Abstract
Multiple methods exist that can reprogram differentiated cells to a pluripotent state similar to that of embryonic stem cells (ESCs). These include somatic cell nuclear transfer (SCNT), fusion-mediated reprogramming (FMR) of somatic cells with ESCs, and the production of induced pluripotent stem cells (iPSCs). All of these methods yield cells in which the endogenous Oct4 gene is reactivated. We were interested in comparing the activity of the Oct4 promoter in three different classes of pluripotent cells, including normal ESCs, FMR cells (FMRCs), and iPSCs. We prepared cells of all three types that harbor a transgene composed of the mouse Oct4 promoter driving green fluorescent protein (Oct4-GFP). All cell derivations started with a characterized transgenic Oct4-GFP mouse, and from this we derived ESCs, FMRCs, and iPSCs with the Oct4-GFP transgene present in an identical genomic integration site in all three cell types. Using flow cytometry we assessed Oct4 promoter expression, cell cycle behavior, and differentiation kinetics. We found similar levels of GFP expression in all three cell types and no significant alterations in pluripotency or differentiation. Our results suggest that the pluripotent condition is a potent “local attractor” state, because it can be achieved through three vastly different avenues.
Introduction
Although acquisition of pluripotency is critically dependent on the co-expression of the pluripotency factors Oct4, Sox2, and Nanog (Boyer et al., 2005; Hanna et al., 2009), mounting evidence suggests that the simple presence of these transcription factors in somatic cells is not sufficient to control artificial reprogramming with an accuracy equal to natural reprogramming during embryogenesis (Shi et al., 2003). In somatic cell nuclear transfer (SCNT), for instance, key obstacles to high efficiency reprogramming include aberrant DNA methylation (Bourc'his et al., 2001; Dean et al., 2001), X chromosome inactivation (Xue et al., 2002), telomere restoration, imprinting, and chromatin remodeling (Xu et al., 2005), leading to low efficiencies in animal cloning. Similar observations have been obtained in an increasing number of recent studies using induced pluripotent stem cells (iPSCs), indicating that reprogrammed pluripotent stem cells frequently retain subsets of epigenetic marks specific to the ancestral somatic epigenome (Kim et al., 2010; Kim et al., 2011; Seiler et al., 2011; Sullivan et al., 2010) and that the iPSC genome contains novel mutations not detected in the ancestral somatic DNA (Krueger et al., 2010; Pasi et al., 2011). Such alterations may raise the probability for immunological incompatibility, tumorigenicity, and limited pluripotency, potentially limiting the clinical utility of iPSCs.
Previously, we reprogrammed mouse embryonic fibroblasts derived from chimeric mice by both fusing them with embryonic stem cells (ESCs), in a process that we call fusion-mediated reprogramming (FMR) (Ambrosi et al., 2007). In the context of increased spontaneous differentiation into adipocytes after partial shRNA knockdown of Oct4 (Hannan and Wolvetang, 2009), we reasoned that the increased rates of spontaneous differentiation might be due to incomplete epigenetic reprogramming or de novo mutations that affect the kinetics and genetic order of reprogramming, leading to differences in the expression of key pluripotency markers that are difficult to detect and difficult to study in mixed populations of cells. One possible explanation for this observation results from the method used for reprogramming; it is likely that the number and concentration of reprogramming components varies from one reprogramming method to another. Thus, it is possible that natural fusion-mediated and transcription factor-induced reprogramming produce small variations in the expression levels of pluripotency factors that subsequently may cause an incomplete reset and/or facilitate increased epigenetic drift of the reprogrammed genome.
Small variations in Oct4 expression levels represent a key candidate for reprogramming method–dependent differences, given the fine-tuned balance of Oct4 levels for maintenance of the pluripotent state and its underlying long-range epigenetic effects. Thus, we surmised that simple variations in Oct4 expression levels alone might be sufficient to trigger increased rates of spontaneous differentiation in single cells. We assessed this hypothesis by using flow cytometry (fluorescence-activated cell sorting [FACS] analysis) to compare green fluorescent protein (GFP) expression levels during proliferation and differentiation of murine (m) ESCs generated from a mouse strain harboring a GFP transgene under the control of the mouse Oct4 promoter with that in FMR and iPSC-derived pluripotent stem cells (PSCs) generated from embryonic fibroblasts derived from the same mouse strain. Here we show that Oct4 expression levels are remarkably similar in pluripotent cells, regardless of their means of derivation or reprogramming.
Materials and Methods
Cell culture
Mouse ESCs expressing enhanced (e) GFP under control of the mouse Oct4 promoter were derived from C57BL/6 mice harboring an Oct4–promoter–eGFP transgene (Boiani et al., 2002; Szabo et al., 2002). Mouse iPSCs were generated as described (Takahashi and Yamanaka, 2006) using lentiviral particles expressing Oct4, Sox2, Klf4, and c-Myc under the control of the human EF1α promoter and mouse embryonic fibroblasts derived from above Oct4-promoter eGFP mice (Boiani et al., 2002; Szabo et al., 2002). Fusion-mediated reprogrammed mouse ESC/fibroblast hybrids (FMRCs) expressing eGFP under control of the mouse Oct4 promoter were generated as described previously (Ambrosi et al., 2007). For analysis of Oct4 expression during differentiation induced by all-trans-retinoic acid (ATRA), cells were plated onto gelatin-coated dishes at a concentration of 10,000 cells/cm2 and exposed to 0.1 μM ATRA for the times indicated. For generation of embryoid bodies (EBs), cells were plated onto low-adhesion plates at a concentration of 20,000 cells/cm2, grown in ESC medium devoid of leukemia inhibitory factor (LIF), and harvested at the times indicated.
FACS analysis
Pluripotent stem cells, grown under feeder-free conditions in the presence of 2000 U/mL LIF, were trypsinized, and both the distribution and intensity of Oct4-expressing cells were analyzed using a FACSCalibur flow cytometer (BD Biosciences Inc.). Live cells were identified either by propidium iodide (PI) exclusion or on the basis of their forward- and side-scatter characteristics. For cell cycle analysis, cells were fixed in ethanol and stained with PI prior to FACS analysis.
RNA extraction
RNA was prepared in triplicate from biologically independent samples. Cells were lysed in RLT buffer (Qiagen Inc.) containing 1% β-mercaptoethanol and processed using RNeasy ion-exchange column chromatography according to the manufacturer's instructions (Qiagen Inc.). Purified RNA was quantified by ultraviolet (UV) spectroscopy.
RT- and qRT-PCR
cDNA was synthesized from 500 ng of total RNA using the iScript™ cDNA synthesis kit (BioRad). Quantitative real-time PCR (qRT-PCR) analyses were performed using the iTaq Fast SYBR Green Supermix with ROX (BioRad) and a 7500 Fast Real Time PCR system (Applied Biosystems). Data were analyzed using the 2−ΔCt method for relative quantification (Livak and Schmittgen, 2001), using β-actin mRNA levels as an endogenous reference. Primers were designed using the Primer Express software (Applied Biosystems) and are listed in Table S1 (for qRT-PCR) and Table S2 (for RT-PCR). (Supplementary Data are available at www.liebertpub.com/cell/.)
Results
Oct4 promoter activity in PSCs
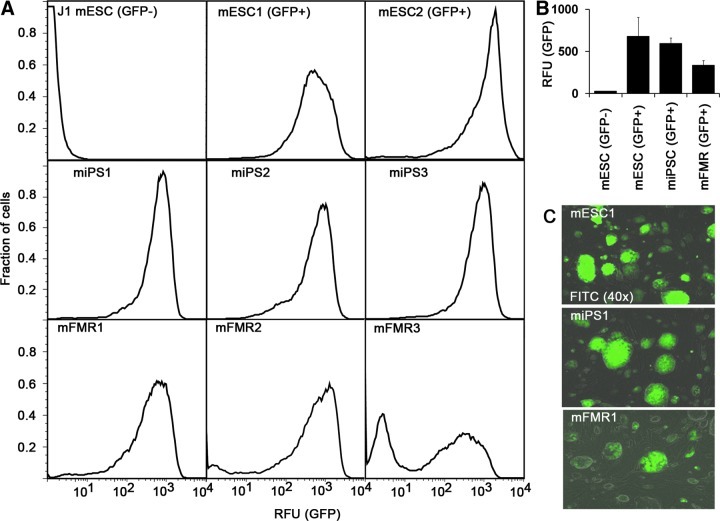
Pluripotent cells can be produced by derivation of ESCs from blastocysts, iPSCs, and by fusion of ESCs with fibroblasts resulting in near-tetraploid FMRCs. We obtained a transgenic mouse strain harboring a transgene in which the Oct4 promoter and enhancers drive eGFP (Boiani et al., 2002; Szabo et al., 2002). This transgene has been previously shown to drive the expression of GFP in mouse ESCs and preimplantation embryos. From this mouse strain, we derived ESCs, iPSCs, and FMRCs, resulting in three different pluripotent cell types in which the transgene resides in an identical genomic location. We used these cell lines to see if Oct4 expression was similar or divergent in the three cell types, in which pluripotency was achieved by three differing routes (i.e., by endogenous regulation in the case of ESCs, and by reprogramming in the case of iPSCs and FMRCs). We used flow cytometry to determine GFP fluorescence as an indicator of Oct4 expression in these cells (Fig. 1A). In the presence of LIF, both mESC cell lines (mESC1 and mESC2) and iPSCs cell lines (miPSC1, miPS2, and miPS3) gave rise to robust GFP signals that were very similar to each other (Fig 1A, B). However, FMRCs (cell lines mFMR1, mFMR2, and mFMR3) expressed only about two-thirds as much Oct4-controlled GFP fluorescence as compared to ESCs and iPSCs (Fig. 1B). Moreover, both FACS analysis (Fig. 1A) and epifluorescence combined with phase-contrast microscopy (Fig. 1C) indicated that cultures of FMRCs contained a significant number of live cells harboring low amounts of GFP fluorescence, whereas similar cells in cultures of ESCs and iPSCs were nearly absent. Although colonies containing cells with low GFP amounts were found in cultures from all three cell lines derived by FMR, we observed significant differences in numbers of cells with low GFP fluorescence, ranging from only a few in the FMR1 line to approximately one-third in the FMR3 line. If we restricted our attention to the high-GFP populations of the FMRCs, two of the lines (mFMR1 and mFMR2) expressed GFP at levels comparable to other pluripotent cells analyzed in this study. FMRCs maintain only a roughly tetraploid complement of chromosomes (Ambrosi et al., 2007). Therefore, it is possible that the GFP-negative cells observed in lines mFMR2 and mFMR3 have lost the chromosome containing the Oct4 promoter–GFP transgene.
FIG. 1.
Oct4 promoter activity in blastocyst, FMR, and iPSC-derived pluripotent stem cells. (A) Histograms of GFP fluorescence emanating from the Oct4-GFP transgene were assayed by FACS for 9 cell lines. Proportions of cells with similar fluorescence (y axis) were plotted as a function of the relative fluorescence units from GFP containing cells (x axis). Only live cells, identified by their forward and side-scatter characteristics were gated and validated by PI exclusion staining (not shown). Nontransgenic J1 mouse ESCs were used to establish the gate for GFP− cells, and this gate was applied to all other populations to identify GFP+ cells. For each assay, 50,000 GFP+ cells were counted using the FL1-H(PE) detector. (B) Graphical representation of the mean GFP fluorescence contents per cell for GFP+ mESCs, iPSCs, and FMR cells. For GFP− cells, only a single mESC line was assayed, and the mean for GFP+ mESCs was derived from two stem cell lines. Note that the average GFP contents value is given as the geometric means from each three independent cell lines (iPSCs and FMRCs) or two independent cell lines (mESCs). (C) Morphology of ESCs, iPSCs, and FMR cell colonies (visible) overlaid with Oct4-GFP fluorescence. Images of three representative cell lines used in this study are shown. Of note is the greater heterogeneity in Oct4 promoter-driven GFP fluorescence observed in the FMR-derived cell line.
Cell cycle kinetics in mouse PSCs
It is well established that the cell cycle in pluripotent and differentiated cells differs with respect to the presence of checkpoints, length, and presence of the G0 stage. In human ESCs, Oct4 together with Sox2 controls expression of essential cell cycle regulators, including cyclin D1 via regulation of the microRNA miR302a, to produce the characteristic stem cell cell cycle profile (Card et al., 2008). Moreover, in mESCs, Oct4 amounts are fined-tuned for the maintenance of the pluripotent state; even two-fold variations in either direction induce differentiation into endoderm or trophectoderm, respectively (Niwa et al., 2000), and this exit from pluripotency induces differentiation-associated alterations in the cell cycle profile. Intriguingly, we observed modest but detectable variations of the Oct4 amounts in iPSCs and FMRCs compared to mESCs and sought to determine if these changes affected the cell cycle profile in the different cell lines (Table 1). Overall, the cell cycle characteristics were relatively similar for all three classes of cells. Nevertheless, we observed an increase of cells in G1 and G2/M for iPSCs and FMRCs, but this effect was only very modest, and the variation among the cell lines within each group of ESCs, iPSCs and FMRCs did not correlate with Oct4 amounts detected (Fig. 1).
Table 1.
Comparative Cell Cycle Analysis of Pluripotent Stem Cells
| Cell line | % G1 | % S | % G2 |
|---|---|---|---|
| J1 | 17.6 | 60.1 | 17.1 |
| mESC1 | 11.1 | 65.2 | 16.5 |
| mESC2 | 8.3 | 63.7 | 15.2 |
| Mean±SEM | 12.3±2.75 | 63.0±1.51 | 16.3±0.56 |
| miPS1 | 22.2 | 45.8 | 28.9 |
| miPS2 | 16.5 | 57.1 | 23 |
| miPS3 | 19.7 | 53.1 | 25.0 |
| Mean±SEM | 19.5±1.65 | 52.0±3.31 | 25.6±1.73 |
| FMR1 | 23.1 | 44.7 | 27.9 |
| FMR2 | 17.4 | 60.0 | 19.8 |
| FMR3 | 15.2 | 58.6 | 21.9 |
| Mean±SEM | 18.6±2.35 | 54.4±4.88 | 21.9±2.43 |
Gating for GFP+ cells was as described. Propidium iodide–based fluorescence was measured after compensation for GFP fluorescence spillover in the Fl2-A (APC) channel. A total of 50,000 events were recorded. The population sizes in the respective cell cycle stages were estimated using the Jett–Dean algorithm for cell cycle modeling.
ESC, embryonic stem cells; SEM, standard error of the mean; FMR, fusion-mediated reprogramming; GFP, green fluorescent protein.
Pluripotency marker expression during EB-based and ATRA-directed differentiation
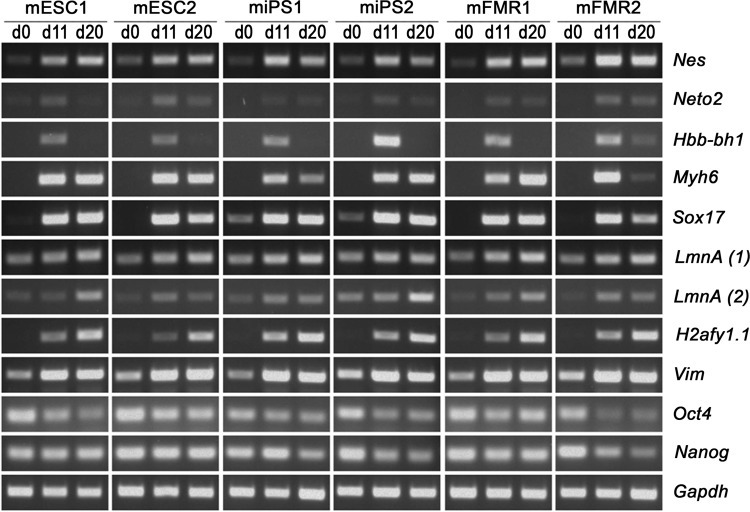
The pluripotency of all cell lines in this study was confirmed by semiquantitative PCR using mRNA derived from EBs after 11 and 20 days of differentiation, respectively (Fig. 2). Expression of the neuronal differentiation markers Nes and Neto2 (Fig. 2) was concordant with their expression in cells induced to neuronal differentiation by ATRA (Fig. 2), as determined by qRT-PCR. We also observed the upregulation of the mesoderm markers Hbb-bh1 and Myh6 in all ESCs, iPSCs, and FMRCs in day-11 EBs (Fig. 2). Interestingly, whereas Hbb-bh1expression consistently declined in all cell lines in this study in day 20 EBs, Myh6 expression remained high in all EBs except those derived from mFMR2 cells, in which expression declined at that time. Likewise, the endoderm-specific differentiation marker Sox17, absent in PSCs, was upregulated in both day-11 and day-20 EBs. Remarkably, we also detected the expression of LMNA, an early differentiation marker for ESCs (Mattout and Meshorer, 2009) in undifferentiated PSCs. The later differentiation marker H2afy1.1 was absent in PSCs and induced in differentiated cells at both day 11 and day 20. Vim, a marker for both endoderm and mesoderm differentiation (Page, 1989), demonstrated an expression profile similar to H2afy1.1 and Sox17. Finally, expression of the pluripotency markers Oct4 and Nanog was also concordant with the expression profiles obtained by qRT-PCR with RNA from ATRA-treated cells for neural induction. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a loading control. Together, these data indicate the ready ability of the cell lines used in this study to differentiate into all three germ layers.
FIG. 2.
Analysis of mRNAs for pluripotency and differentiation markers during EB-based differentiation of mPSCs by semiquantitative RT-PCR. mRNA amounts were determined at day 0, day 11, and day 20 of the differentiation time course. GAPDH mRNA amounts were used as loading control.
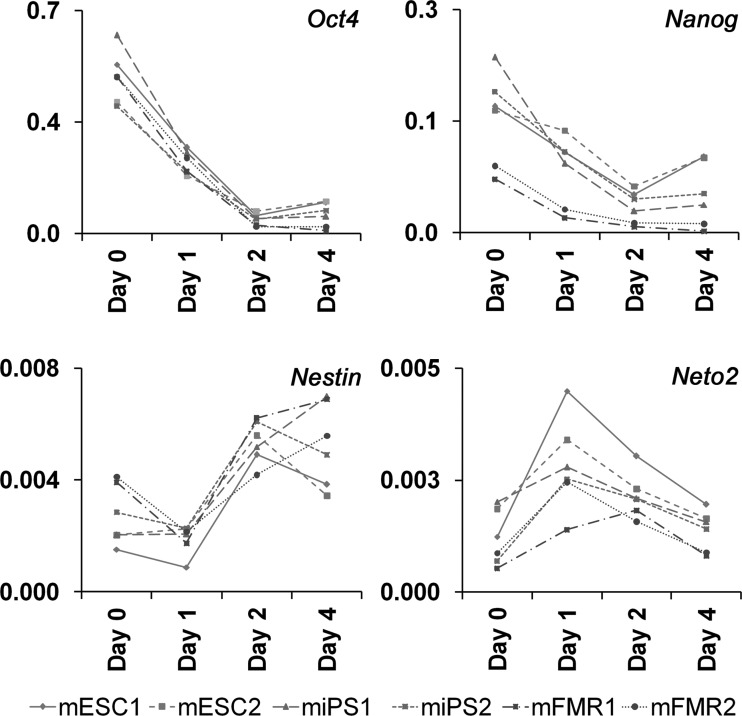
We next looked at the behavior of two pluripotency markers (Oct4 and Nanog) and the ectoderm markers Nestin (Nes) and Neuropilin (Neto) at the level of transcription by qRT-PCR during the course of differentiation induced with ATRA (Fig. 3). For all cell lines in this study, the drop in Oct4 mRNA amounts paralleled differentiation with similar kinetics for the decrease. Similarly, the expression profile for Nanog paralleled the one for Oct4 mRNA (Fig. 3). Likewise, ATRA treatment yielded the upregulation of the neural progenitor marker Nes within 48 h, but Nes mRNA amounts declined thereafter in both mESCs and iPSCs. Interestingly, in FMRCs the Nes mRNA amounts continued to increase beyond 2 days (Fig. 3). Finally, Neto2 mRNA was upregulated within 24 h and declined thereafter in all cell lines employed in this analysis (Fig. 3). We conclude that Oct4 and Nanog are downregulated during the course of differentiation for all cell lines tested, and that all lines have roughly similar patterns of neuroectodermal induction, although Nes behaved slightly differently in FMRCs.
FIG. 3.
Analysis of Oct4, Nanog, Nes, and Neto2 mRNA expression during ATRA-directed differentiation of PSCs into neuronal progenitors by qRT-PCR. Expression levels are given for each individual cell line over a 4-day ATRA time course. Of note is the absence of upregulation for endogenous Oct4 expression both in ESCs and FMRCs. Respective primers are listed in Table S1.
Oct4 promoter activity in mouse PSCs during the exit from pluripotency
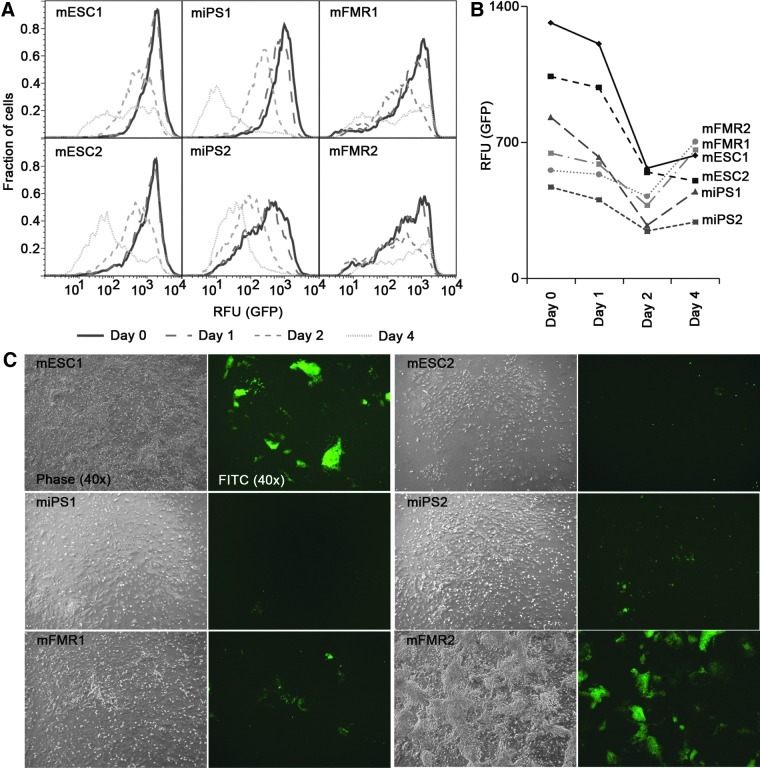
We next investigated the behavior of GFP expression in a similar 4-day ATRA differentiation experiment. In ATRA-treated cultures, we observed the downregulation of Oct4 promoter activity (as judged by GFP fluorescence assessed by flow cytometry) over the course of exposure to the differentiation agent (Fig. 4A, B). All cell lines showed a progressive loss of GFP fluorescence over the first 3 days of the differentiation time course. However, by day 4, we observed for cell lines mESC1, mFMR1, and mFMR2 that the Oct4 transgene was reactivated (Fig. 4A–C). In contrast, no promoter reactivation was seen in cells of iPSC origin (Fig. 4A, B). Quantitative flow cytometric analysis also demonstrated that the maximum decline in GFP amounts occurred within 48 h. The transgene reactivation was approximately 50% in FMRCs (Fig. 4B). In the same cells that reactivated the transgene, endogenous Oct4 mRNA levels declined to low points at day 4 (Fig. 4). Therefore, we conclude that the Oct4-GFP transgene does not faithfully indicate the behavior of the endogenous promoter at later time points during ATRA differentiation. However, the initial rate for the exit from pluripotency appeared highly similar in all PSCs, irrespective of the method by which they were generated (Fig. 4B). Together, these data indicate the pluripotency for all cell lines used in this study.
FIG. 4.
Kinetic analysis of marker expression during ATRA-induced differentiation of pluripotent stem cells into neuronal progenitor cells. (A) Loss of Oct4 expression during ATRA-induced differentiation by FACS. GFP+ cells were quantified as described above, and a total of 10,000 cells were counted. For comparison, the fraction of GFP+ cells was normalized to the number of GFP+ cells at day 0 and plotted. Note the surprising reactivation of Oct4 promoter activity at day 4 of ATRA induced differentiation in both ESCs and FMR cells. (B) Graphical representation of the mean GFP fluorescence contents per cell for GFP+ mESCs, iPSCs, and FMRCs during ATRA-induced differentiation. Note that GFP levels are represented as the geometric mean GFP contents per cell. (C) Assessment of Oct4 promoter activity by phase and fluorescence microscopy, respectively, at day 4 of ATRA-induced differentiation for the cell lines used in this study. All panel pairs show residual Oct4-driven GRP levels after 4 days of ATRA treatment. Note the presence of numerous GFP+ colonies in mESC1, mFMR, and mFMR2 cells as compared to iPSCs.
Discussion
No study has yet been conducted comparing the pluripotency of ESCs, iPSCs, and FMRCs. The results of such a comparison could demarcate whether simple expression of pluripotency factors and subsequent reprogramming through a stochastic process (Yamanaka, 2009) is sufficient for the faithful reset of the somatic genome to an embryonic ground state or if the more ordered epigenetic rearrangement occurring during gametogenesis (Lees-Murdock and Walsh, 2008) and early embryogenesis (Corry et al., 2009; Santos et al., 2005; van der Heijden et al., 2005) is essential for the generation of the epigenetic state specific to inner cell mass cells. For instance, the amounts of reprogramming factors available for somatic reprogramming through cell fusion might be lower than for natural reprogramming or reprogramming through overexpression of Oct4, Sox2, Klf4, and c-Myc, and the PSCs resulting from FMR may have then maintained a larger proportion of the somatic epigenome. The increased retention of epigenetic memory could interfere with the pluripotency of the stem cells by fine tuning amounts of Oct4 and other key pluripotency factors, thereby increasing the chance for spontaneous exit from pluripotency. Examples of genes that affect Oct4 amounts in PSCs include Brg-1 (Kidder et al., 2009), Npr-A (Abdelalim and Tooyama, 2011), and Geminin (Yang et al., 2011b). Support for the hypothesis that variations in Oct4 amounts are pivotal for the spontaneous exit from pluripotency comes from the observation that small changes in the amounts of pluripotency genes may be sufficient to permit progression of differentiation (Zhu et al., 2007). Moreover, in SCNT, evidence exists that the levels of Oct4 appear to be of particular importance for faithful reprogramming and may serve as an indicator for the failed reset of the genetic program leading to a low frequency in ESC derivation from somatic cells (Boiani et al., 2002). It is noteworthy in this context that that Oct4 levels alone direct the cell fate of mESCs and its approximately two-fold up- or downregulation is linked to induction of differentiation to either endoderm and mesoderm or trophoectoderm, respectively (Niwa et al., 2000).
Among the cell lines employed in this study, we did observe some variations in Oct4 amounts when assayed by FACS for Oct4-dependent GFP expression (Fig. 1). However, the differences among the cell lines were much smaller when endogenous Oct4 amounts were determined by population-wide qRT-PCR (Fig. 3). Moreover, these differences were unrelated to a particular reprogramming route and occurred among clones derived by the same procedure. Therefore, it is likely that these variations reflect the extent of somatic reprogramming within each individual cell line. Nevertheless, the minor variations observed did not seem to interfere with the execution of differentiation in EBs because the differentiation markers for all three germ layers were equally upregulated. Moreover, the associated downregulation of pluripotency markers during EB formation equally did not reveal any differences among mESCs, miPSCs, and mPSCs generated by FMR. Similarly, the downregulation of Oct4 expression during ATRA-induced neuronal differentiation followed a similar kinetic for all cell lines employed in this study.
In our cell cycle analysis, we observed that artificially reprogrammed cells, in comparison to naturally reprogrammed ESCs, were retained in G2/M for a longer period of time, and this was compensated for by generally shorter times in S phase. Remarkably, this effect was inversely proportional to the amounts of Oct4 expressed in the cell lines analyzed. The shorter retention time in S phase may reflect that the cell cycle in PSCs is also subject to control by additional regulators that act in parallel to the indirect regulation of the cell cycle regulator cyclin D1 (Card et al., 2008) by Oct4. Examples include Nucleolin (Yang et al., 2011a), Aire, and Deaf1 (Gu et al., 2010), which exert their control independently of Oct4 in mESCs. Further research is necessary to illuminate the significance and causes for the prolonged retention time in G2.
Overall, we found that Oct4 expression, as measured by GFP and directly by qRT-PCR, was rather similar in cells of all three types (i.e., normal ESCs, iPSCs, and FRMCs). All three cell types are reasonably pluripotent as judged by EB formation (this study) and other research (Ambrosi et al., 2007; Battulin et al., 2012; Gridina and Serov, 2010). FMRCs performed well in EB differentiation, but are not as pluripotent as the other two cell types because FMRCs cannot yield mice by tetraploid embryo complementation, although they can contribute to chimeras (Battulin et al., 2012). Upon differentiation to EBs, we found that Oct4 was effectively silenced, as judged by both GFP expression and qRT-PCR for the endogenous Oct4 gene. We cannot rule out the formal possibility that the Oct4 promoter driving the GFP reporter gene was silenced due to methylation, although this seems unlikely because the endogenous Oct4 gene was downregulated with similar kinetics upon differentiation (Fig. 3). Of all of the three cell types, FMRCs were the least stable in terms of their Oct4 expression, and some colonies became negative for Oct4-driven GFP. This could be either due to silencing of the transgene, or loss of the chromosome carrying the transgene, which is a distinct possibility due to karyotypic instability in these cells (Ambrosi et al., 2007).
We conclude that all three reprogramming methods delivered PSCs that, on the single-cell level as assayed by FACS analysis, performed similarly in the regulation of Oct4, exit from pluripotency, and expression of differentiation markers during EB formation, suggesting that the cells produced are of similar quality. Therefore, we propose the inclusion of FACS-based cell cycle analyses for the validation of PSCs. Finally, because pluripotent cells of all three origins (ESC, iPSC, and FMRC) behaved similarly, we surmise that the pluripotent state is a “local attractor” state for gene expression because it can be readily achieved by three very different routes.
Supplementary Material
Acknowledgments
This material is based upon work supported by the State of Connecticut under the Connecticut Stem Cell Research Grants Program grant 09-SCB-UCON-18. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the State of Connecticut, the Department of Public Health of the State of Connecticut or Connecticut Innovations, Inc.
Author Disclosure Statement
The authors declare that no competing financial interests exist.
References
- Abdelalim E.M. Tooyama I. NPR-A regulates self-renewal and pluripotency of embryonic stem cells. Cell Death Dis. 2011;2:e127. doi: 10.1038/cddis.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosi D.J. Tanasijevic B. Kaur A. Obergfell C. O'Neill R.J. Krueger W. Rasmussen T.P. Genome-wide reprogramming in hybrids of somatic cells and embryonic stem cells. Stem Cells. 2007;25:1104–1113. doi: 10.1634/stemcells.2006-0532. [DOI] [PubMed] [Google Scholar]
- Battulin N.R. Khabarova A.A. Boyarskikh U.A. Menzorov A.G. Filipenko M.L. Serov O.L. Reprogramming somatic cells by fusion with embryonic stem cells does not cause silencing of the Dlk1-Dio3 region in mice. World J. Stem Cells. 2012;4:87–93. doi: 10.4252/wjsc.v4.i8.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiani M. Eckardt S. Scholer H.R. McLaughlin K.J. Oct4 distribution and level in mouse clones: Consequences for pluripotency. Genes Dev. 2002;16:1209–1219. doi: 10.1101/gad.966002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourc'his D. Le Bourhis D. Patin D. Niveleau A. Comizzoli P. Renard J.P. Viegas-Pequignot E. Delayed and incomplete reprogramming of chromosome methylation patterns in bovine cloned embryos. Curr. Biol. 2001;11:1542–1546. doi: 10.1016/s0960-9822(01)00480-8. [DOI] [PubMed] [Google Scholar]
- Boyer L.A. Lee T.I. Cole M.F. Johnstone S.E. Levine S.S. Zucker J.P. Guenther M.G. Kumar R.M. Murray H.L. Jenner R.G., et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Card D.A. Hebbar P.B. Li L. Trotter K.W. Komatsu Y. Mishina Y. Archer T.K. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol. Cell. Biol. 2008;28:6426–6438. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corry G.N. Tanasijevic B. Barry E.R. Krueger W. Rasmussen T.P. Epigenetic regulatory mechanisms during preimplantation development. Birth Defects Res. C Embryo Today. 2009;87:297–313. doi: 10.1002/bdrc.20165. [DOI] [PubMed] [Google Scholar]
- Dean W. Santos F. Stojkovic M. Zakhartchenko V. Walter J. Wolf E. Reik W. Conservation of methylation reprogramming in mammalian development: Aberrant reprogramming in cloned embryos. Proc. Natl. Acad. Sci. USA. 2001;98:13734–13738. doi: 10.1073/pnas.241522698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gridina M.M. Serov O.L. Bidirectional reprogramming of mouse embryonic stem cell/fibroblast hybrid cells is initiated at the heterokaryon stage. Cell Tiss. Res. 2010;342:377–389. doi: 10.1007/s00441-010-1085-2. [DOI] [PubMed] [Google Scholar]
- Gu B. Zhang J. Chen Q. Tao B. Wang W. Zhou Y. Chen L. Liu Y. Zhang M. Aire regulates the expression of differentiation-associated genes and self-renewal of embryonic stem cells. Biochem. Biophys. Res. Commun. 2010;394:418–423. doi: 10.1016/j.bbrc.2010.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J. Saha K. Pando B. van Zon J. Lengner C.J. Creyghton M.P. van Oudenaarden A. Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan N.R. Wolvetang E.J. Adipocyte differentiation in human embryonic stem cells transduced with Oct4 shRNA lentivirus. Stem Cells Dev. 2009;18:653–660. doi: 10.1089/scd.2008.0160. [DOI] [PubMed] [Google Scholar]
- Kidder B.L. Palmer S. Knott J.G. SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells. 2009;27:317–328. doi: 10.1634/stemcells.2008-0710. [DOI] [PubMed] [Google Scholar]
- Kim K. Doi A. Wen B. Ng K. Zhao R. Cahan P. Kim J. Aryee M.J. Ji H. Ehrlich L.I., et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. Zhao R. Doi A. Ng K. Unternaehrer J. Cahan P. Huo H. Loh Y.H. Aryee M.J. Lensch M.W., et al. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat. Biotechnol. 2011;29:1117–1119. doi: 10.1038/nbt.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger W.H. Swanson L.C. Tanasijevic B. Rasmussen T.P. Natural and artificial routes to pluripotency. Int. J. Dev. Biol. 2010;54:1545–1564. doi: 10.1387/ijdb.103199wk. [DOI] [PubMed] [Google Scholar]
- Lees-Murdock D.J. Walsh C.P. DNA methylation reprogramming in the germ line. Adv. Exp. Med. Biol. 2008;626:1–15. doi: 10.1007/978-0-387-77576-0_1. [DOI] [PubMed] [Google Scholar]
- Livak K.J. Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mattout A. Meshorer Chromatin plasticity and genome organization in pluripotent embryonic stem cells. Curr. Opin. Cell Biol. 2010;22:334–341. doi: 10.1016/j.ceb.2010.02.001. [DOI] [PubMed] [Google Scholar]
- Niwa H. Miyazaki J. Smith A.G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Page M. Changing patterns of cytokeratins and vimentin in the early chick embryo. Development. 1989;105:97–107. doi: 10.1242/dev.105.1.97. [DOI] [PubMed] [Google Scholar]
- Pasi C.E. Dereli-Oz A. Negrini S. Friedli M. Fragola G. Lombardo A. Van Houwe G. Naldini L. Casola S. Testa G., et al. Genomic instability in induced stem cells. Cell Death Differ. 2011;18:745–753. doi: 10.1038/cdd.2011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos F. Peters A.H. Otte A.P. Reik W. Dean W. Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Dev. Biol. 2005;280:225–236. doi: 10.1016/j.ydbio.2005.01.025. [DOI] [PubMed] [Google Scholar]
- Seiler K. Soroush Noghabi M. Karjalainen K. Hummel M. Melchers F. Tsuneto M. Induced pluripotent stem cells expressing elevated levels of sox-2, oct-4, and klf-4 are severely reduced in their differentiation from mesodermal to hematopoietic progenitor cells. Stem Cells Dev. 2011;20:1131–1142. doi: 10.1089/scd.2010.0391. [DOI] [PubMed] [Google Scholar]
- Shi W. Zakhartchenko V. Wolf E. Epigenetic reprogramming in mammalian nuclear transfer. Differentiation. 2003;71:91–113. doi: 10.1046/j.1432-0436.2003.710201.x. [DOI] [PubMed] [Google Scholar]
- Sullivan G.J. Bai Y. Fletcher J. Wilmut I. Induced pluripotent stem cells: Epigenetic memories and practical implications. Mol. Hum. Reprod. 2010;16:880–885. doi: 10.1093/molehr/gaq091. [DOI] [PubMed] [Google Scholar]
- Szabo P.E. Hubner K. Scholer H. Mann J.R. Allele-specific expression of imprinted genes in mouse migratory primordial germ cells. Mech. Dev. 2002;115:157–160. doi: 10.1016/s0925-4773(02)00087-4. [DOI] [PubMed] [Google Scholar]
- Takahashi K. Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- van der Heijden G.W. Dieker J.W. Derijck A.A. Muller S. Berden J.H. Braat D.D. van der Vlag J. de Boer P. Asymmetry in histone H3 variants and lysine methylation between paternal and maternal chromatin of the early mouse zygote. Mech. Dev. 2005;122:1008–1022. doi: 10.1016/j.mod.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Xu S.J. Xu Y.X. Liu H.L. [Mechanisms of epigenetic reprogramming in Mammalian resonstructed embryos produced by nuclear transfer.] Yi Chuan. 2005;27:473–480. [PubMed] [Google Scholar]
- Xue F. Tian X.C. Du F. Kubota C. Taneja M. Dinnyes A. Dai Y. Levine H. Pereira L.V. Yang X. Aberrant patterns of X chromosome inactivation in bovine clones. Nat. Genet. 2002;31:216–220. doi: 10.1038/ng900. [DOI] [PubMed] [Google Scholar]
- Yamanaka S. Elite and stochastic models for induced pluripotent stem cell generation. Nature. 2009;460:49–52. doi: 10.1038/nature08180. [DOI] [PubMed] [Google Scholar]
- Yang A. Shi G. Zhou C. Lu R. Li H. Sun L. Jin Y. Nucleolin maintains embryonic stem cell self-renewal by suppression of p53 protein-dependent pathway. J. Biol. Chem. 2011a;286:43370–43382. doi: 10.1074/jbc.M111.225185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang V.S. Carter S.A. Hyland S.J. Tachibana-Konwalski K. Laskey R.A. Gonzalez M.A. Geminin escapes degradation in G1 of mouse pluripotent cells and mediates the expression of Oct4, Sox2, and Nanog. Curr. Biol. 2011b;21:692–699. doi: 10.1016/j.cub.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H. Yang H. Owen M.R. Combined microarray analysis uncovers self-renewal related signaling in mouse embryonic stem cells. Syst. Synth. Biol. 2007;1:171–181. doi: 10.1007/s11693-008-9015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.