Abstract
As the usage of fluorescence microscopy as a tool to study biological systems continues to grow, so does the need for additional tools that permit the selective detection of proteins of interest. Existing selective and well-characterized kinase inhibitors may be exploited to develop novel small molecule probes useful in imaging kinases by fluorescence microscopy.
Fluorescence imaging is a powerful tool that permits visualization of specific cell states within a population; however, existing methods for fluorescence labeling cannot be easily applied in many biological systems. Immunofluorescence methods can be limiting due to the lack of appropriate antibodies and/or because the intracellular localization of the protein of interest necessitates fixation of the cells. Expression of the protein of interest as a fusion with a fluorescent protein circumvents the need for fixation but requires prior genetic manipulation that is not compatible with analysis of clinical samples and primary cells. Fluorescent small molecule probes that bind specifically to a protein of interest may offer some advantages over these conventional methods. Unlike antibodies, small molecule probes can be cell permeable and may therefore be useful in live-cell and in vivo imaging experiments; moreover, unlike the expression of EGFP-fusion proteins, small molecule probes do not require genetic manipulation of cells.
Protein kinases are in many ways ideal targets for the development of selective fluorescent small molecule probes. This is because protein kinases are involved in most cellular processes and changes in their localization, accessibility, and abundance are associated with changes in cellular state. Protein kinases have been used as biomarkers in cancer biology because the loss of endogenous kinase regulatory mechanisms by point mutations, gene deletions, gene amplifications, and chromosomal rearrangements has been well-established as a crucial event in many cancers. In addition, drug discovery and chemical biology efforts have in recent decades produced many selective, cell-permeable small molecule ligands of specific cellular kinases. Here we describe our initial attempts to leverage existing, well-characterized kinase inhibitors to develop fluorescent small molecule probes for use as imaging tools in cancer biology.
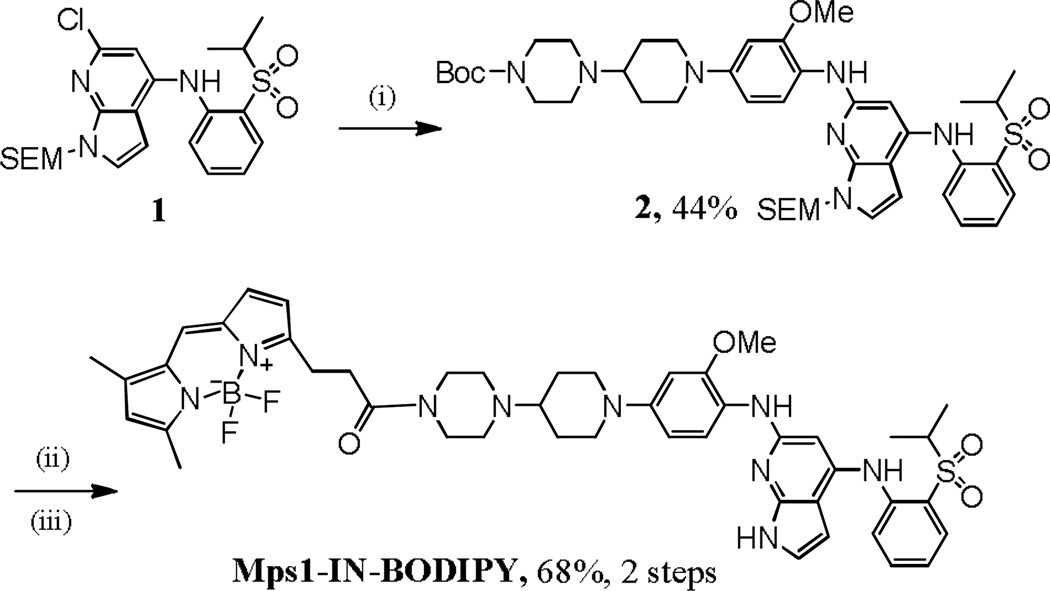
For this effort, we focused on Mps1-IN-1, a recently described inhibitor of the monopolar spindle 1 kinase (Mps1),1 and BI2536, a potent inhibitor of polo-like kinases (PLK1, PLK2, and PLK3).2, 3 These inhibitors target kinases that regulate cell cycle progression and that localize to distinct subcellular structures during mitosis. Mps1 is a dual-specificity kinase whose activity is essential for the establishment and sustained activity of the spindle checkpoint.4–6 The polo-like kinases (PLKs) comprise a family of conserved serine/threonine kinases that are highly conserved from yeast to humans7 and that are known to regulate cell cycle progression.2, 3, 8, 9 We reasoned that the synthesis and use of Mps1- and PLK-selective small molecules probes could facilitate the study of these kinases in different phases of the cell cycle. In addition, such probes might be able to elucidate deregulated states and find use as cancer diagnostics. Towards this end, we endeavoured to convert Mps1-IN-1 and BI2536 to fluorescent probes of their respective kinase targets by conjugation to a cell-permeable fluorophore. 4,4-Difluoro-4-bora-3a,4a-diaza-s-indacene (BODIPY)10 was selected as the fluorescent dye because it has a high quantum yield, it is cell permeable, and its derivatives have been widely used to label proteins11–16 and DNA.17, 18 Synthesis of BODIPY-conjugated derivatives of Mps1-IN-1 and BI2536 (Mps1-IN-BODIPY, and BI-BODIPY, respectively) were adapted from published syntheses of the parent compounds1, 19 and additional steps to conjugate BODIPY are highlighted in Schemes 1 and 2, respectively.
Scheme 1.
Synthesis of Mps1-IN-BODIPY. Reaction and conditions: (i) t-butyl 4-(1-(4-amino-3-methoxyphenyl)piperidin-4-yl)piperazine-1-carboxylate K2CO3, Pd2(dba)3, X-Phos, t-BuOH, 100 °C; (ii) TFA, DCM, 0 °C; LiOH•H2O, 25 °C; (iii) 3-(2-carboxyethyl)-5,5-difluoro-7,9-dimethyl-5H-dipyrrolo[1,2-c:2',1'-f][1,3,2]diazaborinin-4-ium-5-uide (BODIPY acid), DCC, DMAP, THF, 0–25 °C.
Scheme 2.
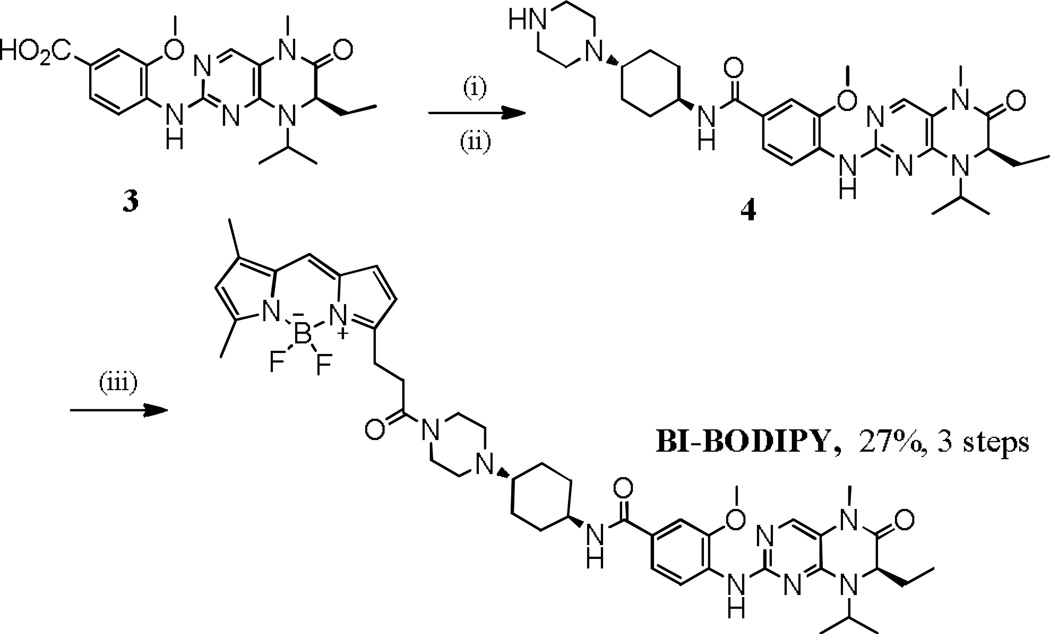
Synthesis of BI-BODIPY. Reaction and conditions: (i) t-butyl 4-((1R,4R)-4-aminocyclohexyl)piperazine-1-carboxylate, HBTU, DIEA, DMF, 25 °C; (ii) TFA, DCM, 25 °C; (iii) 3-(2-carboxyethyl)-5,5-difluoro-7,9-dimethyl-5H-dipyrrolo[1,2-c:2',1'-f][1,3,2]diazaborinin-4-ium-5-uide (BODIPY-acid), DCC, DMAP, THF, 0–25 °C.
For Mps1-IN-BODIPY, compound 1, which was prepared by following the reported procedure1, was reacted with tert-butyl 4-(1-(4-amino-3-methoxyphenyl)piperidin-4-yl)piperazine-1-carboxylate to give 2. Deprotection of 2 followed by amide coupling with the BODIPY acid produced the target Mps1-IN-BODIPY. For BI-BODIPY, compound 3, prepared according to previously reported procedures,19 was coupled with t-butyl 4-((1R,4R)-4-aminocyclohexyl)piperazine-1-carboxylate to afford 4.19 Acid mediated removal of the t-butoxycarbonyl group of 4 followed by amide coupling with BODIPY acid produced the target, BI-BODIPY.
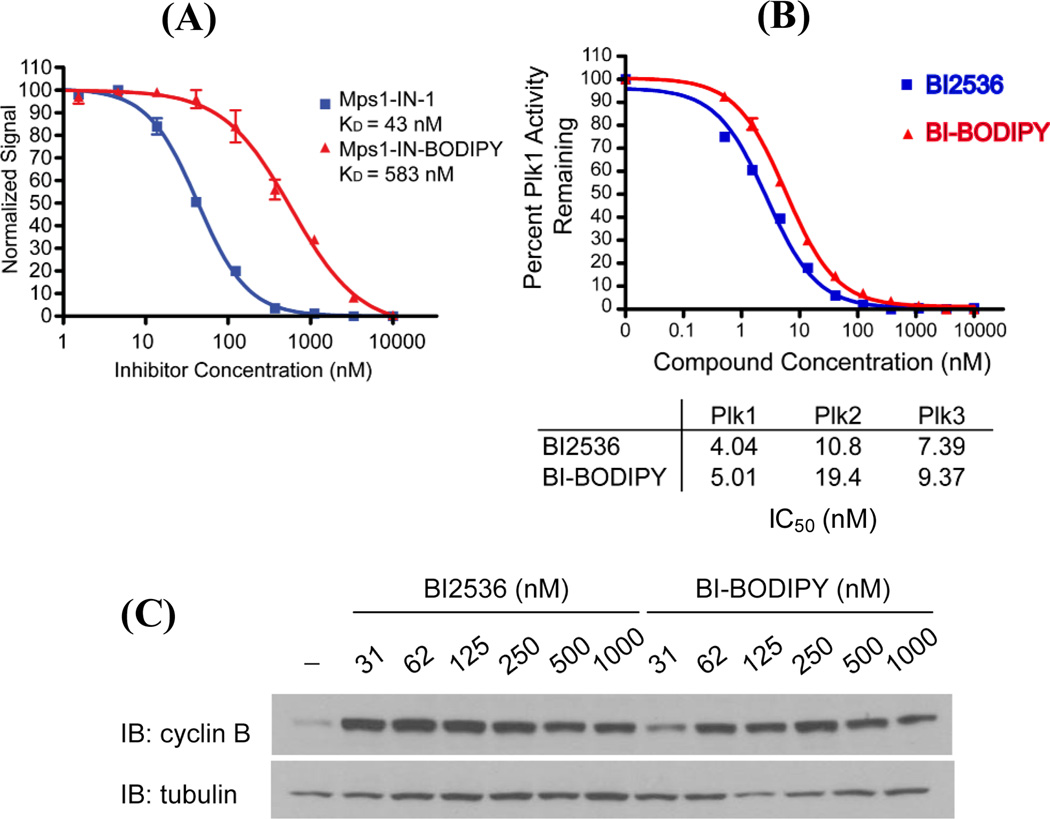
With Mps1-IN-BODIPY and BI-BODIPY in hand, we next performed dose-response experiments using a combination of biochemical and cellular assays to assess whether conjugation to BODIPY negatively affected the binding of the compounds to their kinase targets. Conjugation to BODIPY appeared to significantly reduce the activity of Mps-1-IN, as evidenced by a greater than ten-fold decrease in its binding affinity for recombinant Mps1 (Fig. 1A). Based on this result, we decided not to continue development of this compound as a fluorescent probe of Mps1. In contrast, BI-BODIPY inhibited PLK1, PLK2, and PLK3 in biochemical kinase assays (Fig. 1B) with activities comparable to that of BI25363, demonstrating that linker-modification and conjugation to BODIPY did not significantly alter the biochemical activity of BI2536. To determine if BI-BODIPY elicits a cellular phenotype similar to that of the parent compound, we analysed its effects on cell cycle progression. PLK1 is a critical regulator of cell cycle progression, and both pharmacological inhibition of PLK1 and RNAi-mediated depletion of PLK1 lead to mitotic arrest.2, 3, 8, 9 Cells were treated with BI-BODIPY or BI2536 for 24 hours followed by analysis of the cellular lysates for cyclin B, a protein whose expression peaks during mitosis and which is commonly used as a mitotic marker. Cells treated with BI-BODIPY exhibited a strong mitotic arrest, as exemplified by high and sustained cyclin B levels (compared to DMSO control) (Fig. 1C).2, 3, 8, 9 Importantly, BI-BODIPY was highly potent with only a marginal decrease in activity relative to that observed with BI2536, demonstrating that the BODIPY fluorophore did not adversely affect the cellular activity of the parent compound.
Fig. 1.
Evaluation of biochemical and cellular activities of Mps1-IN-BODIPY and BI-BODIPY against their respective kinase targets. (A) Conjugation to BODIPY reduces the affinity of Mps1-IN for Mps1. Inhibition of Mps1 by Mps1-IN-1 and Mps1-IN-BODIPY was measured by LanthaScreen™ Kinase assay (LifeTechnologies). (B) BI2536 and BI-BODIPY exhibit similar inhibitory activities against recombinant PLK1, PLK2, and PLK3 kinases. Inhibition of PLK1, PLK2, and PLK3 by BI2536 and BI-BODIPY were measured by Z´-Lyte™ kinase assay (Life Technologies). (C) Western blot for cyclin B indicates that BI-BODIPY induces cell cycle arrest at concentrations comparable to the parent inhibitor, BI2536.
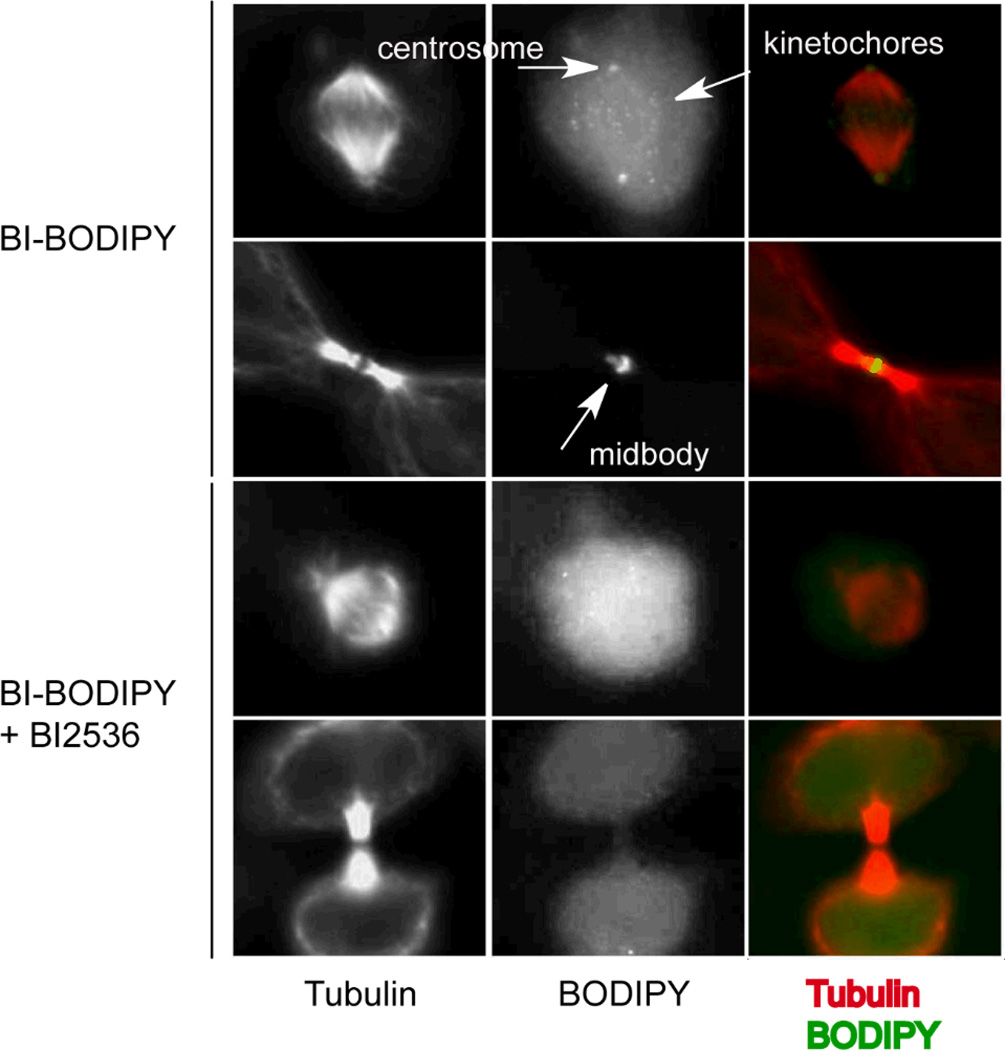
We next evaluated the use of BI-BODIPY as a probe to monitor the localization of PLKs by fluorescence microscopy. Localization of PLKs is controlled by one or more C-terminal polo-box domains (PBDs) that bind a variety of phosphoprotein epitopes, many of which are regulated in a cell cycle-dependent manner.20, 21 The transient interaction of PLKs with different binding partners facilitates the directed localization of PLKs to discrete cellular compartments and enables PLKs to phosphorylate an array of substrates in a cell-cycle dependent manner.22, 23 In particular, PLK1 and other isoforms localize to centrosomes during G2/M, where they regulate that structure’s maturation and maintenance;24–26 to kinetochores during prometaphase/metaphase, where they regulate chromosome segregation and bipolar spindle assembly;24, 27 and to the mid-body during telophase.28–30 We tested whether BI-BODIPY labels these structures during the appropriate cell cycle phases using immunofluorescence staining of tubulin to visualize these subcellular structures. HeLa cells that had been synchronized by 24-hour treatment with thymidine were released from G1/S arrest by washout of thymidine, and then treated 8 hours later with a sub-inhibitory concentration of BI-BODIPY. Cells were then fixed to permit immunofluorescence staining for tubulin. Using tubulin as a structural marker, we were able to localize the BODIPY signal to kinetochores, centrosomes, and the mid-body (Fig. 2, upper panels), consistent with the known localization of PLK1 during mitosis.24–30 Importantly, this staining pattern was disrupted by co-treatment with BI2536, which can compete with BI-BODIPY for binding to PLK1 (Fig. 2, lower panels). Collectively, these data demonstrate that BI-BODIPY can be used to track the localization of PLKs and illustrate proof of concept that existing, well-characterized, selective inhibitors of kinases may be useful in the development of tools for tracking the localization and distribution of specific kinases by fluorescence microscopy.
Fig. 2.
BI-BODIPY localizes to known sites of PLK localization. Hela S3 cells were arrested at the G1/S transition by 24-hour treatment with thymidine. Cells were released by thymidine washout and after 8 hours cells were treated with BI-BODIPY (100 nM) with and without competing unlabeled BI-2536 parent compound (1 µM) for 2 hours. Samples were fixed with 4% paraformaldehyde in PBS at room temperature for 20 minutes, washed with PBS, and then permeabilized with 0.1% Triton-X in PBS. Immunofluorescence staining for tubulin was performed to permit visualization of tubulin and centrosome, midbody, and kinetochore structures. Localization of BI-BODIPY to these structures was prevented in the presence of excess BI2536 as competitor.
Conclusions
The existing collection of highly characterized small molecule kinase inhibitors have the potential to be leveraged in the development of fluorescent, cell permeable, small molecule probes for use in fluorescence imaging. To test the feasibility of this concept, we synthesized fluorophore-conjugated derivatives of the PLK inhibitor BI2536 and the Mps1 inhibitor Mps1-IN-1. Conjugation of Mps1-IN-1 to BODIPY led to a significant perturbation in the interaction of this inhibitor with its kinase target; studies to understand why this modification had a large effect on the inhibitory activity of the compound are on-going. In contrast, BI-BODIPY exhibited biochemical and cellular characteristics that are similar to parent PLK inhibitor BI2536. We demonstrated the utility of BI-BODIPY as a cell permeable probe for monitoring PLK localization. This result serves as the foundation for more sophisticated live-cell and in vivo imaging experiments that we are currently pursuing. This study also provides proof of concept for extension of this strategy to convert other small molecule kinase inhibitors to probes that can analogously be used to monitor localization of their respective kinases.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health / National Cancer Institute grant U54 CA156732 to the University of Massachusetts Boston – Dana-Farber Harvard Cancer Center (UMB-DFHCC) U54 Comprehensive Partnership to Reduce Cancer Health Disparities (Project 3, Co-PIs: N. S. Gray, W. Zhang, and P. L. Yang) and by NIH/NIAID AI076442 (PI: P.L. Yang).
Abbreviations used in the text
- AcOH
acetic acid
- BODIPY
4,4-difluoro-4-bora-3a,4a-diaza-s-indacene
- DCC
N,N'-dicyclohexylcarbodiimide
- DCM
dichloromethane
- DIEA
N,N-diisopropylethylamine
- DMA
dimethylaniline
- DMAP
4-dimethylaminopyridine
- DMF
dimethylformamide
- EGFP
enhanced green fluorescent protein
- EtOH
ethanol
- HBTU
O-(Benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphatemethanol
- MeOH
methanol
- MeI
methyl iodide
- Mps1
monopolar spindle 1 kinase
- PLK
polo-like kinase
- NaOAc
sodium acetate
- Na(OAc)3BH
sodiumtricetoxyborohydride
- NaH
sodium hydride
- TFA
trifluoroacetic acid
- THF
tetrahydrofuran
- t-BuOH
tert-butanol
Footnotes
Published as part of a themed issue dedicated to Emerging Investigators.
Electronic Supplementary Information (ESI) available: Additional experimental details, including analytical data, are provided in a Materials and Methods section. See DOI: 10.1039/b000000x/
Notes and references
- 1.Kwiatkowski N, Jelluma N, Filippakopoulos P, Soundararajan M, Manak MS, Kwon M, Choi HG, Sim T, Deveraux QL, Rottmann S, Pellman D, Shah JV, Kops GJ, Knapp S, Gray NS. Nat Chem Biol. 2010;6:359–368. doi: 10.1038/nchembio.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lenart P, Petronczki M, Steegmaier M, Di Fiore B, Lipp JJ, Hoffmann M, Rettig WJ, Kraut N, Peters JM. Curr Biol. 2007;17:304–315. doi: 10.1016/j.cub.2006.12.046. [DOI] [PubMed] [Google Scholar]
- 3.Steegmaier M, Hoffmann M, Baum A, Lenart P, Petronczki M, Krssak M, Gurtler U, Garin-Chesa P, Lieb S, Quant J, Grauert M, Adolf GR, Kraut N, Peters JM, Rettig WJ. Curr Biol. 2007;17:316–322. doi: 10.1016/j.cub.2006.12.037. [DOI] [PubMed] [Google Scholar]
- 4.Abrieu A, Magnaghi-Jaulin L, Kahana JA, Peter M, Castro A, Vigneron S, Lorca T, Cleveland DW, Labbe JC. Cell. 2001;106:83–93. doi: 10.1016/s0092-8674(01)00410-x. [DOI] [PubMed] [Google Scholar]
- 5.Liu ST, Chan GK, Hittle JC, Fujii G, Lees E, Yen TJ. Mol Biol Cell. 2003;14:1638–1651. doi: 10.1091/mbc.02-05-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vigneron S, Prieto S, Bernis C, Labbe JC, Castro A, Lorca T. Mol Biol Cell. 2004;15:4584–4596. doi: 10.1091/mbc.E04-01-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Carcer G, Manning G, Malumbres M. Cell Cycle. 2011;10:2255–2262. doi: 10.4161/cc.10.14.16494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hansen DV, Loktev AV, Ban KH, Jackson PK. Mol Biol Cell. 2004;15:5623–5634. doi: 10.1091/mbc.E04-07-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moshe Y, Boulaire J, Pagano M, Hershko A. Proc Natl Acad Sci U S A. 2004;101:7937–7942. doi: 10.1073/pnas.0402442101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bergstrom F, Mikhalyov I, Hagglof P, Wortmann R, Ny T, Johansson LB. J Am Chem Soc. 2002;124:196–204. doi: 10.1021/ja010983f. [DOI] [PubMed] [Google Scholar]
- 11.Strandberg L, Karolin J, Johansson LB, Fa M, Aleshkov S, Ny T. Thromb Res. 1994;76:253–267. doi: 10.1016/0049-3848(94)90197-x. [DOI] [PubMed] [Google Scholar]
- 12.Haugland RP. Handbook of Fluorescent Probes and Research Chemicals, 6th ed. edn., Molecular Probes. 1996. [Google Scholar]
- 13.Reis RC, Sorgine MH, Coelho-Sampaio T. Eur J Cell Biol. 1998;75:192–197. doi: 10.1016/S0171-9335(98)80061-7. [DOI] [PubMed] [Google Scholar]
- 14.Chen CS, Chen WN, Zhou M, Arttamangkul S, Haugland RP. J Biochem Biophys Methods. 2000;42:137–151. doi: 10.1016/s0165-022x(00)00048-8. [DOI] [PubMed] [Google Scholar]
- 15.Yee MC, Fas SC, Stohlmeyer MM, Wandless TJ, Cimprich KA. J Biol Chem. 2005;280:29053–29059. doi: 10.1074/jbc.M504730200. [DOI] [PubMed] [Google Scholar]
- 16.Loudet A, Burgess K. Chem Rev. 2007;107:4891–4932. doi: 10.1021/cr078381n. [DOI] [PubMed] [Google Scholar]
- 17.Li X, Traganos F, Melamed MR, Darzynkiewicz Z. Cytometry. 1995;20:172–180. doi: 10.1002/cyto.990200210. [DOI] [PubMed] [Google Scholar]
- 18.Li X, Melamed MR, Darzynkiewicz Z. Exp Cell Res. 1996;222:28–37. doi: 10.1006/excr.1996.0004. [DOI] [PubMed] [Google Scholar]
- 19.EP1599478 and WO2003EP01935. Germany Pat. 2004
- 20.Lowery DM, Lim D, Yaffe MB. Oncogene. 2005;24:248–259. doi: 10.1038/sj.onc.1208280. [DOI] [PubMed] [Google Scholar]
- 21.Barr FA, Sillje HH, Nigg EA. Nat Rev Mol Cell Biol. 2004;5:429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- 22.Elia AE, Rellos P, Haire LF, Chao JW, Ivins FJ, Hoepker K, Mohammad D, Cantley LC, Smerdon SJ, Yaffe MB. Cell. 2003;115:83–95. doi: 10.1016/s0092-8674(03)00725-6. [DOI] [PubMed] [Google Scholar]
- 23.Elia AE, Cantley LC, Yaffe MB. Science. 2003;299:1228–1231. doi: 10.1126/science.1079079. [DOI] [PubMed] [Google Scholar]
- 24.Nigg EA, Blangy A, Lane HA. Exp Cell Res. 1996;229:174–180. doi: 10.1006/excr.1996.0356. [DOI] [PubMed] [Google Scholar]
- 25.Oshimori N, Ohsugi M, Yamamoto T. Nat Cell Biol. 2006;8:1095–1101. doi: 10.1038/ncb1474. [DOI] [PubMed] [Google Scholar]
- 26.Fry AM, Baxter JE. Dev Cell. 2006;11:431–432. doi: 10.1016/j.devcel.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 27.Arnaud L, Pines J, Nigg EA. Chromosoma. 1998;107:424–429. doi: 10.1007/s004120050326. [DOI] [PubMed] [Google Scholar]
- 28.Seong YS, Kamijo K, Lee JS, Fernandez E, Kuriyama R, Miki T, Lee KS. J Biol Chem. 2002;277:32282–32293. doi: 10.1074/jbc.M202602200. [DOI] [PubMed] [Google Scholar]
- 29.Neef R, Preisinger C, Sutcliffe J, Kopajtich R, Nigg EA, Mayer TU, Barr FA. J Cell Biol. 2003;162:863–875. doi: 10.1083/jcb.200306009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou T, Aumais JP, Liu X, Yu-Lee LY, Erikson RL. Dev Cell. 2003;5:127–138. doi: 10.1016/s1534-5807(03)00186-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.