Abstract
Hepatitis B X-interacting protein (HBXIP) is an important oncoprotein that plays critical role in the development of cancer. In this study, we report that HBXIP activates LIM-only protein 4 (LMO4), a transcriptional coregulatory protein, in promotion of cell proliferation. We observed that the messenger RNA (mRNA) expression levels of HBXIP were positively associated with those of LMO4 in clinical breast cancer tissues. We further identified that HBXIP upregulated LMO4 at the levels of promoter, mRNA and protein in MCF-7 and LM-MCF-7 breast cancer cell lines. The expression of cyclin D1 and cyclin E, downstream effectors of LMO4, could be upregulated by HBXIP through LMO4. Then, chromatin immunoprecipitation (ChIP) assay revealed that HBXIP was able to interact with the promoter region of LMO4. Electrophoretic mobility shift assay showed that HBXIP occupied the -237/-206 region of LMO4 promoter containing Sp1 binding element. The mutant of Sp1 binding site in the LMO4 promoter impeded the interaction of HBXIP with the promoter. Co-immunoprecipitation, ChIP and luciferase reporter gene assays showed that HBXIP activated LMO4 promoter through binding to Sp1. In function, flow cytometry, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, 5-ethynyl-2'-deoxyuridine (EdU) incorporation assays and animal transplantation assays demonstrated that HBXIP-enhanced cell proliferation of breast cancer through upregulating LMO4 in vitro and in vivo. Thus, we concluded that oncoprotein HBXIP is able to activate the transcriptional coregulatory protein LMO4 through transcription factor Sp1 in promotion of proliferation of breast cancer cells. HBXIP may serve as a driver gene to activate transcription in the development of cancer.
Introduction
Hepatitis B X-interacting protein (HBXIP), a conserved 19kDa protein, was originally identified by its interaction with hepatitis B virus X protein (1). HBXIP can collaborate with survivin, a baculovirus IAP (inhibitor of apoptosis) repeat (BIR) family chromosomal passenger protein, involved in the control of cell division (2) and lead to the suppression of cell apoptosis (3). Furthermore, HBXIP has been demonstrated to interact with hSuv3, an ATP-dependent DNA/RNA DExH [peptide motif composed of an aspartic acid (D), a glutamic acid (E), a random amino acid (x), and a histidine (H)] box helicase predominantly localized in mitochondria, which exhibits typical characteristics for a nuclear-encoded mitochondrial gene (4). It has been reported that HBXIP plays a critical role in mitosis, particularly in regulating centrosome dynamics, mitotic spindles dynamics as well as cytokinesis (5,6). Our previous study indicated that HBXIP was involved in the regulation of cell proliferation, which was related to cell cycle progression (7,8). Recently, we have reported that HBXIP upregulates S100A4 through activating S100A4 promoter involving STAT4 and inducing PTEN/PI3K/AKT signaling to promote growth and migration of breast cancer cells (9). However, the underlying mechanism of HBXIP in carcinogenesis has not been well defined.
LIM-only protein 4 (LMO4) belongs to the LIM-only family of transcriptional coregulatory proteins characterized by the presence of two tandem LIM domains (10). LIM domains are involved in protein–protein interactions, allowing LIM-only proteins to serve as linker proteins in multiprotein complexes (11–13). In human tissues, LMO4 has a very broad spectrum of expression (14). Increased LMO4 expression is observed in several epithelial cancers including squamous cell carcinomas of oral cavity, prostate, pancreas and breast cancer (10,15). LMO4 is overexpressed in primary breast tumors and its expression is correlated to a worse prognosis (16,17). Significantly, high levels of LMO4 in cell nuclear may be independent predictors of death from breast cancer. And deregulation of LMO4 in breast epithelium of transgenic mice can directly lead to breast neoplasia (11). Moreover, LMO4 has been implicated in the pathogenesis of breast cancer through regulating the rate of cellular proliferation and invasion (11,17). It has been reported that LMO4 plays a crucial role in the centrosome cycle as well as cell cycle progression in breast cancer cells. It can regulate several cell cycle-related genes, including cyclin D1, cyclin E, p21, p27 and BRCA1 (11,18). Strikingly, growing evidence indicates that LMO4 can modulate progression of breast cancer cell cycle by indirectly enhancing the expression of G1 cyclins, such as cyclin D1 and cyclin E, and contributes to the growth of multiple breast cancer cells (19–21). However, whether LMO4 is involved in the carcinogenesis mediated by HBXIP remains unclear.
In this study, we further investigated the mechanism of HBXIP in carcinogenesis. Interestingly, we found that HBXIP was able to activate transcriptional coregulatory protein LMO4 through Sp1 in the cells. Our finding provides new insight into the mechanism of HBXIP in promotion of proliferation of breast cancer cells.
Materials and methods
Cell lines and cell transduction
The breast cancer cell lines, such as MCF-7, T47D and LM-MCF-7 (a metastatic subclone of MCF-7 breast cancer cell line), were maintained in RPMI 1640 (Gibco, Carlsbad, CA) medium containing 10% fetal bovine serum (Gibco) (22). The engineered, stable HBXIP-transfected MCF-7 cell line, termed MCF-7-HBXIP, was maintained in above condition (23).
RNA interference
Small interfering RNAs (siRNAs) targeting the messenger RNAs (mRNAs) of HBXIP (GenBank accession No.NM_006402.2), LMO4 (GenBank accession No.NM_006769.3) and Sp1 were synthesized by Riobio (Guangzhou, China) and transfected into MCF-7 (or T47D) cells according to the manufacturer’s protocols. The sequences of each siRNA were described previously (5,24,25).
Immunohistochemistry
The tissue arrays with breast cancer tissues and normal breast tissues were obtained from Xi’an Aomei Biotechnology Co., (Xi’an, China), which included duplicate core biopsies (1mm in diameter) from fixed, paraffin-embedded tumors. Immunohistochemical staining of samples was performed as described previously (26) with rabbit antihuman HBXIP antibody (Sigma–Aldrich, St Louis, MO) and rabbit antihuman LMO4 antibody (Santa Cruz). The staining level of HBXIP (or LMO4) was scored by using a modified scoring method based on the intensity of staining (0 = negative; 1 = low; 2 = high) and the percentage of stained cells (0 = 0% stained; 1 = 1–49% stained; 2 = 50–100% stained). A multiplied score (intensity score × percentage score) <1 was considered to be negative staining (-), 1 and 2 were considered to be moderate staining (+), 3 and 4 were considered to be intense staining (++).
Plasmid construction
The complete human LMO4 complementary DNA was subcloned into pCMV-tag2B vector to generate the pCMV-LMO4 construct. The upstream region (from -1300 to +35 nt) of LMO4 gene was amplified by PCR from MCF-7 cells using specific primers (27) and was inserted into the KpnI/XhoI site in the pGL3-basic vector. The resulting plasmid was named pGL3-WT-LMO4 (pGL3-LMO4). Mutant construction of -1300/+35 region of LMO4 promoter, named pGL3-mu-Sp1, carried a substitution of three nucleotides within the binding site of Sp1. Mutagenesis primers used were as follows: 5′-GGT CCC CGG CCC CAG GCT AAC GGG TCA CTT CAC CCC A-3′ and 5′-TGG GGT GAA GTG ACC CGT TAG CCT GGG GCC GGG GAC C-3′.
Luciferase reporter gene assay
MCF-7 or LM-MCF-7 cells (2×104 cells per well) grown in 24-well plates were co-transfected with LMO4 luciferase reporter plasmid (0.2 μg) and pRL-TK normalization construct (0.1 μg) using Lipofectamine 2000 (Invitrogen). The pCMV-HBXIP plasmid (0.1–0.3 μg) was co-transfected with reporter plasmids to overexpress HBXIP. siRNAs targeting HBXIP or Sp1 (30–100nM) and the reporter plasmids were co-transfected into the cells. Cells were harvested 48h after transfection and luciferase reporter gene assay was implemented using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions (28,29). All experiments were performed at least three times.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assay was performed using ChIP kit from Epigentek Group (Brooklyn, NY) according to the manufacturer’s instructions. The DNA pulled down by anti-HBXIP antibodies was amplified by PCR (30,31). The negative control primers were described previously (31). DNA from these samples was subjected to PCR analyses with primers sets for LMO4 promoter: 5′-CAG TCA TCC CTT TGT CCT TCC-3′ and 5′-TGA CAG AGC AAA ATC CCA ACT A-3′; the negative control primers were glyceraldehyde 3-phosphate dehydrogenase-1: 5′-GTA TTC CCC CAG GTT TAC AT-3′ and glyceraldehyde 3-phosphate dehydrogenase-2: 5′-TTC TGT CTT CCA CTC ACT CCT-3′, followed by sequencing.
Western blot analysis
The protocol was described previously (32–34). Primary antibodies used were rabbit anti-HBXIP (Santa Cruz), rabbit anti-LMO4 (Santa Cruz), rabbit anti-Sp1 (Epitomics) and mouse anti-β-actin antibodies (Sigma).
Immunofluorescence staining
Cells plated on cover slips in six-well plates were fixed in 4% paraformaldehyde, permeabilized in 0.1% Triton X-100 and blocked in 5% bovine serum albumin. Costaining for HBXIP or LMO4 was performed by incubating with the primary antibodies, such as rabbit anti-LMO4 and rabbit anti-HBXIP, for 2h and with fluorophore-conjugated secondary antibody (1:100) as well as 4′,6-diamidino-2-phenylindole (1:1000) for 1h. The stained cells were observed with Nikon TE200 inverted fluorescence microscope.
Electrophoretic mobility shift assay
Electrophoretic mobility shift assay (EMSA) was performed as described previously (35). Probes were generated by annealing single-strand oligonucleotides covering the LMO4 promoter and labeling the ends with [γ-32P] adenosine triphosphate using T4 polynucleotide kinase (TaKaRa Bio). Nuclear extract (1 μg) of MCF-7-pCMV or MCF-7-HBXIP cells and 15fmol 32P-labeled DNA, with or without 1 μg HBXIP antibody, were mixed in binding buffer (1% NP-40, 20 mmol/l Hepes pH 8.0, 0.5 mmol/l dithiothreitol, 50 mmol/l KCl, 0.05 mmol/l ethylenediaminetetraacetic acid, 5% glycerol, 0.05 μg/μl poly (dI/dC) and 1 mmol/l MgCl2). Samples were incubated on ice for 1h and then were resolved at 4°C using a native 6% polyacrylamide gel in 0.5× tris-borate- EDTA (TBE) buffer. The gel was dried and subjected to autoradiography.
Co-immunoprecipitation
The co-immunoprecipitation assay was performed as described previously (36). To detect the interaction of endogenous proteins, the cell lysis was immunoprecipitated with rabbit anti-HBXIP or anti-Sp1 antibody and protein G-Sepharose. The precipitates were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (12% total acrylamide) followed by immunoblotting.
Flow cytometry analysis
After 48h of transfection, cells (1×106) were harvested and washed with cold phosphate-buffered saline. Washed cells were fixed in 75% ethanol at 4°C overnight. The fixed cells were rinsed twice with phosphate-buffered saline and treated with propidium iodine solution including 50 µg/ml propidium iodine (Sigma) and 50 µg/ml RNaseA (Sigma) at 37°C for 30min. Stained cells were analyzed by a FACScan flow cytometer (Becton Dickinson, Bedford, MA), followed by the analysis using CellQuest software (Becton Dickinson). Cell proliferation index (PI) was calculated by the formula: PI = (G2/M + S) ÷ (G0/G1 + S + G2/M) × 100%, as the sum of the S and G2/M phase cells, expressed as a fraction of the total cell population (37).
Analysis of cell proliferation
For 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays, MCF-7 or T47D cells were seeded onto 96-well plates (3×103cells per well) for 12h before transfection. MTT (Sigma) assays were performed to assess cell proliferation every day for 4 days after transfection. The protocol was described previously (22). In addition, the 5-ethynyl-2'-deoxyuridine (EdU) incorporation assays were also carried out to measure cell proliferation using the Cell-Light TM EdU imaging detecting kit (RiboBio) according to the manufacturer’s instructions.
Xenografts
MCF-7 cells pre-treated with pCMV plasmid [or pCMV-HBXIP plasmid, control siRNA (si-Control), LMO4 siRNA] were harvested and 5×106 cells in 200 μl of phosphate-buffered saline were subcutaneously injected at the shoulder of 4-week-old female BALB-c mice (each group, n = 6). Tumor size was measured in two dimension with calipers every 3 days, up to 30 days after injection. Tumor volume (v) was calculated according to the formula: 0.5 × (length × width2). All experiments were approved by the animal care committee of Nankai University.
Statistical analysis
Statistical significance was assessed by comparing mean values (±standard deviation) using the Student’s t-test for independent groups and was assumed for *P < 0.05 and **P < 0.01. Each experiment was repeated at least three times.
Results
The expression levels of HBXIP are positively associated with those of LMO4 in clinical breast cancer tissues
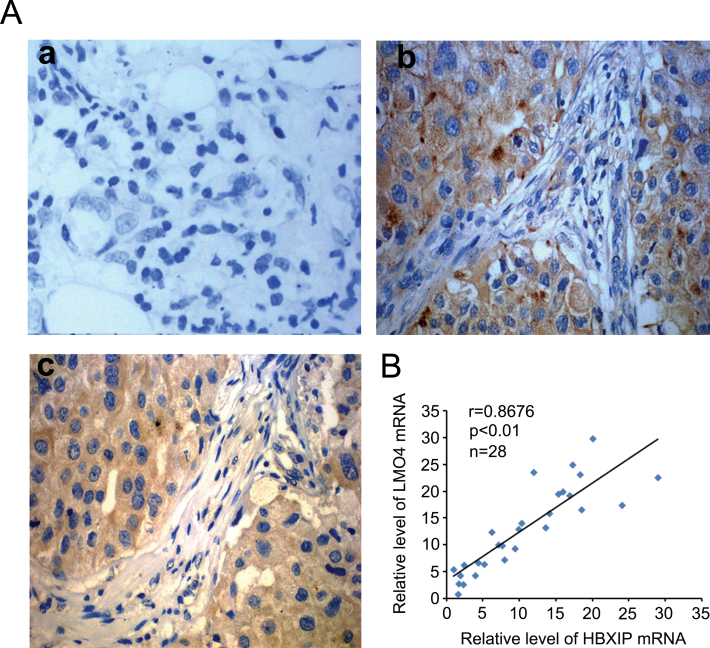
Our previous studies showed that HBXIP was upregulated in breast cancer tissues (23) and it was able to enhance the proliferation of breast cancer cells (7). Meanwhile, LMO4 contributes to breast oncogenesis by regulating cell proliferation (11). Thus, we are interested in the relationship between HBXIP and LMO4 in breast cancer tissues. Immunohistochemistry staining showed that the positive rate of HBXIP was 85.1% (74/87) in clinical breast cancer tissue samples, in which the positive rate of LMO4 was 80.5% (70/87) in the HBXIP-positive specimens (Figure 1A and Supplementary Table 1, available at Carcinogenesis Online). Meanwhile, real-time PCR assays showed that the mRNA expression levels of HBXIP were positively associated with those of LMO4 in fresh breast tumor tissue samples from 28 patients (Figure 1B). Therefore, we conclude that the expression levels of HBXIP are positively associated with those of LMO4 in clinical breast cancer tissues.
Fig. 1.
The expression levels of HBXIP are positively associated with those of LMO4 in clinical breast cancer tissues. (A) Immunohistochemistry showed the expression correlation of HBXIP and LMO4 expression in clinical breast cancer tissues using tissue arrays. Imaged at ×400 magnification. Panel a: negative control; panel b: HBXIP-positive staining; panel c: LMO4-positive staining. (B) The mRNA relative levels of HBXIP and LMO4 in tumor tissues were examined by real-time PCR.
HBXIP is able to upregulate LMO4 in breast cancer cells
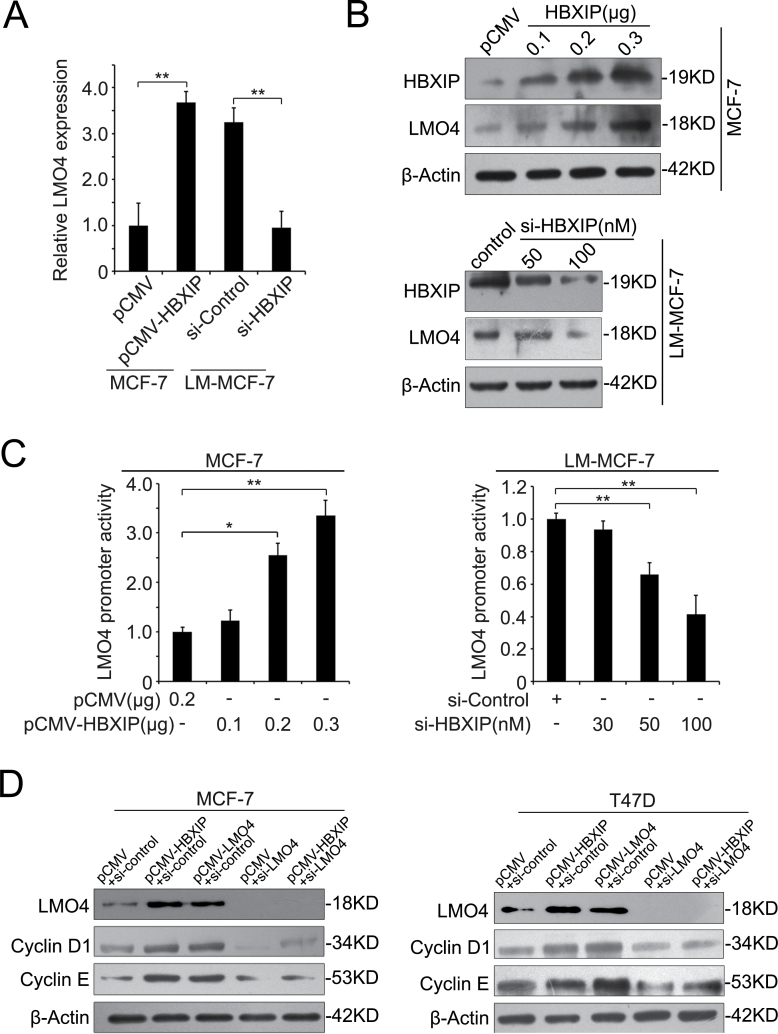
Next, we tested whether HBXIP is able to upregulate LMO4. As shown in Figure 2A and B, we observed that the overexpression of HBXIP enhanced the expression of LMO4 at the levels of mRNA and protein in MCF-7 cells, which expresses low levels of endogenous HBXIP, whereas the knockdown of HBXIP by siRNA decreased the expression of LMO4 in LM-MCF-7 cells with high levels of endogenous HBXIP (9). However, the overexpression or downregulation of LMO4 failed to affect the expression of HBXIP in MCF-7 and LM-MCF-7 cells (data not shown). To assess the effect of HBXIP on LMO4 promoter activity, we cloned the promoter region of LMO4 (-1300/+35) into the pGL3-basic plasmid according to the report (27). Luciferase reporter gene assays showed that the overexpression or knockdown of HBXIP could increase or decrease the promoter activity of LMO4 in a dose-dependent manner (Figure 2C). It has been reported that LMO4 is required for cell proliferation of breast cancer and sustained expression of cell cycle genes, such as cyclin D1 and cyclin E (21). To further validate that HBXIP upregulates LMO4 in breast cancer cells, we next detected the effect of HBXIP on cyclin D1 and cyclin E, two downstream effectors of LMO4. Western blot analysis revealed that the overexpression of HBXIP could upregulate the expression of cyclin D1 and cyclin E in MCF-7 and T47D cells, respectively. Meanwhile, we validated that the overexpression of LMO4 upregulated cyclin D1 and cyclin E in the cells. Inversely, the silence of LMO4 by siLMO4 abolished the upregulation of cyclin D1 and cyclin E mediated by HBXIP (Figure 2D), supporting that HBXIP is able to upregulate LMO4. Thus, we concluded that HBXIP is able to upregulate LMO4 in breast cancer cells.
Fig. 2.
HBXIP is able to upregulate LMO4 in breast cancer cells. (A and B) The expression levels of LMO4 were determined by quantitative real-time PCR and western blot analysis, respectively (*P < 0.05, **P < 0.01; Student’s t-test). MCF-7 cells were transfected with pCMV-HBXIP vector (or control). LM-MCF-7 cells were transfected with HBXIP siRNAs (or control). (C) The activity of LMO4 promoter was examined by luciferase reporter gene assay in MCF-7 and LM-MCF-7 cells (*P < 0.05; Student’s t-test). (D) The expression of cyclin D1 and cyclin E were examined by western blot in MCF-7 and T47D cells.
HBXIP is able to bind to the region of -237 ~ -206 in LMO4 promoter
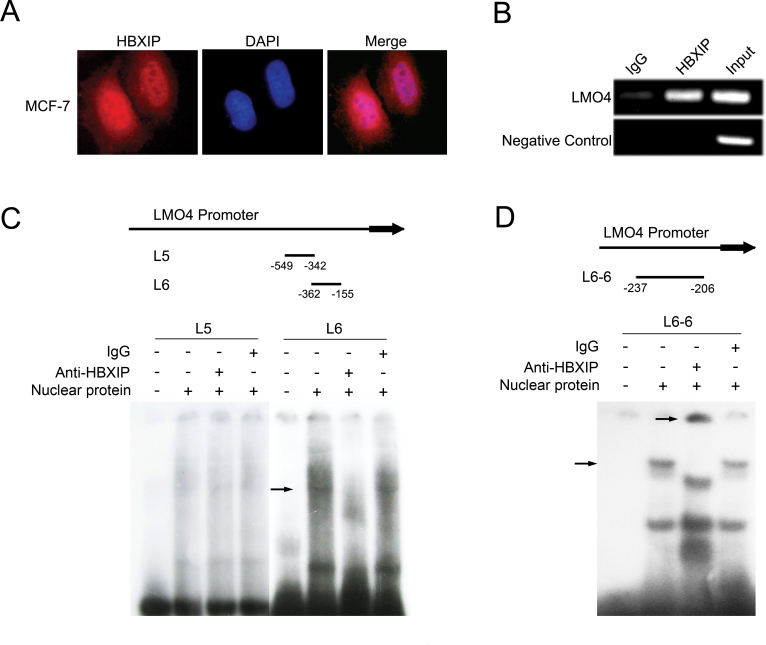
Next, we tried to elucidate the underlying mechanism by which HBXIP upregulates LMO4. Immunofluorescence assay showed that HBXIP was able to localize in the nucleus of MCF-7 cells (Figure 3A). Therefore, we speculated that HBXIP might be involved in the transcriptional regulation of LMO4. Interestingly, ChIP assays revealed that HBXIP was able to bind to the LMO4 promoter (Figure 3B). To map the HBXIP binding sites in LMO4 promoter, we performed EMSA. We used probes to search putative HBXIP binding sites within a 1.3kb region upstream of the LMO4 coding sequence including seven ~200bp DNA fragments, such as L1 (-1300/-1093), L2 (-1112/-905), L3 (-924/-717), L4 (-736/-529), L5 (-549/-342), L6 (-362/-155) and L7 (-175/+35), as shown in Supplementary Figure 1A, available at Carcinogenesis Online. EMSA showed that the overexpression of HBXIP was able to increase the interaction of HBXIP with the L6 fragment (-362/-155) in MCF-7 cells, in which the transfection efficiency was confirmed by western blot analysis (Supplementary Figure 1A, available at Carcinogenesis Online). Furthermore, EMSA validated that the interaction of HBXIP with L6 fragment could be blocked when the anti-HBXIP antibody was added (Figure 3C), suggesting that HBXIP is able to bind to the fragment -362/-155 of LMO4 promoter. To minimize the interaction site between HBXIP and LMO4 promoter, we further divided the L6 fragment into eight ~30bp DNA fragments, such as L6-1~ L6-8 (-362/-332, -337/-307, -312/-282, -287/-257, -262/-232, -237/-206, -211/-181 and -186/-155). We identified that HBXIP could interact with the fragment L6-6 (-237/-206) (Figure 3D and Supplementary Figure 1B, available at Carcinogenesis Online). Thus, we concluded that HBXIP is able to bind to the -237 ~ -206 region in LMO4 promoter.
Fig. 3.
HBXIP is able to bind to LMO4 promoter in the region of -237 ~ -206. (A) The nuclear localization of HBXIP was detected by immunofluorescence staining in MCF-7 cells. The images of HBXIP (red) were shown. 4′,6-Diamidino-2-phenylindole staining (blue) was included to visualize the nucleus. (B) The interaction of HBXIP with LMO4 promoter was determined by ChIP assay. (C) EMSA was used to identify the binding site of HBXIP to LMO4 promoter. DNA fragment L5 (-549/-342) and L6 (-362/-155) from the LMO4 promoter were used as DNA probes, respectively. The shift (arrow) of the fragment L6 was abolished by addition of anti-HBXIP antibody. IgG was used as a control antibody. (D) EMSA assay showed that adding HBXIP antibody led to the supershift (arrow) of the fragment L6-6 (-237/-206).
HBXIP activates LMO4 promoter through transcription factor Sp1
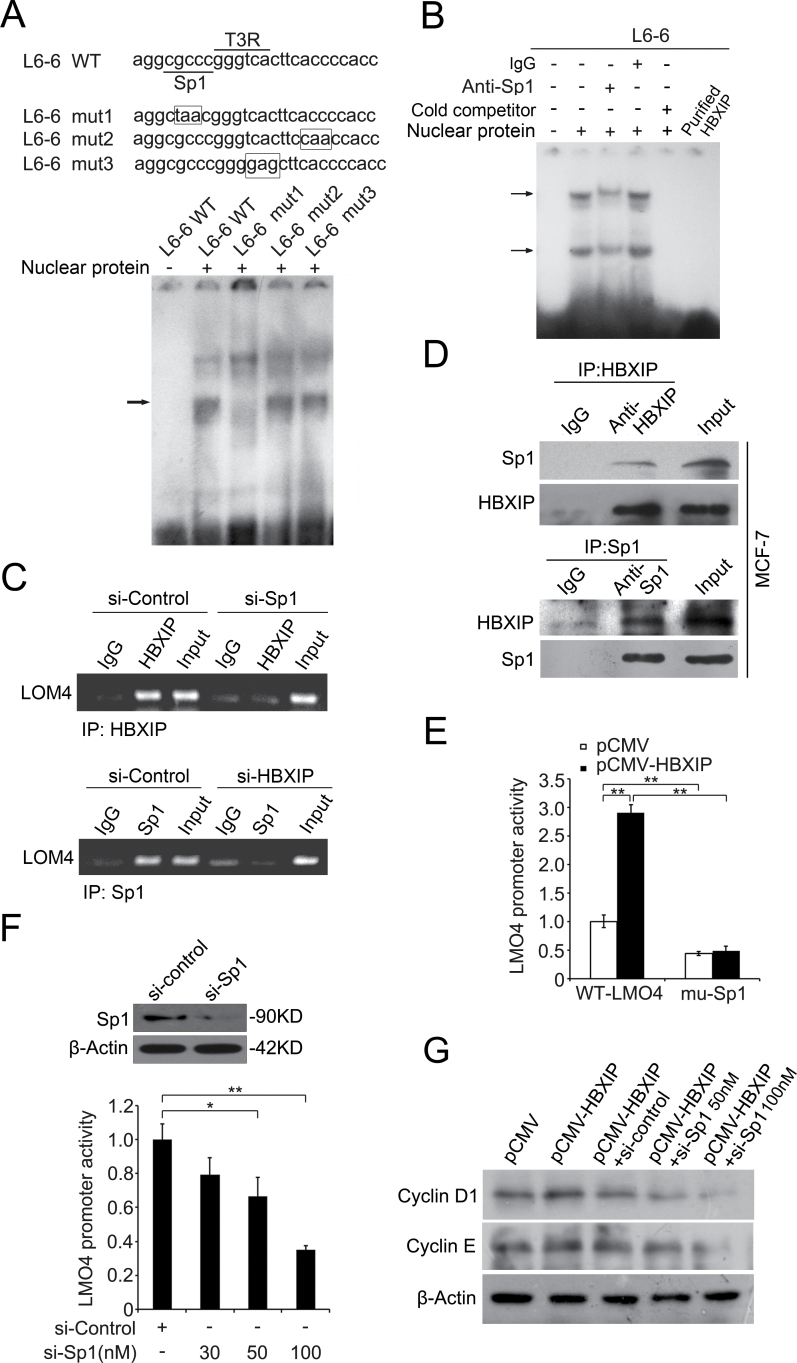
Strikingly, we observed two putative transcription factor binding sites in the -237/-206 region of LMO4 promoter, such as Sp1 and T3R, through prediction using the online promoter analysis tool Gene Regulation (http://www.gene-regulation.com). Next, we examined whether the binding of HBXIP to LMO4 promoter is related to these transcription factors. EMSA demonstrated that only the mutant of Sp1 binding site impeded the HBXIP–DNA interaction (Figure 4A). Meanwhile, the HBXIP–DNA interaction could be strikingly diminished by the addition of anti-Sp1 antibodies. However, the purified recombinant HBXIP alone was unavailable to bind to the -237/-206 DNA oligos by EMSA (Figure 4B), suggesting that Sp1 is involved in the interaction of HBXIP with LMO4 promoter. Moreover, ChIP assay revealed that the knockdown of Sp1 was able to dismiss the interaction of HBXIP with LMO4 promoter. In addition, we also observed the occupancy of Sp1 in LMO4 promoter was disturbed when HBXIP was silenced in the cells (Figure 4C), which supported the above EMSA data. Then, co-immunoprecipitation assay further demonstrated that HBXIP was able to interact with Sp1 in MCF-7 cells (Figure 4D). It suggests that HBXIP indirectly binds to LMO4 promoter via interacting with Sp1. Luciferase reporter gene assays revealed that HBXIP failed to increase the activity of LMO4 promoter when the Sp1 binding site was mutated (Figure 4E). Meanwhile, the knockdown of Sp1 by siRNA led to a significant decrease of LMO4 promoter activity in stably HBXIP-transfected cell lines (MCF-7-HBXIP) in a dose-dependent manner. The silence efficiency of Sp1 was confirmed by western blot analysis (Figure 4F). Moreover, the silence of Sp1 was able to block the upregulation of cyclin D1 and cyclin E mediated by HBXIP in MCF-7 cells (Figure 4G). Thus, we concluded that HBXIP is able to activate LMO4 transcription through interacting with transcription factor Sp1 in breast cancer cells.
Fig. 4.
HBXIP activates LMO4 promoter through transcription factor Sp1. (A) A model of wild and mutation type analysis for the fragments of L6-6 (-237/-206). The binding sites of Sp1and T3R were underlined. The mutated nucleotides of both transcription factor recognizing sequences as well as the mutation of 3'-flank region of L6-6 fragment were highlighted with rectangles. EMSA showed the interaction of the above DNA probes with nucleus proteins of MCF-7 cells. The L6-6 mut 1 dismissed the interaction. (B) EMSA with the addition of anti-Sp1 antibodies was performed to examine the binding of nucleus proteins to fragment L6-6. (C) ChIP showed the interaction of HBXIP with LMO4 promoter in MCF-7 cells when Sp1 was knockdown, and vice versa. (D) The interaction between HBXIP and Sp1 was determined by co-immunoprecipitation in MCF-7 cells. The immunoprecipitates were analyzed by western blot. (E) The promoter activity of LMO4 mediated by HBXIP was measured by luciferase reporter gene assay when Sp1 binding site was mutated. MCF-7-pCMV and MCF-7-HBXIP cells were transiently transfected with pGL3-WT-LMO4 or pGL3-mu-Sp1 plasmids (**P < 0.01; Student’s t-test). (F) The promoter activity of LMO4 was examined by luciferase reporter gene assay in MCF-7-HBXIP cells when the cells were treated with Sp1 siRNA (30, 50 or 100nM) (*P < 0.05, **P < 0.01; Student’s t-test), in which the siRNA efficiency of Sp1 was determined by western blot analysis. (G) The expression levels of cyclin D1 and cyclin E were examined by western blot in MCF-7 cells transiently transfected with pCMV-HBXIP (or pCMV-tag2B), pCMV-HBXIP and Sp1 siRNA (or control siRNA), respectively.
HBXIP enhances the proliferation of breast cancer cells through LMO4 in vitro and in vivo
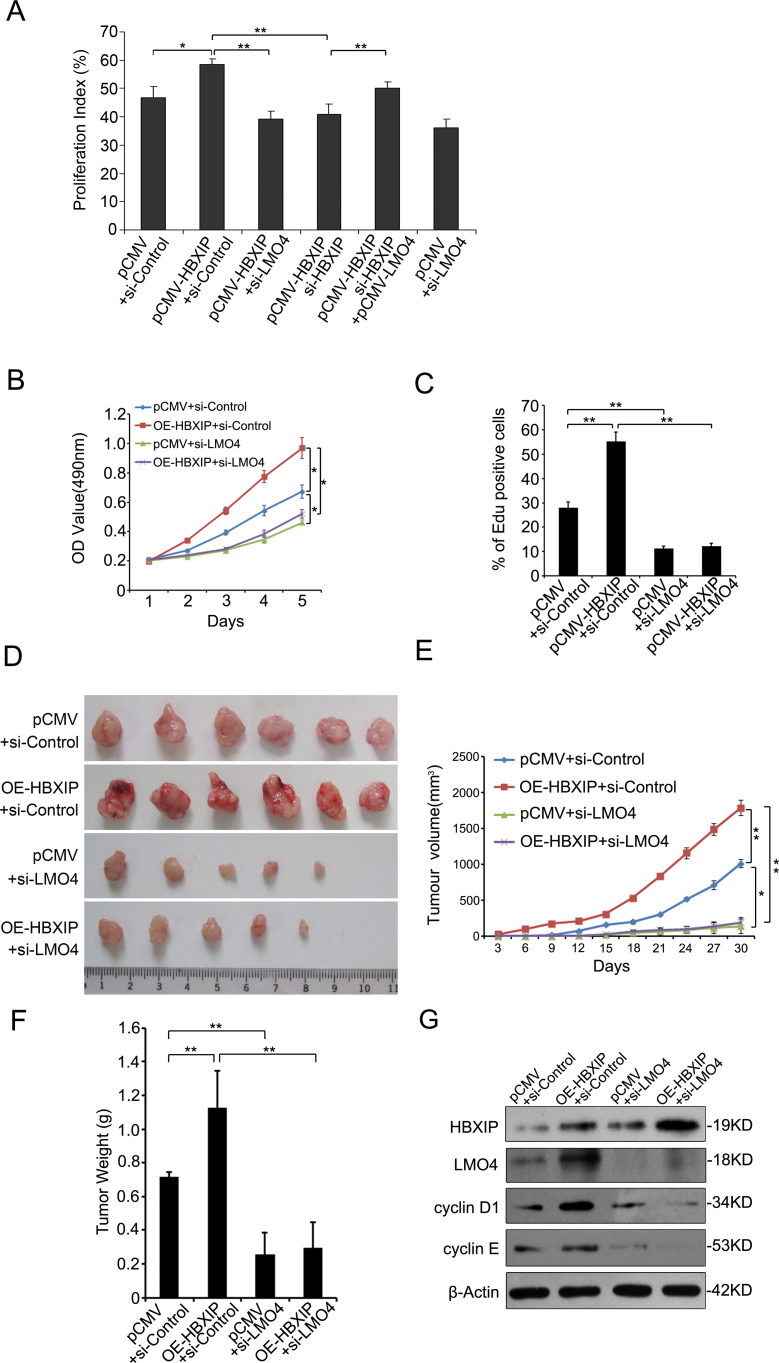
Next, we examined whether LMO4 was involved in the promotion of proliferation of breast cancer cells mediated by HBXIP. Flow cytometry analysis showed that the overexpression of HBXIP resulted in the increase of S phage cells. Interestingly, the knockdown of LMO4 reduced S phage cells in stably HBXIP-transfected MCF-7 cells. Of note, the overexpression of LMO4 could rescue the decrease of S phage cells in MCF-7-HBXIP cells when HBXIP was downregulated by siRNA (Figure 5A and Supplementary Figure 2, available at Carcinogenesis Online). Furthermore, MTT assays showed that the silence of LMO4 abolished HBXIP-enhanced proliferation of MCF-7 cells (Figure 5B). In addition, EdU assays revealed the similar results (Figure 5C). These data suggest that LMO4 is involved in the promotion of proliferation of breast cancer cells mediated by HBXIP. The animal transplantation assay showed that the overexpression of HBXIP enhanced the growth of MCF-7 cells in animal. Importantly, the knockdown of LMO4 dismissed the growth of breast cancer cells in mice. Interestingly, the knockdown of LMO4 was able to block HBXIP-enhanced growth of breast cancer in mice (Figure 5D–F). Meanwhile, western blot analysis showed the mean expression levels of HBXIP, LMO4, cyclin D1 and cyclin E in each group of tumor tissue samples from mice (Figure 5G), which were correspondence to the function of growth of cells. Therefore, we concluded that HBXIP promotes the proliferation of breast cancer cells through LMO4 in vitro and in vivo.
Fig. 5.
HBXIP enhances the proliferation of breast cancer cells through LMO4 in vitro and in vivo. (A) The PIs of MCF-7 cells with different treatments were examined by flow cytometry analysis. The PI values of each group were from three independent experiments, respectively (*P < 0.05, **P < 0.01; Student’st-test). (B and C) The proliferation of MCF-7 cells transfected with pCMV vector, pCMV-HBXIP expression vector, HBXIP or LMO4 siRNA and pCMV-HBXIP/LMO4 siRNA was measured by MTT and EdU assays, respectively (*P < 0.05, **P < 0.01; Student’s t-test). (D) Four groups, such as control, HBXIP overexpression, LMO4 knockdown and HBXIP overexpression/LMO4 knockdown, of MCF-7 cells were transplanted into athymic mice (BALB/c). After 30 days, the tumors were dissected out from mice. (E) The growth curve of tumor. Tumor size was measured every 3 days. Each point showed the mean ± SEM (n = 6). (F) The average weight of tumors. (G) The mean expression levels of HBXIP, LMO4, cyclin D1 and cyclin E in the tumor tissues from each group mice were detected by western blot. The protein samples were from mixture extract of six tumor tissues in each group.
Discussion
Growing evidence indicates that HBXIP is an important oncoprotein that dramatically enhances the proliferation of breast cancer cells (8). However, the underlying mechanisms by which HBXIP contributes to the proliferation of breast cancer cells have not been fully elucidated. LMO4 has been implicated in the pathogenesis of breast cancer by regulating the cellular proliferation and invasion (11–21). Accordingly, we are interested in whether LMO4 is involved in the aggressive accumulation of breast cancer cells mediated by HBXIP.
In this study, we discovered that there is a significant positive correlation between HBXIP and LMO4 expression in breast tumor tissue samples. Then, we found that HBXIP was able to dramatically upregulate the expression levels of LMO4 in breast cancer cells. LMO4, as a transcription regulator, is required for cell cycle progression and sustained expression of cyclin D1 and cyclin E (21). We validated that HBXIP was able to elevate the expression of cyclin D1 and cyclin E through LMO4. However, the underlying mechanism of upregulation of cyclin D1/E mediated by LMO4 is poorly understood. Next, we investigated the mechanisms by which HBXIP upregulates LMO4 expression. Notably, we observed the nucleus localization of HBXIP in MCF-7 cells, implying that HBXIP may exert nuclear functions. Endogenous HBXIP contains the functional domain of leucine zipper, implying its involvements in gene transcription regulations (38,39). Accordingly, we tried to identify whether HBXIP is able to interact with specific transcriptional factors involved in the transcription of LMO4 gene. Of note, we observed the occupancy of HBXIP in the -237/-206 region of LMO4 promoter by EMSA. Furthermore, we predicted two putative binding sites for transcription factor Sp1 and T3R within this promoter region. And further investigations identified that Sp1 was involved in the interaction between HBXIP and LMO4 promoter. Sp1, the prototype of a family of zinc finger (Cys2/Hys2) DNA binding transcription factors, binds to GC-rich sequences such as GC box (40). Sp1 is an ubiquitous transcription factor that activates a broad and diverse spectrum of mammalian genes (41). Moreover, Sp1 plays a critical role in the growth and metastasis of many tumor types including breast cancer by regulating several genes associated with cell growth. Many studies characterized the interactions of Sp1 with several nuclear cofactors including TBP, p53, HDACs, Rb, NF-YA, c-jun, myc, BRCA1 and other proteins (42). In this study, we first report that HBXIP is able to interact with Sp1. We found that the transcription factor Sp1 is required for the binding of HBXIP to the LMO4 promoter and involves the transcriptional activation of LMO4 gene mediated by HBXIP. More importantly, our finding suggests that HBXIP may serve as a transactivator to play crucial roles in the development of breast cancer. It is reported that LMO4 plays critical roles in the pathogenesis of breast cancer through the regulation of cell proliferation. We found that HBXIP promotes the proliferation of breast cancer cells through LMO4 in vitro and in vivo. Our previous report revealed that HBXIP accelerated cell proliferation by modulating cell cycle-related proteins (8). Our findings support the involvement of HBXIP in regulation of cell cycle by targeting cyclin D1 and cyclin E. Our observations imply that the abnormality of HBXIP expression is oncogenic and may facilitate carcinogenesis and tumor progression owing to its contribution to the acceleration of cell cycle process in breast cancer.
Collectively, we report that the oncoprotein HBXIP promotes breast tumor progression through upregulating LMO4 involving the transcription factor Sp1. Our data suggest that HBXIP may serve as a driver gene in the development of cancer. This finding provides new insight into the mechanism of HBXIP in promotion of breast cancer cell proliferation. Therapeutically, HBXIP may act as a new target for breast cancer.
Supplementary material
Supplementary Table 1 and Figures 1 and 2 can be found at http://carcin.oxfordjournals.org/
Funding
National Basic Research Program of China (973 Program: 2011CB512113 and 2009CB521702); Natural Scientific Foundation of China (81071623, 81071624 and 81000870).
Conflict of Interest Statement: These authors declare no conflict of interest.
Supplementary Material
Glossary
Abbreviations:
- ChIP
chromatin immunoprecipitation
- EdU
5-ethynyl-2′-deoxyuridine
- EMSA
electrophoretic mobility shift assay
- HBXIP
hepatitis B X-interacting protein
- LMO4
LIM-only protein 4;
- mRNA
messenger RNA
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PI
proliferation index
- siRNAs
small interfering RNAs.
References
- 1. Melegari M, et al. (1998). Cloning and characterization of a novel hepatitis B virus x binding protein that inhibits viral replication. J. Virol., 72, 1737–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Adams R.R, et al. (2001). Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol., 11, 49–54 [DOI] [PubMed] [Google Scholar]
- 3. Zangemeister-Wittke U, et al. (2004). An IAP in action: the multiple roles of survivin in differentiation, immunity and malignancy. Cell Cycle, 3, 1121–1123 [PubMed] [Google Scholar]
- 4. Minczuk M, et al. (2005). Human ATP-dependent RNA/DNA helicase hSuv3p interacts with the cofactor of survivin HBXIP. FEBS J., 272, 5008–5019 [DOI] [PubMed] [Google Scholar]
- 5. Fujii R, et al. (2006). HBXIP, cellular target of hepatitis B virus oncoprotein, is a regulator of centrosome dynamics and cytokinesis. Cancer Res., 66, 9099–9107 [DOI] [PubMed] [Google Scholar]
- 6. Wen Y, et al. (2008). Interaction of hepatitis B viral oncoprotein with cellular target HBXIP dysregulates centrosome dynamics and mitotic spindle formation. J. Biol. Chem., 283, 2793–2803 [DOI] [PubMed] [Google Scholar]
- 7. Wang F.Z, et al. (2008). Promotion of cell proliferation by HBXIP via upregulation of human telomerase reverse transcriptase in human mesenchymal stem cells. Acta Pharmacol. Sin., 29, 83–89 [DOI] [PubMed] [Google Scholar]
- 8. Wang F.Z, et al. (2007). Involvement of hepatitis B X-interacting protein (HBXIP) in proliferation regulation of cells. Acta Pharmacol. Sin., 28, 431–438 [DOI] [PubMed] [Google Scholar]
- 9. Liu S, et al. (2012). The oncoprotein HBXIP uses two pathways to up- regulate S100A4 in promotion of growth and migration of breast cancer cells. J. Biol. Chem.,; 287, 30228–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yu J, et al. (2008). LIM only 4 is overexpressed in late stage pancreas cancer. Mol. Cancer., 7, 93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sum E.Y, et al. (2005). Overexpression of LMO4 induces mammary hyperplasia, promotes cell invasion, and is a predictor of poor outcome in breast cancer. Proc. Natl. Acad. Sci. U.S.A., 102, 7659–7664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Singh R.R, et al. (2005). Negative regulation of estrogen receptor alpha transactivation functions by LIM domain only 4 protein. Cancer Res., 65, 10594–10601 [DOI] [PubMed] [Google Scholar]
- 13. Kashani A.H, et al. (2006). Calcium activation of the LMO4 transcription complex and its role in the patterning of thalamocortical connections. J. Neurosci., 26, 8398–8408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Murphy N.C, et al. (2008). Expression of LMO4 and outcome in pancreatic ductal adenocarcinoma. Br. J. Cancer., 98, 537–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mousses S, et al. (2002). Clinical validation of candidate genes associated with prostate cancer progression in the CWR22 model system using tissue microarrays. Cancer Res., 62, 1256–1260 [PubMed] [Google Scholar]
- 16. Wang N, et al. (2007). The LIM-only factor LMO4 regulates expression of the BMP7 gene through an HDAC2-dependent mechanism, and controls cell proliferation and apoptosis of mammary epithelial cells. Oncogene., 26, 6431–6441 [DOI] [PubMed] [Google Scholar]
- 17. Tian Y, et al. (2010). Repression of Lim only protein 4-activated transcription inhibits proliferation and induces apoptosis of normal mammary epithelial cells and breast cancer cells. Clin. Exp. Metastasis., 27, 455–463 [DOI] [PubMed] [Google Scholar]
- 18. Montañez-Wiscovich M.E, et al. (2010). Aberrant expression of LMO4 induces centrosome amplification and mitotic spindle abnormalities in breast cancer cells. J. Pathol., 222, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Resnitzky D, et al. (1994). Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. Mol. Cell. Biol., 14, 1669–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee R.J, et al. (2000). Cyclin D1 is required for transformation by activated Neu and is induced through an E2F-dependent signaling pathway. Mol. Cell. Biol., 20, 672–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Montañez-Wiscovich M.E, et al. (2009). LMO4 is an essential mediator of ErbB2/HER2/Neu-induced breast cancer cell cycle progression. Oncogene., 28, 3608–3618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ye L.H, et al. (2006). Screening of a sub-clone of human breast cancer cells with high metastasis potential. Zhonghua Yi Xue Za Zhi., 86, 61–65 [PubMed] [Google Scholar]
- 23. Hu N, et al. (2011). miR-520b regulates migration of breast cancer cells by targeting hepatitis B X-interacting protein and interleukin-8. J. Biol. Chem., 286, 13714–13722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lu Z, et al. (2006). LMO4 can interact with Smad proteins and modulate transforming growth factor-beta signaling in epithelial cells. Oncogene., 25, 2920–2930 [DOI] [PubMed] [Google Scholar]
- 25. Lu S, et al. (2010). Sp1 coordinately regulates de novo lipogenesis and proliferation in cancer cells. Int. J. Cancer., 126, 416–425 [DOI] [PubMed] [Google Scholar]
- 26. Liu B.Y, et al. (2012). Mammary tumor regression elicited by Wnt signaling inhibitor requires IGFBP5. Cancer Res., 72, 1568–1578 [DOI] [PubMed] [Google Scholar]
- 27. Wittlin S, et al. (2003). Two promoters within the human LMO4 gene contribute to its overexpression in breast cancer cells. Genomics., 82, 280–287 [DOI] [PubMed] [Google Scholar]
- 28. Shan C, et al. (2010). Hepatitis B virus X protein promotes liver cell proliferation via a positive cascade loop involving arachidonic acid metabolism and p-ERK1/2. Cell Res., 20, 563–575 [DOI] [PubMed] [Google Scholar]
- 29. Wang W, et al. (2012). MiR-138 induces cell cycle arrest by targeting cyclin D3 in hepatocellular carcinoma. Carcinogenesis., 33, 1113–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dif N, et al. (2006). Insulin activates human sterol-regulatory-element-binding protein-1c (SREBP-1c) promoter through SRE motifs. Biochem. J., 400, 179–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Furuta E, et al. (2008). Fatty acid synthase gene is up-regulated by hypoxia via activation of Akt and sterol regulatory element binding protein-1. Cancer Res., 68, 1003–1011 [DOI] [PubMed] [Google Scholar]
- 32. Jung J.K, et al. (2007). Expression of DNA methyltransferase 1 is activated by hepatitis B virus X protein via a regulatory circuit involving the p16INK4a-cyclin D1-CDK 4/6-pRb-E2F1 pathway. Cancer Res., 67, 5771–5778 [DOI] [PubMed] [Google Scholar]
- 33. Kong G, et al. (2011). Upregulated microRNA-29a by hepatitis B virus X protein enhances hepatoma cell migration by targeting PTEN in cell culture model. PLoS ONE., 6, e19518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Deeb D, et al. (2011). Synthetic triterpenoid CDDO prevents the progression and metastasis of prostate cancer in TRAMP mice by inhibiting survival signaling. Carcinogenesis., 32, 757–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen L, et al. (2009). Cross-regulation of the Nanog and Cdx2 promoters. Cell Res., 19, 1052–1061 [DOI] [PubMed] [Google Scholar]
- 36. Wang Y, et al. (2009). LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell., 138, 660–672 [DOI] [PubMed] [Google Scholar]
- 37. Zhang X, et al. (2005). Hepatitis B virus X protein upregulates survivin expression in hepatoma tissues. J. Med. Virol., 77, 374–381 [DOI] [PubMed] [Google Scholar]
- 38. Garcia-Saez I, et al. (2011). Structural characterization of HBXIP: the protein that interacts with the anti-apoptotic protein survivin and the oncogenic viral protein HBx. J. Mol. Biol., 405, 331–340 [DOI] [PubMed] [Google Scholar]
- 39. Marusawa H, et al. (2003). HBXIP functions as a cofactor of survivin in apoptosis suppression. EMBO J., 22, 2729–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ben-Shachar D, et al. (2007). Sp1 expression is disrupted in schizophrenia; a possible mechanism for the abnormal expression of mitochondrial complex I genes, NDUFV1 and NDUFV2. PLoS ONE., 2, e817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li L, et al. (2010). The role of Sp1 and Sp3 in normal and cancer cell biology. Ann. Anat., 192, 275–283 [DOI] [PubMed] [Google Scholar]
- 42. Safe S, et al. (2005). Sp transcription factor family and its role in cancer. Eur. J. Cancer., 41, 2438–2448 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.