Abstract
HIV-1 subtype C (HIV-1C) CXCR4-using virus is isolated infrequently and is poorly characterized. Understanding HIV-1C env characteristics has implications for the clinical use of antiretrovirals that target viral entry. A total of 209 env clones derived from 10 samples with mixed CCR5-(R5), CXCR4-using (X4) or dual-tropic HIV-1C were phenotyped for coreceptor usage. Intra-patient X4 and R5 variants generally formed distinct monophyletic phylogenetic clusters. X4 compared to R5 envs had significantly greater amino acid variability and insertions, higher net positive charge, fewer glycosylation sites and increased basic amino acid substitutions in the GPGQ crown. Basic amino acid substitution and/or insertion prior to the crown are highly sensitive characteristics for predicting X4 viruses. Chimeric env functional studies suggest that the V3 loop is necessary but often not sufficient to impart CXCR4 utilization. Our studies provide insights into the unique genotypic characteristics of X4 variants in HIV-1C.
Keywords: HIV-1, Subtype C, CCR5, CXCR4, Phylogenetic, Envelope
Introductions
Human immunodeficiency virus type-1 (HIV-1) envelope (env) mediates viral entry by interacting with the primary receptor (CD4) and a coreceptor (CCR5 or CXCR4) on target cells (Choe et al., 1996, 2003;; Feng et al., 1996; Maddon et al., 1986; McDougal et al., 1986; Shioda et al., 1991). HIV transmission predominantly occurs with CCR5-using viruses (R5), and cross-sectional studies show that 70–80% of patients with early-stage disease continue to harbor only R5 variants. In contrast, with advanced disease nearly half of patients have dual/mixed (DM) viruses containing both R5 and variants that can either utilize both coreceptors (dual) and/or CXCR4 using HIV-1 (X4), and only a few patients harbor purely X4 virus even in advanced disease (Melby et al., 2006). The presence of DM or X4 virus is an independent risk factor for accelerated disease progression (Japour et al., 1994; Koot et al., 1993; Richman and Bozzette, 1994).
Most of our current understanding of env interactions with the host cell coreceptor is based on studies on subtype B viruses (HIV-1B). However, the majority of infections worldwide are due to HIV-1 subtype C (HIV-1C) (Osmanov et al., 2002; UNAIDS, 2009). Earlier studies of subtype C infected individuals showed a predominance of R5 viruses regardless of disease stage, suggesting that unique HIV-1C env characteristics potentially limit the emergence of CXCR4-using viruses (Abebe et al., 1999; Bjorndal et al., 1999; Cecilia et al., 2000). However, a recent study from our group showed that around 15% of approximately 150 women in Botswana with advanced disease harbored CXCR4-using viruses (Lin et al., 2011), confirming more recent findings among other smaller HIV-1C cohorts (Cilliers et al., 2003; Connell et al., 2008; Johnston et al., 2003; Michler et al., 2008; Papathanasopoulos et al., 2002; Tien et al., 1999; Tscherning et al., 1998; van Rensburg et al., 2002; Zhang et al., 1996). The small number of CXCR4-using HIV-1C viruses studied to-date has limited the understanding of the determinants of HIV-1 C env coreceptor usage. Previous studies were also limited by the analysis of consensus or population-based viral sequences and comparison of R5 and X4 sequences isolated from different individuals. More precise characterization of the genotypic determinants of HIV-1C coreceptor usage can be achieved by analyzing co-circulating viral clones that have different coreceptor usage (as determined by phenotypic assay), which has been reported for only a few HIV-1C isolates (Zhang et al., 2010). Such analyses are necessary to improve the accuracy of genotypic algorithms for predicting coreceptor usage of HIV-1C (Jensen et al., 2006). This is a high priority because CCR5 antagonists are available as an option for first-line antiretroviral therapy (ART), and CCR5 antagonists are being considered as prophylaxis against HIV-1 transmission (Osmanov et al., 2002). Predicting coreceptor usage by an accurate, timely, and cost-effective test will be important as more infected individuals in developing countries are considered for starting or switching to a CCR5 antagonist. In the current study we performed an in-depth genotypic and phenotypic analysis of clonal HIV-1C envelopes and present data which better define the genotypic characteristics of CXCR4-using HIV-1C.
Results
Co-receptor usage of clonal envelopes from DM HIV-1C viruses
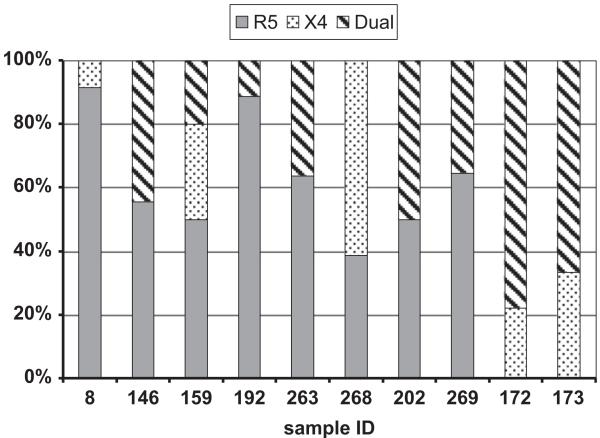
We previously determined that 22 of 148 individuals HIV-1C-infected women harbored DM viruses; none harbored exclusively X4 virus (Lin et al., 2011). In the current study we examined 10 DM samples from 9 of those subjects; 6 samples were obtained prior to ART and 4 samples were collected after virologic failure on ART (Table 1). These samples were selected based on relatively high levels of CXCR4 usage determined by the phenotypic assay. The CD4 cell count level between DM samples which were used in this study (median 201 cells/mm3, range 43–375) and the ones not used for further clonal analysis (median 128.5 cells/mm3, range 5–201) were not significantly different (p=0.050) suggesting that we did not exclude subjects with less progressive disease and potentially recent coreceptor switching. Env sequences were also isolated from an additional 7 subjects harboring exclusively R5 virus (Table 1). The R5 samples, selected based on high levels of CCR5 usage detected by the coreceptor usage assay, tended to be from women without ART exposure although viral levels and CD4 counts were not significantly different compared to DM samples. We isolated a median of 18 env clones (range 11–36) from each sample. Env clones that yielded an infectious pseudo-virus were subsequently sequenced. A total of 209 infectious env clones were sequenced and phenotyped, with a similar number of clones from R5 and DM samples (median 13 [range 7–19] and 12 [9–18] clones per sample, respectively) (Tables 2 and 3). Of the 123 clones isolated from DM samples, 64 were R5, 39 were dual-tropic, and 20 were X4. The proportion of R5 (median 52.8% [range 0–92%]), X4 (median 72.2% [range 0–94%]), and dual-tropic (median 26.1% [range 0–97%]) viruses varied between subjects (Fig. 1). Additionally, 86 env clones were obtained from the 7 subjects infected with R5 virus; all of these clones were confirmed as R5 by phenotyping. A neighbor-joining (NJ) tree of all 164 full-length envs and reference sequences of different subtypes obtained from the Los Alamos National Laboratory (LANL) HIV database confirmed that all isolates were subtype C with bootstrap values >90% (data not shown). Envs from each subject clustered together arguing against any sample contamination or mislabeling.
Table 1.
Clinical characteristics of study participants infected with HIV-1C.
| Sample ID |
Population tropism |
Age | cART Exposure |
CD4 counta (cells/uL) |
Viral loada (copies/mL) |
Infant infected |
|---|---|---|---|---|---|---|
| 8 | DM | 29 | N | 43 | 522,000 | N |
| 146 | DM | 29 | N | 172 | 653,000 | Y |
| 159 | DM | 33 | N | 116 | 154,000 | N |
| 192 | DM | 43 | N | 92 | 121,000 | Y |
| 202 | DM | 31 | N | 147 | 273,056 | N |
| 263 | DM | 29 | N | 201 | 230,000 | N |
| 172 | DM | 30 | Y | 309 | 1060 | N |
| 173 | DM | 25 | Y | 316 | 1290 | Y |
| 268 | DM | 23 | Y | 375 | 333,000 | Y |
| 269 | DM | N/A | Y | 319 | 10,600 | N |
| 19 | R5 | 37 | N | 5 | 399,000 | N |
| 20 | R5 | 37 | N | 83 | 67,600 | N |
| 77 | R5 | 21 | N | 123 | 112,000 | N |
| 115 | R5 | 40 | N | 180 | 741,000 | N |
| 149 | R5 | 35 | N | 65 | 4690 | N |
| 161 | R5 | 34 | N | 50 | 8550 | N |
| 170 | R5 | 30 | N | 165 | 227,000 | N |
Values at or closest to time of sample collection.
Table 2.
HIV-1C V3 sequence characteristics.
| ID | Pheno- type |
# of clones |
V3 loopa | V3 length |
Net charge |
N- glyco |
11/25 Prediction |
C-PSSM prediction |
Change in GPGQb |
|---|---|---|---|---|---|---|---|---|---|
| DM8 | MRCA | CTRPGNNTGRSVRIGPRKAFYTTRKIIGDIRAAHC | |||||||
| R5 | 10 | ------R-----GQ---GE-------- | 35 | 4 | 1 | R5 | X4 | 0 | |
| R5 | 1 | ------R-----GQ---GE---V---- | 35 | 4 | 1 | R5 | X4 | 0 | |
| X4 | 1 | -------------LRT--~-------- | 34 | 7 | 1 | X4 | X4 | 1 | |
| DM146 | MRCA | CTRPDNNTRRSVRMGIGPGQVFYTNDIIGDIRQAHC | |||||||
| Dual | 8 | --------R-------T-------R--- | 36 | 5 | 1 | X4 | X4 | 0 | |
| R5 | 5 | -----------~~----------R--- | 34 | 4 | 1 | R5 | X4 | 0 | |
| R5 | 2 | -----------~~-------------- | 34 | 3 | 1 | R5 | X4 | 0 | |
| R5 | 1 | -----------~~-----------Y-- | 34 | 2 | 1 | R5 | X4 | 0 | |
| R5 | 1 | -----------~~--A----------- | 34 | 3 | 1 | R5 | X4 | 0 | |
| R5 | 1 | -I-------I---~~----A-----E-S- | 34 | 1 | 1 | R5 | X4 | 0 | |
| DM159 | MRCA | CTRPNNNTRKSVRIGPGQTFYATGEIIGDIRRAHC | |||||||
| R5 | 2 | -I------R-M----ALFT---N-Q--- | 35 | 5 | 1 | R5 | X4 | 0 | |
| R5 | 2 | -I------R-M----AYFT---N-Q--- | 35 | 5 | 1 | R5 | X4 | 0 | |
| R5 | 1 | -------------------------- | 35 | 5 | 1 | R5 | R5 | 0 | |
| Dual | 1 | ---I-RERK----RA-FR-QMT---Q-S- | 35 | 6 | 0 | R5 | X4 | 1 | |
| Dual | 1 | ---I-RERK----RA-FR-QMT---Q-S- | 35 | 5 | 0 | R5 | X4 | 1 | |
| X4 | 1 | ---I-RERK----RA-FR-QMT---Q-S- | 35 | 5 | 0 | X4 | X4 | 1 | |
| X4 | 1 | ---I-RERK----RA-FR-QMT---Q-S- | 35 | 6 | 0 | X4 | X4 | 1 | |
| X4 | 1 | ---I-NVRN----RALFK-KMT---Q-S- | 35 | 6 | 0 | X4 | X4 | 1 | |
| DM173 | MRCA | CVRPNNNTRKSVRIGIGRGQTFYANRIIGDIRQAHC | |||||||
| Dual | 6 | --------------------------- | 36 | 7 | 1 | R5 | X4 | 6 | |
| X4 | 3 | --------------------------- | 36 | 7 | 1 | R5 | X4 | 3 | |
| DM172 | MRCA | CIRPGNNTRRRVRMGIGPGQTFYATGNIIGDIRQAHC | |||||||
| Dual | 6 | -M------------------------- | 37 | 6 | 1 | X4 | X4 | 0 | |
| Dual | 1 | --------------------------- | 37 | 6 | 1 | X4 | X4 | 0 | |
| X4 | 1 | --------------------------- | 37 | 6 | 1 | X4 | X4 | 0 | |
| X4 | 1 | -M------------------------- | 37 | 6 | 1 | X4 | X4 | 0 | |
| DM192 | MRCA | CTRPNNNTSQSIRIGPGQTFYAMGRIIGDIRQAHC | |||||||
| R5 | 5 | -------------------------- | 35 | 4 | 2 | X4 | X4 | 0 | |
| R5 | 1 | --------------Y----------- | 35 | 4 | 2 | X4 | X4 | 0 | |
| R5 | 1 | ------RE-------Y--D-------- | 35 | 2 | 1 | R5 | R5 | 0 | |
| R5 | 1 | ------RE-------Y--D-------- | 35 | 2 | 1 | R5 | R5 | 0 | |
| Dual | 1 | -----RRK---------I-GV-----Y- | 35 | 5 | 0 | R5 | X4 | 0 | |
| DM202 | MRCA | CTRPGNNTSRSIRIGPGQTFYATGRITGNIRQAHC | |||||||
| R5 | 1 | -I-----------------V-D--L-- | 35 | 5 | 2 | X4 | X4 | 0 | |
| R5 | 3 | --------------------D----- | 35 | 5 | 2 | X4 | X4 | 0 | |
| R5 | 1 | -I-----------------V-D----- | 35 | 6 | 2 | X4 | X4 | 0 | |
| Dual | 2 | -I-----------------V-D----- | 35 | 5 | 2 | X4 | X4 | 0 | |
| Dual | 1 | -M----K------R-----V-D--K-- | 35 | 8 | 1 | X4 | X4 | 1 | |
| Dual | 1 | -M----R------R-----V-D--K-- | 35 | 8 | 1 | X4 | X4 | 1 | |
| Dual | 1 | -M----K------R-----V-D----- | 35 | 7 | 1 | X4 | X4 | 1 | |
| DM263 | MRCA | CIRPGNNTRKSMRIGPGQTFYATGDIIGDIRKAHC | |||||||
| R5 | 3 | -------------------------- | 37 | 5 | 1 | R5 | R5 | 0 | |
| R5 | 3 | ------------------E-------- | 37 | 5 | 1 | R5 | R5 | 0 | |
| R5 | 1 | -T-------------------N-Q--- | 37 | 5 | 1 | R5 | R5 | 0 | |
| Dual | 1 | ------------------E-------- | 37 | 5 | 1 | R5 | R5 | 0 | |
| Dual | 1 | ----K------I-H-----K-------- | 37 | 9 | 1 | X4 | X4 | 1 | |
| Dual | 1 | ----K------I-H-----K-------- | 37 | 9 | 1 | X4 | X4 | 1 | |
| Dual | 1 | ----K------I-H-----K-------- | 37 | 9 | 1 | X4 | X4 | 1 | |
| DM268 | MRCA | CTRPSNNTRRRVRIGIGPGQAFDATGEIIGDIRQAHC | |||||||
| X4 | 11 | ------------R------Q------- | 37 | 6 | 1 | X4 | X4 | 11 | |
| R5 | 5 | --------S-~~---YT---------- | 35 | 4 | 1 | R5 | R5 | 0 | |
| R5 | 2 | --------S-~~---YT---------- | 35 | 4 | 1 | R5 | R5 | 0 | |
| DM269 | MRCA | CIRPGNNTSRSIRIGPGQAFYATGRIVGDIRQAHC | |||||||
| R5 | 10 | -------------T------------ | 35 | 5 | 2 | X4 | X4 | 0 | |
| R5 | 1 | -------------------------- | 35 | 5 | 2 | X4 | X4 | 0 | |
| Dual | 6 | -M----K------RT---K----K--- | 35 | 8 | 1 | X4 | X4 | 6 |
V3 crow n motif in bold.
Number of clones with substitution in GPGQ crow n.
Table 3.
Concordance of genotypic methods in determining HIV-1C phenotype.
| Phenotype of enva | No. of sequences | 11/25 Rule | %correct | C-PSSM | %correct | GPGQ substitution and/or insertionc |
%correct |
|---|---|---|---|---|---|---|---|
| DM samples | |||||||
| X4 | 20 | 16 | 80.0 | 20 | 100.0 | 20 | 100 |
| Unique X4 V3b | 7 | 6 | 85.7 | 7 | 100.0 | 7 | 100 |
| Dual | 39 | 30 | 76.9 | 38 | 97.4 | 35 | 89.7 |
| Unique Dual V3b | 13 | 9 | 69.2 | 12 | 92.3 | 10 | 76.9 |
| X4+Dual | 59 | 46 | 78.0 | 58 | 98.3 | 55 | 93.2 |
| Unique (X4+Dual) V3b | 20 | 15 | 75.0 | 19 | 95.0 | 17 | 85.0 |
| R5 | 64 | 42 | 65.6 | 18 | 28.1 | 64 | 100 |
| Unique R5 V3b | 23 | 15 | 65.2 | 7 | 30.4 | 23 | 100 |
| R5 samples | |||||||
| R5 | 86 | 86 | 100 | 78 | 90.7 | 86 | 100 |
| Unique R5 V3a | 20 | 20 | 100 | 19 | 95.0 | 20 | 100 |
| ALL samples | |||||||
| ALL R5 | 150 | 128 | 85.3 | 96 | 64.0 | 150 | 100 |
| ALL unique R5 V3a | 43 | 35 | 81.4 | 26 | 60.5 | 43 | 100 |
Coreceptor usage phenotype determined for each clonal env sequence.
Unique V3 loop sequences of corresponding phenotype.
GPGQ substitution and/or insertion around crown predicts CXCR4-usage, absence of both predicts R5 virus.
Fig. 1.
Composition of clones from dual/mixed (DM) env populations. Individual sample identification number are on the x-axis while the y-axis shows the percentage of CCR5 using (R5), exclusive CXCR4 utilizing (X4), and dual-tropic strains (dual).
Phylogenetic relationship of co-circulating R5 and X4 envs
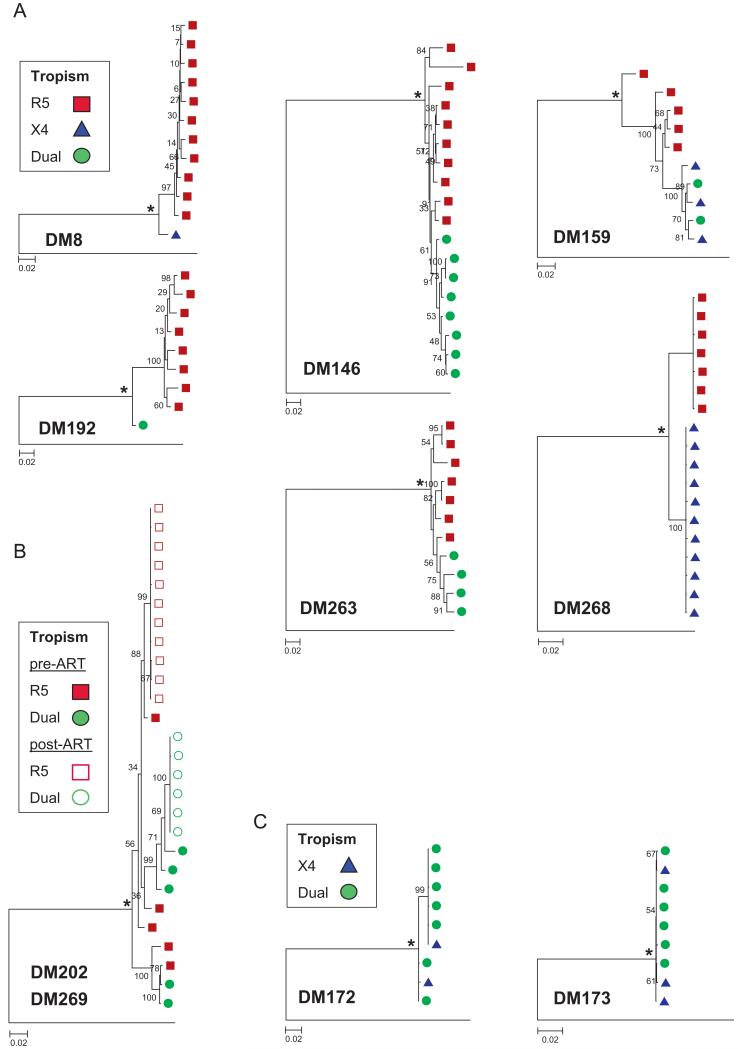
Env sequences obtained from different subjects but with the same coreceptor phenotype did not cluster together, suggesting that intra-subject envs shared greater genotypic similarity than inter-subject envs that used the same coreceptor. Maximum likelihood phylogenies showed that in 7 of the 10 DM samples (DM8, DM146, DM159, DM192, DM263, DM268, and DM269), R5 and CXCR4-using env sequences clustered separately; the segregation was supported with high bootstrap values (≥75%) (Fig. 2A). One individual had two samples collected: DM202 was collected prior to ART whereas DM269 was collected at virological failure after 44 months of ART (Fig. 2B). Samples from both the time-points yielded only R5 and dual-tropic pseudo-viruses. Separate clustering of the R5 and dual-tropic envs was evident in the post-ART but not the pre-ART sequences. The Slatkin–Maddison test, which assesses compartmentalization (i.e. restriction of gene flow) between two populations based on the topology of the phylogenetic trees (Nickle et al., 2003), further confirmed the separation of intra-subject CXCR4 using and R5 sequences. In DM146, DM159, DM263, DM268, and DM269, separating X4 and R5 sequences required more than 3 steps in greater than 95% of the 1000 randomly generated trees, suggesting significant compartmentalization between envs with different coreceptor usage (p<0.05). Within the same five samples, the mean R5-to-X4 pairwise maximum composite likelihood distances were significantly greater than between co-circulating CXCR4-using or R5 sequences (p<0.01). The Slatkin–Maddison and pairwise-distance analyses could not be performed in 2 samples (DM8 and DM192) because only one CXCR4-using env clone was isolated. In DM172 and DM173, which contained no R5 sequences, there was no phylogenetic separation between X4 and dual-tropic clones by ML tree bootstrap support value, Slatkin–Maddison test, and distance comparisons (Fig. 2C). In general, phylogenetic analyses showed that intra-subject R5 sequences clustered independently from X4 envs, whereas dual-tropic envs were often intermixed with either X4 or R5 viruses.
Fig. 2.
Maximum likelihood (ML) tree analysis of co-circulating viral clones among dual/mixed (DM) env populations. Different shapes are used to represent the tropism of individual env clones: square, R5 clones; triangle, X4 clones; and circle, dual-tropic clones. Asterisk (*) indicates predicted MRCA. Bootstrap values from 1000 replications are shown at nodes and rooted to HXB2 env sequence. (A) ML phylogenies of clones isolated from the individuals in which CXCR4-using envs were segregated from R5 variants. (B) All clones are from the same individual but at different time points. Filled shapes indicate clones isolated from sample DM202, which was collected prior to antiretroviral therapy (ART). Open shapes are clones isolated from sample DM269 after virological failure with ART. (C) Two samples (DM172 and DM173) from 2 separate individual harbored only X4 and dual-tropic clones, which were closely related.
Divergence of co-circulating R5 and CXCR4-using sequences from the calculated most recent common ancestor (MRCA) (Fig. 2, asterisk) was used to estimate intra-subject evolution. In samples with more than one of either CXCR4-using or R5 clones, the median distance from the MRCA was greater for the CXCR4-using rather than for the R5 sequences (p<0.05, Wilcoxon rank sum test), except in DM268 (p=0.35). There were also no significant differences in distance from MRCA between X4 and dual-tropic sequences in DM172 and DM173. All the generated MRCA env sequences had the typical HIV-1C R5 V3 crown motifs of GPGQ with no substitutions, except for in DM8 and DM173 (Table 2). The predominance of the R5 V3 loop crown GPGQ motif and the greater distances of CXCR4-using sequences from the predicted MRCA support the observation that R5 viruses predominate early after infection, and CXCR4-utilizing variants typically emerge later in disease.
X4, dual-tropic, and R5 sequences have unique characteristics
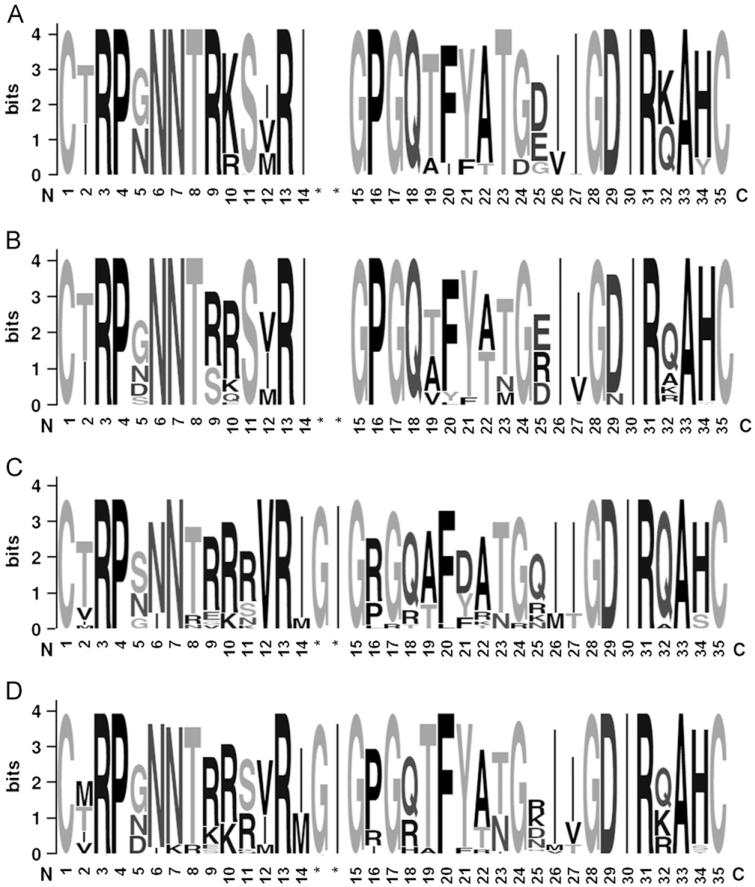
Intra-subject pairwise genetic distances between R5 sequences isolated from the seven subjects with strictly R5 virus (median 0.0096 [range 0.0007–0.016]) were significantly lower compared to the genetic diversity observed among R5 envs isolated from women with DM viruses (median 0.03 [range 0.007–0.05]; p=0.05, 2-tail unpaired t-test). Similarly, the R5 V3 loops cloned from each woman with exclusively R5 virus had lower mean site-specific Shannon Entropy scores compared to R5 viruses cloned from women with DM viruses (mean 0.21 [range 0–1.08] versus 0.31 [0–1.23], respectively), but this difference was not statistically significant (p=0.2). The inter-subject amino acid entropy was significantly lower among R5 V3 loops (median 0 [range 0–1.08]) as compared to X4 (median 0.42 [range 0–1.025]) or dual-tropic (median 0.32 [range 0–1.669]) V3 loops (p=0.03 for each comparison, Wilcoxon rank sum test) (Fig. 3). The GPGQ crown motif was completely conserved in all the isolated R5 sequences (entropy of 0) with no insertions around the crown, while X4 and dual-tropic sequences had much greater variability (median entropy 0.5 and 0.69, respectively) and frequent substitutions around the motif. Interestingly, R5 sequences showed increased variability outside of the V3 loop in the region between C3–C5 (median entropy of 0.262 and 0.236 for clones isolated from R5 and DM samples, respectively) compared to the X4 (median 0, p<0.001) and dual-tropic (median 0, p=0.03) sequences, which were highly conserved.
Fig. 3.
V3 loop sequences with the character and size of each logo representing the proportion of an amino acid at the specific site, and number based on position of alignment with HXB2. The variability at each amino acid position was calculated using the Shannon entropy score and schematically represented by the number of characters at each position, with a single character presenting 0 entropy score. R5 env sequences obtained from individuals infected with a population of exclusive R5 tropic (A) or DM (B) virus. Env sequences of X4 (C) and dual-tropic clones (B) isolated from individuals infected with a population of DM viruses. Asterisk (*) indicates position of amino acid insertions. (A) R5 from R5 quasispecies, (B) R5 from DM quasispecies, (C) X4 from DM quasispecies and (D) Dual-tropic from DM quasispecies.
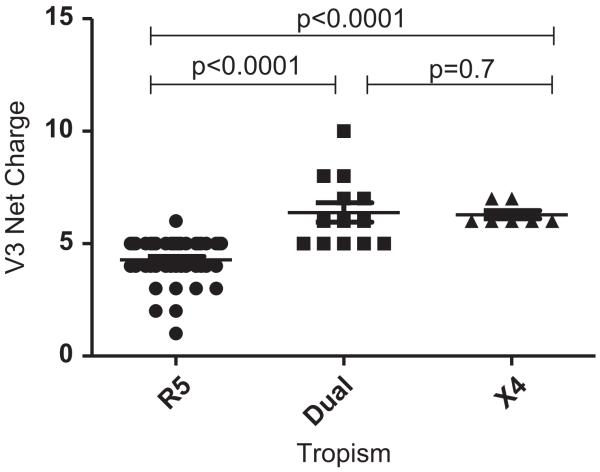
None of the 204 full-length env sequences were identical although several intra-patient sequences had identical V3 loop sequences. Because the number of clones isolated from each subject varied, only unique V3 loop sequences were used for analyses to identify distinct HIV-1C X4 env characteristics. A total of 7 unique X4, 13 unique dual-tropic, and 43 unique R5 V3 loop sequences were compiled from all the clones (Table 3). There was no significant difference in the median length of V3 loops between the unique X4 and R5 sequences (median 36 amino acids (aa) [range 34–37] versus 35 aa [34-36]; p=0.10). The net V3 loop charge was significantly higher among X4 (median+6 [range 6–7]) and dual-tropic viruses (+6 [range 5–10]) as compared to R5 viruses (+5 [range 2–6], both p-values <0.0001) sequences (Fig. 4). The R5 V3 loops had significantly more predicted N-linked glycosylation sites (PNGS) compared to the X4 (p=0.012) and dual-tropic (p=0.025) sequences. Specifically, all unique R5 sequences contained1/4a PNGS at position 6 of the V3 loop. Only some of the unique X4 (5 of 7) and dual-tropic (10 of 13) variants contained a PNGS at position 6, and only 1 of the CXCR4 using variants contained a PNGS at position 7 (Table 2). Interestingly, 8 out of 43 R5 V3 sequences had a second PNGS at position 7, which were all isolated from DM and not R5 quasispecies (Fig. 3A and B).
Fig. 4.
Greater V3 net charge in X4 and dual-tropic sequences compared to R5 env sequences. Each dot represents the net charge of a unique env V3 loop sequence, black lines are medians. x-Axis shows the env tropism, and y-axis displays the calculated charge. P-values from 2-tailed Wilcoxon rank-sum test for pairwise comparison are indicated.
One of the most distinctive properties of the unique CXCR4-using V3 sequences was changes in and around the crown motif. Among unique V3 loop sequences the GPGQ crown displayed an arginine (R) or histidine (H) substitution in 71.4% of the unique X4 (n=7) and 53.8% of dual-tropic envs (n=13) but was conserved in all the unique R5 sequences (n=43) (Fig. 3 and Table 2). The two X4 variants without any amino acid substitution in the GPGQ crown, both isolated from DM172, instead possessed a GI amino acid insertion prior to the crown. This insertion was observed in 45% of the unique X4/dual-tropic V3 loop sequences but was absent in all of the R5 sequences. The prevalence of crown substitutions in other HIV-1C sequences was further investigated in CXCR4-using HIV-1C envs in the Los Alamos National Laboratories (LANL) HIV sequence database, which revealed a total of 42 unique X4 isolates. The coreceptor phenotype for these isolates had been determined by various methods, including the MT2 cell assay and assays using engineered cell lines, such as U87 and Ghost cells. Thirty-seven of the 42 (88.1%) X4 sequences isolated from 20 individual subjects had an R substitution at position 16 and/or R/H at position 18 within the V3 loop crown. Similar to our observation, 4 of the 5 sequences contained a conserved GPGQ crown, of which 3 V3s, isolated from the same patient, had instead a GI insertion prior to the crown and one had an unusual R substitution at position 21. ZAM20 is the only HIV-1C clone in the LANL database that did not contain a positive amino acid substitution in the GPGQ crown or surrounding region (Louwagie et al., 1995; Trkola et al., 1998). In comparison, a search of phenotyped HIV-1C R5 sequences in the LANL database yielded a total of 148 sequences, all of which retained the GPGQ crown, except one with an unusual RPGQ crown motif (accession no. AF391249). Among 15 unique dual-tropic sequences in the database, 9 had a substitution in the crown, 2 had a GI insertion, and only 4 sequences retained the GPGQ crown without any substitutions at the surrounding residues.
Performance of common genotypic predictive algorithms
The ability of commonly used algorithms, such as the 11/25 rule and C-PSSM, to predict coreceptor usage based on the V3 sequence was assessed (Table 3). The 11/25 rule was relatively sensitive in predicting CXCR4 usage among X4 sequences, correctly predicting X4 phenotype in 85.7% of unique sequences and 80% of all 20X4 clones, but with less accuracy in assessing dual-tropic and R5 V3 loop sequences. The 11/25 rule incorrectly predicted CXCR4 usage for R5 clones isolated from 3 DM samples (DM192, 269 and 202) because of the presence of an arginine at position 25 in the V3 loop. In contrast, predictions made by the 11/25 rule were 100% concordant with the phenotype for R5 clones isolated from a population of entirely R5 viruses (86 clones, 20 unique V3).
We also evaluated the predictive value of C-PSSM, an algorithm based on known HIV-1C env sequences and coreceptor phenotype (Table 3). The C-PSSM accurately predicted CXCR4 usage for 100% of all unique X4, and 92% of unique dual-tropic sequences (Table 3). In contrast, C-PSSM accurately predicted CCR5 usage for only 61% of the 43 unique R5 sequences. This decreased accuracy in predicting R5 phenotype was largely influenced by R5 sequences isolated from DM samples with co-circulating X4/dual-tropic viruses. Among R5 clones isolated from samples that harbored exclusively R5 virus, the accuracy was 95% among unique V3 loops. In contrast, among the 23 unique R5 sequences isolated from DM samples CCR5 usage was predicted correctly in only 30%. In comparison to the C-PSSM and 11/25 rule, the presence of one of the two unique X4 V3 loop characteristics we found, either an insertion prior to the crown or substitutions in the GPGQ crown, was 100% predictive for CXCR4-usage among the unique isolated X4 clones. Presence and absence of these two features correctly predicted CXCR4 usage in 10 out of 13 (76.9%) unique dual-tropic and in all of the 43 (100%) unique R5 sequences respectively, regardless of whether the env was isolated from a DM or R5 sample.
Signature amino acids differences between R5 and X4/dual-tropic viruses
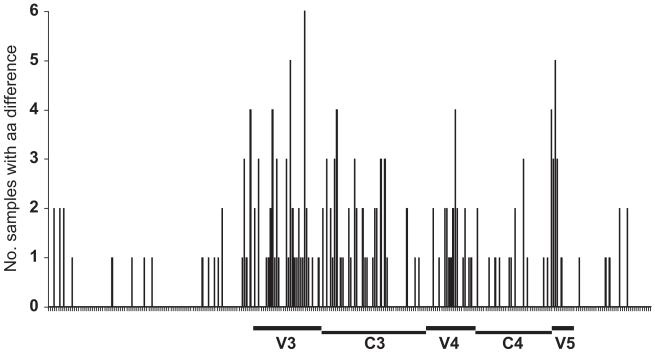
Significant amino acid differences between co-circulating CXCR4-using and R5 sequences isolated from the same subject were identified using VESPA. The 8 intra-subject comparisons were then combined to identify signature env sequence differences across all patients. Although no consistent amino acid was identified, the analysis showed that signature amino acid differences were concentrated in specific regions. The greatest amino acid differences between R5 and X4/dual-tropic sequences were found in the V3 loop region, with some differences in the post-V3 and V5 regions (Fig. 5). Although the V1–V2 region is thought to play a role in determining coreceptor usage (Chohan et al., 2005; Coetzer et al., 2007, 2006; Pollakis et al., 2001), abundant insertions and deletions in this region made alignment of these segments difficult and ambiguous, and thus, many V1–V2 sequences were omitted from this analysis.
Fig. 5.
All env sequences were aligned, and V1–V2, V3, V4–V5 segments corresponding to HXB2 env amino acids were defined. Signature amino acid differences between intra-patient R5 and CXCR4-using clones were mapped relative to the HXB2 env numbering. Signature amino acids were determined using the VESPA program with threshold of 0 and comparison of R5 and CXCR4-using sequences from within each of the 8 subjects. Two DM samples which were not composed of R5 clones, with only X4 and dual-tropic clones, were not included in the analysis.
Determinant of coreceptor usage encoded in V3.
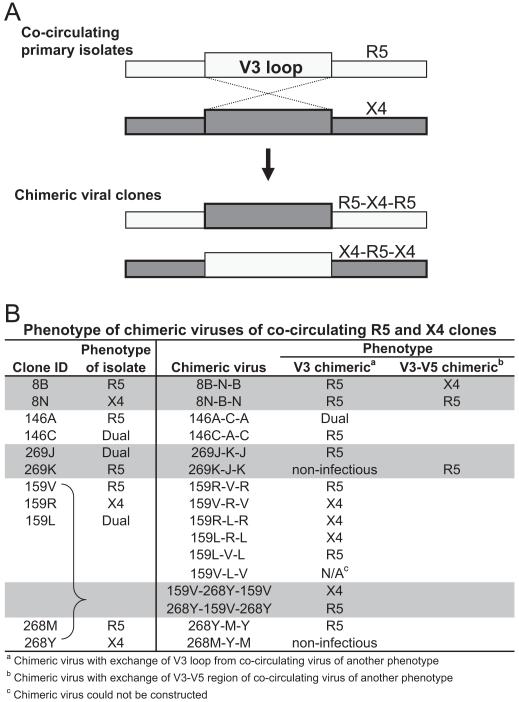
Because amino acid differences between R5 and X4/dual-tropic HIV-1C sequences were concentrated primarily in the V3 loop we exchanged V3 sequences among X4 and R5 clones to examine if coreceptor usage in subtype C depends on the V3 loop. We constructed chimeric viruses by swapping V3 sequences between co-circulating R5 and X4/dual-tropic clones. Chimeric envs were made from isolates from five subjects (Fig. 6A). Exchange of the V3 loop resulted in 13 functional V3 chimeric viruses. In all V3 chimeras, except for 8B-N-B and 159R-L-R, the inserted V3 loop changed the coreceptor usage of the chimeric env to the coreceptor phenotype of the parental V3 loop clone (Fig. 6B). Interestingly, chimeras with V3 loops from original dual-tropic envs were able to use CCR5 and CXCR4 in only one of the four cases (146 A-C-A); two chimeras (159 V-L-V and 269 K-J-K) could not be tested because they yielded env which did not produce infectious pseudovirions, and the fourth chimera (159 R-L-R) in which V3 from a dual-tropic clone was inserted into an X4 background yielded an X4 virus. Similarly, the chimeric 8 B-N-B env, which carried a V3 loop from the X4 8N clone inserted into the R5 8B env backbone, resulted in an R5 phenotype. Because amino acid differences were also concentrated beyond V3, we constructed a few V3–V5 chimeras. Insertion of the X4 V3–V5 region from 8N into the R5 8B env yielded a virus capable of utilizing CXCR4. By contrast, insertion of the V3–V5 segment from the 269J dual-tropic env into a 269 K R5 env resulted in an envelope unable to use CXCR4 for cell entry. In one case, V3 loop exchanges between envs from two different subjects resulted in infectious viruses and a corresponding change in coreceptor phenotype; the V3 loop from subject 268Y X4 env was inserted into the 159 V R5 env and yielded a chimeric env that used the CXCR4 receptor. Conversely, the 159 V R5 V3 loop inserted within the 269Y X4 backbone resulted in an R5 phenotype.
Fig. 6.
Co-circulating viral clones with different coreceptor usage were used as templates for the construction of chimeric env which contained segments of both env. (A) Initially, V3 loop from a R5 env was swapped with a CXCR4-using env using overlapping PCR method, and for some samples a larger region was exchanged. (B) Phenotype of the chimeric envs determined on a phenotypic assay using U87-CD4-CCR5 and U87-CD4-CXCR4 cells.
Discussion
To our knowledge this study is the largest and most comprehensive analysis of CXCR4-using HIV-1C envelope clones to date. We found that in general X4 envs are distinct from R5 variants, while dual-tropic clones often share genetic features with the co-circulating X4 and R5 HIV-1Cs. The majority of distinguishing genotypic characteristics among X4 and R5 strains reside in the env V3 loop. Specifically, all HIV-1C X4 variants had either a two residue insertion prior to the V3 loop crown or basic amino acid substitutions within the generally invariant GPGQ crown motif. Similar to X4 variants of other HIV-1 subtypes, such as clade B, we found that X4 V3 loops of HIV-1C have significantly greater net positive charge and loss of glycosylation sites compared to R5 clones (De Jong et al., 1992; Hoffman et al., 2002; Pollakis et al., 2001). Our chimeric envelope data confirmed that the HIV-1C V3 loop was the primary determinant for coreceptor usage, although in some isolates env regions outside of V3 were also important for conferring CXCR4 usage. Interestingly, we found that R5 clones from subjects with DM viruses demonstrated greater diversity in the V3 loop compared to R5 env sequences from individuals harboring exclusively R5 viruses. This observation has potential implications for understanding the relatively low rate of emergence of CXCR4-using HIV-1C viruses.
We examined a large number of individual env clones (n=209) from 17 different samples with a DM or R5 phenotype (Lin et al., 2011). This approach allowed us to compare both intra- and inter-subject characteristics of X4, R5, and dual-tropic envs. We found that HIV-1C DM viruses compromised varying proportions of X4, R5, and dual-tropic sequences. This observation contrasts with a previous study showing that R5 clones are completely absent among HIV-1C DM isolates (Singh et al., 2009). However, in that earlier study env clones were isolated from proviral DNA after in vitro passage of peripheral blood mononuclear cells, which may have selected for X4 and dual-tropic viruses (Singh et al., 2009). This study is also unique in that most previous studies have compared HIV-1C envelopes of differing coreceptor phenotypes isolated from different subjects and used population-based sequencing strategies to determine env sequences. We directly isolated virus sequences from plasma and examined clonal sequences to overcome some of the limitations inherent in these previous studies. Our intra- and inter-subject comparisons also allowed us to demonstrate that in the majority of cases X4 viruses are phylogenetically distinct from co-circulating R5 strains, even though the diverse X4 variants did not form a unique cluster when we examined viruses from different subjects. On the other hand, dual-tropic strains clustered with either co-circulating R5 or X4 strains in the intra-subject analyses. Together, these observations suggest that as with other subtypes, dual-tropic HIV-1C variants may be evolutionary intermediates between R5 and X4 viruses.
Dual-tropic viruses likely emerge from R5 viruses, and additional selective mutations are required for conformational changes in the coreceptor binding site that presumably increase affinity for CXCR4 while completely restricting binding to CCR5 coreceptor. It has been suggested that dual-tropic viruses have more flexibility in displaying different structural confirmations and binding sites that potentially can allow interactions with both CCR5 and CXCR4 (Cardozo et al., 2007; Hartley et al., 2005; Nolan et al., 2008). The genetic modifications in env that prevent CCR5 use and enhance CXCR4-usage likely result in the phylogenetic separation of R5 and X4 variants within a subject. Future analysis of longitudinal samples may provide additional insight into the emergence and evolution of X4 viruses from dual-tropic and R5 strains.
Our sequence analysis also demonstrated that the V3 loop contained the primary distinguishing features separating X4 and R5 variants. Interestingly, although X4 and R5 envelope V3 loops had distinctive signature motifs, the V3 sequences among co-circulating viruses were still more closely related to each other than to X4 and R5 variants, respectively, from different subjects. Among all unique R5 sequences, the net V3 loop charge was significantly lower than X4 or dual-tropic sequences, which is consistent with earlier observations in smaller numbers of isolates (Coetzer et al., 2006; Johnston et al., 2003). Although we did not find any significant differences between lengths of V3 loop we did find a significant difference in the number of PNGS among R5 compared to X4 or dual-tropic V3 loops, with a conserved site at position 6 among all R5 clones and in many a second prominent glycosylation site. This glycosylation pattern may facilitate the virus-CCR5 interaction (Pollakis et al., 2004, 2001; Polzer et al., 2002). Similar glycosylation changes have been observed in the subtype B and D comparisons between CXCR4-using and R5 variants (Clevestig et al., 2006; Fenyo et al., 1988; Huang et al., 2007). We also found that positive amino acid substitutions or two-amino acid insertions were highly specific for subtype C X4 virus, consistent with previous observations of crown substitution at the 4th position with an R, Y, K or H amino acids in CXCR4-using HIV-1C isolates (Abebe et al., 1999; Batra et al., 2000; Bjorndal et al., 1999; Coetzer et al., 2006; Ping et al., 1999). Interestingly, while crown substitutions and pre-crown insertions were observed among the majority (90%) of X4 envs, these characteristics were present in some but were not a necessary and dominating characteristic of the isolated dual-tropic clones. The crown motif forms the beta turn in the env secondary structure, and basic amino acid substitutions or the two residue insertion likely resulted in conformational changes that allow for better CXCR4 versus CCR5 receptor interactions (Hartley et al., 2005; Hu et al., 2000; Pollakis et al., 2004; Suphaphiphat et al., 2007). Besides changing coreceptor usage these V3 loop modifications may also change V3 presentation and subsequently result in differences in antibody reactivity against X4 versus R5 isolates. Antibodies against the HIV-1C V3 domain are rarely elicited early in infection (Bou-Habib et al., 1994; Li et al., 2006), and it remains unclear if they appear with greater frequency against X4 variants, potentially explaining the limited emergence of CXCR4 using viruses during HIV-1C infection.
We further confirmed the importance of the envelope V3 loop as a determinant for coreceptor usage by constructing and examining chimeric envelopes. Few previous studies have done similar functional analyses to confirm whether certain env regions influence coreceptor usage among HIV-1C viruses. One study constructed chimeric envelopes from a single subject infected with HIV-1 subtype C and showed that the principal determinant for coreceptor usage was in the V3 loop although changes in the V4 regions were speculated as compensatory or stabilizing mutations (Zhang et al., 2010). We observed that sequences outside the V3 loop are likely involved in determining coreceptor usage because some dual-tropic clones had identical V3 loops as co-circulating R5 or X4 viruses. This finding may also explain the inability of some previously described genotypic methods to accurately predict coreceptor usage in all cases solely based on V3 loop sequences. For example, some dual-tropic V3 loops from DM159, DM173, and DM263 were identical to the related X4 or R5 variants. These findings also suggest that V3 loop mutations that distinguish pure X4 and R5 variants are only established after modifications outside the V3 loop that allow for both CXCR4 and CCR5 usage among HIV-1C. Our chimeric envelope constructions incorporating V3–V5 env regions confirm that non-V3 segments are often necessary elements in dictating coreceptor usage among HIV-1C. Envelope V1–V2 loop modifications potentially also contribute to coreceptor switching (Hoffman et al., 2002; Labrosse et al., 2001), but we did not examine this issue in our sequence or chimera analysis. Extensive V1–V2 variability makes alignments and phylogenic interpretations unreliable. Our chimera studies clearly show that in the majority of cases coreceptor usage is determined by the V3 loop. Exchanging V3 loop segments between two different subjects demonstrates that changes in the V3 loop can confer coreceptor switching even when other portions of the envelope are not highly homologous. Some sequences, such as DM8, require V3–V5 elements for changing coreceptor usage. And in subject DM269, we were unable to generate a dual-tropic variant within a R5 envelope backbone suggesting that it may require other segments beyond V3–V5, such as the V1–V2 loops.
Our large genotype-phenotype dataset allowed us to test the accuracy of the two most commonly used genotypic methods for determining coreceptor usage at a clonal level. Both C-PSSM and the 11/25 rule had high sensitivity for predicting X4 usage among HIV-1C CXCR4-using clones, but the algorithms were associated with high false positive rates among co-circulating R5 clones. False positive identification of CXCR4-usage may have occurred because the C-PSSM algorithm was based on a training set of 279 subtype C sequences derived predominantly from population sequencing, and coreceptor usage phenotype was determined using a variety of assays (Jensen et al., 2006). In addition, a previous study using HIV-1C isolates from a similar population of subjects from Botswana found that the 11/25 rule predicted CXCR4 use only 39% of the time (Ndung’u et al., 2006). Sequences in that study also were obtained by population sequencing and likely failed to detect genotypic differences in minority CXCR4-using species. These observations suggest that population sequences of R5 and X4 variants may inadequately identify salient genotypic changes necessary for CXCR4 usage in subtype C. Our analyses also highlight the high prevalence of pre-crown insertions or basic amino substitutions within the GPGQ motif specifically in X4 variants. This genotype signature (basic amino acid insertion or substitution in the GPGQ crown) was present in 41 of 42 unique HIV-1C X4 sequences present in the LANL database. Unfortunately, this signature sequence motif fails to detect CXCR4 usage in all dual-tropic strains, thus the presence of this signature motif can only be used to exclude candidates for CCR5 antagonist therapy in the absence of phenotypic data. Identifying appropriate HIV-1C infected candidates for CCR5 inhibitors will still require improvement in the accuracy of genotypic algorithms or phenotypic data.
Interestingly, we found that the 11/25 rule and C-PSSM algorithm were better at predicting CCR5 usage in clones isolated from subjects with exclusively R5 virus as opposed to clones isolated from individuals with DM viruses. In addition, we found that R5 clones from subjects with strictly R5 viruses had less env amino acid variability compared to R5 envs from subjects with a mixture of viruses using both the CCR5 and CXCR4 coreceptor. It is possible that the selective pressures on a quasispecies with X4 variants affect co-circulating R5 viruses in ways which give them less “typical” R5 features, leading to the poor performance of genotypic algorithms. Differences in selective pressure may also explain differences in diversity observed among R5 env sequences isolated from subjects with R5 versus DM quasispecies. On the other hand, CXCR4-using viruses in individuals with DM quasispecies may emerge only after some threshold level of env sequence variability is achieved in the infected host. Only longitudinal analysis can help determine whether HIV-1C CXCR4 emergence depends on a pre-existing level of env sequence diversity or whether presence of dual-tropic and X4 viruses generate more virus divergence.
This study had several limitations to consider. We did not employ single genome amplification (SGA), which may limit interpretation of some of the results from the study because of possibility of resampling and PCR-mediated recombination (Fang et al., 1998; Liu et al., 1996; Meyerhans et al., 1990). The env genes were initially amplified in an earlier study to determine the coreceptor usage (Lin et al., 2011). We did employ PCR conditions to minimize founder effects and recombination. Analyses of our env sequence datasets using the Recombinant Identification Program (RIP) (http://www.hiv.lanl.gov) and Genetic Algorithm for Recombination Detection (GARD) (Kosakovsky Pond et al., 2006) suggest that some isolated sequences could be recombinants although the programs failed to identify a consistent breakpoint among all the clones within a subject. It remains unclear, however, if potential recombination occurred in vivo or during PCR amplification. Failure to observe env sequences which were phylogenetically in-between the X4 and R5 variants suggests that distinct clustering among the exclusively CCR5 and CXCR4-using viruses was likely not an artifact of bulk PCR, and in fact represent sequences circulating in vivo. We potentially could have missed phylogenetically intermediate recombinant sequences because of limited sequence sampling during the clonal analysis. However, a recent study comparing traditional PCR cloning with SGA did not find that one method was more biased than the other in isolating certain species, and the level of diversity of an HIV quasispecies obtained from both methods was similar (Jordan et al., 2010). Even though this is one of the largest sets of env clones with associated phenotypic data on coreceptor usage, we still had a limited number of DM samples because of the small number of HIV-1C individuals with CXCR4-using viruses. Lastly, the CXCR4-using isolates in this study were not isolated at the initial time of coreceptor switching, during which we could most likely identify the minimal genetic changes necessary for CXCR4 usage. This was not possible as this cohort study did not have such extensive longitudinal samples.
Overall, this study adds to the limited genetic characterization of coreceptor usage in HIV-1C, which has previously been restricted by the small number of HIV-1C CXCR4-using isolates available, and the heterogeneity of the sequence and phenotype information. To our knowledge this is the first study to decipher the genetic determinants of CXCR4 usage in HIV-1C at a clonal level, among co-circulating clones of different coreceptor phenotype, and in a large panel of subjects harboring DM and strictly R5 HIV-1C. The observations made in this study extend our current knowledge about HIV-1C coreceptor usage, and provide one of the largest datasets of env clonal sequence and corresponding coreceptor phenotype in HIV-1C isolates. Our findings are relevant for effective therapeutics and coreceptor based prevention strategies against the large and rapidly spreading HIV-1C epidemic.
Materials and methods
Study subjects, cloning and phenotypic coreceptor testing
HIV-1 envelope clones were isolated from plasma from nine women (ten different plasma samples) previously shown to harbor DM virus and seven women harboring exclusively R5 virus (Lin et al., 2011). Samples were from selected women in the Mashi study, which compared different strategies for prevention of mother-to-child transmission in Botswana (clinicaltrials.gov identifier: NCT00197587) (Lockman et al., 2007; Thior et al., 2006). The use of samples was approved by the institutional review boards in both United States institutions (Harvard School of Public Health and Partners Healthcare Systems) and by the Botswana Ministry of Health.
Co-receptor usage was determined as previously described (Lin et al., 2010). Briefly, env amplicons were isolated from plasma by pooling independent nested RT-PCR reactions. The resulting amplicons were then cloned into the TOPO-TA vector (Invitrogen). The CMV promoter was attached to individual full-length env genes using overlapping PCR. Pseudotyped viruses were generated by co-transfecting each env clone with the CMV promoter and an env deficient HIV-1 backbone (pNL4-3R-E-) plasmid expressing luciferase. Coreceptor usage was determined by assessing entry capacity in the presence and absence of a coreceptor antagonist on U87 cells expressing CD4 and either CXCR4 or CCR5.
Sequence analyses
Full-length bidirectional sequencing of env clones which produced viable virions for coreceptor determination was performed using standard primers (Sanders-Buell and McCutchan, 1995). All env sequences will be deposited in Genbank (accession numbers pending). FindModel (http://www.hiv.lanl.gov) was employed to determine the parameters for a best-fit evolutionary model, which were used to construct maximum likelihood trees using PhyML v3.0, with stability of the nodes assessed by bootstrap resampling at 100 iterations. NJ trees were constructed and edited using MEGA (Kumar et al., 1994) using all isolated env sequences and reference sequences of different subtypes obtained from the LANL HIV sequence database (http://www.hiv.lanl.gov). Segregation among different sequences was examined using the Slatkin–Maddison test as implemented on MacCLade, version 4.01, and maximum composite likelihood distances as calculated using MEGA. Viral diversity was estimated as the average pairwise genetic distance using PAUP version 4.02b2a using the previously estimated maximum likelihood model. The divergence of each sequence from the MRCA was calculated in DIVEIN (Deng et al., 2010). Sequence variability at each amino acid position was determined by calculating Shannon entropy scores using the LANL Entropy-one tool (Korber et al., 1993) with comparisons made using the two-tailed paired t-test. N-linked glycosylation sites were predicted using the N-Glycosite tool from the LANL database (Zhang et al., 2004). Signature amino acid differences were identified using the VESPA program from the LANL database, and histograms displaying the number of differences at each site were constructed using the Microsoft Excel program.
Construction of chimeric viruses
Chimeric env sequences were constructed by swapping a region from a R5 clone with the corresponding region from a X4 or dual-tropic env using overlapping PCR (Kirchherr et al., 2007); specific primer sequences are available upon request. Chimeras were confirmed by sequence analysis, and the coreceptor usage of these chimeric envelopes was determined as previously detailed (Lin et al., 2010).
Statistical analysis
P-values were determined by 2-sided tests, with values less than or equal to 0.05 considered significant. All statistical tests were performed with GraphPad Prism software version 5.0 (Graph Pad Software Inc., San Diego, CA) and Stata version 8.0 (Stata Corporation).
Acknowledgments
Dr. Lin was supported by the Harvard Center for AIDS Research (CFAR) Scholar Award, Hearst Fund and Burroughs-Wellcome ASTMH Research Fellowship. This work was supported by NIH Grants RR016482, R37 AI055357 and U01 AI068636 (an ACTG Virology Support contract) to D.R.K. and AI1077473 to M.S.
References
- Abebe A, Demissie D, Goudsmit J, Brouwer M, Kuiken CL, Pollakis G, Schuitemaker H, Fontanet AL, Rinke de Wit TF. HIV-1 subtype C syncytium- and non-syncytium-inducing phenotypes and coreceptor usage among Ethiopian patients with AIDS. AIDS. 1999;13(11):1305–1311. doi: 10.1097/00002030-199907300-00006. [DOI] [PubMed] [Google Scholar]
- Batra M, Tien PC, Shafer RW, Contag CH, Katzenstein DA. HIV type 1 envelope subtype C sequences from recent seroconverters in Zimbabwe. AIDS Res. Hum. Retroviruses. 2000;16(10):973–979. doi: 10.1089/08892220050058399. [DOI] [PubMed] [Google Scholar]
- Bjorndal A, Sonnerborg A, Tscherning C, Albert J, Fenyo EM. Phenotypic characteristics of human immunodeficiency virus type 1 subtype C isolates of Ethiopian AIDS patients. AIDS Res. Hum. Retroviruses. 1999;15(7):647–653. doi: 10.1089/088922299310944. [DOI] [PubMed] [Google Scholar]
- Bou-Habib DC, Roderiquez G, Oravecz T, Berman PW, Lusso P, Norcross MA. Cryptic nature of envelope V3 region epitopes protects primary monocytotropic human immunodeficiency virus type 1 from antibody neutralization. J. Virol. 1994;68(9):6006–6013. doi: 10.1128/jvi.68.9.6006-6013.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo T, Kimura T, Philpott S, Weiser B, Burger H, Zolla-Pazner S. Structural basis for coreceptor selectivity by the HIV type 1 V3 loop. AIDS Res. Hum. Retroviruses. 2007;23(3):415–426. doi: 10.1089/aid.2006.0130. [DOI] [PubMed] [Google Scholar]
- Cecilia D, Kulkarni SS, Tripathy SP, Gangakhedkar RR, Paranjape RS, Gadkari DA. Absence of coreceptor switch with disease progression in human immunodeficiency virus infections in India. Virology. 2000;271(2):253–258. doi: 10.1006/viro.2000.0297. [DOI] [PubMed] [Google Scholar]
- Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath PD, Wu L, Mackay CR, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85(7):1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- Choe H, Li W, Wright PL, Vasilieva N, Venturi M, Huang CC, Grundner C, Dorfman T, Zwick MB, Wang L, Rosenberg ES, Kwong PD, Burton DR, Robinson JE, Sodroski JG, Farzan M. Tyrosine sulfation of human antibodies contributes to recognition of the CCR5 binding region of HIV-1 gp120. Cell. 2003;114(2):161–170. doi: 10.1016/s0092-8674(03)00508-7. [DOI] [PubMed] [Google Scholar]
- Chohan B, Lang D, Sagar M, Korber B, Lavreys L, Richardson B, Overbaugh J. Selection for human immunodeficiency virus type 1 envelope glycosylation variants with shorter V1–V2 loop sequences occurs during transmission of certain genetic subtypes and may impact viral RNA levels. J. Virol. 2005;79(10):6528–6531. doi: 10.1128/JVI.79.10.6528-6531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cilliers T, Nhlapo J, Coetzer M, Orlovic D, Ketas T, Olson WC, Moore JP, Trkola A, Morris L. The CCR5 and CXCR4 coreceptors are both used byhuman immunodeficiency virus type 1 primary isolates from subtype C. J. Virol. 2003;77(7):4449–4456. doi: 10.1128/JVI.77.7.4449-4456.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevestig P, Pramanik L, Leitner T, Ehrnst A. CCR5 use by human immunodeficiency virus type 1 is associated closely with the gp120 V3 loop N-linked glycosylation site. J. Gen. Virol. 2006;87(3):607–612. doi: 10.1099/vir.0.81510-0. [DOI] [PubMed] [Google Scholar]
- Coetzer M, Cilliers T, Papathanasopoulos M, Ramjee G, Karim SA, Williamson C, Morris L. Longitudinal analysis of HIV type 1 subtype C envelope sequences from South Africa. AIDS Res. Hum. Retroviruses. 2007;23(2):316–321. doi: 10.1089/aid.2006.0207. [DOI] [PubMed] [Google Scholar]
- Coetzer M, Cilliers T, Ping LH, Swanstrom R, Morris L. Genetic characteristics of the V3 region associated with CXCR4 usage in HIV-1 subtype C isolates. Virology. 2006;356(1-2):95–105. doi: 10.1016/j.virol.2006.07.030. [DOI] [PubMed] [Google Scholar]
- Connell BJ, Michler K, Capovilla A, Venter WD, Stevens WS, Papathanasopoulos MA. Emergence of X4 usage among HIV-1 subtype C: evidence for an evolving epidemic in South Africa. AIDS. 2008;22(7):896–899. doi: 10.1097/QAD.0b013e3282f57f7a. [DOI] [PubMed] [Google Scholar]
- De Jong JJ, De Ronde A, Keulen W, Tersmette M, Goudsmit J. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J. Virol. 1992;66(11):6777–6780. doi: 10.1128/jvi.66.11.6777-6780.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Maust BS, Nickle DC, Learn GH, Liu Y, Heath L, Kosakovsky Pond SL, Mullins JI. DIVEIN: a web server to analyze phylogenies, sequence divergence, diversity, and informative sites. Biotechniques. 2010;48(5):405–408. doi: 10.2144/000113370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G, Zhu G, Burger H, Keithly JS, Weiser B. Minimizing DNA recombination during long RT-PCR. J. Virol. Methods. 1998;76(1-2):139–148. doi: 10.1016/s0166-0934(98)00133-5. [DOI] [PubMed] [Google Scholar]
- Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272(5263):872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- Fenyo EM, Morfeldt-Manson L, Chiodi F, Lind B, von Gegerfelt A, Albert J, Olausson E, Asjo B. Distinct replicative and cytopathic characteristics of human immunodeficiency virus isolates. J. Virol. 1988;62(11):4414–4419. doi: 10.1128/jvi.62.11.4414-4419.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley O, Klasse PJ, Sattentau QJ, Moore JP. V3: HIV’s switch-hitter. AIDS Res. Hum. Retroviruses. 2005;21(2):171–189. doi: 10.1089/aid.2005.21.171. [DOI] [PubMed] [Google Scholar]
- Hoffman NG, Seillier-Moiseiwitsch F, Ahn J, Walker JM, Swanstrom R. Variability in the human immunodeficiency virus type 1 gp120 Env protein linked to phenotype-associated changes in the V3 loop. J. Virol. 2002;76(8):3852–3864. doi: 10.1128/JVI.76.8.3852-3864.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu QX, Barry AP, Wang ZX, Connolly SM, Peiper SC, Greenberg ML. Evolution of the human immunodeficiency virus type 1 envelope during infection reveals molecular corollaries of specificity for coreceptor utilization and AIDS pathogenesis. J. Virol. 2000;74(24):11858–11872. doi: 10.1128/jvi.74.24.11858-11872.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Eshleman SH, Toma J, Fransen S, Stawiski E, Paxinos EE, Whitcomb JM, Young AM, Donnell D, Mmiro F, Musoke P, Guay LA, Jackson JB, Parkin NT, Petropoulos CJ. Coreceptor tropism in human immunodeficiency virus type 1 subtype D: high prevalence of CXCR4 tropism and heterogeneous composition of viral populations. J. Virol. 2007;81(15):7885–7893. doi: 10.1128/JVI.00218-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Japour AJ, Fiscus SA, Arduino JM, Mayers DL, Reichelderfer PS, Kuritzkes DR. Standardized microtiter assay for determination of syncytium-inducing phenotypes of clinical human immunodeficiency virus type 1 isolates. J. Clin. Microbiol. 1994;32(9):2291–2294. doi: 10.1128/jcm.32.9.2291-2294.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen MA, Coetzer M, van’t Wout AB, Morris L, Mullins JI. A reliable phenotype predictor for human immunodeficiency virus type 1 subtype C based on envelope V3 sequences. J. Virol. 2006;80(10):4698–4704. doi: 10.1128/JVI.80.10.4698-4704.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston ER, Zijenah LS, Mutetwa S, Kantor R, Kittinunvorakoon C, Katzenstein DA. High frequency of syncytium-inducing and CXCR4-tropic viruses among human immunodeficiency virus type 1 subtype C-infected patients receiving antiretroviral treatment. J. Virol. 2003;77(13):7682–7688. doi: 10.1128/JVI.77.13.7682-7688.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan MR, Kearney M, Palmer S, Shao W, Maldarelli F, Coakley EP, Chappey C, Wanke C, Coffin JM. Comparison of standard PCR/cloning to single genome sequencing for analysis of HIV-1 populations. J. Virol. Methods. 2010;168(1-2):114–120. doi: 10.1016/j.jviromet.2010.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchherr JL, Lu X, Kasongo W, Chalwe V, Mwananyanda L, Musonda RM, Xia SM, Scearce RM, Liao HX, Montefiori DC, Haynes BF, Gao F. High throughput functional analysis of HIV-1 env genes without cloning. J. Virol. Methods. 2007;143(1):104–111. doi: 10.1016/j.jviromet.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koot M, Keet IP, Vos AH, de Goede RE, Roos MT, Coutinho RA, Miedema F, Schellekens PT, Tersmette M. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann. Intern. Med. 1993;118(9):681–688. doi: 10.7326/0003-4819-118-9-199305010-00004. [DOI] [PubMed] [Google Scholar]
- Korber BT, Farber RM, Wolpert DH, Lapedes AS. Covariation of mutations in the V3 loop of human immunodeficiency virus type 1 envelope protein: an information theoretic analysis. Proc. Natl. Acad. Sci. USA. 1993;90(15):7176–7180. doi: 10.1073/pnas.90.15.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. GARD: a genetic algorithm for recombination detection. Bioinformatics. 2006;22(24):3096–3098. doi: 10.1093/bioinformatics/btl474. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA: Molecular Evolutionary Genetics Analysis software for microcomputers. Comput. Appl. Biosci. 1994;10(2):189–191. doi: 10.1093/bioinformatics/10.2.189. [DOI] [PubMed] [Google Scholar]
- Labrosse B, Treboute C, Brelot A, Alizon M. Cooperation of the V1/V2 and V3 domains of human immunodeficiency virus type 1 gp120 for interaction with the CXCR4 receptor. J. Virol. 2001;75(12):5457–5464. doi: 10.1128/JVI.75.12.5457-5464.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Salazar-Gonzalez JF, Derdeyn CA, Morris L, Williamson C, Robinson JE, Decker JM, Li Y, Salazar MG, Polonis VR, Mlisana K, Karim SA, Hong K, Greene KM, Bilska M, Zhou J, Allen S, Chomba E, Mulenga J, Vwalika C, Gao F, Zhang M, Korber BT, Hunter E, Hahn BH, Montefiori DC. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa. J. Virol. 2006;80(23):11776–11790. doi: 10.1128/JVI.01730-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin NH, Negusse DM, Beroukhim R, Giguel F, Lockman S, Essex M, Kuritzkes DR. The design and validation of a novel phenotypic assay to determine HIV-1 coreceptor usage of clinical isolates. J. Virol. Methods. 2010;169(1):39–46. doi: 10.1016/j.jviromet.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin NH, Smeaton LM, Giguel F, Novitsky V, Moyo S, Mitchell RM, Makhema J, Essex M, Lockman S, Kuritzkes DR. Prevalence and clinical associations of CXCR4-using HIV-1 among treatment-naive subtype C-infected women in Botswana. J. Acquir. Immune Defic. Syndr. 2011;57(1):46–50. doi: 10.1097/QAI.0b013e318214fe27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SL, Rodrigo AG, Shankarappa R, Learn GH, Hsu L, Davidov O, Zhao LP, Mullins JI. HIV quasispecies and resampling. Science. 1996;273(5274):415–416. doi: 10.1126/science.273.5274.415. [DOI] [PubMed] [Google Scholar]
- Lockman S, Shapiro RL, Smeaton LM, Wester C, Thior I, Stevens L, Chand F, Makhema J, Moffat C, Asmelash A, Ndase P, Arimi P, van Widenfelt E, Mazhani L, Novitsky V, Lagakos S, Essex M. Response to antiretro-viral therapy after a single, peripartum dose of nevirapine. N. Engl. J. Med. 2007;356(2):135–147. doi: 10.1056/NEJMoa062876. [DOI] [PubMed] [Google Scholar]
- Louwagie J, Janssens W, Mascola J, Heyndrickx L, Hegerich P, van der Groen G, McCutchan FE, Burke DS. Genetic diversity of the envelope glycoprotein from human immunodeficiency virus type 1 isolates of African origin. J. Virol. 1995;69(1):263–271. doi: 10.1128/jvi.69.1.263-271.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddon PJ, Dalgleish AG, McDougal JS, Clapham PR, Weiss RA, Axel R. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell. 1986;47(3):333–348. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- McDougal JS, Kennedy MS, Sligh JM, Cort SP, Mawle A, Nicholson JK. Binding of HTLV-III/LAV to T4+ T cells by a complex of the 110K viral protein and the T4 molecule. Science. 1986;231(4736):382–385. doi: 10.1126/science.3001934. [DOI] [PubMed] [Google Scholar]
- Melby T, Despirito M, Demasi R, Heilek-Snyder G, Greenberg ML, Graham N. HIV-1 coreceptor use in triple-class treatment-experienced patients: baseline prevalence, correlates, and relationship to enfuvirtide response. J. Infect. Dis. 2006;194(2):238–246. doi: 10.1086/504693. [DOI] [PubMed] [Google Scholar]
- Meyerhans A, Vartanian JP, Wain-Hobson S. DNA recombination during PCR. Nucleic Acids Res. 1990;18(7):1687–1691. doi: 10.1093/nar/18.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michler K, Connell BJ, Venter WD, Stevens WS, Capovilla A, Papathanasopoulos MA. Genotypic characterization and comparison of full-length envelope glycoproteins from South African HIV type 1 subtype C primary isolates that utilize CCR5 and/or CXCR4. AIDS Res. Hum. Retroviruses. 2008;24(5):743–751. doi: 10.1089/aid.2007.0304. [DOI] [PubMed] [Google Scholar]
- Ndung’u T, Sepako E, McLane MF, Chand F, Bedi K, Gaseitsiwe S, Doualla-Bell F, Peter T, Thior I, Moyo SM, Gilbert PB, Novitsky VA, Essex M. HIV-1 subtype C in vitro growth and coreceptor utilization. Virology. 2006;347(2):247–260. doi: 10.1016/j.virol.2005.11.047. [DOI] [PubMed] [Google Scholar]
- Nickle DC, Shriner D, Mittler JE, Frenkel LM, Mullins JI. Importance and detection of virus reservoirs and compartments of HIV infection. Curr. Opin. Microbiol. 2003;6(4):410–416. doi: 10.1016/s1369-5274(03)00096-1. [DOI] [PubMed] [Google Scholar]
- Nolan KM, Jordan AP, Hoxie JA. Effects of partial deletions within the human immunodeficiency virus type 1 V3 loop on coreceptor tropism and sensitivity to entry inhibitors. J. Virol. 2008;82(2):664–673. doi: 10.1128/JVI.01793-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmanov S, Pattou C, Walker N, Schwardlander B, Esparza J. Estimated global distribution and regional spread of HIV-1 genetic subtypes in the year 2000. J. Acquir. Immune Defic. Syndr. 2002;29(2):184–190. doi: 10.1097/00042560-200202010-00013. [DOI] [PubMed] [Google Scholar]
- Papathanasopoulos MA, Cilliers T, Morris L, Mokili JL, Dowling W, Birx DL, McCutchan FE. Full-length genome analysis of HIV-1 subtype C utilizing CXCR4 and intersubtype recombinants isolated in South Africa. AIDS Res. Hum. Retroviruses. 2002;18(12):879–886. doi: 10.1089/08892220260190362. [DOI] [PubMed] [Google Scholar]
- Ping LH, Nelson JA, Hoffman IF, Schock J, Lamers SL, Goodman M, Vernazza P, Kazembe P, Maida M, Zimba D, Goodenow MM, Eron JJ, Jr., Fiscus SA, Cohen MS, Swanstrom R. Characterization of V3 sequence heterogeneity in subtype C human immunodeficiency virus type 1 isolates from Malawi: underrepresentation of X4 variants. J. Virol. 1999;73(8):6271–6281. doi: 10.1128/jvi.73.8.6271-6281.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollakis G, Abebe A, Kliphuis A, Chalaby MI, Bakker M, Mengistu Y, Brouwer M, Goudsmit J, Schuitemaker H, Paxton WA. Phenotypic and genotypic comparisons of CCR5- and CXCR4-tropic human immunodeficiency virus type 1 biological clones isolated from subtype C-infected individuals. J. Virol. 2004;78(6):2841–2852. doi: 10.1128/JVI.78.6.2841-2852.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollakis G, Kang S, Kliphuis A, Chalaby MI, Goudsmit J, Paxton WA. N-linked glycosylation of the HIV type-1 gp120 envelope glycoprotein as a major determinant of CCR5 and CXCR4 coreceptor utilization. J. Biol. Chem. 2001;276(16):13433–13441. doi: 10.1074/jbc.M009779200. [DOI] [PubMed] [Google Scholar]
- Polzer S, Dittmar MT, Schmitz H, Schreiber M. The N-linked glycan g15 within the V3 loop of the HIV-1 external glycoprotein gp120 affects coreceptor usage, cellular tropism, and neutralization. Virology. 2002;304(1):70–80. doi: 10.1006/viro.2002.1760. [DOI] [PubMed] [Google Scholar]
- Richman DD, Bozzette SA. The impact of the syncytium-inducing phenotype of human immunodeficiency virus on disease progression. J. Infect. Dis. 1994;169(5):968–974. doi: 10.1093/infdis/169.5.968. [DOI] [PubMed] [Google Scholar]
- Sanders-Buell ES, M O, McCutchan FE. A Compilation and analysis of nucleic acid and amino acid sequences. Human Retroviruses and AIDS. 1995 Part III-15. [Google Scholar]
- Shioda T, Levy JA, Cheng-Mayer C. Macrophage and T cell-line tropisms of HIV-1 are determined by specific regions of the envelope gp120 gene. Nature. 1991;349(6305):167–169. doi: 10.1038/349167a0. [DOI] [PubMed] [Google Scholar]
- Singh A, Page T, Moore PL, Allgaier RL, Hiramen K, Coovadia HM, Walker BD, Morris L, Ndung’u T. Functional and genetic analysis of coreceptor usage by dualtropic HIV-1 subtype C isolates. Virology. 2009;393(1):56–67. doi: 10.1016/j.virol.2009.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suphaphiphat P, Essex M, Lee TH. Mutations in the V3 stem versus the V3 crown and C4 region have different effects on the binding and fusion steps of human immunodeficiency virus type 1 gp120 interaction with the CCR5 coreceptor. Virology. 2007;360(1):182–190. doi: 10.1016/j.virol.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thior I, Lockman S, Smeaton LM, Shapiro RL, Wester C, Heymann SJ, Gilbert PB, Stevens L, Peter T, Kim S, van Widenfelt E, Moffat C, Ndase P, Arimi P, Kebaabetswe P, Mazonde P, Makhema J, McIntosh K, Novitsky V, Lee TH, Marlink R, Lagakos S, Essex M. Breastfeeding plus infant zidovudine prophylaxis for 6 months vs. formula feeding plus infant zidovudine for 1 month to reduce mother-to-child HIV transmission in Botswana: a randomized trial: the Mashi Study. J. Am. Med. Assoc. 2006;296(7):794–805. doi: 10.1001/jama.296.7.794. [DOI] [PubMed] [Google Scholar]
- Tien PC, Chiu T, Latif A, Ray S, Batra M, Contag CH, Zejena L, Mbizvo M, Delwart EL, Mullins JI, Katzenstein DA. Primary subtype C HIV-1 infection in Harare, Zimbabwe. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 1999;20(2):147–153. doi: 10.1097/00042560-199902010-00006. [DOI] [PubMed] [Google Scholar]
- Trkola A, Paxton WA, Monard SP, Hoxie JA, Siani MA, Thompson DA, Wu L, Mackay CR, Horuk R, Moore JP. Genetic subtype-independent inhibition of human immunodeficiency virus type 1 replication by CC and CXC chemokines. J. Virol. 1998;72(1):396–404. doi: 10.1128/jvi.72.1.396-404.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tscherning C, Alaeus A, Fredriksson R, Bjorndal A, Deng H, Littman DR, Fenyo EM, Albert J. Differences in chemokine coreceptor usage between genetic subtypes of HIV-1. Virology. 1998;241(2):181–188. doi: 10.1006/viro.1997.8980. [DOI] [PubMed] [Google Scholar]
- UNAIDS UNAIDS: AIDS Epidemic Update. UNAIDS, Ed. 2009 [Google Scholar]
- van Rensburg EJ, Smith TL, Zeier M, Robson B, Sampson C, Treurnicht F, Engelbrecht S. Change in co-receptor usage of current South African HIV-1 subtype C primary isolates. AIDS. 2002;16(18):2479–2480. doi: 10.1097/00002030-200212060-00015. [DOI] [PubMed] [Google Scholar]
- Zhang H, Tully DC, Zhang T, Moriyama H, Thompson J, Wood C. Molecular determinants of HIV-1 subtype C coreceptor transition from R5 to R5 4. Virology. 2010;407(1):68–79. doi: 10.1016/j.virol.2010.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Huang Y, He T, Cao Y, Ho DD. HIV-1 subtype and second-receptor use. Nature. 1996;383(6603):768. doi: 10.1038/383768a0. [DOI] [PubMed] [Google Scholar]
- Zhang M, Gaschen B, Blay W, Foley B, Haigwood N, Kuiken C, Korber B. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology. 2004;14(12):1229–1246. doi: 10.1093/glycob/cwh106. [DOI] [PubMed] [Google Scholar]