Abstract
We previously dissected the components of the innate immune response to Helper-dependent adenoviral vectors (HDAds) using genetic models, and demonstrated that multiple pattern recognition receptor signaling pathways contribute to this host response to HDAds in vivo. Based on analysis of cytokine expression profiles, type I interferon (IFN) mRNA is induced in host mouse livers at 1 hour post-injection. This type I IFN signaling amplifies cytokine expression in liver independent of the nature of vector DNA sequences after 3 hours post-injection. This type I IFN signaling in response to HDAds administration contributes to transcriptional silencing of both HDAd prokaryotic and eukaryotic DNA in liver. This silencing occurs early and is mediated by epigenetic modification as shown by in vivo chromatin immunoprecipitation (ChIP) with anti-histone deacetylase (HDAC) and promyelocytic leukemia protein (PML). In contrast, self-complementary adeno-associated viral vectors (scAAVs) showed significantly lower induction of type I IFN mRNA in liver compared to HDAds at both early and late time points. These results show that the type I IFN signaling dependent transgene silencing differs between AAV and HDAd vectors after liver-directed gene transfer.
Introduction
Adenoviral vectors (Ads) have been extensively used for gene therapy because of their ability to efficiently transduce a wide variety of cell types, independently of the cell cycle while directing high levels of transgene expression without integration into the host genome. The development of helper-dependent Ads (HDAds), devoid of viral coding genes, has markedly diminished the acquired cellular immune response and chronic toxicity associated with first-generation Ads (FGAds). Furthermore, viral gene deletion facilitates the insertion of large transgenes or combinatorial therapy using multiple transgenes. Several groups have now reported long-term persistence of vector, transgene expression, and minimal chronic toxicity in multiple small and large animal models treated with HDAd.1,2,3,4,5,6,7
However, systemic administration of HDAd induces a rapid host innate immune response similar to that seen with FGAds.8,9,10 This acute response represents a serious obstacle to clinical translation of Ad-based vectors.11 We and others have thus begun to dissect mechanisms responsible for induction of the innate response by combining in vitro and in vivo strategies using genetic mouse models. Multiple pattern recognition receptor signaling pathways (e.g., Toll-like receptors (TLRs)/myeloid differentiation primary response gene 88 (MyD88) and Nod-like receptors) have been found to contribute to the induction of an acute inflammatory response following the administration of Ad-based vectors in vivo, leading to the production of cytokines and chemokines.12,13,14,15,16 Given that over 80% of Ad5-based vectors localize to livers of mice after systemic administration,17 we next addressed how host tissues/cells amplify the innate response to HDAd in the liver following intravenous injection of HDAd.
In the present study, we found that type I IFN (but not type II IFN) mRNA is induced in livers at 1-hour post-injection of HDAd and amplifies cytokine expression in the liver after 3 hours. This induction was independent of the nature of the transgene or promoter sequences within the HDAd. Type I interferons (IFNs) are a family of cytokines that constitute 13 and 17 IFNα subtypes in mice and humans, respectively, and one IFNβ in both species. Upon recognition of type I IFN, IFNα receptor triggers a signaling cascade leading to the transcription of over 100 IFN-stimulated genes essential for antiviral immunity.18 This type I IFN-dependent innate immune response was confirmed by systemic administration of a DNA viral vector (HSV amplicon vector) and RNA viral vector (lentiviral vector) in vivo. This led to silencing of transgene expression from these viral vectors in vivo.19,20 Type I IFN signaling induced by infection of FGAd contributes to infiltration of immune cells (e.g., NK cells) in livers of mice at 3 days post-injection and development of neutralizing antibody to Ad viral particles (Vps) at 10 days. These immune responses dependent on type I IFN signaling ultimately lead to loss of vector-transduced cells.21,22
The objective of this study is to evaluate the role of type I IFN signaling in liver at an early time point following intravenous injection of HDAd. Interestingly, both prokaryotic and eukaryotic transgene expression of HDAd are suppressed at the transcriptional level in the liver due to type I IFN signaling at 24 hours post-injection. Based on in vivo chromatin immunoprecipitation (ChIP), we demonstrated that type I IFN signaling induces epigenetic modification of both prokaryotic and eukaryotic sequences in HDAd vector DNA. In contrast, self-complementary adeno-associated viral vector (scAAV), an alternative DNA viral vector showed significantly lower induction of type I IFN mRNA in liver. This attenuated type I IFN signaling was not associated with transgene silencing for scAAV at both early and late time points. Thus, these results suggest that type I IFN signaling dependent transgene silencing in vivo is not driven by the nature of vector DNA, but rather by host responses to the vector components (e.g., viral particle).
Results
Type I IFN mRNA is induced at 1 hour post-injection of HDAd and amplifies cytokine expression in host animals
Previously, we and other groups observed that host animals induce cytokine expression at both the mRNA level in liver and protein level in blood depending on MyD88 signaling at 1 hour post-injection with 5 × 1011 Vps/kg HDAds.23,24 To characterize how host animals amplify the innate response, we quantified type I IFN (IFNβ) and type II IFN (IFNγ) mRNA expression in livers of wild-type (WT) C57Bl/6 mice at 1 hour after systemic administration of 5 × 1011 Vp/kg HDAdLacZ25 (Figure 1a), since over 80% of Ad5-based vectors localize to mouse liver within 30 minutes of post-injection.17 Although there was no difference in IFNγ mRNA levels between control [phosphate-buffered saline (PBS) injected] and HDAd-injected mice, HDAd-injected mice showed tenfold higher IFNβ mRNA expression compared to control mice. To test whether this type I IFN signaling contributes to an innate response to HDAd (amplification of proinflammatory cytokine and chemokine expression), we compared IFNβ, interleukin 6 (IL-6), and monocyte chemoattractant protein-1 (MCP-1) mRNA levels in livers between WT C56Bl/6 mice and IFNα receptor deficient (IFNαR−/−) mice at various time points (1, 3, 6, 12, and 24 hours) following systemic administration of 5 × 1011 Vp/kg HDAdLacZ (Figure 1b). IL-6 is known to be highly induced in the blood of small and large animal models after systemic administration of Ad-based vectors.16,23,26,27,28 MCP-1 was highly upregulated in the blood of mice compared to other chemokines (e.g., KC, RANTES) at 6 hours post-injection with HDAds.25 We also evaluated IL-6 and MCP-1 expression in the blood at the same time points (Figure 1c). There was no significant difference in IFNβ, IL-6, and MCP-1 liver mRNA expression or in blood levels of IL-6 and MCP-1 between WT and IFNαR−/− mice at 1 hour post-injection, indicating that induction of inflammatory cytokines at 1 hour is independent of type I IFN signaling.23 IFNαR−/− mice showed an over 80% reduction of IFNβ, IL-6, and MCP-1 mRNA in livers compared to WT mice after 3 hours post-injection, suggesting that type I IFN signaling via IFNαR contributes to induction of these cytokine and chemokine mRNA in livers after 3 hours post-injection. This result could also suggest that upregulation of these cytokine and chemokine mRNAs in WT livers could be dependent on an influx of immune cells (e.g., Natural killer cells, dendritic cells) that endogenously express these mRNAs (e.g., type I IFN).21,22 IFNαR−/− mice still showed residual expression of these cytokine and chemokine in livers after 3 hours post-injection, suggesting that residual expression in IFNαR−/− livers after 3 hours could be dependent on pattern recognition receptor signaling pathways (e.g., TLRs or Nod-like receptors).12,25 Both IL-6 and MCP-1 protein levels in the blood of IFNαR−/− mice showed significantly lower levels compared with WT mice after 6 hours post-injection of HDAd. Interestingly, IL-6 liver mRNA returned to a basal level in IFNαR−/− mice at 6 hours post-injection, although IL-6 in blood of IFNαR−/− mice at 6 hours still showed significantly higher expression compared to the basal level of IFNαR−/− mice. In contrast, MCP-1 in the blood of IFNαR−/− mice exhibited an ~99% reduction compared to WT mice at 6 hours post-injection. However, MCP-1 liver mRNA in IFNαR−/− mice still showed significantly higher expression compared to the basal level in IFNαR−/− mice. These results suggest that cytokine and chemokine levels in the blood may be regulated not only in the liver, but also other tissues (e.g., spleen).23,24
Figure 1.
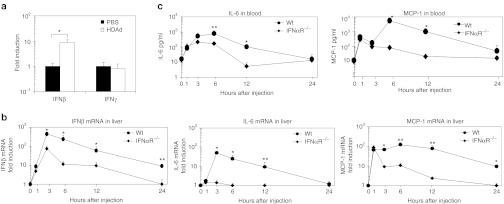
Type I interferon (IFN) mRNA is induced in livers at 1 hour post-injection of HDAdLacZand amplifies cytokine expression in livers and blood of mice. Wild-type (WT) C57Bl/6 mice were injected with 5 × 1011 viral particle (Vp)/kg of HDAdLacZ into tail veins. (a) Total RNA was extracted from livers at 1 hour post-injection, and the levels of IFNβ and IFNγ mRNA were determined by quantitative real-time PCR. Each value was calculated relative to that of control mice injected with phosphate-buffered saline (PBS). Data are presented as means ± SD (n = 3). *P < 0.005. The experiment was repeated with similar results. WT C57Bl/6 and IFNαR−/− mice were injected with 5 × 1011 Vp/kg of HDAdLacZ into tail veins. (b) Total RNA was extracted from livers at 1, 3, 6, 12, and 24 hours post-injection, and the levels of IFNβ, IL-6, and MCP-1 mRNA were determined by quantitative real-time PCR. Each value was calculated relative to that of control mice (untreated). Data are presented as means ± SD (n = 3). *P < 0.002 and **P < 0.01 for IFNβ. *P < 0.001 and **P < 0.01 for IL-6. *P < 0.003 and **P < 0.001 for MCP-1. (c) Plasma samples were collected after 1, 3, 6, 12, and 24 hours post-injection. IL-6 and MCP-1 levels were measured by using BD cytokine multiplex bead array system. Data are presented as means ± SD (n = 5). *P < 0.01 and **P < 0.05 for IL-6. *P < 0.001 for MCP-1. HDAd, Helper-dependent adenoviral vector.
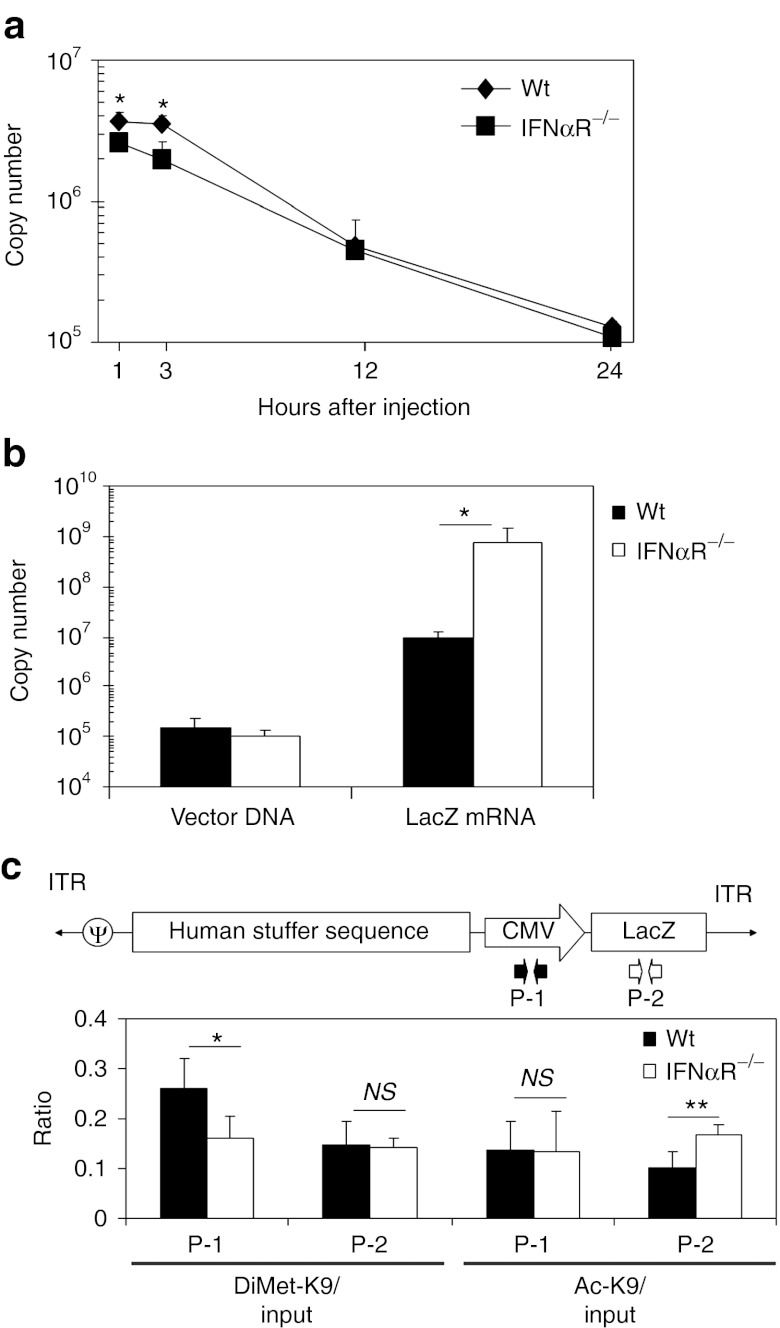
Type I IFN signaling suppresses transgene transcription of HDAd but does not contribute to vector elimination in livers at 24 hours
Type I IFN signaling induced by FGAd injection contributes to infiltration of NK cells and eliminates Ad transduced cells at 3 days post-injection.22 To test whether type I IFN signaling contributes to the elimination of HDAd vector DNA in liver within 24 hours, we quantified vector copies in WT and IFNαR−/− mouse livers at various time points (1, 3, 12, and 24 hours) after systemic administration of 5 × 1011 Vp/kg HDAdLacZ (Figure 2a). Vector copy numbers in WT and IFNαR−/− mouse livers showed an ~95% reduction from 1 hour to 24 hours post-injection. Vector copies normalized to a reference gene (GAPDH), also showed a similar reduction from 1 hour to 24 hours post-injection in both WT and IFNαR−/− mouse livers (Supplementary Figure S1). These results suggest that the elimination of HDAd vector DNA in liver within 24 hours is independent of type I IFN signaling similar to that observed with FGAd.22 To test whether type I IFN signaling affects transgene expression from HDAd at this early time point, we quantified transgene mRNA (LacZ mRNA) in WT and IFNαR−/− mouse livers at 24 hours post-injection (Figure 2b). IFNαR−/− mice showed a 50-fold higher level of LacZ mRNA levels compared to WT mouse livers, indicating that type I IFN signaling suppresses transgene expression from HDAd. Previously, we and other groups reported that an innate immune response from an infection of viral vectors induces epigenetic modification of prokaryotic transgene expression cassettes in vector DNA to suppress transgene expression in vitro.12,29 To test whether type I IFN signaling induces epigenetic modification of prokaryotic sequences in HDAd vector DNA in vivo, we performed in vivo ChIP of liver samples collected at 24 hours post-injection of 5 × 1011 Vp/kg HDAdLacZ. After immunoprecipitation, samples were subjected to quantitative PCR (Q-PCR) with primer sets (Table 1) designed specifically for different regions of the transgene. Anti-histone H3 antibody was used to confirm whether type I IFN signaling affects chromatin formation of HDAd vector DNA in liver (Supplementary Figure S2). No difference in the association ratio of histone H3 to input in the bacterial LacZ gene region was seen between WT and IFNαR−/− liver samples, suggesting that chromatin formation of HDAd vector DNA in the liver is independent of type I IFN signaling. We next quantified the chromatin status of vector DNA in WT and IFNαR−/− mouse liver samples using anti-Dimethyl Lys9 histone H3 (DiMet-K9 H3) antibody as a heterochromatin marker (inactive form marker) and anti-acetyl Lys9 histone H3 (Ac-K9 H3) antibody as an euchromatin marker (active form marker) (Figure 2c). These antibodies have been previously used to correlate both epigenetic status and transgene expression in vitro.29,30,31,32 The cytomegalovirus promoter of vector DNA in WT liver samples showed a significantly higher association ratio of DiMet-K9 H3 compared to IFNαR−/− mouse liver samples, and the LacZ of vector DNA in WT liver samples showed a significantly lower association ratio of Ac-K9 H3 compared to that in IFNαR−/− liver samples. These results suggest that the prokaryotic transgene expression cassette in HDAd vector DNA exists in an epigenetic state that is silenced in a type I IFN-dependent fashion.12,29 There was no significant difference in the control GAPDH levels between WT and IFNαR−/− mouse liver samples after immunoprecipitation with these antibodies (Supplementary Figure S3).
Figure 2.
Type I interferon (IFN) signaling suppresses transgene expression of Helper-dependent adenoviral vector (HDAd) at transcriptional level in livers of mice. Wild-type (WT) C57Bl/6 and IFNαR−/− mice were injected with 5 × 1011 viral particle (Vp)/kg of HDAdLacZ into tail veins. (a) Total DNA was extracted from livers at 1, 3, 12, and 24 hours post-injection, and vector copy number in total DNA was determined by quantitative PCR. Data are presented as means ± SD (n = 3). *P < 0.01. (b) Total DNA and total RNA were extracted from their livers at 24 hours post-injection, and vector copy number in total DNA and LacZ mRNA copy number in total RNA were determined by quantitative PCR. Data are presented as means ± SD (n = 3). *P < 0.001. The experiment was repeated with similar results. (c) Chromatin immunoprecipitation (ChIP) analysis of liver samples at 24 hours post-injection. After immunoprecipitation with anti-DiMet K9 H3 (heterochromatin marker) and anti-Ac K9 H3 (euchromatin marker) antibodies, vector copy numbers of each region were determined by quantitative PCR with specific primer sets (P-1 for cytomegalovirus (CMV) promoter, P-2 for LacZ). Data are presented as means ± SD (n = 3). *P < 0.05 and **P < 0.02. The experiment was repeated with similar results.
Table 1. Oligonucleotides used for quantitative PCR of ChIP products.

PML mRNA is induced by type I IFN signaling in liver and associates with HDAd vector DNA
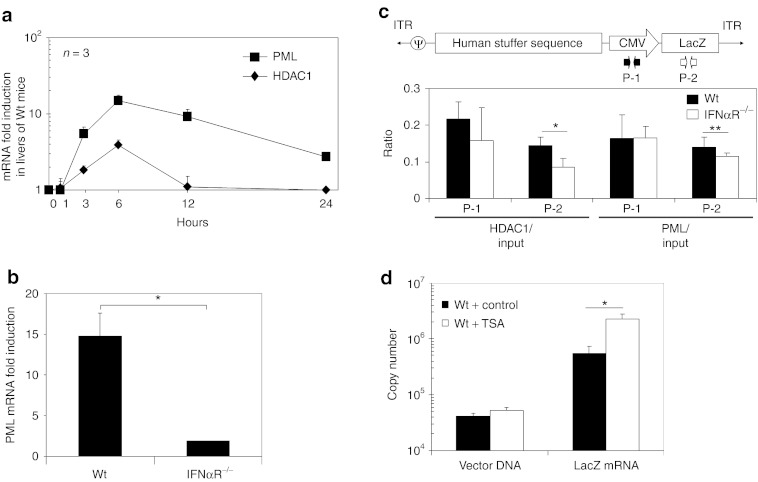
Our results suggest that type I IFN signaling contributes to the epigenetic modification of HDAd vector DNA in vivo. Expression of promyelocytic leukemia protein (PML) is induced by type I IFN signaling and results in the formation of PML nuclear bodies (PML-NBs, also known as ND10).33,34 PML-NBs were reported to associate with the viral DNA of simian virus 40, herpes simplex virus, and cytomegalovirus in nuclei and to recruit the histone deacetylase (HDAC) complex to silence viral gene expression during natural infection by these viruses.35 To test whether this mechanism might be induced in livers of mice after infection with HDAd, we quantified PML and HDAC1 mRNA in livers of WT mice at various time points (1, 3, 6, 12, and 24 hours) after systemic administration of 5 × 1011 Vp/kg HDAdLacZ (Figure 3a). Both mRNAs were confirmed to be increased in livers after 3 hours post-injection of HDAd and peaked at 6 hours post-injection. To confirm whether this PML mRNA induction was dependent on type I IFN signaling, we compared PML mRNA levels between WT and IFNαR−/− mouse livers at 6 hours post-injection (Figure 3b). There was no induction of PML mRNA in IFNαR−/− mouse livers at 6 hours post-injection of HDAds, suggesting that upregulation of PML mRNA in the liver is dependent on type I IFN signaling.
Figure 3.
Promyelocytic leukemia protein (PML) mRNA is induced dependently on type I interferon (IFN) signaling and associated with Helper-dependent adenoviral vector (HDAd) DNA in livers of wild-type (WT) mice. WT C57Bl/6 mice were injected with 5 × 1011 viral particle (Vp)/kg of HDAdLacZ into tail veins. (a) Total RNA was extracted from livers at 1, 3, 6, 12, and 24 hours post-injection, and the levels of PML and HDAC1 mRNA were determined by quantitative real-time PCR. Each value was calculated relative to that of control mice (untreated). Data are presented as means ± SD (n = 3). WT C57Bl/6 and IFNαR−/− mice were injected with 5 × 1011 Vp/kg of HDAdLacZ into tail veins. (b) Total RNA was extracted from livers at 6 hours post-injection, and the levels of PML mRNA were determined by quantitative real-time PCR. Each value was calculated relative to that of control mice (untreated). Data are presented as means ± SD (n = 3). *P < 0.005. (c) Chromatin immunoprecipitation (ChIP) analysis of liver samples at 24 hours post-injection. After immunoprecipitation with anti-HDAC1 and anti-PML antibodies, vector copy numbers of each region were determined by quantitative PCR with specific primer sets (P-1 for cytomegalovirus (CMV) promoter, P-2 for LacZ). Data are presented as means ± SD (n = 3). *P < 0.05 and **P < 0.02. The experiment was repeated with similar results. (d) WT C57Bl/6 mice were coinjected TSA (control: buffer alone) with 5 × 1011 Vp/kg of HDAdLacZ into tail veins. Total DNA and total RNA were extracted from their livers at 24 hours post-injection, and vector copy number in total DNA and LacZ mRNA copy number in total RNA were determined by quantitative PCR. Data are presented as means ± SD (n = 3). *P < 0.01.
To test whether PML-NBs and/or the HDAC complex tend to associate with HDAd vector DNA in a type I IFN-dependent fashion, we performed in vivo ChIP of HDAd from injected WT and IFNαR−/− mouse liver samples using anti-HDAC1 and anti-PML antibodies (Figure 3c). Vector DNA in WT liver samples showed a significantly higher association ratio with HDAC1 and PML compared to IFNαR−/− mouse liver samples, suggesting that PML may associate with HDAd vector DNA in the nuclei (as PML-NBs) and recruit the HDAC complex for transgene silencing. To test whether this HDAC1 association contributes to transgene silencing of HDAd, we coinjected HDAC inhibitor (TSA) with 5 × 1011 Vp/kg of HDAdLacZ into WT mice, and quantified vector DNA copy and LacZ mRNA copy numbers in livers at 24 hours post-injection (Figure 3d). There was no difference in inflammatory cytokine levels (e.g., IL-6, MCP-1) in the blood between untreated and TSA-treated mice at 6 hours post-injection (data not shown). There was no difference in the reference/housekeeping gene expression levels between TSA-treated livers and untreated livers at 24 hours post-injection (Supplementary Figure S4) suggesting that the TSA treatment protocol did not significantly affect endogenous gene expression at this time point. Although there was no significant difference in vector DNA copy number between control and TSA-treated mice, TSA-treated mice showed a sevenfold higher level of LacZ mRNA compared to livers of control mice. These results suggest that association of HDAC with vector DNA contributes to transgene silencing after HDAd infection.
Induction of type I IFN mRNA in the liver is independent of vector DNA sequences and induces epigenetic modification of eukaryotic sequences
Previous in vitro ChIP with DNA viral vectors showed that epigenetic modification originated in prokaryotic sequences of vector DNA in transduced cells.30,36,37 However, our in vivo ChIP data revealed that eukaryotic, human genomic stuffer sequence within HDAdLacZ also associated with PML and HDAC in a type I IFN-dependent epigenetic modification similar to what we have seen with prokaryotic sequence (bacterial LacZ sequence) (Supplementary Figure S5). To assess this phenomenon within the context of a therapeutic transgene, we quantified IFNβ mRNA in livers of WT mice at 3 hours post-injection receiving either 5 × 1011 Vp/kg of HDAd0 or HDAdhFVIII (Figure 4a). HDAd0 contains only packaging signal sequences and human genomic stuffer sequences,13 and HDAdhFVIII contains a human FVIII expression cassette. The hFVIII-coding DNA is driven by liver-restricted phosphoenolpyruvate carboxykinase (PEPCK) promoter. There was no significant difference in IFNβ mRNA levels in livers of WT mice injected with HDAd0, HDAdhFVIII, or HDAdLacZ, indicating that IFNβ mRNA induction is independent of the nature of the sequence at 3 hours. hFVIII transcription was suppressed in a type I IFN-dependent fashion (Supplementary Figure S6). We performed in vivo ChIP of WT and IFNαR−/− mouse liver samples after systemic injection of 5 × 1011 Vp/kg HDAd0 (Figure 4b,c). After immunoprecipitation of HDAd0 injected liver samples, we quantified the chromatin status of vector DNA (DiMet-K9 H3 and Ac-K9 H3) at three different regions of the human stuffer sequence in the vector DNA (Figure 4b). Although HDAd0 vector DNA contains only packaging signal sequences and human genomic stuffer sequences, vector DNA in WT liver samples trended toward a higher ratio of DiMet-K9 H3 and a lower ratio of Ac-K9 H3 compared to vector DNA in IFNαR−/− mouse liver samples (some regions were significantly different). To test whether this chromatin was also associated with HDAC1 and PML, we performed in vivo ChIP with anti-HDAC1 and anti-PML antibodies and quantified the same regions by Q-PCR (Figure 4c). Vector DNA in WT liver samples exhibited a higher association ratio of HDAC1 and PML compared to vector DNA in IFNαR−/− mouse liver samples, suggesting that type I IFN signaling dependent PML-NBs also sense HDAd vector DNA encoding eukaryotic sequences.
Figure 4.
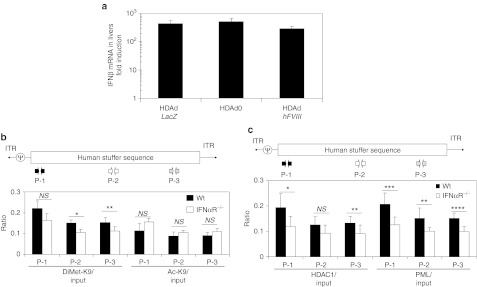
Helper-dependent adenoviral vector (HDAd) coding eukaryotic sequences induces similar level of interferon β (IFNβ) mRNA in livers of wild-type (WT) mice and is epigenetically modified dependently on type I interferon (IFN) signaling in livers of mice. (a) WT C57Bl/6 mice were injected with 5 × 1011 viral particle (Vp)/kg of HDAdLacZ, HDAd0, or HDAdhFVIII into tail veins. Total RNA was extracted from livers at 3 hours post-injection, and the levels of IFNβ mRNA were determined by quantitative real-time PCR. Each value was calculated relative to that of control mice injected with phosphate-buffered saline (PBS). Data are presented as means ± SD (n = 3). The experiment was repeated with similar results. WT C57Bl/6 and IFNαR−/− mice were injected with 5 × 1011 viral particle (Vp)/kg of HDAd0 into tail veins. Liver samples at 24 hours post-injection were collected and subjected to Chromatin immunoprecipitation (ChIP) analysis. After immunoprecipitation with anti-DiMet K9 H3 and anti-Ac K9 H3 antibodies (b), anti-HDAC1 and anti-PML antibodies (c), vector copy numbers of each region were determined by quantitative PCR with specific primer sets and calculated the ratio to input. Data are presented as means ± SD (n = 3). *P < 0.02 and **P < 0.01 for (b). *P < 0.03, **P < 0.04, ***P < 0.01, and ****P < 0.005 for (c). The experiment was repeated with similar results. PML, promyelocytic leukemia protein (PML).
scAAV2/8-mediated gene expression is not silenced by type I IFN dependent signaling in livers either at early or late time points
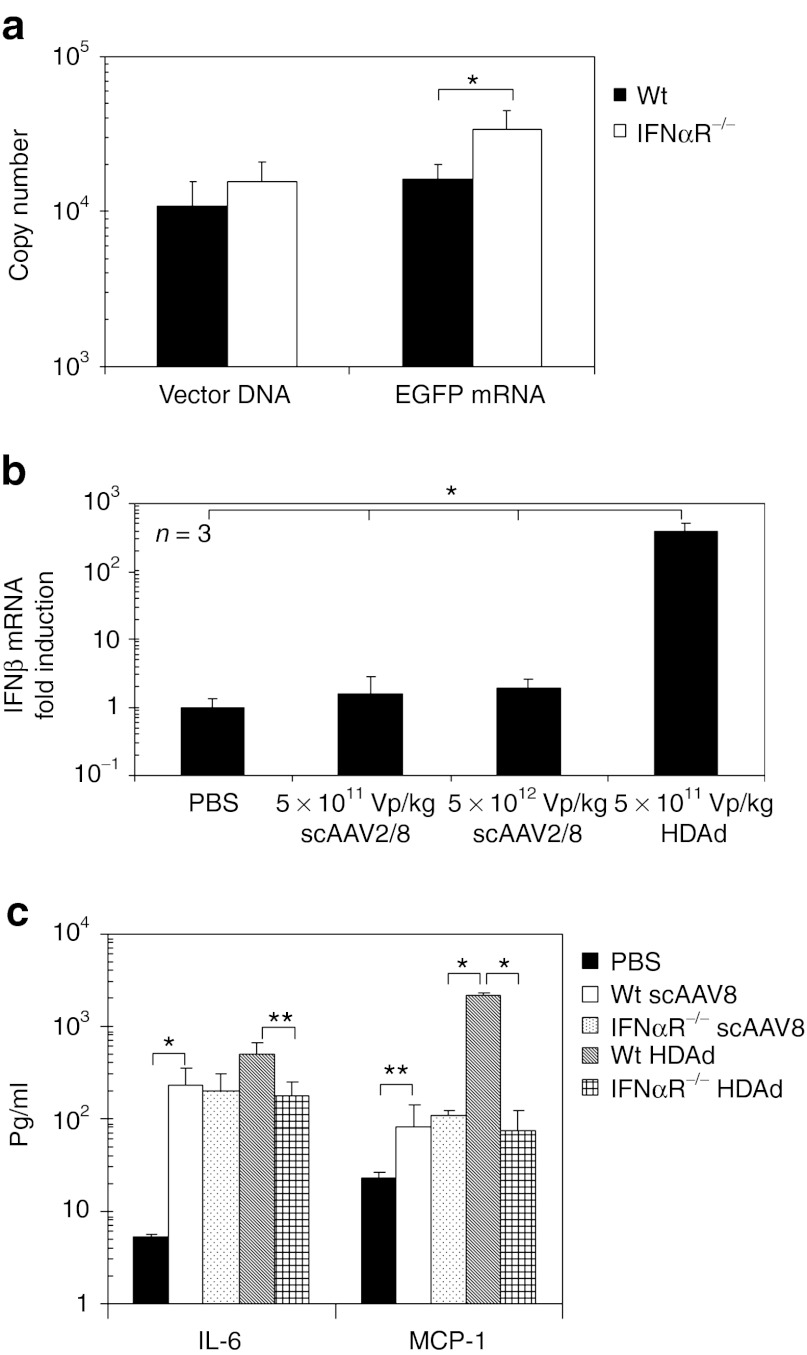
Previously, we showed that host animals induce the expression of inflammatory cytokine mRNAs (including type I IFN) to scAAVs via TLR9 in livers of mice.38 To test whether transgene expression from scAAVs is also silenced in type I IFN-dependent fashion, we quantified copies of vector DNA and transgene mRNA (enhanced green fluorescent protein (EGFP) mRNA) in livers of WT and IFNαR−/− mice at 24 hours post-injection of 5 × 1011 Vg/kg scAAV2/8EGFP (Figure 5a). ScAAV2/8 is known to have a strong tropism to the liver after systemic administration.39 While HDAd-injected IFNαR−/− mice showed an over 50-fold higher transgene level of mRNA compared to WT mouse livers at an identical vector dose, scAAV-injected IFNαR−/− mice actually had a 1.5-fold higher EGFP mRNA level compared to WT mouse livers. For scAAV2/8, no difference in the ratio of EGFP mRNA to vector DNA was observed between WT and IFNαR−/− mice (data not shown). These results show that transgene expression from scAAV2/8 vector was not inhibited by type I IFN signaling at 24 hours post-injection.
Figure 5.
ScAAV2/8 attenuates type I interferon (IFN) signaling dependent transgene silencing at 24 hours post-injection. (a) Wild-type (WT) C57Bl/6 and IFNαR−/− mice were injected with 5 × 1011 Vg/kg of scAAV2/8EGFP into tail veins. Total DNA and total RNA were extracted from their livers at 24 hours post-injection, and vector copy number in total DNA and EGFP mRNA copy number in total RNA were determined by quantitative PCR. Data are presented as means ± SD (n = 3). *P < 0.04. (b) WT C57Bl/6 mice were injected with 5 × 1011 Vg/kg or 5 × 1012 Vg/kg of scAAV2/8EGFP or 5 × 1011 viral particle (Vp)/kg of HDAdLacZ into tail veins. Total RNA was extracted from livers at 3 hours post-injection, and the levels of IFNβ mRNA were determined by quantitative real-time PCR. Each value was calculated relative to that of control mice injected with phosphate-buffered saline (PBS). Data are presented as means ± SD (n = 3). *P < 0.001. (c) WT C57Bl/6 and IFNαR−/− mice were injected with 5 × 1011 Vg/kg of scAAV2/8EGFP or 5 × 1011 Vp/kg of HDAdLacZ into tail veins. Plasma samples were collected at 6 hours post-injection. IL-6 and MCP-1 levels were measured by using BD cytokine multiplex bead array system. Data are presented as means ± SD (n = 3). *P < 0.001 and **P < 0.05. The experiment was repeated with similar results. HDAd, Helper-dependent adenoviral vector.
We also quantified the induction level of IFNβ mRNA in livers of WT mice at 3 hours post-injection of 5 × 1011 Vg/kg or 5 × 1012 Vg/kg of scAAV2/8EGFP vs. 5 × 1011 Vp/kg of HDAdLacZ (Figure 5b). Although WT mice injected with 5 × 1012 Vg/kg scAAVs showed an approximately tenfold higher type I IFN mRNA level compared to control mice (PBS injection) at 2 hours post-injection,38 there was no significant difference of IFNβ mRNA between PBS injected livers and scAAV2/8 injected livers (both dosages) at 3 hours post-injection. WT mice injected with other scAAV vectors (scAAV2, scAAV2/5) also failed to show a significant difference of IFNβ mRNA compared to PBS injected livers at 3 hours post-injection (data not shown). We next measured the cytokine levels in the blood of WT and IFNαR−/− mice at 6 hours post-injection of 5 × 1011 Vg/kg of scAAV2/8EGFP or 5 × 1011 Vp/kg of HDAdLacZ (Figure 5c), because HDAd injected mice showed contribution of IFNαR-dependent cytokine expression at 6 hours post-injection (Figure 1c). Although IFNαR−/− mice injected with HDAd showed significant induction of both IL-6 and MCP-1 compared to WT mice, there was no difference in the levels of these cytokines between WT and IFNαR−/− mice after systemic administration of scAAV2/8. These results suggest that scAAV2/8 does not amplify proinflammatory innate immune responses through type I IFN signaling via IFNαR.
Ad-based vector injected mice showed type I IFN signaling dependent development of an acquired response to Ad Vps and transgene expression was significantly reduced in livers of WT mice compared to IFNαR−/− mice at late time points (Figure 6a).21,40 To test whether type I IFN signaling contributes to transgene silencing of scAAV at a later time points, we measured human factor IX (hFIX) levels in the blood of WT and IFNαR−/− mice at 1, 2, 3, 4, and 5 weeks post-injection of 5 × 1012 Vg/kg of scAAV2hFIX (Figure 6b). No difference in hFIX levels in the blood of WT and IFNαR−/− mice was seen. These results also suggest that type I IFN signaling minimally affects the transgene expression of scAAV at a later time points.
Figure 6.
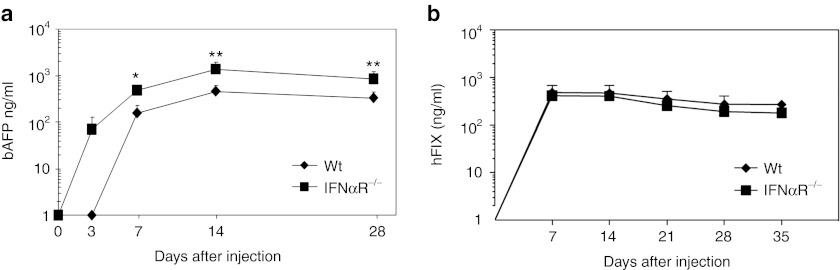
Contribution of type I interferon (IFN) signaling to transgene expression of Helper-dependent adenoviral vectors (HDAds) or scAAV2 at long-time points. (a) Wild-type (WT) C57Bl/6 and IFNαR−/− mice were injected with 5 × 1011 viral particle (Vp)/kg of HDAdbAFP into tail veins. Serum samples were collected at 3, 7, 14, and 28 days post-injection. Baboon AFP levels were measured by using ELISA. Data are presented as means ± SD (n = 5). *P < 0.001 and **P < 0.02. (b) WT C57Bl/6 and IFNαR−/− mice were injected with 5 × 1011 Vg/kg of scAAV2hFIX or into tail veins. Plasma samples were collected at 7, 14, 21, and 28 days post-injection. The hFIX levels were measured by using ELISA. Data are presented as means ± SD (n = 5).
Discussion
The host innate immune response is complex and involves many cellular and humoral factors. Hence, it is likely that no single pathway accounts for the overall immune response to Ad-based or AAV-based vectors. The development of HDAd, devoid of all functional viral genes, has largely, but not entirely, overcome the cell mediated host immune response against antigenic epitopes associated with viral proteins expressed by FGAd.9 However, the major obstacle for clinical translation in humans remains the dose-related acute toxicity associated with systemic delivery of Ad-based vectors.26 We and other groups have previously reported that multiple pattern recognition receptor signaling pathways contribute to the innate response to Ad-based vectors in vivo.12,13,14,15,16,23,24,41 In this study, we found that type I IFN induced at 1 hour contributes by amplifying the innate response to HDAd in livers of mice. Another DNA viral vector, HSV-1-based vector, is also known to induce the expression of type I IFN mRNA (but not type II IFN) in livers of WT mice at 1-hour post-injection after systemic administration.29 HSV-1-based vector administered liver mRNA profiles showed that the expression of multiple cytokine and chemokine mRNAs in the liver is primarily regulated by type I IFN/STAT1 signaling. In that study, cytokine and chemokine mRNAs were significantly decreased in livers of STAT1−/− mice after 3 hours post-injection of HSV-1 based vector (a finding consistent with ours). These results suggest that proinflammatory cytokines and chemokines contributed to an antiviral vector response that could be primarily amplified through type I IFN signaling.
We also found that an innate response to HDAds (dependent on type I IFN signaling) contributes to epigenetic silencing of both eukaryotic and prokaryotic vector DNA. However, type I IFN signaling does not appear to contribute to loss of HDAd vector DNA in the liver at 24 hours similar to that seen with FGAd.22 We and others have shown that negatively charged circulating Ad Vps in the blood may be captured by Kupffer cells via scavenger receptors in the liver leading to necrotic loss of Kupffer cells (and vector DNA).42,43 We found that type I IFN signaling regulates transgene transcription of HDAd in liver at 24 hours post-injection. In both prokaryotic (bacterial LacZ) and eukaryotic (human FVIII) genes, transcription from vector was observed to be suppressed. This suggests that type I IFN signaling induced by HDAd administration in livers of mice contributes to suppression of transgene expression from HDAd at the transcriptional level and this is independent of transgene or promoter sequences.
We found that type I IFN signaling dependent epigenetic modification contributes to silencing transgene expression of HDAd at the transcriptional level in liver. Induction of PML mRNA is dependent on type I IFN signaling, and PML and HDAC1 associate with both prokaryotic and eukaryotic sequences of HDAd vector DNA in liver cells. Other groups have previously reported that the prokaryotic sequence of vector DNA was first sensed in transduced cells and epigenetically modified toward transgene silencing (heterochromatic formation). That transcriptionally inactive, heterochromatic region spreads over the entire vector DNA in vitro.30,36 Epigenetic modification of the prokaryotic sequence in vector DNA directed toward transgene silencing occurred independently of methylation in the CpG motif of vector DNA sequences and independently of gene delivery methods (both nonviral vector and viral vectors).32,36,44 Flanking the prokaryotic sequence with insulator sequences partially attenuated epigenetic modification of other vector DNA regions in vitro.36 However, our in vivo ChIP results indicated that eukaryotic sequences in HDAd vector DNA were also epigenetically modified toward gene silencing similar to prokaryotic sequences by type I IFN-dependent signaling in livers of mice.
In contrast, scAAVs (scAAV2/8), which are currently in a clinical trial for treatment of hemophilia by hepatic gene transfer,39 were not silenced by the type I IFN signaling dependent processes. There was also no difference seen in long-term expression of the scAAV2 transgene between WT and IFNαR−/− mice. Previously, we found that scAAVs induced the expression of type I IFN mRNA in mouse liver within 2 hours post-injection, a response that was TLR9 and Kupffer cell-dependent was seen to subside by 6 hours.38 Here, we found minimal induction of IFNβ mRNA by scAAV2/8 in the liver at 3 hours post-injection. This could be why systemically administered scAAVs show attenuated type I IFN signaling dependent transgene silencing over the short- and long-term relative to other DNA and RNA viral vectors.20,21,22,29 However, recent reports showed that type I IFN induced by TLR9/MyD88 signaling has an essential role in the development of both cellular and humoral acquired responses to AAV viral particle (and transgene product) after intramuscular injection of AAVs.45 These adaptive responses can influence transgene expression from AAV vectors at late time points.45 While AAV vectors have been used to treat multiple genetic diseases,46 some patients showed a transient therapeutic effect, likely due to the development of B- or T-cell responses to viral proteins after systemic administration.47 Although our data suggest that systemically administered scAAVs attenuate type I IFN-dependent transgene silencing, their administration may still contribute to the development of an acquired immune response in humans. AAV vectors activate inflammatory genes within 2 hours after gene transfer via the NF-κB pathway, which may be caused by TLR2 or TLR9 sensing of capsid or vector DNA.38,48,49 Interestingly, here we found a minimal effect of type I IFN signaling on the induction of IL-6 and MCP-1 responses to scAAVs in blood, suggesting that the attenuated type I IFN response fails to amplify a broader inflammatory response.
It is necessary to note that rodents are extremely tolerant to systemically delivered high doses of Ad-based vectors, unlike primates.26 The reasons for this difference are not known but likely involve species to species differences in the quality of the innate immune response or sensitivities of the end organs to pathologic sequelae.26 Thus, additional studies in large animal models may be helpful to assess the contribution of type I IFN signaling to systemic administered AAV-based vectors and to define the full-molecular mechanisms that specify the innate and acquired immune responses to scAAVs in vivo for development/improvement of human gene therapy.
Materials and Methods
Viral vectors (HDAds and scAAVs). HDAd HDΔ28E4 (HDAd0) and HDAd HDΔ28E4LacZ constructs containing the β-galactosidase transgene driven by the cytomegalovirus promoter (HDAdLacZ) were produced as described elsewhere.50 HDAd HDΔ25.3E4PEPCK-hFVIII constructs containing the human FVIII cDNA transgene driven by the liver-restricted PEPCK promoter (HDAdhFVIII) were produced as described elsewhere.51,52 HDAd HDΔ21.7E4PEPCK-bAFP-WL constructs containing the baboon AFP cDNA transgene driven by the liver-restricted PEPCK promoter (HDAdbAFP) were produced as described elsewhere.52 Helper-virus contamination in viral preparations was assessed by Southern blot and Phosphor Imager analysis and was estimated to be <0.05% as described elsewhere.22,50
Various AAV vector genome constructs and serotypes were used: AAV2, AAV8. For scAAV vectors, hFIX was driven by the liver-restricted transthyretin promoter, and GFP vectors contained the cytomegalovirus enhancer/chicken β-actin promoter as described elsewhere.38
Animals and injections. IFNαR−/− mice (back-crossed with C57BL/6) were provided by W. Yokoyama (Washington University). WT C57BL/6 mice were purchased from Jackson Laboratories (Bar Harbor, ME). All of the mice were housed under pathogen-free conditions; food and water were provided ad libitum. Mice used in all experiments were males and between 7–10 weeks of age. All experimental procedures were conducted in accordance with institutional guidelines for animal care and use. HDAds or scAAVs were diluted in PBS and injected into tail veins. The injections were performed in a total volume of 200 µL. Blood was collected retro-orbitally for analyses. Plasma and serum samples were frozen immediately and stored at −80 °C until analysis. Upon sacrifice, livers were harvested and kept at −80 °C until analysis.
Chemical and antibodies. HDAC inhibitor TSA was purchased from Sigma-Aldrich Inc. (St. Louis, MO) and dissolved to ethanol to a stock. To evaluate the role of HDACs, 5 µg/kg of TSA was coinjected with HDAd injection.53
Anti-Histone H3 (ab1791), anti-Dimethyl Lysine 9 histone H3 (ab1220), anti-Acetyl Lysine 9 histone H3 (ab4441), anti-HDAC1 (ab51846), and anti-PML (ab50637) antibodies were purchased from Abcam (Cambridge, MA).
Cytokine analysis. Mouse IL-6 and MCP-1 in plasma were assayed by using the BD cytokine multiplex bead array system (BD Biosciences, San Jose, CA), and analyzed using a BD FacsArray instrument (BD Biosciences) according to manufacturer's instructions.
Quantitative real-time RT-PCR analysis of cytokine expression. Mice were injected with 5 × 1011 Vp/kg of HDAd0, HDAdLacZ or HDAdhFVIII as described above. The animals were sacrificed at stated time points, and total RNA was extracted from the liver of each animal using TRIzol reagent (Invitrogen, Carlsbad, CA). First-strand cDNA was synthesized from RNA samples using Superscript III with oligo dT priming (Invitrogen) and analyzed by SYBR green quantitative real-time PCR analysis (10 minutes at 95°C and then 45 cycles of 10 seconds at 95°C, 7 seconds at 58°C, and 30 seconds at 72°C) using a Roche LightCycler 1.1, and Roche master mix (Roche) according to the manufacturer's protocol. The following primer sequences were designed and used for the analysis of mouse IFNγ: Forward: 5′-GTGCTGCTGATGGGAGGAGATGTCTA-3′ and Reverse: 5′-GTTATTTGTCATTCGGGTGTA-3′, mouse IFNβ: Forward: 5′-TGCG TTCCTGCTGTGCTTCTC-3′ and Reverse: 5′-GCATCTTCTCCGTCATCT CCATAG-3′. To control for template variation among samples, the level of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA expression levels was determined for each sample using specific primers Forward: (5′-GCAAGAGAGGCCCTATCCCAA-3′ and Reverse: 5′-CTCCCTAGG CCCCTCCTGTTATT-3′). To evaluate the impact of TSA treatment to a reference/housekeeping gene, the level of β-2-microglobulin mRNA expression levels was also determined for each sample (TSA treated or untreated) using specific primers Forward: (5′-GGTCTTTCTGGTGCTTGTC-3′ and Reverse: 5′-CGTATGTATCAGTCTCAGT-3′).
Vector genome DNA and transgene mRNA in livers of mice. Mice were injected with 5 × 1011 Vp/kg of HDAd0, HDAdLacZ, HDAdhFVIII or 5 × 1011 Vg/kg of scAAV2/8EGFP as described above. Total DNA was extracted from livers using a DNeasy Blood and Tissue Kit (QIAGEN, Valencia, CA). Total RNA was extracted from the liver of each animal, and first-strand cDNA was synthesized from RNA samples as described above. DNA and cDNA samples prepared from each liver were analyzed by quantitative real-time PCR (10 minutes at 95°C and then 45 cycles of 10 seconds at 95°C, 7 seconds at 60°C, and 30 seconds at 72°C) using a Roche LightCycler 1.1, and Roche master mix (Roche) and Human stuffer gene-specific primers Forward: 5′-TCTGAATAATTTTGTGTTACTCATAGCGCG-3′ and Reverse: 5′-CCCATAAGCTCCTTTTAACTTGTTAAAGTC-3′, LacZ-specific primers Forward: 5′-ATACTGTCGTCGTCCCCTCAAACT-3′ and Reverse: 5′-CCTCCAGATAACTGCCGTCACTC-3′, hFVIII-specific primers Forward: 5′-TCCTTACTGCTCAAACACTC-3′ and Reverse: 5′-TCCGTGAGGGTAGATGTTAT-3′ and EGFP-specific primers Forward: 5′-CCTGGCCCACCCTCGTGAC-3′ and Reverse: 5′-GCGCT CCTGGACGTAGCCTTC-3′. Vector copy numbers per microgram of total DNA or cDNA were calculated from the standard curve generated by Q-PCR spanning nine orders of magnitude by serial dilution of the original plasmid DNA of pHDAdLacZ, pHDAdhFVIII or pHDAdEGFP.12,13 For quantification of vector copy numbers to genomic DNA, we analyzed liver DNA samples by quantitative real-time PCR using mouse genomic GAPDH specific primers Forward: 5′-TAGGCCAGGATGTAAAGGTCATTAAG-3′ and Reverse: 5′-CCAGAAAGGTCACACGGCTAAA-3′.
In vivo ChIP analysis (mouse liver ChIP analysis). We followed previous reported mouse liver ChIP analysis protocol29,30,32,54 with modifications. Briefly, mice were sacrificed at 24 hours post-injection of 5 × 1011 Vp/kg of HDAd0, HDAdLacZ or HDAdhFVIII as described above. Livers were dissected and Dounce homogenized in 10 mL of Dulbecco's modified Eagle's medium supplemented with protease inhibitor (Complete, EDTA-Free; Roche). Homogenized samples were fixed with 1% formaldehyde (Sigma-Aldrich Inc.) for 15 minutes at room temperature. The reaction was quenched by the addition of 125 mM Glycine. After centrifugation (2,000 rpm for 5 minutes), samples were washed two times with PBS supplemented protease inhibitor and resuspended with 10 mL PBS. One milliliter samples were centrifuged (2,000 rpm for 5 minutes), cells were resuspended in 300 µL SDS-lysis buffer (Millipore, Billerica, CA). Samples were incubated 30 minutes on ice and sonicated with a Bioruptor XL (Diagnode Inc., Denville, NJ) for ten times (30-second pulses at 1-minute interval). After centrifugation (13,000 rpm for 10 minutes), supernatants were collected and diluted with 3 mL ChIP dilution buffer (Millipore). Part of each sample was tested to confirm that cross-linked DNA was sheared to 200–1,000 base pairs in length (Supplementary Figure S7). The diluted samples were pre-cleared by the addition of Protein G agarose with salmon sperm DNA (Millipore) and rotated at 4 °C for 2 hours. After centrifugation (1,000g for 2 minutes), supernatants were distributed into six aliquots. One aliquot was used as “Input. ” Other aliquots received 2 µg of antibodies and rotated overnight at 4 °C. After incubation, 60 µL Protein G agarose were added to each sample and rotated for 2 hours at 4 °C. Each sample was washed with ChIP wash buffers according to manufacturer's instructions (Millipore). Samples were eluted with 400 µL of elution buffer (100 mM NaHCO3, 1% sodium dodecyl sulfate). 16 µL of 5 M NaCl (final concentration of 0.2 M) was added, and samples including “Input ” samples were reverse cross-linked by incubation at 65 °C for overnight. After incubation, samples were treated with proteinase K at 45 °C for 1 hour, and DNA was extracted with Phenol:Chloroform, and ethanol-precipitated in the presence of glycogen (Invitrogen). The final DNA samples were resuspended in nuclease-free water, and DNA concentration was measured. These samples were quantified by real-time PCR (for 10 minutes at 95 °C and then 45 cycles of 10 seconds at 95 °C, 30 seconds at 55 °C, and 30 seconds at 72 °C) using a Roche LightCycler 1.1, and Roche master mix (Roche) with specifically designed primer sets (Table 1). Vector copy numbers per microgram of total DNA were calculated from the standard curve generated by Q-PCR spanning nine orders of magnitude by serial dilution of the original plasmid DNA. The ratio of each sample to Input was calculated by dividing vector copy numbers of each sample to that in “Input”.
AFP expression in blood of mice. Baboon AFP (bAFP) in serum was assayed by using the Alpha-Fetoprotein (AFP) Elisa kit (CALBIOTECH, Spring Valley, CA) according to manufacturer's instructions.
Systemic hFIX levels. ELISA for hFIX levels in plasma samples was as described elsewhere.38
Statistical analysis. Data were analyzed by t-test or one-way ANOVA analysis of variance followed by Shapiro–Wilk's protected least significant difference test (SigmaPlot; Systat Software, San Jose, CA).
SUPPLEMENTARY MATERIAL Figure S1. Type I IFN signaling minimally affects vector DNA clearance in livers of mice at 24 hours post-injection. Figure S2. Type I IFN signaling does not affect the chromatin formation of HDAd vector DNA in livers. Figure S3. There is no difference of the ratio of GAPDH between WT and IFNαR−/− samples after immunoprecipitation. Figure S4. TSA treatment minimally affects housekeeping gene expression in WT livers at 24 hours post-injection. Figure S5. Eukaryotic sequence of vector DNA in HDAdLacZ is also epigenetically modified due to type I IFN signaling. Figure S6. Mammalian transgene expression cassette is also suppressed at transcriptional level and epigenetically modified due to type I IFN signaling. Figure S7. Liver DNA samples were mainly shared to 200–1,000 base pairs in length after sonication.
Acknowledgments
This work was supported by the National Institutes of Health (R01HL87836) to B.L., National Institutes of Health (K99HL098692) to M.S., National Institutes of Health (R01AI51390) to R.W.H., National Institutes of Health (R01DK067324) to P.N., and the Baylor College of Medicine Intellectual and Developmental Disability Research Center and the Texas Gulf Coast Digestive Disease Center.
Supplementary Material
Type I IFN signaling minimally affects vector DNA clearance in livers of mice at 24 hours post-injection.
Type I IFN signaling does not affect the chromatin formation of HDAd vector DNA in livers.
There is no difference of the ratio of GAPDH between WT and IFNαR−/− samples after immunoprecipitation.
TSA treatment minimally affects housekeeping gene expression in WT livers at 24 hours post-injection.
Eukaryotic sequence of vector DNA in HDAdLacZ is also epigenetically modified due to type I IFN signaling.
Mammalian transgene expression cassette is also suppressed at transcriptional level and epigenetically modified due to type I IFN signaling.
Liver DNA samples were mainly shared to 200–1,000 base pairs in length after sonication.
REFERENCES
- Brunetti-Pierri N, Stapleton GE, Law M, Breinholt J, Palmer DJ, Zuo Y.et al. (2009Efficient, long-term hepatic gene transfer using clinically relevant HDAd doses by balloon occlusion catheter delivery in nonhuman primates Mol Ther 17327–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerullo V, McCormack W, Seiler M, Mane V, Cela R, Clarke C.et al. (2007Antigen-specific tolerance of human alpha1-antitrypsin induced by helper-dependent adenovirus Hum Gene Ther 181215–1224. [DOI] [PubMed] [Google Scholar]
- McCormack WM, Jr, Seiler MP, Bertin TK, Ubhayakar K, Palmer DJ, Ng P.et al. (2006Helper-dependent adenoviral gene therapy mediates long-term correction of the clotting defect in the canine hemophilia A model J Thromb Haemost 41218–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mian A, McCormack WM, Jr, Mane V, Kleppe S, Ng P, Finegold M.et al. (2004Long-term correction of ornithine transcarbamylase deficiency by WPRE-mediated overexpression using a helper-dependent adenovirus Mol Ther 10492–499. [DOI] [PubMed] [Google Scholar]
- Toietta G, Mane VP, Norona WS, Finegold MJ, Ng P, McDonagh AF.et al. (2005Lifelong elimination of hyperbilirubinemia in the Gunn rat with a single injection of helper-dependent adenoviral vector Proc Natl Acad Sci USA 1023930–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunetti-Pierri N, Stapleton GE, Palmer DJ, Zuo Y, Mane VP, Finegold MJ.et al. (2007Pseudo-hydrodynamic delivery of helper-dependent adenoviral vectors into non-human primates for liver-directed gene therapy Mol Ther 15732–740. [DOI] [PubMed] [Google Scholar]
- Brunetti-Pierri N, Stapleton GE, Law M, Breinholt J, Palmer DJ, Zuo Y.et al. (2009Efficient, long-term hepatic gene transfer using clinically relevant HDAd doses by balloon occlusion catheter delivery in nonhuman primates Mol Ther 17327–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey AP, Fawcett P, Nakai H, McCaffrey RL, Ehrhardt A, Pham TT.et al. (2008The host response to adenovirus, helper-dependent adenovirus, and adeno-associated virus in mouse liver Mol Ther 16931–941. [DOI] [PubMed] [Google Scholar]
- Muruve DA, Cotter MJ, Zaiss AK, White LR, Liu Q, Chan T.et al. (2004Helper-dependent adenovirus vectors elicit intact innate but attenuated adaptive host immune responses in vivo J Virol 785966–5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mane VP, Toietta G, McCormack WM, Conde I, Clarke C, Palmer D.et al. (2006Modulation of TNFalpha, a determinant of acute toxicity associated with systemic delivery of first-generation and helper-dependent adenoviral vectors Gene Ther 131272–1280. [DOI] [PubMed] [Google Scholar]
- Raper SE, Yudkoff M, Chirmule N, Gao GP, Nunes F, Haskal ZJ.et al. (2002A pilot study of in vivo liver-directed gene transfer with an adenoviral vector in partial ornithine transcarbamylase deficiency Hum Gene Ther 13163–175. [DOI] [PubMed] [Google Scholar]
- Suzuki, M, Cerullo, V, Bertin, TK, Cela, R, Clarke, C, Guenther, M.et al. (2009MyD88-dependent silencing of transgene expression during the innate and adaptive immune response to Helper-dependent adenovirus Hum Gene Ther 21325–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Cela R, Bertin TK, Sule G, Cerullo V, Rodgers JR.et al. (2011NOD2 signaling contributes to the innate immune response against helper-dependent adenovirus vectors independently of MyD88 in vivo Hum Gene Ther 221071–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muruve DA, Pétrilli V, Zaiss AK, White LR, Clark SA, Ross PJ.et al. (2008The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response Nature 452103–107. [DOI] [PubMed] [Google Scholar]
- Cerullo V, Seiler MP, Mane V, Brunetti-Pierri N, Clarke C, Bertin TK.et al. (2007Toll-like receptor 9 triggers an innate immune response to helper-dependent adenoviral vectors Mol Ther 15378–385. [DOI] [PubMed] [Google Scholar]
- Appledorn, DM, Patial, S, McBride, A, Godbehere, S, Van Rooijen, N, Parameswaran N.et al. (2008Adenovirus vector-induced innate inflammatory mediators, MAPK signaling, as well as adaptive immune responses are dependent upon both TLR2 and TLR9 in vivo J Immunol 1812134–2144. [DOI] [PubMed] [Google Scholar]
- Sakurai F, Mizuguchi H, Yamaguchi T., and, Hayakawa T. Characterization of in vitro and in vivo gene transfer properties of adenovirus serotype 35 vector. Mol Ther. 2003;8:813–821. doi: 10.1016/s1525-0016(03)00243-0. [DOI] [PubMed] [Google Scholar]
- Sadler AJ., and, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi O, Kawai T, Takeda K, Matsumoto M, Tsutsui H, Sakagami M.et al. (1998Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function Immunity 9143–150. [DOI] [PubMed] [Google Scholar]
- Brown BD, Sitia G, Annoni A, Hauben E, Sergi LS, Zingale A.et al. (2007In vivo administration of lentiviral vectors triggers a type I interferon response that restricts hepatocyte gene transfer and promotes vector clearance Blood 1092797–2805. [DOI] [PubMed] [Google Scholar]
- Zhu J, Huang X., and, Yang Y. A critical role for type I IFN-dependent NK cell activation in innate immune elimination of adenoviral vectors in vivo. Mol Ther. 2008;16:1300–1307. doi: 10.1038/mt.2008.88. [DOI] [PubMed] [Google Scholar]
- Zhu J, Huang X., and, Yang Y. Innate immune response to adenoviral vectors is mediated by both Toll-like receptor-dependent and -independent pathways. J Virol. 2007;81:3170–3180. doi: 10.1128/JVI.02192-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo NC, Miao EA, Iwakura Y, Murali-Krishna K, Aderem A, Flavell RA.et al. (2009Virus binding to a plasma membrane receptor triggers interleukin-1 alpha-mediated proinflammatory macrophage response in vivo Immunity 31110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doronin K, Flatt JW, Di Paolo NC, Khare R, Kalyuzhniy O, Acchione M.et al. (2012Coagulation factor X activates innate immunity to human species C adenovirus Science 338795–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M.et al. NOD2 signaling contributes to the innate immune response against helper-dependent adenovirus vectors independently of MyD88 in vivo Hum Gene Ther 221071–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell MA, Zhang Y, Tazelaar J, Gao GP, Yu QC, Qian R.et al. (2001Activation of innate immunity in nonhuman primates following intraportal administration of adenoviral vectors Mol Ther 35 Pt 1708–722. [DOI] [PubMed] [Google Scholar]
- Zaiss AK, Liu Q, Bowen GP, Wong NC, Bartlett JS., and, Muruve DA. Differential activation of innate immune responses by adenovirus and adeno-associated virus vectors. J Virol. 2002;76:4580–4590. doi: 10.1128/JVI.76.9.4580-4590.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wonganan P, Clemens CC, Brasky K, Pastore L., and, Croyle MA. Species differences in the pharmacology and toxicology of PEGylated helper-dependent adenovirus. Mol Pharm. 2011;8:78–92. doi: 10.1021/mp100216h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Chiocca EA., and, Saeki Y. Early STAT1 activation after systemic delivery of HSV amplicon vectors suppresses transcription of the vector-encoded transgene. Mol Ther. 2007;15:2017–2026. doi: 10.1038/sj.mt.6300273. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Kasai K., and, Saeki Y. Plasmid DNA sequences present in conventional herpes simplex virus amplicon vectors cause rapid transgene silencing by forming inactive chromatin. J Virol. 2006;80:3293–3300. doi: 10.1128/JVI.80.7.3293-3300.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muruve DA, Barnes MJ, Stillman IE., and, Libermann TA. Adenoviral gene therapy leads to rapid induction of multiple chemokines and acute neutrophil-dependent hepatic injury in vivo. Hum Gene Ther. 1999;10:965–976. doi: 10.1089/10430349950018364. [DOI] [PubMed] [Google Scholar]
- Riu E, Chen ZY, Xu H, He CY., and, Kay MA. Histone modifications are associated with the persistence or silencing of vector-mediated transgene expression in vivo. Mol Ther. 2007;15:1348–1355. doi: 10.1038/sj.mt.6300177. [DOI] [PubMed] [Google Scholar]
- Everett RD., and, Chelbi-Alix MK. PML and PML nuclear bodies: implications in antiviral defence. Biochimie. 2007;89:819–830. doi: 10.1016/j.biochi.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Regad T., and, Chelbi-Alix MK. Role and fate of PML nuclear bodies in response to interferon and viral infections. Oncogene. 2001;20:7274–7286. doi: 10.1038/sj.onc.1204854. [DOI] [PubMed] [Google Scholar]
- Bishop CL, Ramalho M, Nadkarni N, May Kong W, Higgins CF., and, Krauzewicz N. Role for centromeric heterochromatin and PML nuclear bodies in the cellular response to foreign DNA. Mol Cell Biol. 2006;26:2583–2594. doi: 10.1128/MCB.26.7.2583-2594.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross PJ, Kennedy MA., and, Parks RJ. Host cell detection of noncoding stuffer DNA contained in helper-dependent adenovirus vectors leads to epigenetic repression of transgene expression. J Virol. 2009;83:8409–8417. doi: 10.1128/JVI.00796-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbalat R, Lau L, Locksley RM., and, Barton GM. Toll-like receptor 2 on inflammatory monocytes induces type I interferon in response to viral but not bacterial ligands. Nat Immunol. 2009;10:1200–1207. doi: 10.1038/ni.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino AT, Suzuki M, Markusic DM, Zolotukhin I, Ryals RC, Moghimi B.et al. (2011The genome of self-complementary adeno-associated viral vectors increases Toll-like receptor 9-dependent innate immune responses in the liver Blood 1176459–6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathwani AC, Tuddenham EG, Rangarajan S, Rosales C, McIntosh J, Linch DC.et al. (2011Adenovirus-associated virus vector-mediated gene transfer in hemophilia B N Engl J Med 3652357–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Huang X., and, Yang Y. Type I IFN signaling on both B and CD4 T cells is required for protective antibody response to adenovirus. J Immunol. 2007;178:3505–3510. doi: 10.4049/jimmunol.178.6.3505. [DOI] [PubMed] [Google Scholar]
- Chiu YH, Macmillan JB., and, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Xu Z., and, Byrnes AP. A quantitative assay for measuring clearance of adenovirus vectors by Kupffer cells. J Virol Methods. 2008;147:54–60. doi: 10.1016/j.jviromet.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Xu Z, Tian J, Smith JS., and, Byrnes AP. Clearance of adenovirus by Kupffer cells is mediated by scavenger receptors, natural antibodies, and complement. J Virol. 2008;82:11705–11713. doi: 10.1128/JVI.01320-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZY, Riu E, He CY, Xu H., and, Kay MA. Silencing of episomal transgene expression in liver by plasmid bacterial backbone DNA is independent of CpG methylation. Mol Ther. 2008;16:548–556. doi: 10.1038/sj.mt.6300399. [DOI] [PubMed] [Google Scholar]
- Zhu J, Huang X., and, Yang Y. The TLR9-MyD88 pathway is critical for adaptive immune responses to adeno-associated virus gene therapy vectors in mice. J Clin Invest. 2009;119:2388–2398. doi: 10.1172/JCI37607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingozzi F., and, High KA. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat Rev Genet. 2011;12:341–355. doi: 10.1038/nrg2988. [DOI] [PubMed] [Google Scholar]
- Mingozzi F., and, High KA. Immune responses to AAV in clinical trials. Curr Gene Ther. 2011;11:321–330. doi: 10.2174/156652311796150354. [DOI] [PubMed] [Google Scholar]
- Jayandharan GR, Aslanidi G, Martino AT, Jahn SC, Perrin GQ, Herzog RW.et al. (2011Activation of the NF-kappaB pathway by adeno-associated virus (AAV) vectors and its implications in immune response and gene therapy Proc Natl Acad Sci USA 1083743–3748. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hösel M, Broxtermann M, Janicki H, Esser K, Arzberger S, Hartmann P.et al. (2012Toll-like receptor 2-mediated innate immune response in human nonparenchymal liver cells toward adeno-associated viral vectors Hepatology 55287–297. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Cela R, Clarke C, Bertin TK, Mourino S, Lee B. Large-scale production of high-quality helper-dependent adenoviral vectors using adherent cells in cell factories. Human Gene Therapy. 2009. [DOI] [PMC free article] [PubMed]
- Cerullo V, Seiler MP, Mane V, Cela R, Clarke C, Kaufman RJ.et al. (2007Correction of murine hemophilia A and immunological differences of factor VIII variants delivered by helper-dependent adenoviral vectors Mol Ther 152080–2087. [DOI] [PubMed] [Google Scholar]
- Brunetti-Pierri N, Ng T, Iannitti DA, Palmer DJ, Beaudet AL, Finegold MJ.et al. (2006Improved hepatic transduction, reduced systemic vector dissemination, and long-term transgene expression by delivering helper-dependent adenoviral vectors into the surgically isolated liver of nonhuman primates Hum Gene Ther 17391–404. [DOI] [PubMed] [Google Scholar]
- Gaetano C, Catalano A, Palumbo R, Illi B, Orlando G, Ventoruzzo G.et al. (2000Transcriptionally active drugs improve adenovirus vector performance in vitro and in vivo Gene Ther 71624–1630. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Kasai K, Ohtsuki A, Godlewski J, Nowicki MO, Chiocca EA.et al. (2009ICP0 inhibits the decrease of HSV amplicon-mediated transgene expression Mol Ther 17707–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Type I IFN signaling minimally affects vector DNA clearance in livers of mice at 24 hours post-injection.
Type I IFN signaling does not affect the chromatin formation of HDAd vector DNA in livers.
There is no difference of the ratio of GAPDH between WT and IFNαR−/− samples after immunoprecipitation.
TSA treatment minimally affects housekeeping gene expression in WT livers at 24 hours post-injection.
Eukaryotic sequence of vector DNA in HDAdLacZ is also epigenetically modified due to type I IFN signaling.
Mammalian transgene expression cassette is also suppressed at transcriptional level and epigenetically modified due to type I IFN signaling.
Liver DNA samples were mainly shared to 200–1,000 base pairs in length after sonication.