Abstract
Helper-dependent adenoviral (HDAd) vectors can mediate long-term, high-level transgene expression from transduced hepatocytes with no chronic toxicity. However, a toxic acute response with potentially lethal consequences has hindered their clinical applications. Liver sinusoidal endothelial cells (LSECs) and Kupffer cells are major barriers to efficient hepatocyte transduction. Understanding the mechanisms of adenoviral vector uptake by non-parenchymal cells may allow the development of strategies aimed at overcoming these important barriers and to achieve preferential hepatocyte gene transfer with reduced toxicity. Scavenger receptors on Kupffer cells bind adenoviral particles and remove them from the circulation, thus preventing hepatocyte transduction. In the present study, we show that HDAd particles interact in vitro and in vivo with scavenger receptor-A (SR-A) and with scavenger receptor expressed on endothelial cells-I (SREC-I) and we exploited this knowledge to increase the efficiency of hepatocyte transduction by HDAd vectors in vivo through blocking of SR-A and SREC-I with specific fragments antigen-binding (Fabs).
Introduction
Helper-dependent adenoviral (HDAd) vectors hold tremendous potential for liver-directed gene therapy because they can mediate long-term, high-level transgene expression from transduced hepatocytes with no chronic toxicity.1 Following systemic administration in mice and large animal models, Ad vector particles reach the liver through the hepatic artery; in thin-wall liver sinusoids viral particles (vp) come in contact with fenestrated liver sinusoidal endothelial cells (LSEC) and with Kupffer cells, that are liver resident macrophages protruding into the vascular space and rapidly remove blood-borne Ad particles.2,3,4,5 This uptake of vector by the reticuloendothelial system appears to contribute to Ad-induced innate immune responses.5,6,7 A thorough understanding of the molecular events and players involved in recognition and uptake of intravenously injected Ad particles by the reticuloendothelial system is an important first step for overcoming the obstacles to successful HDAd-mediated gene therapy. Such efforts are required to achieve preferential hepatocyte gene transfer and to minimize Ad-mediated acute toxic response.8,9
Ad vectors have shown great potential for cancer treatment and have been extensively studied in clinical trials involving over 15,000 patients.10,11,12,13 Although some effects on tumor growth have been reported, the efficacy of intravenous administration of Ad vectors in patients with metastatic cancers has been limited, likely because of rapid vector clearance from blood by Kupffer cells.13,14 Therefore, understanding of the receptors involved in Ad vector uptake and strategies to avoid these receptors have potential to increase tumor transduction by Ad vectors and therefore, to improve therapeutic efficacy against cancer cells.15
Ad infection occurs through different routes, with a variety of receptors on the plasma membrane of the host cells that interact with Ad capsid proteins. Serotype 5 Ad (Ad5) binds to coxsackie and adenovirus receptor,16,17 integrins,16,17 and scavenger receptors.18,19 Moreover, blood-borne Ad5 are opsonized by coagulation factors, especially factor X, complement, and natural antibodies, thus affecting vector tropism.20 In the liver, hepatocytes are transduced via heparan sulfate proteoglycan-mediated FX binding.17,21,22 In contrast, the mechanism by which Kupffer cells take up Ad particles does not require coxsackie and adenovirus receptor or integrins.23 Ad uptake by LSEC is almost unknown and their role in Ad liver sequestration is controversial.2,24,25 Recently, it has been reported that administration of polyinosine (poly[I]), as well as other polyanionic ligands, into mice before Ad injection drastically reduces Ad accumulation in Kupffer cells and increases hepatocyte gene transfer.18,26 This observation has led to the hypothesis that scavenger receptor-A (SR-A), and possibly other scavenger receptors, bind intravenously injected Ad particles.19,26 Experiments in Chinese hamster ovary cells suggested that also SREC-I bind Ad5 particles.27 Pre-incubation of Kupffer cells with knob 5 resulted in strong inhibition of Ad5 infection thus suggesting a role of the knob in virus uptake by SR-A.19 However, direct evidence of in vivo interaction of the Ad vector with SR-A and SREC-I have not been provided yet.
In this study, we show that HDAd particles interact in vivo with SR-A and we identified the scavenger receptor expressed on endothelial cells-I (SREC-I) as a new player involved in HDAd vector binding and uptake. In addition, we show that blocking in vivo these receptors can be exploited to increase hepatocyte transduction efficiency.
Results
Poly[I] increases HDAd-mediated hepatocyte transduction but lowers the threshold of HDAd toxicity
Improving the therapeutic index of HDAd vector by increasing hepatocyte transduction efficiency has potential for clinical translation of liver-directed gene therapy. Towards this goal, we have investigated the effect of poly[I] at increasing hepatocyte gene transfer. With the notion that poly[I] pre-treatment improves the therapeutic index of HDAd, we have determined the minimal poly[I] dose able to increase HDAd-mediated liver expression. Previous studies have used 0.2 mg of poly[I] by intravenous injection to increase Ad-mediated liver transduction.19,26,28 We have tested 0.4, 0.1, 0.05, and 0.025 mg of poly[I] in comparison to 0.2 mg of poly[I] (at least n = 3 per group), injected intravenously 5 minutes before injection of 5 × 1011 viral particle (vp)/kg of an HDAd vector expressing the baboon α-fetoprotein (AFP) reporter gene under the control of a liver-specific promoter (HDAd-AFP)29,30 in wild-type C57BL/6 mice. The serum levels of AFP measured at 7 days post-injection were similar in mice pre-dosed with 0.2 or 0.05 mg of poly[I], both being approximately fivefold higher than serum AFP measured in control mice receiving saline as pre-dosing (one-way analysis of variance (ANOVA) and post-hoc Tukey's test: P < 0.01) (Figure 1a). Therefore, we concluded that the minimal effective dose of poly[I] that increases HDAd-mediated hepatocyte transduction in mice is 0.05 mg. The dose of 0.05 mg of poly[I] is also associated with long-term transgene expression following the injection of 1 × 1011 vp/kg of HDAd-AFP (n = 3 per group; Figure 1b). Therefore, poly[I] pre-treatment does not compromise HDAd-mediated long-term transgene expression.
Figure 1.
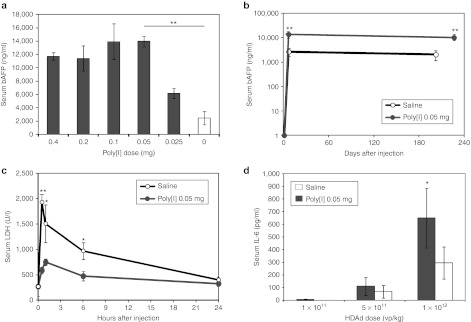
Polyinosine increases hepatocyte transduction, results in long-term transgene expression, reduces serum LDH a marker of Kupffer cell death, but it lowers threshold of HDAd acute toxicity. (a) Serum AFP levels 7 days post-injection in C57BL/6 wild-type mice pre-treated with different doses of polyinosine (poly[I]) (gray bars), or saline (white bar), 5 minutes before the injection of 5 × 1011 vp/kg of HDAd-AFP. The dose of 0.05 mg poly[I] is the minimal effective dose that increased liver transduction (at least n = 3 per group; one-way ANOVA and post-hoc Tukey's test: **P < 0.01). (b) Serum AFP in wild-type C57BL/6 mice injected with poly[I] before the injection of 1 × 1011 vp/kg of HDAd-AFP showed stable and higher levels of transgene expression compared to mice treated with saline before the injection of the same vector dose (n = 3 per group; **P < 0.01). (c) Serum LDH in wild-type C57BL/6 mice injected with either saline or poly[I] before the injection of 5 × 1011 vp/kg of HDAd-AFP. Early (30 minutes, 2 hours, 6 hours) increase in serum LDH post-HDAd vector administration, a marker of Kupffer cell killing, is reduced in poly[I] pre-treated wild-type C57BL/6 mice (at least n = 3 per group; *P < 0.05, **P < 0.01). (d) Serum IL-6 in wild-type C57BL/6 mice treated with 0.05 mg of poly[I] or saline 5 minutes before the injection of different doses of HDAd-AFP. The dose of 1 × 1012 vp/kg resulted in significantly higher levels of serum IL-6 at 6 hours post-injection compared with saline pre-treated mice (n = 5 per group; *P < 0.05). At the highest dose used (5 × 1012 vp/kg) pre-treatment with poly[I] resulted in 100% mortality within 1 hour post-injection and serum IL-6 levels could not be determined. AFP, α-fetoprotein; ANOVA, analysis of variance; HDAd, helper-dependent adenoviral vector; IL, interleukin; LDH, lactate dehydrogenase; Poly[I], polyinosine; vp, viral particle.
The increased hepatocyte transduction observed as a consequence of poly[I] pre-treatment is explained by a reduction of vp uptake by Kupffer cells.18 To confirm this, we determined the hepatic clearance rate of vector genomes between poly[I] and saline pre-treated mice. We found that hepatic HDAd copy number in animals pre-treated with poly[I] was the same as saline pre-treated mice at 1 hour post-injection but it was approximately 2.5-fold higher (t-test: P < 0.05) at 24 and 48 hours post-injection in poly[I] pre-treated compared with saline pre-treated animals (n = 3 per group; Supplementary Figure S1). The reduced hepatic HDAd genome clearance in poly[I] pre-treated mice is consistent with increased hepatocyte transduction and reduced HDAd absorption by Kupffer cells, because while HDAd transduction does not kill hepatocytes, Kupffer cells that take up Ad are killed and quickly removed from the liver.31 This finding is also consistent with immunostaining data showing less Kupffer cell uptake and killing in poly[I]-treated mice.18,19 Acute elevation in serum lactate dehydrogenase (LDH) is a marker of Kupffer cell killing following uptake of Ad.31 Thus, we measured serum LDH shortly (30 minutes) after HDAd injection and found reduced serum LDH in poly[I] pre-treated mice compared with saline pre-treated mice, a result consistent with reduced Kupffer cell killing by Ad in poly[I] pre-treated mice (n = 3 per group; Figure 1c).
Although effective at improving hepatocyte gene transfer efficiency, poly[I] lowered the threshold of HDAd-mediated acute toxicity. The increase in serum interleukin-6 was higher in animals receiving 1 × 1012 vp/kg of HDAd following pre-treatment with poly[I] compared with saline pre-treated mice (n = 5 per group; one-way ANOVA and post-hoc Tukey's test: P < 0.001) (Figure 1d). At a higher dose of 5 × 1012 vp/kg, 100% mortality occurred within the first hour post-injection for mice (n = 5) pre-treated with poly[I], while no mortality was observed in mice pre-treated with saline (n = 5). Mortality was not observed neither in mice injected with 5 × 1012 vp/kg alone nor with 0.05 mg poly[I] alone. These data are consistent with previous observations of increased mortality in mice receiving poly[I] at the higher dose of 0.2 mg in combination with an early generation Ad vector.26 The reasons for the increased toxicity of poly[I] and HDAd are unknown. Taken together, these data indicate that poly[I] increases acute toxicity of HDAd vectors and thus, poly[I] has limited potential for clinical application aiming at improving Ad-mediated liver transduction. Nevertheless, poly[I] remains a useful tool to understand mechanisms leading to improved hepatocyte transduction and to acute toxicity. Understanding the mechanisms involved in poly[I]-mediated increase in hepatocyte transduction by Ad vectors and avoidance of Kupffer cell uptake may allow the development of strategies to achieve preferential hepatocyte gene transfer.
HDAd hepatocyte transduction is paradoxically reduced in SR-A−/− mice
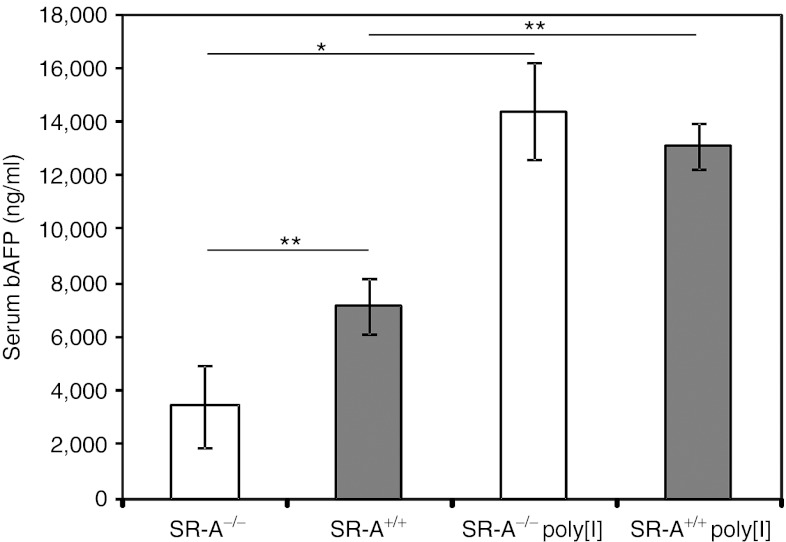
Poly[I] is thought to block the uptake of Ad particles mediated by SR-A expressed on the Kupffer cells.19 To investigate the role of SR-A in improving transduction and acute toxicity, we injected wild-type C57BL/6 mice and SR-A knockout (SR-A−/−) mice in C57BL/6 background,32 with 1 × 1012 vp/kg of HDAd-AFP (n = 3 per group). We observed a significant 2.1-fold reduction of hepatocyte transduction in SR-A−/− mice compared with wild-type mice (SR-A+/+) at 7 days post-injections (one-way ANOVA and post-hoc Tukey's test: P < 0.01; Figure 2). These results were unexpected because poly[I] is a known inhibitor of SR-A18,28 and knockout of this gene was expected to mimic poly[I]-mediated inhibition of SR-A, thus resulting in increased hepatocyte transduction. We next investigated whether poly[I] would increase hepatocyte transduction in SR-A−/− mice. HDAd-mediated hepatocyte transduction in SR-A−/− mice pre-treated with 0.05 mg of poly[I] was still significantly (4.1-fold) higher than saline pre-treated SR-A−/− mice (P < 0.05; Figure 2). Based on these findings, we hypothesized that other scavenger receptor(s) besides SR-A could be inhibited by poly[I] and therefore, are involved in Ad vector uptake. This hypothesis was supported by previous data showing that Ad vector DNA content following intravenous injection of Ad is similar in the livers of wild-type and SR-A−/− mice.17,26
Figure 2.
SR-A−/− mice showed unexpected reduction in HDAd-mediated hepatocyte transduction and poly[I] retains its effect of increasing hepatocyte transduction in SR-A−/− mice. Wild-type (SR-A+/+) and SR-A−/− mice were treated with 0.05 mg of poly[I] or saline 5 minutes prior the injection of 5 × 1011 vp/kg of HDAd-AFP. Serum AFP levels measured at 7 days post-injections were higher in both wild-type and SR-A−/− mice treated with poly[I] compared to saline pre-treated controls (at least n = 3 per group; one-way ANOVA and post-hoc Tukey's test: *P < 0.05; **P < 0.01). A 2.1-fold reduction in hepatocyte transduction was observed in SR-A−/− mice compared with wild-type mice (at least n = 3 per group; **P < 0.01). AFP, α-fetoprotein; ANOVA, analysis of variance; HDAd, helper-dependent adenoviral vector; Poly[I], polyinosine; vp, viral particle.
SREC-I is involved in HDAd uptake in vitro
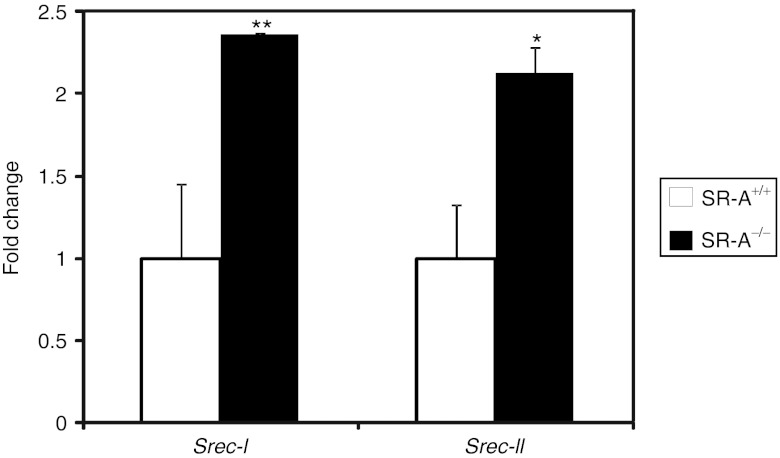
SR-A and SREC-I have been previously shown to cooperate in recognition of hepatitis C virus NS3 protein and TLR2-mediated activation of myeloid cells.33 Moreover, SREC-I is expressed on both macrophages and endothelial cells34,35 which are barriers to Ad-mediated hepatocyte gene transfer.1 Remarkably, SREC-I was also demonstrated to increase Ad5 transduction when stably transfected in Chinese hamster ovary cells.27 Therefore, we measured the levels of expression of Srec-I and II in livers of SR-A−/− mice by real-time PCR and found that their expression was indeed increased compared with wild-type C57BL/6 mice (t-test: P < 0.01 and P < 0.05, respectively; Figure 3). As a control, SR-A levels were also measured in SR-A−/− mice and were not detectable (data not shown). Therefore, SR-A−/− mice exhibit a compensatory increase in expression of SREC-I and II. Based on these findings, we hypothesized that SREC-I and SREC-II are involved in Ad uptake in vivo and their upregulation might contribute to the reduced levels of hepatic transduction observed in SR-A−/− mice.
Figure 3.
SR-A−/− mice have a compensatory increase in Srec-I and Srec-II mRNA levels. By real-time PCR, the levels of expression of Srec-I and II in livers of SR-A−/− mice were increased compared with wild-type C57BL/6 mice (SR-A+/+). Data are expressed as expression fold change calibrated to SR-A+/+ expression (n = 3 per group; t-test: *P < 0.05, **P < 0.01).
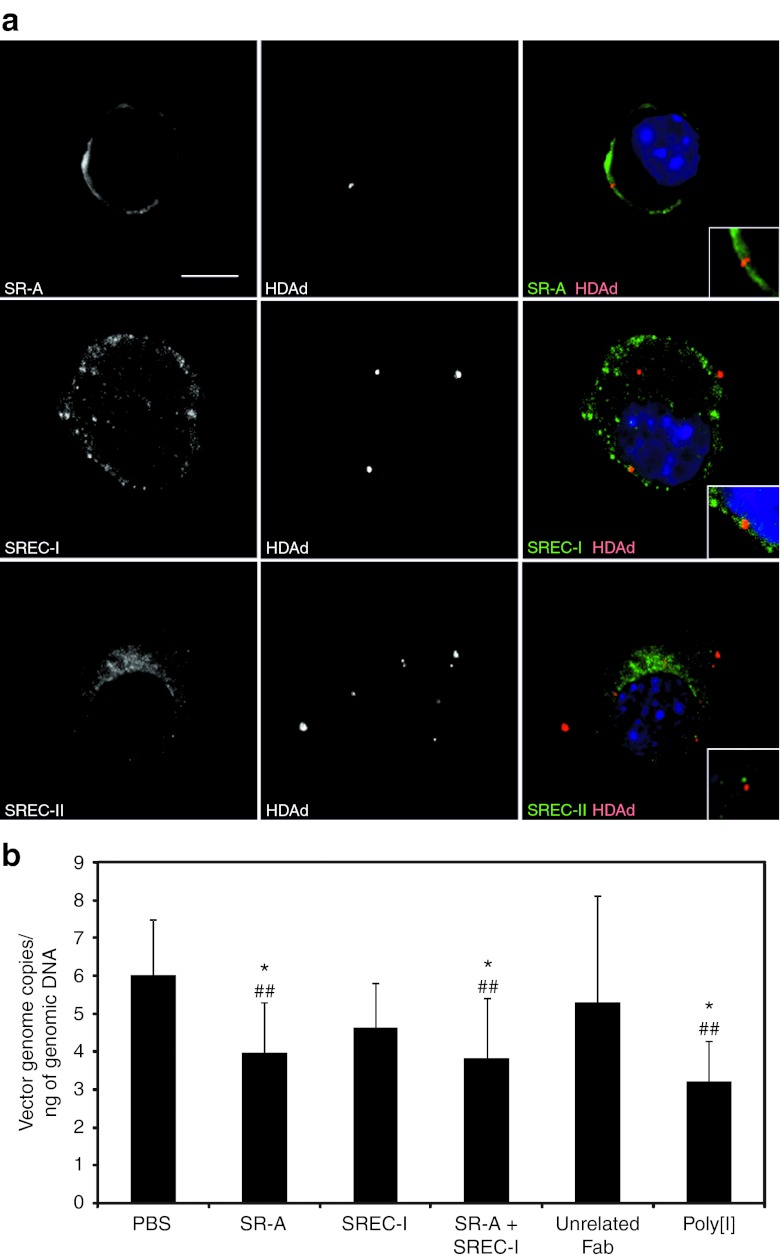
To test the hypothesis that SRECs are involved in HDAd binding, we infected J774A.1 cells, a murine monocyte-macrophage derived cell line, with AlexaFluor-555–labeled HDAd and performed immunofluorescence staining against SR-A, SREC-I, and SREC-II followed by confocal image analysis. As shown in Figure 4a, SR-A and SREC-I were found to colocalize with fluorescent HDAd vector at the plasma membrane level in 36 and 28% of analyzed cells, respectively. No colocalization was detected with SREC-II. To rule out that vector infectivity was affected by the fluorescent labeling procedure, we measured vector genome copy number by quantitative PCR (qPCR) in J774A.1 cells infected with either AlexaFluor-555–labeled HDAd or with an unlabeled HDAd, as a control, at different multiplicity of infections (MOIs). Although the infectivity of the AlexaFluor-555–labeled HDAd was slightly reduced at higher MOIs (ranging from 27 to 35%) compared with the unlabeled HDAd (t-test: P < 0.05; Supplementary Figure S2), the reduction falls within the range of variability in infectivity among vector lots.36
Figure 4.
HDAd vector binds to SR-A and SREC-I and blocking of SR-A and SREC-I reduces HDAd uptake in J774A.1 cells. (a) Immunofluorescence showed colocalization of SR-A (top panel) (green) and SREC-I (middle panel) (green) with AlexaFluor-555–labeled-HDAd (red) at the plasma membrane of J774A.1 cells, while no colocalization was detected with SREC-II (bottom panel) (green). Nuclei are counterstained with DAPI (blue). Bar: 10 µm. (b) HDAd genome copy number quantification by real-time PCR showed reduced viral uptake in J774A.1 cells pre-treated with anti-SR-A Fab compared with control treated with PBS or unrelated Fab. Cells were first pre-incubated with poly[I] (30 µg/ml), anti-SR-A Fab (1 ng/ml), anti-SREC-I Fab (1 ng/ml), anti-SR-A Fab + anti-SREC-I Fab (1 ng/ml each), unrelated IgG Fab (1 µg/ml), or left untreated (PBS) and then infected with HDAd-LacZ at MOI of 100 vp/cell. Results are expressed as vector genome copies per ng of total genomic DNA (n = 3 at least per treatment group; one-way ANOVA and post-hoc multicomparison Tukey's test: *P < 0.05 compared with PBS group; ##P < 0.01 compared with unrelated Fab). ANOVA, analysis of variance; DAPI, 4′,6-diamidino-2-phenylindole; HDAd, helper-dependent adenoviral vector; MOI, multiplicity of infection; PBS, phosphate-buffered saline; Poly[I], polyinosine; vp, viral particle.
We next evaluated whether SREC-I is involved in HDAd uptake and infected J774A.1 cells with HDAd expressing LacZ (HDAd-LacZ)8 in the presence of fragments antigen-binding (Fabs) purified from anti-SR-A antibody, anti-SREC-I antibody, and an unrelated goat IgG. As controls, J774A.1 cells were infected with either HDAd-LacZ alone or in the presence of poly[I]. We found that pre-incubation with anti-SR-A blocking Fabs resulted in a reduction of virion uptake compared with cells pre-incubated with the unrelated Fab or with phosphate-buffered saline (PBS) (one-way ANOVA and post-hoc Tukey's test: P < 0.01 versus unrelated Fab and P < 0.05 versus PBS), as shown by qPCR for HDAd vector genomes (Figure 4b). Anti-SREC-I Fab also reduced HDAd uptake, even though the results did not reach statistical significance, while the combination of anti-SR-A and anti-SREC-I Fabs showed a statistically significant inhibitory effect on HDAd uptake (P < 0.01 versus unrelated Fab and P < 0.05 versus PBS) (Figure 4b). It should be pointed out that SREC-I expression in macrophages is lower compared with SR-A, and therefore these cells are not a good model to investigate SREC-I functions.35 Pre-incubation with Fab from an unrelated goat IgG did not affect HDAd transduction (Figure 4b).
SR-A and SREC-I are receptors for HDAd
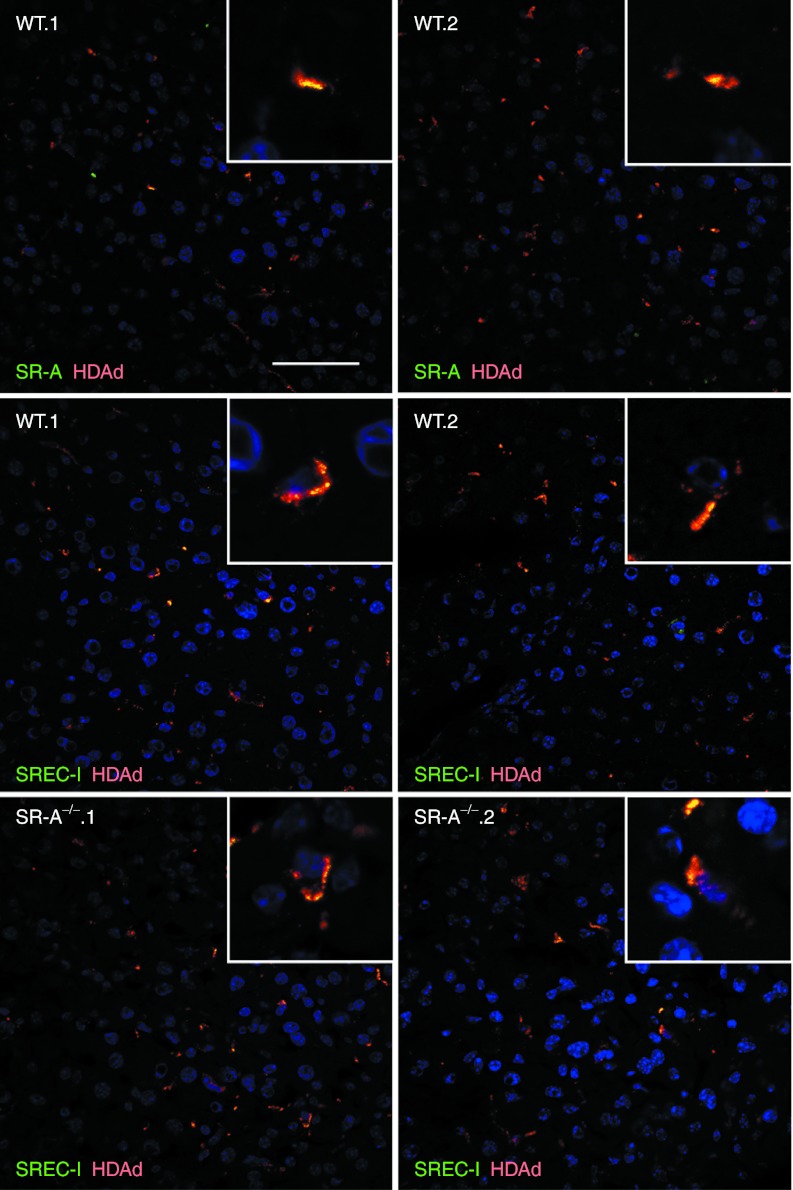
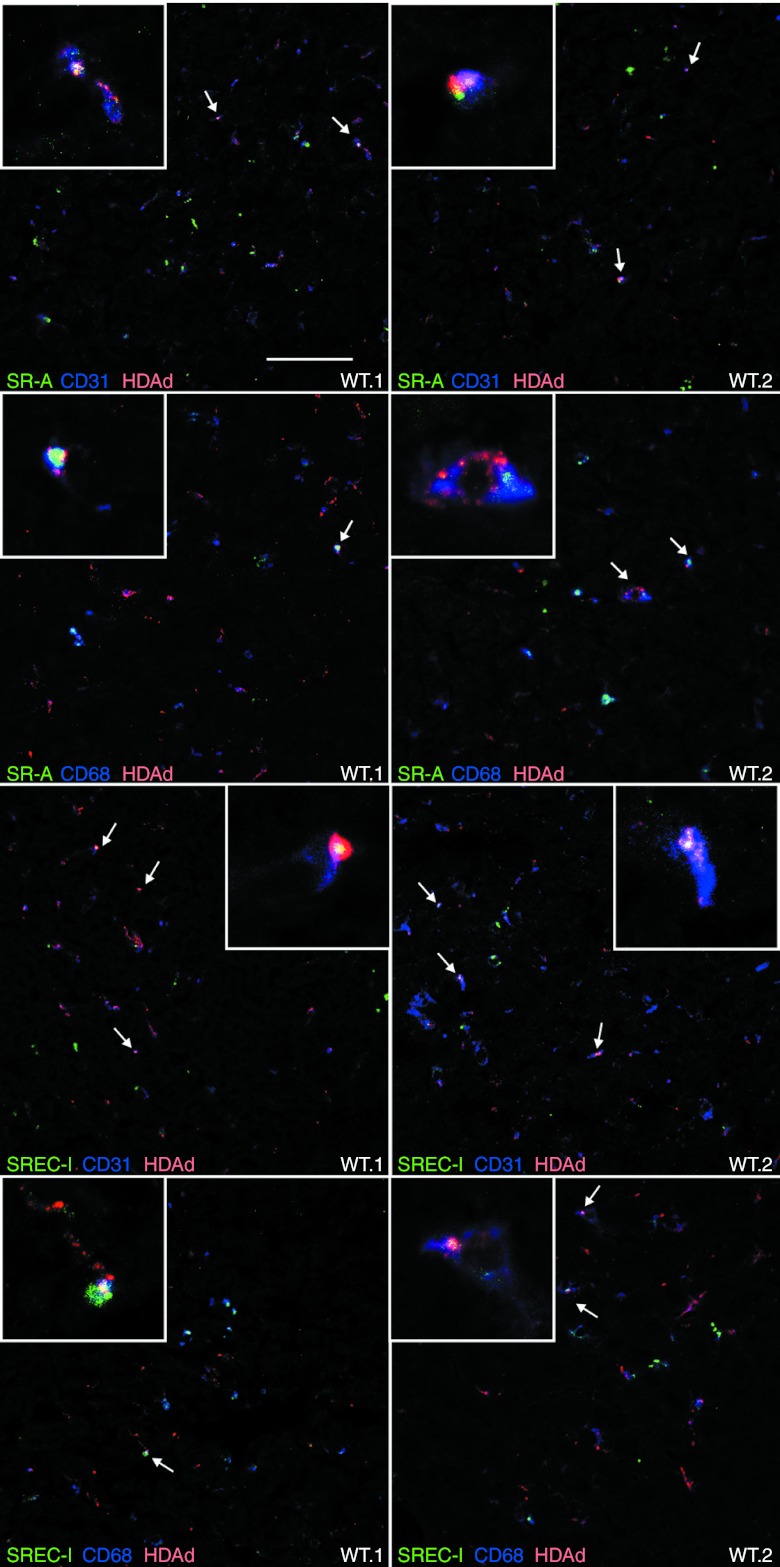
C57BL/6 mice were injected intravenously with 5 × 1011 vp/kg of AlexaFluor-555–labeled HDAd (n = 4) or saline (n = 2) and livers, harvested 30 minutes after injection, showed colocalization of SREC-I and HDAd vector, thus suggesting in vivo interaction of HDAd particles with SREC-I receptor, both in wild-type and SR-A−/− mice (Pearson's correlation coefficient (RP) = 0.70 and 0.56, respectively; Figure 5, Supplementary Figures S3 and S4). Interaction of SR-A with fluorescent-labeled HDAd was also detected (RP = 0.55; Figure 5, Supplementary Figures S3 and S4). Co-staining with CD31 and CD68 suggested that in vivo HDAd interactions with SR-A and SREC-I occur in both LSECs and Kupffer cells, respectively (Figure 6, Supplementary Figures S5 and S6).
Figure 5.
SR-A and SREC-I colocalize with HDAd vector particles in mouse livers. Livers from AlexaFluor-555–labeled-HDAd (red) injected wild-type C57BL/6 (WT.1 and WT.2) and SR-A−/− mice (SR-A−/−.1 and SR-A−/−.2) were stained for SR-A (green) and SREC-I (green) for confocal analyses. Both SR-A and SREC-I colocalize with HDAd in wild-type livers, while SREC-I-HDAd double-positive signals are increased in SR-A−/− livers (bottom panels). Nuclei were counterstained with DAPI (blue). Bar: 50 µm. DAPI, 4′,6-diamidino-2-phenylindole; HDAd, helper-dependent adenoviral vector.
Figure 6.
SR-A and SREC-I colocalize with HDAd particles in both Kupffer and endothelial cells in the liver. Confocal analysis of livers from AlexaFluor-555–labeled HDAd-injected wild-type C57BL/6 mice (WT.1 and WT.2) stained for SR-A (green) or SREC-I (green) and CD31 (blue) or CD68 (blue), as markers of endothelial and Kupffer cells, respectively. SR-A-HDAd and SREC-I-HDAd double-positive signals were detected in both CD31+ and CD68+ cells. Triple-positive cells are indicated by the arrows. Bar: 50 µm. HDAd, helper-dependent adenoviral vector.
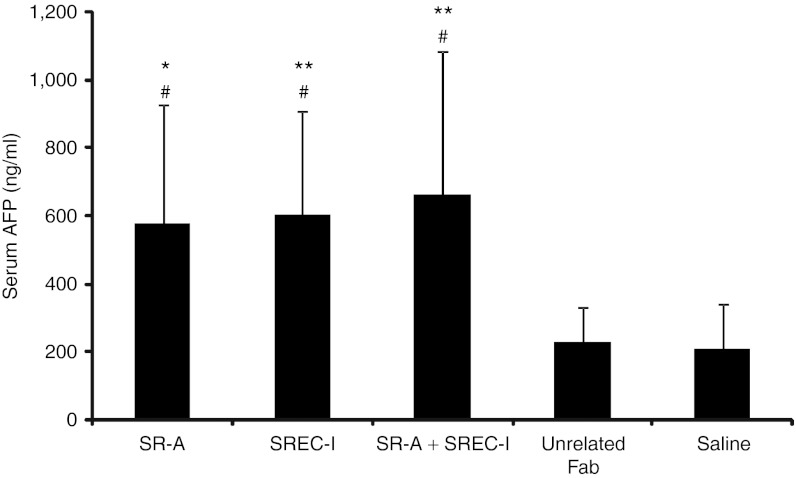
To investigate whether blocking in vivo of SR-A and SREC-I increases hepatocyte transduction efficiency, we pre-treated BALB/c mice with anti-SR-A and anti-SREC-I Fabs before the intravenous injection of 5 × 1011 vp/kg of HDAd-AFP vector. BALB/c mice were used for these studies because available blocking anti-SR-A 2F8 antibody was developed from the BALB/c strain-derived macrophage cell line, RAW264.7 and does not cross-react with cells and tissues from C57BL/6 mice.37 As previously reported,38 the efficiency of HDAd-mediated liver transduction in BALB/c mice is reduced compared with C57BL/6 (Figures 1 and 7). Serum AFP levels at 7 days post-injection were found to be significantly increased in mice pre-treated with anti-SR-A (one-way ANOVA and post-hoc Tukey's test: P < 0.05) or anti-SREC-I (P < 0.01 and P < 0.05) Fabs, or with a combination of the two (P < 0.01 and P < 0.05) compared with saline and urelated Fab pre-treated animals, respectively (Figure 7). Pre-administration of Fab from unrelated IgG instead did not result in any significant change of liver transduction efficiency compared with saline-injected controls (Figure 7).
Figure 7.
Pre-treatment with anti-SR-A and anti-SREC-I blocking antibodies enhances hepatocyte transduction efficiency. Wild-type BALB/c mice were injected with anti-SR-A Fab (SR-A), anti-SREC-I Fab (SREC-I), a combination of both (SR-A + SREC-I), unrelated IgG Fab (unrelated Fab), or saline 5 minutes before the injection of 5 × 1011 vp/kg of HDAd-AFP. Pre-treatments with anti-SR-A or anti-SREC-I Fabs and with a combination of both resulted in increased serum AFP levels at 7 days post-injection (at least n = 7 per treatment group; one-way ANOVA and post-hoc multicomparison Tukey's test: *P < 0.05; **P < 0.01 compared with saline group; #P < 0.05 compared with unrelated Fab). AFP, α-fetoprotein; ANOVA, analysis of variance; HdAd, helper-dependent adenoviral vector; vp, viral particle.
Discussion
Intravenously injected Ad particles reach the liver through the portal vein and contact most hepatocytes only after passing through the liver sinusoids, the walls of which are formed by endothelial cells. Within the liver sinusoids are located the Kupffer cells, which are non-parenchymal macrophages that avidly take up blood-borne Ad particles. In the present study, we used poly[I], a polyanionic compound, that is a well-established inhibitor of scavenger receptors on non-parenchymal liver cells,39 as a tool to investigate the molecular players involved in Ad uptake, which are important obstacles for efficient hepatocyte transduction by HDAd vectors. We have shown that poly[I], at the minimal dose of 0.05 mg per mouse before HDAd injection, increases hepatocyte transduction at 7 days post-injection without affecting long-term transgene expression for up to 226 days post-injection, reduces serum LDH elevation and HDAd vector genome hepatic clearance (Figure 1 and Supplementary Figure S1). Delineating the mechanisms for poly[I]-mediated increase of hepatic transduction efficiency by Ad vectors is important for improving the vector therapeutic index. The poly[I]-mediated increase in hepatocyte transduction by Ad vectors has been attributed to blockade of Ad vector uptake by Kupffer cells which are hypothesized to bind Ad particles through scavenger receptors.26 More recently, the SR-A has been suggested to play a major role in this process.19 However, evidence of Ad uptake by Kupffer cells via SR-A has not been demonstrated in vivo so far.
Scavenger receptors are a highly heterogenic group of membrane receptors sharing little sequence homology with high redundancy of ligand binding.40 They bind modified low-density lipoproteins and modified albumin but are also involved in recognition and uptake of blood-borne pathogens. Scavenger receptors recognize negatively charged materials, without any need for opsonization by plasma proteins.41 Interestingly, the Ad capsid bears an overall negative charge, with the amounts of negative charge differing among serotypes due primarily to differences in the hypervariable region of hexon loops.42 Ad5 and the closely related Ad2, which are the most commonly used serotypes in gene therapy studies, are among the most negatively charged serotypes.42 Moreover, it has been demonstrated that some hypervariable regions of Ad5 are able to bind scavenger receptors.27 Unexpectedly, we found that hepatocyte transduction is reduced in mice lacking SR-A (Figure 2). This finding led us to hypothesize the involvement of other SRs in Ad uptake in vivo. We selected SREC-I as a candidate because: (i) we found SREC-I to be upregulated in SR-A−/− mice (Figure 3), (ii) it has been previously shown that SREC-I cooperates with SR-A in recognition of the hepatitis C virus,33 and (iii) SREC-I is expressed on both macrophages and endothelial cells.34,35 We indeed confirmed both in vitro and in vivo that HDAd particles colocalize with SREC-I, which is upregulated in SR-A−/− mice (Figures 4 and 5). Moreover, we provide for the first time evidence of colocalization of HDAd with SR-A in vivo (Figure 5) and demonstrated that this colocalization occurs in both Kupffer cells and LSECs (Figure 6). The role of LSECs in the clearance of blood-borne Ad has recently emerged.2 Our study supports this concept and provides the molecular receptors involved in such important interaction. The role of these receptors in Ad uptake is supported by pre-treatment of mice with anti-SR-A and anti-SREC-I Fabs, purified from blocking antibodies, that resulted in an increased hepatocyte transduction efficiency (Figure 7).
Scavenger receptors are important players in Ad vector clearance and modulation of their interactions with blood-borne Ad particles has great potential to improve systemic gene delivery in vivo. However, Ad interactions in vivo involve multiple processes and cell types. Kupffer cells and endothelial cells clearly play an important role in such interactions. Ad particles are opsonized by natural IgM antibodies and by mouse complement components C3 and C4.26 Whether Kupffer cells can recognize Ad particles opsonized with IgM and complement remains to be clarified. Clearly, Kupffer cells take up Ad particles from the bloodstream via scavenger receptors without any need for opsonization by plasma proteins.
The role of scavenger receptors in innate immune defense as pattern recognition receptors is of a growing interest.43 Interestingly, SR-A and SREC-I have previously been shown to share common ligands: Hsp110 and Grp170, members of the Hsp70 superfamily,44 and the hepatitis C virus NS3 protein.33
Because scavenger receptors recognize negatively charged materials, less negatively charged Ad serotypes might be more successful at evading Kupffer cells. Serotype 6 Ad (Ad6) vectors which bear less net negative charge in hypervariable regions are less efficiently phagocytosed by Kupffer cells compared with Ad5 and chimeric Ad5/Ad6 viruses.45 Modification of Ad vector capsids with non-reactive polymers, such as polymers of poly[N-(2-hydroxypropyl)methacrylamide] or polyethylene glycol, may be another method for evading Kupffer cell uptake.46,47 Alternative strategies to improve the HDAd vector therapeutic index include blockade of scavenger receptors before HDAd administration. Selective blockade of SR-A and SREC-I, as performed in this study by blocking Fabs before vector administration, may be a safe and effective approach to obtain efficient hepatocyte transduction.
In conclusion, this study shows SR-A and SREC-I are both involved in vivo in Ad vector uptake and are potential targets to improve vector therapeutic index by reducing reticuloendothelial uptake that may allow the use of lower, less toxic doses.
Materials and Methods
Vectors. HDAd-AFP and HDAd-LacZ vectors bear a PEPCK-WL-bAFP and a MCMV-LacZ expression cassette, respectively.29,48 HDAd was produced in 116 cells with the helper virus AdNG163 as described previously.36,48 Helper virus contamination levels were determined and were found to be <0.05%. DNA analyses of HDAd genomic structure was confirmed as described elsewhere.48 Fluorescent-labeled HDAd was conjugated with AlexaFluor-555 carboxylic acid, succinimidyl ester (Invitrogen Life Technologies, Carlsbad, CA), as previously described with some modifications.49 Briefly, 100 µl of dimethyl sulfoxide 99% anhydrous (Sigma Aldrich, St Louis, MO) was added to 1 mg of the lyophilized Alexa Fluor dye. The HDAd vector stock was diluted to 5 × 1011 vp/ml in 2 ml of 0.1 mol/l sodium bicarbonate buffer, pH 8.5 and the entire 100 µl of dye was added to 2 ml of diluted virus stock while the solution was mixed by vortexing. The reagents were mixed continuously by vortex for 1 hour at room temperature in a foil-wrapped 15 ml tube. After the 1 hour incubation, the entire 2 ml volume was injected into a 10 kDa MWCO Slide-A-Lyzer dialysis cassette (Pierce Thermo Scientific, Rockford, IL) and dialyzed overnight at 4 °C against a total of three changes of 0.1 mol/l Tris-HCl pH 7.8, 0.1 mol/l MgCl2, 1.5 mol/l NaCl. The dialyzed AlexaFluor-555–labeled HDAd was adjusted to a final concentration of 10% glycerol and stored at −80 °C.
Mice and injections. Nine- to twelve-weeks-old male C57BL/6 (The Jackson Laboratory, Bar Harbor, ME) or BALB/c mice (Charles River Laboratories International, Wilmington, MA) were used for all the experiments. SR-A−/− mice were purchased from The Jackson Laboratory. All the injections were performed retro-orbitally in a volume of 200 µl. Polyinosinic acid potassium salt (Sigma Aldrich) was dissolved in PBS. Fabs from anti-SR-A (HM1061; HyCult Biotechnology, Montrouge, France), anti-SREC-I (AF2409; R&D, Minneapolis, MN), and goat anti-rat IgG (Pierce Thermo Scientific) were generated by Antibody Research (St Charles, MO) by papain digestion and protein A purification (Pierce Thermo Scientific) and were injected at the dose of 500 µg/kg. Poly[I], Fabs, or saline pre-treatments were followed after 5 minutes by vector injection. Serum AFP and mouse interleukin-6 levels were measured as previously reported.29,50 Serum LDH was measured by the chemistry laboratory of the Center for Comparative Medicine (Baylor College of Medicine, Houston, TX). One-way ANOVA followed by post-hoc multicomparison Tukey's test was performed for comparison of experimental groups.
Reverse transcription and real-time qPCR. Complementary DNA was synthesized from 1 µg of total RNA extracted from snap-frozen mouse livers by reverse transcription using Superscript II according to the manufacturer's instruction (Invitrogen Life Technologies). Samples were then diluted 1:10 in nuclease-free water and used as template for real-time qPCR. Real-time qPCR was performed using the Light Cycler Faststart DNA Master SYBR Green I (Roche, Indianapolis, IN) in a total volume of 20 µl with 2 µl of template DNA, 4 mmol/l MgCl2, and 5 µmol/l of each specific primer. Sequences of the mouse primer used in real-time PCR were as follows: Srec-I forward: 5′-GGTCCTGTCTGGTCTGTCTTTCGTCGTT-3′, reverse: 5′-CGCAGAGGCTTAGGGATAGCACTCTTT-3′ Srec-II forward: 5′-CTCCAGGGCCTCCTTCTCATCATTCGACA3′, reverse: 5′-CAGTCGCTTCCTCGTGGGGCACACAGTACA-3′ SRA1 forward: 5′-CTCAGACTGAAGGACTGGGAACACTCAC-3′, reverse 5′-TCACCTTTAACACCTGGAATACCTCTTA-3′ β-actin forward: 5′-TGTTTTGTTTTGGCGCTTTTGACTC-3′, reverse: 5′-TTGTAGAACTTTGGGGGATGTTTGCT-3′.
Cycling conditions consisted of 95 °C for 10 seconds, 50–65 °C for 7 seconds, and 72 °C for 20 seconds. Raw data were analyzed with the 2−ΔΔCT method, normalized to β-actin as housekeeping gene and calibrated to SR-A+/+ expression, using Light Cycler software version 3.5 (Roche).
In vitro infection and vector genome copy analysis. J774A.1 cells were purchased from European Collection of Cell Culture (Salisbury, UK) and cultured according to supplier's instructions. Cells were pre-treated with anti-SR-A, anti-SREC-I, unrelated IgG Fabs, or poly[I] 30 minutes before HDAd infection or left untreated. Cells were infected with HDAd-LacZ at an MOI of 100 vp/cell for 1 hour, infection media were then removed and replaced with fresh regular maintenance media. Total DNA was extracted after 24 hours using standard phenol-chloroform extraction and quantitated by absorbance at 260 nm. At least three different experiments were performed per treatment group.
Quantitative real-time PCR was performed in duplicate for each experiment using the LightCycler FastStart DNA Master SYBER Green I (Roche) in a total volume of 20 ml with 100 ng of template DNA, 1 mmol/l of each HDAd-specific primers (5′-TCTGAATAA TTTTGTGTTACTCATAGCGCG-3′ and 5′-CCCATAAGCTCCTTTT AACTTGTTAAAGTC-3′). Cycling conditions consisted of 95 °C for 10 minutes followed by 45 cycles at 95 °C for 10 seconds, 60 °C for 7 seconds, and 72 °C for 20 seconds. Serial dilutions of a plasmid bearing the PCR target sequence were used as a control to determine the amounts of HDAd. Results were analyzed with Light Cycler software version 3.5 (Roche). One-way ANOVA followed by post-hoc multicomparison Tukey's test was performed as statistical analysis.
Immunofluorescence studies. J774A.1 cells were infected with AlexaFluor-555–labeled HDAd at an MOI of 3 × 104 vp/cell for 15 minutes at 37 °C, and then fixed in 4% paraformaldehyde/PBS pH 7.4. Anti-SR-A and anti-SREC-I (sc-11298; Santa Cruz Biotechnology) were used as primary antibodies. Secondary antibodies were anti-rat AlexaFluor-488 for SR-A and anti-goat AlexaFluor-488 for SREC-I (Invitrogen Life Technologies). Nuclei were counterstained with DAPI (4′,6-diamidino-2-phenylindole) (Invitrogen Life Technologies). Each experiment was performed in triplicate and 20 randomly chosen images were analyzed for each staining.
Four AlexaFluor-555–labeled HDAd (dose 5 × 1011 vp/kg) and two saline-injected animals were perfused with PBS pH 7.4 and harvested livers were fixed with 1% paraformaldehyde/PBS pH 7.4 for 5 minutes. After immersion, post-fixation in 1% paraformaldehyde and 0.5% glutaraldehyde solution for 3 hours followed by overnight immersion in 30% sucrose, livers were included in OCT compound (Tissue-Tek; Sakura Finetek, Torrance, CA). Embedded livers were cryosectioned at 4 µm and fixed in 4% paraformaldehyde. Sections were then permeabilized in PBS, 0.2% Triton and blocked in 5% BSA for SR-A staining and in 3% BSA, 5% donkey serum, 0.3% Tween20, 20 mmol/l MgCl2 for SREC-I staining. Primary antibodies used were anti-SR-A (NBP1-00092; Novus Biologicals, Cambridge, UK), anti-CD31 (550274-BD; Pharmingen, Oxford, UK), anti-CD68 (MCA1957; BD Serotec, Kidlington, UK), and anti-SREC-I (AF2409; R&D). Secondary antibodies were anti-rabbit AlexaFluor-488 for SR-A, anti-goat AlexaFluor-488 for SREC-I, and anti-rat AlexaFluor-647 for CD31 and CD68 (Invitrogen Life Technologies). Confocal images were obtained using LSM 710 microscope (Plan-Apochromat 63X/1.40oil DIC M27 objective; lasers excitation wavelength: 488, 561, 633, and 405 nm; filters: ChS1-494-552 for 488, ChS2-562-630 for 555, Ch2-637-655 for 647, Ch1-409-495 for DAPI; zoom was ×2.0 for cell studies and ×0.6 for tissue studies) and ZEN 2008 software (Carl Zeiss, Oberkochen, Germany). At least five images per animal were analyzed for each staining. Image Correlation Analysis tool of ImageJ software (NIH, Bethesda, MD) was used for quantitative analysis of colocalization, with at least three images from three different animals analyzed per group.
SUPPLEMENTARY MATERIAL Figure S1. Clearance of HDAd in saline or poly[I] pre-treated wild-type C57BL/6 mice. Figure S2. Alexa555-HDAd infectivity. Figure S3. SREC-I and SR-A colocalize with HDAd vector in mouse livers. Figure S4. Controls for SR-A and SREC-I staining and Alexa555-HDAd autofluorescence. Figure S5. SR-A colocalizes with HDAd particles in both endothelial and Kupffer cells in the liver. Figure S6. SREC-I colocalizes with HDAd particles in both endothelial and Kupffer cells in the liver.
Acknowledgments
We thank Edoardo Nusco for assistance with mouse studies and TIGEM Bioinformatic Core for statistical analyses. This work was supported by the Fondazione Telethon, (TCBP37TELC and TCBMT3TELD to N.B.-P.), by a research grant of The Hyperoxaluria and Oxalosis Foundation to N.B.-P., by the Italian Ministry of Health (GR-2009-1594913 to N.B.-P.), and by National Institutes of Health grant R01DK067324 to P.N. The authors declared no conflict of interest.
Supplementary Material
REFERENCES
- Brunetti-Pierri N., and, Ng P. Helper-dependent adenoviral vectors for liver-directed gene therapy. Hum Mol Genet. 2011;20 R1:R7–13. doi: 10.1093/hmg/ddr143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan LP, Mohanty S, Kim J, Clark KR, Robinson JM., and, Anderson CL. Rapid and efficient clearance of blood-borne virus by liver sinusoidal endothelium. PLoS Pathog. 2011;7:e1002281. doi: 10.1371/journal.ppat.1002281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alemany R, Suzuki K., and, Curiel DT. Blood clearance rates of adenovirus type 5 in mice. J Gen Virol. 2000;81 Pt 11:2605–2609. doi: 10.1099/0022-1317-81-11-2605. [DOI] [PubMed] [Google Scholar]
- Tao N, Gao GP, Parr M, Johnston J, Baradet T, Wilson JM.et al. (2001Sequestration of adenoviral vector by Kupffer cells leads to a nonlinear dose response of transduction in liver Mol Ther 328–35. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chirmule N, Gao GP, Qian R, Croyle M, Joshi B.et al. (2001Acute cytokine response to systemic adenoviral vectors in mice is mediated by dendritic cells and macrophages Mol Ther 35 Pt 1697–707. [DOI] [PubMed] [Google Scholar]
- Smith JS, Tian J, Muller J., and, Byrnes AP. Unexpected pulmonary uptake of adenovirus vectors in animals with chronic liver disease. Gene Ther. 2004;11:431–438. doi: 10.1038/sj.gt.3302149. [DOI] [PubMed] [Google Scholar]
- Lieber A, He CY, Meuse L, Schowalter D, Kirillova I, Winther B.et al. (1997The role of Kupffer cell activation and viral gene expression in early liver toxicity after infusion of recombinant adenovirus vectors J Virol 718798–8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunetti-Pierri N, Palmer DJ, Beaudet AL, Carey KD, Finegold M., and, Ng P. Acute toxicity after high-dose systemic injection of helper-dependent adenoviral vectors into nonhuman primates. Hum Gene Ther. 2004;15:35–46. doi: 10.1089/10430340460732445. [DOI] [PubMed] [Google Scholar]
- Morral N, O'Neal WK, Rice K, Leland MM, Piedra PA, Aguilar-Córdova E.et al. (2002Lethal toxicity, severe endothelial injury, and a threshold effect with high doses of an adenoviral vector in baboons Hum Gene Ther 13143–154. [DOI] [PubMed] [Google Scholar]
- Immonen A, Vapalahti M, Tyynelä K, Hurskainen H, Sandmair A, Vanninen R.et al. (2004AdvHSV-tk gene therapy with intravenous ganciclovir improves survival in human malignant glioma: a randomised, controlled study Mol Ther 10967–972. [DOI] [PubMed] [Google Scholar]
- Kirn D. Clinical research results with dl1520 (Onyx-015), a replication-selective adenovirus for the treatment of cancer: what have we learned. Gene Ther. 2001;8:89–98. doi: 10.1038/sj.gt.3301377. [DOI] [PubMed] [Google Scholar]
- Peng Z. Current status of gendicine in China: recombinant human Ad-p53 agent for treatment of cancers. Hum Gene Ther. 2005;16:1016–1027. doi: 10.1089/hum.2005.16.1016. [DOI] [PubMed] [Google Scholar]
- Nemunaitis J, Cunningham C, Buchanan A, Blackburn A, Edelman G, Maples P.et al. (2001Intravenous infusion of a replication-selective adenovirus (ONYX-015) in cancer patients: safety, feasibility and biological activity Gene Ther 8746–759. [DOI] [PubMed] [Google Scholar]
- Nemunaitis J, Senzer N, Sarmiento S, Zhang YA, Arzaga R, Sands B.et al. (2007A phase I trial of intravenous infusion of ONYX-015 and enbrel in solid tumor patients Cancer Gene Ther 14885–893. [DOI] [PubMed] [Google Scholar]
- Koski A, Rajecki M, Guse K, Kanerva A, Ristimäki A, Pesonen S.et al. (2009Systemic adenoviral gene delivery to orthotopic murine breast tumors with ablation of coagulation factors, thrombocytes and Kupffer cells J Gene Med 11966–977. [DOI] [PubMed] [Google Scholar]
- Shayakhmetov DM, Di Paolo NC., and, Mossman KL. Recognition of virus infection and innate host responses to viral gene therapy vectors. Mol Ther. 2010;18:1422–1429. doi: 10.1038/mt.2010.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo NC, van Rooijen N., and, Shayakhmetov DM. Redundant and synergistic mechanisms control the sequestration of blood-born adenovirus in the liver. Mol Ther. 2009;17:675–684. doi: 10.1038/mt.2008.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haisma HJ, Kamps JA, Kamps GK, Plantinga JA, Rots MG., and, Bellu AR. Polyinosinic acid enhances delivery of adenovirus vectors in vivo by preventing sequestration in liver macrophages. J Gen Virol. 2008;89 Pt 5:1097–1105. doi: 10.1099/vir.0.83495-0. [DOI] [PubMed] [Google Scholar]
- Haisma HJ, Boesjes M, Beerens AM, van der Strate BW, Curiel DT, Plüddemann A.et al. (2009Scavenger receptor A: a new route for adenovirus 5 Mol Pharm 6366–374. [DOI] [PubMed] [Google Scholar]
- Khare R, Chen CY, Weaver EA., and, Barry MA. Advances and future challenges in adenoviral vector pharmacology and targeting. Curr Gene Ther. 2011;11:241–258. doi: 10.2174/156652311796150363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shayakhmetov DM, Gaggar A, Ni S, Li ZY., and, Lieber A. Adenovirus binding to blood factors results in liver cell infection and hepatotoxicity. J Virol. 2005;79:7478–7491. doi: 10.1128/JVI.79.12.7478-7491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyuzhniy O, Di Paolo NC, Silvestry M, Hofherr SE, Barry MA, Stewart PL.et al. (2008Adenovirus serotype 5 hexon is critical for virus infection of hepatocytes in vivo Proc Natl Acad Sci USA 1055483–5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Xu Z, Tian J, Stevenson SC., and, Byrnes AP. Interaction of systemically delivered adenovirus vectors with Kupffer cells in mouse liver. Hum Gene Ther. 2008;19:547–554. doi: 10.1089/hum.2008.004. [DOI] [PubMed] [Google Scholar]
- Merrick AF, Shewring LD, Sawyer GJ, Gustafsson KT., and, Fabre JW. Comparison of adenovirus gene transfer to vascular endothelial cells in cell culture, organ culture, and in vivo. Transplantation. 1996;62:1085–1089. doi: 10.1097/00007890-199610270-00011. [DOI] [PubMed] [Google Scholar]
- Hegenbarth S, Gerolami R, Protzer U, Tran PL, Brechot C, Gerken G.et al. (2000Liver sinusoidal endothelial cells are not permissive for adenovirus type 5 Hum Gene Ther 11481–486. [DOI] [PubMed] [Google Scholar]
- Xu Z, Tian J, Smith JS., and, Byrnes AP. Clearance of adenovirus by Kupffer cells is mediated by scavenger receptors, natural antibodies, and complement. J Virol. 2008;82:11705–11713. doi: 10.1128/JVI.01320-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare R, Reddy VS, Nemerow GR., and, Barry MA. Identification of adenovirus serotype 5 hexon regions that interact with scavenger receptors. J Virol. 2012;86:2293–2301. doi: 10.1128/JVI.05760-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Xu Z., and, Byrnes AP. A quantitative assay for measuring clearance of adenovirus vectors by Kupffer cells. J Virol Methods. 2008;147:54–60. doi: 10.1016/j.jviromet.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Brunetti-Pierri N, Ng T, Iannitti DA, Palmer DJ, Beaudet AL, Finegold MJ.et al. (2006Improved hepatic transduction, reduced systemic vector dissemination, and long-term transgene expression by delivering helper-dependent adenoviral vectors into the surgically isolated liver of nonhuman primates Hum Gene Ther 17391–404. [DOI] [PubMed] [Google Scholar]
- Brunetti-Pierri N, Stapleton GE, Palmer DJ, Zuo Y, Mane VP, Finegold MJ.et al. (2007Pseudo-hydrodynamic delivery of helper-dependent adenoviral vectors into non-human primates for liver-directed gene therapy Mol Ther 15732–740. [DOI] [PubMed] [Google Scholar]
- Manickan E, Smith JS, Tian J, Eggerman TL, Lozier JN, Muller J.et al. (2006Rapid Kupffer cell death after intravenous injection of adenovirus vectors Mol Ther 13108–117. [DOI] [PubMed] [Google Scholar]
- de Winther MP, Gijbels MJ, van Dijk KW, van Gorp PJ, suzuki H, Kodama T.et al. (1999Scavenger receptor deficiency leads to more complex atherosclerotic lesions in APOE3Leiden transgenic mice Atherosclerosis 144315–321. [DOI] [PubMed] [Google Scholar]
- Beauvillain C, Meloni F, Sirard JC, Blanchard S, Jarry U, Scotet M.et al. (2010The scavenger receptors SRA-1 and SREC-I cooperate with TLR2 in the recognition of the hepatitis C virus non-structural protein 3 by dendritic cells J Hepatol 52644–651. [DOI] [PubMed] [Google Scholar]
- Hölzl MA, Hofer J, Kovarik JJ, Roggenbuck D, Reinhold D, Goihl A.et al. (2011The zymogen granule protein 2 (GP2) binds to scavenger receptor expressed on endothelial cells I (SREC-I) Cell Immunol 26788–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y, Osuga J, Adachi H, Tozawa R, Takanezawa Y, Ohashi K.et al. (2004Scavenger receptor expressed by endothelial cells I (SREC-I) mediates the uptake of acetylated low density lipoproteins by macrophages stimulated with lipopolysaccharide J Biol Chem 27930938–30944. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Cela R, Clarke C, Bertin TK, Mouriño S., and, Lee B. Large-scale production of high-quality helper-dependent adenoviral vectors using adherent cells in cell factories. Hum Gene Ther. 2010;21:120–126. doi: 10.1089/hum.2009.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty A, Whitman SC, Block AE., and, Rateri DL. Polymorphism of class A scavenger receptors in C57BL/6 mice. J Lipid Res. 2000;41:1568–1577. [PubMed] [Google Scholar]
- Snoeys J, Mertens G, Lievens J, van Berkel T, Collen D, Biessen EA.et al. (2006Lipid emulsions potently increase transgene expression in hepatocytes after adenoviral transfer Mol Ther 1398–107. [DOI] [PubMed] [Google Scholar]
- Kawabata K, Takakura Y., and, Hashida M. The fate of plasmid DNA after intravenous injection in mice: involvement of scavenger receptors in its hepatic uptake. Pharm Res. 1995;12:825–830. doi: 10.1023/a:1016248701505. [DOI] [PubMed] [Google Scholar]
- Platt N., and, Gordon S. Is the class A macrophage scavenger receptor (SR-A) multifunctional? – The mouse's tale. J Clin Invest. 2001;108:649–654. doi: 10.1172/JCI13903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy JE, Tedbury PR, Homer-Vanniasinkam S, Walker JH., and, Ponnambalam S. Biochemistry and cell biology of mammalian scavenger receptors. Atherosclerosis. 2005;182:1–15. doi: 10.1016/j.atherosclerosis.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Konz JO, Livingood RC, Bett AJ, Goerke AR, Laska ME., and, Sagar SL. Serotype specificity of adenovirus purification using anion-exchange chromatography. Hum Gene Ther. 2005;16:1346–1353. doi: 10.1089/hum.2005.16.1346. [DOI] [PubMed] [Google Scholar]
- Areschoug T., and, Gordon S. Scavenger receptors: role in innate immunity and microbial pathogenesis. Cell Microbiol. 2009;11:1160–1169. doi: 10.1111/j.1462-5822.2009.01326.x. [DOI] [PubMed] [Google Scholar]
- Facciponte JG, Wang XY., and, Subjeck JR. Hsp110 and Grp170, members of the Hsp70 superfamily, bind to scavenger receptor-A and scavenger receptor expressed by endothelial cells-I. Eur J Immunol. 2007;37:2268–2279. doi: 10.1002/eji.200737127. [DOI] [PubMed] [Google Scholar]
- Khare R, May SM, Vetrini F, Weaver EA, Palmer D, Rosewell A.et al. (2011Generation of a Kupffer cell-evading adenovirus for systemic and liver-directed gene transfer Mol Ther 191254–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok H, Palmer DJ, Ng P., and, Barry MA. Evaluation of polyethylene glycol modification of first-generation and helper-dependent adenoviral vectors to reduce innate immune responses. Mol Ther. 2005;11:66–79. doi: 10.1016/j.ymthe.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Green NK, Herbert CW, Hale SJ, Hale AB, Mautner V, Harkins R.et al. (2004Extended plasma circulation time and decreased toxicity of polymer-coated adenovirus Gene Ther 111256–1263. [DOI] [PubMed] [Google Scholar]
- Palmer D., and, Ng P. Improved system for helper-dependent adenoviral vector production. Mol Ther. 2003;8:846–852. doi: 10.1016/j.ymthe.2003.08.014. [DOI] [PubMed] [Google Scholar]
- Warren JC, Rutkowski A., and, Cassimeris L. Infection with replication-deficient adenovirus induces changes in the dynamic instability of host cell microtubules. Mol Biol Cell. 2006;17:3557–3568. doi: 10.1091/mbc.E05-09-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunetti-Pierri N, Palmer DJ, Mane V, Finegold M, Beaudet AL., and, Ng P. Increased hepatic transduction with reduced systemic dissemination and proinflammatory cytokines following hydrodynamic injection of helper-dependent adenoviral vectors. Mol Ther. 2005;12:99–106. doi: 10.1016/j.ymthe.2005.03.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.