Abstract
CuA is a dinuclear mixed-valence center located in subunit 2 of the ba3 type cytochrome oxidase from Thermus thermophilus. The assembly of this site within the periplasmic membrane is believed to be mediated by the copper chaperones Sco and/or PCuAC, but the biological mechanisms are still poorly understood, thereby stimulating interest in the mechanisms of CuA formation from inorganic ions. The formulation of the CuA center as an electron-delocalized Cu1.5 – Cu1.5 system, implicates both Cu(II) and Cu(I) states in the metalation process. In earlier work we showed that selenomethionine (SeM) substitution of the coordinated M160 residue provided a ligand-directed probe for studying the copper coordination environment via the Se XAS signal, which was particularly useful for interrogating the Cu(I) states where other spectroscopic probes are absent. In the present study we have investigated the formation of mixed-valence CuA and its M160SeM derivative by stopped-flow UV-vis, EPR, and XAS at both Cu and Se edges, while the formation of fully reduced di-Cu(I) CuA has been studied by XAS alone. Our results establish the presence of previously undetected mononuclear intermediates, and show important differences from the metalation reactions of purple CuA azurin. XAS spectroscopy at Cu and Se edges has allowed us to extend mechanistic inferences to formation of the di-Cu(I) state which may be more relevant to biological CuA assembly. In particular, we find that T. thermophilus CuA assembles more rapidly than reported for other CuA systems, and that the dominant intermediate along the pathway to mixed-valence is a new green species with λmax = 460 nm. This intermediate has been isolated in a homogeneous state, and shown to be a mononuclear Cu(II)-(His)(Cys)2 species with no observable Cu(II)-(Met) interaction. Reduction with dithionite generates its Cu(I) homologue which is again mononuclear, but now shows a strong interaction with the Met160 thioether. The results are discussed within the framework of (i) the “coupled distortion” model for Cu(II) thiolates, and (ii) their relevance to biological metalation reactions of the CuA center.
Introduction
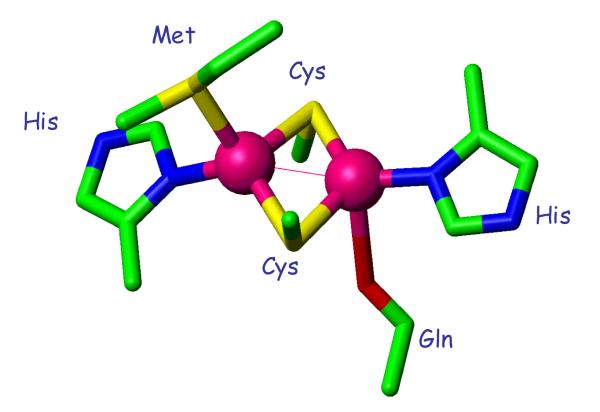
Cytochrome oxidases are heme-copper proteins found ubiquitously in both eukaryotic and prokaryotic systems. They contain a catalytic binuclear Fe – CuB center at which molecular oxygen undergoes a 4-electron reduction to water coupled to the creation of the membrane proton gradient required for the synthesis of ATP 1-5. The reducing electrons are shuttled into the catalytic center by additional cofactors including the 6-coordinate heme a in subunit 1, and in the a3-type oxidases by a unique dinuclear electron delocalized mixed-valence copper center in subunit 2 termed CuA (Fig. 1) 6. Electron transfer into and out of the CuA center is remarkable efficient due to unique coordination features which include a pair of copper atoms bridged by two cysteine residues and a short Cu-Cu distance of 2.4 – 2.5 Å 7-10. Direct bonding between the Cu atoms leads to complete delocalization of the single unpaired electron over both Cu nuclei within a stable Cu2S2 diamond core11-13. The terminal ligands coordinated to each Cu are different, with one Cu ligated by His and a more distant methionine, and the other ligated by His and the amide O of glutamine 10. Cu A centers are also found in nitrous oxide reductase enzymes. CuA assembly therefore imposes an important requirement for cellular copper, with the added complexity of ensuring selective loading into a heterogeneous dinuclear environment.
Figure 1.
Structure of the dinuclear CuA center (taken from pdb file 2CUA).
The assembly and metalation of cytochrome oxidase is a complex process requiring a large number of accessory proteins. In particular, assembly of the CuA center appears to be mediated by Sco, a mitochondrial inner membrane red copper protein which binds both Cu(II) and Cu(I) in a (Cys)2His ligand environment. In yeast and humans, Sco1 has been implicated in CuA assembly while a paralog Sco2 plays an additional poorly understood role 14-16. Deletion of Sco1 but not Sco2 in yeast induces a respiratory phenotype, whereas in humans mutations in either Sco1 or Sco2 result in severe Cox deficiency and early onset fatal clinical outcome. Perhaps the strongest evidence for the direct association of Sco with the CuA center has come from studies in B. subtilis where the scoΔ mutant eliminates the oxidation of the dye TMPD, a phenotype directly associated with a defect in electron transfer to the Cu center of the aa3 oxidase 17-19. Despite this genetic evidence, direct transfer from Cu-loaded Sco to apo-CuA has not been demonstrated. Rather, in Thermus thermophilus NMR studies have shown that the bis-thiol form of apo-Sco is capable of reducing the disulfide form of apo-CuA, while a separate Cu(I)-binding transporter PCuAC completes the transfer of Cu(I) into the resulting bis-thiolate CuA metal-binding site 20. Although elegant and compelling with respect to the Thermus system, these observations do not resolve the questions of CuA metalation in other systems, since PCuAC homologues are absent from eukaryotes and from many CuA-containing prokaryotes, including B. subtilis.
The complexity of in vivo CuA assembly emphasizes the need to gain a better understanding of the metalation process in model systems where CuA can be metalated using inorganic ions. Model CuA centers have been introduced into protein scaffolds 21,22 using protein engineering, an important system being the purple CuA-azurin in which the C-terminal loop of azurin has been replaced with a 17 amino-acid sequence that provides the ligand side chains and the additional cysteine residue necessary to construct the CuA center 6. Recent studies by Lu and coworkers on the metalation of this center from inorganic copper has provided useful insights into the mechanism of the process, including the detection of mononuclear intermediates 23. At low Cu(II) to protein ratios, three mononuclear species were detected appearing sequentially along the pathway to dinuclear CuA assembly. Early in the process a “red” (Sco-like) type 2 Cu(II)-thiolate species with absorption maxima at 390 nm was observed, which rapidly converted to a novel intermediate Ix with λmax 410 nm and then to a “blue” copper center with λmax 640 nm before converting into the final purple CuA product. These species were similar to those reported earlier for the metalation of the CuA center of nitrous oxide reductase 24, and suggested a common pathway of copper incorporation. However, neither the red intermediate nor the intermediate Ix could be isolated as homogeneous entities frustrating attempts to accurately determine their identity.
Thermus thermophilus (Tt) cytochrome ba3 contains a CuA center in subunit 2 which can be isolated as a soluble domain in which the N-terminal transmembrane anchor has been cleaved 10,25. When expressed as an apo protein, this construct rapidly reacts with aqueous Cu(II) to generate the purple mixed-valence CuA center which can be reduced with dithionite to form the di-Cu(I) form. In previous work we used EXAFS spectroscopy to characterize the uniquely short Cu-Cu distances in the mixed-valence and di-Cu(I) forms 9, and the interaction of the M160 thioether with the Cu center in the selenomethionine (SeM) -substituted form 8. In the present paper we have extended these studies to explore the mechanism of copper addition to the apo protein to form either the mixed-valence or di-Cu(I) species. We have again used the SeM substituted protein, where the Se atom provides a ligand-directed spectroscopic probe of coordination via XAS at the Se edge 26-28. The formation of mixed-valence CuA has been studied by stopped-flow UV-vis, EPR, and XAS at both Cu and Se edges, while the formation of fully reduced di-Cu(I) CuA has been studied by XAS alone. Our results establish important differences between the metalation reactions of T. thermophilus and purple CuA azurin, and allow us to extend mechanistic inferences to formation of the di-Cu(I) state which may be more relevant to biological CuA assembly. In particular, we find that T. thermophilus CuA assembles more rapidly than reported for other CuA systems, and that the dominant intermediate along the pathway to mixed-valence is a new green species with λmax = 460 nm. This intermediate has been isolated in a homogeneous state, and shown to be a mononuclear Cu(II)-(His)(Cys)2 species with no observable Cu(II)-(Met) interaction. Reduction with dithionite generates its Cu(I) homologue which is again mononuclear, but now shows a strong interaction with the Met160 thioether. The results suggest different pathways for the mixed-valence and di-Cu(I) assembly, and lead to important inferences with respect to chaperone-mediated copper transfer to CuA.
Materials and methods
Expression of wild-type His6-Tt CuA
Ten milliliters of culture medium (LB and 50mg/mL kanamycin) was inoculated from a freshly streaked plate of BL21(DE3) cells containing the wild-type (WT) His6-Tt CuA plasmid. After overnight incubation, this culture was used to inoculate a 1L flask of culture medium. After approximately 3-4 hours of incubation at 37° C with shaking, the OD600 of the cell culture reached 0.6 - 0.8. At this point, the cell culture was induced by addition of IPTG to a final concentration of 0.4 mM and incubated for 4 hours at 37° C. Cells were harvested by pelleting via centrifugation at 5000 × g for 15 minutes. Typically, 1L of culture produced 8-10 grams of cells.
Expression of selenomethionine (SeM) His6-Tt CuA
Ten milliliters of culture medium (LB and 50mg/mL kanamycin) were inoculated from a freshly streaked plate of BL21(DE3) Met auxotrophic cells containing the His6-Tt CuA plasmid. After overnight incubation at 37° C, 100 microliters of this culture was used to inoculate a 10 milliliter flask of minimal culture medium which included L-methionine. After overnight incubation at 37° C with shaking, this culture was used to inoculate a 1L flask of minimal medium that contained SeM in place of methionine. After approximately 8 hours, the OD600 of the cell culture reached 0.6 - 0.8. At this point, the cell culture was induced by addition of IPTG to a final concentration of 0.4 mM, and incubated for 6 hours at 30° C. Cells were harvested by pelleting via centrifugation at 5000 × g for 15 minutes. Typically, 1L of culture produced 4 grams of cells.
Purification of wild-type and selenomethionine Tt CuA
Pelleted cells were resuspended in lysis buffer, lysed using a French Press, and the cell debris pelleted by centrifugation. The lysate was passed through a 0.45 micron syringe filter. Tt CuA was purified from the lysate by chromatography on a nickel-NTA affinity column. The His6 tag was cleaved by overnight incubation with r-TEV protease followed by dialysis against imidazole-free buffer, and a second metal affinity chromatography step was used to remove the cleaved His6 tag. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE) (8-12% gradient stained with Coomassie brilliant blue R-250) showed a single band. Following purification, the protein was dialyzed into 50mM phosphate buffer, pH 7.0 with 10% glycerol and immediately stored at 80° C. Both the WT and SeM Tt CuA protein concentrations were routinely determined by the bicinchoninic acid (BCA) assay, and Inductively Coupled Plasma Optical Emission Spectroscopy (ICP-OES) was used to accurately determine selenium content in the SeM Tt CuA. Protein yield of the WT Tt CuA was ~25 mg per liter of culture medium, and yield of the SeM variant was generally ~12 mg per liter. Before use, apo-Tt CuA samples were thawed, dialyzed into the desired buffer, and then incubated anaerobically with 3 mM tris(2-carboxyethyl)phosphine (TCEP) for up to 1 hour to reduce any disulfide crosslinks at the CuA site. The reduced protein was then dialyzed into excess anaerobic phosphate buffer (50 mM, pH 7) overnight to remove the TCEP.
Reconstitution of wild-type and SeM apo-Tt CuA with Cu(I) and Cu(II)
Reconstitution was carried out in a Coy anaerobic chamber to prevent oxidation of the cysteine residues. Reduced and dialyzed protein was incubated with up to a 3-fold excess of Cu(II) using an aqueous Cu(SO4) solution, or up to a 3-fold excess of Cu(I) using tetrakis acetonitrile copper (I) hexafluorophosphate ([Cu(CH3CN)4]PF6) in acetonitrile solvent. The protein was stirred while using a syringe pump which was set to deliver the given metal ion solution at a rate of 1 microliter per minute. The final concentration of acetonitrile did not exceed 10% of the total volume of protein solution. The reconstituted protein was then dialyzed exhaustively with metal-free anaerobic buffer. Upon reconstitution with Cu(II), both wild-type and SeM apo-Tt CuA exhibited the expected purple color and remained stable indefinitely. Reconstitution with Cu(I) gave colorless solutions which were found to oxidize back to the purple mixed-valence species on removal of excess Cu(I) by dialysis. Therefore, for Cu(I) reconstitution, excess Cu(I) was removed by passing through three successive desalting columns equilibrated with buffer that contained decreasing concentrations of acetonitrile. The resulting samples were then flash-frozen in liquid N2 to prevent oxidation. Bound metal concentrations were determined by ICP-OES. Partial reconstitution of WT and SeM-Tt CuA with Cu (II) was performed on reduced and dialyzed proteins, by slow, anaerobic addition of metal solution using a syringe pump set to deliver a pre-determined molarity of metal ions at 0.8-0.9:1 (SeM) or 0.5:1 (WT) copper to protein mole ratios.
UV-vis and EPR Spectroscopic measurements
Electronic spectra were recorded on a Cary 50 spectrophotometer. Electron paramagnetic resonance measurements were carried out on a Bruker ER085CS spectrometer under the following conditions: 9.4 Ghz frequency, 120 K, 2mW power, 25 db gain, 4 G modulation amplitude and a sweep time of 42 s. To determine the relative concentrations of paramagnetic copper in a given sample, a series of standard solutions containing 100-600 μM Cu(II)-EDTA in 50 mM phosphate buffer at pH 7.0 were measured in the same tubes as were used for protein samples. The concentrations of paramagnetic copper were determined by double integration. EPR spectra were simulated using SIMPIP as previously described 17,18,29,30.
X-ray absorption data
X-ray absorption data were collected at the Stanford Synchrotron Radiation Lightsource. The extended X-ray absorption fine structure (EXAFS) and X-ray absorption near-edge structure (XANES) of Cu (9.8 keV) and Se (12.6 keV) were measured on beamline 9-3 and 7-3 using a Si 220 monochromator with crystal orientation φ = 90°, and a Rh-coated mirror located upstream of the monochromator set to 13 keV (Cu) or 15 keV (Se) cutoff to reject harmonics. Samples were measure as frozen aqueous glasses in 20-25% ethylene glycol at temperatures between 7 and 15 K, and the XAS was detected as Kα fluorescence using either a 100-element (beamline 9-3) or 30-element (beamline 7-3) Canberra Ge array detector. A Z-1 metal oxide filter (Ni, As) and Soller slit assembly was placed in front of the detector to attenuate the elastic scatter peak. Four to six scans of a buffer blank were measured at each absorption edge and subtracted from the raw data to produce a flat pre-edge and eliminate residual Ni/As Kβ fluorescence of the metal oxide filter. Energy calibration was achieved by placing a Cu or Se metal foil between the second and third ionization chamber. Data reduction and background subtraction were performed using EXAFSPAK 31. The data from each detector channel were inspected for drop outs and glitches before being included into the final average. Spectral simulation was carried out using the program EXCURVE 9.2 as previously described 8,9,32,33.
Stopped-flow spectrophotometry
SeM-Tt CuA Cu(II) binding kinetics were followed by stopped-flow using an Applied Photophysics SX-20 stopped flow spectrometer. In the first series of experiments, TCEP-reduced anaerobic apo protein was rapidly mixed with an excess of anaerobic Cu(SO4)(aq) to form the SeM Tt CuA-Cu(II) intermediates and final mixed valence form. In a second series of experiments TCEP-reduced apo protein was rapidly mixed with 0.4 molar equivalents Cu(II) with the purpose of following the formation and decay of the green mononuclear intermediate under copper limiting conditions which precluded binding of a second Cu atom in the dinuclear site. Kinetic data were analyzed using the program DYNAFIT 34 and by the Pro-K II global analysis software package (Applied Photophysics).
Results
Identification of a green copper intermediate in Tt CuA
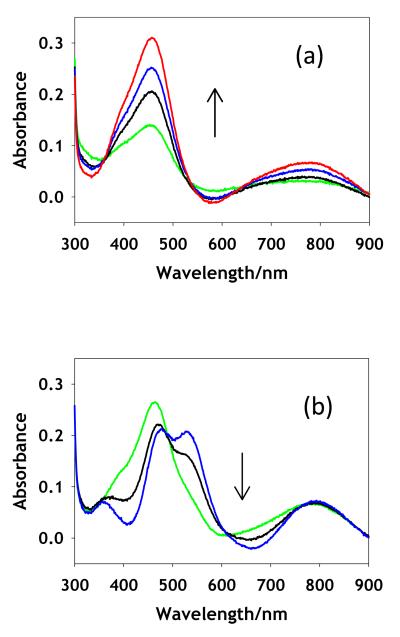
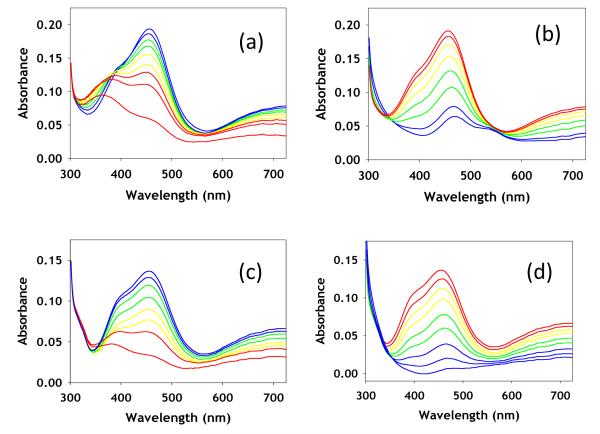
Slow addition of Cu(II) ions to TCEP-reduced, anaerobic apo CuA led to a rapid conversion to an intense green species absorbing at 460 nm with a shoulder at ~410 nm, and less intense absorptions at 660 and 790 nm. This green species was stable at copper to protein ratios at or below 0.5:1 under anaerobic conditions for the WT protein, but showed greater stability in the M160SeM derivative where the 460 nm species persisted to 0.8-0.9 Cu:P. For both derivatives, further addition of Cu(II) ions converted the green species to the native purple of the mixed valence state, with the characteristic bands at 360, 480, 530, and 790 nm. Fig. 2(a) shows the UV/vis spectrum due to the formation of the green intermediate from WT, while Fig. 2(b) shows its subsequent conversion to purple product. The UV/vis spectral bands attributable to the green species increase monotonically in the formation phase reaching a maximum, and thereafter convert to purple CuA with isosbestic behavior. This suggests that the green species in Fig. 2(a) corresponds to an intermediate which can be isolated as a single species at molar ratios of ~0.5 or below for WT or 0.9 or below for the SeM derivative.
Figure 2.
Titration of apo WT (M160S(Met)) CuA with increasing equivalents of aqueous Cu(II) sulfate. The spectra in panel (a) represent increase in formation of the green intermediates with Cu(II) to protein values (mM) of 0.2 (green), 0.3 (black), 0.4 (blue) and 0.5 (red). The spectra in panel (b) represent conversion of the intermediate to mixed-valence product with Cu(II) to protein values (mM) of 0.7 (green), 0.9 (black) and 1.25 (blue).
Electron Paramagnetic Resonance Spectra
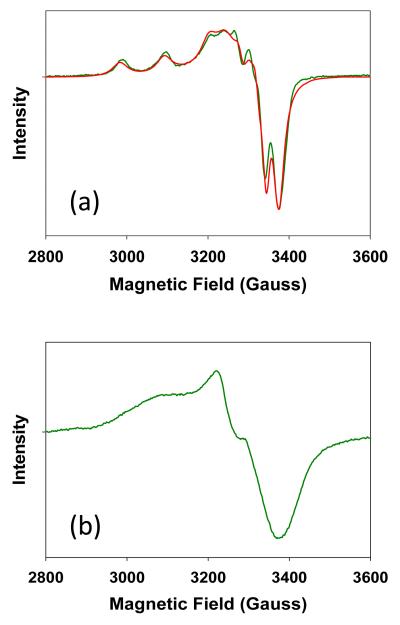
To investigate the nature of the green species, an EPR sample of apo SeM CuA was prepared by incubation with Cu(II) at 0.5 coppers to protein and then flash frozen to ensure stability in the open atmosphere. The resulting spectrum is shown in Fig. 3(a). A copper concentration that was less than stoichiometric was selected in order to minimize the possible presence of the mixed valence species, which would complicate extraction of the EPR signal of the intermediate. The spectrum is characteristic of a mononuclear type 2 cupric center with well-resolved hyperfine in both the parallel and perpendicular regions of the spectrum. Double integration of the signal versus Cu(II)-EDTA as standard resulted in an unpaired spin concentration of 98 per cent of the total copper, indicating that the intermediate was fully in the cupric state. The assignment as a mononuclear species was confirmed by simulation of the spectrum using the program SIMPIP which resulted in spin Hamiltonian parameters listed in the legend to Fig. 3. The spectrum is axial, with gx = gy < gz, corresponding to a mononuclear Cu(II) species and differs significantly from the mixed valence CuA spectrum, particularly with respect to the partially resolved seven-line parallel hyperfine splitting pattern observed in the M160SeM CuA center (Fig.3 (b) and 8) which results from coupling of the single unpaired spin over two copper nuclei. Instead, a tetragonally distorted four-line splitting pattern is observed with gx = 2.0127, gy = 2.0521, and gz = 2.1344. The anisotropy of the hyperfine interaction is calculated to be Ax = Ay = 47 MHz and Az = 326 MHz (116 G), respectively. The gz value is similar to that reported recently for red Cu(II)-Sco (gz = 2.1501, Az = 572 MHz) 29 which is classified as a type II copper-thiolate ligated by two cysteines and one histidine residue 35,36. The value of Az in the green intermediate places its coordination between type 1 and type 2, and differentiates the spectrum from both the Sco-like red copper thiolates and the “green” cupredoxin sites such as that found in nitrite reductase (A = 188 MHz (67 G)) 37.
Figure 3.
(a) Experimental and simulated X-band EPR spectrum of the green intermediate. The spectrum was simulated with the following Hamiltonian parameters: gx = 2.0127, gy = 2.0521, gz = 2.1344, Ax =Ay = 47 MHz, and Az = 326 MHz (116 G). (b) X-band EPR spectrum of purple mixed-valence CuA for comparison. Experimental conditions were as follows: frequency 9.400 GHz, temperature 120K, microwave power 2mW, modulation amplitude 4 G, instrument gain 25 db, sweep time 42 s.
X-ray Absorption Spectroscopy
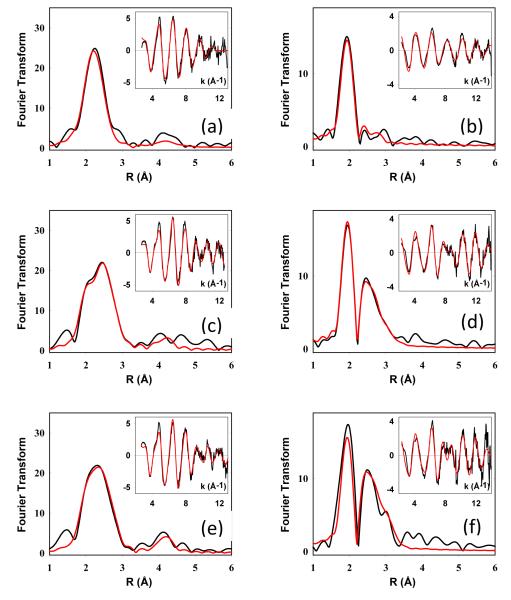
The nature of the intermediates formed in the metalation process was probed by X-ray absorption spectroscopy. To calibrate the system, we first remeasured and simulated EXAFS spectra for the mixed-valence and di-Cu(I) forms of the WT, and the SeM160 Tt CuA at both the Cu and Se edges. The results of these studies were similar to those previously reported 8,9, and are shown for reference in Figure S1 and Table S1 of the Supplementary Information. The coordinate structure of the green intermediate formed in both WT and SeM derivatives was also probed by X-ray absorption spectroscopy at both the Cu and Se edges. Experimental and simulated data are shown in Figure 4 with the parameters used in the fits listed in Table 1. Cu K-edge spectra for the SeM intermediate (Fig. 4 (a)) could be simulated by 1 Cu-His ligated at 1.98 Å together with 2 Cu.S(Cys) ligated at 2.22 Å. This coordination suggests that the SeM160 ligand is not coordinated in the mononuclear Cu(II) intermediate. A maximum of 0.15 Cu-Se could be tolerated by the simulation accompanied by ~0.2-0.3 Cu-Cu suggesting a small contaminating fraction of the mixed-valence product. Measurements at the Se edge confirmed this result as shown in Fig. 4(b), where the Se EXAFS consists entirely of a single Se-C shell from the methyl and methylene C atoms covalently bound to Se in the selenomethionine residue. Consistent with the Cu data, a small component (~0.2) of Se-Cu could be fitted to the data again suggesting the presence of a small percentage of mixed-valence product. Studies on the WT green intermediate (at the Cu edge only) were also consistent with these conclusions, refining to 1 Cu-His at 1.98 Å, and 2 Cu-S at 2.27 Å, although in this case the S shell cannot be unambiguously assigned to the Cys residues such that Met coordination is not excluded by the data (Table 1).
Figure 4.
Experimental and simulated Fourier transforms and EXAFS (insets) for the Cu and Se EXAFS of mononuclear intermediates in the metalation of M160SeM CuA. Spectra (a), (c) and (e) on the left are Cu EXAFS and spectra (b), (d) and (f) on the right are Se EXAFS. Spectra represent the following samples top (a) and (b), green mononuclear Cu(II) intermediate; middle (c) and (d), mononuclear Cu(I) intermediate prepared by dithionite reduction of the green intermediate; bottom (e) and (f), mononuclear Cu(I) intermediate prepared by reconstitution of apo bis-thiol CuA with [Cu(I)(CH3CN)4]PF6. Parameters used in the fits are listed in Table 1.
Table 1.
Fits obtained to the EXAFS of mononuclear intermediates formed during the metalation of the M160SeM variant of T. thermophilus CuA
| Sample/Fit | Fa | C-N(His)b | Cu-Se | Cu-S | -E0 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Noc | R (Å)d |
DW (Å2) |
Noc | R (Å)d |
DW (Å2) |
Noc | R (Å)d |
DW (Å2) |
|||
| Cu edge (S-Met) | |||||||||||
| Cu(II) | 0.540 | 1 | 1.98 | 0.003 | 2 | 2.27 | 0.013 | 1.1 | |||
| Cu(I)-Dithio Fit A | 0.514 | 1 | 2.10 | 0.030 | 2 | 2.20 | 0.015 | 0.3 | |||
| 1 | 2.33 | 0.003 | |||||||||
| Cu(I)-Dithio Fit B | 0.478 | 1 | 1.99 | 0.012 | 1 | 2.21 | 0.007 | 1.7 | |||
| 1 | 2.33 | 0.002 | |||||||||
| Cu edge (Se-Met) | |||||||||||
| Cu(II) | 0.615 | 1 | 1.99 | 0.020 | 0.15 | 2.48 | 0.012e | 2 | 2.22 | 0.013 | 1.0 |
| Cu(I)-Dithio Fit A | 0.443 | 1 | 1.98 | 0.030 | 1 | 2.47 | 0.007 | 2 | 2.21 | 0.014 | −1.9 |
| Cu(I)-Dithio Fit B | 0.465 | 1 | 1.96 | 0.004 | 1 | 2.47 | 0.006 | 1 | 2.23 | 0.002 | −0.4 |
| Cu(I)-ACN Fit A | 0.442 | 1 | 1.93 | 0.025 | 1 | 2.46 | 0.007 | 2 | 2.21 | 0.011 | −3.0 |
| Cu(I)-ACN Fit B | 0.408 | 1 | 1.97 | 0.003 | 1 | 2.47 | 0.008 | 1 | 2.23 | 0.002 | 0.2 |
| Se edge (Se-Met) | Se-C(met) | Se-Cuf | Se-S | ||||||||
| Cu(II) | 2 | 1.96 | 0.006 | 0.2e | 2.49 | 0.012 | |||||
| Cu(I)-Dithio | 0.703 | 2 | 1.97 | 0.004 | 0.9 | 2.48 | 0.007 | 1 | 3.09 | 0.025 | 5.1 |
| Cu(I)-ACN | 1.15 | 2 | 1.96 | 0.005 | 0.9 | 2.49 | 0.008 | 1 | 3.07 | 0.014 | 4.2 |
F is a least-squares fitting parameter defined as ( Data - Model )2
Fits modeled histidine coordination by an imidazole ring, which included single and multiple scattering contributions from the second shell (C2/C5) and third shell (C3/N4) atoms respectively. The Cu-N-Cx angles were as follows: Cu-N-C2 126°, Cu-N-C3 −126°, Cu-N-N4 163°, Cu-N-C5 −163°.
Coordination numbers are generally considered accurate to ± 25%
In any one fit, the statistical error in bond-lengths is ±0.005 Å. However, when errors due to imperfect background subtraction, phase-shift calculations, and noise in the data are compounded, the actual error is probably closer to ±0.02 Å.
This shell is accompanied by ~0.3 Cu-Cu at 2.48 Å, and arises from a small proportion of mixed-valence dinuclear species
Samples were prepared at Cu to protein ratios below 1 to protect against conversion of the Cu(II) monomer to the dinuclear species which takes place at Cu:P < 1. Such samples will therefore have Se:Cu ratios less than 1, with some of the Se-containing sites in the apo form. The values of the Se-Cu coordination numbers determined from EXAFS simulation have therefor been renormalized by dividing by the Se:Cu ratio in the protein sample. This process renders the Cu-Se and Se-Cu coordination numbers determined from the Cu and the Se edges comparable.
We next inquired as to the whether a similar mononuclear intermediate could form from apo-enzyme and Cu(I). Cu(I) species were prepared by (i) reduction of the green Cu(II) mononuclear species with sodium dithionite and (ii) by reconstitution of apo protein with sub-stoichiometric amounts of tetrakis acetonitrile Cu(I) hexafluorophosphate [Cu(MeCN)4.PF6], using established protocols 38. For WT where where Se XAS is absent, the intermediate was formed using 0.5 mole equivalents. For the SeM derivative, mole ratios of 0.9:1 were employed so as to maximize the formation of the intermediate and avoid contribution from apo SeM protein at the Se edge. Cu K edge Fourier transform and EXAFS data for the Cu(I) species formed from the SeM derivative by each of these methods are shown in Fig.4(c) and (e) with Se K edge data shown in Fig 3 (d) and (f). Parameters used in the simulations are listed in Table 1. Inspection of the Se data show a clear difference relative to the Cu(II) intermediates, namely that a strong Se-Cu interaction is now visible as an intense second-shell peak at ~2.5 Å in the FTs. This provides unambiguous evidence that Cu(I) recruits the Met ligand. The Cu data confirm the presence of a Cu-Se interaction for both dithionite reduction and Cu(MeCN)4 reconstitution at the same (2.46-2.49 Å) distance.
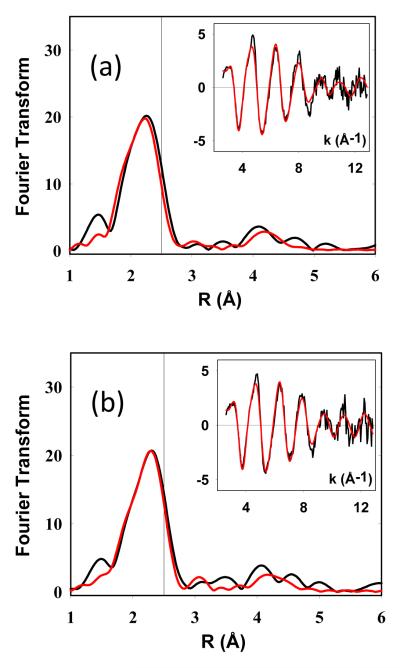
Whereas the green Cu(II) intermediate is clearly mononuclear both from the EPR and XAS data, a similar assignment for the putative Cu(I) mononuclear species is not as definitive. Dithionite reduction of the Cu(II) intermediate would be expected to generate a mononuclear Cu(I) species, but the coordination is different for the two oxidation states, since the Se edge data clearly shows Cu(I) coordinated by the M160 ligand. However, detailed analysis of the XAS data reveals ambiguities in distinguishing between two possible structural models for the Cu(I) coordination. The first of these involves a simple Cu(I) mononuclear species in which the Cu(I) ion adopts a similar structure to the Cu(II) mononuclear species but increases its coordination by binding the M160 S or Se donor. In this scenario, it we expect the Se edge data to show 1 Se-Cu interaction, and the Cu-edge data to show 1 Cu-Se (at the same distance) and no Cu-Cu interaction. This is modeled in Table 1 for both the dithionite reduced form and the Cu(I) reconstituted form and is consistent with a mononuclear assignment. The second possibility is that the Cu(I) mononuclear species is unstable relative to formation of the di-Cu(I) product, such that at 1:1 Cu(I) to P, equal amounts of di-Cu(I) and apo protein are present. Here we expect the Se data to show 0.5 Se-Cu (since half the Se in the sample is apo), while the Cu data should show 0.5 Cu-Se and 1 Cu-Cu, similar to the fit observed for the di-Cu(I) (Supporting Info, Figs. S1 and Table S1). Put in another way, comparison of the Se edge data for 1:1 with the fully formed 2:1 Cu(I):P species should show equal intensity of the Se-Cu peaks in the FT if the 1:1 species is mononuclear, but be half as intense if the species is a mixture of 2:1 and apo protein. Fig 5(a) compares Se edge FTs for the Cu(I) species generated by dithionite reduction of the green intermediate (Cu(I):P = 0.9:1) and purple mixed valence (Cu(I):P = 2:1), from which it can be seen that the Se-Cu peaks are (i) of comparable intensity and (ii) have very different shapes, suggesting differences in the identity of the species and favoring the mononuclear model. Additional evidence comes from analysis of the Cu EXAFS data, where good fits are obtained with 1 Cu-Se and no Cu-Cu. However this latter result is subject to uncertainty since the Cu-Se and Cu-Cu interactions have the same distance and similar phase shifts, making them difficult to distinguish and quantify.
Figure 5.
Experimental and simulated Fourier transforms and EXAFS (insets) for the Cu EXAFS of mononuclear intermediates in the metalation of the WT (M160S(Met) CuA. Top spectra, mononuclear Cu(I) intermediate prepared by dithionite reduction of the green intermediate; bottom, mononuclear Cu(I) intermediate prepared by reconstitution of apo bis-thiol CuA with [Cu(I)(CH3CN)4]PF6. Parameters used in the fits are listed in Table 1. The vertical reference lines in each FT are drawn to illustrate the absence of a strong Cu-Cu interaction at 2.5 Å.
Paradoxically, distinguishing between mononuclear and dinuclear species is easier for the WT (S-Met) forms of CuA, since the species now differ only in the presence of the Cu-Cu. Figure 6 compares the Cu FTs and EXAFS for M160M (S-met) CuA prepared by dithionite reduction of its green mononuclear intermediate, and by reconstitution with 0.5:1 [Cu(MeCN)4]+. The data are well simulated by a mononuclear species with Cu(I) bound by 1 His, 1-2 Cys residues (vide infra) and 1 S(Met). The vertical line at ~2.5 Å represents where Cu-Cu is expected, and the lack of intensity at this distance indicates Cu-Cu scattering at 2.5 is absent from the spectra. These data confirm the assignment as mononuclear species.
Figure 6.
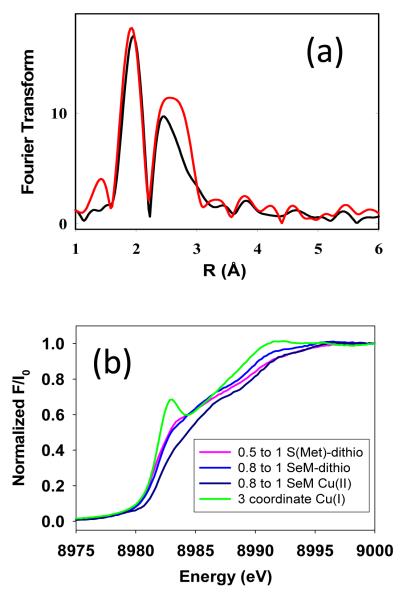
Evidence that 1:1 Cu(I) to protein species are mononuclear. Top panel, comparison of Fourier transforms of the Se EXAFS of the M160SeM species at Cu(I) to protein ratios of 1:1 and 2:1 showing near equivalency of the intensity of the Se-Cu peaks in the FT expected for a mononuclear Cu(I) species. Bottom panel, absorption edges of Cu(I) mononuclear species compared with the 3-coordinate Cu(I) derivative of the H135A variant of B. subtilis Sco (where Cu(I) is bound by 2 Cys and 1 Met residue). The low intensity of the 8983 eV peaks of the mononuclear Cu(I) CuA intermediates suggest 4-coordination.
A final ambiguity arises with respect to the number of coordinated cysteine residues in the mononuclear Cu(I) entities. Comparable fits (as judged by the value of F) can be obtained for S-Met and SeM derivatives using either 1 or 2 Cys residues, and are labeled Fits A and B for each species in Table 1. For the SeM derivatives where the Met donor atom is Se, Fit A (2 Cys residues per Cu) produces a very large and chemically unreasonable Debye-Waller factor for the Cu-His shell, whereas in Fit B (1 Cys) the DW values for both Cu-N and Cu-S are smaller and well within the expected limits. We also examined the absorption edges where the 8983 eV edge feature is often used as an indicator of coordination number. 3-coordinate species (corresponding to the 1 Cys model) have well resolved low-intensity peaks on the rising edge, whereas 4-coordinate species have featureless edges. Figure 5(b) compares the absorption edges of the mononuclear Cu(I) species with that of a genuine 3-coordinate Cu(I) species, the H135M variant of B. subtilis Sco which is coordinated to 2 Cys and 1 Met residue 39. The lack of 8983 eV peaks on the mononuclear CuA(I) are more suggestive of 4-coordination favoring the 2-cys model. Although this observation seems to contradict the modeling, one possibility is that the mononuclear Cu(I) may sample all the ligands in the coordination environment in a fluxional fashion, or be held in a pseudo tetrahedral structure by the influence of the fourth ligand.
Stopped Flow Kinetic Analysis of CuA Assembly
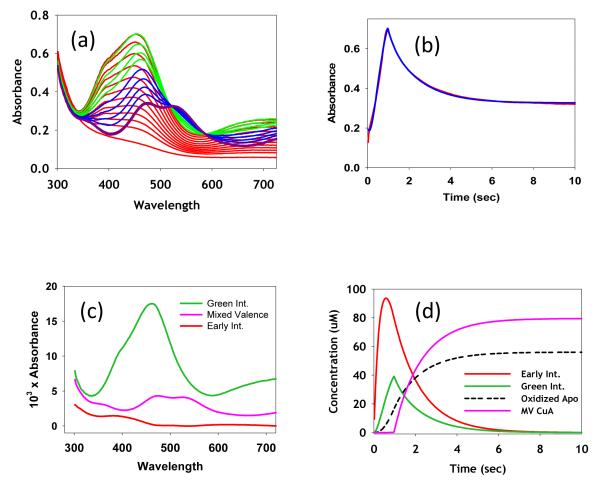
To gain further insight into the mechanism of CuA assembly, we studied the kinetics of the metalation process using stopped flow spectrophotometry. The rates of formation of the mixed-valence product and its mononuclear intermediates were determined, starting from the bis-thiolate apo protein and aqueous Cu(II) ion. The results of a typical experiment starting from 170 μM apo protein and 500 μM Cu(II) are shown in Figure 7(a) with the complete kinetic analysis given in Fig. 7(b)-(d). Fig. 7(a) shows the time course of the reaction on a time scale of 0 -10 s (10-104 ms) where the evolution of the spectra is color coded from red (0.04 – 84 ms) to green (1 – 1.64 s) to blue (2.84 – 9.64 s) to purple (final products above 10 s). It can be seen that a spectrum similar to the green mononuclear intermediate with λmax= 460 forms initially and is then cleanly converted to the mixed-valence product. Fig. 7(b) shows the absorbance versus time kinetic trace at 462 nm which is characterized by an increase to a sharp maximum followed by a slower decrease, corresponding to the respective formation and decay of the intermediate species. Absorbance versus time data was fit to trial mechanistic models using the program DYNAFIT 34 which solves the series of differential and mass balance equations which define any particular mechanism for the associated rate constants. First we tested a simple A → I → P reaction scheme, where apo protein (A) reacts with Cu(II) to form intermediate I, which subsequently converts into the mixed-valence product P, but this reaction sequence failed to reproduce the sharpness of the maximum. The fact that the green mononuclear Cu(II) intermediate was converted into the mixed-valence product suggested that the latter was formed by addition of Cu(I) to I. This prompted us to include steps that could generate Cu(I) including reaction of Cu(II) with the bis-thiolate apo protein to form Cu(I) and oxidized apo protein A(S-S),
or disproportionation of I,
Figure 7.
Stopped-flow kinetic analysis of the reaction of the bis-thiol apo M160SeM CuA with aqueous Cu(II) sulfate. (a) Transient spectra collected at increasing time intervals color coded as follows: red (0.04 – 84 ms), green (1 – 1.64 s), blue (2.84 – 9.64 s), purple (final products above 10 s). (b) Fit to the absorbance versus time data collected at 462 nm. (c) Absorption spectra of colored species (including intermediates) derived from a global kinetic analysis of data at all wavelengths, using the Pro-K II software package. (d) Time dependence of the formation and decay of colored species during the reaction. Fits, spectra of intermediates, and formation-decay curves were derived using the mechanism and rate constants listed in Table 2. Concentrations were apo protein, 170 μM and Cu(II) 500 μM
However, neither set of Cu(I) generating reactions improved the fit. The solution to the problem came from two experimental observations, first that very early in the reaction an additional species (the early intermediate) was present (Figure 8, vide infra), and second that the subsequent green intermediate was stable at sub-stoichiometric ratios of Cu(II), but converted to purple mixed-valence when the molar equivalents of Cu(II) rose above a threshold (Fig. 2). This latter result implied that Cu(I) is formed when the green intermediate reacts with Cu(II) and this Cu(I) may either react further with the green intermediate to form purple product or react with apo protein to form the Cu(I) mononuclear species. A mechanism incorporating all of these observations
was tested, and after refinement of the rate constants for each step, gave an excellent fit to the data as shown in Fig 7(b).
Figure 8.
Stopped-flow data at substoichiometric Cu to protein (0.4 : 1) for WT (left) and M160SeM (right). Top spectra (a) and (b) show increase of the green intermediate at 460 nm while bottom spectra (c) and (d) show the decay of the 460 nm green intermediate.
The PRO-K II software of the Applied Photophysics stopped flow instrument was used to complete the kinetic analysis. The mechanism determined above was used to undertake global fitting of the complete data set at all wavelengths and time points. This analysis generated the set of rate constants listed in Table 2, together with the spectra of each intermediate (Fig 7(c)) and the time course of their formation and decay (Fig 7(d)). Fits to the absorbance versus time curves for a selected number of wavelengths are given in Figure S2. The excellent quality of the fits over the complete wavelength range gives a high level of confidence in the postulated mechanism.
Table 2.
Rate constants for the individual reaction steps leading to the formation of the mixed-valence form of M160SeM T. thermophilus CuA.
| Reaction Stepa | Rate Constant |
|---|---|
| A + Cu(II) → Iearly | 7.5±1.1 × 103 mol−1s−1 |
| Iearly → I | 0.81±0.18 s−1 |
| I + Cu(II) → A(S-S) + 2Cu(I) | 3.0±0.31 × 103 mol−1s−1 |
| I + Cu(I) → P | 11.0±3.8 × 106 mol−1s−1 |
| A + Cu(I) → I-Cu(I) | indeterminate |
A is the bis-thiol apo protein, A(S-S) is the disulfide bridged apo protein, P is the mixed valence product, Iearly is the red copper intermediate, I is the green copper intermediate, and I-Cu(I) the mononuclear Cu(I) intermediate. I-Cu(I) may react with Cu(I) to form the di-Cu(I) product but no information on this step could be extracted from the data. The rate constant for reaction of A with Cu(I) is also not well defined.
As a further test of this mechanism, we followed the time course of the reaction under conditions of limiting Cu(II) at low temperature (10°C). These data (Fig. 8) confirmed the presence of the early intermediate as a species with λmax in the 350-400 nm range. Significantly, the S(Met) and SeM derivatives give rise to early intermediates with different absorption maxima at λmax = 360 and 380 nm respectively. In contrast, the absorption maxima of the green intermediates do not change although small differences in the ratio of the 460 nm main absorption to the 400 - 410 nm shoulder are apparent. As fully predicted by the mechanism, when Cu(II) is limiting, the mixed-valence product does not form (except in very low amounts for the S(Met) derivative late in the reaction), confirming that major route to its formation is by addition of Cu(II) to the green intermediate. We may speculate that addition of cupric ion to the green intermediate initially forms an unstable di-Cu(II) species which then rapidly decays to Cu(I) and oxidized apo protein, perhaps via the intermediacy of Cu(I)-thiyl radicals. However in the absence of excess Cu(II) this reaction cannot proceed: instead, the green intermediate loses intensity over longer time periods, suggesting that it undergoes slow autoreduction.
Discussion
Biological mechanisms of metal transport have been the subject of intense investigation. In particular, the metalation reactions of the CuA center of cytochrome oxidase have generated special interest because of (i) the uniqueness of its mixed-valence Cu1.5 – Cu1.5 electronic structure 13,40, and (ii) the role played by the putative chaperone Sco in its assembly 17. Whereas Sco has been shown to be essential for CuA assembly in vivo19,41,42, direct transfer between copper-loaded Sco and apo CuA has never been observed. On the other hand, transfer of Cu(I) center from the mononuclear periplasmic protein PCuAC has been shown to generate the dinuclear di-Cu(I) CuA 20. We undertook a detailed study of the metalation reactions of the T. thermophilus CuA with the goal of answering fundamental questions relating to the chemical mechanism, for example is CuA metalated via mononuclear intermediates, which redox state of copper binds preferentially, and what are the structures of the putative mononuclear intermediates in each oxidation state.
The dinuclear mixed-valence end product could be generated from the reaction of apo protein with aqueous Cu(II). Slow titration of sub-stoichiometric amounts of Cu(II) formed a green species with λmax=460 nm, which was clearly a mononuclear entity as judged by its UV-vis, EPR, and XAS spectral properties. However, stopped-flow kinetics showed that this was not the first intermediate to form. Early in the reaction, a species with an absorption maximum between 360 and 380 nm was observed which rapidly converted into the green intermediate. This early intermediate has also been observed in the metalation of nitrous oxide reductase 24 and purple copper azurin 23, where it has been ascribed to a “red copper” center 17,43-45, typical of a cupric thiolato species coordinated in a tetragonally distorted environment.
Nature of the intermediates
The binding of Cu(II) in both the intermediates is best discussed within the framework of the “coupled distortion” model for understanding the electronic structure in type 1 and type 2 copper thiolates, of the kind found in blue, green and red cupredoxin sites 46,47. Blue copper centers of the type found in azurin 48 or plastocyanin 49 are characterized by intense visible absorption around 600 nm, a weaker band at around 400 nm, and narrow A∥ hyperfine splittings, assigned to a trigonal planar coordination of two His residues and a Cys residue, with weak axial interaction to a Met and/or a carbonyl main chain O. The spectroscopic features have been assigned to a strong S p(π) interaction with the Cu dx2-y2 giving rise to the intense Cu-S CT band at 600 nm, and a weaker S p(σ) interaction of lower intensity at higher energy. The π-interaction results in a highly covalent Cu-S(Cys) interaction corresponding to 38% S(Cys) 3p in the Cu 3dx2-y2 orbital for plastocyanin 50,51. As the axial interaction strengthens, the site undergoes an increase in ligand field strength together with tetragonal distortion which is coupled to a rotation of the dx2-y2 orbital. The coupled distortion causes the π-interaction to weaken and shift to higher energy while the σ-interaction achieves better overlap and gains intensity resulting in the green T1 center found in nitrite reductase with two absorptions of almost equal intensity at 460 and 495 nm 46. In the limit of strong axial interaction, a 5-coordinate pseudo square pyramidal (type 2 copper) structure is formed, as exemplified by the red copper sites of nitrosocyanin where four ligands (2 His, 1 Cys and a water) occupy the equatorial plane with a strong 2.1 Å interaction to an axial Glu residue 43,44,52. Another example is found in Sco where 1 His, 2 Cys, and an endogenous 0/N (non-His) act as equatorial ligands 17,29. In these systems the Cu-S p(σ) interaction is now the dominant absorption at 350 -400 nm with much weaker S p(π) around 450 nm and much lower Cu-S covalency (20% for nitrosocyanin 44, 22% over both Cu-S bonds in Sco 17. As further confirmation of the validity of the coupled distortion model, it has been possible to convert green copper into blue copper by mutation of the axial Met in NIR to threonine 53, and a blue site into a red site via increasing the axial ligand field strength in azurin from Met to homocysteine 45.
The protein derived ligand set available for binding Cu(II) in the early intermediate of CuA is one or two His residues, one or two cysteines and the Met or Gln residues which act as axial ligands in the mixed-valence product. The absorption maxima of 360 nm for the S(Met) and 380 nm for the SeM derivative places the species in the category of a red copper center, where the axial interaction is strong enough to induce a 5-coordinate tetragonally distorted structure. An important observation is the fact that the absorption maxima are dependent on the donor atom in the M160 residue, undergoing a red shift when S(Met) is substituted by SeM. This effect has been reported previously in the H135M and H135SeM variants of B. subtilis Sco, where Se edge XAS confirmed the coordination of the SeM ligand to Cu(II) 39. Therefor it is likely that the early intermediate coordinates the two Cys residues in the equatorial plane with a strong axial interaction to M160. The identity of the remaining equatorial ligands is unclear but since this is an early capture complex, two water ligands would not be unreasonable.
In the Sco system, the coordinated H135 ligand has been shown to stabilize the Cu(II) center 18 and its mutation to either Ala or Met results in autoreduction of the Cu(II) and concomitant oxidation of the two thiolates to a disulfide. This chemistry suggests that the initial red copper center would be unstable but could stabilize itself via a rearrangement to bind the His residue and generate the green intermediate, which we have shown by EXAFS to have a N(His)+S(Cys)2 ligand set. While this coordination is similar to the WT Sco, the UV-vis spectrum is quite different, resembling more closely the green copper center of NIR. This suggests that the rearrangement that results from binding of the His residue may induce a tetragonal to trigonal perturbation which would increase the amount of S p(π) interaction, and increase the Cu-S covalency, and it is notable in this regard that the mixed-valence product is also highly covalent 13,54. A∥ values from EPR spectra provide a further point of comparison with values for the green intermediate and NIR of 326 and 188 MHz respectively 37, relative to the red copper sites of BSco (533 MHz) and nitrosocyanin (387 MHz 43). From this comparison, the green intermediate lies between the cupredoxin-like NIR spectrum and the tetragonally distorted nitrosocyanin, suggesting that the rearrangement induced by His binding decreases the axial interaction below that found in typical red copper centers. The absence of observable Cu-Se interactions in the XAS supports this view, since T1 sites with axial Met ligands seldom show any contribution from the Met S in the EXAFS 55. However, the value of A∥ is at best an approximate indicator of axial ligand strength since the WT NIR (green) has an identical A∥ to that of its M182T variant (blue) in which the moderately strong axial interaction with S(Met) has been eliminated 37.
The structure of the Cu(I) mononuclear intermediate is easier to formulate. Here EXAFS at both the Cu and Se edges provide good evidence for thioether coordination from M160 together with one of the available His residues. The coordination sphere is completed by addition of 1 or both of the Cys residues. Interestingly, detailed analysis of the XAS parameters suggested that the Cu(I) may sample all of its available ligands in a fluxional fashion, perhaps suggesting that the metal is “rattling around” in the site. This may reflect one reason why addition of a second Cu(I) to form the di-Cu(I) species is a facile reaction, whereas larger ions such as Hg(II) appear to form mononuclear sites preferentially 56.
Reaction Mechanism
Stopped-flow kinetic studies have suggested a mechanism in which the early red intermediate converts to the green intermediate, the fate of which is dependent on the availability of additional copper in the system. Excess copper in either oxidation state converts the green intermediate into mixed-valence. Addition of Cu(I) is straightforward mechanistically as the excess metal will populate the additional binding site and generate the mixed-valence product. Addition of excess Cu(II) also forms mixed-valence, and our kinetic analysis shows that this step is obligatory in the conversion of green to purple product. This suggests that Cu(II) can bind in the second site to form a transient di-Cu(II) species which is unstable with respect to disproportionation to form oxidized disulfide, and Cu(I). The Cu(I) can then rapidly react with residual green mononuclear species to form the purple product. The different stoichiometry at which the green intermediate is maximally populated varies between the S(Met) and SeM derivatives, and we believe that this reflects differences in the rates at which the S(Met) and SeM green mononuclear species react with excess copper. The rate of this reaction is likely to be influenced by the presence or absence of a coordinated S- or Se- methionine residue, since the latter will have differing stabilizing influence on the Cu(II) monomer. It is unclear why the di-Cu(II) species is so unstable, but Hay and coworkers have argued that the site is destabilized if the overall charge exceeds +3 56. This empirical observation explains the formation of Hg(II)Ag(I) derivatives which appear to be stable, and is consistent with the formation of mononuclear but not dinuclear Co(II) derivatives in CuA-azurin at high pH.
Comparisons with other CuA systems
The intermediates characterized in the present study show both similarities and differences from those described for other CuA systems such as purple CuA azurin and N2O reductase 23. The early intermediate appears to be similar, and is formulated as a red copper species. In CuA azurin under conditions of excess copper, the red copper species converts to a blue type 1 copper species with λmax = 640 nm, which clearly differs from the green intermediate of T.t CuA with λmax = 460 nm. This may be the result of small differences in the copper sites imposed by the different protein matrices, and is not inconsistent with the observed differences in Cu-Cu distances, where the azurin model exhibits a shorter Cu-Cu in the mixed-valence state than T.t CuA (2.39 versus 2.44 Å respectively 9,57). At low copper, a new species Ix is observed in the azurin model, which is only transiently stable, and converts either aerobically to the T1 site or anaerobically to a Cu(I) species. The spectral properties of Ix show similarities to the T.t green intermediate, with absorptions at 410, ~460 and 760, but the relative intensities of the 410 and 460 nm bands are reversed. However, whereas the A∥ values are similar (326 versus 345 MHz; 116 versus 123 G) the gz values differ significantly (2.13 versus 2.25) for the green intermediate and Ix respectively. Wilson and coworkers propose a structure for Ix based on similarities to imidazole and pyrazole adducts of horse liver alcohol dehydrogenase with a suggested (His)(Cys)2 ligand set similar to that of the green intermediate presented above. However, the large differences in gz values suggest differences in electronic structure which are clearly manifest in the decreased stability of Ix over the green intermediate.
Biological Implications
The pathway for CuA metalation in vivo is still poorly understood. Whereas Sco-type proteins appear to be essential, direct transfer from Sco to CuA has not been demonstrated. The periplasmic Cu(I)-binding protein PCuAC remains the only protein copper donor shown to successfully metalate CuA 20. The reaction of T. thermophilus Cu(I)-loaded PCuAC with T. thermophilus apo CuA generated a NMR spectrum characteristic of the fully metalated di-Cu(I) CuA species, but the study did not detect mononuclear intermediates, nor any protein-protein interactions. PCuAC homologues are not universally present in prokaryotes, and are absent from eukaryotes, calling into question a direct role in CuA metalation. A recent study of CuA assembly in Rhodobacter spheroides which expresses both PCuAC and Sco (PrrC) homologues showed that deletion strains of either or both proteins lowered the accumulation of the CuA-containing aa3-type oxidase but showed a stronger phenotype for PrrC (96%) than for PCuAC (86%) 58. The data indicated distinct roles for each chaperone, as overexpression of PCuAC in a ΔPrrC background did not compensate for the defect. The defects in both deletion strains could be partially restored in cells grown at high (1.6 μM) added Cu(II) which mitigated against the role suggested for PrrC/Sco by Abriata et al. 20 as a disulfide reductase. The two scenarios most consistent with the findings on R. spheroides CuA assembly were (i) each chaperone could add one copper or (ii) PrrC adds both coppers but is itself metalated by PCuAC. In mitochondria where PCuAC is absent, it has been shown that Sco1 is metalated by Cox17 (absent in bacteria) which would explain the lack of a universal requirement for PCuAC in CuA assembly 59,60.
While the evidence is compelling for metalation of both copper sites by Sco/PrrC, the inability to demonstrate this chemistry in vitro is worrisome. It is possible that removal of the membrane anchor to produce soluble Sco constructs perturbs its transfer chemistry, or destabilizes mechanistically-relevant oligomers, but in the absence of such effects, the lack of transfer chemistry suggests a missing component, and argues in favor of a mechanism in which each of the two copper ions is added sequentially by a separate chaperone. In previous work, we showed that Bacillus subtilis Sco variants that lack the coordinating His residue (H135) are inactive in assembly of the aa3 oxidase 17,18,39. This His residue stabilizes the Cu(II) state as shown by the large increase in reduction rate for the H135A variant. The Cu(I) state is stabilized in the H135M variant, which has properties similar to those of the reduced WT protein, but this derivative is completely inactive. These data suggest that the Cu(II) state of Sco is important for CuA assembly, and when this oxidation state is inaccessible or unstable as in H135A or H135M variants, the transfer is impeded. These observations support the idea that Sco may provide a cupric ion to generate the green mononuclear intermediate, but the assembly of the site by Cu(I) addition may occur via a different pathway. Like those of others, our own efforts to metalate CuA in either oxidation state using copper loaded WT Sco or its H135A variant have been unsuccessful, leading instead to a dead-end complex with unusual UV-vis properties. Given the facile metalation chemistry of CuA from inorganic ions, and the weakened binding affinity of the H135A variant of Sco, these finding seem difficult to reconcile. Further work is underway to use the techniques described herein to probe the transfer chemistry of CuA with Sco and PCuAC with the goal of resolving these issues.
Supplementary Material
Acknowledgement
This work was funded by a grant from the National Institute of Health R01 GM054803 to NJB. KNC is supported by National Science Foundation Graduate Research Fellowship DGE-0925180. We gratefully acknowledge the use of facilities at the Stanford Synchrotron Radiation Lightsource which is supported by the National Institutes of Health Biomedical Research and Technology Program Division of Research Resources, and by the US Department of Energy Office of Biological and Environmental Research.
Footnotes
Supporting Information Two figures describing fits to the Cu and Se EXAFS of the fully metalated dicopper forms of T. thermophilus M160SeM of CuA in the mixed-valence and fully reduced oxidation states, and multiwavelength fits to kinetics of formation of the mixed-valence M160SeM derivative; one table of EXAFS parameters for simulation of mixed-valence and di-Cu(I) forms of WT and M160SeM CuA. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaona R, Yoshikawa S. Science. 1996;272:1136. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- (2).Ostermeir C, Harrenga A, Ermler U, Michel H. Proc. Natl. Acad. Sci. USA. 1997;94:10547. doi: 10.1073/pnas.94.20.10547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Soulimane T, Buse G, Bourenkov GP, Bartunik HD, Huber R, Than ME. EMBO J. 2000;19:1766. doi: 10.1093/emboj/19.8.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Svensson-Ek M, Abramson J, Larsson G, Tornroth S, Brzezinski P, Iwata S. J. Mol. Biol. 2002;321:329. doi: 10.1016/s0022-2836(02)00619-8. [DOI] [PubMed] [Google Scholar]
- (5).Liu B, Chen Y, Doukov T, Soltis SM, Stout CD, Fee JA. Biochemistry. 2009;48:820. doi: 10.1021/bi801759a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Savelieff M, Lu Y. J. Biol. Inorg. Chem. 2010;15:461. doi: 10.1007/s00775-010-0625-2. [DOI] [PubMed] [Google Scholar]
- (7).Blackburn NJ, Barr ME, Woodruff WH, van der Oost J, de Vries S. Biochemistry. 1994;33:10401. doi: 10.1021/bi00200a022. [DOI] [PubMed] [Google Scholar]
- (8).Blackburn NJ, Ralle M, Gomez E, Hill MG, Patsuszyn A, Sanders D, Fee JA. Biochemistry. 1999;38:7075. doi: 10.1021/bi982500z. [DOI] [PubMed] [Google Scholar]
- (9).Blackburn NJ, de Vries S, Barr ME, Houser RP, Tolman WB, Sanders D, Fee JA. J. Am. Chem. Soc. 1997;119:6135. [Google Scholar]
- (10).Williams PA, Blackburn NJ, Sanders D, Bellamy H, Stura EA, Fee JA, McRee DE. Nat. Struct. Biol. 1999;6:509. doi: 10.1038/9274. [DOI] [PubMed] [Google Scholar]
- (11).Glaser T, Hedman B, Hodgson KO, Solomon EI. Acc. Chem. Res. 2000;33:859. doi: 10.1021/ar990125c. [DOI] [PubMed] [Google Scholar]
- (12).Solomon EI, Hedman B, Hodgson KO, Dey A, Szilagyi RK. Coord. Chem. Rev. 2005;249:97. [Google Scholar]
- (13).Williams KR, Gamelin DR, Lacroix LB, Houser RP, Tolman WB, Mulder MC, de Vries S, Hedman B, Hodgson KO, Solomon EI. J. Am. Chem. Soc. 1997;119:613. [Google Scholar]
- (14).Leary SC, Cobine PA, Kaufman BA, Guercin GH, Mattman A, Palaty J, Lockitch G, Winge DR, Rustin P, Horvath R, Shoubridge EA. Cell Metab. 2007;5:9. doi: 10.1016/j.cmet.2006.12.001. [DOI] [PubMed] [Google Scholar]
- (15).Leary SC, Kaufman BA, Pellecchia G, Guercin GH, Mattman A, Jaksch M, Shoubridge EA. Hum. Mol. Genet. 2004;13:1839. doi: 10.1093/hmg/ddh197. [DOI] [PubMed] [Google Scholar]
- (16).Leary SC, Sasarman F, Nishimura T, Shoubridge EA. Hum Mol Genet. 2009;18:2230. doi: 10.1093/hmg/ddp158. [DOI] [PubMed] [Google Scholar]
- (17).Siluvai GS, Mayfield M, Nilges MJ, DeBeer George S, Blackburn NJ. J. Am. Chem. Soc. 2010;132:5215. doi: 10.1021/ja910759v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Siluvai GS, Nakano M, Mayfield M, Nilges MJ, Blackburn NJ. Biochemistry. 2009;48:12133. doi: 10.1021/bi901480g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Mattatall NR, Jazairi J, Hill BC. J. Biol. Chem. 2000;275:28802. doi: 10.1074/jbc.M002741200. [DOI] [PubMed] [Google Scholar]
- (20).Abriata LA, Banci L, Bertini I, Ciofi-Baffoni S, Gkazonis P, Spyroulias GA, Vila AJ, Wang S. Nat Chem Biol. 2008;4:599. doi: 10.1038/nchembio.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Andrew CR, Lappalainen P, Saraste M, Hay MT, Lu Y, Dennison C, Canters GW, Fee JA, Slutter CE, Nakamura N, Sanders-Loehr J. J. Am. Chem. Soc. 1995;117:10759. [Google Scholar]
- (22).Wilmanns M, Lapplalainen P, Kelly M, Sauer-Eriksson E, Saraste M. Proc. Natl. Acad. Sci. U.S.A. 1995;92:11955. doi: 10.1073/pnas.92.26.11955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Wilson TD, Savelieff MG, Nilges MJ, Marshall NM, Lu Y. J. Am. Chem. Soc. 2011;133:20778. doi: 10.1021/ja205281t. [DOI] [PubMed] [Google Scholar]
- (24).Savelieff MG, Wilson TD, Elias Y, Nilges MJ, Garner DK, Lu Y. Proc. Natl. Acad. Sci. USA. 2008;105:7919. doi: 10.1073/pnas.0711316105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Slutter CE, Sanders D, Wittung P, Malmstrom BG, Aasa R, Richards JH, Gray H, Fee JA. Biochemistry. 1996;35:3387. doi: 10.1021/bi9525839. [DOI] [PubMed] [Google Scholar]
- (26).Bagai I, Rensing C, Blackburn NJ, McEvoy MM. Biochemistry. 2008;47:11408. doi: 10.1021/bi801638m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Barry AN, Blackburn NJ. Biochemistry. 2008;49:4916. doi: 10.1021/bi8001049. [DOI] [PubMed] [Google Scholar]
- (28).Barry AN, Clark KM, Otoikhian A, van der Donk WA, Blackburn NJ. Biochemistry. 2008;47:13074. doi: 10.1021/bi801438g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Andruzzi L, Nakano M, Nilges MJ, Blackburn NJ. J. Am. Chem. Soc. 2005;127:16548. doi: 10.1021/ja0529539. [DOI] [PubMed] [Google Scholar]
- (30).Nilges MJ. Illinois EPR Research Center (IERC), University of Illinois; Urbana-Champaign: 1979. [Google Scholar]
- (31).George GN. Menlo Park, CA; Stanford Synchrotron Radiation Laboratory: 1995. [Google Scholar]
- (32).Gurman SJ, Binsted N, Ross I. J. Phys. C. 1984;17:143. [Google Scholar]
- (33).Gurman SJ, Binsted N, Ross I. J. Phys. C. 1986;19:1845. [Google Scholar]
- (34).Kuzmic P. Anal. Biochem. 1996;237:260. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- (35).Balatri E, Banci L, Bertini I, Cantini F, Ciofi-Baffoni S. Structure. 2003;11:1431. doi: 10.1016/j.str.2003.10.004. [DOI] [PubMed] [Google Scholar]
- (36).Williams JC, Sue C, Banting GS, Yang H, Glerum DM, Hendrickson WA, Schon EA. J. Biol. Chem. 2005;280:15202. doi: 10.1074/jbc.M410705200. [DOI] [PubMed] [Google Scholar]
- (37).Olesen K, Veselov A, Zhao Y, Wang Y, Danner B, Scholes CP, Shapleigh JP. Biochemistry. 1998;37:6086. doi: 10.1021/bi971603z. [DOI] [PubMed] [Google Scholar]
- (38).Ralle M, Lutsenko S, Blackburn NJ. J. Biol. Chem. 2003;278:23163. doi: 10.1074/jbc.M303474200. [DOI] [PubMed] [Google Scholar]
- (39).Siluvai GS, Nakano M, Mayfield M, Blackburn NJ. J. Biol. Inorg. Chem. 2011;16:285. doi: 10.1007/s00775-010-0725-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Gamelin DR, Randall DW, Hay MT, Houser RP, Mulder TC, Canters GW, de Vries S, Tolman WB, Lu Y, Solomon EI. J. Am. Chem. Soc. 1998;120:5246. [Google Scholar]
- (41).Horng YC, Leary SC, Cobine PA, Young FB, George GN, Shoubridge EA, Winge DR. J. Biol. Chem. 2005;280:34113. doi: 10.1074/jbc.M506801200. [DOI] [PubMed] [Google Scholar]
- (42).Nittis T, George GN, Winge DR. J. Biol. Chem. 2001;276:42520. doi: 10.1074/jbc.M107077200. [DOI] [PubMed] [Google Scholar]
- (43).Arciero DM, Pierce BS, Hendrich MP, Hooper AB. Biochemistry. 2002;41:1703. doi: 10.1021/bi015908w. [DOI] [PubMed] [Google Scholar]
- (44).Basumallick L, Sarangi R, DeBeer George S, Elmore B, Hooper AB, Hedman B, Hodgson KO, Solomon EI. J. Am. Chem. Soc. 2005;127:3531. doi: 10.1021/ja044412+. [DOI] [PubMed] [Google Scholar]
- (45).Clark KM, Yu Y, Marshall NM, Sieracki NA, Nilges MJ, Blackburn NJ, van der Donk WA, Lu Y. J. Am. Chem. Soc. 2010;132:10093. doi: 10.1021/ja102632p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).LaCroix LB, Shadle SE, Wang Y, Averill BA, Hedman B, Hodgson KO, Solomon EI. J. Am. Chem. Soc. 1996;118:7755. [Google Scholar]
- (47).Solomon EI. Inorg. Chem. 2006;45:8012. doi: 10.1021/ic060450d. [DOI] [PubMed] [Google Scholar]
- (48).Adman ET. Adv. Protein Chem. 1991;42:145. doi: 10.1016/s0065-3233(08)60536-7. [DOI] [PubMed] [Google Scholar]
- (49).Penfield KW, Gewirth AA, Solomon EI. J. Am. Chem. Soc. 1985;107:4519. [Google Scholar]
- (50).Solomon EI, Baldwin MJ, Lowery ML. Chem. Rev. 1992:521. [Google Scholar]
- (51).Shadle SE, Penner-Hahn JE, Schugar HJ, Hedman B, Hodgson KO, Solomon EI. J. Am. Chem. Soc. 1993;115:767. [Google Scholar]
- (52).Lieberman RL, Arciero DM, Hooper AB, Rosenzweig AC. Biochemistry. 2001;40:5674. doi: 10.1021/bi0102611. [DOI] [PubMed] [Google Scholar]
- (53).Basumallick L, Szilagyi RK, Zhao Y, Shapleigh JP, Scholes CP, Solomon EI. J. Am. Chem. Soc. 2003;125:14784. doi: 10.1021/ja037232t. [DOI] [PubMed] [Google Scholar]
- (54).DeBeer George S, Metz M, Szilagyi RK, Wang H, Cramer SP, Lu Y, Tolman WB, Hedman B, Hodgson KO, Solomon EI. J. Am. Chem. Soc. 2001;123:5757. doi: 10.1021/ja004109i. [DOI] [PubMed] [Google Scholar]
- (55).Berry SM, Ralle M, Low DW, Blackburn NJ, Lu Y. J. Am. Chem. Soc. 2003;125:8760. doi: 10.1021/ja029699u. [DOI] [PubMed] [Google Scholar]
- (56).Hay MT, Lu Y. J. Biol. Inorg. Chem. 2000;5:699. doi: 10.1007/s007750000158. [DOI] [PubMed] [Google Scholar]
- (57).Hay MT, Ang MC, Gamelin DR, Solomon EI, Antholine WE, Ralle M, Blackburn NJ, Massey PD, Wang X, Kwon AH, Lu Y. Inorg. Chem. 1998;37:191. [Google Scholar]
- (58).Thompson AK, Gray J, Liu A, Hosler JP. Biochim Biophys Acta. 2012;1817:955. doi: 10.1016/j.bbabio.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Banci L, Bertini I, Ciofi-Baffoni S, Katsari E, Katsaros N, Kubicek K, Mangani S. Proc. Natl. Acad. Sci. U S A. 2005;102:3994. doi: 10.1073/pnas.0406150102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Horng YC, Cobine PA, Maxfield AB, Carr HS, Winge DR. J. Biol. Chem. 2004;279:35334. doi: 10.1074/jbc.M404747200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.