SUMMARY
The endocycle is a variant cell cycle consisting of successive DNA synthesis and Gap phases that yield highly polyploid cells. Although essential for metazoan development, relatively little is known about its control or physiologic role in mammals. Using novel lineage-specific cre mice we identified two opposing arms of the E2F program, one driven by canonical transcription activation (E2F1, E2F2 and E2F3) and the other by atypical repression (E2F7 and E2F8), that converge on the regulation of endocycles in vivo. Ablation of canonical activators in the two endocycling tissues of mammals, trophoblast giant cells in the placenta and hepatocytes in the liver, augmented genome ploidy, whereas ablation of atypical repressors diminished ploidy. These two antagonistic arms coordinate the expression of a unique G2/M transcriptional program that is critical for mitosis, karyokinesis and cytokinesis. These results provide in vivo evidence for a direct role of E2F family members in regulating non-traditional cell cycles in mammals.
The archetypal cell division cycle culminates in the generation of genetically identical diploid progenies. However, a number of cell cycle variations in nature exist, with the endocycle being a remarkable variant utilized by a wide range of organisms in diverse environmental contexts1. This modified cell cycle is characterized by alternating DNA synthesis (S) and Gap (G) phases in the absence of intervening mitosis (M), karyokinesis and cytokinesis. As a result of successive rounds of whole genome duplication, cells may achieve genome polyploidy that exceed 1000C2,3. The biological advantages of polyploidy have been proposed to include an augmented response to strenuous cellular metabolic demands; dampened sensitivity to apoptotic or checkpoint stimuli4; and buffering against deleterious mutations that may cause cancer5,6.
The molecular mechanisms that control mitotic and variant cell cycles among species are presumed to be similar. In the textbook view of cell cycle control, E2F transcriptional activators and repressors are portrayed to orchestrate a gene expression program essential for mitotic cell cycle progression7, with E2F activators and repressors oppositely regulating a common set of target genes. However, careful analyses of mouse knockout tissues demonstrate that E2F function in vivo do not strictly adhere to this elegant dichotomous paradigm8–16. This is perhaps not surprising given the large number of genes in the mammalian E2F family, which consists of eight distinct genes encoding at least nine structurally related protein products that can be divided into activator or repressor subclasses17. Canonical E2F activators (E2F1, E2F2, E2F3a and E2F3b) have a single DNA binding domain (DBD), heterodimerize with DP1/DP2 proteins and associate with co-activator proteins to robustly induce RNA polymerase II-dependent gene expression18. Canonical repressors (E2F4, E2F5 and E2F6) also have a single DBD, heterodimerize with DP1/DP2 proteins and associate with chromatin modifying proteins to repress gene expression18. The most recently identified family members encode the atypical repressors E2F7 and E2F8, which have two tandem DBDs and repress gene expression independent of DP1/DP2 heterodimerization19.
While flies and plants use endocycles as a mechanism for cell growth in a variety of tissue types1,20, endocycles in mammals are mainly restricted to trophoblast giant cells (TGCs) in the placenta and hepatocytes in the liver1,5. Endocycles in these mammalian tissues are believed to be integral to organ physiology1,5, however, surprisingly little evidence exists to support this contention. Here we developed novel genetic tools in mice and show by depletion of either canonical E2F activators or atypical E2F repressors that these two separate E2F transcriptional programs converge to control the endocycle through the regulation of cellular events important for mitosis, karyokinesis and cytokinesis. Surprisingly, placentas and livers with severely restricted polyploidization retained sufficient physiological function to carry them through apparently normal development.
RESULTS
E2f1-3 Ablation Results in Hyperploidy
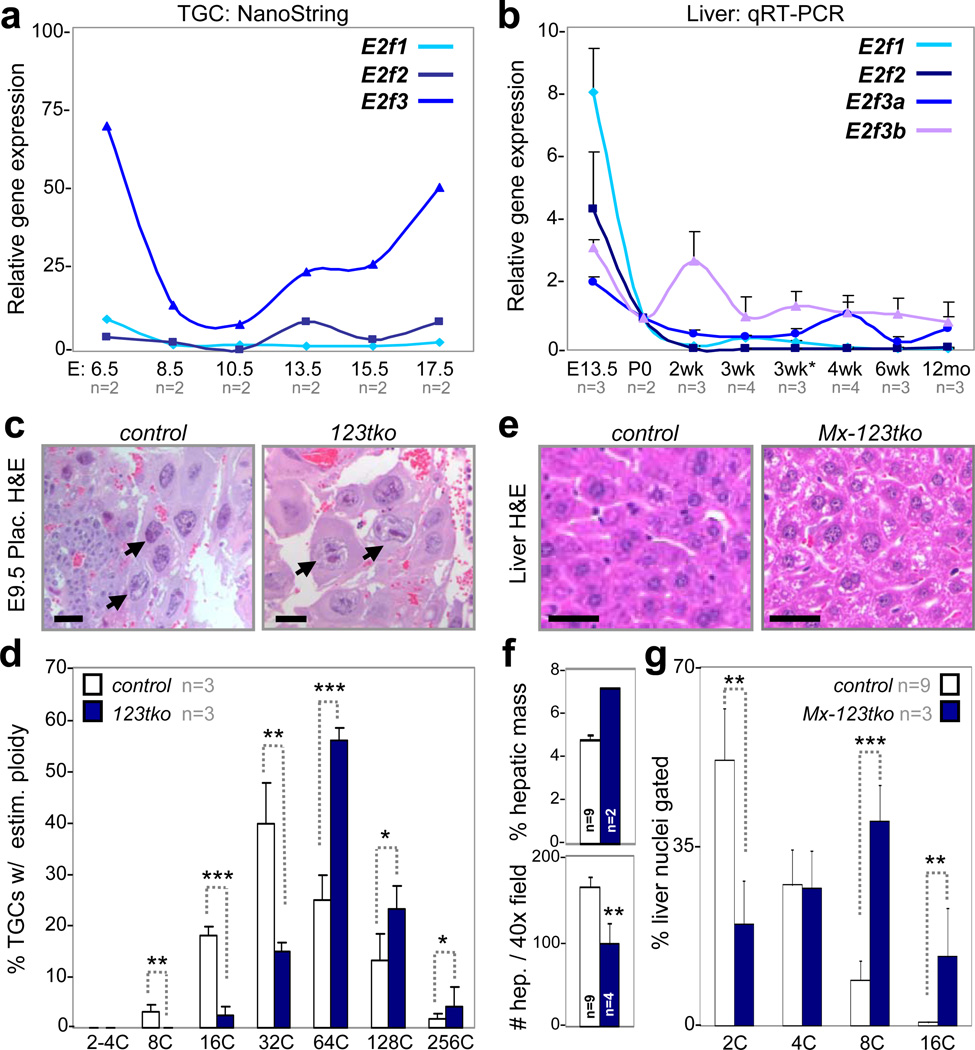
E2F mediated transcription contributes to endocycle control in flies and plants1,5,19–21, but its role in mammals has not been thoroughly evaluated. Whole genome duplication in mammalian endocycling cells is temporally regulated during development with major ploidy increases in TGCs during mid gestation and in hepatocytes following weaning at around three weeks of age22,23. The levels of E2f1 and E2f2 mRNAs in laser capture microdissected TGCs from wildtype placentas, as measured by NanoString technology (Fig. 1a), were low throughout placental development, whereas the level of E2f3 mRNA was high prior to E8.5 and following E13.5. Expression analysis in livers by quantitative real-time polymerase chain reaction (qRT-PCR) showed relatively high levels of E2f1 and E2f2 during fetal development and a precipitous decrease following birth (P0), whereas expression of E2f3 (E2f3a and E2f3b) decreased gradually following birth and was almost undetectable post-weaning (Fig. 1b). Consistent with the expression pattern of their mRNAs, E2F3a and E2F3b protein levels decreased in newborn pups post weaning (Supplementary Fig. 1a), while E2F1 and E2F2 proteins could not be detected beyond the intrauterine period (data not shown). Thus, it would appear that total E2F1-3 activator levels are minimized when TGCs and hepatocytes are most actively endocycling.
Figure 1. Loss of E2f activators promotes TGC and hepatocyte endocycles.
a, NanoString analysis of TGC-specific E2f1-3 expression in laser capture microdissected wild type cells, n=2 placentas analyzed per time point. b, Quantitative RT-PCR analysis of E2f activators in pre- (E13.5) and post-natal (P0-12mo) wild type livers, with n per time point as indicated. 3wk*, 1 day post weaning (3wk). c, Representative E9.5 H&E placenta sections showing control and E2f1−/−;E2f2−/−;E2f3−/− (123tko) TGCs. Arrows point to selected TGCs. Scale bar, 12.5µm. d, Feulgen quantification of ploidy in E9.5 control and 123tko TGCs, n=3 per genetic group. One-way ANOVA, * p≤0.05, ** p≤0.01, *** p≤0.001. e, Representative H&E sections showing control and Mx-cre;E2f1−/−;E2f2−/−;E2f3f/f (Mx-123tko) livers of 8-week-old mice 10 days after pIpC injection. Scale bar, 10µm. f, Measurements showing enlarged Mx-123tko livers (top graph) and hepatocytes (bottom graph) relative to control tissues and hepatocytes, with n per genetic group as indicated. Two-tailed Student T-test, ** p≤0.01. g, Flow cytometry of control and Mx-123tko liver nuclei from 8-week-old mice, with n per genetic group as indicated. One-way ANOVA, ** p≤0.01, *** p≤0.001. c control, wild type TGCs; e control, pIpC injected Mx-cre;E2f1−/−;E2f2−/−;E2f3+/+ livers; f-g control, wild type livers. Data in a, b, d, f and g reported as average values, ± SD are included when n>2 samples were analyzed.
We utilized targeted gene inactivation strategies in mice to rigorously evaluate the role of E2F activators in endocycling TGCs and hepatocytes. To avoid functional compensation24, we evaluated TGCs in embryos deficient for the entire E2F activator subclass (E2f1-3). Visual inspection of H&E stained E9.5 E2f1−/−;E2f2−/−;E2f3−/− (123tko) placentas revealed the presence of TGCs with abnormally large nuclei (Fig. 1c). Feulgen staining and quantification of genome ploidy by standardized methods (Supplementary Fig. 1b) showed a further shift of the 123tko TGC population towards higher ploidy (Fig. 1d and Supplementary Fig. 1c). Analysis of 123tko TGCs at later stages of development was precluded due to embryo lethality at E10.525,26.
To evaluate the role of the three activator E2Fs in hepatocytes we used pIpC-inducible Mx-cre mice27 and conditional alleles of E2f3 (Mx-cre;E2f1−/−;E2f2−/−;E2f3f/f; Mx-123tko) to examine the consequences of their combined ablation. Injection of pIpC in six-week-old Mx-123tko mice resulted in increased liver weights (Fig. 1f) and proportionately larger hepatocytes (Fig. 1e, f) than age-matched controls. Loss of E2f1 alone or in combination with E2f2 and E2f3 resulted in a gene dose-dependent increase in hepatocyte ploidy (Fig. 1g and Supplementary Fig. 1d). The expression of E2f repressors (E2f4-8) was not affected by loss of E2f1-3 (Supplementary Fig. 1e). Together, these findings suggest that mammalian E2F activators have a physiologic role in suppressing endocycles.
E2f7/E2f8 Ablation Reduces TGC Ploidy
Next, we explored the role of E2F repressors in endocycle control. Expression analysis of the canonical repressors (E2f4-6) showed relatively high levels of E2f4 in TGCs of E6.5 placentas that decreased with time, whereas E2f5-6 expression was relatively constant throughout placental development (Fig. 2a, left panel); expression of E2f4-6 was relatively high in fetal livers and their expression decreased after birth (Supplementary Fig. 2a). Inspection of E2f4−/− or E2f5−/− embryos and mice revealed normal placentas and livers (Supplementary Fig. 2b, c); E2f6−/− placentas and livers were not evaluated.
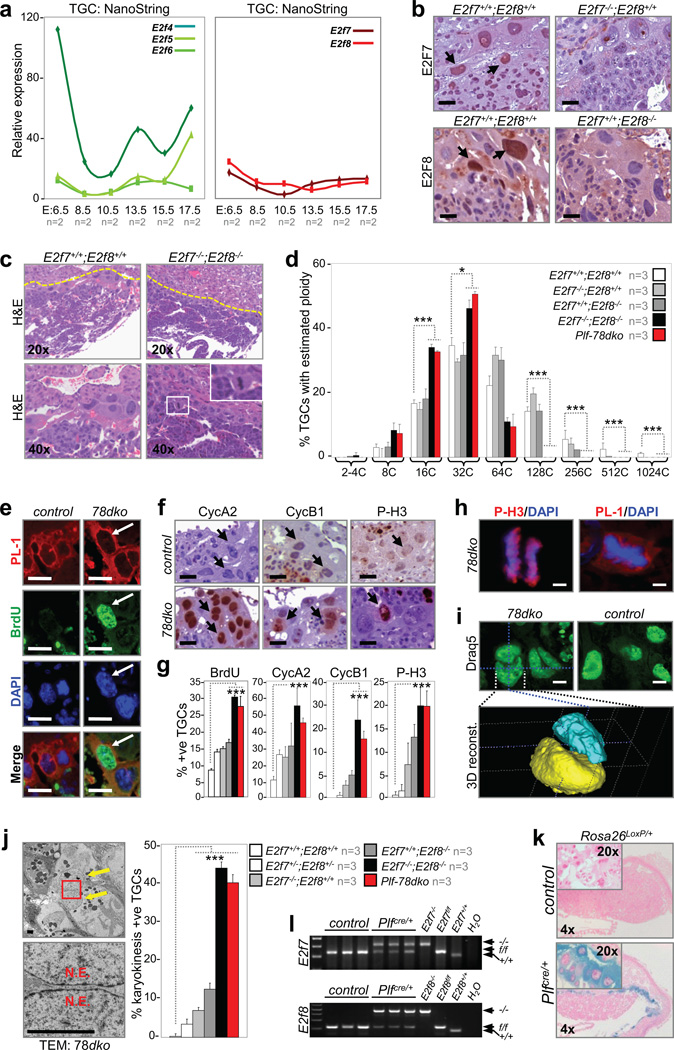
Figure 2. E2f7 and E2f8 promote TGC endocycles.
a, NanoString analysis of TGC-specific E2f4-6 (left panel) and E2f7/E2f8 (right panel) expression in laser capture microdissected wild type cells, n=2 placentas analyzed per time point. b, Immunohistochemistry demonstrating E2F7 (top left) and E2F8 (bottom left) expression in wild type E10.5 TGCs but not mutant controls. Arrows point to selected TGCs. Scale bar, 10µm. c, Representative H&E sections of E10.5 control and E2f7−/−;E2f8−/− (78dko) placentas. Inset, a 78dko TGC in metaphase. d, Feulgen quantification of genome ploidy in E10.5 TGCs, n=3 placentas analyzed per genetic group. e–g, Immunostaining and quantification of S and M phase proteins in E10.5 control and 78dko TGCs. Arrows point to TGCs. n=3 placentas analyzed per genetic group. Scale bar in e–f, 12.5µm. h, Co-immunofluorescence showing E10.5 78dko TGCs in anaphase (left, P-H3) and metaphase (right, PL-1). DAPI stained total DNA. Scale bar, 5µm. i, Representative confocal images of nuclei in E10.5 control and 78dko TGCs (top) and 3D reconstruction of a binucleated 78dko TGC (bottom). Draq5 stained total DNA pseudocolored in green. Scale bar, 10µm. j, Left, transmission electron micrograph of a 78dko E10.5 TGC (left top, arrows indicate two nuclei; left bottom, enlarged view of boxed area showing separation between nuclear envelopes). Right, quantification of binucleated E10.5 TGCs, n=3 placentas analyzed per genetic group. k, X-gal staining of E10.5 control and Plfcre/+ placentas carrying the reporter allele Rosa26LoxP. l, PCR genotyping of genomic DNA isolated from laser capture microdissected E10.5 control and Plfcre/+ TGCs. e, f and i control, E2f7+/+;E2f8+/+; e, f and h–j 78dko, E2f7−/−;E2f8−/−; k control, Plf+/+;Rosa26LoxP/+; l control, Plf+/+;E2f7f/f;E2f8f/f; d, g, and j Plf-78dko, Plfcre/+;E2f7f/f;E2f8f/f. PL-1, placental lactogen 1; N.E., nuclear envelope. Data in a, d, g and j reported as average values, ± SD are included when n>2 samples were analyzed. One-way ANOVA, * p≤0.05; ** p≤0.01; ***p≤0.001.
We then focused our attention on the atypical repressors, E2F7 and E2F8. These E2Fs are expressed in TGCs throughout placental development (Fig. 2a, right panel). Immunohistochemistry (IHC) showed robust staining of E2F7 and E2F8 proteins in a portion of TGCs (Fig. 2b, arrows), consistent with their established cell cycle dependent expression in late S and G2 phases28–31. Visual inspection of E2f7−/−;E2f8−/− placentas at E10.5, one day prior to their embryonic lethality16, revealed a surprising number of TGCs at various stages of mitosis (Fig. 2c), implying an interruption of normal endocycles. Indeed, the ploidy of E2f7−/−;E2f8−/− TGCs never exceeded 64C (Fig. 2d, black bars), whereas wildtype TGCs with genomes >1000C were readily detected (Fig. 2d, white bars). The combined inactivation of E2f7 and E2f8 led to ectopic BrdU incorporation (Fig. 2e and 2g), increased levels of G2/M (cyclin A2) and M phase-specific proteins (cyclin B1 and phosphorylated-H3, P-H3) (Fig. 2f, g) and the appearance of numerous TGCs in metaphase and anaphase (Fig. 2h). Confocal microscopy and 3D reconstruction of serially imaged E2f7−/−;E2f8−/− placentas showed that approximately 40% of TGCs contained at least two nuclei (Fig. 2i), which was confirmed by transmission electron microscopy (Fig. 2j). Placentas lacking E2f7, E2f8, or one allele of each (E2f7+/−;E2f8+/−) exhibited TGCs with an intermediate reduction in ploidy (Fig. 2d and Supplementary Fig. 2d, e) and a proportionate increase in G2/M and M phase related events (Fig. 2g and 2j), once again suggesting a gene dosage effect.
Because expression of trophoblast cell lineage markers was altered in placentas globally deleted for E2f7 and E2f8 (TGCs, labyrinth trophoblasts and spongiotrophoblasts)32, we could not formally rule out a general disruption of differentiation as a possible cause for the endocycle defects observed in mutant TGCs. To specifically ablate E2f7 and E2f8 in TGCs of placentas we crossed Plfcre/+ knockin mice (Fig. 2k and data not shown)32 with E2f7f/f;E2f8f/f mice to yield Plf-78dko embryos. We confirmed the TGC-specific ablation of conditional alleles by PCR genotyping of laser capture microdissected samples (Fig. 2l). The same abnormalities were observed in Plf-78dko TGCs as in E2f7−/−;E2f8−/− TGCs, including the reduced ploidy, increased cyclin A2, cyclin B1 and P-H3 expression levels, and ectopic karyokinesis (Fig. 2d, 2g and 2j; red bars). In summary, these data demonstrate cell autonomous functions for E2F7 and E2F8 in facilitating TGC endocycles by repressing key molecular events required for mitosis and karyokinesis.
E2f7/E2f8 Ablation Prevents Hepatocyte Endocycles
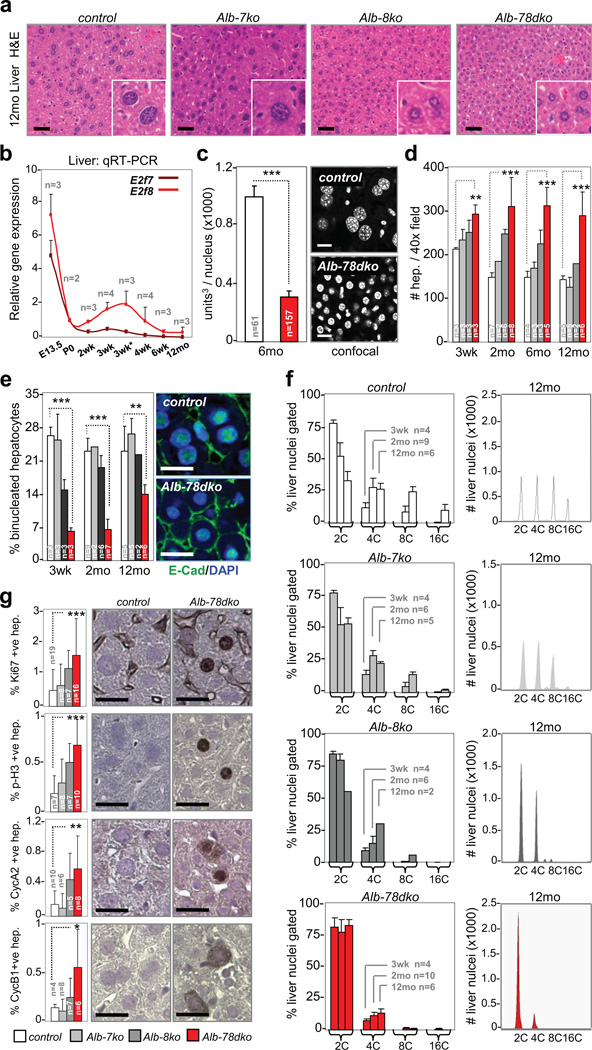
The expression of E2f7 and E2f8 in fetal livers was high but decreased to almost undetectable levels by weaning age (Fig. 3b and Supplementary Fig. 3a). To examine their roles during postnatal liver development we crossed E2f7f/f and E2f8f/f mice with Albumin-cre mice (Alb-cre)33, which express cre in hepatocytes of late term embryos and newborn pups. The conditional inactivation of E2f7 (Alb-7ko), E2f8 (Alb-7ko) or both (Alb-78dko) did not significantly affect hepatic mass (Supplementary Fig. 3b). However, evaluation of H&E and DAPI stained liver sections from Alb-8ko and Alb-78dko mice revealed markedly smaller hepatocytes with smaller nuclei than in control animals (Fig. 3a and 3c). This reduction in cell size was compensated by an increase in the total number of hepatocytes (Fig. 3d). Moreover, Alb-8ko and Alb-78dko livers had a decreased proportion of binucleated hepatocytes (Fig. 3e), which is an early event believed to precede the onset of polyploidization34. Strikingly, flow cytometry showed that hepatocytes in Alb-8ko and Alb-78dko livers remained diploid over the entire lifetime of the mouse, whereas hepatocytes in Alb-7ko livers had only a modest reduction in genome ploidy (Fig. 3f). The percentages of Ki67-, cyclin A2-, cyclin B1- and P-H3-positive hepatocytes were significantly elevated in 2-month-old Alb-8ko mice and even more so in similarly aged Alb-78dko mice (Fig. 3g). Analysis of mice globally deleted for E2f7 (E2f7−/−) or E2f8 (E2f8−/−) yielded identical phenotypes as conditionally deleted Alb-7ko and Alb-8ko mice (Supplementary Fig. 3c, d). Surprisingly, Alb-78dko mice survived to old age in apparent good health and retained the capacity to regenerate their livers upon chemical (carbon tetrachloride) or physical (partial hepatectomy) injury (Supplementary Fig. 3e-h and data not shown). From these results, we conclude that E2F8 is the principal atypical E2F that functions to promote hepatocyte endocycles in mice.
Figure 3. E2f8 is sufficient to promote hepatocyte endocycles.
a, Representative H&E sections of aged (12mo) livers. Scale bar, 12.5µm. b, Quantitative RT-PCR analysis of E2f7 and E2f8 in pre- (E13.5) and post-natal (P0-12mo) wild type livers, with n per time point analyzed as indicated. 3wk*, 1 day post weaning (3wk). c, Left graph, assessment of nuclei volume by confocal imaging and 3-D reconstruction in 6-month-old control and Alb-78dko hepatocytes. n, number of nuclei evaluated per genotype. Right panels, representative confocal images of DAPI-stained liver sections. Scale bar, 10µm. d, Assessment of hepatocyte size in young (3wk), adult (2-6mo) and aged (12mo) livers by quantifying number of hepatocytes per image field, with n per genetic group analyzed as indicated. e, Left graph, quantification of binucleated hepatocytes in young, adult and aged livers, with n per genetic group analyzed as indicated. Right panels, immunofluoresence with anti-E-cadherin marking the boundary of mono- or binucleated hepatocytes. Scale bar, 10µm. f, Left column, flow cytometry of liver nuclei in young, adult and aged livers. n, number of livers analyzed per genotype and age group. Right column, representative FACS profiles of liver nuclei from aged (12mo) mice. g, Immunohistochemistry and quantification of hepatocyte proliferation in 2-month-old livers, with n per genetic group analyzed as indicated. Scale bar, 10µm. control, E2f7f/f;E2f8f/f; Alb-7ko, Alb-cre;E2f7f/f; Alb-8ko, Alb-cre;E2f8f/f; Alb-78dko, Alb-cre;E2f7f/f;E2f8f/f. FACS, fluorescence activated cell sorting. Data in b–g reported as average values, ± SD are included when n>2 samples were analyzed. One-way ANOVA, * p≤0.05; ** p≤0.01; ***p≤0.001.
The onset of genome endoreduplication in hepatocytes coincides with the weaning of pups, but DNA content continues to increase through adulthood even though very little E2f8 is expressed at this time (see Fig. 3b). These observations raised the possibility that E2F8 might only be required for the programmed onset and not the continuation of endocycles. We thus used a tamoxifen-inducible system (SA-creERTT2) to express cre in hepatocytes following birth and/or the weaning of pups35. As determined by qRT-PCR, supplying SA-creERTT2;E2f8f/f mice with a tamoxifen diet beginning at either 7 days (two weeks before weaning, pre-weaning) or 28 days of age (one week following weaning, post-weaning) resulted in the tissue-specific efficient deletion of E2f8 (data not shown). Ablation of E2f8 pre-weaning prevented efficient polyploidization of hepatocytes (Supplementary Fig. 4a-c), whereas ablation post-weaning failed to impact hepatocyte size, binucleation and DNA content (data not shown and Supplementary Fig. 4a, 4b and 4d). These results suggest that E2F8 is largely dispensable for the execution of endocycles following the weaning of pups.
E2F7/E2F8 Regulate Endocycle-related Genes
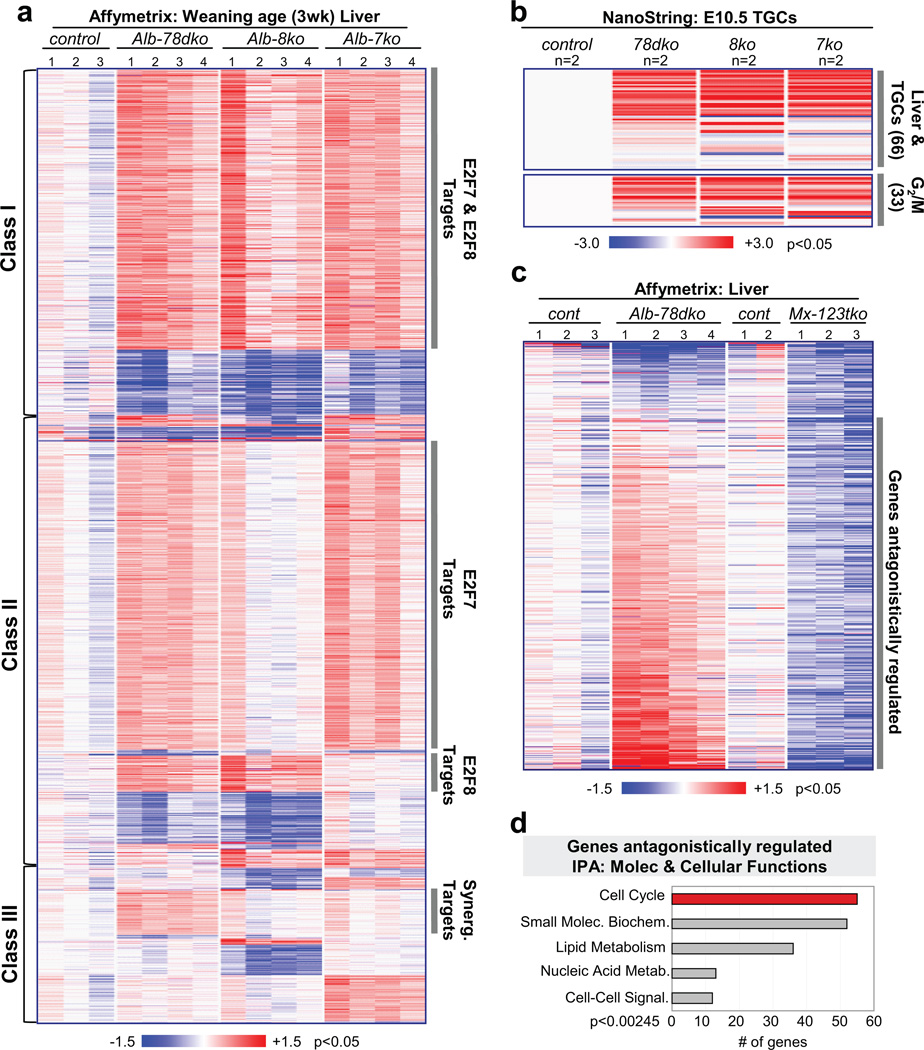
Given the role of E2F7 and E2F8 in transcriptional repression, we anticipated changes in gene expression as the principal underlying cause for the observed endocycle defects in E2f7/E2f8 deficient tissues. Gene expression profiling (Affymetrix Mouse Genome 430 2.0) using RNA purified from livers of 3-week-old wildtype, Alb-7ko, Alb-8ko and Alb-78dko mice showed that there were significantly more upregulated than downregulated genes in mutant tissues, consistent with their roles as repressors (Fig. 4a). Expression changes were confirmed by NanoString technology (Supplementary Fig. 5a). Supervised clustering methods identified three main classes of differentially expressed genes (Fig. 4a). Class I includes genes differentially expressed in all three mutant genetic groups relative to wildtype controls. These genes may represent targets regulated by both E2F7 and E2F8 since loss of either member led to their derepression. Class II represent genes differentially expressed in two of the three mutant genetic groups and include target genes that may be uniquely repressed by either E2F7 or E2F8. Finally, class III represent genes differentially expressed in only one mutant genetic group, and include target genes that may be synergistically repressed by E2F7 and E2F8.
Figure 4. Canonical activator and atypical repressor E2Fs regulate key transcriptional networks coordinating endocycles.
a, Heatmap of approximately 4,500 differentially expressed genes in weaning age (3wk) livers. Class I: genes regulated by E2F7 and E2F8. Class II: genes regulated by either E2F7 or E2F8. Class III: genes synergistically regulated by E2F7/E2F8. n=3–4 livers analyzed per genetic group. control, E2f7f/f;E2f8f/f; Alb-78dko, Alb-cre;E2f7f/f;E2f8f/f; Alb-8ko, Alb-cre;E2f8f/f; Alb-7ko, Alb-cre;E2f7f/f. b, Top, heatmap of TGC gene expression in a custom NanoString mRNA codeset. RNA was isolated from laser capture microdissected E10.5 TGCs in frozen placental tissues. Bottom, G2/M-related genes in NanoString codeset with deregulated expression in TGCs. n=2 placentas analyzed per genetic group. control, E2f7+/+;E2f8+/+; 78dko, E2f7−/−;E2f8−/−; 8ko, E2f7+/+;E2f8−/−; 7ko, E2f7−/−;E2f8+/+. c, Heatmap of downregulated genes in Mx-123tko livers with significantly deregulated expression in Alb-78dko livers. n=3–4 livers analyzed per genetic group. Alb-78dko, Alb-cre;E2f7f/f;E2f8f/f; Mx-123tko, Mx-cre;E2f1−/−;E2f2−/−;E2f3f/f. d, Top five Molecular & Cellular Functions revealed through Ingenuity Pathway Analysis (IPA) of genes antagonistically regulated by E2F1-3 and E2F7/E2F8.
Further analysis focused on genes upregulated in both Alb-8ko and Alb-78dko livers (Class II), since hepatocytes in these two genetic groups displayed the most pronounced reduction in genome ploidy, as well as genes upregulated in all three mutant cohorts (in Class I), since these could also contribute to the observed defect in polyploidization. Closer inspection of these genes by Ingenuity Pathway Analysis (IPA) revealed a striking bias for functions related to cytokinesis, G2/M progression and various stages of mitosis, such as chromosome condensation, stabilization and segregation (Supplementary Fig. 5b, c). By manual annotation we also noted an increase in gene products with GTPase-activating (GAP) or GTP exchange factor (GEF) functions (data not shown), which regulate Rho family of small GTPases essential for successful cytokinesis36. NanoString technology was then used to interrogate the expression of a panel of genes with cell cycle and G2/M related functions in TGCs from E10.5 wildtype, E2f7−/−, E2f8−/− and E2f7−/−;E2f8−/− placentas. As shown in Figure 4b, 66 of the 99 genes interrogated had predominantly increased expression in E2f7/E2f8 deficient TGCs, suggesting a common mechanism for how E2F7 and E2F8 may regulate endocycles in TGCs and hepatocytes.
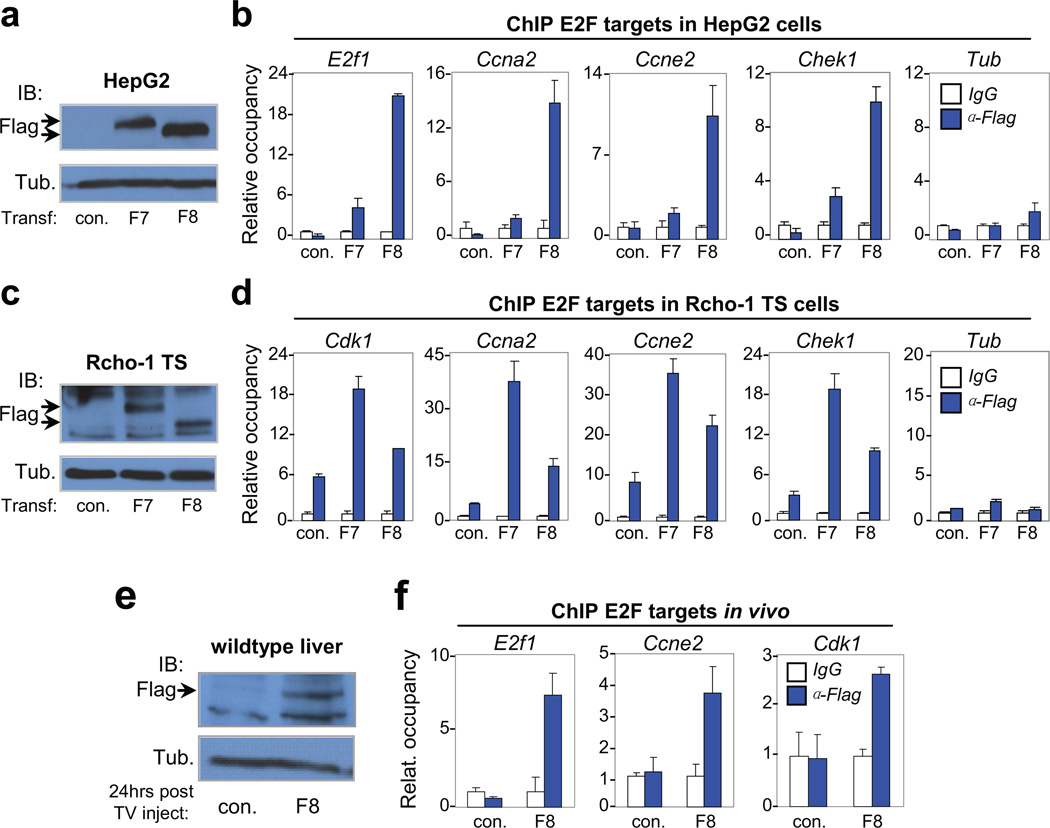
A significant portion of common upregulated genes in E2f7/E2f8 deficient TGCs and hepatocytes have E2F binding elements on their promoters, raising the possibility that these may represent direct targets of E2F7/E2F8. To test this hypothesis, we transfected human HepG2 cells and rat Rcho-1 trophoblast stem (TS) cells with plasmids expressing Flag-tagged E2F7 (F7) or E2F8 (F8) and performed chromatin immunoprecipitation (ChIP) assays with anti-Flag antibodies. As shown in Figures 5b and 5d, Flag-specific antibodies, but not control IgG, co-immunoprecipitated promoter sequences containing E2F binding elements, but not irrelevant sequences lacking E2F binding sites (Tub). The above results were corroborated by additional ChIP assays in human embryonic kidney (HEK) 293 cells transfected with either F7 or F8 (Supplementary Fig. 5d) and in wildtype liver tissues transfected by hydrodynamic-based methods with plasmids expressing F8 (Fig. 5e, f). Together, these results suggest that E2F7 and E2F8 repress a core group of genes with prominent involvement in the regulation of G2/M related events.
Figure 5. Atypical repressors E2F7/E2F8 directly bind gene targets involved in endocycle control.
a, Immunoblot of transfected HepG2 cells showing exogenous expression of flag-tagged E2F7 (F7) and E2F8 (F8) proteins with Tubulin as control. Arrows indicate tagged protein. b, Representative chromatin immunoprecipitation (ChIP) assay in transfected HepG2 cells with anti-Flag antibodies demonstrating enhanced occupancy of F7 and F8 proteins on E2F binding sites in the promoter of G1/S (E2f1 and Ccne2) and G2/M (Ccna2 and Chek1) genes. The Tubulin (Tub) gene, a non-E2F target, demonstrates specific recruitment of F7/F8 to target promoters containing consensus E2F binding sequences. n=3 independent transfection-ChIP experiments performed. c, Immunoblot of transfected Rcho-1 TS cells showing exogenous expression of F7 and F8 proteins with Tubulin as control. Arrows indicate tagged protein. d, Representative ChIP assays in transfected Rcho-1 TS cells of G1/S and G2/M genes as in HepG2 cells. n=3 independent transfection-ChIP experiments performed. e, Immunoblot of liver lysates from 6-week-old mice demonstrating expression of tagged E2F8 protein. A total of 10µg of plasmid DNA was delivered through tail vein injection. Lysates were prepared from livers 24 hours after injection. Tubulin served as loading control. n=3 and n=5 wild type mice were injected with control (empty) plasmid and plasmid expressing Flag-E2F8, respectively. One out of 5 had detectable expression of F8 as shown by Western blot (Fig. 5e). f, ChIP assay in liver lysates with anti-Flag antibodies demonstrating enhanced occupancy of F8 on E2F binding sites in the promoter of G1/S (E2f1 and Ccne2) and G2/M (Cdk1) genes. a–f con., empty plasmid; F7, plasmid expressing flag-tagged E2F7; F8, plasmid expressing flag-tagged E2F8. Data in b, d and f from the representative experiment are reported as average values from triplicate quantitative RT-PCR reactions ± SD.
E2F1 Functionally Opposes Atypical E2F7/E2F8
Given that ablation of E2f1-3 and E2f7/E2f8 led to opposite effects on hepatocyte ploidy, we explored the possibility of cross-regulation between these two E2F subclasses. Loss of E2f1-3 did not impact the expression of other E2Fs (E2f4-8; see Supplementary Fig. 1e), however, loss of E2f7/E2f8 resulted in a transient increase of E2f1 expression soon after birth in the liver (Supplementary Fig. 5e). Interestingly, loss of E2f7/E2f8 had no significant effect on the expression of E2f1-6 in TGCs (Supplementary Fig. 5f). We then profiled global gene expression in hepatocytes lacking the three E2F activators (Mx-123tko). Strikingly, this analysis showed that the majority of genes upregulated in E2f7/E2f8 deficient hepatocytes were downregulated in E2f1-3 deficient hepatocytes (Fig. 4c), and many of these genes had annotated ‘cell cycle’ functions related to G2/M transitions or mitoses and had E2F binding sites in their promoters (Fig. 4d and Supplementary Fig. 5g, h).
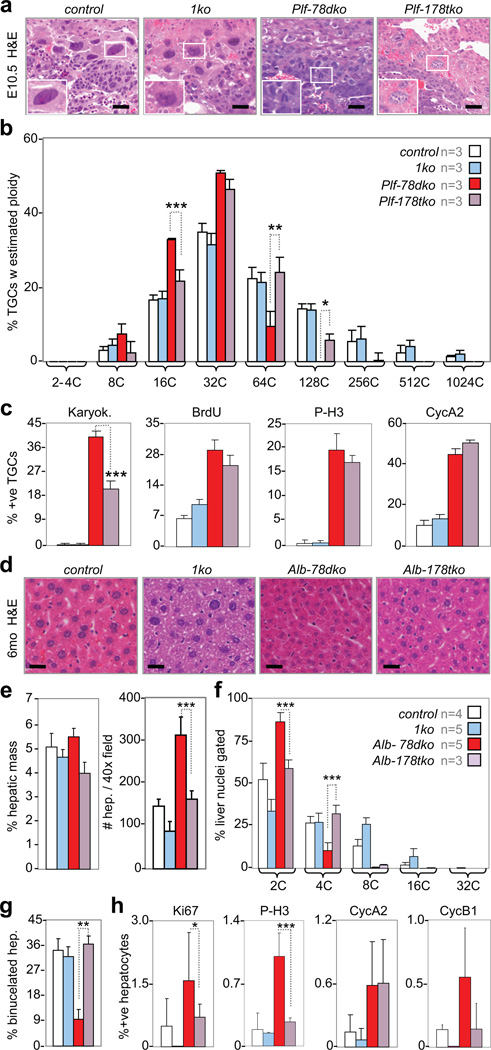
Based on the above results we entertained the hypothesis that canonical E2F activators may functionally antagonize atypical E2F repressors in the control of endocycles. To rigorously test this possibility we generated and analyzed mice with TGCs or hepatocytes deficient for E2F7, E2F8 and E2F1 (E2f1−/−). The ablation of E2f7f/f and E2f8f/f alleles in TGCs was achieved by Plfcre/+ (Plf-178tko) and their ablation in hepatocytes was achieved by Alb-cre (Alb-178tko). Placenta sections from E10.5 Plf-178tko embryos exhibited TGCs with large nuclei having increased ploidy (Fig. 6a), with some TGCs having 128C and 256C genomes (Fig. 6b), which was never observed in Plf-78dko TGCs. This increase in ploidy was accompanied by a significant decrease in the number of multinucleated TGCs and a slight decrease in BrdU- and P-H3-positive cells, even though the number of cyclin A2-positive TGCs was not reduced (Fig. 6c). Similar results were obtained when placentas globally deleted for E2f1, E2f7 and E2f8 (178tko) were examined (Supplementary Fig. 6a-c). Evaluation of Alb-178tko livers revealed larger hepatocytes containing greater DNA content than in Alb-78dko livers, with some reaching 8C ploidy (Fig. 6d-f). The Alb-178tko livers also had many more binucleated hepatocytes (Fig. 6g), and importantly, had fewer Ki67-, P-H3- and cyclin B1-positive hepatocytes, even though again, the percentage of cyclin A2-positive cells was not reduced (Fig. 6h). Thus it would appear that loss of E2f1 suppressed some but not all mitotic defects in TGCs and hepatocytes caused by E2f7/E2f8 deficiency.
Figure 6. Loss of E2f1 restores endocycles in E2f7/E2f8 deficient TGCs and hepatocytes.
a, Representative H&E sections of E10.5 TGCs. Scale bar, 12.5µm. b, Feulgen quantification of genome ploidy in E10.5 TGCs, n=3 placentas per genetic group analyzed. Note significant increase in proportions of 64C and 128C Plf-178tko TGCs relative to Plf-78dko TGCs. c, Quantification of E10.5 TGCs undergoing karyokinesis and expressing S/M phase markers, n=3 placentas per genetic group analyzed. Note significant reduction in proportion of Plf-178tko TGCs undergoing karyokinesis relative to Plf-78dko TGCs. d, Representative H&E liver sections from 6-month-old mice. Scale bar, 10µm. e, Measurements showing no significant change in percent hepatic mass in 6-month-old mice and a significant decrease in number of hepatocytes per image field in Alb-178tko relative to Alb-78dko livers, with n per genetic group analyzed as indicated in Fig. 6f. f, Flow cytometry of liver nuclei in 6-month-old mice, with n per genetic group analyzed as indicated in legend. Note significant increase and decrease in proportion of 4C and 2C nuclei, respectively, in Alb-178tko livers relative to Alb-78dko livers. g, Quantification of binucleated hepatocytes in 2-month-old livers, with n per genetic group analyzed as indicated in Fig. 6f. h, Quantification of hepatocyte proliferation in 2-month-old livers, with n per genetic group analyzed as indicated in Fig. 6f. a–c control, E2f7+/+;E2f8+/+; d-h control, E2f7f/f;E2f8f/f; 1ko, E2f1−/−; Plf-178tko, Plfcre/+;E2f1−/−;E2f7f/f;E2f8f/f; Alb-178tko, Alb-cre;E2f1−/−;E2f7f/f;E2f8f/f. Data in b, c and e-h reported as average ± SD. b and f One-way ANOVA, * p≤0.05; ** p≤0.01; ***p≤0.001. c, e, g and h, Two-tailed Student T-test, * p≤0.05; ** p≤0.01; ***p≤0.001.
Loss of Cyclin A2 Reinstates G2/M Block and Polyploidy in E2f7/E2f8 Deficient Cells
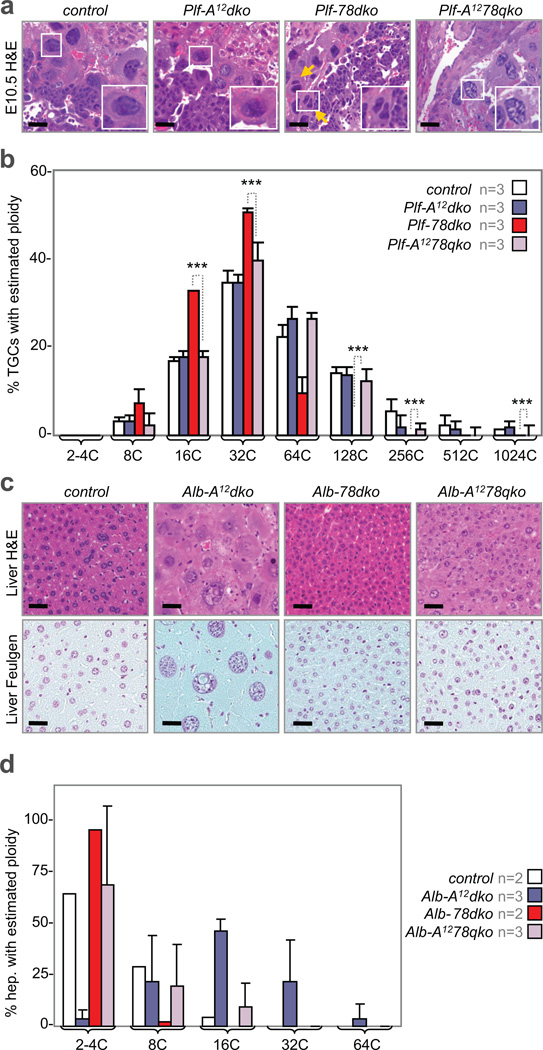
Because the inactivation of E2f7/E2f8 results in ectopic mitoses without inhibiting DNA replication, we predicted that re-establishing a mitotic block would restore polyploidy in E2f7/E2f8 deficient cells. Previous analysis of TS cell cultures showed that p57KIP2 activity participates in driving TGC differentiation and endocycles by inhibiting Cdk1 function37. However, we failed to detect any measureable change in p57KIP2 mRNA or protein levels that might underlie the phenotype of E2f7/E2f8 deficient TGCs or hepatocytes (Supplementary Fig. 6d). Given that cyclin A activity is critical for G2/M progression by regulating the APC/Cyclosome38, we sought to evaluate the consequence of inactivating cyclin A1/A2 in E2f7/E2f8 deficient TGCs and hepatocytes. We thus intercrossed Ccna1−/−;Ccna2f/f mice39 with Plfcre/+;E2f7f/f;E2f8f/f and generated embryos with quadruply deficient TGCs (Plf-A1278qko). Remarkably, TGCs in E10.5 Plf-A1278qko embryos had larger nuclei with greater DNA content than TGCs in Plf-78dko embryos, with some reaching 1024C (Fig. 7a, b). We also intercrossed Ccna1−/−;Ccna2f/f mice with Alb-cre;E2f7f/f;E2f8f/f mice to generate offspring with quadruply deficient hepatocytes (Alb-A1278qko). As shown in Figure 7d, hepatocytes in 3-month-old Alb-A1278qko mice had similar ploidy levels as in age-matched wildtype mice. Interestingly, the loss of Ccna1/Ccna2 alone (Alb-A12dko) in hepatocytes resulted in hyperploidy (Fig. 7c, d). In summary, inhibiting the transcriptional network that signals G2/M progression (by E2f1 inactivation) or interfering with the mitotic machinery (by Ccna1/Ccna2 inactivation) re-established a mitotic block and reinstated higher levels of polyploidy in E2f7/E2f8 deficient cells.
Figure 7. Cyclin A ablation reinstates genome ploidy of E2f7/E2f8 deficient TGCs and hepatocytes.
a, H&E images of E10.5 TGCs. The combined loss of Ccna1 and Ccna2 (Plf-A12dko) led to TGCs similar in appearance to control TGCs. The additional loss of Ccna1 and Ccna2 in E2f7/E2f8 deficient TGCs (Plf-A1278qko) led to TGCs with normal (wildtype-like) appearance, in contrast to Plf-78dko TGCs that appeared small or were binucleated (yellow arrows). Scale bar, 12.5µm. b, Feulgen quantification of genome ploidy in E10.5 TGCs. The intensities (i.e. estimated genome content) of 120–140 TGCs were quantified per placenta sample, with n placenta samples analyzed per genetic group as indicated. A number of TGCs quadruply deficient for Ccna1, Ccna2, E2f7 and E2f8 reach ploidy levels of 128C and 256C. control, E2f7f/f;E2f8f/f; Plf-A12, Plfcre/+;Ccna1−/−;Ccna2f/f; Plf-78dko, Plfcre/+;E2f7f/f;E2f8f/f; Plf-A1278qko, Plfcre/+;Ccna1−/−;Ccna2f/f;E2f7f/f;E2f8f/f. c, H&E images of 3-month-old liver sections, which were stained with the Feulgen technique to facilitate quantification of DNA content, showing an increase in both cellular and nuclear size of hepatocytes lacking Ccna1 and Ccna2 (Alb-A12dko) when compared with control hepatocytes. Hepatocytes quadruply deficient for Ccna1, Ccna2, E2f7 and E2f8 (Alb-A1278qko) had nuclei resembling control hepatocytes. Scale bar, 12.5µm. d, Feulgen quantification of genome ploidy in livers of 3-month-old mice. The intensities (i.e. estimated genome content) of 100 hepatocyte nuclei were quantified per liver sample, with n liver samples analyzed per genetic group as indicated. Ccna1/Ccna2 deficient livers had increased proportion of hepatocytes with higher ploidy levels (16C, 32C and 64C) relative to control livers. Quadruple deficient livers had significantly elevated number of hepatocytes with 16C genomes relative to Alb-78dko livers. control, E2f7f/f;E2f8f/f; Alb-A12, Alb-cre;Ccna1−/−;Ccna2f/f; Alb-78dko, Alb-cre;E2f7f/f;E2f8f/f; Alb-A1278qko, Alb-cre;Ccna1−/−;Ccna2f/f;E2f7f/f;E2f8f/f. Data in b and d reported as average values, ± SD are included when n>2 samples were analyzed. One-way ANOVA, * p≤0.05; ***p≤0.001.
DISCUSSION
In multi-cellular organisms there exists diversity in the kinetics and composition of cell cycles and presumably, in the mechanisms that regulate them. The endocycle is commonly utilized in flies and plants and hence, more is known about how they are controlled in these organisms than in mammals40–45. The balanced action of the E2F activator (dE2f1) and repressor (dE2f2) in Drosophila is critical for the tight regulation of cyclin E expression and successful completion of S phase in mitotic and endocycling cells21,42,43. While E2Fs are also involved in controlling endocycles in plants, their role appears to be different than in mammals. For example the atypical E2Fe repressor in Arabidopsis is required to prevent endocycles44, while we show that the atypical E2F7 and E2F8 family members in mice promote endocycles. A comparison of the regulation of how atypical E2Fs are expressed during development and identification of their direct targets might shed light on these differences. In summary, our current work exposes an intricate E2F network involving balanced and antagonistic activities of canonical E2F activators (E2F1-3) and atypical E2F repressors (E2F7-8) that controls mammalian endocycles. Perhaps the most dramatic manifestation of altering the balance in the E2F network is the ectopic mitosis and karyokinesis observed in E2f7/E2f8 deficient TGCs.
The results presented here expose two distinct arms of the E2F network, one arm involving canonical activators (E2F1-3) and a second involving atypical repressors (E2F7/E2F8), that converge to antagonistically regulate a common gene expression program critical for mammalian endocycles in vivo. A similar antagonistic role for E2F activators and atypical repressors appears to be at play in the control of a distinct program that is critical for mitotic cell cycles during placental development32. Because neither loss of E2f4 or E2f5 in mice12–14 appeared to affect polyploidization of TGCs and hepatocytes (Supplementary Fig. 2c and data not shown) we propose that the most ancient E2F repressor arm consisting of atypical E2f7 and E2f8 evolved to specifically regulate variant cell cycles in metazoans in a manner distinct from the function of canonical E2F repressor proteins. One reason for this might be to keep E2F7/E2F8 mediated repression outside the influence of the cyclin-CDK axis. Because E2F7/E2F8 DNA binding activity is independent of DP proteins, atypical repressors would remain unresponsive to CDK-mediated phosphorylation of DP, which negatively regulates the DNA binding and functions of canonical repressors45–47, allowing repression at a time (S/G2) when CDK activity is high. However, the exact molecular mechanism of how canonical activators and atypical repressors coordinate gene expression in endocycling tissues remains obscure.
It has been assumed that polyploidy in mammals, as in flies and plants, is integral to the physiologic functions of placentas and livers. Here we generated the first mouse models with significantly reduced TGC and hepatocyte ploidy to assess this hypothesis. Surprisingly, TGCs and hepatocytes having significantly restricted ploidy levels appeared to impart placentas and livers, respectively, with adequate organ function to carry animals through embryonic and adult development. Whether altered liver ploidy in mice impacts aging-related processes such as tumor development remains to be evaluated. In summary, genetic, systems-based and biochemical approaches revealed a fundamental role for the canonical activator and atypical repressor arms of the E2F network in orchestrating mammalian endocycles in vivo.
METHODS
Mouse husbandry and genotyping
All mice were housed in barrier conditions with a 12-hour light/dark cycle and had access to food/water ad libitum. Genotyping primers are provided in Supplementary Table 1.
Feulgen analysis
5µm sections of placentas/livers were treated with 1N hydrochloric acid and stained with Schiff reagent. Cytoplasm was counterstained with aqueous light green (1%). Nuclear intensity was quantified using ImageJ (http://rsbweb.nih.gov/ij/)48. TGC ploidy was expressed relative to 2C-4C labrynthine trophoblast cells. For binucleated TGCs, ploidy levels of individual nuclei were summed to derive cumulative ploidy. Hepatocyte ploidy was expressed relative to 2C–4C hepatocytes deficient for E2f7/EfF8. Hepatocyte ploidy was expressed on a per nucleus basis.
Flow cytometry analysis
Nuclei suspensions were obtained from frozen liver tissue as previously described49. Total DNA content of minimum 30,000 nuclei per liver sample were analyzed using LSR II (BD Biosciences). Cell cycle profiles were generated using FlowJo (Tree Star).
Affymetrix analyses
Total RNA was extracted from minimum 3 livers per genetic group using TRIzol (Invitrogen). RNA quality was verified using Agilent 2100 Bioanalyzer, and samples submitted to OSUCCC Microarray Shared Resource facility for biotinylated cRNA generation and MOE4302.0 hybridization. GeneChip Scanner 3000 (Affymetrix) was used for data acquisition. Pre-processing of raw signal intensities was performed using the Robust Multi-chip Average (RMA) method50. BRB-Array Tools 3.8.1 was then used to identify differentially expressed genes51. For heatmap images, geometric mean of genes after RMA normalization from control liver samples was set as reference, with colors corresponding to the ratio between expression of genes in mutant samples and the reference. Functional annotation was performed using Ingenuity Pathway Analysis (Ingenuity Systems).
NanoString analyses
From each tissue sample, total RNA (200ng) was profiled on a custom codeset containing 200 selected genes following manufacturer’s instructions (Supplementary Table 3). For TGC gene expression analysis, we isolated TGCs from frozen placenta sections using laser capture microdissection (PALM MicroLaser system). Total RNA was then extracted using Arcturus PicoPure RNA Isolation Kit (Applied Biosystems 12204-01). For validation of gene expression changes in the liver (Supplementary Fig. 4a), the same set of TRIzol-isolated RNA was utilized in both Affymetrix and NanoString assays. The NanoString data is first normalized to the geometric mean of positive control spike counts in each lane of the cartridge, followed by subtracting the minimum negative control spike counts in each lane to remove the background. Finally, expression of each gene in the codeset is normalized with respect to the geometric mean of the housekeeping genes. Heatmaps were generated in the same manner as for Affymetrix gene expression data.
Confocal microscopy-3D reconstruction
70µm section of tissues were stained with 1mM Draq5 (Biostatus) at 4°C overnight. Sections were examined by confocal laser scanning microscopy (Zeiss LSM 510) at 63X and 0.7X scan zoom. Optical sections with between-plane-plane resolution of 0.40µm and axial resolution of 0.42µm were acquired. 4 to 5 non-overlapping regions per placenta were imaged with a field view of 207µm × 207µm and depth of 40 ± 2µm, resulting in a stack of 100 images for each region. This process was identically repeated for liver samples. For 3D reconstruction, the image stacks were processed using Insight Toolkit and ITK-SNAP52. Each nucleus was segmented by active contour segmentation using region competition as the stopping criterion53. Nuclei of binucleated TGCs were constructed separately and assigned different colors. Only the volumes of individual hepatocyte nuclei were measured.
Transmission Electron Microscopy
Placentas were fixed/processed following a standard protocol (http://cmif.osu.edu/419.cfm) and embedded in Eponate resin. 70nm placental cross sections were mounted on copper grids, double stained with uranyl acetate and lead citrate, and observed with a Technai G2 Spirit transmission electron microscope (FEI).
Transient transfections
For HepG2, subconfluent cells were transfected with 1:3 ratio of plasmid DNA:reagent [X-tremeGENE HP DNA Transfection Reagent (Roche)] in 10% FBS RPMI-1640. Cells were collected 48 hours after transfection. In 1 experiment, 3 100cm plates of transfected cells were combined for lysis and sonication as 1 sample in the chromatin immunoprecipitation (ChIP) assay. For Rcho-1, transfection conditions were same as described for HepG2 cells. Rcho-1 cells were maintained as previously described54. In 1 experiment, two 150cm plates were combined as one sample utilized in the ChIP. A total of 3 independent experiments for each cell line was performed. For HEK293 cells, subconfluent cells in 10% FBS DMEM were transfected using calcium phosphate with 25µg of plasmid DNA expressing wildtype flag-E2F7/flag-E2F8 or mutant flag-E2F7/flag-E2F8.
Systemic administration of plasmid DNA
Six-week-old wildtype male mice were injected with either sterile PBS or sterile PBS containing 10µg plasmid DNA through the tail vein following a published protocol55. Mice were sacrificed by CO2 asphyxiation 24 hours after TV injection. Whole body perfusion with warm saline was performed using the intracardiac method prior to excision of the liver, subsequent derivation of single-celled suspension by passing perfused tissue through 40µm mesh and fixation/processing for chromatin immunoprecipitation assays (next section).
Chromatin immunoprecipitation assays
For HepG2 and Rcho-1 TS cell lines, transfected cells were cross-linked in growth media containing 1% formaldehyde followed by neutralization with 1.25M glycine. The cytoplasmic fraction was discarded (cytoplasmic lysis buffer: 5mM PIPES pH 8.0, 85mM KCl, 0.5% NP40 and fresh protease inhibitors). Nuclear lysates were collected by incubating the nuclei pellet in lysis buffer containing 50mM Tris pH 8.0, 10mM EDTA, 0.3% SDS and fresh cocktail of protease/phosphatase inhibitors. Sonicated chromatin (~100–300bp) was incubated with 10µg of anti-Flag mouse monoclonal antibodies (Sigma F1804) or normal mouse IgG (Santa Cruz sc-2025) at 4°C overnight. Antibody-protein-DNA complexes were recovered by addition of 50% Protein G agarose slurry (Millipore 16–266) pre-cleared with 10mg/ml tRNA and 10mg/ml BSA. De-crosslinking was performed at 65°C overnight with periodic agitation. Recovered DNA was purified using Qiagen PCR extraction kit. 5% input and 2µl of eluted DNA was used for quantitative Real-Time PCR (RT-PCR) with gene-specific primers and SYBR Green (BioRad 170–8880). For HEK293 cells, the EZ-CHIP assay kit (Millipore 17–371) was used following manufacturer’s instructions. Sonicated chromatin was incubated with 2µg of anti-Flag antibodies (Sigma F1804) or normal mouse IgG (Santa Cruz sc-2025) at 4°C overnight. Antibody-protein-DNA complexes were recovered by addition of Salmon Sperm DNA/Protein G agarose slurry (Millipore 16–201). Immunoprecipitated DNA fractions were de-crosslinked at 65°C overnight and purified using QIAquick columns (QIAGEN). Quantitative RT-PCR was performed as above except 1% input was used. All quantitative RT-PCRs reactions were performed in triplicate using the following conditions: 1 cycle at 95°C 5 min, followed by 20–40 cycles of 95°C 15 sec, 60–64°C 30 sec and 72°C 30 sec.
Immunoblot assays
Whole cell lysates were prepared from lysis of HepG2 and Rcho-1 TS cells in a standard radioimmunoprecipitation (RIPA) buffer containing freshly diluted cocktail of protease and phosphatase inhibitors. For preparation of liver lysates, frozen tissue (~100mg) was thawed on ice followed by homogenization in RIPA buffer and then centrifugation at maximum speed for 15 minutes in 4°C. Total protein concentration was measured using the Bio-Rad Protein Assay kit (BioRad 500-0006). Denatured samples were separated in 8–10% SDS-polyacrylamide gels, transferred onto PDVF membranes, blocked in 5% milk-PBS solution and incubated overnight in 4°C with the following primary antibodies: anti-E2F3 1:5000 (Santa Cruz sc-878x), anti-Flag 1:1000 (Sigma F1804), anti-Tubulin 1:5000 (Sigma T6199) and anti-HSP70 1:5000 (BD Transduction Laboratories 610608). HRP-conjugated species-specific secondary antibodies were applied to membranes followed by detection with SuperSignal West Femto kit (Thermo Scientific 34095).
Immunohistochemistry and immunofluoresence
After antigen retrieval using Target Retrieval Solution (DAKO S1699), 5µm deparaffinized sections of placenta or liver tissues were incubated with primary antibodies at 4°C overnight against E2F7 1:100 (Abcam ab56022), E2F8 1:50 (polyclonal against residues 576–595 of murine E2F8), BrdU 1:100 (DAKO MO-0744), Ki67 1:100 (BD Pharmingen 550609), phospho-Histone 3 (Ser10) 1:100 (Millipore 06-570), Cyclin A2 1:100 (Santa Cruz sc-596), Cyclin B1 1:100 (Santa Cruz sc-752), E-Cadherin 1:100 (Abcam ab53033), p57Kip2 1:100 (Santa Cruz sc-8298)56, 57 and placental lactogen 1:200 (PL-1, F. Talamantes). TGCs in M phase (Fig. 2h) were identified by detecting P-H3 or the lineage-specific PL-1 protein. For IF detection, fluorophore-conjugated secondary antibodies were applied to the slides. For IHC, the VECTASTAIN ABC system was utilized (Vector Laboratories).
BrdU incorporation assays
Pregnant females (E10.5) received a single intraperitoneal injection of bromodeoxyuridine (BrdU, Sigma B5002) in sterile PBS at 100µg/g body weight 1 hour prior to sacrifice for collection of placentas. 5µm deparaffinized tissue sections were used for BrdU detection after incubation in 2N HCl and neutralization with 10mM sodium borate (pH 8.0). Primary antibody was then applied overnight.
pIpC injections
6-week-old mice received 5 intraperitoneal injections of 250µg of polyinosine-polycytidine (pIpC, Sigma P1530) dissolved in sterile PBS every alternate day. Mice were sacrificed 24 hours after the last injection, and their livers harvested for flow cytometry and microarray analyses.
Carbon tetrachloride injection
6-month-old mice received a single intraperitoneal injection of 10% carbon tetrachloride (Sigma 319961) dissolved in corn oil. Mice were sacrificed at 0, 24, 48, 60 and 168 hours (when liver regeneration is complete in rodents58) post-injection. Mice were administered an intraperitoneal injection of BrdU solution (100µg/g body weight) to label hepatocytes in S phase of the cell cycle 1 hour prior to sacrifice.
Tamoxifen induction of cre
Tamoxifen chow (Teklad Lab Animal Diets, Harlan Laboratories) was prepared by mixing 3kg of irradiated standard chow with 1g tamoxifen (Sigma T5648). For pre-weaning induction of cre, 7-day-old pups and their mother were switched from regular to tamoxifen chow for 1 week. After 1 week, they were again fed regular chow until weaning and up to time of sacrifice (age 2 months). For post-weaning induction of cre, pups were weaned from their mother at age 21 days and maintained on tamoxifen chow starting from age 28 days up to age 2 months. Tamoxifen chow was stored in the dark at 4°C and changed weekly. It was provided to mice at 40–80mg/kg body weight59.
X-gal staining
OCT-embedded placentas were sectioned and fixed in 0.2% PBS-glutaraldehyde solution (1.25mM EGTA pH 7.3 and 2mM MgCl2). Sections were rinsed with wash buffer [2mM MgCl2, 0.01% sodium deoxycholate and 0.02% IGEPAL CA-630 (Sigma I8896)] and stained in LacZ solution (4mM potassium ferricyanide, 4mM potassium ferrocyanide and 1mg/ml X-gal in wash buffer) at 37°C overnight with protection from light. Nuclear Fast Red was used as counterstain.
E2F Binding site analysis
Promoter sequences (−3000bp ~ +2000bp) of genes antagonistically regulated by E2F1-3 and E2F7/E2F8 were obtained from the UCSC genome browser (http://genome.ucsc.edu/). E2F consensus binding sites were identified using TFSearch (http://www.cbrc.jp/research/db/TFSEARCH.html).
Statistical analyses
Binucleated TGCs, as a percentage of minimum 143 total TGCs assessed in each genetic group, was quantified in confocal images using the LSM image browser (Zeiss). All other IHC/IF-associated quantification of TGCs/hepatocytes was performed using Metamorph 6.1 in photomerged images (placenta) or 5–7 random images (liver) acquired at 40× from minimum of 3 sections per sample. Results were reported as an average percentage ± S.D. of positive cells in number (n) of control and mutant samples analyzed. Flow cytometry profiles (e.g. Fig. 3f) are representative of minimum 3 independent experiments per genotype and age group. Flow cytometry results were reported as an average percentage ± S.D. of nuclei with certain DNA content out of minimum 30,000 nuclei gated per sample in n of control and mutant samples analyzed. Pairwise comparisons were evaluated by 2-tailed Student’s T-test, while multiple comparisons were evaluated by One-way ANOVA. When appropriate, p-values were adjusted by the Holm’s method within each ploidy category.
Supplementary Material
ACKNOWLEDGEMENTS
We thank L. Rawahneh, J. Moffitt and N. Lovett for excellent technical assistance with histology, and P. Wenzel for generating and harvesting 123tko placentas. We also thank these individuals from OSUCCC Shared Resources: T. Wise, J. Palatini, H. Alders, P. Yan, P. Fada and B. Rodriguez (Microarray and Nucleic Acid Shared Resources); B. McElwain (Analytic Cytometry); R. Burry, K. Wolken, and B. Kemmenoe (Microscopy and Imaging); and K. La Perle (Comparative Pathology and Mouse Phenotyping). We are gracious for PL-1 antibodies provided by F. Talamantes, Rcho-1 TS cells from M. Soares and HepG2 cells from S. Jacobs. This work was funded by NIH grants to G.L. (R01CA85619, R01CA82259, R01HD047470, P01CA097189) and P.S. (R01CA132740). G.L. is the recipient of The Pew Charitable Trust Scholar Award and the Leukemia & Lymphoma Society Scholar Award. H.-Z. Chen, M. Mair, S. Senepati, S. Raman and T. Pécot are recipients of the Pelotonia Fellowship Award.
Footnotes
AUTHOR CONTRIBUTIONS
H.-Z.C., M.M.O., J.L., and G.L. designed the experiments. H.-Z.C., M.M.O., J.L., T.P., V.C., L.K. and S.B. performed experiments. H.-Z.C. and M.M.O. co-wrote the paper with G.L.; all other authors listed helped perform experiments. Specifically, J.C.T., S.R., S.S., M.L., R.M. and K.H. helped with experiments relating to confocal microscopy and 3D reconstruction. X.M. and S.F. helped analyze data and performed statistics. I.K., D.J.W. and P.S. provided mice for the study and reviewed drafts of the manuscript. T.H., B.L. and V.J. contributed to discussion relating to Affymetrix/NanoString analyses.
The authors declare no competing financial interests
REFERENCES
- 1.Lilly MA, Duronio RJ. New insights into cell cycle control from the Drosophila endocycle. Oncogene. 2005;24:2765–2775. doi: 10.1038/sj.onc.1208610. [DOI] [PubMed] [Google Scholar]
- 2.Varmuza S, Prideaux V, Kothary R, Rossant J. Polytene chromosomes in mouse trophoblast giant cells. Development. 1988;102:127–134. doi: 10.1242/dev.102.1.127. [DOI] [PubMed] [Google Scholar]
- 3.Cross J. How to make a placenta: mechanisms of trophoblast cell differentiation in mice – a review. Placenta. 2005;26:S3–S9. doi: 10.1016/j.placenta.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 4.Mehrotra S, Maqbool SB, Kolpakas A, Murnen K, Calvi BR. Endocycling cells do not apoptose in response to DNA rereplication genotoxic stress. Genes Dev. 2008;22:3158–3171. doi: 10.1101/gad.1710208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee HO, Davidson JM, Duronio RJ. Endoreplication: polyploidy with purpose. Genes Dev. 2009;23:2461–2477. doi: 10.1101/gad.1829209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Storchova Z, Pellman D. From polyploidy to aneuploidy, genome instability and cancer. Nature Rev. Mol. Cell. Biol. 2004;5:45–54. doi: 10.1038/nrm1276. [DOI] [PubMed] [Google Scholar]
- 7.Harbour JW, Dean DC. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 2000;14:2393–2409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- 8.Yamasaki L, et al. Tumor induction and tissue atrophy in mice lacking E2F-1. Cell. 1996;85:537–548. doi: 10.1016/s0092-8674(00)81254-4. [DOI] [PubMed] [Google Scholar]
- 9.Field SJ, et al. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell. 1996;85:549–561. doi: 10.1016/s0092-8674(00)81255-6. [DOI] [PubMed] [Google Scholar]
- 10.Humbert PO, et al. E2f3 is critical for normal cellular proliferation. Genes Dev. 2000;14:690–703. [PMC free article] [PubMed] [Google Scholar]
- 11.Murga M, et al. Mutation of E2F2 in mice causes enhanced T lymphocyte proliferation, leading to the development of autoimmunity. Immunity. 2001;15:959–970. doi: 10.1016/s1074-7613(01)00254-0. [DOI] [PubMed] [Google Scholar]
- 12.Humbert PO, et al. E2F4 is essential for normal erythrocyte maturation and neonatal viability. Mol. Cell. 2000;6:281–91. doi: 10.1016/s1097-2765(00)00029-0. [DOI] [PubMed] [Google Scholar]
- 13.Rempel RE, et al. Loss of E2F4 activity leads to abnormal development of multiple cellular lineages. Mol. Cell. 2000;6:293–306. doi: 10.1016/s1097-2765(00)00030-7. [DOI] [PubMed] [Google Scholar]
- 14.Lindeman GJ, et al. A specific, nonproliferative role for E2F-5 in choroid plexus function revealed by gene targeting. Genes Dev. 1998;12:1092–1098. doi: 10.1101/gad.12.8.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pohlers M, et al. A role for E2F6 in the restriction of male-germ-cell-specific gene expression. Curr. Biol. 2005;15:1051–1057. doi: 10.1016/j.cub.2005.04.060. [DOI] [PubMed] [Google Scholar]
- 16.Li J, et al. Synergistic function of E2F7 and E2F8 is essential for cell survival and embryonic development. Dev. Cell. 2008;14:62–75. doi: 10.1016/j.devcel.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen H-Z, Tsai S-Y, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nature Rev. Cancer. 2009;9:785–797. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trimarchi JM, Lees JA. Sibling rivalry in the E2F family. Nature Rev. Mol. Cell Biol. 2002;3:11–20. doi: 10.1038/nrm714. [DOI] [PubMed] [Google Scholar]
- 19.Lammens T, Li J, Leone G, De Veylder L. Atypical E2Fs: new players in the E2F transcription factor family. Trends Cell Biol. 2009;19:111–118. doi: 10.1016/j.tcb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Breuer C, Ishida T, Sugimoto K. Developmental control of endocycles and cell growth in plants. Curr. Opin. Plant Biol. 2010;13:654–660. doi: 10.1016/j.pbi.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 21.Zielke N, et al. Control of Drosophila endocycles by E2F and CRL4(CDT2) Nature. 2011;480:123–127. doi: 10.1038/nature10579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barbason H, Van Cantfort J, Houbrechts N. Correlation between tissular and division functions in the liver of young rats. Cell Tissue Kinet. 1974;7:319–326. doi: 10.1111/j.1365-2184.1974.tb00414.x. [DOI] [PubMed] [Google Scholar]
- 23.Dallman PR, Spirito RA, Siimes MA. Diurnal patterns of DNA synthesis in the rat: modification by diet and feeding schedule. J. Nutr. 1974;104:1234–1241. doi: 10.1093/jn/104.10.1234. [DOI] [PubMed] [Google Scholar]
- 24.Tsai S-Y, et al. Mouse development with a single E2F activator. Nature. 2008;454:1137–1141. doi: 10.1038/nature07066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chong J-L, et al. E2f1-3 switch from activators in progenitor cells to repressors in differentiating cells. Nature. 2009;462:930–934. doi: 10.1038/nature08677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wenzel P, et al. Cell proliferation in the absence of E2F1-3. Dev. Biol. 2010;351:35–45. doi: 10.1016/j.ydbio.2010.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;8:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 28.de Bruin A, et al. Identification and characterization of E2F7, a novel mammalian E2F family member capable of blocking cellular proliferation. J. Biol. Chem. 2003;278:42041–42049. doi: 10.1074/jbc.M308105200. [DOI] [PubMed] [Google Scholar]
- 29.Di Stefano L, et al. E2F7, a novel E2F featuring DP-independent repression of a subset of E2F-regulated genes. EMBO J. 2003;22:6289–6298. doi: 10.1093/emboj/cdg613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maiti B, et al. Cloning and characterization of mouse E2F8, a novel mammalian E2F family member capable of blocking cellular proliferation. J. Biol. Chem. 2005;280:18211–18220. doi: 10.1074/jbc.M501410200. [DOI] [PubMed] [Google Scholar]
- 31.Christensen J, et al. Characterization of E2F8, a novel E2F-like cell-cycle regulated repressor of E2F-activated transcription. Nucleic Acids Res. 2005;33:5458–5470. doi: 10.1093/nar/gki855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ouseph MM, et al. Atypical E2F repressors and activators coordinate placental development. Dev. Cell. 2012;22:849–862. doi: 10.1016/j.devcel.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Postic C, Magnuson MA. DNA excision in liver by an albumin-cre transgene occurs progressively with age. Genesis. 2000;26:149–150. doi: 10.1002/(sici)1526-968x(200002)26:2<149::aid-gene16>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 34.Margall-Ducos G, Celton-Morizur S, Couton D, Bregerie O, Desdouets C. Liver tetraploidization is controlled by a new process of incomplete cytokinesis. J. Cell Sci. 2007;120:3633–3639. doi: 10.1242/jcs.016907. [DOI] [PubMed] [Google Scholar]
- 35.Schuler M, Dierich A, Chambon P, Metzger D. Efficient temporally controlled targeted somatic mutagenesis in hepatocytes of the mouse. Genesis. 2004;39:167–172. doi: 10.1002/gene.20039. [DOI] [PubMed] [Google Scholar]
- 36.Normand G, King RW. Understanding cytokinesis failure. Adv. Exp. Med. Biol. 2010;676:27–55. doi: 10.1007/978-1-4419-6199-0_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ullah Z, Lee CY, Lilly MA, DePamphilis ML. Developmentally programmed endoreduplication in animals. Cell Cycle. 2009;8:1501–1509. doi: 10.4161/cc.8.10.8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sorensen CS, et al. A conserved cyclin-binding domain determines functional interplay between anaphase-promoting complex-Cdh1 and cyclin A-Cdk2 during cell cycle progression. Mol. Cell. Biol. 2001;21:3692–3703. doi: 10.1128/MCB.21.11.3692-3703.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalaszczynska I, et al. Cyclin A is redundant in fibroblasts but essential in hematopoietic and embryonic stem cells. Cell. 2009;138:352–365. doi: 10.1016/j.cell.2009.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edgar BA, Orr-Weaver TL. Endoreplication cell cycles: more for less. Cell. 2001;105:297–306. doi: 10.1016/s0092-8674(01)00334-8. [DOI] [PubMed] [Google Scholar]
- 41.Inzé D, De Veylder L. Cell cycle regulation in plant development. Annu. Rev. Genet. 2006;40:77–105. doi: 10.1146/annurev.genet.40.110405.090431. [DOI] [PubMed] [Google Scholar]
- 42.Duronio RJ, O’Farrell PH, Xie JE, Brook A, Dyson N. The transcription factor E2F is required for S phase during Drosophila embryogenesis. Genes Dev. 1995;9:1445–1455. doi: 10.1101/gad.9.12.1445. [DOI] [PubMed] [Google Scholar]
- 43.Weng L, Zhu C, Xu J, Du W. Critical role of active repression by E2F and Rb proteins in endoreplication during Drosophila development. EMBO J. 2003;22:3865–3875. doi: 10.1093/emboj/cdg373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lammens T, et al. Atypical E2F activity restrains APC/CCCS52A2 function obligatory for endocycle onset. Proc. Natl. Acac. Sci U.S.A. 2008;105:14721–14726. doi: 10.1073/pnas.0806510105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kohn MJ, Bronson RT, Harlow E, Dyson NJ, Yamasaki L. Dp1 is required for extra-embryonic development. Development. 2003;130:1295–1305. doi: 10.1242/dev.00355. [DOI] [PubMed] [Google Scholar]
- 46.Krek W, et al. Negative regulation of the growth-promoting transcription factor E2F-1 by a stably bound cyclin A-dependent protein kinase. Cell. 1994;78:161–172. doi: 10.1016/0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 47.Krek W, et al. Cyclin A-kinase regulation of E2F-1 DNA binding function underlies suppression of an S phase checkpoint. Cell. 1995;83:1149–1158. doi: 10.1016/0092-8674(95)90141-8. [DOI] [PubMed] [Google Scholar]
- 48.Hardie DC, Gregory TR, Hebert PD. From pixels to picograms: a beginners' guide to genome quantification by Feulgen image analysis densitometry. J. Histochem. Cytochem. 2002;50:735–749. doi: 10.1177/002215540205000601. [DOI] [PubMed] [Google Scholar]
- 49.Mayhew CN, et al. Liver-specific pRB loss results in ectopic cell cycle entry and aberrant ploidy. Cancer Res. 2005;65:4568–4577. doi: 10.1158/0008-5472.CAN-04-4221. [DOI] [PubMed] [Google Scholar]
- 50.Irizarry RA, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A Comparison of normalization methods for high density oligonucleotide array data based on bias and variance. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 52.Yushkevich PA, et al. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. NeuroImage. 2006;31:1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 53.Zhu SC, Yuille A. Region competition: unifying snakes, region growing, and bayes/mdl for multiband image segmentation. IEEE Trans. Pattern Anal. 1996;18:884–900. [Google Scholar]
- 54.Sahgal N, Canham LN, Canham B, Soares MJ. Rcho-1 trophoblast stem cells: a model system for studying trophoblast cell differentiation. Methods Mol. Med. 2006;121:159–178. [PubMed] [Google Scholar]
- 55.Liu F, Song YK, Liu D. Hydrodynamic-based transfection in animal by systemic administration of plasmid DNA. Gene Therapy. 1999;6:1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- 56.Ullah Z, Kohn MJ, Yagi R, Vassilev LT, DePamphilis ML. Differentiation of trophoblast stem cells into giant cells is triggered by p57/Kip2 inhibition of CDK1 activity. Genes Dev. 2008;22:3204–3236. doi: 10.1101/gad.1718108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nagahama H, et al. Spatial and temporal expression patterns of the cyclin-dependent kinase (CDK) inhibitors p27Kip1 and p57Kip2 during mouse development. Anat. Embryol. 2001;203:77–87. doi: 10.1007/s004290000146. [DOI] [PubMed] [Google Scholar]
- 58.Schultze B, Gerhard H, Maurer W. Liver Regeneration after Experimental Injury. New York: Grune and Stratton; 1975. A quantitative model of liver regeneration in the mouse after CCl4 intoxification. [Google Scholar]
- 59.Kiermayer C, Conrad M, Schneider M, Schmidt J, Brielmeier M. Optimization of spatiotemporal gene inactivation in mouse heart by oral application of tamoxifen citrate. Genesis. 2007;45:11–16. doi: 10.1002/dvg.20244. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.