Abstract
Trinucleotide repeat expansions cause 17 heritable human neurological disorders. In some diseases, somatic expansions occur in non-proliferating tissues such as brain where DNA replication is limited. This finding stimulated significant interest in replication-independent expansion mechanisms. Aberrant DNA repair is a likely source, based in part on mouse studies showing that somatic expansions are provoked by the DNA repair protein MutSβ (Msh2-Msh3 complex). Biochemical studies to date used cell-free extracts or purified DNA repair proteins to yield partial reactions at triplet repeats. The findings included expansions on one strand but not the other, or processing of DNA hairpin structures thought to be important intermediates in the expansion process. However, it has been difficult to recapitulate complete expansions in vitro, and the biochemical role of MutSβ remains controversial. Here, we use a novel in vitro assay to show that human cell-free extracts catalyze expansions and contractions of trinucleotide repeats without the requirement for DNA replication. The extract promotes a size range of expansions that is similar to certain diseases, and triplet repeat length and sequence govern expansions in vitro as in vivo. MutSβ stimulates expansions in the extract, consistent with aberrant repair of endogenous DNA damage as a source of expansions. Overall, this biochemical system retains the key characteristics of somatic expansions in humans and mice, suggesting that this important mutagenic process can be restored in the test tube.
Keywords: expansion, mutagenesis, neurological disease, cell-free extract, DNA repair, MutSβ
Introduction
Trinucleotide repeat (TNR) expansions cause at least 17 inherited neurological diseases, including Huntington's disease (HD) and myotonic dystrophy type 1 (DM1). It is generally accepted that the TNR contributes to its own expansion through the tendency to form non-B-DNA structures such as hairpins and slipouts, which arise during aberrant replication or misrepair of damage1,2,3. These hairpins/slipouts are likely the mutagenic precursors to the expansion itself. Thus, cellular proteins that either help create hairpins/slipouts or process them further are expected to be key components of the expansion process1,2,3. The mismatch recognition protein MutSβ was identified as one important causative factor for expansions in mouse models of HD or DM1. Most expansions, both inherited and somatic, are blocked by knockout of Msh2 or Msh34,5,6,7,8,9,10, which encode the two subunits of MutSβ. This evidence supports the hypothesis that misrepair of DNA damage is one important source of expansions, especially for somatic events in non-proliferating tissues where replication is limited.
Biochemical studies of expansions have largely focused on partial reactions. In one approach11,12, CAG repeats containing an 8-oxo-guanine lesion were treated with purified Ogg1 glycosylase and apurinic endonuclease to remove the damage and leave a one-nucleotide gap. Repair synthesis by purified DNA polymerase β and flap endonuclease created excess CAG repeats, thus expanding the damaged DNA strand. Expansion of the undamaged CTG strand was not documented, although it was proposed to occur through a subsequent misrepair event11. Another approach13,14,15,16,17 started with preformed CTG hairpins or CAG slipouts and monitored their processing in cell-free extracts. In some cases, the excess repeats were precisely excised and repaired, consistent with expansion prevention. Other substrates were either unrepaired or misrepaired to give products retaining extra repeats on one strand. A third approach used human cell extracts to drive SV40 DNA replication of plasmid templates with (CAG•CTG)79 repeats18. A few large expansions in the replicated DNA could be distinguished from background events. The authors suggested that replication helps create mutagenic intermediates that are then somehow misrepaired to give expansions18. The biochemistry of MutSβ with respect to expansions has been examined due to the genetic results cited above. MutSβ binds to CAG hairpins9,19 and it is required for repair of short slipouts16, but the biochemical role of MutSβ in expansions remains controversial9,19,20,21.
The aim of the current study was to establish an in vitro system for generating the complete expansion process. As described below, human cell-free extracts catalyze expansions in a manner consistent with the key genetic characteristics from in vivo studies in humans and mice, including MutSβ stimulation of expansions. This extract system provides the ability to assess directly the roles of DNA replication, repair and transcription in driving expansions.
Results
Expansions occur in vitro
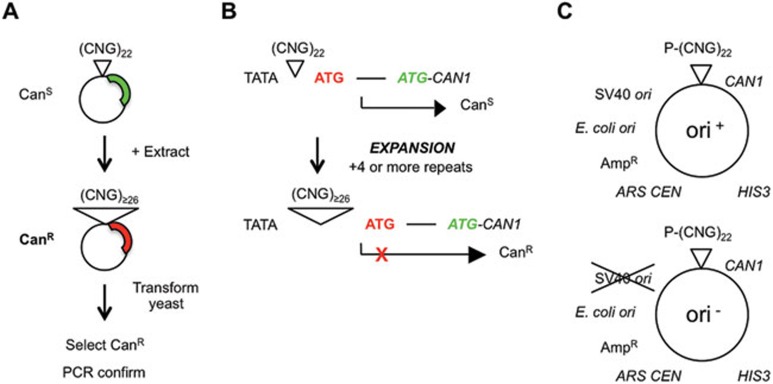
Our objective was to recapitulate the complete expansion reaction in vitro. A novel but simple cell-free system that generates expansions was developed. A 22-repeat plasmid is incubated with human cell extract to generate expansions in the test tube. Expansions are detected following DNA recovery and transformation into yeast (Figure 1A). Control samples without extract indicate the background level of expansions, such as any arising from pre-existing expansions in the substrate preparation or expansion after transformation into yeast. Most experiments used HeLa extracts, which support many DNA metabolic processes in vitro, such as replication, repair and transcription. Furthermore, use of HeLa allows comparison with previous studies of TNR hairpin processing in vitro13,14,15,16,17. Using yeast as a biosensor improves the sensitivity for detecting expansions compared with most biochemical methods. The assay is also quantitative, and PCR is employed to verify expansions and to measure their sizes. A drug resistance assay is used to detect TNR expansions to final lengths of ≥ 26 repeats22 (Figure 1B). The plasmid substrates are amenable to insertion of different TNR lengths and sequences (Figure 1C). In addition, plasmids either contain the SV40 origin of DNA replication (ori+) or not (ori−) to assess whether replication is required for any expansions observed (Figure 1C). The extract can be supplemented as desired with SV40 large T antigen (Tag). Experiments with the ori− plasmid and the omission of Tag therefore preclude nearly all DNA replication.
Figure 1.
Key features of the in vitro assay for expansions. (A) Assay procedure. A 22-repeat shuttle vector was incubated with cell-free extract, then recovered and transformed into yeast. Plasmids containing an expanded (≥ 26) repeat confer canavanine resistance. The repeat region is amplified by PCR and the expansion is confirmed on polyacrylamide gels. (B) Expansion selection22. At 22 repeats, CAN1 is expressed, causing canavanine sensitivity. Expansions adding ≥ 4 repeats cause a change in the transcriptional start site, such that an out-of-frame initiator codon is incorporated into the mRNA. Translation from this initiator codon is out-of-frame with respect to the CAN1 structural gene, causing canavanine resistance22. (C) Shuttle vectors. Plasmids are based on those used by Claassen and Lahue35, modified with CAN1 selection cassette22. Briefly, each plasmid is a three-way shuttle vector capable of replicating in Saccharomyces cerevisiae, E. coli, and human cells or cell-free extracts supplemented with SV40 large T antigen. The TNR is usually (CTG)22, although other sequences can be substituted. P, adh1 promoter for expression in yeast.
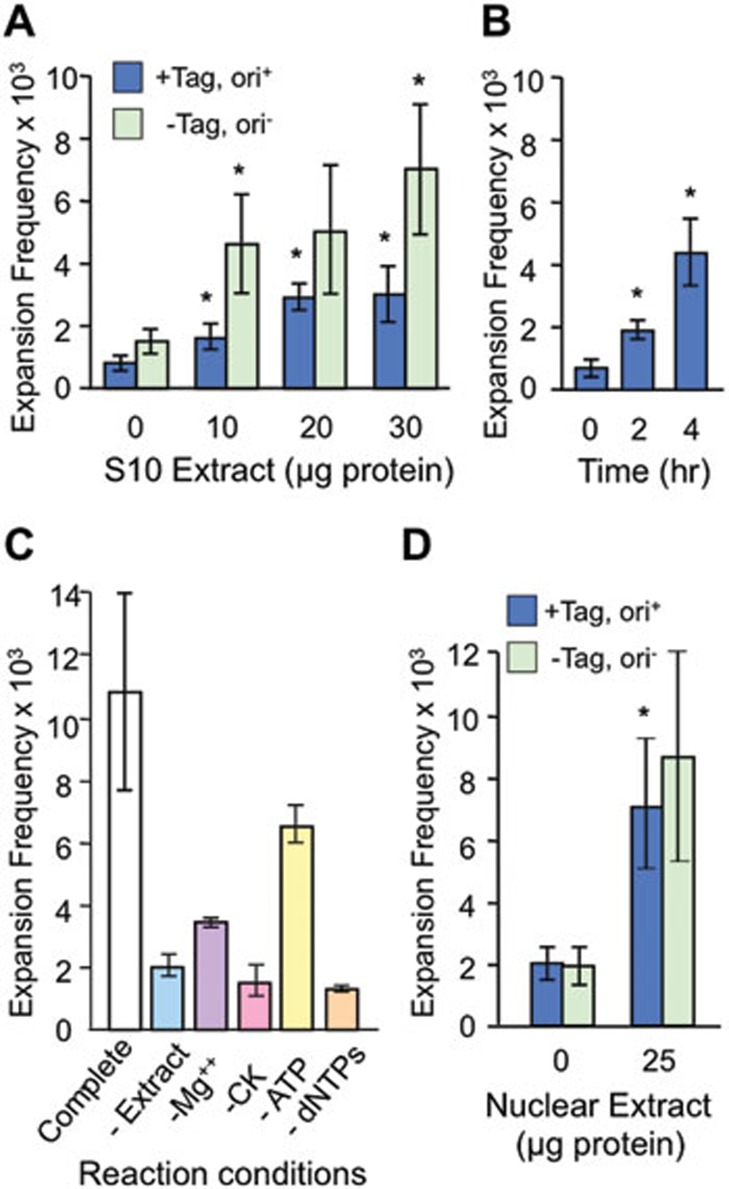
If expansions are generated in vitro, they should accumulate in an extract-dependent manner. Expansions clearly increased with addition of HeLa whole-cell extract (Figure 2A). The majority of expansions, typically 80% or more, were extract dependent compared with the no extract control. The reaction is reasonably effective, catalyzing expansion at frequencies that approach 1%. Expansions did not require DNA replication; unexpectedly, more expansions were observed when replication was blocked (–Tag, ori−) than when it was permitted (+Tag, ori+; Figure 2A). There was an atypical trend in Figure 2A towards more expansions when replication was blocked; in most experiments there was no negative or positive effect of replication on expansion frequencies (see below). Expansions were also dependent on reaction time (Figure 2B), consistent with the prediction that the extract processed more DNA molecules over time and therefore should generate more expansions. In further support of this prediction, expansions required supplementation of the reaction with Mg2+, creatine kinase and dNTPs (Figure 2C). The dependence on dNTPs is consistent with the requirement for DNA repair synthesis to increase the length of the TNR tract. The partial dependence on ATP is presumably due to ATP from the extract or from the regenerating system. Expansions also occurred at similar levels in HeLa nuclear extract, and preventing DNA replication did not alter the frequency of expansions (Figure 2D). In summary, these data indicate that expansions are catalyzed in vitro by HeLa cell-free extracts, with no detectable requirement for DNA replication.
Figure 2.
Expansions require extract, time and cofactors. The (CTG)22 starting plasmid was tested in all panels. (A) Extract-dependent expansion frequencies for ori+ and ori− plasmids incubated with HeLa whole-cell extract for 4 h. +Tag, supplemented with 2 μg SV40 large T antigen. (B) Time course of expansions for ori+ plasmid treated with 30 μg whole-cell extract plus 2 μg SV40 T-antigen. (C) Cofactor requirements for expansions of ori− plasmid in 30 μg whole-cell extract for 4 h. CK, creatine kinase. (D) Expansion frequencies for ori+ and ori− plasmids in HeLa nuclear extract for 4 h. In all panels, error bars are ± SEM except panel C, error bars = range. *P < 0.05 compared with 0 extract control in panels (A and D), or compared with 0 time control in panel (C).
The size of expansions in vitro and the sequence requirements for generating them accurately reflect in vivo expansions
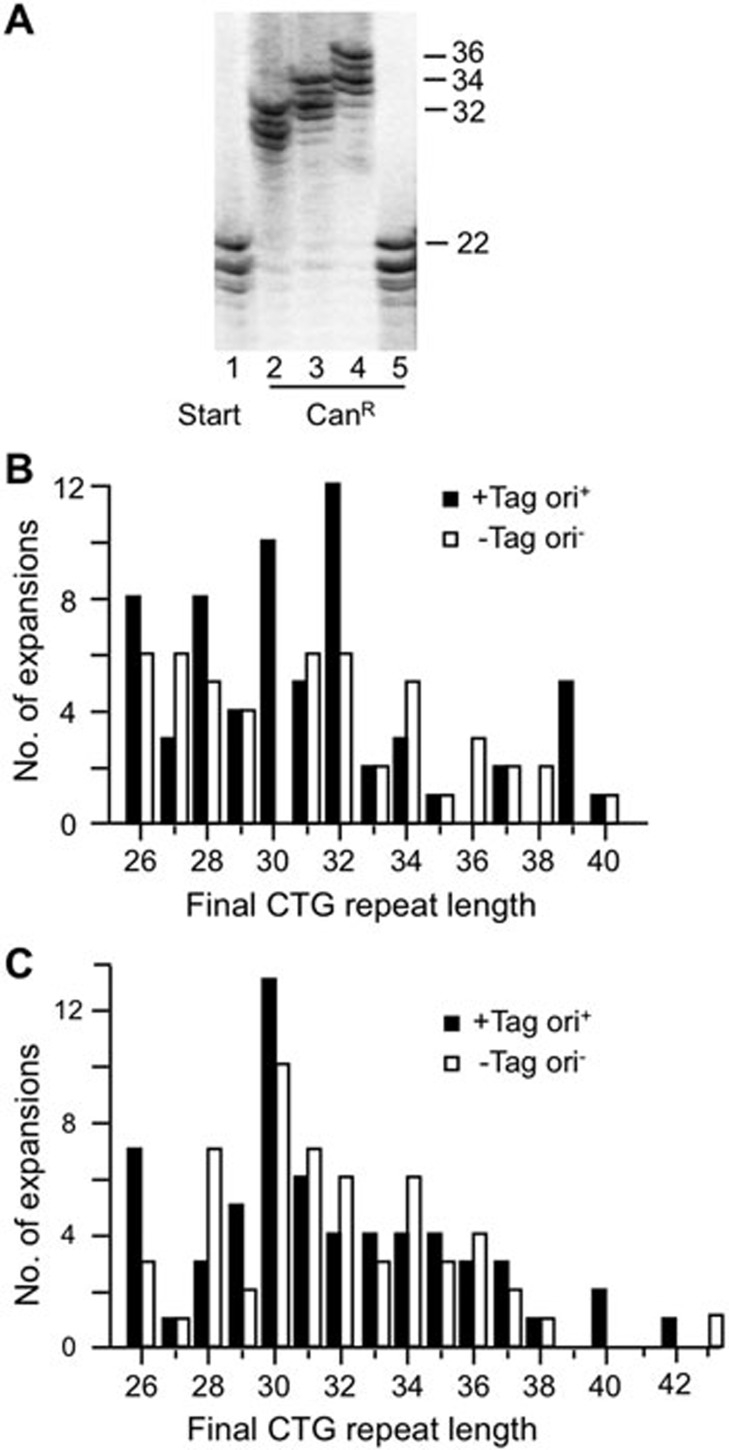
A crucial test of in vitro expansions is whether they are consistent with known characteristics from genetic studies. We first examined expansion sizes as one indicator of disease relevance. PCR analysis showed that expansions catalyzed by extracts occurred as jumps rather than recurrent small gains (Figure 3A). The size distribution of expansions in HeLa whole-cell extracts ranged from +4 to +18 repeats, with no detectable difference regardless of whether replication was permitted or not (Figure 3B). Expansions catalyzed by HeLa nuclear extracts gave a similar size distribution, with the addition of one expansion each of +20 and +21 to final repeat lengths of 42 and 43 (Figure 3C). Both the single jumps and the size range results are consistent with expansions seen in male germline from HD individuals23,24. The largest expansions seen in vitro, up to a final length of 40-43 repeats, correspond to disease-causing alleles of a number of polyglutamine diseases including HD25 and SCA126. In summary, expansions occurring in cell-free extracts overlap in size with those seen in a number of TNR expansion diseases.
Figure 3.
Analysis of expansion sizes. (A) Representative polyacrylamide gel showing PCR product from starting repeat (22 repeats; lane 1), three confirmed expansions (lanes 2-4) and one false positive (lane 5). Sizes are expressed as TNR length. (B) Expansion spectrum showing final repeat sizes from (CTG)22 ori+ and ori− plasmids in 30 μg whole-cell extract. n, 64 for +Tag ori+; 49 for −Tag ori−. (C) Expansions of (CTG)22 treated for 4 h with HeLa nuclear extract. Spectra are pooled from experiments using either 25, 50 or 100 μg extract. +Tag, ori+, extract supplemented with 2 μg large T antigen, substrate contained SV40 origin, n = 62; −Tag, ori−, no large T antigen was added, substrate without SV40 origin, n = 56.
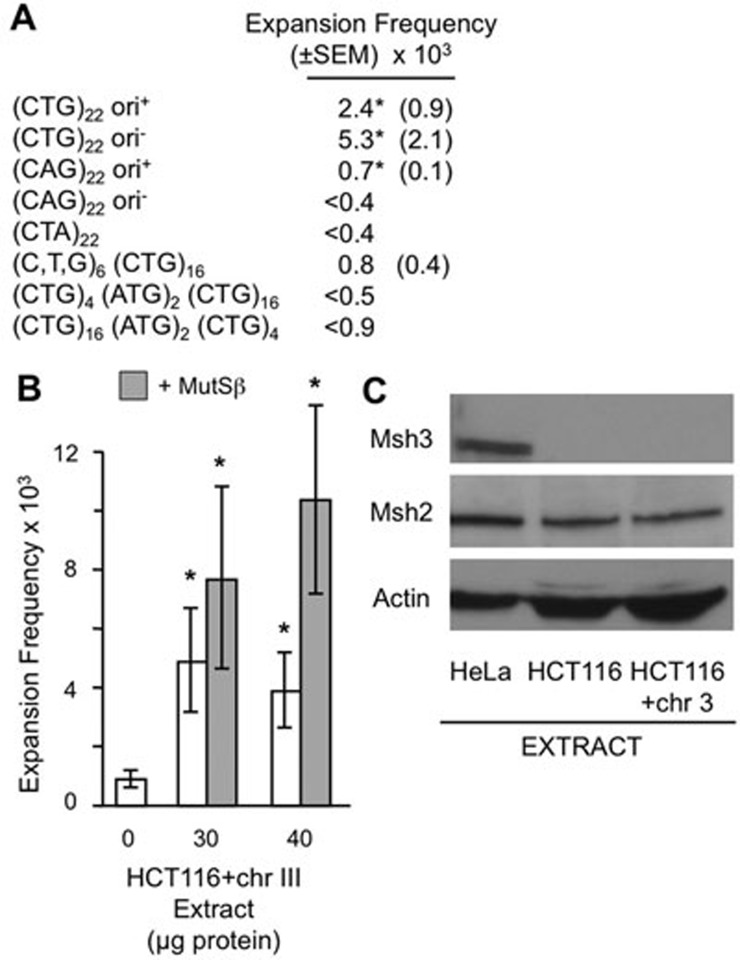
Disease-causing expansions in humans and mice occur in CNG repeats, where N denotes any nucleotide, with the sole exception of a GAA repeat that is unstable in Friedreich's ataxia1,2,3. Additional sequence features that favor expansions include longer repeats and the absence of any base pair changes within the tract ('interruptions'). We utilized the versatility of our plasmid substrates to examine to what extent in vitroexpansions mimic the in vivosequence requirements (Figure 4A). Compared with the (CTG)22 reference plasmid, expansions of (CAG)22 substrates under replication-permissive conditions occurred about 30% as efficiently. Thus both CTG and CAG repeats expand in vitro. The non-structure-forming sequence (CTA)2227 gave no detectable expansions, suggesting the structure-forming capacity of the TNR is important for reactivity. Shortening the CTG tract from 22 to 16 repeats reduced expansions by two-thirds relative to (CTG)22 (Figure 4A) and also showed a more constrained size distribution of +6 to +14 repeats. The inclusion of two ATG interruptions within the CTG tract stabilized the repeat to the point where no expansions were detected (Figure 4A). Together, this analysis supports the hypothesis that DNA elements of the TNR govern expansions similarly in vitro and in vivo.
Figure 4.
DNA and protein requirements for expansions. (A) Expansion frequencies for plasmid substrates incubated with 30 μg whole-cell extract for 4 h. All plasmids were identical except for the indicated changes to TNR tract and the presence or absence of SV40 origin. Zero extract control values were subtracted out. <, upper limits, as PCR analysis showed all false positives. *P < 0.05 with extract versus zero extract. Additional details are provided in Supplementary information, Table S1. (B) Expansion frequencies for (CTG)22 ori+ plasmid with HCT116+chr3 whole-cell extract with or without 400 ng MutSβ for 4 h. Error bars are ± 1 SEM; *P < 0.05 versus zero extract. P is also < 0.05 comparing 40 μg extract with MutSβ to 40 μg extract without MutSβ. (C) Immunoblot for Msh3, Msh2 and actin (loading control) of 30 μg of protein from HeLa, HCT116 or HCT116+chr3 extracts.
Stimulation of in vitro expansions by MutSβ
In mouse models of HD and DM1, most expansions require the presence of MutSβ (Msh2-Msh3 complex)4,5,6,7,8,9. To test whether MutSβ is required for expansions in vitro, extracts were prepared from human HCT116 cells complemented with chromosome 3 ('HCT116+chr3'). HCT116 cells are defective in both Mlh1 and Msh328, the latter causing a deficiency in MutSβ. Stable complementation with chromosome 3 corrects the MLH1 defect but not MSH3, making HCT116+chr3 cells proficient for mismatch repair28,29 but still deficient in MutSβ28. Immunoblot analysis (Figure 4C) verified that HCT116+chr3 cells were deficient in Msh3, whereas HeLa cells expressed Msh3. Both cell lines expressed Msh2 at similar levels. Expansions were measured in 30-40 μg extract from HCT116+chr3 cells, with or without addition of purified MutSβ (Figure 4B). The addition of MutSβ stimulated expansions by about two-fold, suggesting that about one-half of the expansions are dependent on MutSβ under these conditions. The enhancement in expansion activity upon addition of MutSβ achieved statistical significance with 40 μg extract. To our knowledge, this is the first biochemical demonstration that MutSβ facilitates expansions.
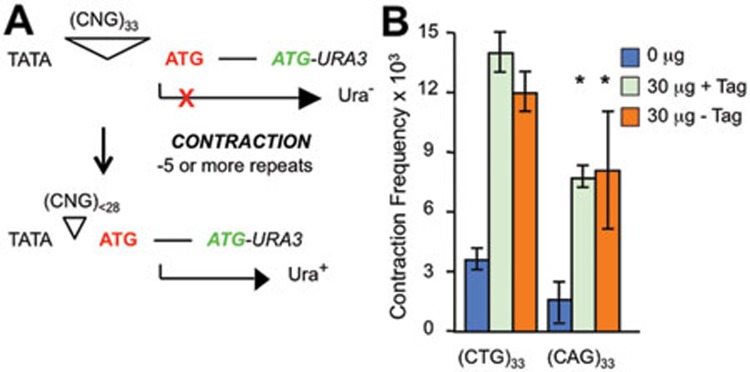
Contractions of triplet repeats are also catalyzed by human cell-free extracts
In addition to expansions, triplet repeats in humans and mice occasionally contract (shorten). Although infrequent in most TNR expansion families, contractions are beneficial mutations that can reduce the risk of disease or delay its onset2. We modified the in vitro assay slightly to start with tract lengths of (CTG)33 and (CAG)33 and then measured contractions of 5 repeats or more (Figure 5A). HeLa whole-cell extracts also catalyzed contractions of both sequences (Figure 5B). Contraction frequencies were roughly two-fold higher than expansion frequencies for CTG tracts, and ∼10-fold greater for CAG repeats (compare Figure 5B with Figures 2A and 4A). The higher frequency of contractions could be due to the longer starting tract for contractions than expansions, 33 versus 22 repeats. Similar to what was seen for expansions, preventing DNA replication did not alter contraction frequencies in vitro (Figure 5B). The results presented here show that human cell-free extracts are active in catalyzing both expansions and contractions of triplet repeats.
Figure 5.
Contractions catalyzed by HeLa cell extract. (A) Overview of contractions based on published work34. Plasmids with 33 repeats do not express the reporter gene URA3 due to a block in translation (red X), thus yeast cells require uracil for growth (Ura–). A TNR contraction removing 5 or more repeats allows expression of URA3 in yeast, thus providing a Ura+ phenotype. Subsequent PCR analysis of the repeat region verifies the contraction. (B) Contraction frequencies of (CTG)33 and (CAG)33 ori+ plasmids incubated for 4 h with 30 μg HeLa whole-cell extract with or without supplementation with 2 μg SV40 large T antigen. (CTG)33 results are from two independent experiments; error bars display the range of frequency values. (CAG)33 results are from three independent experiments; error bars are ± SEM, *P < 0.05 versus no extract control.
Discussion
The biochemical system described here retains the key characteristics of somatic expansions from non-proliferating tissues in humans and mice. There is no requirement for DNA replication, expansion frequencies are governed by the TNR length, sequence and purity, expansion sizes overlap with those seen in polyglutamine diseases such as HD, and MutSβ stimulates expansions. Transcription-mediated instability is unlikely in our system as there is no obvious human promoter in the substrate plasmid (Figure 1C), and addition of the transcription inhibitor α-amanitin showed no effect on expansion frequencies (data not shown). As expansions are replication independent and can be stimulated by MutSβ, we postulate that expansions occur through misrepair of endogenous DNA damage in this system. Human cell-free extracts also catalyzed contractions in a replication-independent manner. The most likely mechanism of contraction is also misrepair, consistent with a previous observation that contractions are the predominant in vitro event after repair of a double-strand break within a CTG•CTG tract30.
This in vitro expansion system has the virtues of simplicity, sensitivity and versatility, but its efficiency of ∼1% is lower than in vivo. One major difference is the TNR length. We start with 22 repeats and monitor expansions that approach and sometimes cross the crucial threshold of ∼30-40 repeats where instability becomes apparent in humans1,3,31 and where diseases such as HD and SCA1 first express a phenotype25,26. The expansion efficiency seen in vitro for 22-repeat substrates is in line with in vivo results. In sperm from normal and HD individuals, expansions first become apparent for starting alleles between 30 and 36 repeats, and expansion frequencies exceed 90% for TNR lengths of 38-51 repeats23. Overall, the in vitro system generates expansions that mirror disease-initiating mutations and at frequencies in line with expectations.
MutSβ stimulates expansions by about two-fold in vitro, suggesting that MutSβ functions on short, threshold-length TNR tracts. This is consistent with recent work in cultured human astrocytes showing that MutSβ promotes most expansions of (CTG)22 tracts, and that both Msh2 and Msh3 are specifically enriched at the TNR, compared with a randomized control sequence32. Together, these findings extend the range of repeat lengths at which MutSβ stimulates expansions down to 22 repeats in both extracts and cell culture. Previous studies in mice showed that MutSβ is required for most inherited and somatic expansions of long TNR tracts from 84 to > 300 repeat units in length4,5,6,7,8,9,10. Thus MutSβ promotes expansions over more than a 10-fold range in repeat length. Although the precise mechanism by which MutSβ functions remains to be solved, this protein exhibits its effects over an impressive range of TNR sizes.
Materials and Methods
Shuttle vectors
CAN1 shuttle vector plasmids were created by modification of pBL245-based shuttle vectors described previously33,34,35. These are pRS313-based plasmids36 containing the SV40 origin of replication cloned into the ApaI site. To create the plasmids used in this study, the promoter-TNR-URA3 region of pBL25235 was removed by BamHI and XhoI digestion, and the backbone was religated with a BamHI-XhoI promoter-TNR-CAN1 fragment. To create the derivatives lacking the SV40 origin of replication, XhoI digestion was followed by partial KpnI digestion to remove the SV40 sequences, the ends were then blunted with Klenow fragment and ligated together. For all plasmids, the named repeat (e.g., (CTG)22) refers to a plasmid where the CTG repeats are on the non-coding strand relative to the reporter gene CAN1 or URA3.
Preparation of human cell extracts
Frozen HeLa cells were obtained from Cilbiotech (Mons, Belgium). HCT116 and HCT116+chr3 cells were a gift from Dr Alan Clark (National Institute of Environmental Health Sciences, Research Park, NC, USA). Whole-cell S10 extracts were prepared, essentially, according to Li and Kelly37. Frozen cell pellets were thawed on ice, and resuspended in an equal volume of chilled hypotonic lysis buffer and sucrose. Cells were pelleted at 1 000× g for 10 min at 4 °C and supernatant was removed. Cells were resuspended in an equal volume of chilled hypotonic lysis buffer, pelleted at 1 000× g for 10 min at 4 °C, and supernatant was removed. Cell pellet was resuspended in an equal volume of chilled hypotonic lysis buffer (including DTT, PMSF, protease inhibitors and phosphatase inhibitors), and incubated for 10 min on ice. Cell suspension was transferred into prechilled Dounce homogenizer and cells were lysed on ice by 20-30 strokes using a loose-fitting pestle. Extract was incubated on ice for 30 min, inverting tube occasionally to mix, then centrifuged at 10 000× g for 30 min at 4 °C. 5 M NaCl was added to the supernatant to a final concentration of 0.1 M NaCl, and then centrifuged at 10 000× g for 30 min at 4 °C. The supernatant was aliquoted and stored at −80 °C.
HeLa cell nuclear extracts were prepared essentially as described38. Cells (2-3 × 109) were harvested at 4 000× g for 5 min and nuclei were obtained from lysed cells by centrifugation (9 000× g for 5 min). Nuclear pellets were resuspended in extraction buffer (50 mM Hepes, pH 7.5, 0.5 mM DTT, 0.1% PMSF, 10% (w/v) sucrose, 150 mM NaCl), and incubated with shaking at 4 °C for 1 h. After centrifugation (15 000× g for 20 min), the supernatant was concentrated by ammonium sulfate precipitation (0.42 g/ml). The ammonium sulfate pellet was then dialyzed in a buffer containing 25 mM Hepes-KOH (pH 7.5), 0.1 mM EDTA and 50 mM KCl. After clarification by centrifugation, the resulting nuclear extract was frozen in small aliquots in liquid nitrogen and stored at −80 °C.
Immunoblot
30 μg of protein extract from HeLa, HCT116 or HCT116+chr3 cells was separated by denaturing polyacrylamide gel electrophoresis and transferred to Immobilon membrane (Millipore). Primary antibodies were against Msh2 (Calbiochem #NA26), Msh3 (Professor Glenn Morris, Wolfson Centre for inherited Neuromuscular Disease, UK;39) or the loading control, actin (Sigma A2066). Horseradish peroxidase-conjugated secondary antibodies from Jackson ImmunoResearch Labs were anti-rabbit 711-035-152 for Msh2 and 115-035-003 for Msh3. Visualization was achieved with Western Lightning Plus-ECL (Perkin Elmer).
Cell-free assay to measure triplet repeat expansions
Assays were performed essentially as described18. Each 100 μl reaction contained 300 ng of plasmid template, 20 μl 5× buffer (500 μM each dATP, dCTP, dGTP and dTTP, 1 mM each CTP, GTP and UTP, 20 mM ATP, 200 mM creatine phosphate, 35 mM magnesium acetate, 150 mM HEPES pH 7.8, 2.5 mM DTT), 16 μl creatine kinase (6 μg/ml), and 10-100 μg protein extract (S10 or nuclear). dNTPs, rNTPs and magnesium were included to replace any cofactors lost or diluted during extract preparation. Some reactions contained 2 μg SV40 large T-antigen (CHIMERx, Milwaukee, WI, USA), or as described40. Assays were incubated at 37 °C for 4 h, except where otherwise noted. Reactions were terminated with 100 μl stop solution (2 mg/ml proteinase K, 2% SDS, 50 mM EDTA pH 8.0) at 37 °C for 30 min. Plasmid DNA was then purified using a Qiagen miniprep kit, and eluted in 80 μl buffer EB.
Genetic assays and analysis of expanded TNR alleles
Contraction assays using the URA3 reporter have been described previously33. Triplet repeat expansion assays using the CAN1reporter were performed as described22 with the following modifications. Recovered plasmids were transformed into yeast; 1/40 of the total was plated on a SC-His (synthetic complete media lacking histidine) plate to assess total plasmid population and the remainder was plated on a separate SC-His plate. After two days, the large-volume plates were replica-plated to SC-His-Arg+Can (synthetic complete media lacking histidine and arginine and supplemented with 120 μg/ml canavanine) to assess growth for potential expansions. All expansions were verified by single-colony PCR across the repeat tract followed by analysis on high-resolution polyacrylamide gels22. Expansion frequencies are expressed as (CanR colonies/total colonies) × (% authentic expansions), where expansions are authenticated by PCR as in Figure 2A. For some TNR sequences, no authentic expansions were detected out of at least 30 CanR colonies tested by PCR. These data are presented as upper limits, denoted by “<” symbol.
Recombinant MutSβ protein production
Recombinant human MutSβ was purified from insect cells using the baculovirus expression system as described41. Briefly, insect cells (HighFive) co-infected with MSH2- and MSH3-baculoviruses were lysed in buffer A (25 mM HEPES, pH 7.6, 150 mM NaCl, 4 mM β-mercaptoethanol, 100 mM PMSF, 190 mM benzamidine, 0.05 μg/ml protease inhibitors (pepstatin A, leupeptin and antipain). After centrifugation, the supernatant was used for MutSβ purification through three columns (GE Healthcare), a HisTrap HP column, a Mono S column, and a Superdex 200 column. MutSβ was eluted in the HisTrap HP column and the Mono S column with a 50-ml imidazole gradient (from 20 mM to 300 mM) and a NaCl gradient (150 mM to 500 mM) in buffer A, respectively. The Superdex 200™ column was developed with buffer A containing 150 mM NaCl. The protein eluted from the last column was near homogeneity (∼99% pure), and was aliquoted, quickly frozen in liquid nitrogen and stored at −80 °C.
Acknowledgments
This study was supported by Science Foundation Ireland (10/IN.1/B2973) and Irish Health Research Board (RP/2008/12) to RSL, and US National Institutes of Health (GM089684) to G-M L. We thank Heinz-Peter Nasheuer and Irina Tikhanovich for assistance with preparing and assaying the T antigen.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Material
Expansion frequency data for Fig 4A
References
- Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- Lopez Castel A, Cleary JD, Pearson CE. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat Rev Mol Cell Biol. 2010;11:165–170. doi: 10.1038/nrm2854. [DOI] [PubMed] [Google Scholar]
- McMurray CT. Mechanisms of trinucleotide repeat instability during human development. Nat Rev Genet. 2010;11:786–799. doi: 10.1038/nrg2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley K, Shirley TL, Flaherty L Messer A. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat Genet. 1999;23:471–473. doi: 10.1038/70598. [DOI] [PubMed] [Google Scholar]
- Kovtun IV, McMurray CT. Trinucleotide expansion in haploid germ cells by gap repair. Nat Genet. 2001;27:407–411. doi: 10.1038/86906. [DOI] [PubMed] [Google Scholar]
- van den Broek WJAA, Nelen MR, Wansink DG, et al. Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum Mol Genet. 2002;11:191–198. doi: 10.1093/hmg/11.2.191. [DOI] [PubMed] [Google Scholar]
- Savouret C, Brisson E, Essers J, et al. CTG repeat instability and size variation timing in DNA repair-deficient mice. EMBO J. 2003;22:2264–2273. doi: 10.1093/emboj/cdg202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler VC, Lebel L-A, Vrbanac V, Teed A, te Riele H, MacDonald ME. Mismatch repair gene Msh2 modifies the timing of early disease in HdhQ111 striatum. Hum Mol Genet. 2003;12:273–281. doi: 10.1093/hmg/ddg056. [DOI] [PubMed] [Google Scholar]
- Owen BAL, Yang Z, Lai M, et al. (CAG)n-hairpin DNA binds to Msh2-Msh3 and changes properties of mismatch recognition. Nat Struct Mol Biol. 2005;12:663–670. doi: 10.1038/nsmb965. [DOI] [PubMed] [Google Scholar]
- Foiry L, Dong L, Savouret C, Hubert L, et al. Msh3 is a limiting factor in the formation of intergenerational CTG expansions in DM1 transgenic mice. Hum Genet. 2006;119:520–526. doi: 10.1007/s00439-006-0164-7. [DOI] [PubMed] [Google Scholar]
- Kovtun IV, Liu Y, Bjoras M, Klungland A, Wilson SH, McMurray CT. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007;447:447–452. doi: 10.1038/nature05778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Prasad R, Beard WA, et al. Coordination between polymerase beta and FEN1 can modulate CAG repeat expansion. J Biol Chem. 2009;284:28352–28366. doi: 10.1074/jbc.M109.050286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi GB, Lau R, Montgomery SE, Leonard MR Pearson CE. Slipped (CAG)•(CTG) repeats can be correctly repaired, escape repair or undergo error-prone repair. Nat Struct Mol Biol. 2005;12:654–662. doi: 10.1038/nsmb959. [DOI] [PubMed] [Google Scholar]
- Hou C, Chan NLS, Gu L, Li G-M. Incision-dependent and error-free repair of (CAG)n/(CTG)n hairpins in human cell extracts. Nat Struct Mol Biol. 2009;16:869–876. doi: 10.1038/nsmb.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel AL, Tomkinson AE, Pearson CE. CTG/CAG repeat instability is modulated by the levels of human DNA ligase I and its interaction with proliferating cell nuclear antigen: a distinction between replication and slipped-DNA repair. J Biol Chem. 2009;284:26631–26645. doi: 10.1074/jbc.M109.034405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi GB, Slean MM, Simard JP, Gileadi O, Pearson CE. Isolated short CTG/CAG slip-outs are repaired efficiently by hMutSbeta, but clustered slip-outs are poorly repaired. Proc Natl Acad Sci USA. 2010;107:12593–12598. doi: 10.1073/pnas.0909087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Huang J, Gu L, Li G-M. In vitro repair of DNA hairpins containing various numbers of CAG/CTG trinucleotide repeats. DNA Repair. 2012;11:201–209. doi: 10.1016/j.dnarep.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi GB, Cleary JD, Pearson CE. In vitro (CTG)•(CAG) expansions and deletions by human cell extracts. J Biol Chem. 2002;277:13926–13934. doi: 10.1074/jbc.M109761200. [DOI] [PubMed] [Google Scholar]
- Lang WH, Coats JE, Majka J, et al. Conformational trapping of mismatch recognition complex MSH2/MSH3 on repair-resistant DNA loops. Proc Natl Acad Sci USA. 2011;108:E837–E844. doi: 10.1073/pnas.1105461108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Hou C, Tian K, Holcomb NC, Gu L, Li G-M. Mismatch recognition protein MutSβ does not hijack (CAG)n hairpin repair in vitro. J Biol Chem. 2009;284:20452–20456. doi: 10.1074/jbc.C109.014977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tome S, Holt I, Edelmann W, et al. MSH2 ATPase domain mutation affects CTG•CAG repeat instability in transgenic mice. PLoS Genet. 2009;5:e1000482. doi: 10.1371/journal.pgen.1000482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debacker K, Frizzell A, Gleeson O, Kirkham-McCarthy L, Mertz T, Lahue RS. Histone deacetylase complexes promote trinucleotide repeat expansions. PLoS Biol. 2012;10:e1001257. doi: 10.1371/journal.pbio.1001257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeflang EP, Zhang L, Tavare S, et al. Single sperm analysis of the trinucleotide repeats in the Huntington's disease gene: quantification of the mutation frequency spectrum. Hum Mol Genet. 1995;4:1519–1526. doi: 10.1093/hmg/4.9.1519. [DOI] [PubMed] [Google Scholar]
- Leeflang EP, Tavare S, Marjoram P, et al. Analysis of germline mutation spectra at the Huntington's disease locus supports a mitotic mutation mechanism. Hum Mol Genet. 1999;8:173–183. doi: 10.1093/hmg/8.2.173. [DOI] [PubMed] [Google Scholar]
- Group T.H.s.D.C.R. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes Cell 199372971–983. [DOI] [PubMed] [Google Scholar]
- Orr HT, Chung M, Banfi S, et al. Expansion of an unstable trinucleotide (CAG) repeat in spinocerebellar ataxia type 1. Nat Genet. 1993;4:221–226. doi: 10.1038/ng0793-221. [DOI] [PubMed] [Google Scholar]
- Gacy AM, Goellner G, Juranic N, Macura S, McMurray CT. Trinucleotide repeats that expand in human disease form hairpin structure in vitro. Cell. 1995;81:533–540. doi: 10.1016/0092-8674(95)90074-8. [DOI] [PubMed] [Google Scholar]
- Boland CR, Koi M, Chang DK, Carethers JM. The biochemical basis of microsatellite instability and abnormal immunohistochemistry and clinical behavior in Lynch Syndrome: from bench to bedside. Fam Cancer. 2008;7:41–52. doi: 10.1007/s10689-007-9145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koi M, Umar A, Chauhan DP, Cherian SP, et al. Human chromosome 3 corrects mismatch repair deficiency and microsatellite instability and reduces N-methyl-N'-nitro-N-nitrosoguanidine tolerance in colon tumor cells with homozygous hMLH1 mutation. Cancer Res. 1994;54:4308–4312. [PubMed] [Google Scholar]
- Marcadier JL, Pearson CE. Fidelity of primate cell repair of a double-strand break within a (CTG)•(CAG) tract. Effect of slipped DNA structures. J Biol Chem. 2003;278:33848–33856. doi: 10.1074/jbc.M304284200. [DOI] [PubMed] [Google Scholar]
- Paulson HL, Fischbeck KH. Trinucleotide repeats in neurogenetic disorders. Annu Rev Neurosci. 1996;19:79–107. doi: 10.1146/annurev.ne.19.030196.000455. [DOI] [PubMed] [Google Scholar]
- Gannon A-MM, Frizzell A, Healy E, Lahue RS. MutSβ and histone deacetylase complexes promote expansions of trinucleotide repeats in human cells. Nucleic Acids Res. 2012;40:10324–10333. doi: 10.1093/nar/gks810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier R, Farrell BT, Miret JJ, Lahue RS. Mechanistic features of CAG•CTG repeat contractions in cultured cells revealed by a novel genetic assay. Nucleic Acids Res. 2005;33:5667–5676. doi: 10.1093/nar/gki880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell BT, Lahue RS. CAG•CTG repeat instability in cultured human astrocytes. Nucleic Acids Res. 2006;34:4495–4505. doi: 10.1093/nar/gkl614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claassen DA, Lahue RS. Expansions of CAG•CTG repeats in immortalized human astrocytes. Hum Mol Genet. 2007;16:3088–3096. doi: 10.1093/hmg/ddm270. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JJ, Kelly TJ. Simian virus 40 DNA replication in vitro: specificity of initiation and evidence for bidirectional replication. Mol Cell Biol. 1985;5:1238–1246. doi: 10.1128/mcb.5.6.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes J, Clark S, Modrich P. Strand-specific mismatch correction in nuclear extracts of human and Drosophila melanogaster cell lines. Proc Natl Acad Sci USA. 1990;87:5837–5841. doi: 10.1073/pnas.87.15.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt I, Lam LT, Tome S, et al. The mouse mismatch repair protein, MSH3, is a nucleoplasmic protein that aggregates into denser nuclear bodies under conditions of stress. J Cell Biochem. 2011;112:1612–1621. doi: 10.1002/jcb.23075. [DOI] [PubMed] [Google Scholar]
- Mahon C, Liang B, Tikhanovich I, et al. Restriction of human polyomaevirus BK virus DNA replication in murine cells and extracts. J Virol. 2009;83:5708–5717. doi: 10.1128/JVI.00300-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yuan F, Presnell SR, et al. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell. 2005;122:693–705. doi: 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expansion frequency data for Fig 4A