Abstract
Purpose
To identify single nucleotide polymorphisms (SNPs) associated with development of erectile dysfunction (ED) among prostate cancer patients treated with radiotherapy.
Methods and Materials
A two-stage genome-wide association study (GWAS) was performed. Patients were split randomly into a stage I discovery cohort (132 cases, 103 controls) and a stage II replication cohort (128 cases, 102 controls). The discovery cohort was genotyped using Affymetrix 6.0 genome-wide arrays. The 940 top ranking SNPs selected from the discovery cohort were genotyped in the replication cohort using Illumina iSelect custom SNP arrays.
Results
12 SNPs identified in the discovery cohort and validated in the replication cohort were associated with development of ED following radiotherapy (Fisher combined p-values 2.1×10−5 to 6.2×10−4). Notably, these 12 SNPs lie in or near genes involved in erectile function or other normal cellular functions (adhesion and signaling) rather than DNA damage repair. In a multivariable model including non-genetic risk factors, the odds ratios for these SNPs ranged from 1.6 to 5.6 in the pooled cohort. There was a striking relationship between the cumulative number of SNP risk alleles an individual possessed and ED status (Sommers’ D p-value = 1.7×10−29). A one-allele increase in cumulative SNP score increased the odds for developing ED by a factor of 2.2 (p-value = 2.1×10−19). The cumulative SNP score model had a sensitivity of 84% and specificity of 75% for prediction of developing ED at the radiotherapy planning stage.
Conclusions
This GWAS identified a set of SNPs that are associated with development of ED following radiotherapy. These candidate genetic predictors warrant more definitive validation in an independent cohort.
Keywords: prostate cancer, genetic predictors, late effects, brachytherapy
Introduction
Prostatectomy, brachytherapy, and external beam radiation therapy (EBRT) as treatments of prostate adenocarcinoma offer excellent rates of long-term disease-free survival1. Thus, patients and clinicians consider risk of short and long term side effects for choice of treatment. These toxicities include genitourinary, gastrointestinal, and erectile dysfunction (ED).
Radiation therapy results in favorable erectile function preservation. Brachytherapy with or without EBRT has a 57.9% overall potency preservation rate at 10 years and up to 80% in selected men2. Similar results have been reported for brachytherapy3–4. Treatment-related factors that increase the risk for ED include age, pretreatment potency, use of androgen deprivation therapy (ADT), radiation type, dose, and comorbidities2,5–6. Even when controlling for these factors, significant variation exists for developing ED suggesting an intrinsic radiation sensitivity genetic risk. This hypothesis is supported by SNP association studies performed by ourselves and others7–8.
To broaden the search for genetic factors predictive for ED development in men treated with brachytherapy and/or EBRT, we performed a two-stage GWAS. This analysis identified 12 new candidate SNPs.
Methods and Materials
Patient Characteristics
Men treated with definitive radiation for biopsy proven adenocarcinoma of the prostate were recruited from XXX and XXX. This study was approved by the Institutional Review Board of XXX. All patients provided informed consent. Radiation consisted of brachytherapy alone with a full 125Iodine implant prescribed to 160Gy (TG43), a partial 103Palladium implant prescribed to 100Gy followed in 6–8 weeks by external beam irradiation (3D conformal or image guided intensity modulated radiation therapy, IMRT) to 24 to 50Gy (median 45Gy), or EBRT alone to 66.6–81Gy (IMRT). Approximately 95% of implants were performed by or under the direct supervision of a single physician (RS). The remaining 5% were performed by providers at FROG using the same treatment algorithm (1.8 Gy fractions to 45 Gy using image guided IMRT). Delivered radiation doses were converted to the biologically effective dose (BED) as described previously9.
Patients were followed prospectively every 3–6 months with the patient-administered Sexual Health Inventory for Men (SHIM) questionnaire10. Patients treated prior to introduction of the SHIM questionnaire (42%), were assessed using the Mount Sinai Erectile Function (MSEF) score (0=no erections; 1=erections but insufficient for intercourse; 2=erections suboptimal but sufficient for intercourse; 3=optimal erections), which correlates with SHIM 11. Included patients (N=593) were potent before treatment (SHIM ≥ 16 or MSEF ≥ 2), and have ≥1 year follow up. ADT was administered for 3–6 months in low risk patients with prostate volumes >50cc, in intermediate risk patients for 6 months with brachytherapy monotherapy or for 9–24 months in high risk patients with combination brachytherapy and EBRT. Patients with testosterone levels <50ng/dl at one-year post treatment were excluded (n=5). ED was assessed using SHIM scores reported between 1 and 5 years post-treatment. Cases were defined by at least one post-treatment SHIM ≤ 7 (“severe ED”), and controls were defined by all post-treatment SHIM ≥ 16 (“no” or “mild” ED) with or without use of a phosphodiesterase type 5 inhibitor.
260 patients met the ED case definition and 205 met the ED control definition, and were included in the GWAS. Cases were significantly older than controls at time of treatment (65.7 years vs. 60.3 years, p<0.001), were significantly more likely to have received ADT (60.8% vs. 31.5%, p<0.001), had significantly larger prostate volume at time of treatment (48.1mm3 vs. 44.4 mm3; p=0.048), and were significantly more likely to have been treated with combination implant + EBRT (53.5% vs. 35.1%, p<0.001) (Table 1). Gleason score was higher in cases than controls, but because Gleason score correlates with treatment modality, it was omitted from analyses.
Table 1.
Patient characteristics. Bold values are significantly different between cases and controls (p-value <0.05). Ethnicity, smoking status, diabetes, and hypertension are patient-reported.
| Cases N = 260 |
Controls N = 205 |
Intermediate N = 128 |
Total N = 593 |
|
|---|---|---|---|---|
| Age (yrs), mean(sd) | 66.5 (6.7) | 60.5 (6.9) | 62.6 (6.1) | 63.7 (7.1) |
| Follow-up (months), mean(sd) | 45.1 (14.8) | 45.1 (14.4) | 47.3 (13.0) | 45.6 (14.3) |
| Hormones, N(%) | 158 (60.8%) | 72 (35.1%) | 61 (47.7%) | 306 (49.0%) |
| Treatment, N(%) | ||||
| Implant Only | 114 (43.8%) | 128 (62.4%) | 79 (61.7%) | 346 (55.4%) |
| Implant + EBRT | 139 (53.5%) | 72 (35.1%) | 48 (37.5%) | 266 (42.6%) |
| EBRT Only | 7 (2.7%) | 5 (2.4%) | 1 (0.8%) | 13 (2.1%) |
| Total BED (Gy2), mean(sd) Gleason, N(%) | 205.5 (21.1) | 202.8 (22.8) | 203.1 (21.4) | 202.5 (24.5) |
| ≤= 6 | 141 (54.2%) | 146 (71.2%) | 90 (70.3%) | 398 (63.7%) |
| 7 | 76 (29.2%) | 47 (22.9%) | 31 (24.2%) | 165 (26.4%) |
| ≥ 8 | 43 (16.5%) | 12 (5.9%) | 7 (5.5%) | 62 (9.9%) |
| Prostate Volume at time of diagnosis (mm3), mean(sd) Self-reported ethnicity, N(%) | 48.1 (21.3) | 44.4 (14.5) | 47.1 (17.5) | 46.6 (18.3) |
| European American | 197 (76.4%) | 161 (80.5%) | 99 (77.3%) | 482 (78.9%) |
| African American | 27 (10.5%) | 25 (12.5%) | 16 (12.5%) | 74 (12.1%) |
| Hispanic | 25 (9.7%) | 10 (5.0%) | 10 (7.8%) | 46 (7.5%) |
| Asian | 6 (2.3%) | 2 (1.0%) | 1 (0.8%) | 9 (1.5%) |
| Smoker, N(%) | 113 (43.5%) | 78 (38.0%) | 55 (43.0%) | 255 (40.8%) |
| Diabetes, N(%) | 15 (5.8%) | 6 (2.9%) | 12 (9.4%) | 34 (5.4%) |
| Hypertension, N(%) | 89 (34.2%) | 58 (28.3%) | 39 (30.5%) | 190 (30.4%) |
A small group of prostate cancer patients (n=55) was recruited from the Maastrict Radiation Oncology clinic and utilized as a test cohort. Patients were treated with EBRT alone prescribed to 68–72Gy in 2Gy fractions or with brachytherapy alone prescribed to 145Gy. A unique questionnaire for measuring ED was utilized in this group. The 4 elements of this questionnaire which correlated directly to the SHIM were utilized to define cases and controls based on post-treatment score after excluding patients who had a low pre-treatment score.
Genotyping
Genomic DNA was isolated from lymphocytes. Discovery cohort DNA samples were genotyped for ~909,000 SNPs using Affymetrix v6.0 arrays (Affymetrix, Santa Clara, CA), and genotypes were called using Genotyping Console. Replication cohort DNA was genotyped using Illumina iSelect custom SNP arrays (Illumina, San Diego, CA), and genotypes were called by GenomeStudio. SNPs were excluded from analysis if missing in >5% of samples (137,589 SNPs in the discovery dataset; 22 SNPs in the replication dataset) or if they had minor allele frequency <5% (157,580 SNPs in the discovery dataset). Individuals were excluded if they showed cryptic relatedness as assessed by pairwise identity-by-state (8 pairs) or if they had call rate <90% (N=2). Duplicate control samples included in both rounds of genotyping showed >95% concordance. Three control samples (a trio of two parents and an offspring) were included on both the Affymetrix and Illumina genotyping platforms and showed >99% concordance across platforms. PLINK was used to perform QC and association tests12.
Principal components analysis (PCA) was performed using 11 reference populations from the International HapMap Project13. PCA was performed using a random 100,000 SNPs in the discovery study and 860 ancestry-informative SNPs in the replication study. SNP data processing and PCA was carried out using R 2.13.114.
Statistical Analysis
Association between non-genetic variables and ED was assessed using chi-square tests or analysis of variance. SNP association tests were carried out using multivariable logistic regression. Combined p-values for the discovery and replication cohorts were calculated using Fisher’s method after filtering for agreement in odds ratio direction and inheritance model (out of additive, genotypic, dominant and recessive). Top SNPs were examined for their utility in predicting ED following radiotherapy using multivariate logistic regression models inclusive of age, ADT, treatment, and PCs. The area under the ROC curve was used as a measure of the discriminatory power. IBM SPSS Statistics 19 was used for univariate and multivariate analysis.
Results
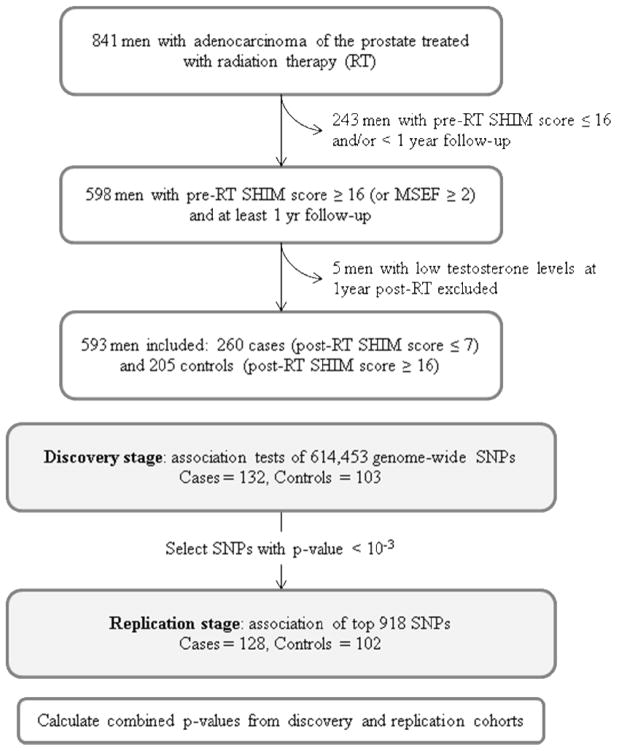
A two-stage GWAS was performed to identify SNPs associated with ED (Figure 1). After QC, the discovery cohort included 132 cases and 103 controls genotyped for 614,453 SNPs with a call rate of ~99%. In agreement with self-reported race/ethnicity (Table 1), PCA confirmed that the majority of the patients were of European ancestry, with approximately 25% of individuals showing African, Hispanic and/or Asian ancestry. Inclusion of the first five PCs in association tests adequately controlled for ancestry as evidenced by a low genomic inflation factor (1.02 after correction compared to 1.11 before correction). Multivariable logistic regression was performed to test the association for each of the 614,453 SNPs with ED status while controlling for PCs and non-genetic factors associated with ED on univariate analysis (age, ADT, and treatment; Table 1). 940 SNPs from ~550 genomic regions had p-values from 8×10−4 to 2×10−6 and were genotyped in the replication cohort.
Figure 1.
Study design.
The replication cohort included 128 cases and 102 controls genotyped successfully (call rate >99%) for 918 SNPs. After adjusting for age, ADT, treatment, and PCs, 21 SNPs were associated with ED in the replication cohort (p-value < 0.10 and agreement in inheritance model and odds ratio direction). These 21 SNPs had Fisher-combined p-values between 9×10−4 and 2×10−5 (Table 2). Four additional SNPs had combined p-value between 1×10−4 and 2×10−5 but replication p-values >0.1. These SNPs are clustered within one genomic region suggesting a true association, and were included in downstream analysis. We ran a re-sampling procedure to reduce the list to the most robustly associated SNPs. We randomly split the pooled cohorts into two equal groups and calculated separate p-values and Fisher combined p-values. After 1,000 iterations, we identified 12 of the 25 SNPs with median Fisher p-value <1×10−3 (Table 2). In the case where multiple SNPs were in linkage disequilibrium, we selected one from the cluster.
Table 2.
SNPs associated with ED following radiation therapy for treatment of prostate cancer. Results are adjusted for age, ADT, treatment, and PCs. Genotypes are reported with the risk allele listed first. Alleles are shown as risk/non-risk.
| Discovery Cohort | Replication Cohort | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||
| dbSNPrsID | Location | Alleles | Nearest Gene(s) | Genotype Cases | Genotype Controls | OR (95%CI) | p-value | Genotype Cases | Genotype Controls | OR (95%CI) | p-value | Combined p-value | Inheritance Model |
| rs322895* | 1q32.1 | C/T | NR5A2/PTPRC | 45/70/14 | 28/38/33 | 4.7 (2.1,10.6) | 2.2×10−4 | 49/64/15 | 35/47/20 | 2.2 (1.0,5.2) | 6.0×10−2 | 1.6×10−4 | Recessive |
| rs1779969 | 1q44 | G/A | AHCTF1 | 25/82/25 | 19/47/37 | 3.4 (1.6,6.9) | 8.8×10−4 | 34/70/24 | 27/45/30 | 1.9 (0.9,3.8) | 8.6×10−2 | 7.9×10−4 | Recessive |
| rs13389878 | 2p25.3 | G/A | MYT1L | 116/14/2 | 72/30/1 | 4.3 (1.9,9.4) | 3.7×10−4 | 104/22/2 | 72/29/1 | 2.2 (1.1,4.5) | 2.8×10−2 | 1.3×10−4 | Dominant |
| rs11693002* | 2p25.3 | C/G | MYT1L | 114/15/2 | 70/32/1 | 4.2 (1.9,9.2) | 2.8×10−4 | 101/25/2 | 67/34/1 | 2.5 (1.3,5.1) | 7.9×10−3 | 3.1×10−5 | Dominant |
| rs1563740 | 2p13.2 | A/T | DYSF/CYP26B1 | 2/37/91 | 1/15/86 | 3.8 (1.7,8.5) | 9.7×10−4 | 1/28/99 | 2/18/82 | 2.0 (0.9,4.4) | 9.6×10−2 | 9.6×10−4 | Dominant |
| rs3749191* | 3p21.31 | G/A | CDCP1 | 75/49/7 | 14/59/30 | 3.8 (2.0,7.2) | 4.3×10−5 | 70/46/12 | 46/46/10 | 2.0 (1.1,3.7) | 3.3×10−2 | 2.1×10−5 | Dominant |
| rs11747037* | 5q22.3 | A/G | KCNN2/YTHDC2 | 14/42/76 | 37/48/18 | 2.1 (1.4,3.3) | 6.9×10−4 | 57/58/13 | 41/45/16 | 1.5 (0.9,2.4) | 8.3×10−2 | 6.2×10−4 | Additive |
| rs1896974 | 6p12.2 | C/T | PKHD1 | 45/59/27 | 12/70/20 | 4.1 (1.4,12.6) | 3.1×10−4 | 39/67/22 | 23/51/28 | 2.9 (1.1,5.4) | 4.5×10−2 | 1.7×10−4 | Genotypic^ |
| rs9382062 | 6p12.2 | G/A | PKHD1 | 46/59/26 | 15/68/20 | 2.6 (1.3,5.0) | 1.4×10−3 | 40/66/22 | 23/51/28 | 3.0 (1.2,7.8) | 4.2×10−2 | 6.4×10−4 | Genotypic^ |
| rs1548335 | 6q13 | A/G | RIMS1/KCNQ5 | 81/47/4 | 44/44/15 | 2.6 (1.6,4.4) | 2.9×10−4 | 76/41/11 | 54/36/12 | 1.5 (0.9,2.3) | 1.1×10−1 | 3.5×10−4 | Additive |
| rs6557362* | 6q25.2 | T/A | CNKSR3 | 37/59/36 | 12/44/47 | 2.2 (1.4,3.5) | 3.7×10−4 | 22/64/42 | 11/53/38 | 1.5 (0.9,2.4) | 7.9×10−2 | 3.3×10−4 | Additive |
| rs766838* | 7p15.2 | G/A | NFE2L3/NPVF | 45/70/13 | 29/39/31 | 4.4 (1.9,10.2) | 4.2×10−4 | 43/65/20 | 28/47/26 | 1.9 (0.9,4.1) | 9.4×10−2 | 4.4×10−4 | Recessive |
| rs1486147* | 7p12.3 | C/T | TNS3/IGFBP3 | 27/46/59 | 4/42/56 | 8.3 (2.4,28.3) | 7.1×10−4 | 16/49/63 | 8/38/56 | 2.7 (0.9,7.8) | 7.3×10−2 | 5.7×10−4 | Recessive |
| rs1397294 | 8p22 | G/C | SGCZ | 52/67/12 | 39/40/24 | 4.3 (1.8,10.3) | 8.3×10−4 | 52/63/13 | 27/55/20 | 2.5 (1.1,6.2) | 4.0×10−2 | 3.7×10−4 | Recessive |
| rs16902486 | 8q24.21 | G/C | PVT1 | 124/6/2 | 87/15/0 | 7.2 (2.1,23.9) | 1.4×10−3 | 120/7/1 | 88/14/0 | 3.8 (1.2,12.1) | 2.2×10−2 | 3.4×10−4 | Dominant |
| rs7866508 | 9q31.3 | G/A | PTPN3 | 120/9/0 | 77/21/0 | 5.2 (2.0,13.9) | 9.1×10−4 | 118/9/1 | 86/15/1 | 2.6 (1.0,7.1) | 6.1×10−2 | 6.0×10−4 | Dominant |
| rs11017104 | 10q26.3 | A/G | GLRX3 | 116/13/1 | 64/36/2 | 5.9 (2.7,13.1) | 1.1×10−5 | 101/25/2 | 74/27/1 | 1.5 (0.7,3.0) | 2.5×10−1 | 3.6×10−5 | Dominant |
| rs10829695 | 10q26.3 | G/A | GLRX3 | 118/13/1 | 66/35/2 | 5.8 (2.6,12.8) | 1.4×10−5 | 101/25/1 | 74/27/1 | 1.5 (0.8,3.1) | 2.2×10−1 | 4.1×10−5 | Dominant |
| rs10764930* | 10q26.3 | G/A | GLRX3 | 118/13/1 | 65/36/2 | 5.9 (2.7,13.1) | 9.3×10−6 | 101/25/2 | 74/27/1 | 1.5 (0.7,3.0) | 2.5×10−1 | 3.2×10−5 | Dominant |
| rs10829696 | 10q26.3 | G/A | GLRX3 | 118/13/1 | 65/36/1 | 5.7 (2.6,12.7) | 1.5×10−5 | 101/25/2 | 74/27/1 | 1.5 (0.7,3.0) | 2.5×10−1 | 5.0×10−5 | Dominant |
| rs895255* | 11q14.1 | C/A | SYTL2/CCDC83 | 68/57/7 | 47/35/21 | 5.8 (2.1,15.0) | 5.7×10−4 | 64/55/9 | 45/40/17 | 2.8 (1.0,7.8) | 4.6×10−2 | 3.0×10−4 | Recessive |
| rs9595967* | 13q14.2 | C/G | CYSLTR2 | 49/65/16 | 24/49/26 | 5.0 (2.1,11.8) | 3.0×10−4 | 48/61/19 | 28/47/27 | 1.9 (0.9,4.2) | 8.5×10−2 | 2.9×10−4 | Recessive |
| rs11648233* | 16q23.3 | C/A | HSD17B2 | 50/58/19 | 30/43/29 | 2.2 (1.4,3.6) | 9.2×10−4 | 49/58/21 | 30/42/30 | 1.8 (1.2,2.8) | 7.7×10−3 | 9.1×10−5 | Additive |
| rs4794940 | 17q11.2 | A/G | ACCN1 | 23/67/39 | 9/45/48 | 2.4 (1.5,4.0) | 4.3×10−4 | 24/60/44 | 13/49/40 | 1.5 (0.9,2.3) | 9.2×10−2 | 4.4×10−4 | Additive |
| rs7245988* | 19q13.43 | G/A | NLRP11 | 1/41/89 | 0/12/90 | 4.65 (2.1,10.5) | 2.2×10−4 | 1/31/96 | 2/12/88 | 2.0 (0.9,4.4) | 1.0×10−1 | 2.6×10−4 | Dominant |
SNP included in multivariable model;
OR is for the homozygous risk genotype relative to the homozygous non-risk genotype.
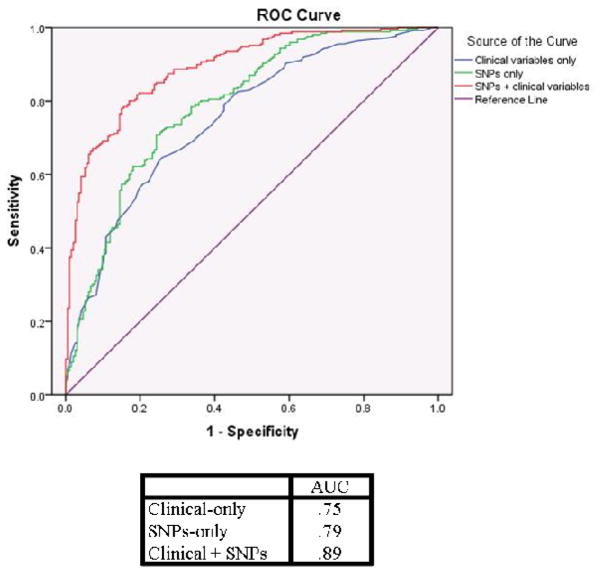
Table 3A shows the odds ratios associated with each of these 12 SNPs in the full cohort (N = 465) in a multivariable model with age, ADT, treatment and PCs. The odds ratios for the individual SNPs ranges from 1.6 to 5.6 indicating that after controlling for clinical factors, the SNPs represent clinically relevant predictors of ED. Though this was a multi-ethnic cohort, it is important to note that PCs were not significantly associated with ED, suggesting that ancestry was not a major confounder. The model including SNPs and non-genetic variables was a better predictor of ED compared to the model with age, ADT and treatment alone (AUCs of 0.89 and 0.75 respectively) (Figure 2). The AUC in the full cohort is likely an over-estimate because the model was being tested in the same patients from which it was developed. The AUC from the replication cohort alone (0.85) is likely closer to the true value, but the model should be tested further in additional large, independent cohorts.
Table 3.
Multivariable logistic regression models with (A) individual SNPs, (B) and (C) cumulative SNP score. Odds ratios for SNPs and SNP score are adjusted for non-genetic factors listed and PCs.
| A.
| ||
|---|---|---|
| Variable | OR (95%CI) | p-value |
| rs11693002 | 2.6 (1.4,5.0) | 2.8×10−3 |
| rs3749191 | 2.5 (1.5,4.3) | 7.8×10−4 |
| rs10764930 | 2.0 (1.1,3.7) | 3.4×10−2 |
| rs11648233 | 2.0 (1.4,3.0) | 2.7×10−4 |
| rs322895 | 2.8 (1.4,5.6) | 4.3×10−3 |
| rs895255 | 5.6 (2.3,14.0) | 1.9×10−4 |
| rs1486147 | 5.5 (2.0,15.2) | 1.1×10−3 |
| rs7245988 | 3.4 (1.7,6.8) | 5.7×10−4 |
| rs9595967 | 2.3 (1.2,4.6) | 1.2×10−2 |
| rs6557362 | 1.6 (1.1,2.4) | 1.1×10−2 |
| rs766838 | 2.2 (1.2,4.3) | 1.6×10−2 |
| rs11747037 | 1.8 (1.2,2.6) | 4.5×10−3 |
| Age | 1.2 (1.1,1.2) | 8.4×10−15 |
| ADT | 1.9 (1.0,3.4) | 3.7×10−2 |
| EBRT Only* | 0.3 (0.0,1.3) | 0.11 |
| Implant + EBRT* | 1.9 (1.0,3.4) | 4.1×10−2 |
| B.
| ||
|---|---|---|
| Variable | OR (95%CI) | p-value |
| Cumulative SNP score | 2.2 (1.8,2.6) | 2.1×10−19 |
| Age | 1.2 (1.1,1.2) | 3.1×10−15 |
| ADT | 2.0 (1.1,3.5) | 0.02 |
| EBRT Only* | 0.2 (0.04,0.9) | 0.03 |
| Implant + EBRT* | 1.8 (1.0,3.1) | 0.05 |
| C.
| ||
|---|---|---|
| Variable | OR (95%CI) | p-value |
| Cumulative SNP score | 2.3 (1.9,2.8) | 2.2×10−19 |
| Age | 1.2 (1.1,1.2) | 2.3×10−13 |
| ADT | 2.9 (1.7,5.0) | 1.1×10−4 |
| BED (10Gy2) | 1.1 (0.9,1.2) | 0.29 |
| Prostate Volume (10mm3) | 1.1 (0.9,1.2) | 0.09 |
Reference category is “implant only”.
Figure 2.
ROC curves from multivariable logistic regression models. Clinical-only model contains age, ADT, and treatment; SNP-only model contains 12 SNPs and PCs; Clinical+SNP model contains age, ADT, treatment, 12 SNPs and PCs.
When these 12 SNPs were combined into a cumulative SNP score representing a quantitative measure of risk alleles possessed by each individual, a one-allele increase in cumulative SNP score increased the odds of developing ED by a factor of 2.2 (95% CI 1.9,2.6; p-value = 2.1×10−19) after controlling for clinical factors and ancestry (Table 3B). We investigated the effect of including BED and prostate volume in the multi-variable model rather than treatment modality. These factors did not have a substantial effect on the odds ratios for the cumulative SNP score (Table 3C). Using ordered logistic regression, the cumulative SNP score was also a significant predictor of SHIM category15 (OR=1.5, 95%CI 1.4–1.7; Supplemental Table 1). The ordered logistic regression model correctly assigned 52.4% of the patients to their SHIM category; just 4.3% of individuals were grossly misclassified (i.e. group 1 classified as group 5 and vice versa). Furthermore, there was a striking dose-dependent relationship between number of risk alleles and ED case status (Sommers’ D p-value for directional association = 1.7×10−29) (Table 4A). The multivariable model including cumulative SNP score, age, ADT, treatment, and PCs has sensitivity of 83.7% and specificity of 74.6%.
Table 4.
A. Rank-correlation of cumulative SNP score and ED. SNPs with an additive inheritance pattern were counted as 0, 1, or 2 risk alleles; SNPs with a dominant or recessive inheritance pattern were counted as either 0 or 1 risk alleles with the appropriate genotypes collapsed into a single group. Sensitivity and specificity were calculated using predicted outcomes from the model including SNP score, age, ADT, treatment and PCs. Each row shows the sensitivity and specificity achieved if the SNP score was dichotomized at the number shown for that row. B. Relationship with cumulative SNP score and age. Percentages are based on row totals.
| A.
| ||||
|---|---|---|---|---|
| Cumulative SNP score | Cases, N(%) | Controls, N(%) | Sensitivity | Specificity |
| 3 | 0 | 2 (1.0%) | 77.6% | 58.5% |
| 4 | 0 | 4 (2.1%) | 77.6% | 60.1% |
| 5 | 1 (0.4%) | 14 (7.3%) | 79.3% | 64.2% |
| 6 | 4 (1.6%) | 28 (14.5%) | 82.1% | 65.8% |
| 7 | 17 (6.9%) | 37 (19.2%) | 84.6% | 72.0% |
| 8 | 38 (15.4%) | 38 (19.7%) | 83.3% | 71.0% |
| 9 | 48 (19.5%) | 27 (14.0%) | 80.9% | 73.1% |
| 10 | 56 (22.8%) | 23 (11.9%) | 78.5% | 71.5% |
| 11 | 45 (18.3%) | 15 (7.8%) | 77.2% | 66.3% |
| 12 | 21 (8.5%) | 4 (2.1%) | 77.2% | 62.7% |
| 13 | 12 (4.9%) | 1 (0.5%) | 77.6% | 59.6% |
| 14 | 3 (1.2%) | 0 | 77.6% | 59.1% |
| 15 | 1 (0.4%) | 0 | 76.2% | 60.5% |
|
| ||||
| Total | 246 | 193 | 83.7% | 74.6% |
| B.
| |||
|---|---|---|---|
| Case, N(%) | Control, N(%) | Total | |
| ≤ 62 years | |||
| 3–9 risk alleles | 22 (21.8%) | 79 (78.2%) | 101 |
| 10–15 risk alleles | 39 (52.7%) | 35 (47.3%) | 74 |
|
| |||
| > 62 years | |||
| 3–9 risk alleles | 86 (54.8%) | 71 (45.2%) | 157 |
| 10–15 risk alleles | 99 (92.5%) | 8 (7.5%) | 107 |
Alemozaffar et al. recently developed validated models to predict erectile function at 2 years following radiation therapy16. Among patients who selected brachytherapy for treatment, age, pre-treatment function, race/ethnicity and body-mass index were significant predictors of erectile function (AUC=0.90). Among patients who selected EBRT, PSA level, pre-treatment function and ADT were significant predictors of erectile function (AUC=0.87). We sought to determine how well these models fit our cohort, and whether adding the SNP score improves the AUC. Since our cohort includes a large proportion of patients treated with combination brachytherapy with EBRT, we combined the elements of the two models from Alemozaffar et al, excluding BMI, which was not available. This model achieved an AUC of 0.80 in our cohort. Addition of the SNP score (dichotomozied at the median of 9 risk alleles) improved the AUC to 0.85. These AUCs would likely increase with addition of BMI.
To show a simplified understanding of what the SNPs could mean in the context of age alone, we stratified the pooled cohort by age (using the median, 62, as a cut-off) and cumulative SNP score (using the median, 9, as a cut-off). Approximately 21.8% of younger men with ≤ 9 risk alleles developed ED compared to 52.7% of younger men with > 9 risk alleles (Table 4B). Conversely, 54.8% of older men with ≤ 9 risk alleles developed ED compared to 92.5% of older men with > 9 risk alleles. Stated more generally, in this cohort, a younger man with more risk alleles has roughly the same likelihood of developing ED as an older man with fewer risk alleles.
We tested the model in two independent, small cohorts of patients treated with radiotherapy and followed for development of ED. The XXX cohort was comprised of 55 individuals (38 cases and 17 controls) treated with either EBRT alone (59%) or brachytherapy alone (41%). The AUC for the model including SNPs and non-genetic variables in this cohort was 0.79. The second test cohort comprised 66 individuals of African American ancestry from our previously published GWAS8. In this cohort, the model produced an AUC of 0.81. Though these cohorts were under-powered to expect significant SNP-phenotype associations, they provide an indicator of overall model performance.
Finally, we investigated the top SNPs from the previously published GWAS of post-radiotherapy ED in an African American cohort. Seven of the top 31 SNPs from that study have p-values < 0.05 in the cohort from the current study, though none of the SNPs reached genome-wide significance (Supplementary Table 2). This is not surprising since the SNPs identified in the African American study are rare in European Americans8. To better attempt to replicate the regions tagged by these SNPs in the current study, we scanned 150kb upstream and downstream of each SNP identified in the African American GWAS, and identified 5 regions that contain at least one SNP with a low p-value (<1×10−2) (Supplementary Table 3). Because we searched a limited number of regions with a priori evidence for association with ED, a genome-wide level of significance may be too strict, and some of these regions may represent true positives.
Discussion
We identified 25 SNPs that showed low p-values, with effects of similar magnitude and in the same direction in both discovery and replication cohorts. Using re-sampling, we reduced the complexity of this list to 12 SNPs. Although none of these SNPs met a strict genome-wide significance p-value threshold of 10−7 to 10−8, they may represent true associations as evidenced by their increased accuracy for risk prediction compared with non-genetic variables alone for the case, control and intermediate groups. The AUC achieved by the model developed from this study is likely to be an overestimate as it was tested on the pooled discovery and replication cohorts from which it was developed. Still, this increased accuracy was observed in two small independent groups of cases and controls from the XXX cohort and the African American cohort previously studied. The utility of these SNPs for predicting risks was shown not only in the multi-SNP model, but also in a model of additive allelic risk.
There are several limitations to note. First, none of the individual SNPs reached genome-wide significance. Other studies that did not achieve genome-wide significance have observed replication of suggestive SNPs however17. Thus, it will be important to investigate the SNPs identified in this study further in a large, independent cohort of similarly treated patients. A second limitation is that data on co-morbidities was limited. While we found no significant association between ED and smoking or diabetes as assessed at time of diagnosis, these factors could have changed over the course of follow-up. It is possible that some proportion of the risk for ED is due to worsening co-morbid conditions, which could modify the effects of the SNP-phenotype association. A third limitation is that more extensive analysis of dose-volume parameters could not be adequately assessed in this cohort because of the relatively uniform treatment received by the patients.
Although this study and our first GWAS on ED following radiation therapy for prostate cancer were similar, the ethnic make-up of the two cohorts was quite different8. The first study was comprised of African-American men, whereas this study was composed predominantly of men of European ancestry. When comparing the results of these two studies, most of the top 31 SNPs from the first study did not have low p-values. This result was expected, because the SNPs we identified as associated with ED in men of African ancestry are almost universally rare in European populations. Yet, part of the same repertoire of genes may be involved, because 11 out of the top 31 genes in the current study had at least one SNP within 150kb of the SNP associated with ED in African-Americans with p-values < 10−2.
As with the previous study, some of the SNPs identified in this study lie in or near genes that may encode biological activities involved in erectile function rather than DNA damage repair – the focus of earlier studies. For example, the 17-beta-hydroxysteroid dehydrogenase II gene (HSD17B2) catalyzes the oxidative metabolism of androgens and estrogens in human peripheral tissues18. Other physiological functions included cell adhesion and cell matrix association (CDCP1)19, glutaredoxin oxidoreductase enzyme activity (GLRX3)20, and mineralocorticoid receptor signaling in response to aldosterone (CNKSR3)21. The Ingenuity Pathway Analysis software (Ingenuity Systems, Inc., Redwood City, CA) grouped 11 of these 12 genes into a common pathway containing direct or indirect connections between each other. This pathway contains genes involved in cellular growth and proliferation, cell-cell signaling and tissue development. The SNPs associated with ED may encode sensitivity to radiation injury through these various pathways.
It is important to note that the markers identified in this study are tag SNPs indirectly associated with ED. Higher density genotyping and/or sequencing could identify more predictive SNPs or rare variants that are in linkage disequilibrium and may produce direct effects on protein expression, modification, or transcriptional regulation.
Supplementary Material
Summary.
Through a two-stage genome wide association study, 12 SNPs were identified that were associated with the development of ED following radiation treatment for prostate cancer. If validated further, these SNPs could provide the basis for an assay that would enable radiation oncologists to more accurately predict which men are most likely to develop ED following prostate cancer radiotherapy.
Acknowledgments
This research was supported by grants RSGT-05-200-01-CCE from the American Cancer Society, PC074201 from the Department of Defense and 1R01CA134444 from the National Institutes of Health.
Footnotes
Conflict of interest: N.S. has received consulting fees or honorarium from Amgen, Ferring, Janssen, Diversified Conference Management, Prologics LLC, and Nihon MediPhysics. RS has received fees for lectures and development of educational presentations from Bard.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Grimm P, Billiet I, Bostwick D, et al. Comparative analysis of prostate-specific antigen free survival outcomes for patients with low, intermediate and high risk prostate cancer treatment by radical therapy. Results from the Prostate Cancer Results Study Group. BJU Int. 2012 Feb;109 (Suppl 1):22–29. doi: 10.1111/j.1464-410X.2011.10827.x. [DOI] [PubMed] [Google Scholar]
- 2.Snyder KM, Stock, Richard G, Buckstein, Michael, Stone, Nelson N. Long-term potency preservation following brachytherapy for prostate cancer. BJU Int. 2012 doi: 10.1111/j.1464-410X.2011.10800.x. [DOI] [PubMed] [Google Scholar]
- 3.Merrick GS, Butler WM, Wallner KE, et al. Erectile function after prostate brachytherapy. International journal of radiation oncology, biology, physics. 2005 Jun 1;62(2):437–447. doi: 10.1016/j.ijrobp.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Potters L, Torre T, Fearn PA, Leibel SA, Kattan MW. Potency after permanent prostate brachytherapy for localized prostate cancer. International journal of radiation oncology, biology, physics. 2001 Aug 1;50(5):1235–1242. doi: 10.1016/s0360-3016(01)01578-4. [DOI] [PubMed] [Google Scholar]
- 5.Robinson JW, Moritz S, Fung T. Meta-analysis of rates of erectile function after treatment of localized prostate carcinoma. International journal of radiation oncology, biology, physics. 2002 Nov 15;54(4):1063–1068. doi: 10.1016/s0360-3016(02)03030-4. [DOI] [PubMed] [Google Scholar]
- 6.Talcott JA, Manola J, Clark JA, et al. Time course and predictors of symptoms after primary prostate cancer therapy. J Clin Oncol. 2003 Nov 1;21(21):3979–3986. doi: 10.1200/JCO.2003.01.199. [DOI] [PubMed] [Google Scholar]
- 7.Rosenstein BS. Identification of SNPs associated with susceptibility for development of adverse reactions to radiotherapy. Pharmacogenomics. 2011 Feb;12(2):267–275. doi: 10.2217/pgs.10.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kerns SL, Ostrer H, Stock R, et al. Genome-wide association study to identify single nucleotide polymorphisms (SNPs) associated with the development of erectile dysfunction in African-American men after radiotherapy for prostate cancer. International journal of radiation oncology, biology, physics. 2010 Dec 1;78(5):1292–1300. doi: 10.1016/j.ijrobp.2010.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stock RG, Stone NN, Cesaretti JA, Rosenstein BS. Biologically effective dose values for prostate brachytherapy: effects on PSA failure and posttreatment biopsy results. International journal of radiation oncology, biology, physics. 2006 Feb 1;64(2):527–533. doi: 10.1016/j.ijrobp.2005.07.981. [DOI] [PubMed] [Google Scholar]
- 10.Rosen RC, Cappelleri JC, Smith MD, Lipsky J, Pena BM. Development and evaluation of an abridged, 5-item version of the International Index of Erectile Function (IIEF-5) as a diagnostic tool for erectile dysfunction. Int J Impot Res. 1999 Dec;11(6):319–326. doi: 10.1038/sj.ijir.3900472. [DOI] [PubMed] [Google Scholar]
- 11.Zagar TM, Stock RG, Cesaretti JA, Stone NN. Assessment of postbrachytherapy sexual function: a comparison of the IIEF-5 and the MSEFS. Brachytherapy. 2007 Jan-Mar;6(1):26–33. doi: 10.1016/j.brachy.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 12.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007 Sep;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Consortium IH. The International HapMap Project. Nature. 2003 Dec 18;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 14.R: A language and environment for statistical computing [computer program] Version 2.13.1. Vienna, Austria: 2011. [Google Scholar]
- 15.Wheat JC, Hedgepeth RC, He C, Zhang L, Wood DP., Jr Clinical interpretation of the Expanded Prostate Cancer Index Composite-Short Form sexual summary score. J Urol. 2009 Dec;182(6):2844–2849. doi: 10.1016/j.juro.2009.08.088. [DOI] [PubMed] [Google Scholar]
- 16.Alemozaffar M, Regan MM, Cooperberg MR, et al. Prediction of erectile function following treatment for prostate cancer. JAMA. 2011 Sep 21;306(11):1205–1214. doi: 10.1001/jama.2011.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panagiotou OA, Ioannidis JP. What should the genome-wide significance threshold be? Empirical replication of borderline genetic associations. Int J Epidemiol. 2012 Feb;41(1):273–286. doi: 10.1093/ije/dyr178. [DOI] [PubMed] [Google Scholar]
- 18.Wu L, Einstein M, Geissler WM, Chan HK, Elliston KO, Andersson S. Expression cloning and characterization of human 17 beta-hydroxysteroid dehydrogenase type 2, a microsomal enzyme possessing 20 alpha-hydroxysteroid dehydrogenase activity. J Biol Chem. 1993 Jun 15;268(17):12964–12969. [PubMed] [Google Scholar]
- 19.Brown TA, Yang TM, Zaitsevskaia T, et al. Adhesion or plasmin regulates tyrosine phosphorylation of a novel membrane glycoprotein p80/gp140/CUB domain-containing protein 1 in epithelia. J Biol Chem. 2004 Apr 9;279(15):14772–14783. doi: 10.1074/jbc.M309678200. [DOI] [PubMed] [Google Scholar]
- 20.Witte S, Villalba M, Bi K, Liu Y, Isakov N, Altman A. Inhibition of the c-Jun N-terminal kinase/AP-1 and NF-kappaB pathways by PICOT, a novel protein kinase C-interacting protein with a thioredoxin homology domain. J Biol Chem. 2000 Jan 21;275(3):1902–1909. doi: 10.1074/jbc.275.3.1902. [DOI] [PubMed] [Google Scholar]
- 21.Ziera T, Irlbacher H, Fromm A, et al. Cnksr3 is a direct mineralocorticoid receptor target gene and plays a key role in the regulation of the epithelial sodium channel. FASEB J. 2009 Nov;23(11):3936–3946. doi: 10.1096/fj.09-134759. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.