There is an exciting new player in the ever-expanding field of genome editing. In a study reported in the January 2013 issue of Science, two groups—Cong et al.1 and Mali et al.2—explored the limits and adaptability of a prokaryotic RNA-based system for mammalian genome-wide editing. This new method of genome engineering is derived from an adaptive immune system known as CRISPR (Clustered Regulatory Interspaced Short Palindromic Repeats) that bacteria and archaea use as a means to protect themselves against foreign invasive elements. These two studies show that the CRISPR system is an efficient method to alter mammalian genomes. At present, four types of discrete systems have been shown to generate, to different degrees of specificity and efficiency, genome-wide editing: three distinct protein-based nuclease systems,3,4,5 a chemical-based nuclease system,6 an adeno-associated virus (AAV)–based system,7 and now a protein RNA–based system.1,2
The homing endonucleases (HEs), such as I-SceI, were the first of these systems shown to be able to modify the mammalian genome in a precise fashion.8 HEs are naturally occurring nucleases that have specific long recognition sites (more than 14 base pairs). Although progress has been made in modifying HEs to recognize new target sequences, this protein-engineering problem has remained a tough nut to crack, and the use of modified HEs has yet to spread widely among researchers. Russell and Hirata later showed that recombinant single-stranded AAV could be used for highly efficient genome editing without the use of nucleases,9 with absolute editing efficiencies of 1% being possible under certain conditions. Although a few investigators have used the AAV approach effectively and Horizon Discovery has adopted this strategy commercially, it has not been widely adopted.
Zinc-finger nucleases (ZFNs), the next system to be described, were critical in demonstrating the broad feasibility of precise genome editing in vertebrate cells.10,11,12 ZFNs are artificial proteins that fuse a zinc-finger DNA-binding domain with a nonspecific nuclease domain derived from the type II restriction enzyme FokI. The nuclease domain requires dimerization to be active, and thus efficient genome modification occurs only when a pair of ZFNs is engineered to recognize specific sites that facilitate dimerization of the nuclease domain. A decade later, ZFNs have been used in clinical trials as a strategy to engineer an HIV-resistant immune system.13 Several methods to engineer ZFNs exist, but they all require substantial technical expertise to engineer high-quality ZFNs; this has limited their use. In the 2000s, a peptide–nucleic acid–based chemical system to modify genomes was developed, but this strategy has not been widely used by researchers other than those who first reported it.14
In the past two years, the genome-editing field has further expanded with the development of TAL effector nucleases (TALENs).15 TALENs are similar to ZFNs except for the substitution of a TAL effector DNA-binding domain for the zinc-finger DNA-binding domain in conferring sequence specificity. The ease of engineering TALENs for a wide variety of target binding sites, their high success rate in genome editing, and the lower cellular toxicity of TALENs as compared with ZFNs have all contributed to the rapid expansion of their use.16 For these reasons, TALENs have supplanted ZFNs as the most useful nuclease-based platform for genome editing. The new CRISPR system, however, may soon prove to be a viable alternative to TALENs, and the recent articles in Science describe a critical early step in its development.1,2,17
The type II CRISPR system is used by bacteria and archaea to provide immunological memory against subsequent invasions of foreign DNA. It works by incorporating short exogenous DNA sequences from the invading pathogen into specific loci of the host genome. Upon transcription, these sequences are processed into pre−CRISPR RNAs (crRNAs) and further into crRNAs, following maturation, which then function as detectors of foreign DNA. The crRNA guides the Cas9 nuclease machinery, the sole effector enzyme, to the foreign DNA, where the Cas9 nuclease cleaves and thereby inactivates the foreign DNA. Two additional players are required to complete the system in prokaryotes: a trans-activating crRNA (tracrRNA) that base-pairs with the crRNA to provide the substrate for host ribonuclease RNase III. When this system comes together, it can identify DNA sequences complementary to the crRNA and degrade them (Figure 1c). Cong et al.1 showed that a three-component system comprising (i) a guide RNA that hybridizes to the target DNA (the crRNA), (ii) a protein nuclease that cleaves the target DNA (bacterial Cas9, not to be confused with caspase 9), and (iii) a linker RNA that brings the nuclease to the guide RNA (the tracrRNA) was sufficient to mediate efficient genome editing in human cells. Interestingly, Mali et al.2 showed that a two-component system comprising (i) the Cas9 protein and (ii) a guide RNA consisting of a crRNA–tracrRNA hybrid molecule was sufficient.
Figure 1.
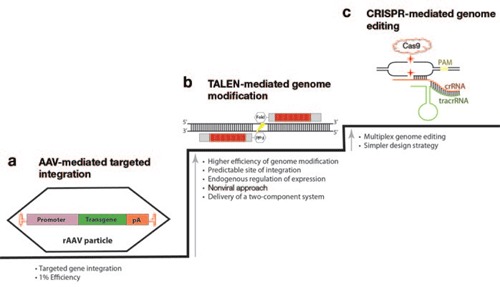
Evolution of tools designed for genome editing. (a) Adeno-associated virus (AAV)-based integration was the first strategy for targeted modification of any genomic site in mammalian somatic cells in a highly efficient manner (up to 1%). It provided an alternative to the unpredictable insertion of integrating viral systems (e.g., retroviruses, lentiviruses, foamy viruses). (b) Zinc-finger nucleases (ZFNs), homing endonucleases, and TAL effector nucleases (TALENs) are nonviral protein–based strategies for inducing precise genomic modifications by either mutagenic nonhomologous end joining or homologous recombination. These methods provide higher genome-editing frequencies than AAV and have entered clinical trials. The ease of engineering highly active TALENs has increased the popularity of the protein-based strategy for genome editing. Depicted is a schematic of a pair of TALENs binding to a target sequence in which the TAL effector DNA-binding domain is depicted by the red boxes and the FokI nuclease domain is labeled. (c) Protein RNA–based tools, the CRISPR system, are the latest strategy for genome engineering. The ease of assembly, high efficiency of genome editing, and the ability to be used for multiplex engineering open exciting new avenues in the field. crRNA, CRISPR RNA; pA, polyadenylation signal sequence; PAM, protospacer-adjacent motif; rAAV, recombinant adeno-associated virus; tracrRNA, trans-activating crRNA.
There are several key findings in these two studies. One is that the CRISPR components can be transfected into cells in the form of plasmids that include appropriate promoter elements for expression in the target cells (a mammalian protein expression promoter such as cytomegalovirus or elongation factor 1a to drive Cas9 expression and U6 to drive RNA expression), and the components can assemble in the transfected cells. Another finding is that the CRISPR system can efficiently modify the genome in a site-specific fashion both at endogenous loci and at reporter genes by either mutagenic nonhomologous end joining (NHEJ) or homologous recombination (HR) pathways. Cong et al.,1 for example, mutated up to 19% of EMX1 alleles in HEK-293T cells using their CRISPR system, and Mali et al.2 stimulated gene correction in ~7% of cells in an integrated reporter gene. The latter group also demonstrated allelic mutation frequency up to 37% in an endogenous gene in K562 cells and up to 4% in an induced pluripotent stem cell line.2 Whether the reduced efficiency in induced pluripotent stem cells relative to K562 cells reflects differential activity of the CRISPR system or a fundamental difference in mutagenic repair between these cell types remains to be determined. Third, both groups demonstrated the feasibility of multiplex genome engineering. Cong et al.,1 for example, showed that they could simultaneously modify two genes; Mali et al.2 describe the range of genomic targets that are accessible to the CRISPR system (40% of known exons are potential targets). However, the large number of potential CRISPR target sites may still fall substantially short of the range of target sites that can be modified by TALENs. Fourth, both groups found that the efficiency of genome modification using the CRISPR system is greater than that reported for TALENs, although the frequency of genome modification by the TALENs used by both groups falls short of what has been reported elsewhere. Future studies that compare the CRISPR system with more active TALENs will be important in establishing the relative efficiency of the two systems.
Finally, a version of Cas9 that creates nicks rather than breaks had previously been described, and both groups studied the genome-editing capability of the CRISPR-nicking system. In prior studies examining nicking nucleases vs. breaking nucleases, nicking systems showed higher relative rates of HR-based repair as compared with mutagenic NHEJ-based repair. As expected, both groups found a low rate of insertions or deletions at the target site using the nicking system, with a relative bias toward HR-mediated repair. In contrast to recombination activating gene (RAG)–based18 and ZFN-based19,20 nicking studies in which HR frequency was often 10-fold or more lower than that generated by double-strand breaks, the CRISPR nicking system resulted in only a twofold decrease in HR. Confirmation and further study of this finding will be of great interest because minimizing unwanted NHEJ mutations without compromising HR-mediated genome editing is a desired property in terms of improving the safety of a nuclease-mediated HR-based approach to gene therapy.
A limitation of the TALEN system is that TALENs cannot be easily packaged into some of the more common virus-based delivery systems. The large size of TALENs, for example, means that they cannot be easily packaged into AAV vectors, particularly self-complementary AAV vectors. The highly repetitive nature of TALENs means that they cannot be easily packaged into retroviral and lentiviral vectors that do not tolerate repetitive elements. The CRISPR–Cas system is smaller and lacks repetitive elements, and therefore might be incorporated into AAV and retroviral or lentiviral delivery systems.
With all nuclease-based genome-editing systems, off-target effects are a concern because off-target activity could lead to genome changes such as small mutations or gross chromosomal rearrangements that would predispose the cell to transformation. Interestingly, Cong et al.1 demonstrated that, whereas their system could tolerate certain single-base-pair changes between the guide and target sequences, other single-base-pair changes, particularly in the 12-base-pair “seed” region, resulted in complete abrogation of activity. These results suggest that the specificity of the CRISPR system may not be conferred by the entire length of the guide RNA but may instead be determined by a 12- to 14-base-pair core that consists of a “seed” plus “PAM,” protospacer-adjacent motif, sequence. This length has important implications regarding the genome-wide specificity of the CRISPR system: the shorter the DNA recognition site of the CRISPR system, the higher the probability that the system will recognize other genomic sites, thereby increasing the probability of off-target genetic changes.
In addition to elucidating the issue of off-target activity, it is important to understand how a prokaryotic-based system will react in a mammalian environment. The system requires pairing of the guide RNA with single-stranded DNA. Although there is some “breathing” of double-stranded DNA, most genomic DNA is in a double-stranded form. It will be interesting to determine whether the CRISPR system requires transcription or replication by the host machinery to create single-stranded DNA to which the guide RNA can pair or whether the intrinsic helicase activity of Cas9 is capable of creating the RNA–DNA hybrid duplex without the need for host proteins. If Cas9 is not able to create such hybrids, it suggests that areas of the genome that are not transcribed in cell types that are not dividing would be resistant to CRISPR-mediated modification.
In summary, the CRISPR system is a new and exciting approach to nuclease-mediated genome editing. The ease of engineering TALENs relative to ZFNs has been a critical factor in the recent spread in genome editing as a research tool. Of the available methods for engineering TALENs relatively simply, the CRISPR strategy offers the potential to be particularly straightforward because it requires only the identification of a short guide-RNA sequence that can be incorporated into a simple expression vector. Thus, the near future is likely to be an exciting time as TALENs and CRISPR battle for the hearts and minds of researchers interested in the research and clinical applications of genome editing.
References
- Cong L, Ran FA, Cox D, Lin S, Barretto L, Habib N.et al. (2013Multiplex genome engineering using CRISPR/Cas Systems Science e-pub ahead of print 3 January 2013 [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE.et al. (2013RNA-guided human genome engineering via Cas9 Science e-pub ahead of print 3 January 2013; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard BL. Homing endonucleases: from microbial genetic invaders to reagents for targeted DNA modification. Structure. 2011;19:7–15. doi: 10.1016/j.str.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S.et al. (2005Highly efficient endogenous human gene correction using designed zinc-finger nucleases Nature 435646–651. [DOI] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF.et al. (2011A TALE nuclease architecture for efficient genome editing Nat Biotechnol 29143–150. [DOI] [PubMed] [Google Scholar]
- Aiba Y, Sumaoka J., and, Komiyama M. Artificial DNA cutters for DNA manipulation and genome engineering. Chem Soc Rev. 2011;40:5657–5668. doi: 10.1039/c1cs15039a. [DOI] [PubMed] [Google Scholar]
- Hendrie PC. Hirata RK and Russell, DW (2003). Chromosomal integration and homologous gene targeting by replication-incompetent vectors based on the autonomous parvovirus minute virus of mice. J Virol. 77:13136–13145. doi: 10.1128/JVI.77.24.13136-13145.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica MS., and, Soddard BL. Homing endonucleases: structure, function and evolution. Cell Mol Life Sci. 1999;10:1304–1326. doi: 10.1007/s000180050372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DW., and, Hirata RK. Human gene targeting by viral vectors. Nat Genet. 1998;4:325–330. doi: 10.1038/ng0498-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YG, Cha Y., and, Chandrasegaram S. Hybrid restriction enzymes: zinc finger fusion to FokI cleavage domain. Proc Natl Acad Sci USA. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Beumer K, Trautman JK., and, Carroll D.2003Enhancing gene targeting with designed zinc finger nucleases Science 300764. [DOI] [PubMed] [Google Scholar]
- Porteus MH., and, Baltimore D.2003Chimeric nucleases stimulate gene targeting in human cells Science 300763 [DOI] [PubMed] [Google Scholar]
- Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O.et al. (2008Establishing of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases Nat Biotechnol 26808–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiyama M, Aiba Y, Yamamoto Y., and, Sumaoka J. Artificial restriction DNA cutter for site-selective scission of double-stranded DNA with tunable scission site and specificity. Nat Protoc. 2008;4:655–662. doi: 10.1038/nprot.2008.7. [DOI] [PubMed] [Google Scholar]
- Bogdanove AJ., and, Voytas DF. TAL effectors: customizable proteins for DNA targeting. Science. 2011;6051:1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- Pruett-Miller SM, Connelly JP, Maeder ML, Joung JK., and, Porteus MH. Comparison of zinc finger nucleases for use in gene targeting in mammalian cells. Mol Ther. 2008;16:707–717. doi: 10.1038/mt.2008.20. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA., and, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock DM, Nakanishi K, Helgadottir HR., and, Jasin M. Assaying double-strand break repair pathway choice in mammalian cells using a targeted endonuclease or the RAG recombinase. Methods Enzymol. 2006;409:524–540. doi: 10.1016/S0076-6879(05)09031-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez CL, Certo MT, Mussolino C, Goodwin MJ, Cradick TJ, McCaffrey AP.et al. (2012Engineered zinc finger nickases induce homologous-direct repair with reduced mutagenic effects Nucleic Acids Res 405560–5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Friedman G, Doyon Y, Wang NS, Li CJ, Miller JC.et al. (2012Targeted gene addition to a predetermined site in the human genome using ZFN-based nicking enzyme Genome Res 71316–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]


