Abstract
The centromere is the fundamental unit for insuring chromosome inheritance. This complex region has a distinct type of chromatin in which histone H3 is replaced by a structurally different homologue identified in humans as CENP-A. In metazoans, specific DNA sequences are neither required nor sufficient for centromere identity. Rather, an epigenetic mark comprised of CENP-A containing chromatin is thought to be the major determinant of centromere identity. In this view, CENP-A deposition and chromatin assembly are fundamental processes for the maintenance of centromeric identity across mitotic and meiotic divisions. Several lines of evidence support CENP-A deposition in metazoans occurring at only one time in the cell cycle. Such cell cycle-dependent loading of CENP-A is found in divergent species from human to fission yeast, albeit with differences in the cell cycle point at which CENP-A is assembled. Cell cycle dependent CENP-A deposition requires multiple assembly factors for its deposition and maintenance. This review discusses the regulation of new CENP-A deposition and its relevance to centromere identity and inheritance.
Introduction
The cell genetic material in eukaryotic cells is organized in a packed nucleoprotein complex, chromatin. Most chromatin is comprised of nucleosomes with ~147 bp of DNA wrapped around a histone octamer that contains two molecules of H3, H4, H2A and H2B. This nucleosome organization is thought to be present along the entire chromosome, with the most conspicuous exception at the centromere, a specialized chromosomal domain necessary for the correct segregation of eukaryotic chromosomes prior to cell division (Cleveland et al., 2003). This region is a complex DNA domain that in human contains extensive tandemly repeated arrays of a 171 bp DNA sequence element called α-satellite (Schueler et al., 2001). Remarkably, although centromeric DNA is the element that insures chromosome inheritance, it has no sequence conservation species to species. A common feature of centromeres, despite size and sequence divergence across species, is that in centromeric chromatin from yeast to man the canonical nucleosome histone H3 is replaced by a centromere specific variant initially identified in humans (Palmer et al., 1987) and named CENP-A (Earnshaw et al., 1986). Homologues have now been identified in many other species (e.g., CID in Drosophila melanogaster, Cnp1 in Schizosaccharomyces pombe, and Cse4 in Saccharomyces cerevisiae).
Not surprisingly, CENP-A incorporation at centromeric nucleosomes is tightly controlled. Artificially targeting of CENP-ACID to a new location in Drosophila cells correlates with ectopic centromere deposition and kinetochore assembly, including microtubule-binding proteins and spindle assembly checkpoint protein (Ndc80 and Mad2); this leads to formation of multicentric chromosomes that are consequently mis-segregated at high frequency (Mendiburo et al., 2011; Olszak et al., 2011). Together with the discovery in humans of neocentromeres (the stable acquisition of a new centromere at a new chromosomal site without any DNA sequence change) (Amor et al., 2004; Warburton, 2004) and stably bound by CENP-A (Bassett et al., 2010), the current evidence supports a model in which CENP-A chromatin rather than DNA sequence is the major determinant of mammalian centromere identity.
The question of how CENP-A can epigenetically define centromeres remains unsolved. One initial proposal is that it can induce a conformationally more constricted nucleosome structure that serves to template its own replication and nucleate recruitment of the ~50 other centromeric components. Evidence for this initially came from deuterium exchange/mass spectrometric analysis, in which exchange of the amide proton on each peptide bond with protons in solution provides a direct measure of accessibility to solvent. Buried residues that are inaccessible to solvent – and which remain inaccessible despite transient flexing driven by thermal energy - are easily detected by absence of (or very slow) exchange onto them from tritium in solution. This approach revealed that CENP-A nucleosomes are more rigid than their counterpart histone H3-containing nucleosomes – with many positions exchanging amide protons >10 fold more slowly. This rigidity/conformational inflexibility initiates within the CENP-A/histone H4 pre-nucleosomal heterotetramer (Black et al., 2007a; Black et al., 2004; Sekulic et al., 2010) and can be transmitted into histone H3 by a 22 amino acid exchange of the helix α1 and α2 of CENP-A for the corresponding domain of histone H3 (Black et al., 2007a; Black et al., 2004). This short domain has been called the CENP-A targeting domain (CATD) because when substituted into histone H3 (Black et al., 2004) it is sufficient to convert H3 into a centromere targeting histone where it can maintain centromere function when CENP-A levels are lowered (Black et al., 2007b).
A crystal structure of the CENP-A/histone H4 heterotetramer confirmed its structural compactness (Sekulic et al., 2010), with a 9–14° skewing of the CENP-A dimer (relative to the corresponding domains of histone H3 in the H3/H4 heterotetramer). Nevertheless, a subsequent crystal structure of the octameric CENP-A nucleosome did not reveal the same compactness of the CENP-A nucleosome relatively to the H3 when assembled into the full nucleosome (Tachiwana et al., 2011). The crystal structure is, of course, a static view that does not assay conformational dynamics. Indeed, despite the similarity in final structure, it should be emphasized that in solution the CENP-A-containing nucleosomes are substantially more conformationally constrained – especially in the α–helix 1 and 2, as revealed by deuterium exchange/ mass spectrometry (Black et al., 2007a). Lastly, an insight from the crystal structure is that Loop1 in the CATD domain protrudes from the CENP-A nucleosome (and heterotetramer), positioned appropriately for it to be able attract additional centromeric components (Sekulic et al., 2010; Tachiwana et al., 2011).
Beyond a structurally altered nucleosome – with the exposed Loop 1 – an alternative, but not mutually exclusive, possibility for CENP-A chromatin is that CENP-A induces changes in the higher-order chromatin structure as it modifies the entry/exit of the DNA from the nucleosome (Conde e Silva et al., 2007), potentially producing a more condensed chromatin state (Panchenko et al., 2011; Tachiwana et al., 2011).
CENP-A chromatin may also be marked by a structure that differs from the classical canonical octameric H3-nucleosomes. Recent studies offer conflicting evidence for the structure of CENP-A-containing chromatin (Black and Cleveland, 2011). The different proposals include, among others, a classical octameric CENP-A nucleosome with two copies of each histone, a tetrasome lacking H2A:H2B dimers, a hemisome with one copy of each histone, and involvement of the budding yeast Scm3 in the final structure either as a hexasome or as a trisome of CENP-ACse4, H4, and Scm3. These different structures of CENP-A chromatin may represent a process of cell-cycle-dependent maturation of centromeric chromatin that is a result of the separation between centromeric DNA replication and CENP-A deposition (discussed below). Whether this is indeed the case is yet to be established. However, such a model of chromatin maturation could provide a basis for the stable inheritance of centromere identity.
CENP-A deposition: only once at the right time
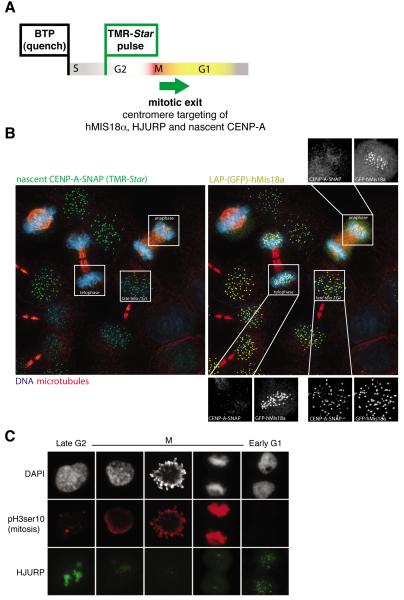
A very surprising feature of centromeric chromatin is that its replication in metazoans is uncoupled from centromeric DNA replication. Indeed, once per cell cycle assembly of CENP-A at active centromeres is a highly conserved feature of centromeres in species spanning from fission yeast to man, albeit the cell cycle position during which centromeric chromatin is replicated different in different species (Table 1). In human cells, centromeric DNA is replicated in mid-to-late S-phase (Ten Hagen et al., 1990), while synthesis of CENP-A increases in G2 (Shelby et al., 2000; Shelby et al., 1997). Pulse-chase fluorescent labeling based on SNAP-tagging has demonstrated that new CENP-A loading occurs only once per cell cycle in telophase/early G1 and requires passage through mitosis (Figure 1A, B) (Jansen et al., 2007), a result confirmed by photobleaching experiments (Hemmerich et al., 2008). Similar to the human findings, in chicken DT40 cells new CENP-A loading is prevented by blocking advance past anaphase with microtubule drugs (Silva et al., 2012) (Table 1). So, too, for frogs, as has been shown with Xenopus egg extracts in which CENP-A assembly occurs in a short time window following mitotic exit (Table 1)(Bernad et al., 2011; Moree et al., 2011).
TABLE 1.
| Organism | CENP-A cell cycle-dependent deposition | References |
|---|---|---|
| Human cells | Late Telophase-early G11 | Jansen et al., 2007 |
| Drosophila melanogaster embryos/S2 cells | Anaphase/Metaphase2 |
Schuh et al., 2007
Mellone et al., 2011 |
| Xenopus Laevis | Mitotic exit-early interphase3 |
Bernard et al., 2011
Moree et al., 2011 |
| DT4O chicken cells | Mitotic exit | Silva et al., 2012 |
| S. pombe | S-phase (and G2 phase)4 | Takayama et al., 2008 |
| S. cerevisiae | S-phase5 | Pearson et al., 2004 |
| Arabidopsis thaliana | G2 phase | Lermontova et al., 2006 |
HJURP localizes to centromere in telophase just prior CENP-A assembly, hMIS18α in anaphase (Dunleavy et al., 2009; Foltz et al., 2009; Fujita et al., 2007; Silva and jansen, 2009)
CAL1 is recruited to centromere during prophase (Mellone et al., 2011)
xHJURP localizes at centromere in mitosis, M18BP1 during metaphase (Bernard et al, 2011; Moree et al., 2011)
Scm3 and Mis18 complex localize to centromere at all cell cycle phase except from metaphase to mid-anaphase (Hayashi et al., 2004; Pidoux et al., 2009; Williams et al., 2009)
Scm3 is present at centromere at all cell cycle phase (Xiao et al., 2011)
Figure 1.
hMis18α, HJURP and CENP-A assemble at centromeres in a cell cycle-dependent manner. (A). Outline of the SNAP tagging experiments in (B) to establish temporal order of Mis18 recruitment to centromeres at mitotic exit and CENP-A loading. (B) Mis18 recruitment to centromeres precedes new CENP-A assembly at mitotic exit, as demonstrated with synchronized HeLa cells stably expressing CENP-A-SNAP. LAP-hMis18α is targeted to centromere in anaphase, prior to the late telophase/G1-CENP-A-SNAP assembly [adapted from (Silva and Jansen, 2009)]. (C) HJURP localization at centromere occurs after mitotic exit, as determined with indirect immunofluorescence for phospho-histone H3 and HJURP [adapted from (Dunleavy et al., 2009)].
Quantitative fluorescence measurements in Drosophila syncytial embryos revealed that GFP-CID is rapidly assembled at centromeres during or after anaphase (Schuh et al., 2007). More recent, higher resolution quench-chase-pulse experiments using SNAP-tagging in a pair of Drosophila cell lines (S2 and Kc167 cells) have revealed that new CENP-ACID incorporation at centromeres initiates during metaphase (Table 1) (Mellone et al., 2011). Loading may require degradation of cyclin A, since blocking the proteasome by MG132 treatment or using a non-degradable form of cyclin A reduced CENP-ACID loading prior to anaphase onset (Mellone et al., 2011).
Overall, while the precise cell cycle position of loading differs between flies and the other animal species studied, the common feature for CENP-A loading across different species of higher eukaryotes is that it occurs independent of DNA replication. Moreover, in these species, cells undergo mitosis with only half the maximal CENP-A content loaded at centromeres. All these finding suggest that passage through mitosis is a prerequisite for new CENP-A loading and assembly at the centromere.
Contrary to the animal species described above, CENP-ACENH3 loading in Arabidopsis thalianato has been reported to occur in late G2 (Table 1) (Lermontova et al., 2006), as demonstrated by measuring the relative proportions of CENP-ACENH3 immunostaining in different phases of the cell cycle. In single cell eukaryotes, the timing of assembly of the CENP-A homolog apparently correlates with the timing of DNA replication as it does for the canonical histones. Photobleaching experiments in budding yeast are consistent with Cse4-GFP assembly occurring mainly early during DNA replication (Pearson et al., 2004). A similar result was observed in fission yeast where incorporation of CENP-ACnp1 occurs predominantly during S-phase (although a 25% of new CENP-ACnp1 loading occurs during late G2) and the timing of this replication-dependent deposition requires the cell cycle regulated GATA-type transcription factor Ams2 (Takayama et al., 2008). Indeed, in Δams2 deficient cells the cell cycle-dependent loading of CENP-ACnp1 shifts almost completely to G2 (Takayama et al., 2008) (summarized in Table 1). This difference between lower and higher eukaryotes in timing of CENP-A incorporation correlates with a shift in centromere DNA replication timing in S-phase, since yeast centromeres are replicated very early during S-phase (Kim et al., 2003; Raghuraman et al., 2001) while human and Drosophila centromeres are replicated at mid-late S-phase (Sullivan albicans and Karpen, 2001; Ten Hagen et al., 1990). Indeed, in the yeast Candida albicans stable and heritable neocentromeres are associated with a shift in replication timing (from late to early in S-phase) (Koren et al., 2010).
The CENP-A's assembly factors
Similar to the canonical histone H3 that is deposited on chromatin by a series of chaperones, CENP-A loading at centromeres is mediated by factors that chaperone the loading process. Two independent affinity purification studies in human identified a pre-nucleosomal (non-chromatin associated) CENP-A complex that consists of CENP-A, HJURP, histone H4 and nucleophosmin 1 (NPM1) (Dunleavy et al., 2009; Foltz et al., 2009). In one of those efforts (Dunleavy et al., 2009), an additional component was also present - RbAp48, a previously known component of both the CAF-1 complex responsible for loading the replication dependent histone H3.1 and the HIRA complex responsible for facilitating loading of histone H3.3. The role for NPM1 in CENP-A loading is not established and unlikely to be unique for CENP-A, as it binds ATP and has been implicated as an ATP-dependent chromatin remodeler/chaperone for both H3:H4 and H2A:H2B and multiple other cellular roles (Frehlick et al., 2007; Grisendi et al., 2006).
HJURP (Holliday junction recognizing protein) was demonstrated to act as the CENP-A chaperone protein that binds directly to soluble CENP-A and is required for its chromatin assembly (Dunleavy et al., 2009; Foltz et al., 2009). Xenopus HJURP was also shown to have characteristics of a CENP-A loader, as adding exogenous xHJURP to egg extracts results in substantial assembly of new CENP-A into centromeric chromatin (Moree et al., 2011). HJURP contains a region of homology to the yeast Scm3 (Sanchez-Pulido et al., 2009) and both appear to function similarly; depletion of HJURP from human cells (Dunleavy et al., 2009; Foltz et al., 2009) or from Xenopous egg extracts (Bernad et al., 2011) causes defects in CENP-A assembly similar to the defects resulting from Scm3 mutation in yeast (Bernad et al., 2011; Dunleavy et al., 2009; Foltz et al., 2009; Pidoux et al., 2009). Furthermore, tethering HJURP to an ectopic non-centromeric locus is sufficient to induce incorporation of CENP-A into the chromatin at this site and results in creating a functional de novo kinetochore (Barnhart et al., 2011).
The targeting of CENP-A and HJURP to centromeres is dependent on another group of proteins, the Mis18 complex, initially identified in a study of chromosome mis-segregation mutants in fission yeast and which when mutated lead to the loss of CENP-ACnp1 from centromeres (Hayashi et al., 2004). The human homologue for the fission yeast mis18 is a complex comprised of three components, Mis18-α, Mis18-β and M18BP1. Depletion of any of the three subunits by RNAi rapidly abolishes the centromere recruitment of newly synthesized CENP-A, followed by defects such as misaligned chromosomes, anaphase mis-segregation and interphase micronuclei (Fujita et al., 2007). Depletion of C. elegans M18BP1KNL-2 from C. elegans embryos (Maddox et al., 2007) or xM18BP1 from Xenopus egg extracts (Moree et al., 2011) also prevents assembly of CENP-A at centromere. Furthermore, in cells depleted of the Mis18 complex, HJURP fails to localize to centromeres (Barnhart et al., 2011).
Since both the Mis18 complex and HJURP are necessary to achieve spatial and temporal control of CENP-A nucleosome assembly, a key goal ahead is to decipher the regulatory signals that control their centromeric localization. In human cells, HJURP is localized to centromeres during late anaphase/telophase (Figure 1C) and remains associated with centromeres during G1, the time window during which new CENP-A assembly occurs (Dunleavy et al., 2009; Foltz et al., 2009). The human M18BP1 complex localizes to centromeres at anaphase, just before HJURP, and remains associated until mid G1 (Figure 1A, B) (Fujita et al., 2007) (Silva and Jansen, 2009). Similar to human cells, in X. laevis xHJURP localizes at centromere in mitosis (Bernad et al., 2011) and M18BP1 during metaphase just prior to CENP-A loading (Bernad et al., 2011; Moree et al., 2011). In Drosophila, CAL1 has been hypothesized to have similar function to HJURP and M18BP1, despite the lack of sequence homology. Accordingly, similar to HJURP/M18BP1 cell cycle-dependent loading, CAL1 associates with centromeres in prophase just before CENP-ACID loading (Mellone et al., 2011).
These results led to the proposal that the Mis18 complex “primes” the centromere and prepares it for the loading of new CENP-A (Barnhart et al., 2011), perhaps through histone H4 acetylation (Barnhart et al., 2011; Fujita et al., 2007). Surprisingly, in yeast, Scm3 and the Mis18 complex seem to remain associated during all the stages of the cell cycle, excluding a short time from metaphase to mid-anaphase in fission yeast (Hayashi et al., 2004; Pidoux et al., 2009; Williams et al., 2009; Xiao et al., 2011). Despite their suggestive localization pattern and clear role in CENP-A assembly, none of the yeast or human Mis18 proteins appears to bind CENP-A directly (Fujita et al., 2007; Hayashi et al., 2004) and the process by which M18BP1 is recruited to centromeres during mitosis is unknown.
Regulation, stabilization and maintenance of newly incorporated CENP-A
Cell cycle-dependent loading of CENP-A at centromeres is now recognized to require the constitutive centromere protein CENP-C. Straight and colleagues have recently shown that CENP-C directly interacts with M18BP1 in both Xenopus and human (Moree et al., 2011). Further, since CENP-C binds CENP-A nucleosomes directly (Carroll et al., 2010) and is required for new CENP-A assembly (Carroll et al., 2010; Erhardt et al., 2008), the interaction between CENP-C and M18BP1 creates a molecular bridge between CENP-A and M18BP1. Furthermore, depletion of CENP-C resulted in disruption of centromere targeting of both M18BP1 and HJURP during late mitosis, leading to inhibition of new CENP-A assembly in G1 (Moree et al., 2011). Thus, a key role of CENP-C is the recruitment of M18BP1 to centromeres as a second means to promote CENP-A assembly.
It should be emphasized that it remains unresolved what is the mitotic trigger that initiates centromere propagation. Nevertheless, work from the Jansen group has established that the CENP-A assembly process is inhibited by Cdk activity (Silva et al., 2012). Treatment of G2-synchronized HeLa cells with the pan-Cdk inhibitors roscovitine or purvalanol resulted in premature centromeric CENP-A assembly in 50% of the G2 cell population accompanied by a rapid recruitment of Mis18-α and M18BP1 to centromeres prior to the assembly of CENP-A. Similarly, using induced gene inactivation in DT40 chicken cell lines, cells without both Cdk1 and Cdk2 induce CENP-A assembly during G2, indicating that both Cdk1 and Cdk2 function to inhibit premature CENP-A loading prior to mitotic exit. Moreover, the machinery responsible for CENP-A assembly is present already in S phase but is normally inhibited primarily by Cdk2 activity.
Lastly, premature centromere recruitment of M18BP1 is prevented by its phosphorylation (by a kinase yet to be determined), which also occurs in a cell cycle dependent manner - peaking at mitosis. Upon mitotic exit M18BP1 is dephosphorylated and only the unphosphorylated form of M18BP1 can then be rapidly targeted to centromeres to initiate the process of CENP-A replenishment.
Stabilizing newly replicated centromeric chromatin in G1
What happens after nascent CENP-A is deposited at mitotic exit and early G1? Recent studies have pointed to several factors that may have important roles in centromeric chromatin maturation in G1. Rsf-1 and SNF2h are two subunits of the ATP-dependent chromatin remodeling and spacing factor RSF. These two proteins were found along the components in CENP-A affinity interphase precipitates (Izuta et al., 2006; Obuse et al., 2004). Native chromatin immunoprecipitation (nChIP) and immunoprecipitation experiments by Yoda and colleagues showed that unlike the Mis18 proteins, Rsf-1 and SNF2h associates with CENP-A chromatin at the mononucleosome level (Perpelescu et al., 2009). Rsf-1 is localized at centromeres in mid-G1 and is retained at centromeres for a period of 4 hours. Thus, Rsf-1 centromere localization occurs several hours after the recruitment of the Mis18 complex and the deposition of new CENP-A.
Interestingly, depletion of Rsf-1 using siRNA resulted in normal CENP-A levels at centromeres, but this centromeric CENP-A became sensitive to high salt extraction and was removed easily from the chromatin core following a 0.6M NaCl salt wash. These results suggest a model in which CENP-A is initially targeted to centromeres in late M/early G1 with relatively weak association to the chromatin, after which it is incorporated and assembled into stable core chromatin through the RSF remodeling and spacing function. Future experiments are still needed to further determine whether the salt sensitive pool observed indeed represents newly targeted CENP-A.
Following incorporation into core chromatin, newly assembled CENP-A molecules are apparently further stabilized by the action of MgcRacGAP (Lagana et al., 2010). MgcRacGAP was previously known to exhibit GAP activity for three Rho family small GTPases (RhoA, Rac1 and Cdc42) (Minoshima et al., 2003) and to be a part of the centralspindlin complex that functions during cytokinesis and localizes to the spindle midzone following chromosome segregation (Kawashima et al., 2000; Minoshima et al., 2003). Added to this, MgcRacGAP had been previously shown to co-purify with CENP-A chromatin (Izuta et al., 2006; Perpelescu et al., 2009).
A role for MgcRacGAP in regulation of centromere chromatin has recently emerged. Using immunoprecipitation experiments, Maddox and colleagues have shown that MgcRacGAP co-purifies with M18BP1 (Lagana et al., 2010). Depletion of MgcRacGAP using shRNA resulted in the loss of only the new incorporated-CENP-A at centromeres, without any effect on M18BP1. Quantitative live cell measurements revealed that MgcRacGAP localizes to centromeres only at late G1 (~10h after anaphase) after CENP-A levels at centromeres doubled. Furthermore, the authors suggested that MgcRacGAP functions together with its GEF partner, ECT2, to target the small GTPase Cdc42, since depletion of either one resulted in loss of centromeric CENP-A. It still remains to be demonstrated what are the downstream targets of Cdc42 in regulating centromeric chromatin. Nevertheless, these localization patterns suggest that MgcRacGAP transiently activates a Cdc42-mediated switch at centromeres to maintain newly incorporated CENP-A.
A multi-step model for centromeric chromatin replication
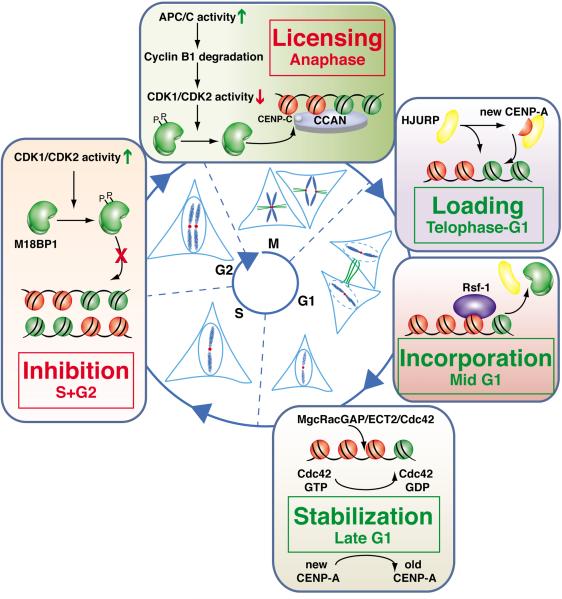
Taken all together, the flurry of recent findings has led to a model in which a multi-step pathway controls the location and timing of human CENP-A nucleosome assembly (Figure 2). During S-phase and G2, Cdk1 and Cdk2 activities are up-regulated and are essential for the initiation of DNA replication and entry into mitosis (Bell and Dutta, 2002). Cdk1/2 activities at these phases lead to phosphorylation of M18BP1 (and likely other proteins as well, yet to be identified) and prevent targeting to centromeres. Mitotic exit is triggered by degradation of cyclin B, the Cdk1 activator, leading to loss of Cdk1 activity, thereby permitting association of unphosphorylated M18BP1 to centromeres as an initiating step in the process of CENP-A replenishment. Recruitment of the Mis18 complex to the centromere requires binding of M18BP1 to CENP-C and provides the licensing step that primes each centromere for CENP-A loading. Following priming and licensing, HJURP is recruited to the centromere while providing chaperone function for soluble CENP-A subunits.
Figure 2.
Model of the multi-step pathway of CENP-A-containing centromeric chromatin replication utilizing sequential, temporally separated steps across the mammalian cell cycle. See full description in the main text.
Loading of new CENP-A subunits by HJURP takes place from late telophase to early-mid G1. Further maturation of this newly assembled centromeric chromatin occurs slowly through the transient recruitment of the chromatin remodeling and spacing factor complex RSF at mid G1, contemporaneous with removal of HJURP and the Mis18 complex. Further on at late G1, a GTPase switch at centromeres by MgcRacGAP-ECT2 is used to further stabilize newly assembled CENP-A -containing chromatin.
Several possibilities may explain the complexity of the CENP-A assembly process. First, following mitotic exit there is a need to epigenetically propagate the centromere before the next round of DNA replication. This process may take time since it involves a large number of proteins and multiple assembly and disassembly events in order to assemble and stabilize CENP-A nucleosomes in a precise manner (Prendergast and Sullivan, 2010). Second, the multi-step pathway may present opportunities for quality control, especially in the stabilization phase by centromeric-localized MgcRacGAP, with the extended time and contributions from multiple components allowing promiscuous CENP-A mis-incorporated at non-centromeric loci to be removed by mechanisms that perhaps recognize weakly associated “new” CENP-A which has not been stabilized by MgcRacGAP (Lagana et al., 2010).
Acknowledgements
This work was supported by a grant from the National Institutes of Health to D.W.C. (GM74150). D.F. has been supported by a postdoctoral fellowship from EMBO. D.W.C. receives salary support from the Ludwig Institute for Cancer Research.
References
- Amor DJ, Bentley K, Ryan J, Perry J, Wong L, Slater H, Choo KH. Human centromere repositioning “in progress”. Proc Natl Acad Sci U S A. 2004;101:6542–6547. doi: 10.1073/pnas.0308637101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart MC, Kuich PH, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol. 2011;194:229–243. doi: 10.1083/jcb.201012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett EA, Wood S, Salimian KJ, Ajith S, Foltz DR, Black BE. Epigenetic centromere specification directs aurora B accumulation but is insufficient to efficiently correct mitotic errors. J Cell Biol. 2010;190:177–185. doi: 10.1083/jcb.201001035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Bernad R, Sanchez P, Rivera T, Rodriguez-Corsino M, Boyarchuk E, Vassias I, Ray-Gallet D, Arnaoutov A, Dasso M, Almouzni G, et al. Xenopus HJURP and condensin II are required for CENP-A assembly. J Cell Biol. 2011;192:569–582. doi: 10.1083/jcb.201005136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Brock MA, Bedard S, Woods VL, Jr., Cleveland DW. An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc Natl Acad Sci U S A. 2007a;104:5008–5013. doi: 10.1073/pnas.0700390104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Jr., Cleveland DW. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- Black BE, Jansen LE, Maddox PS, Foltz DR, Desai AB, Shah JV, Cleveland DW. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol Cell. 2007b;25:309–322. doi: 10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- Carroll CW, Milks KJ, Straight AF. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J Cell Biol. 2010;189:1143–1155. doi: 10.1083/jcb.201001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- Conde e Silva N, Black BE, Sivolob A, Filipski J, Cleveland DW, Prunell A. CENP-A-containing nucleosomes: easier disassembly versus exclusive centromeric localization. J Mol Biol. 2007;370:555–573. doi: 10.1016/j.jmb.2007.04.064. [DOI] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- Earnshaw W, Bordwell B, Marino C, Rothfield N. Three human chromosomal autoantigens are recognized by sera from patients with anti-centromere antibodies. J Clin Invest. 1986;77:426–430. doi: 10.1172/JCI112320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt S, Mellone BG, Betts CM, Zhang W, Karpen GH, Straight AF. Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J Cell Biol. 2008;183:805–818. doi: 10.1083/jcb.200806038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Bailey AO, Yates JR, 3rd, Bassett EA, Wood S, Black BE, Cleveland DW. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frehlick LJ, Eirin-Lopez JM, Ausio J. New insights into the nucleophosmin/nucleoplasmin family of nuclear chaperones. Bioessays. 2007;29:49–59. doi: 10.1002/bies.20512. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev Cell. 2007;12:17–30. doi: 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Grisendi S, Mecucci C, Falini B, Pandolfi PP. Nucleophosmin and cancer. Nat Rev Cancer. 2006;6:493–505. doi: 10.1038/nrc1885. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Hemmerich P, Weidtkamp-Peters S, Hoischen C, Schmiedeberg L, Erliandri I, Diekmann S. Dynamics of inner kinetochore assembly and maintenance in living cells. J Cell Biol. 2008;180:1101–1114. doi: 10.1083/jcb.200710052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izuta H, Ikeno M, Suzuki N, Tomonaga T, Nozaki N, Obuse C, Kisu Y, Goshima N, Nomura F, Nomura N, et al. Comprehensive analysis of the ICEN (Interphase Centromere Complex) components enriched in the CENP-A chromatin of human cells. Genes Cells. 2006;11:673–684. doi: 10.1111/j.1365-2443.2006.00969.x. [DOI] [PubMed] [Google Scholar]
- Jansen LE, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima T, Hirose K, Satoh T, Kaneko A, Ikeda Y, Kaziro Y, Nosaka T, Kitamura T. MgcRacGAP is involved in the control of growth and differentiation of hematopoietic cells. Blood. 2000;96:2116–2124. [PubMed] [Google Scholar]
- Kim SM, Dubey DD, Huberman JA. Early-replicating heterochromatin. Genes Dev. 2003;17:330–335. doi: 10.1101/gad.1046203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren A, Tsai HJ, Tirosh I, Burrack LS, Barkai N, Berman J. Epigenetically-inherited centromere and neocentromere DNA replicates earliest in S-phase. PLoS Genet. 2010;6:e1001068. doi: 10.1371/journal.pgen.1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagana A, Dorn JF, De Rop V, Ladouceur AM, Maddox AS, Maddox PS. A small GTPase molecular switch regulates epigenetic centromere maintenance by stabilizing newly incorporated CENP-A. Nat Cell Biol. 2010;12:1186–1193. doi: 10.1038/ncb2129. [DOI] [PubMed] [Google Scholar]
- Lermontova I, Schubert V, Fuchs J, Klatte S, Macas J, Schubert I. Loading of Arabidopsis centromeric histone CENH3 occurs mainly during G2 and requires the presence of the histone fold domain. Plant Cell. 2006;18:2443–2451. doi: 10.1105/tpc.106.043174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox PS, Hyndman F, Monen J, Oegema K, Desai A. Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J Cell Biol. 2007;176:757–763. doi: 10.1083/jcb.200701065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellone BG, Grive KJ, Shteyn V, Bowers SR, Oderberg I, Karpen GH. Assembly of Drosophila centromeric chromatin proteins during mitosis. PLoS Genet. 2011;7:e1002068. doi: 10.1371/journal.pgen.1002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendiburo MJ, Padeken J, Fulop S, Schepers A, Heun P. Drosophila CENH3 is sufficient for centromere formation. Science. 2011;334:686–690. doi: 10.1126/science.1206880. [DOI] [PubMed] [Google Scholar]
- Minoshima Y, Kawashima T, Hirose K, Tonozuka Y, Kawajiri A, Bao YC, Deng X, Tatsuka M, Narumiya S, May WS, Jr., et al. Phosphorylation by aurora B converts MgcRacGAP to a RhoGAP during cytokinesis. Dev Cell. 2003;4:549–560. doi: 10.1016/s1534-5807(03)00089-3. [DOI] [PubMed] [Google Scholar]
- Moree B, Meyer CB, Fuller CJ, Straight AF. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J Cell Biol. 2011;194:855–871. doi: 10.1083/jcb.201106079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obuse C, Yang H, Nozaki N, Goto S, Okazaki T, Yoda K. Proteomics analysis of the centromere complex from HeLa interphase cells: UV-damaged DNA binding protein 1 (DDB-1) is a component of the CEN-complex, while BMI-1 is transiently co-localized with the centromeric region in interphase. Genes Cells. 2004;9:105–120. doi: 10.1111/j.1365-2443.2004.00705.x. [DOI] [PubMed] [Google Scholar]
- Olszak AM, van Essen D, Pereira AJ, Diehl S, Manke T, Maiato H, Saccani S, Heun P. Heterochromatin boundaries are hotspots for de novo kinetochore formation. Nat Cell Biol. 2011;13:799–808. doi: 10.1038/ncb2272. [DOI] [PubMed] [Google Scholar]
- Palmer DK, O'Day K, Wener MH, Andrews BS, Margol RL. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J Cell Biol. 1987;104:805–815. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchenko T, Sorensen TC, Woodcock CL, Kan ZY, Wood S, Resch MG, Luger K, Englander SW, Hansen JC, Black BE. Replacement of histone H3 with CENP-A directs global nucleosome array condensation and loosening of nucleosome superhelical termini. Proc Natl Acad Sci U S A. 2011;108:16588–16593. doi: 10.1073/pnas.1113621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Yeh E, Gardner M, Odde D, Salmon ED, Bloom K. Stable kinetochore-microtubule attachment constrains centromere positioning in metaphase. Curr Biol. 2004;14:1962–1967. doi: 10.1016/j.cub.2004.09.086. [DOI] [PubMed] [Google Scholar]
- Perpelescu M, Nozaki N, Obuse C, Yang H, Yoda K. Active establishment of centromeric CENP-A chromatin by RSF complex. J Cell Biol. 2009;185:397–407. doi: 10.1083/jcb.200903088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux AL, Choi ES, Abbott JK, Liu X, Kagansky A, Castillo AG, Hamilton GL, Richardson W, Rappsilber J, He X, et al. Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin. Mol Cell. 2009;33:229–311. doi: 10.1016/j.molcel.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast L, Sullivan KF. A GTPase switch maintains CENP-A at centromeric chromatin. Nat Cell Biol. 2010;12:1128–1130. doi: 10.1038/ncb1210-1128. [DOI] [PubMed] [Google Scholar]
- Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, Conway A, Lockhart DJ, Davis RW, Brewer BJ, Fangman WL. Replication dynamics of the yeast genome. Science. 2001;294:115–121. doi: 10.1126/science.294.5540.115. [DOI] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Pidoux AL, Ponting CP, Allshire RC. Common ancestry of the CENP-A chaperones Scm3 and HJURP. Cell. 2009;137:1173–1174. doi: 10.1016/j.cell.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schueler MG, Higgins AW, Rudd MK, Gustashaw K, Willard HF. Genomic and genetic definition of a functional human centromere. Science. 2001;294:109–115. doi: 10.1126/science.1065042. [DOI] [PubMed] [Google Scholar]
- Schuh M, Lehner CF, Heidmann S. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr Biol. 2007;17:237–243. doi: 10.1016/j.cub.2006.11.051. [DOI] [PubMed] [Google Scholar]
- Sekulic N, Bassett EA, Rogers DJ, Black BE. The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres. Nature. 2010;467:347–351. doi: 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby RD, Monier K, Sullivan KF. Chromatin assembly at kinetochores is uncoupled from DNA replication. J Cell Biol. 2000;151:1113–1118. doi: 10.1083/jcb.151.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby RD, Vafa O, Sullivan KF. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva MC, Bodor DL, Stellfox ME, Martins NM, Hochegger H, Foltz DR, Jansen LE. Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev Cell. 2012;22:52–63. doi: 10.1016/j.devcel.2011.10.014. [DOI] [PubMed] [Google Scholar]
- Silva MC, Jansen LE. At the right place at the right time: novel CENP-A binding proteins shed light on centromere assembly. Chromosoma. 2009;118:154–574. doi: 10.1007/s00412-009-0227-3. [DOI] [PubMed] [Google Scholar]
- Sullivan B, Karpen G. Centromere identity in Drosophila is not determined in vivo by replication timing. J Cell Biol. 2001;154:683–690. doi: 10.1083/jcb.200103001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachiwana H, Kagawa W, Shiga T, Osakabe A, Miya Y, Saito K, Hayashi-Takanaka Y, Oda T, Sato M, Park SY, et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476:232–235. doi: 10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- Takayama Y, Sato H, Saitoh S, Ogiyama Y, Masuda F, Takahashi K. Biphasic incorporation of centromeric histone CENP-A in fission yeast. Mol Biol Cell. 2008;19:682–690. doi: 10.1091/mbc.E07-05-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten Hagen KG, Gilbert DM, Willard HF, Cohen SN. Replication timing of DNA sequences associated with human centromeres and telomeres. Mol Cell Biol. 1990;10:6348–6355. doi: 10.1128/mcb.10.12.6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton PE. Chromosomal dynamics of human neocentromere formation. Chromosome Res. 2004;12:617–626. doi: 10.1023/B:CHRO.0000036585.44138.4b. [DOI] [PubMed] [Google Scholar]
- Williams JS, Hayashi T, Yanagida M, Russell P. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol Cell. 2009;33:287–298. doi: 10.1016/j.molcel.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Mizuguchi G, Wisniewski J, Huang Y, Wei D, Wu C. Nonhistone Scm3 binds to AT-rich DNA to organize atypical centromeric nucleosome of budding yeast. Mol Cell. 2011;43:369–380. doi: 10.1016/j.molcel.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]