Abstract
Next-generation DNA sequencing of human tumors has led to discovery of chromoanagenesis, in which large numbers of complex rearrangements occur at one or a few chromosomal loci in a single catastrophic event. Two mechanisms underlie these rearrangements, both of which can be facilitated by a mitotic chromosome segregation error to produce a micronucleus containing the chromosome to undergo rearrangement. In the first, chromosome shattering (called chromothripsis) is produced by mitotic entry before completion of DNA replication within the micronucleus, with failure to disassemble the micronuclear envelope encapsulating the chromosomal fragments for random reassembly in the subsequent interphase. Alternatively, locally defective DNA replication (also potentially within a micronucleus) initiates serial, microhomology-mediated template switching (called chromoanasynthesis) that produces local rearrangements with altered gene copy numbers. Complex, localized rearrangements are present in a broad spectrum of tumors and in individuals with congenital or developmental defects, highlighting the impact of chromoanagenesis in human disease.
Introduction
Karyotype abnormalities take the form of numerical and structural alterations in chromosomes and are a defining feature of the cancer cell genome. Structural rearrangements in chromosomes are caused by erroneous repair of DNA double strand breaks and include deletions, duplications, inversions and translocations. Recurrent translocations are common in hematological malignancies, where they have been shown to drive tumorigenesis through the creation of fusion genes derived from portions of two normal genes joined together1. In addition, rearrangements also contribute to disruption of tumor suppressor genes and amplification of oncogenes.
The advent of high throughput DNA sequencing has enabled the interrogation of the cancer genome in unprecedented detail. Catalogues of the somatic mutations present in cancer cells are rapidly appearing (http://www.sanger.ac.uk/genetics/CGP/Census/). Sequencing of both ends of the same DNA fragment (known as paired-end sequencing) reduces alignment ambiguities when matching short sequence reads to the reference genome. Paired-end sequencing of millions of genomic fragments from a single tumor is able to map genome-wide chromosomal rearrangements. Its use has recently brought considerable attention to the impact of structural chromosomal changes in cancer development2-4 and uncovered an unexpected phenomenon in which tens to hundreds of rearrangements occur within one or a handful of genomic regions5.
Two mechanisms have been proposed to provoke such rearrangements in a single event: 1) a cellular crisis termed chromothripsis5 (from the Greek chromo for chromosomes and thripsis, for shattering into pieces) and 2) local rearrangements with altered gene copy numbers produced by serial, microhomology-mediated template switching during DNA replication, termed chromoanasynthesis6 (from the Greek chromo for chromosomes and anasynthesis, for reconstitution). Chromothripsis is the likely mechanism underlying most of the events identified to date in cancer. Chromoanasynthesis appears to underlie most rearrangements in development. Recognizing that at least two mechanisms produce complex, localized rearrangements, we propose the word chromoanagenesis (from the Greek chromo for chromosomes and anagenesis, to be reborn) as a descriptor of this class of chromosomal rearrangement that is independent of the provoking mechanism.
In this perspective, we discuss the evidence supporting the view that chromoanagenesis occurs as a one-off cellular event that may contribute to initiation and development of human cancer. We outline the mechanisms that have been proposed to create highly localized complex genomic rearrangements, including provocative recent work suggesting chromoanagenesis is initiated by a chromosome missegregation error producing a micronucleus in which the localized shattering and religation take place in two subsequent cell cycles. We also describe how similarly complex rearrangements with copy number changes can be driven by cellular stress during DNA replication that leads to replication fork collapse coupled with microhomology-mediated template switching.
A one-off cellular cataclysm
Three primary lines of evidence indicate that many of the localized chromosomal rearrangements observed in cases of chromoanagenesis do not arise from a progressive series of independent rearrangements, but rather occur in a single catastrophic event5.
First, for the cancer examples now known, the chromosome rearrangements primarily alternate between two copy number states. The lower copy number state represents heterozygous deletion of a DNA fragment and the higher copy state indicates retention of a DNA piece (Figure 1). [The higher copy number state does not always result from two copies of a DNA fragment, as tumors are often aneuploid (containing an abnormal number of chromosomes).] Progressive models with sequential chromosomal translocations would predict substantially more than two copy number states5.
Figure 1. Mechanism for the creation of complex chromosomal rearrangements by non-homologous end joining after chromosome shattering.
Chromothripsis results in the shattering of one or a few chromosomes (or a chromosome arm) leading to the simultaneous creation of many double strand breaks. Most of the shattered fragments are stitched back together though Non-Homologous End Joining (NHEJ) leading to chromoanagenesis: the creation of a chromosome with complex, high-localized chromosomal rearrangements. The rearranged chromosome contains two copy number states: a high copy number state for each religated fragment and a low copy number state for fragments not-reincorporated and therefore lost. Broken DNA fragments may also be joined together to form circular, extrachromosomal double minute chromosomes that often harbor oncogenes and are frequently amplified, resulting in a dramatically increased copy number of DNA fragments on these chromosomes.
Second, heterozygosity is preserved in multiple separate regions with higher copy number states, where DNA fragments have been retained. Regions where heterozygosity is maintained can be encompassed within an area spanned by multiple additional rearrangements that have the orientation of deletions, duplications and inversions5. If a deletion occurred early in a successive series of rearrangements then heterozygosity would be permanently eliminated between the breakpoints. Thus, for a progressive model to explain chromoanagenesis, deletion events could only occur late in the sequence of rearrangements, a scenario that seems unlikely given the number of rearrangements involved in chromoanagenesis5. On the other hand, alternating regions of heterozygosity (retention of a DNA fragment) and loss of heterozygosity (loss of a DNA fragment) inevitably result from rearrangements that are caused by a one-off cataclysmic event proposed to occur during chromothripsis (Figure 1).
Third and finally, the chromosomal breakpoints cluster to a greater degree than expected from sequential independent rearrangements5.
Overall therefore, it is likely that most of the rearrangements present in the chromoanagenesis found in cancer occurs in a single catastrophic event arising from chromosome pulverization followed by rejoining of chromosomal fragments in a random order (Figure 1). The idea that can cancer genomes evolve in “rapid bursts” is in line with the evolutionary theory of “punctuated equilibrium” originally proposed by Eldredge and Gould in 1972, which posits that species undergo little alteration for most of their evolutionary history, with rare events leading to rapid evolutionary shifts that can result in the creation of a new species. Similarly, creating many alterations in a single genomic event increases the probability that large adaptive leaps can be achieved, which may be advantageous in the severe genetic or environmental pressures encountered in tumors.
Even accepting that the multiple complex rearrangements arising can occur in a single event, the high frequency of genome changes in cancer cells firmly suggests additional rearrangements can also be expected before or after chromoanagenesis, consistent with the widely held view that genomic changes in many cancers accumulate in a succession of errors. Indeed, some regions of rearranged chromosomes alternate between two and three copy number states, which suggests that a partial duplication of the rearranged chromosome occurred after chromoanagenesis had taken place5. Alternatively, if an initiating event created massive DNA double strand breaks simultaneously on both genetically identical sister chromatids of a replicated chromosome, then the random stitching together of chromosome fragments could lead to a duplication of specific chromosomal fragments in the rearranged sister chromatids7.
Solitary confinement: locked away in a micronucleus
Since its discovery, the most perplexing feature of chromoanagenesis is how chromosomal rearrangements can be limited to a very small subset of chromosomes, often a single chromosome or chromosome arm. What event(s) causes this massive damage and how can it be highly localized to distinct genomic regions? A very surprising mechanism was identified in early 2012: chromosome shattering may arise as a result of an error in chromosome segregation in mitosis that leads to the production of a micronucleus8.
During normal mitosis, the replicated genetic information is divided equally into the two new daughter nuclei such that each cell receives a single copy of each duplicated chromosome. Errors in chromosome segregation during mitosis result in the production of aneuploid cells. Aneuploidy is a hallmark of cancer and has been widely proposed to play a role in the initiation and development of tumors9,10. While aneuploidy and structural alterations in chromosomes have often been thought to arise independently of one another, recent evidence has shown that these two chromosomal aberrations can be mechanistically linked.
Most tumor cells do not possess a stably aneuploid genome, but rather exhibit a continually changing karyotype driven by high rates of chromosome gain and loss during division, a phenomenon known as chromosomal instability (CIN)11. Live cell imaging experiments have revealed that chromosomally unstable tumor cells exhibit an increase in chromosomes that lag in the middle of the spindle during anaphase12,13. One or both copies of such lagging chromosomes often fail to reach the two major chromosome masses at the poles of the cells before nuclear envelope reassembly, and consequently form a self-contained micronucleus (Figure 2A-D). Surprisingly, newly formed micronuclei frequently possess an inadequate number of nuclear pores (Figure 2E) and consequently exhibit defects in nuclear import of some components in the subsequent interphase8,14.
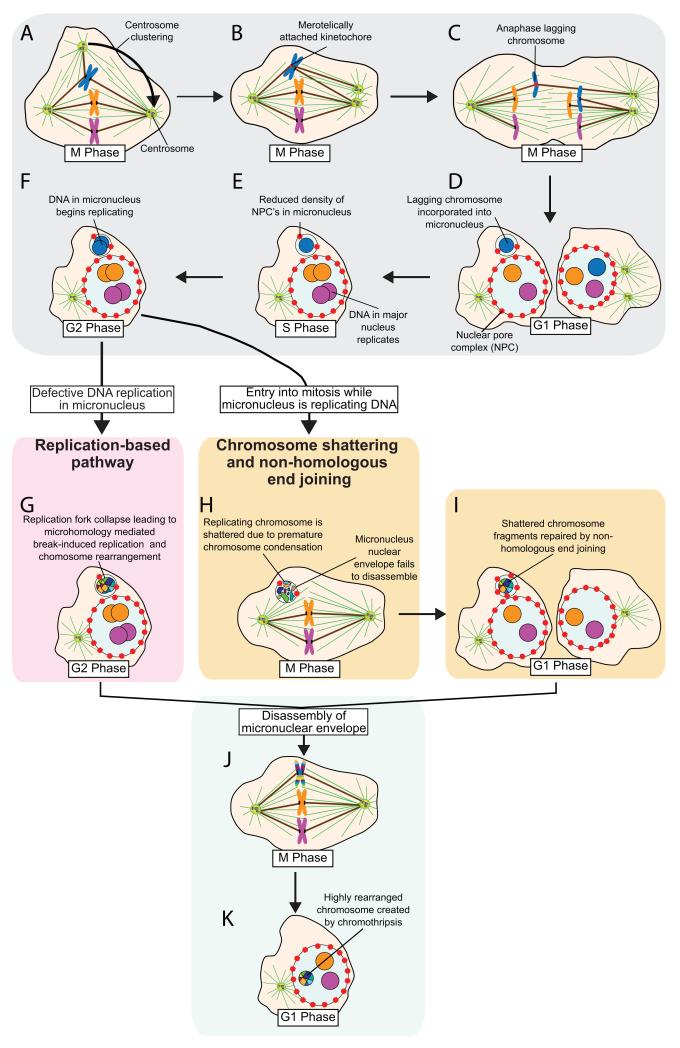
Figure 2. Mitotic errors produce micronuclei and subsequent chromoanagenesis.
(A) Cells with extra centrosomes form multi-polar mitotic spindles. In many instances centrosomes coalesce into two groups prior to anaphase. (B) Centrosome clustering increases the frequency of merotelic attachments, the situation in which a duplicated chromosome attaches through its kinetochore to microtubules arising from both mitotic spindle poles48,49. (C) If not corrected before anaphase merotelically attached chromosomes may lag in the middle of the mitotic spindle. (D) Lagging chromosomes are sometimes excluded from both daughter nuclei and instead form a micronucleus in one of the daughter cells in the subsequent interphase. (E) Micronuclei often contain fewer nuclear pore complexes, impairing nuclear import and (F) delaying DNA replication of chromosome(s) in the micronucleus. Chromoanagensis (the creation of complex, localized chromosomal rearrangements) can arise in micronuclei through two distinct mechanisms. (G) The predominant pathway known as chromothripsis5, involves chromosome shattering following mitotic entry while the micronucleus is still replicating its DNA. The incompletely replicated micronuclear DNA undergoes premature chromosome condensation that results in pulverization of the trapped chromosome(s). Often the nuclear envelope of the micronucleus fails to disassemble during the next mitosis and the intact micronucleus randomly segregates at mitotic exit into one of the daughter cells. (H) During the subsequent interphase, shattered chromosome pieces within the micronucleus are repaired by NHEJ. (I) A second pathway known as chromoanasynthesis6, leads to the creation of complex rearrangements in micronuclei through a replication-based mechanism, such as Microhomology Mediated Break-Induced Replication (MMBIR). In this phenomenon, defective DNA replication in the micronucleus leads to a collapsed replication fork that initiates microhomology-dependent priming of DNA replication and serial template switching. MMBIR can result in chromoanagenesis, with the creation of complex chromosomal rearrangements at genomic regions surrounding the collapsed replication fork. (J) The micronucleus nuclear envelope eventually disassembles during a subsequent mitosis, releasing the rearranged chromosome. (K) The rearranged chromosome is segregated on the mitotic spindle and reincorporated into the major nucleus of the cell.
Reduced nuclear import has a number of consequences for the chromatin sequestered inside micronuclei. First, micronuclei exhibit defective DNA damage response signaling, resulting in defective/delayed repair of induced DNA damage (Figure 2E-F)8,15,16. Second, DNA replication in micronuclei is delayed compared with the major nucleus, with some micronuclei still replicating DNA when the major nucleus is in the G2 phase8. Third, entry into mitosis while the micronucleus is undergoing DNA replication produces massive DNA double strand breaks in the micronuclear DNA (detailed below)8.
Pulverizing chromosomes within a micronucleus
The most plausible mechanism for the observed chromosomal pulverization that characterizes chromothripsis is entry into mitosis prior to completion of DNA replication within a micronucleus, resulting in breaks in the incompletely replicated micronuclear DNA during premature chromosome condensation (PCC). PCC was originally described in classic cell fusion experiments and occurs when cyclin-dependent kinase activity in a mitotic cell induces incompletely replicated chromosomes in S phase nuclei to undergo chromosome condensation and shattering17-19. PCC of an incompletely replicated micronucleus is expected to create focal and catastrophic DNA damage (Figure 2G)20.
While it has been widely assumed that the nuclear envelop will dissemble at each mitosis, thereby allowing the chromosome(s) contained within each micronucleus to spill into the cytoplasm of the next mitosis, this is not what happens for the many micronuclei. Indeed, disassembly of the micronuclear envelope frequently fails at the onset of the subsequent mitosis, with the intact micronucleus randomly segregating at mitotic exit into one of the daughter cells8. This failure of efficient nuclear envelop disassembly is crucial for containing the fragments of pulverized chromosomes in an isolated compartment for their subsequent religation. Persistence of a micronucleus into interphase of the second cell cycle after its initial formation provides a plausible mechanism for repair by ligation (in a random order) of the chromosomal fragments that were initially generated as a result of PCC (Figure 2H). For the subset of micronuclei for which the nuclear membrane does disassemble, the acentric fragments (which lack centromeres and microtubule attachment sites) of pulverized chromosomes will be unable to be segregated and may be lost or form de novo micronuclei at mitotic exit.
How a micronucleus escapes nuclear envelope disassembly during mitosis is unsettled, but the reduced density of nuclear pores may reflect reduced incorporation of several key envelope constituents that are phosphorylated by mitotic cyclin-dependent kinases to promote nuclear envelope breakdown. Eventually, however, further cycling will yield micronuclear envelope disassembly upon entry into a subsequent mitosis, releasing the rearranged chromosome into the mitotic cytoplasm and allowing its conventional mitotic segregation with the main mitotic chromosome mass (Figure 2J-K).
The missegregation of chromosomes into micronuclei provides a plausible route through which whole chromosome missegregation can promote chromosome breaks and subsequent rearrangement, thereby mechanistically coupling events leading to the acquisition of numerical and structural chromosomal alterations. In addition, this pathway also provides an elegant explanation for how the DNA breaks acquired during chromothripsis or chromoanasynthesis may be circumscribed to one or a small number of chromosomes – the one(s) trapped within a micronucleus8. Thus, an initial error in chromosome segregation during mitosis is likely to be one key event in the initiation of chromoanagenesis. As such, it will now be of high interest to examine the tumors formed in mice that have been genetically manipulated to exhibit CIN and aneuploidy for evidence of chromoanagenesis21.
Alternative proposals for chromosome shattering
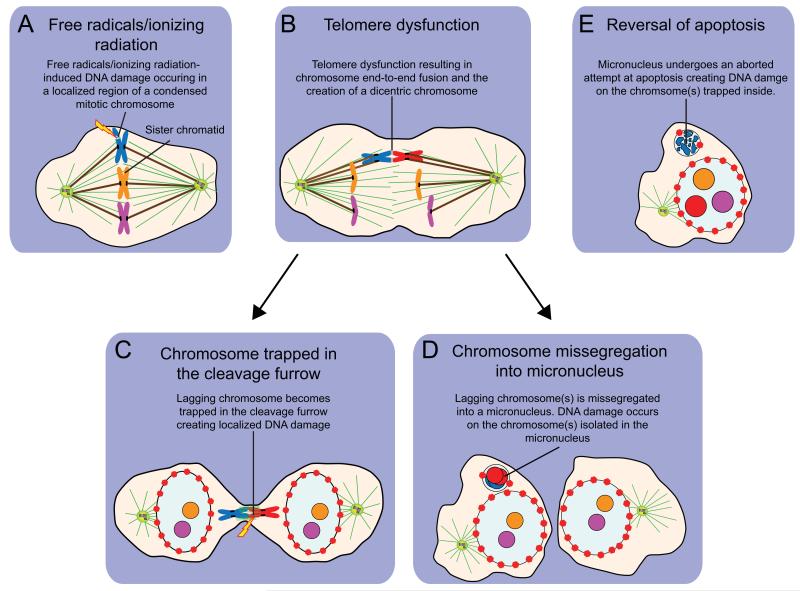
It should be noted that at present the mechanism(s) responsible for chromoanagenesis remains controversial. In addition to shattering from mitotic entry with incompletely replicated DNA as introduced above, three additional proposals have been put forward to explain how localized chromosome shattering may result in complex, localized rearrangements. As detailed below, we propose that each of these mechanisms is made more plausible if one imparts the formation of micronuclei as a means to either spatially localize DNA damage or to contain the chromosome fragments created by this damage so that they may be religated to produce chromosomal rearrangements.
To explain the confined nature of the DNA damage created during chromothripsis, an initial proposal was that localized DNA double strand breaks from free radicals or ionizing radiation may be created during mitosis when chromosomes are highly compacted and DNA damage signaling is suppressed5,22. While it is otherwise unclear how damage could be isolated to just one or a few chromosomes, the formation at mitotic exit of a micronucleus containing the damaged chromosome fragments would provide a means to constrain the fragments produced so as to facilitate their religation into a rearranged chromosome characteristic of chromoanagenesis.
Telomere dysfunction has also been proposed as a cause of chromothripsis5. Continued proliferation of somatic cells in the absence of telomerase activity leads to the progressive attrition of telomeres. Eventually, telomere-shortening exposes uncapped chromosome ends that are prone to fuse, creating a dicentric chromosome with two microtubule attachment sites on each sister chromatid. If these sites attach to opposite poles of the cell during mitosis, the resulting chromosome will become highly stretched during anaphase. Since chromothripsis appears to occur in a single catastrophic cellular event, one possibility is that the bridging chromosome undergoes massive localized genomic damage at the cleavage furrow during cytokinesis23. A more attractive explanation, however, is that the lagging dicentric chromosome fails to incorporate into the major nucleus of either daughter cell and instead forms a micronucleus. Therefore, telomerase deficiency could promote chromothripsis indirectly, through disrupting chromosome segregation leading to the production of micronuclei. Examining telomerase deficient mouse models for evidence of extensive localized genomic rearrangements will form an important test of whether telomere dysfunction can promote chromoanagenesis.
Finally, chromothriptic chromosome shattering has been suggested to result from an aborted attempt at apoptosis24. While apoptosis has traditionally been considered as an irreversible cascade that once initiated, irrevocably leads to cell death, recent evidence has clearly demonstrated that initial apoptotic events can be reversed if the initiating stimulus is removed25. Reversal of apoptosis has been termed “anastasis” (Greek for “rising to life”) and can occur following measurable DNA damage, allowing cells to acquire permanent genetic changes that facilitate transformation25. Anastasis promotes an increase in numerical and structural chromosomal alterations as well as an increase in micronuclei formation25. Reversal of apoptosis after the initiation of DNA damage and chromosome fragmentation may lead to the religation of chromosome fragments and the production of chromosomal rearrangements26,27. In the tumor microenvironment, apoptosis could be initiated by a variety of stresses including chemotherapy, ionizing radiation, hypoxia and nutrient deprivation. This raises the possibility that these transient stress stimuli induce an aborted apoptosis that initiates the DNA damage responsible for chromothripsis. Like the other proposed mechanisms, if anastasis occurred specifically within a micronucleus, DNA damage would be confined to the chromosome(s) trapped inside28. It will be of interest to establish if anastasis can initiate chromothripsis and the development of complex chromosomal rearrangements in cells in culture.
After shattering: weaving together chromosomal fragments
Most of the breakpoints of the reassembled chromosomes created by chromothripsis in human cancers show a lack of homology or only areas of microhomology, pointing towards Non-Homologous End Joining (NHEJ) as the predominant mechanism used to stitch the shattered chromosomes back together following extensive double strand breaks5,29,30. NHEJ can occur at any point in the cell cycle and often occurs at regions of microhomology that are 1-4 nucleotides in length31. NHEJ occurs in a series of steps31. First, the Ku protein heterodimer (Ku70/Ku80) is recruited to both ends of the DNA at the site of a double strand break. Ku recruits a complex of Artemis and the DNA-dependent protein kinase catalytic subunit involved in processing the ends of DNA breaks. As a final step, the Ligase IV complex (comprised of DNA ligase IV and its cofactor XRCC4) is recruited by Ku and ligates the adjacent DNA ends, thereby repairing the double strand break. Ligation of incorrect ends though NHEJ can lead to chromosomal translocations. The random reassembly by NHEJ of many simultaneously created chromosome fragments can explain the vast majority of the chromosomal translocations created during chromoanagenesis.
Constitutional rearrangements from a slip-up during DNA replication not shattering
Unlike the chromosome shattering and NHEJ found in cancer, many (but not all) complex constitutional chromosomal rearrangements carry a signature of microhomologies at the ends of rearranged segments that is indicative of a direct DNA replication-based mechanism6 (right refxx??) (Box 1). These rearrangements are associated with congenital or developmental abnormalities and contain multiple duplications and triplications, neither of which can be readily explained by a mechanism involving the NHEJ-mediated repair of many simultaneously created double strand breaks.
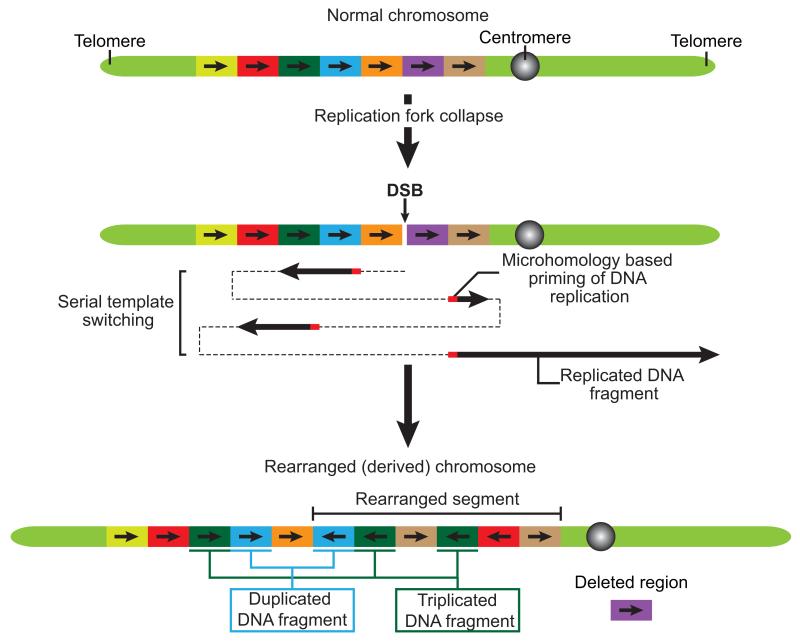
The most persuasive evidence for rearrangements from DNA replication based mechanisms, including Fork-Stalling and Template Switching (FoSTes)32 and Microhomology Mediated Break-Induced Replication (MMBIR)33, is that sequencing of breakpoint junctions reveals areas of microhomologies and templated insertions (54-1542 bp’s)6. MMBIR may occur when a replication fork collapses after encountering a nick in the template strand (Box 2). Breakage of a replication fork then promotes microhomology-dependent priming of DNA replication and serial template switching, a process resulting in complex chromosomal rearrangements surrounding the site of the collapsed fork (Figure 3)6. In FoSTes, rearrangements appear to arise as a result of a stalled replication fork coupled with a consecutive series of long-range replication fork template switches (Figure 3).
Figure 3. Mechanism for complex chromosomal rearrangements as a result of Fork-Stalling and Template Switching (FoSTes) and Microhomology Mediated Break-Induced Replication (MMBIR).
Fork-Stalling and Template Switching (FoSTes) occurs when a replication fork stalls at a DNA lesion while MMBIR is initiated following replication fork collapse. FoSTes and MMBIR lead to microhomology-dependent priming of DNA replication and serial template switching. This leads to chromoanagenesis: the creation of complex chromosomal rearrangements in a genomic region surrounding the collapsed replication fork. In addition to the deletion and retention of DNA fragments, FoSTes and MMBIR can also lead to duplication and triplication of DNA sequences. Therefore, FoSTes and MMBIR can result in more than two copy number states on the rearranged chromosome. Modified from reference50.
Replication-based mechanisms do not necessarily require micronuclei to explain the formation of complex localized chromosomal rearrangements. Indeed, the organization of chromosomes into distinct “territories” within a cell’s nucleus that may help bias microhomology-dependent template switching to sequences on a single, or small subset of, chromosome(s) that are in close proximity in three-dimensional space. Nevertheless, micronuclei often exhibit defective DNA replication8 and the partitioning of a chromosome into a micronucleus would provide an elegant explanation for how aberrant DNA replication could be restricted to one or a few spatially isolated chromosomes (Figure 2I).
While MMBIR offers a plausible route for the generation of complex copy number variations and offers an explanation for how multiple copy number changes may arise in some germline cases of chromoanagenesis (Box 1 and 2)6,34, it should also be borne in mind that inspection of breakpoints in several additional cases of constitutional structural rearrangements bear similarity to what is predicted by local chromosome shattering followed by NHEJ34,35. Thus, while the weight of evidence supports a mechanism of massive chromosomal breakage followed by NHEJ to explain the complex genomic rearrangements observed in many cancers5,29,30, there appear to be at least two distinct mechanisms responsible for the chromoanagenesis-like rearrangements associated with genomic disorders6,35.
Since oncogene activation can cause cellular senescence as a result of DNA damage caused by oncogene-induced activation of DNA replication stress36,37.
Chromoanagenesis in human cancer
The multiple, localized, chromosome rearrangements characteristic of chromoanagenesis occur in many different types of cancers with an overall frequency of ~2-3%5,29,30,36-40. The frequency of chromoanagenesis is elevated in specific tumor types, including ~25% of bone cancers5 and ~18% of high stage neuroblastomas40. Moreover, chromoanagenesis is widespread in primary and metastatic colorectal cancer30 and there is a striking association between mutations in TP53 (encoding the p53 protein) and chromoanagenesis in Sonic-Hedgehog Medulloblastoma (SHH-MB) and Acute Myeloid Leukemia (AML)29. Importantly, chromoanagenesis has been associated with a poor survival in a variety of tumor types29,39,40, but further studies will be required to define the incidence and consequence of chromoanagenesis across a larger set of human cancers.
In the vast majority of cases it is probable that chromoanagenesis creates genomic alterations that fail to produce any significant advantage or lead to a substantial reduction in cellular fitness. As a result the occurrence of chromoanagenesis in cancer is likely to be considerably higher than the ~3% observed, with the majority of cases of chromoanagenesis not providing a selectable advantage and escaping clinical detection. However, in rare circumstances chromoanagenesis may lead to the creation of one or more cancer-causing lesions in a single catastrophic event, thereby providing an advantage for cellular growth.
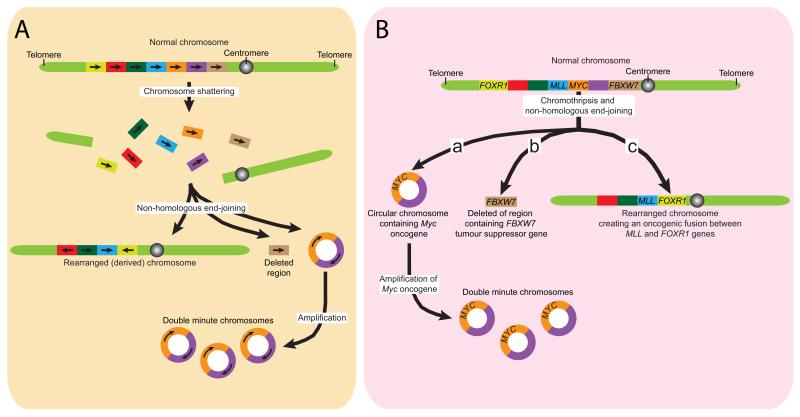
One important route through which chromosome shattering and religation by NHEJ could promote aberrant cellular proliferation is by facilitating oncogene amplification through the creation of small, circular fragments of DNA lacking centromeres and telomeres and frequently harboring oncogenes (Figure 4A). These extrachromosomal fragments are known as double minute chromosomes and are often present at many copies per cell41. During chromothripsis it is postulated that individual chromosomes (or portions of them) are initially broken into many pieces and randomly reassembled by NHEJ. While many of the pieces are stitched back together in random order to produce a highly rearranged chromosome, some fragments may also be joined together to create a circular double minute chromosome (Figure 4A), whose amplification can be selected for if it confers a growth advantage5,29. One example of the latter is exemplified in one small cell lung cancer cell line that has been found to contain a double minute chromosome (carrying the MYC oncogene) created by fusing several segments of chromosome 8 that were absent from a rearranged copy of chromosome 8 generated by chromoanagenesis5.
Figure 4. Chromoanagenesis may create oncogenic lesions.
The complex chromosomal rearrangements created by chromoanagenesis can be oncogenic. (A) Initial chromosomal shattering followed by rejoining by NHEJ may create circular fragments of DNA harboring oncogenes, such as MYC. Amplification of these extrachromosomal “double minute” chromosomes can provide a growth advantage. Other pieces of a shattered chromosome may be joined together to create a highly rearranged chromosome. (B) Chromoanagenesis can lead to the loss or disruption of regions containing tumor suppressor genes, such as FBXW7. (C) Rearrangements may also create oncogenic fusion genes by joining the coding sequence two normal genes together, for example, the fusion of the MLL and the FOXR1 genes.
A second potential route by which chromoanagenesis could create cancer-causing mutations is through loss or disruption of tumor suppressor genes (Figure 4B). For example, in one chordoma, chromosome shattering facilitated the loss of chromosomal fragments containing or led to rearrangements that directly disrupted, each of three tumor suppressor genes (FBXW7, WRN and CDKN2A)5. In addition, in colorectal cancer the breakpoints generated by chromothripsis have been found to affect several known cancer causing genes (NOTCH2, EXO1 and MLL3)30. In many cases, the rearrangements created by chromoanagenesis affect only a single allele of a tumor suppressor gene with the other copy retained, implying that the second intact allele may be inactivated epigenetically. Alternatively, the affected gene may act as a haploinsufficient tumor suppressor, as has been shown for the tumor suppressor FBXW742,43.
Finally, the chromosome shattering and religation of chromoanagenesis can generate oncogenic fusion genes by joining the coding portions of two genes in the same orientation (Figure 4C). For example, chromoanagenesis in medulloblastoma tumors leads to recurrent translocations that fuse PVT1 (a non-coding gene) to the MYC proto-oncogene, resulting in MYC-amplification.
Chromoanagenesis: an early or late even in human tumors?
While chromoanagenesis can sculpt the cancer genome leading to the creation of potentially oncogenic lesions, it should be noted that it has yet to be formally demonstrated that the genetic abnormalities that arise as a consequence of chromoanagenesis act, either individually or in combination, to drive tumorigenesis. The handful of studies reported thus far have analyzed chromoanagenesis by sampling a single and relatively late stage in tumor development and thus, it remains unsettled at which stage during tumor evolution chromoanagenesis occurs. Chromoanagenesis often occurs following TP53 mutations in AML and SHH-MB patients29, while in neuroblastoma chromoanagenesis has been identified in 18% of late stage cancers but is absent in early stage tumors40. This argues that chromoanagenesis may be a relatively late event, at least in the development of these cancers. In the future it will be important to perform longitudinal studies in animals models or human patients to establish whether chromoanagenesis is an early initiating event or a later event that only occurs once additional defects, such as TP53 mutations, have been acquired.
Stayin’ alive
It is remarkable that a cell can survive the catastrophic events of chromoanagenesis that arise either following replication fork collapse in chromoanasynthesis or after tens to hundreds of DNA breaks accompanying chromothripsis. One implication is that acquired defects in DNA damaging signaling cascades may set the stage for tolerating massive DNA damage. Whole genome sequencing coupled with microarray analysis has uncovered a striking association between chromoanagenesis and both germline and somatic inactivation of the TP53 tumor suppressor gene29,37,38. There is a considerable enrichment for chromoanagenesis in TP53 mutated samples of AML29. In one study chromoanagenesis was observed in all ten of the TP53 mutated SHH-MB samples examined, but was not observed in SHH-MBs with an intact TP53 gene29. An independent study found that chromoanagenesis in Group 3 medulloblastomas is associated of loss of the TP53 gene38. Strikingly, other medulloblastoma subtypes rarely display chromoanagenesis (including WNT subtype medulloblastomas harboring mutated TP53), indicating the link between p53 mutation and chromoanagenesis is dependent upon the specific tumor type29,38.
Germline mutations of TP53 in SHH-MBs patients occur prior to chromoanagenesis, suggesting that TP53 mutations may predispose cells to chromothripsis, or allow cellular survival following catastrophic DNA damage. Indeed, p53 plays an important role in promoting cell cycle arrest, apoptosis or senescence in response to DNA damage44. Analysis of early T-cell precursor acute lymphoblastic leukaemia has also hinted at a link between mutation in genes involved in DNA repair genes and chromoanagenesis36. Two neuroblastoma tumors displaying evidence of chromoanagenesis were also found to contain mutations in genes functioning in the Fanconi Anemia-DNA damage response pathway, indicating that lesions which attenuate DNA damage signaling pathways may play a general role in facilitating the survival of cells which undergo events to initiate chromothripsis40.
Clinical implication of chromoanagenesis
It clear that chromoanagenesis has the capacity to create novel genetic alterations that can potentially drive tumor progression. Indeed, chromoanagenesis has been associated with poor survival in AML29, neuroblastoma40 and multiple myeloma patients39. In AML the association of chromoanagenesis with poor survival is independent of patient age and karyotype classification raising the possibility that this distinctive genetic alteration may form a useful prognostic marker to predict disease outcome or therapeutic responsiveness29. However, in neuroblastoma, chromoanagenesis has only been observed in late-stage patients (stage 3-4) which have a poorer outcome40. Therefore, the prognostic value of chromoanagenesis will likely depend upon the cancer type and will be sensitive to when during tumor evolution chromoanagenesis occurs and what additional genetic events (such as TP53 mutations) predispose to chromoanagenesis. In addition to its role in sculpting cancer genomes, chromoanagenesis has also been reported to create complex constitutional genomic rearrangements, which likely contributes to congenital or developmental defects (Box 1). Clearly more work will be needed in a larger patient cohort to determine the full clinical implications arising from chromoanagenesis.
Li-Fraumeni syndrome (LFS) patients with heterozygous germline mutations in the TP53 gene show an increased incidence of chromoanagenesis in SHH-MB and possibly also other LFS-associated malignancies29. The use of DNA damaging agents and ionizing radiation in cancer therapy has the possibility of inducing chromothripsis. Comparing the genome of primary tumors with those that relapse after therapy will provide important insights into whether chromoanagenesis can be induced by specific therapeutic regimes and whether this may contribute to the emergence of resistance in the primary tumor.
Looking forward
Given the large number of genomic alterations occurring in a single event, chromoanagenesis could allow the rapid development of new phenotypes that facilitate tumor initiation, progression and the evolution of resistance to drug therapy. For a more complete understanding of the role of chromoanagenesis in tumorigenesis it will now be important to establish at which point during the clonal evolution of a tumor chromoanagenesis occurs. In addition, establishing which types of tumors display the highest incidence of chromoanagenesis will aid in the discovery of additional co-operating genetic alterations that facilitate the initiation of, or response to, chromoanagenesis. Included here will be determining whether chromoanagenesis is more common in tumors which specific DNA damage signaling defects and establishing if there is tissue-type or tumor-type context specificity in the mechanisms leading to chromoanagenesis.
Along with variation in the genetic makeup of individual tumors, emerging evidence points toward considerable levels of intratumoral genetic heterogeneity45,46. Establishing what fraction of cells in a given tumor possess complex chromosomal rearrangements and how this subpopulation evolves over time is now an important next step in understanding subclonal tumor architecture and the context specific factors that determine tumor development following chromoanagenesis.
Box 1: Germline chromothripsis contributes to human disease.
Complex genomic rearrangements consist of at least two breakpoint junctions and are associated with a variety of congenital or developmental abnormalities. Unlike the rearrangements generated by cancer chromoanagenesis, which arise in differentiated somatic cells, constitutional rearrangements occur in the germline or very early in embryonic development. Several recent studies have revealed that some patients with inherited genetic defects exhibit complex constitutional chromosomal rearrangements that strongly resemble the somatic rearrangements found in cancer chromoanagenesis. Analysis of a family trio identified a complex series of de novo chromosomal rearrangements occurring in a child with congenital abnormalities35. These rearrangements clustered in small genomic regions on three chromosomes (chromosomes 1, 4 and 10) and bore the hallmarks of chromoanagenesis. An additional study of 17 patients with developmental and congenital abnormalities revealed four with inherited chromoanagenesis-like rearrangements in a single chromosome (involving chromosomes 1 or 9 in one patient each and chromosome 22 in two additional patients)6.
High-resolution analysis of the breakpoints in 52 patients with cytogenetically defined chromosomal abnormalities identified at least two additional cases of constitutional complex genomic rearrangements that share similarities with the rearrangements identified in cancer chromoanagenesis47. In these two patients, the genomic rearrangements involved two or three chromosomes (chromosome 5 and X or chromosomes 3, 5 and 7) and showed few losses and gains of DNA segments. The largely dosage balanced state (in which genes or DNA sequences are present in the correct copy number) observed in these multi-chromosome constitutional rearrangements is distinct from more extensive copy number changes frequently observed in cancer chromoanagenesis5,29,35,37,39,40 and some other individuals with constitutional chromoanagenesis involving only a single chromosome6. Dosage alterations may be favored in cancer cells due to loss of tumor suppressor genes, while the more balanced chromosomal translocations observed in complex genomic rearrangements may reflect a selection for rearrangements that are compatible with organismal viability. Indeed, less complex rearrangements are expected for heritable disorders, since massive constitutional rearrangements would be expected to be detrimental during development.
In an effort to determine the mechanism responsible for the creation of constitutional complex chromosomal rearrangements, a recent study analyzed the breakpoints in ten individuals with congenital abnormalities34. The rearrangements consisted of between three and twenty-four inter and inrachromosomal breakpoints with features similar to those observed in cancer chromoanagenesis. Eight of the patients exhibited rearrangements that are most likely to have arisen through the creation of multiple simultaneous DNA double strand breaks followed by non-homologous repair (as originally proposed for chromothripsis in cancer cells5), while two others displayed a distinct signature at the junctions of rearrangements that are most consistent with defective replication leading to serial template switching (chromoanasynthesis) (Box 2)34.
Together these studies highlight the remarkable structural plasticity of the human genome and indicate that chromoanagenesis can produce stable and viable constitutional rearrangements that are likely to contribute to congenital and developmental defects in humans.
Box 2: Replication based mechanims leading to the creation of complex chromosomal rearrangements.
The insertion of short sequences at breakpoint junctions that are “templated’ from nearby genomic regions provides evidence of a DNA replication-associated mechanism for chromosomal rearrangements. Such a mechanism (chrmoanasynthesis) has been proposed to account for some examples of complex constitutional chromosomal rearrangements (Box 1)6,34. Two related mechanisms have been proposed for these genomic alterations. The first, known as Fork-Stalling and Template Switching (FoSTes), occurs when a replication fork stalls at a DNA lesion, allowing the lagging strand of the replication fork to disengage and switch to an area of microhomology on a neighboring replication fork32. The two-replication forks will be in physical proximity, but may be separated by large stretches of DNA sequence. DNA synthesis would initiate temporarily at this second site before the nascent strand disengages again and invades an additional replication fork. This process may repeat multiple times leading to serial template switching before completion of DNA synthesis on the original template.
The second mechanism is known as Microhomology Mediated Break-Induced Replication (MMBIR) and is initiated when a replication fork collapses upon encountering a nick in the template strand33. This process creates a DNA double strand break in one arm of the replication fork; however, since there is not an additional DNA end to be used in double strand break repair, the 5′ end of the broken arm is resected to leave a 3′ single-stranded DNA overhang, which invades a DNA sequence with microhomology to the single stranded 3′ end. The 3′ end primes DNA synthesis and establishes a replication fork. The extended arm eventually separates from the template and the 3′ end reinvades an additional region to repeat the process. Eventually a switch occurs to the original genomic region and replication continues to the chromosome end.
FoSTes and MMBIR can result in complex genomic rearrangements surrounding the site of the original defective replication fork. Serial template switching can lead to the insertion of specific DNA sequence from distinct genomic regions that lie in close proximity in three dimensional space and also explain the increases in copy number (duplications and triplications) observed in some cases of complex constitutional chromosomal rearrangements6. For example, duplication can occur when a template switch occurs to a DNA sequence that lies behind (relative to the direction of the fork) where the replication fork collapsed33.
Acknowledgements
We apologize to all whose work on aneuploidy was not cited because of space restrictions. This work was supported by a grant (GM29513) from the National Institutes of Health to D.W.C, who receives salary support from the Ludwig Institute for Cancer Research. A.J.H. is supported by a Leukemia & Lymphoma Society special fellowship.
References
- 1.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 2.Stephens PJ, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell PJ, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell PJ, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu P, et al. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell. 2011;146:889–903. doi: 10.1016/j.cell.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen JM, Ferec C, Cooper DN. Transient hypermutability, chromothripsis and replication-based mechanisms in the generation of concurrent clustered mutations. Mutat Res. 2011 doi: 10.1016/j.mrrev.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 8.Crasta K, et al. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nat Rev Genet. 2012;13:189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- 10.Holland AJ, Cleveland DW. Losing balance: the origin and impact of aneuploidy in cancer. EMBO Rep. 2012;13:501–514. doi: 10.1038/embor.2012.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 12.Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. The Journal of cell biology. 2008;180:665–672. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gascoigne KE, Taylor SS. Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell. 2008;14:111–122. doi: 10.1016/j.ccr.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 14.Hoffelder DR, et al. Resolution of anaphase bridges in cancer cells. Chromosoma. 2004;112:389–397. doi: 10.1007/s00412-004-0284-6. [DOI] [PubMed] [Google Scholar]
- 15.Terradas M, Martin M, Hernandez L, Tusell L, Genesca A. Nuclear envelope defects impede a proper response to micronuclear DNA lesions. Mutat Res. 2012;729:35–40. doi: 10.1016/j.mrfmmm.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 16.Terradas M, Martin M, Tusell L, Genesca A. DNA lesions sequestered in micronuclei induce a local defective-damage response. DNA Repair (Amst) 2009;8:1225–1234. doi: 10.1016/j.dnarep.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 17.Rao PN, Johnson RT. Mammalian cell fusion: studies on the regulation of DNA synthesis and mitosis. Nature. 1970;225:159–164. doi: 10.1038/225159a0. [DOI] [PubMed] [Google Scholar]
- 18.Johnson RT, Rao PN. Mammalian cell fusion: induction of premature chromosome condensation in interphase nuclei. Nature. 1970;226:717–722. doi: 10.1038/226717a0. [DOI] [PubMed] [Google Scholar]
- 19.Sperling K, Rao PN. The phenomenon of premature chromosome condensation: its relevance to basic and applied research. Humangenetik. 1974;23:235–258. doi: 10.1007/BF00272508. [DOI] [PubMed] [Google Scholar]
- 20.Meyerson M, Pellman D. Cancer genomes evolve by pulverizing single chromosomes. Cell. 2011;144:9–10. doi: 10.1016/j.cell.2010.12.025. [DOI] [PubMed] [Google Scholar]
- 21.Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nature Reviews Molecular Cell Biology. 2009;10:478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang W, Peng G, Lin SY, Zhang P. DNA damage response is suppressed by the high cyclin-dependent kinase 1 activity in mitotic mammalian cells. J Biol Chem. 2011;286:35899–35905. doi: 10.1074/jbc.M111.267690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janssen A, van der Burg M, Szuhai K, Kops GJ, Medema RH. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science. 2011;333:1895–1898. doi: 10.1126/science.1210214. [DOI] [PubMed] [Google Scholar]
- 24.Tubio JM, Estivill X. Cancer: When catastrophe strikes a cell. Nature. 2011;470:476–477. doi: 10.1038/470476a. [DOI] [PubMed] [Google Scholar]
- 25.Tang HL, et al. Cell survival, DNA damage and oncogenic transformation following a transient and reversible apoptotic response. Mol Biol Cell. 2012 doi: 10.1091/mbc.E11-11-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stanulla M, Wang J, Chervinsky DS, Thandla S, Aplan PD. DNA cleavage within the MLL breakpoint cluster region is a specific event which occurs as part of higher-order chromatin fragmentation during the initial stages of apoptosis. Mol Cell Biol. 1997;17:4070–4079. doi: 10.1128/mcb.17.7.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stanulla M, Chhalliyil P, Wang J, Jani-Sait SN, Aplan PD. Mechanisms of MLL gene rearrangement: site-specific DNA cleavage within the breakpoint cluster region is independent of chromosomal context. Hum Mol Genet. 2001;10:2481–2491. doi: 10.1093/hmg/10.22.2481. [DOI] [PubMed] [Google Scholar]
- 28.Terradas M, Martin M, Tusell L, Genesca A. Genetic activities in micronuclei: is the DNA entrapped in micronuclei lost for the cell? Mutat Res. 2010;705:60–67. doi: 10.1016/j.mrrev.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 29.Rausch T, et al. Genome Sequencing of Pediatric Medulloblastoma Links Catastrophic DNA Rearrangements with TP53 Mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kloosterman WP, et al. Chromothripsis is a common mechanism driving genomic rearrangements in primary and metastatic colorectal cancer. Genome Biol. 2011;12:R103. doi: 10.1186/gb-2011-12-10-r103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 33.Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kloosterman WP, et al. Constitutional Chromothripsis Rearrangements Involve Clustered Double-Stranded DNA Breaks and Nonhomologous Repair Mechanisms. Cell Reports. 2012;1:648–655. doi: 10.1016/j.celrep.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 35.Kloosterman WP, et al. Chromothripsis as a mechanism driving complex de novo structural rearrangements in the germline. Hum Mol Genet. 2011;20:1916–1924. doi: 10.1093/hmg/ddr073. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu C, et al. Poly-gene fusion transcripts and chromothripsis in prostate cancer. Genes Chromosomes Cancer. 2012 doi: 10.1002/gcc.21999. [DOI] [PubMed] [Google Scholar]
- 38.Northcott PA, et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature. 2012;488:49–56. doi: 10.1038/nature11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Magrangeas F, Loiseau H, Munshi NC, Minvielle S. Chromothripsis identifies a rare and aggressive entity among newly diagnosed multiple myeloma patients. Blood. 2011;118:675–678. doi: 10.1182/blood-2011-03-344069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Molenaar JJ, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012 doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- 41.Wahl GM. The importance of circular DNA in mammalian gene amplification. Cancer Res. 1989;49:1333–1340. [PubMed] [Google Scholar]
- 42.Mao JH, et al. Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature. 2004;432:775–779. doi: 10.1038/nature03155. [DOI] [PubMed] [Google Scholar]
- 43.Kemp Z, et al. CDC4 mutations occur in a subset of colorectal cancers but are not predicted to cause loss of function and are not associated with chromosomal instability. Cancer Res. 2005;65:11361–11366. doi: 10.1158/0008-5472.CAN-05-2565. [DOI] [PubMed] [Google Scholar]
- 44.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 45.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yap TA, Gerlinger M, Futreal PA, Pusztai L, Swanton C. Intratumor heterogeneity: seeing the wood for the trees. Sci Transl Med. 2012;4:127 ps110. doi: 10.1126/scitranslmed.3003854. [DOI] [PubMed] [Google Scholar]
- 47.Chiang C, et al. Complex reorganization and predominant non-homologous repair following chromosomal breakage in karyotypically balanced germline rearrangements and transgenic integration. Nat Genet. 2012 doi: 10.1038/ng.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Silkworth WT, Nardi IK, Scholl LM, Cimini D. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS ONE. 2009;4:e6564. doi: 10.1371/journal.pone.0006564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maher CA, Wilson RK. Chromothripsis and human disease: piecing together the shattering process. Cell. 2012;148:29–32. doi: 10.1016/j.cell.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]