Abstract
We showed previously that breast cancer chemoprevention with benzyl isothiocyanate (BITC) in MMTV-neu mice was associated with induction of E-cadherin protein in vivo. Loss of E-cadherin expression and induction of mesenchymal markers (e.g. vimentin) are biochemical hallmarks of epithelial–mesenchymal transition (EMT), a developmental process implicated in progression of cancer to aggressive state. This study offers novel insights into the mechanism by which BITC inhibits EMT. Exposure of MDA-MB-231, SUM159 and MDA-MB-468 human breast cancer cells to BITC (2.5 and 5 µM) resulted in transcriptional repression of urokinase-type plasminogen activator (uPA) as well as its receptor (uPAR). However, ectopic expression of uPAR in MDA-MB-468 cells failed to confer protection against induction of E-cadherin and inhibition of cell invasion/migration resulting from BITC treatment. The BITC-mediated induction of E-cadherin and inhibition of cell migration was sustained in MDA-MB-231 and SUM159 cells transiently transfected with an uPAR-targeted small interfering RNA. Overexpression of Forkhead Box Q1 (FOXQ1), whose protein and messenger RNA levels were decreased by BITC treatment in cells and MDA-MB-231 xenografts, conferred marked protection against BITC-mediated inhibition of EMT and cell migration. In conclusion, this study implicates FOXQ1 suppression in BITC-mediated inhibition of EMT in human breast cancer cells.
Introduction
Novel approaches for chemoprevention of breast cancer are clinically attractive because (i) breast cancer continues to be a leading cause of cancer-related deaths in women worldwide despite tremendous advances toward targeted therapies and personalized medicine (1–3), and (ii) many risk factors associated with this disease (4–6) are not easily modifiable. Feasibility of mammary cancer prevention is exemplified by successful clinical application of selective estrogen receptor modulators (e.g. tamoxifen) and aromatase inhibitors (7–10). Unfortunately, these approaches are ineffective against estrogen receptor-negative breast cancers as well as a subset of women with estrogen receptor-positive breast cancer and have rare but unwanted side effects (7–11).
Bioactive food components continue to draw attention for the discovery of cancer chemopreventive agents (12,13). Benzyl isothiocyanate (BITC) is one such naturally occurring constituent of edible cruciferous vegetables (e.g. garden cress) with in vivo preventive efficacy in experimental animals (14–17). For example, BITC treatment prior to the carcinogen challenge inhibited polycyclic aromatic hydrocarbon-induced mammary cancer in rats (14). Likewise, BITC administered during the initiation stage resulted in suppression of pancreatic atypical hyperplasia and adenocarcinoma induced by N-nitrosobis(2-oxopropyl)amine in hamsters (17).
Previous studies from our laboratory have revealed that BITC-mediated prevention of mammary cancer development in mouse mammary tumor virus-neu transgenic mice is associated with induction of E-cadherin protein expression (18). These observations prompted us to explore whether BITC treatment inhibited epithelial–mesenchymal transition (EMT), which is a normal physiological process essential for embryonic development, tissue remodeling and wound healing, and implicated in progression of cancer to invasive state (19–21). Indeed, we found that exposure of MDA-MB-231 human breast cancer cells to BITC resulted in inhibition of EMT characterized by induction of E-cadherin protein level and suppression of vimentin expression (22). Furthermore, the BITC-mediated inhibition of MDA-MB-231 xenograft growth in vivo was accompanied by induction of E-cadherin protein expression and suppression of vimentin protein level in the tumor (22,23). This study builds upon these observations and provides novel insights into the mechanism by which BITC inhibits EMT.
Materials and methods
Ethics statement
Archived tumor tissues from our previously published in vivo study (23) were used to determine the effect of BITC administration on expression of various proteins. Use of mice and their care were in accordance with the University of Pittsburgh Institutional Animal Care and Use Committee guidelines.
Reagents
BITC (purity >98%) was purchased from LKT Laboratories (St Paul, MN). Cell culture reagents were purchased from Invitrogen-Life Technologies (Carlsbad, CA). Anti-E-cadherin antibody was purchased from BD Biosciences (San Diego, CA). Antibodies against vimentin and actin were purchased from Sigma–Aldrich (St Louis, MO). Antibodies against urokinase-type plasminogen activator (uPA) and its receptor (uPAR), slug and Forkhead Box Q1 transcription factor (FOXQ1) were from Santa Cruz Biotechnology (Santa Cruz, CA). Antisnail antibody was purchased from Abgent (San Diego, CA). An antibody against vimentin used for immunofluorescence microscopy was purchased from Santa Cruz Biotechnology. An antibody against copper zinc–superoxide dismutase (Cu,Zn–SOD) was from Calbiochem-EMD Millipore (Billerica, MA). Small interfering RNA (siRNA) targeted against uPAR was purchased from Santa Cruz Biotechnology. A control non-specific siRNA was purchased from Qiagen (Valencia, CA).
Cell lines
The MDA-MB-231, MCF-7 and MDA-MB-468 cells were purchased from the American Type Culture Collection (Manassas, VA) and cultured as described by us previously (24) or recommended by the provider. The SUM159 cell line was purchased from Asterand (Detroit, MI) and maintained as suggested by the supplier. The MDA-MB-468 and MCF-7 cells stably transfected with a plasmid encoding for uPAR (hereafter abbreviated as uPAR cells) and empty-vector (hereafter abbreviated as EV cells) were generously provided by Dr Steven L.Gonias (University of California, San Diego, CA) and maintained as recommended by the provider (25). The human mammary epithelial cell line (HMLE) stably transfected with FOXQ1 (hereafter abbreviated as FOXQ1) and EV (abbreviated as LacZ) were generously provided by Dr Guojun Wu (Karmanos Cancer Institute, Departments of Oncology and Pathology, Wayne State University, Detroit, MI) and maintained as recommended by the provider (26). Details of MDA-MB-231 cell line stably transfected with empty pcDNA3.1 vector and the same vector encoding for Cu,Zn–SOD and their culture conditions have been described by us previously (27). Stock solution of BITC was prepared in dimethyl sulfoxide (DMSO) and an equal volume of DMSO (final concentration <0.15%) was added to the controls.
Western blotting
DMSO-treated control and BITC-treated cells and MDA-MB-231 xenografts from control and BITC-treated mice (7.5 µmol BITC/mouse) (23) were processed for western blotting as described by us previously (28,29).
Luciferase reporter assays
Luciferase reporter assay was performed to determine the effect of BITC treatment on E-cadherin and uPA transcription. Cells (2 × 104 cells per well) were seeded in 12-well plates and allowed to attach by overnight incubation at 37°C. Cells were then co-transfected with 8 μg of pGL2 Basic-EcadK1-Luc plasmid (Addgene, Cambridge, MA; promoter sequence from -108 to +125) and 0.8 μg of pRL-CMV using Fugene6 (Roche Applied Science, Indianapolis, IN). The uPA luciferase plasmid was a generous gift from Dr Shafaat A.Rabbani (McGill University, Montreal, Canada) (30). For uPA luciferase assay, cells were co-transfected with 6 μg of pGL-3 Basic-uPA-Luc plasmid (promoter region -745 to -30) and 0.6 μg of pGL-3 basic plasmid using Fugene6. Twenty-four hours after transfection, the cells were treated with DMSO or BITC for specified time periods. Luciferase activity was determined as described by us previously (22). Luciferase activity was normalized against protein concentration and expressed as a ratio of firefly luciferase to renilla luciferase units.
Immunocytochemical analysis for E-cadherin and vimentin
The SUM159 cells and the MDA-MB-231 cells stably transfected with empty pcDNA3.1 vector or Cu,Zn–SOD plasmid were cultured on cover slips, allowed to attach by overnight incubation and then exposed to DMSO (control) or BITC for 24h. Cells were fixed with 2% paraformaldehyde for 1h, permeabilized with 0.05% Triton X-100 in phosphate-buffered saline (PBS) for 10min and blocked with 0.5% bovine serum albumin (in PBS) for 1h at room temperature. Cells were then incubated overnight with anti-E-cadherin or antivimentin antibody at 4°C, washed with PBS and incubated with Alexa Fluor 568-conjugated or Alexa Fluor 488-conjugated secondary antibody for 1h at room temperature. Cells were washed twice with PBS, mounted and examined under a Leica DC300F microscope.
Reverse transcription–PCR
Total RNA from DMSO-treated control and BITC-treated cells was isolated using RNeasy kit (Invitrogen-Life Technologies) and reverse transcribed with the use of reverse transcriptase and oligo(deoxythymidine)20 to synthesize complementary DNA (cDNA). Reverse transcription–PCR reaction was carried out using GoTaq green master mix (Promega, Madison, WI), gene-specific primers and cDNA. The uPA primers were 5ʹ-CTGTGACTGTCTAAATGGAGG-3ʹ (forward) and 5ʹ-GACGATGTAGTCCTCCTTCTT-3ʹ (reverse). Amplification conditions for uPA were 95ºC for 3min, 30 cycles 95ºC for 30 s, 60ºC for 30 s, 72ºC for 30 s and 72ºC for 5min. The primers used for uPAR were 5ʹ-CATGCAGTGTAAGACCAACG-3ʹ (forward) and 5ʹ-CTCTCACAGCTCATGTCTGATGAGCCAC-3ʹ (reverse). Amplification conditions for uPAR were 95ºC for 5min, 25 cycles 95ºC for 20 s, 56ºC for 60 s, 72ºC for 30 s and 72ºC for 3min. House-keeping gene GAPDH was used as an internal control (22). PCR products were resolved on 1–2% agarose gels pre-stained with ethidium bromide and visualized under an UV illuminator.
uPA ELISA assay
A commercially available kit (IMUBIND uPA ELISA kit) from American Diagnostics (Greenwich, CT) was used for quantitative measurement of uPA in conditioned media of cells and plasma of control and BITC-treated mice.
Migration and invasion assays
Effect of BITC treatment on in vitro migration and invasion of breast cancer cells was determined using Transwell Boyden chamber (Corning, NY) containing a polycarbonate filter with a pore size of 8 µm as described by us previously (22). For invasion assay, the Boyden chamber was coated with 30 μl of Matrigel (1:2 dilutions in medium). Briefly, cells (1×105 cells per well) in 0.2ml of serum-free media were mixed with DMSO (control) or desired concentration of BITC and then placed in the upper compartment of the chamber. Lower compartment of the chamber contained 0.6ml of complete medium containing the same concentrations of BITC or DMSO. Following incubation at 37°C for 24h, non-motile cells from upper membrane were removed gently using a cotton swab. Membrane was washed with PBS and the motile cells on the bottom face of the membrane were fixed with 90% ethanol and stained with hematoxylin and eosin. Number of invaded or migrated cells was counted under a microscope. Quantitation of cell migration and invasion is representative of triplicate measurements in each experiment.
RNA interference of uPAR
MDA-MB-231 or SUM159 cells (0.5–1 × 105 cells per well) were seeded in six-well plates one day before transfection. Cells were washed three times with serum/growth factor-free OptiMEM (Invitrogen-Life Technologies) and transfected with 200nM of uPAR-targeted siRNA or a non-specific control siRNA using OligofectAMINE. Forty-eight hours after transfection, cells were treated with DMSO or BITC for 24h and processed for immunoblotting and in vitro migration assay.
Real-time PCR for FoxQ1 expression
Total RNA from DMSO-treated control and BITC-treated cells was isolated using RNeasy kit (Qiagen). First-strand cDNA was synthesized using Superscript reverse transcriptase (Invitrogen-Life Technologies) with oligo (deoxythymidine)20 primer. The FOXQ1 primers were 5ʹ-CGCGGACTTTGCACTTTGAA-3′ and 5′-AGCTTTAAGGCACGTTTGATGGAG-3′. Quantitative real-time PCR was done using 2× SYBR Green master mix (Applied Biosystems-Life Technologies) with 95°C (30 s) and 60°C (60 s) for 40 cycles. Relative gene expression was calculated using the method described by Livak et al. (31) and normalized against GAPDH.
Results
BITC treatment inhibited EMT in cultured SUM159 cells
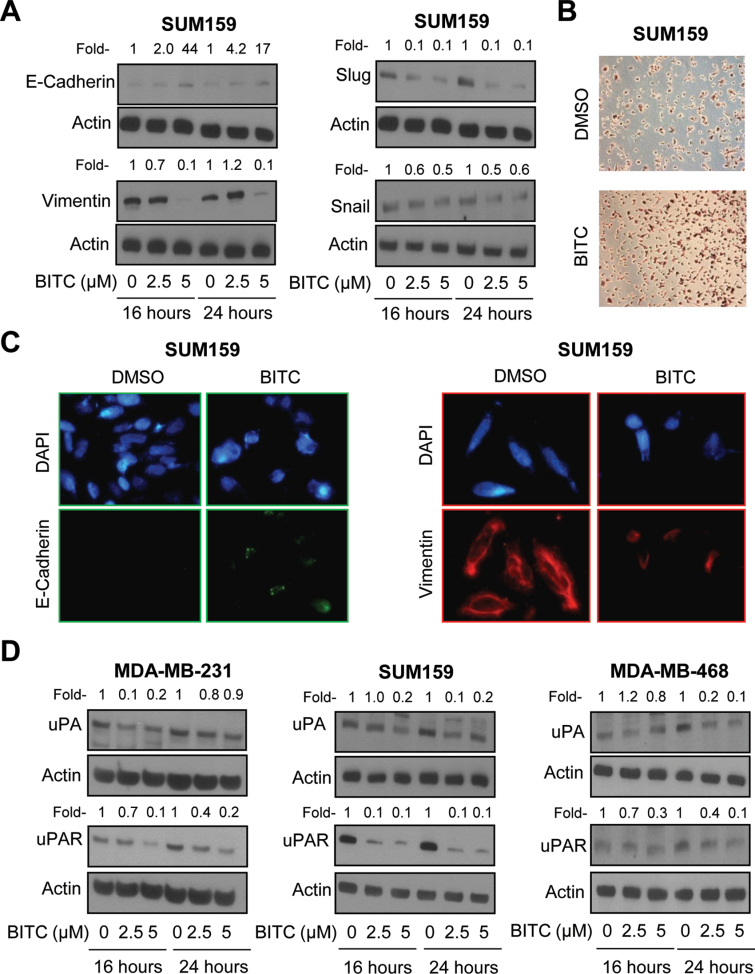
We have shown previously that BITC treatment inhibits EMT in MDA-MB-231 human breast cancer cells (22). Initially, we designed experiments using a triple-negative breast cancer cell line (SUM159) to determine generality of the BITC-mediated inhibition of EMT. Exposure of SUM159 cells to pharmacologically relevant concentrations of BITC (32) resulted in induction of E-cadherin protein expression, which was accompanied by suppression of vimentin protein level especially at the 5 μM dose (Figure 1A). Protein levels of slug and snail, which are transcriptional repressors of E-cadherin (33,34), were also decreased markedly after treatment of SUM159 cells with BITC, but this effect was relatively more pronounced on slug protein compared with snail (Figure 1A). Similar to MDA-MB-231 cells (22), the BITC-mediated induction of E-cadherin protein expression in SUM159 cells was accompanied by its transcriptional upregulation as revealed by E-cadherin luciferase reporter assay (results not shown). Moreover, BITC treatment resulted in restoration of some cell–cell contact, which was rare in DMSO-treated control SUM159 cells (Figure 1B). Immunofluorescence microscopy confirmed induction of E-cadherin and suppression of vimentin protein expression in BITC-treated SUM159 cells (Figure 1C). These results indicated that the BITC-mediated inhibition of EMT was not a cell line-specific phenomenon.
Fig. 1.
BITC treatment suppresses uPA and uPAR protein levels in human breast cancer cells. (A) Western blotting for E-cadherin, vimentin, snail and slug proteins using lysates from SUM159 cells treated for 16–24h with DMSO (control) or the indicated concentrations of BITC. (B) Morphology of SUM159 cells after 6h of treatment with DMSO or 2.5 μM of BITC. (C) Immunofluorescence microscopy for expression of E-cadherin and vimentin in SUM159 cells treated for 24h with DMSO or 5 μM BITC (×200 objective magnification). (D) Immunoblotting for uPA and uPAR using lysates from MDA-MB-231, SUM159 and MDA-MB-468 cells treated for 16–24h with DMSO or the indicated concentrations of BITC. Actin was probed as a loading control. Numbers on top of the immunoreactive bands represent changes in protein levels relative to corresponding DMSO-treated control. Each experiment was done at least twice and representative data from one such experiment are shown.
BITC treatment caused transcriptional repression of uPA and uPAR in breast cancer cells
Recent studies have shown that overexpression of uPAR alone is sufficient to drive EMT in breast cancer cells (25,35). The uPAR together with uPA constitutes an integral component of extracellular matrix proteolysis and cell–extracellular matrix interaction as well as cell signaling involving receptor tyrosine kinases (36). The BITC treatment caused suppression of both uPA and uPAR protein levels in MDA-MB-231, SUM159 and MDA-MB-468 cells, albeit with different kinetics especially for uPA (Figure 1D). For example, the BITC-mediated suppression of uPA protein seemed transient in the MDA-MB-231 cell line, but not in SUM159 or MDA-MB-468 cells (Figure 1D).
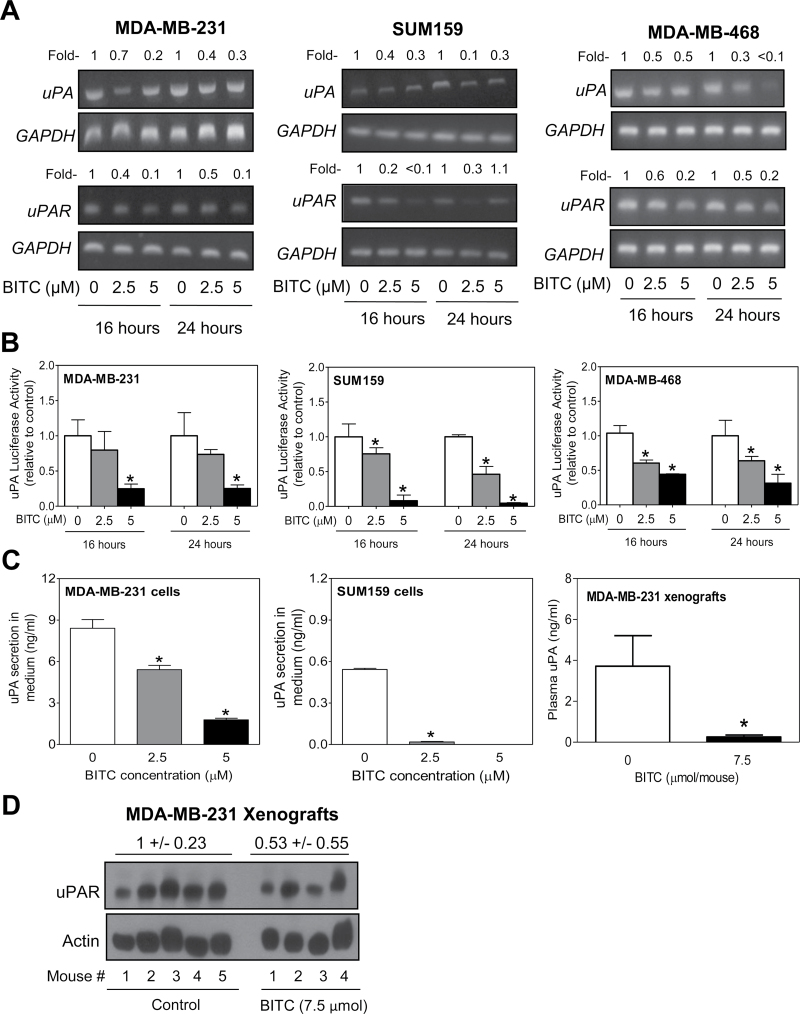
Next, we proceeded to determine the mechanism by which BITC treatment suppressed uPA and uPAR protein levels. The BITC treatment caused suppression of uPA and uPAR messenger RNA (mRNA) levels in MDA-MB-231, SUM159 and MDA-MB-468 cells as revealed by reverse transcription–PCR (Figure 2A). Transcriptional repression of uPA in BITC-treated breast cancer cells was confirmed by uPA luciferase reporter assay (Figure 2B). Moreover, BITC treatment resulted in a significant decrease in secretion of uPA into the culture medium of MDA-MB-231 and SUM159 cells (Figure 2C). Collectively, these results indicated that BITC treatment not only caused transcriptional repression of uPA and uPAR in cells but also inhibited uPA secretion into the conditioned media.
Fig. 2.
BITC treatment causes transcriptional repression of uPA and uPAR in human breast cancer cells. (A) Reverse transcription–PCR for uPA, uPAR and GAPDH mRNA levels in MDA-MB-231, SUM159 and MDA-MB-468 cells treated for 16–24h with DMSO (control) or the indicated concentrations of BITC. Number above band represents change in expression relative to corresponding DMSO-treated control. (B) uPA luciferase activity in MDA-MB-231, SUM159 and MDA-MB-468 cells after 16–24h of treatment with DMSO or the indicated concentrations of BITC. Results shown are mean ± SD (n = 3). *Significantly different (P < 0.05) compared with DMSO-treated control by one-way analysis of variance (ANOVA) with Dunnett’s adjustment. (C) Quantitation of uPA secretion into the culture media of MDA-MB-231 and SUM159 cells (24h treatment), and plasma of control (n = 5) and BITC-treated (n = 4) MDA-MB-231 xenograft bearing female athymic mice. Results for uPA secretion in culture media are shown as mean ± SD (n = 3). *Significantly different compared withcontrol by one-way ANOVA with Dunnett’s adjustment (media of MDA-MB-231 and SUM159 cells) or Student’s t-test (plasma levels). (D) Western blotting for uPAR protein using MDA-MB-231 xenograft supernatants from control (n = 5) and BITC-treated mice (n = 4).
We have shown previously that BITC administration retards in vivo growth of MDA-MB-231 cells implanted in female athymic mice (23). We used tumor tissues and plasma specimens from this study to determine in vivo relevance of the BITC-mediated suppression of uPA and uPAR proteins. The plasma level of uPA (Figure 2C) and expression of uPAR in the tumor tissue (Figure 2D) were markedly reduced upon BITC administration. These observations provided in vivo evidence for the BITC-mediated suppression of uPA and uPAR proteins.
uPAR was dispensable for BITC-mediated inhibition of EMT and cell migration
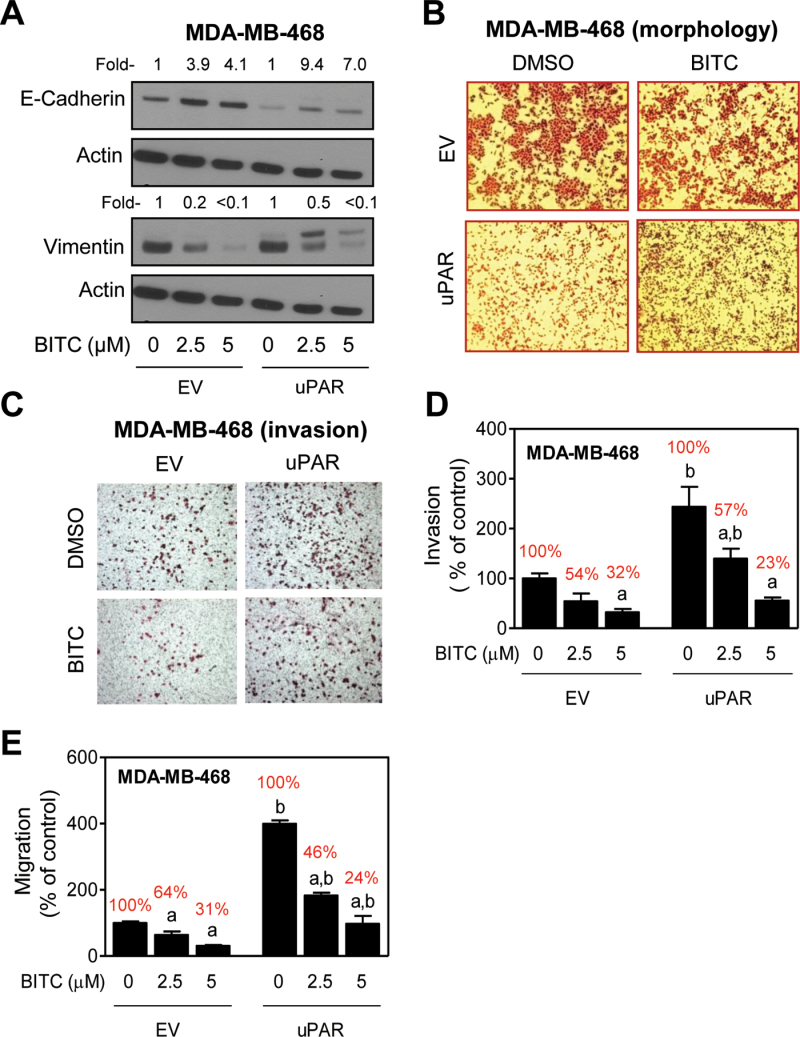
Stable overexpression of uPAR alone has been shown to drive EMT in MDA-MB-468 cells characterized by suppression of E-cadherin and induction of vimentin protein expression (25). We used uPAR overexpressing MDA-MB-468 cells (uPAR overexpression results not shown) and corresponding EV-transfected control cells to study the role of uPAR in BITC-mediated inhibition of EMT. Consistent with the published literature (25), we also noticed suppression of E-cadherin expression by overexpression of uPAR in MDA-MB-468 cells (Figure 3A). Treatment of both EV-transfected control MDA-MB-468 cells and uPAR overexpressing cells with BITC resulted in induction of E-cadherin and suppression of vimentin expression (Figure 3A). However, when the protein change quantitation was normalized against respective DMSO-treated control for each cell line (EV and uPAR cells), the BITC-mediated induction of E-cadherin and suppression of vimentin was clearly visible. It is important to point out that a band with reduced electrophoretic mobility in western blotting for vimentin protein was observed in uPAR overexpressing MDA-MB-468 cells only after treatment with BITC (Figure 3A). Slower migrating band for vimentin protein was not detectable in EV-transfected MDA-MB-468 cells (Figure 3A) or in SUM159 cells (Figure 1A) regardless of the BITC exposure. It is possible that the band with reduced electrophoretic mobility represents posttranslational modification of the vimentin protein. At the same time, the possibility that the slower migrating band is a consequence of non-specific reactivity cannot be excluded.
Fig. 3.
uPAR overexpression is dispensable for BITC-mediated inhibition of EMT and cell invasion/migration in MDA-MB-468 cells. (A) Western blotting for E-cadherin and vimentin proteins using lysates from MDA-MB-468 cells stably transfected with EV or uPAR plasmid (uPAR) (25) and treated for 24h with DMSO or BITC (2.5 and 5 μM). Actin was probed as a loading control. Numbers above the bands represent densitometric quantitation relative to corresponding DMSO-treated control cells. (B) Morphology of EV and uPAR MDA-MD-468 cells after 24h of treatment with DMSO or 1 μM BITC. (C) Microscopic images depicting cell invasion (×100 magnifications) after 24h of treatment with DMSO or 2.5 μM of BITC. (D) Quantitation of cell invasion after 24h of treatment with DMSO or BITC (2.5 or 5 μM). (E) Quantitation of cell migration after 24h of treatment with DMSO or BITC (2.5 or 5 μM). For data in panels D and E, three-four fields on each filter were scored for cell invasion and migration. Results shown are relative to DMSO-treated EV cells (mean ± SD, n = 3). Significantly different (P < 0.05) compared with acorresponding DMSO-treated control and bbetween EV and uPAR cells at each concentration of BITC (0, 2.5 and 5 μM BITC) by one-way ANOVA followed by Bonferroni’s multiple comparison test. Percent values in red font represent cell migration normalized to DMSO-treated control for each cell type. Similar results were observed in replicate experiments.
As shown in Figure 3B, uPAR overexpression resulted in a change in cell morphology characterized by loss of cell–cell contact. Similar to the results in SUM159 cells (Figure 1B), the loss of cell–cell contact resulting from forced expression of uPAR was partially restored after BITC treatment (Figure 3B). The uPAR overexpressing MDA-MB-468 cells exhibited a significantly higher propensity for cell invasion compared with EV-transfected control cells (Figure 3C). However, when the results were normalized to DMSO-treated control for each cell line (percentage values relative to respective control are shown in red font), protection against BITC-mediated inhibition of cell invasion (Figure 3D) or cell migration (Figure 3E) was not evident. Collectively, these results indicated that uPAR overexpression was dispensable for BITC-mediated inhibition of EMT and cell migration. Similarly, uPAR overexpression in MCF-7 cells (uPAR overexpression data not shown) failed to confer protection against BITC-mediated inhibition of cell migration (results not shown). Stable overexpression of uPAR alone does not induce EMT in MCF-7 cells (25), and thus, western blotting for EMT-related proteins was not performed.
Effect of uPAR knockdown on BITC-mediated inhibition of EMT and cell migration
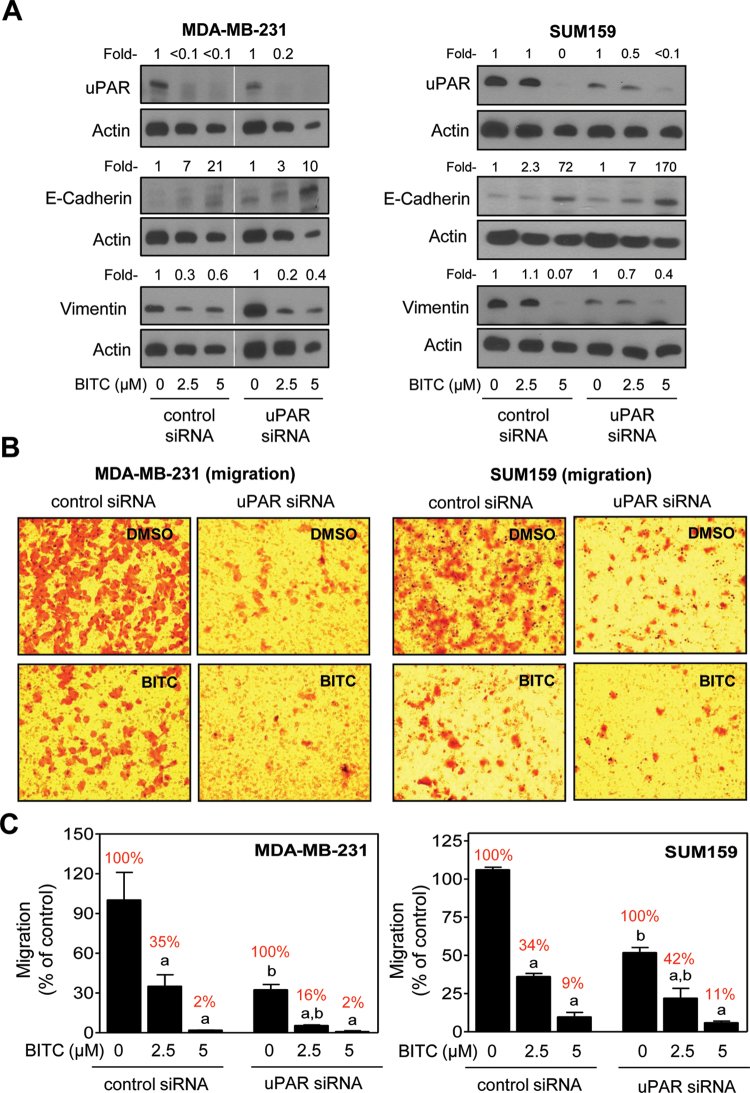
We used siRNA targeted against uPAR to obtain additional evidence for lack of its involvement in BITC-mediated inhibition of EMT. Expression of uPAR was decreased by about 80–90% in MDA-MB-231 and SUM159 cells transiently transfected with the uPAR-targeted siRNA compared with control siRNA transfected cells (Figure 4A). The BITC-mediated induction of E-cadherin and suppression of vimentin protein expression was observed in cells transfected with the non-specific siRNA and uPAR-targeted siRNA (Figure 4A). In agreement with these results, inhibition of MDA-MB-231 and SUM159 cell migration resulting from BITC exposure was observed in cells transfected with the non-specific siRNA and uPAR-targeted siRNA (Figure 4B and C). These observations ruled out involvement of uPAR in BITC-mediated inhibition of EMT at least in MDA-MB-231 and SUM159 cells.
Fig. 4.
Effect of uPAR protein knockdown on BITC-mediated inhibition of EMT and cell migration in MDA-MB-231 and SUM159 cells. (A) Western blotting for uPAR, E-cadherin and vimentin proteins using lysates from MDA-MB-231 and SUM159 cells transiently transfected with a non-specific (control) siRNA or uPAR-targeted siRNA and treated for 24h with DMSO or BITC (2.5 and 5 μM). Numbers above the bands represent densitometric quantitation relative to corresponding DMSO-treated control. (B) Representative microscopic images depicting migration by MDA-MB-231 and SUM159 cells transiently transfected with a non-specific (control) siRNA or uPAR-targeted siRNA and treated for 24h with DMSO or 2.5 μM BITC (×100 magnifications). (C) Quantitation of cell migration from data shown in panel B. Results shown are relative to control siRNA transfected cells treated with DMSO (mean ± SD, n = 3). Percent values in red font represent normalization to corresponding DMSO-treated control for each cell type. Significantly different (P < 0.05) compared with acorresponding DMSO-treated control and bbetween control siRNA and uPAR siRNA transfected cells at each concentration of BITC (0, 2.5 and 5 μM) by one-way ANOVA followed by Bonferroni’s multiple comparison test. Similar results were observed in replicate experiments in each cell line.
BITC treatment caused transcriptional repression of FOXQ1
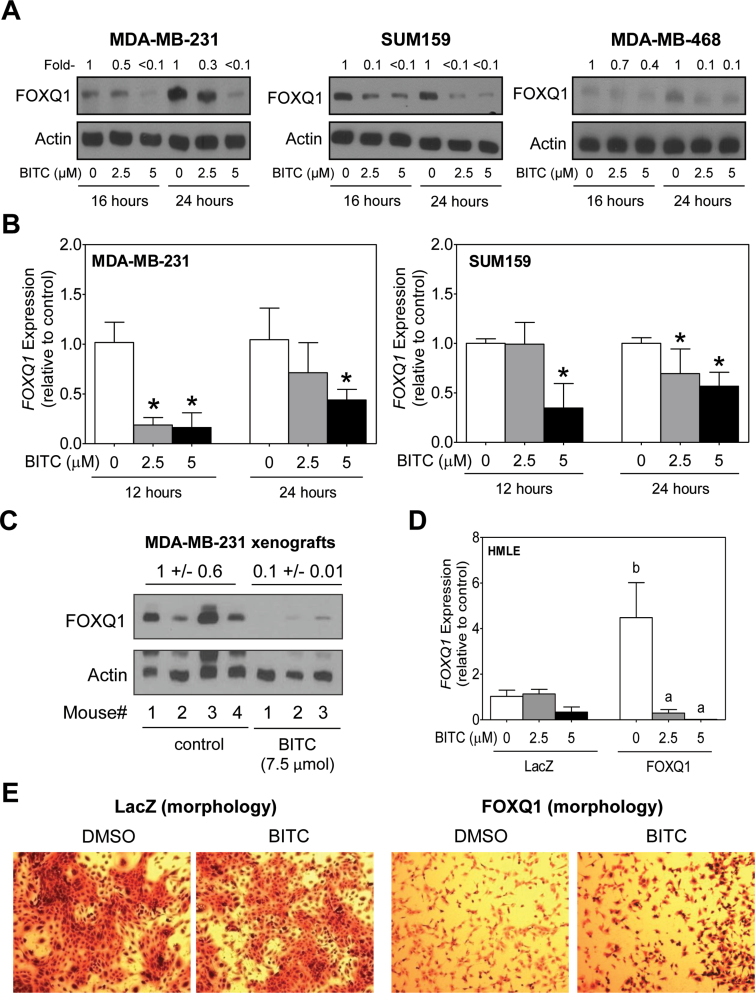
Recent studies have shown that overexpression of FOXQ1 protein leads to EMT induction and promotion of tumor growth (26,37,38). Protein level of FOXQ1 was decreased markedly upon treatment with BITC in MDA-MB-231, SUM159 and MDA-MB-468 cells (Figure 5A), which was accompanied by suppression of FOXQ1 mRNA levels as revealed by quantitative real-time PCR (Figure 5B). Moreover, the MDA-MB-231 xenografts from BITC-treated mice exhibited a marked decrease in the level of FOXQ1 protein compared with tumors from vehicle-treated control mice (Figure 5C). We used HMLE stably transfected with FOXQ1 (abbreviated as FOXQ1 in Figure 5) and EV-transfected control cells (abbreviated as LacZ in Figure 5) to determine the role of FOXQ1 in BITC-mediated inhibition of EMT. Overexpression of FOXQ1 was confirmed by quantitative real-time PCR (Figure 5D). BITC treatment caused suppression of FOXQ1 mRNA level in EV-transfected cells as well as in FOXQ1 overexpressing cells (Figure 5D). Overexpression of FOXQ1 resulted in change in cell morphology when compared with LacZ cells and this effect was partially reversible after treatment with BITC in the cells stably transfected with FOXQ1 (Figure 5E).
Fig. 5.
BITC treatment causes transcriptional repression of FOXQ1 in human breast cancer cells. (A) Western blotting for FOXQ1 protein using lysates from MDA-MB-231, SUM159 and MDA-MB-468 cells after 16–24h of treatment with DMSO (control) or the indicated concentrations of BITC. Actin was probed as a loading control. Numbers on top of the immunoreactive bands represent changes in protein levels relative to corresponding DMSO-treated control. (B) Quantitative real-time PCR for FOXQ1 mRNA levels in MDA-MB-231 and SUM159 cells after 12 and 24h of treatment with DMSO (control) or BITC (2.5 or 5 μM). Results shown are relative to corresponding DMSO-treated control (mean ± SD, n = 6). *Significantly different (P < 0.05) compared with DMSO-treated control by one-way ANOVA followed by Dunnett’s test. (C) Western blotting for FOXQ1 protein using MDA-MB-231 xenograft supernatants from control (n = 4) and BITC-treated mice (n = 3). Actin was probed as a loading control. (D) Quantitative real-time PCR for FOXQ1 mRNA levels in immortalized HMLE cells stably transfected with EV (abbreviated as LacZ) or FOXQ1 plasmid (abbreviated as FOXQ1) and treated for 24h with DMSO or BITC (2.5 or 5 μM). Results shown are relative to DMSO-treated LacZ cells (mean ± SD, n = 3). Significantly different (P < 0.05) compared with acorresponding DMSO-treated control and bbetween LacZ and FOXQ1 cells at each concentration of BITC (0, 2.5 and 5 μM BITC) by one-way ANOVA followed by Bonferroni’s multiple comparison test. (E) Morphology of LacZ and FOXQ1 cells after 24h of treatment with DMSO or 1 μM of BITC. Each experiment was repeated at least twice, and representative data from one such experiment are shown.
FOXQ1 overexpression conferred protection against BITC-mediated induction of E-cadherin and inhibition of cell migration
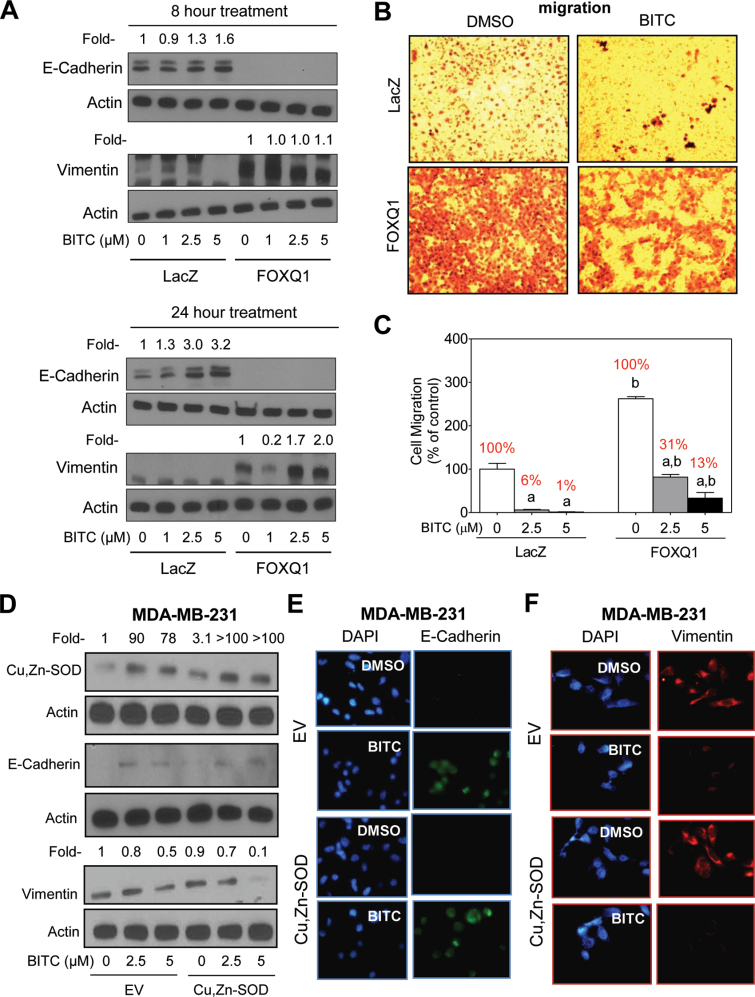
Treatment of LacZ cells with BITC resulted in a dose-dependent increase in protein levels of E-cadherin (Figure 6A). However, the BITC treatment was unable to rescue E-cadherin protein loss resulting from overexpression of FOXQ1 (Figure 6A). In agreement with these results, BITC treatment failed to suppress vimentin protein level in FOXQ1 overexpressing cells (Figure 6A). The BITC-mediated decrease in vimentin protein level observed at 1 µM concentration in Figure 6A was not consistent in different experiments (results not shown). Overexpression of FOXQ1 increased migratory potential of mammary epithelial cells (Figure 6B). In addition, overexpression of FOXQ1 conferred partial but marked protection against BITC-mediated inhibition of cell migration (compare percentage of cell migration normalized against corresponding DMSO-treated control for each cell line; values shown in red font in Figure 6C). For example, 24h exposure of LacZ cells with 2.5 µM BITC resulted in 94% decrease in cell migration (Figure 6C). Migration of FOXQ1 overexpressing cells was reduced by 69% by a similar treatment with BITC. These results indicated that suppression of FOXQ1, at least in part, contributed to BITC-mediated inhibition of cell migration. We attempted to solidify these conclusions using FOXQ1-targeted siRNA and small-hairpin RNA for transient and stable transfection, respectively, in human breast cancer cells. Unfortunately, we were not successful in knockdown of FOXQ1 using these approaches.
Fig. 6.
FOXQ1 overexpression confers protection against BITC-mediated inhibition of EMT and cell migration. (A) Western blotting for E-cadherin and vimentin proteins using lysates from HMLE cells stably transfected with EV (LacZ) or FOXQ1 plasmid (FOXQ1) after 8 and 24h of treatment with DMSO or BITC (1, 2.5 or 5 μM). Numbers on top of the immunoreactive bands represent changes in protein levels relative to corresponding DMSO-treated control. (B) Representative microscopic images depicting cell migration by LacZ and FOXQ1 overexpressing cells following 24h of treatment with DMSO or BITC (2.5 μM) (×100 magnifications). (C) Quantitation of cell migration from data shown in panel B. Results shown are relative to LacZ cells treated with DMSO (mean ± SD, n = 3). Percent values in red font represent normalization to corresponding DMSO-treated control for each cell type. Significantly different (P < 0.05) compared with acorresponding DMSO-treated control, and bbetween LacZ and FOXQ1 cells at each concentration of BITC by one-way ANOVA followed by Bonferroni’s multiple comparison test. (D) Western blotting for Cu,Zn–SOD, E-cadherin and vimentin proteins using lysates from MDA-MB-231 cells stably transfected with empty pcDNA3.1 vector (EV) or the same vector encoding for Cu,Zn–SOD and treated for 24h with DMSO or the indicated concentrations of BITC. Numbers on top of the immunoreactive bands represent changes in protein levels relative to DMSO-treated EV cells. Immunofluorescence microscopy for expression of (E) E-cadherin and (F) vimentin in EV cells and Cu,Zn–SOD overexpressing MDA-MB-231 cells after 24h of treatment with DMSO or 5 μM of BITC. Each experiment was repeated and representative data from one such experiment are shown.
Stable overexpression of Cu,Zn–SOD failed to confer protection against BITC-mediated inhibition of EMT
We have shown previously that BITC-induced apoptotic cell death in human breast cancer cells is associated with production of reactive oxygen species (24). Furthermore, the BITC-induced apoptosis is fully attenuated by ectopic expression of Cu,Zn–SOD (24). We proceeded to determine the association, if any, between BITC-mediated inhibition of EMT and apoptosis induction using MDA-MB-231 cells stably transfected with empty pcDNA3.1 vector or the same vector encoding for Cu,Zn–SOD. As shown in Figure 6D, western blotting revealed BITC-mediated induction of E-cadherin and suppression of vimentin protein level not only in EV-transfected cells but also in Cu,Zn–SOD overexpressing MDA-MB-231 cells. Quantitation for E-cadherin was not possible as an immunoreactive band corresponding to this protein was not detectable in the DMSO-treated control cells (Figure 6D). These results, which were confirmed by immunofluorescence microscopy for E-cadherin (Figure 6E) and vimentin protein (Figure 6F), indicated that the BITC-mediated inhibition of EMT was independent of its proapoptotic effect.
Discussion
This study demonstrates that BITC treatment suppresses uPA and uPAR protein expression and secretion of uPA in cultured and xenografted breast cancer cells. Downregulation of constitutive uPA in cultured HT29 human colon cancer cells and suppression of hepatocyte growth factor-stimulated secretion of uPA in MDA-MB-231 cells upon treatment with BITC has been reported previously (39,40), but this study is the first published report to show uPAR downregulation by this agent. We also provide evidence indicating that uPAR suppression is dispensable for BITC-mediated inhibition of EMT at least in human breast cancer cells. The uPAR can also function as a signaling receptor promoting cell proliferation and survival. Signaling pathways activated through uPAR include mitogen-activated protein kinases, focal adhesion kinase, Src and the Rho family small GTPase Rac (41–44). The BITC-mediated alterations in many of these signaling pathways have been observed in cancer cells. For example, we have shown previously that BITC treatment decreases levels of Tyr416 phosphorylated Src in MDA-MB-231 cells (45). The BITC has been shown to downregulate extracellular signal-regulated kinases and RhoA proteins in gastric cancer AGS cells (46). Further studies are needed to determine if suppression of uPA and uPAR contributes to BITC-mediated inhibition of cell proliferation observed in previous studies (24).
Evidence continues to accumulate to implicate FOXQ1 in regulation of EMT in cancer cells (26,37,38). For example, a cross-species gene expression profiling using metastatic human and mouse breast cancer cell lines revealed a novel function for FOXQ1 in regulation of EMT (26). Ectopic expression of FOXQ1 increased cell migration and invasion in vitro in association with EMT induction and enhanced the lung metastatic capability of mammary epithelial cells in vivo (26). In another study, FOXQ1 expression in high-grade basal-like breast cancer was associated with poor clinical outcomes (37). Forced expression of FOXQ1 in differentiated HMLEs and epithelial cancer cell lines induced an EMT phenotype as well as acquisition of resistance to chemotherapy-induced apoptosis (37). FOXQ1 overexpression was also shown to mediate angiogenic and antiapoptotic effects in vivo in colorectal cancer cells (38). Furthermore, FOXQ1 overexpression was shown to repress expression of E-cadherin by binding to the E-box in its promoter region (26). Expression of FOXQ1 is regulated by transforming growth factor-β1 and BITC treatment inhibits transforming growth factor-β1-induced EMT in breast cancer cells (22). It is possible that the BITC-mediated downregulation of FOXQ1 is mediated by suppression of transforming growth factor-β1 signaling, but further work is needed to explore this possibility.
Notably, FOXQ1 overexpression confers only partial protection against BITC-mediated inhibition of cell migration at least in breast cancer cells. Further studies are needed to shed light on other mechanisms potentially contributing to BITC-mediated inhibition of cell migration. For example, a very recent study has shown that mTORC1 and mTORC2 are involved in regulation of cell motility and metastasis via RhoA and Rac1 signaling pathways (47). Interestingly, BITC-induced autophagy in MDA-MB-231 and MCF-7 cells is associated with inhibition of mTORC1 (48). Although mTORC1 is dispensable for autophagic death by BITC (48), the possibility of its involvement in BITC-mediated inhibition of cell migration cannot be ignored.
Funding
United States Public Health Service (grant RO1 CA129347-05) awarded by the National Cancer Institute; National Cancer Institute at the National Institutes of Health (P30 CA047904).
The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- ANOVA
analysis of variance
- BITC
benzyl isothiocyanate
- cDNA
complementary DNA
- Cu,Zn–SOD
copper zinc–superoxide dismutase
- DMSO
dimethyl sulfoxide
- EMT
epithelial–mesenchymal transition
- EV
empty-vector
- FOXQ1
Forkhead Box Q1 transcription factor
- HMLE
human mammary epithelial cell line
- mRNA
messenger RNA
- PBS
phosphate-buffered saline
- siRNA
small interfering RNA
- uPA
urokinase-type plasminogen activator
- uPAR
uPA receptor.
References
- 1. Jemal A, et al. (2010). Cancer statistics, 2010. CA. Cancer J. Clin., 60, 277–300 [DOI] [PubMed] [Google Scholar]
- 2. Alvarez R.H. (2010). Present and future evolution of advanced breast cancer therapy. Breast Cancer Res., 12 Suppl 2, S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Higgins M.J, et al. (2011). Breast cancer in 2010: novel targets and therapies for a personalized approach. Nat. Rev. Clin. Oncol., 8, 65–66 [DOI] [PubMed] [Google Scholar]
- 4. Hulka B.S, et al. (1995). Breast cancer: cause and prevention. Lancet, 346, 883–887 [DOI] [PubMed] [Google Scholar]
- 5. Zhang Y, et al. (1997). Bone mass and the risk of breast cancer among postmenopausal women. N. Engl. J. Med., 336, 611–617 [DOI] [PubMed] [Google Scholar]
- 6. van Zitteren M, et al. (2011). Genome-based prediction of breast cancer risk in the general population: a modeling study based on meta-analyses of genetic associations. Cancer Epidemiol. Biomarkers Prev., 20, 9–22 [DOI] [PubMed] [Google Scholar]
- 7. Fisher B, et al. (1998). Tamoxifen for prevention of breast cancer: report of the National Surgical Adjuvant Breast and Bowel Project P-1 Study. J. Natl. Cancer Inst., 90, 1371–1388 [DOI] [PubMed] [Google Scholar]
- 8. Cauley J.A, et al. (2001). Continued breast cancer risk reduction in postmenopausal women treated with raloxifene: 4-year results from the MORE trial. Multiple outcomes of raloxifene evaluation. Breast Cancer Res. Treat., 65, 125–134 [DOI] [PubMed] [Google Scholar]
- 9. Goss P.E, et al. ; NCIC CTG MAP.3 Study Investigators (2011). Exemestane for breast-cancer prevention in postmenopausal women. N. Engl. J. Med., 364, 2381–2391 [DOI] [PubMed] [Google Scholar]
- 10. Obiorah I, et al. (2011). Progress in endocrine approaches to the treatment and prevention of breast cancer. Maturitas, 70, 315–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boccardo F, et al. (2006). Switching to anastrozole versus continued tamoxifen treatment of early breast cancer. Updated results of the Italian tamoxifen anastrozole (ITA) trial. Ann. Oncol., 17(suppl. 7),vii10–14 [DOI] [PubMed] [Google Scholar]
- 12. Surh Y.J. (2003). Cancer chemoprevention with dietary phytochemicals. Nat. Rev. Cancer, 3, 768–780 [DOI] [PubMed] [Google Scholar]
- 13. Stan S.D, et al. (2008). Bioactive food components and cancer risk reduction. J. Cell. Biochem., 104, 339–356 [DOI] [PubMed] [Google Scholar]
- 14. Wattenberg L.W. (1977). Inhibition of carcinogenic effects of polycyclic hydrocarbons by benzyl isothiocyanate and related compounds. J. Natl. Cancer Inst., 58, 395–398 [DOI] [PubMed] [Google Scholar]
- 15. Sugie S, et al. (1993). Inhibitory effects of benzyl isothiocyanate and benzyl thiocyanate on diethylnitrosamine-induced hepatocarcinogenesis in rats. Jpn. J. Cancer Res., 84, 865–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Okazaki K, et al. (2002). Simultaneous treatment with benzyl isothiocyanate, a strong bladder promoter, inhibits rat urinary bladder carcinogenesis by N-butyl-N-(4-hydroxybutyl)nitrosamine. Nutr. Cancer, 42, 211–216 [DOI] [PubMed] [Google Scholar]
- 17. Kuroiwa Y, et al. (2006). Protective effects of benzyl isothiocyanate and sulforaphane but not resveratrol against initiation of pancreatic carcinogenesis in hamsters. Cancer Lett., 241, 275–280 [DOI] [PubMed] [Google Scholar]
- 18. Warin R, et al. (2009). Prevention of mammary carcinogenesis in MMTV-neu mice by cruciferous vegetable constituent benzyl isothiocyanate. Cancer Res., 69, 9473–9480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thiery J.P. (2002). Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer, 2, 442–454 [DOI] [PubMed] [Google Scholar]
- 20. Tomaskovic-Crook E, et al. (2009). Epithelial to mesenchymal transition and breast cancer. Breast Cancer Res., 11, 213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hollier B.G, et al. (2009). The epithelial-to-mesenchymal transition and cancer stem cells: a coalition against cancer therapies. J. Mammary Gland Biol. Neoplasia, 14, 29–43 [DOI] [PubMed] [Google Scholar]
- 22. Sehrawat A, et al. (2011). Benzyl isothiocyanate inhibits epithelial-mesenchymal transition in cultured and xenografted human breast cancer cells. Cancer Prev. Res. (Phila)., 4, 1107–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Warin R, et al. (2010). Inhibition of human breast cancer xenograft growth by cruciferous vegetable constituent benzyl isothiocyanate. Mol. Carcinog., 49, 500–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xiao D, et al. (2008). Benzyl isothiocyanate targets mitochondrial respiratory chain to trigger reactive oxygen species-dependent apoptosis in human breast cancer cells. J. Biol. Chem., 283, 30151–30163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jo M, et al. (2010). Cell signaling by urokinase-type plasminogen activator receptor induces stem cell-like properties in breast cancer cells. Cancer Res., 70, 8948–8958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang H, et al. (2011). Forkhead transcription factor foxq1 promotes epithelial-mesenchymal transition and breast cancer metastasis. Cancer Res., 71, 1292–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hahm E.R, et al. (2011). Withaferin A-induced apoptosis in human breast cancer cells is mediated by reactive oxygen species. PLoS ONE, 6, e23354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xiao D, et al. (2003). Allyl isothiocyanate, a constituent of cruciferous vegetables, inhibits proliferation of human prostate cancer cells by causing G2/M arrest and inducing apoptosis. Carcinogenesis, 24, 891–897 [DOI] [PubMed] [Google Scholar]
- 29. Xiao D, et al. (2006). Diallyl trisulfide suppresses growth of PC-3 human prostate cancer xenograft in vivo in association with Bax and Bak induction. Clin. Cancer Res., 12, 6836–6843 [DOI] [PubMed] [Google Scholar]
- 30. Guo Y, et al. (2002). Regulation of DNA methylation in human breast cancer. Effect on the urokinase-type plasminogen activator gene production and tumor invasion. J. Biol. Chem., 277, 41571–41579 [DOI] [PubMed] [Google Scholar]
- 31. Livak K.J, et al. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods, 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 32. Boreddy S.R, et al. (2011). Pancreatic tumor suppression by benzyl isothiocyanate is associated with inhibition of PI3K/AKT/FOXO pathway. Clin. Cancer Res., 17, 1784–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hajra K.M, et al. (2002). The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res., 62, 1613–1618 [PubMed] [Google Scholar]
- 34. Batlle E, et al. (2000). The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat. Cell Biol., 2, 84–89 [DOI] [PubMed] [Google Scholar]
- 35. Lester R.D, et al. (2007). uPAR induces epithelial-mesenchymal transition in hypoxic breast cancer cells. J. Cell Biol., 178, 425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kwaan H.C, et al. (2009). The role of plasminogen-plasmin system in cancer. Cancer Treat. Res., 148, 43–66 [DOI] [PubMed] [Google Scholar]
- 37. Qiao Y, et al. (2011). FOXQ1 regulates epithelial-mesenchymal transition in human cancers. Cancer Res., 71, 3076–3086 [DOI] [PubMed] [Google Scholar]
- 38. Kaneda H, et al. (2010). FOXQ1 is overexpressed in colorectal cancer and enhances tumorigenicity and tumor growth. Cancer Res., 70, 2053–2063 [DOI] [PubMed] [Google Scholar]
- 39. Lai K.C, et al. (2010). Benzyl isothiocyanate (BITC) inhibits migration and invasion of human colon cancer HT29 cells by inhibiting matrix metalloproteinase-2/-9 and urokinase plasminogen (uPA) through PKC and MAPK signaling pathway. J. Agric. Food Chem., 58, 2935–2942 [DOI] [PubMed] [Google Scholar]
- 40. Kim E.J, et al. (2012). Benzyl isothiocyanate inhibits basal and hepatocyte growth factor-stimulated migration of breast cancer cells. Mol. Cell. Biochem., 359, 431–440 [DOI] [PubMed] [Google Scholar]
- 41. Aguirre Ghiso J.A. (2002). Inhibition of FAK signaling activated by urokinase receptor induces dormancy in human carcinoma cells in vivo . Oncogene, 21, 2513–2524 [DOI] [PubMed] [Google Scholar]
- 42. Liu D, et al. (2002). EGFR is a transducer of the urokinase receptor initiated signal that is required for in vivo growth of a human carcinoma. Cancer Cell, 1, 445–457 [DOI] [PubMed] [Google Scholar]
- 43. Kjøller L, et al. (2001). Rac mediates cytoskeletal rearrangements and increased cell motility induced by urokinase-type plasminogen activator receptor binding to vitronectin. J. Cell Biol., 152, 1145–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nusrat A.R, et al. (1991). An autocrine role for urokinase in phorbol ester-mediated differentiation of myeloid cell lines. J. Clin. Invest., 87, 1091–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kim S.H, et al. (2011). Benzyl isothiocyanate inhibits oncogenic actions of leptin in human breast cancer cells by suppressing activation of signal transducer and activator of transcription 3. Carcinogenesis, 32, 359–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ho C.C, et al. (2011). Benzyl isothiocyanate (BITC) inhibits migration and invasion of human gastric cancer AGS cells via suppressing ERK signal pathways. Hum. Exp. Toxicol., 30, 296–306 [DOI] [PubMed] [Google Scholar]
- 47. Gulhati P, et al. (2011). mTORC1 and mTORC2 regulate EMT, motility, and metastasis of colorectal cancer via RhoA and Rac1 signaling pathways. Cancer Res., 71, 3246–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xiao D, et al. (2012). Benzyl isothiocyanate causes FoxO1-mediated autophagic death in human breast cancer cells. PLoS ONE, 7, e32597 [DOI] [PMC free article] [PubMed] [Google Scholar]