Abstract
The deregulation of paxillin (PXN) has been involved in the progression and metastasis of different malignancies including colorectal cancer (CRC). miR-137 is frequently suppressed in CRC. PXN is predicted to be a direct target of miR-137 in CRC cells. On this basis, we hypothesized that overexpression of PXN induced by suppression of miR-137 may promote tumor progression and metastasis and predicts poor prognosis. We detected the expression of PXN and miR-137 in clinical tumor tissues by immunohistochemical analysis and real-time PCR, positive PXN staining was observed in 198 of the 247 (80.1%) cases, whereas no or weak PXN staining was observed in the adjacent non-cancerous area. Higher level of PXN messenger RNA (mRNA) and lower level of miR-137 was observed in cancer tissues than adjacent non-cancerous tissues. High expression of PXN and low expression of miR-137 was associated with aggressive tumor phenotype and adverse prognosis. Moreover, the expression of PXN was negatively correlated with miR-137 expression. A dual-luciferase reporter gene assay validated that PXN was a direct target of miR-137. The use of miR-137 mimics or inhibitor could decrease or increase PXN mRNA and protein levels in CRC cell lines. Knockdown of PXN or ectopic expression of miR-137 could markedly inhibit cell proliferation, migration and invasion in vitro and repress tumor growth and metastasis in vivo. Taken together, these results demonstrated that overexpression of PXN induced by suppression of miR-137 promotes tumor progression and metastasis and could serve as an independent prognostic indicator in CRC patients.
Introduction
Colorectal cancer (CRC) is an important public health problem. It ranks as the third most common malignancy and as the second leading cause of cancer-related deaths in Western countries (1). The pathological process and molecular mechanism of CRC is complicated and is not yet fully understood. Previous studies have indicated that genetic alternations and changes in the transduction pathways are involved in CRC initiation and progression. These include alterations in oncogenes, tumor suppresser genes (KRAS, APC, DCC, Smad-2, Smad-4 and P53), transforming growth factor-β and the WNT signaling pathways (2). However, the precise molecular biomarkers that can predict prognosis and recurrence have not yet been determined.
The cytoskeleton is important for abnormal cell growth, invasion and metastasis, which are frequently observed in malignant tumors (3,4). The paxillin (PXN) gene encodes for a focal adhesion molecule of 68kDa, which is one of the key focal adhesion proteins that contribute to form a structural link between the extracellular matrix and actin cytoskeleton (5). PXN is a multidomain adaptor protein that integrates multiple signals from cell surface receptors, integrins and growth factors (5). Through these protein–protein interactions, PXN functions as a regulator of a variety of physiological process, including cell motility, metastasis, gene expression, matrix organization, tissue remolding, cell proliferation and survival (5–7). Previously, PXN has been found to be upregulated and act as an oncogene in various malignancies, such as non-small cell lung cancer (8), prostate cancer (9) and esophageal squamous cell carcinoma (10). In CRC, Ayaki et al. (11) reported that PXN expression levels were higher in CRC tissues than in paired normal tissues. More recently, Jun et al. (12) demonstrated that overexpression of PXN could stimulate migration, invasion and adhesion abilities of CRC cells, whereas downregulation of PXN by small interfering RNA (siRNA) inhibited these capacities. However, the underlying molecular mechanism and clinicopathological implication of PXN in CRC is not yet known.
MicroRNAs (miRNAs) are a class of small non-coding RNAs (approximately 18–22 nucleotides in length), which function as regulators of the expressions of a wide variety of genes (13–15). By complementary binding to the 3′-untranslated region (3′UTR) of messenger RNA (mRNA), miRNAs enhance their cleavage or inhibit their posttranscriptional translation (16). Evidence is increasing that miRNAs play important roles during the biological processes of carcinogenesis and tumor progression through their regulation of cancer cell proliferation, differentiation, apoptosis and invasion (17–19). miR-137 has been reported to be downregulated in several cancer types, such as melanoma (20), head and neck carcinoma (21) and breast cancer (22). In CRC, epigenetic silencing of miR-137 was found to be a contributor and an early event in tumorigenesis (23).
In this study, we first determined the expression level of PXN and miR-137 in tumor tissues surgically resected from patients with CRC by immunohistochemical analysis and RT-qPCR, and the clinicopathological significance of PXN and miR-137 was evaluated. The regulative relationship of PXN by miR-137 was confirmed in CRC cell lines. We further examined the cell proliferation, migration and invasion abilities in vitro and in vivo to verify whether overexpression of PXN induced by downregulation of miR-137 could stimulate tumor growth and metastasis and thus predict prognosis.
Materials and methods
Human tissue specimens and cell lines
Paraffin-embedded, archived tissue samples were collected from 247 CRC patients who underwent surgery in Sun Yat-sen University Cancer Center (Guangzhou, China) between 2000 and 2004. Fresh CRC tissues and matched adjacent non-cancerous tissues were obtained from 102 of the 247 CRC patients and stored in liquid nitrogen until use. All the patients had a histological diagnosis of CRC. A written informed consent was obtained from all the patients involved in this study and approval was obtained from the ethics committee of Sun Yat-sen University Cancer Center. The stages of all collected specimens were determined according to the World Health Organization’s classification system on TNM staging. All the patients were followed-up, postoperation, at 3 month intervals. The median follow-up period was 51 months (range 3–136 months). All the clinicopathological data including age, gender, tumor size, tumor depth, differentiation status, lymph node invasion, venous invasion, peritoneal dissemination, liver metastasis and TNM stage were obtained from patients’ medical records.
Six CRC cell lines (HT-29, HCT116, SW480, SW620, DLD-1 and LoVo), an embryonic kidney cell line 293Ta and a normal colonic cell line CCD-112-CoN were obtained from the American Type Culture Collection; cells were cultured and stored according to the instructions of American Type Culture Collection. Cells were routinely authenticated every 6 months (last examined in February 2012) by growth curve analysis, cell morphology monitoring and testing for mycoplasma (R&D Systems’ new MycoProbe Mycoplasma Detection Kit).
Immunohistochemistry analysis
The paraffin-embedded tissue blocks were cut into 4 μm slides. A PXN rabbit antibody (CST#2542) was used. Immunohistochemistry (IHC) analysis of PXN was performed according to a previously described method (24). To quantify PXN protein expression, both the extent and intensity of immunoreactivity were assessed and scored. In this study, the scores of the extent of immunoreactivity ranged from 0 to 3 and were according to the percentage of cells that had positive staining in each microscopic field of view (0, <25%; 1, 25–50%; 2, 50–75%; 3, 75–100%). The scores of IHC intensity were as follows: 0, negative staining; 1, weak staining; 2, moderate staining; 3, strong staining. By multiplying the scores for extent and intensity, a total score ranging from 0 to 9 was achieved. The expression level of PXN was considered high when the total scores were ≥4 and low when the total scores were <4.
RNA isolation and real-time quantitative RT-PCR analysis of miR-137 and PXN mRNA expression levels
For detection of PXN mRNA, total RNA was extracted from the tissues and cells with Trizol reagent (Invitrogen). For measurement of miR-137, RNA was extracted using mirVana miRNA isolation kit (Applied Biosystems/Ambion, Austin, TX) according to the manufacturer’s instructions. RNA concentration and quality were measured with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). miR-137 expression was detected with a Bulge-Loop™ miRNA qRT-PCR Primer Set (Ribobio, Guangzhou, China) according to the manufacturer’s instructions; U6 snRNA was used as the endogenous control. PXN mRNA expression was measured using a SYBR Premix Ex Taq™ kit (Takara); β-actin expression was used as a reference. The primers used are listed in Supplementary Table I, available at Carcinogenesis Online. Real-time PCR was carried out with an ABI PRISM® 7500 Sequence Detection System. Data were analyzed using the 2−△△ct method.
Vector constructs, transfections and luciferase activity assay
A miR-137 expression plasmid (pcDNAmiR-137) was constructed with synthetic oligonucleotides and the pcDNA6.2-GW/EmGFP vector. A PXN expression plasmid (pcDNA3.1PXN) containing the coding sequence but lacking the 3′UTR was constructed using PCR-generated fragments and pcDNA3.1 (+) vector. The 3′UTR of PXN mRNA and a mutant variant were generated by q-PCR and cloned to the XbaI site of a pGL3-basic vector (Promega) and termed PXN-wt-3′UTR and PXN-mt-3′UTR, respectively.
For the luciferase assay, HCT116 and SW620 cells were seeded in 6-well plates the day before transfection and were infected with the pGL3 reporter vector (250ng/well), pRL-TK luciferase reporters (25ng/well) and pcDNAmiR-137(750ng/well) or a negative-control (NC) vector using lipofectamine 2000 (Invitrogen). Luciferase activities were measured with a Dual-Luciferase Reporter Assay Kit (Promega) according to the manufacturer’s instructions.
Western blot analysis
For immunoblotting of PXN and α-tubulin, a rabbit PXN antibody (CST#2542) was purchased from Cell Signaling Technology and a mouse α-tubulin antibody was obtained from Santa Cruz Biotechnology. The procedures were as described previously (24).
Oligonucleotide transfection
The hsa-miR-137 mimic and NC oligonucleotides (#miR10000843-1-5), has-miR-137 inhibitor and NC oligonucleotides (#miR20000429-1-5) were obtained from Ribobio. Ribobio also synthesized three siRNA duplex oligonucleotides targeting human PXN mRNA (PXNsi). The targeting sequences were PXNsi-1: GCAGCAACCTTTCTGAACT; PXNsi-2: GTGTGGAGCCTTCTTTGGT; PXNsi-3: GCTGGAACTGAACGCTGTA. In the present study, we used PXNsi-1 because it could effectively inhibited endogenous PXN expression in our preliminary experiments. Cells were plated in a 6-well plate the day before transfection. For PCR, western blot and other functional assays, HCT116 and SW620 were transfected with has-miR-137 mimic (50 nmol/l) or PXNsi (50 nmol/l). HT-29 cells were infected with has-miR-137 inhibitor (100 and 150 nmol/l) or NC with lipofectamine 2000 (Invitrogen).
Cell proliferation assays
A 3-(4,5-dimethylthiazole-2-yl)-2,5-biphenyl tetrazolium bromide (MTT) assay was performed to test cell viability. The spectrophotometric absorbance at 490nm was measured for each sample; all the experiments were repeated three times in triplicate and the mean was calculated.
For the colony formation assay, 24h after infection, 500 cells were placed in a 6-well plate and cultured for 14 days with McCoy’s 5A medium containing 10% fetal bovine serum. Colonies were fixed with methanol and stained with 0.1% crystal violet (1mg/ml).
In vitro invasion and migration assay
The cell invasive and migratory potential was evaluated using transwell chambers (8 μm pore; BD Biosciences). For the invasion assay, 24h after infection, 1×105 cells suspended in 100 μl serum-free medium were added to the upper chamber of the inserts, which were coated with a mitrigel mix; fetal bovine serum (500 μl) was added to the lower chamber as a chemoattractant. After incubation for 24h, non-invading cells on the upper surface were removed and cells that migrated to the lower side of the membrane were fixed with methanol, stained with 0.1% crystal violet, air dried and photographed. For the migration assay, tumor cells (5×104 cells in 100 μl serum-free medium) were placed in the top chamber of each insert without matrix gel, and 500 μl serum was added to the lower compartment. Sixteen hours later, the cells on the upper side were removed, and the cells that migrated to the lower chamber were fixed and stained with crystal violet. The number of invading or migrating cells was determined by microscopically counting five different fields.
Lentivirus production and transduction
The lentiviral plasmid pEZX-MR03 (GeneCopoeia™) expressing miR-137 and NC miRNA precursor sequences were obtained from GeneCopoeia (termed pLV-miR-137, pLV-miR-NC). Lenti-Pac HIV Expression Packaging mix (GeneCopoeia™), pLV-miR-137 or pLV-miR-NC were cotransfected into the 293Ta cells using EndoFectin Lenti transfection reagent and following the manufacturer’s instructions. Forty-eight hours posttransfection, lentiviral particles in the supernatant were harvested and filtered by centrifugation (500g for 10min). HCT116 cells were then transduced with the lentivirus expressing miR-137 or NC miRNA. For the selection of stably transduced cells, 48h after infection, puromycin (2 μg/ml) was added to the culture medium and the cells were cultured for 14 days. Clonal cell populations were selected and cultured for further studies. Cell lines stably expressing miR-137 and the NC vector were termed HCT116-LV-miR-137 and HCT116-LV-miR-NC cells, respectively.
To generate PXN stable knockdown cells, lentivirus containing PXN short hairpin RNA or non-target oligonucleotides were obtained from GenePharma (Shanghai, China). An annealed short interfering RNA (PXNsi-1) for PXN selected from three different target sequences was inserted into the LV-3 (pGLVH1/GFP+Puro) vector. HCT116 cells were transduced with lentivirus and stable cell lines were selected according to the manufacturer’s instructions. Clonal cell populations were selected and cultured for in vivo studies. The cell lines stably expressing PXN short hairpin RNA and non-target oligonucleotides were named HCT116-sh-PXN and HCT116-sh-NT, respectively.
In vivo proliferation and metastasis assays
Male BABL/c athymic nude mice (4 weeks old) were obtained from the Animal Center of Guangdong province (Guangzhou, China). All the animal experiments were conducted according to the National Institutes of Health animal use guidelines on the use of experimental animals.
To evaluate the in vivo proliferative effect of PXN and miR-137, the HCT116-sh-PXN, HCT116-sh-NT, HCT116-LV-miR-137 and HCT116-LV-miR-NC cells (1×106 cells/mouse) were injected subcutaneously into the dorsal flanks of four groups of nude mice (five for each cell group). Tumor size was measured weekly and tumor volume was estimated. After 6 weeks, the mice were killed and the tumors were removed and weighted. To investigate the effect of PXN and miR-137 on metastasis, the HCT116-sh-PXN, HCT116-sh-NT, HCT116-LV-miR-137 and HCT116-LV-miR-NC cells (2×106 cells/mouse) were injected into the tail vein of nude mice (eight for each cell group). Six weeks postinjection, the mice were killed and the lungs and livers were removed and embedded in paraffin. Consecutive sections (4 μm) were made and stained with hematoxylin–eosin. The micrometastases in the lungs and livers were examined and counted under a dissecting microscope as described previously (25).
Statistical analysis
Statistical analysis was performed using the SPSS software package (version 13.0, SPSS). Statistical significance was tested by a Student’s t-test or a chi-square test as appropriate. Survival analysis was performed using the Kaplan–Meier method and log-rank test. Parameters with a P value <0.05 by univariate analysis were included in multivariate analysis. All differences were statistically significant with a P value <0.05.
Results
Overexpression of PXN is correlated with downregulation of miR-137 in CRC tissues
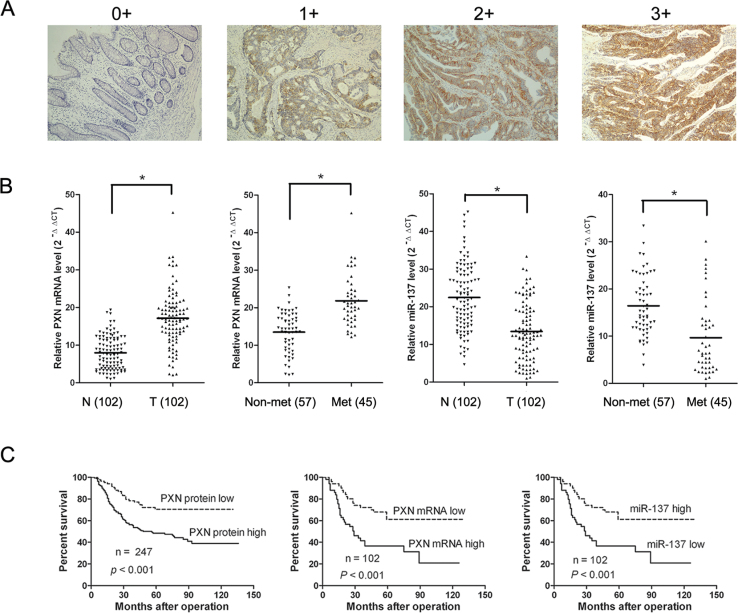
PXN protein was evaluated in 247 paraffin-embedded, archived CRC tissues by IHC. PXN protein was detected in 198 of 247 (80.1%) tumor samples, whereas there was no or weak staining in the adjacent non-cancerous areas in all sections detected. Representative PXN protein expression in CRC tissues is shown in Figure 1A, which shows positive PXN immunoreactivity is mainly located in the cytoplasm of tumor cells. To better understand whether expression of PXN is associated with miR-137, the level of PXN mRNA and miR-137 was detected in 102 paired tumor tissues and adjacent non-cancerous tissues (taken from 102 of the 247 patients) by real-time PCR. PXN mRNA and miR-137 levels were normalized to β-actin and U6, respectively (Supplementary Figure S1, available at Carcinogenesis Online). PXN mRNA level was significantly higher in tumor tissues (mean ± SD, 17.14±7.31) than in non-cancerous tissues (mean ± SD, 7.97±4.40) (Figure 1B). On the contrary, miR-137 expression level was significantly lower in tumor tissues (mean ± SD, 13.44±7.74) than in non-cancerous tissues (mean ± SD, 22.43±8.81) (Figure 1B). In addition, when the patients were divided into two groups based on the presence or absence of tumor metastasis, we found that PXN mRNA levels in patients with metastasis (mean ± SD, 21.83±6.77) were higher than those in patients without metastasis (mean ± SD, 13.50±5.45), whereas miR-137 levels were lower in patients with metastasis (mean ± SD, 9.66±7.76) than those in patients without metastasis (mean ± SD, 16.42±6.33) (Figure 1B). Chi-square test was performed to analyze the association between the expression of PXN and miR-137, which showed that the expression of PXN protein and mRNA in CRC patients was negatively associated with miR-137 expression (PXN protein, P = 0.009; PXN mRNA, P = 0.029, Table I).
Fig. 1.
PXN and miR-137 expression levels in CRC tissues and their prognostic significance. (A) Immunoreactive intensity of PXN in CRC was translated into a four-tier system as negative (score = 0), weak staining (score = 1), moderate staining (score = 2) and strong staining (score = 3). (B) Relative PXN mRNA and miR-137 expression levels in CRC tissue samples. Relative PXN mRNA and miR-137 expression in N (adjacent non-tumorous tissues), T (tumor tissues), Non-met (non-metastatic tumor tissues) and Met (metastatic tumor tissues) was determined by real-time PCR. The line in the middle indicates the mean value. (C) Kaplan–Meier analysis of overall survival based on PXN protein, PXN mRNA and miR-137 levels.
Table I.
The correlation between clinicopathological parameters and miR-137 and PXN expression, and the correlation between miR-137 and PXN expression levels
| Factor | No. | PXN protein | P | No. | miR-137 | P | ||
|---|---|---|---|---|---|---|---|---|
| Low, n (%) | High, n (%) | Low, n (%) | High, n (%) | |||||
| Age | ||||||||
| <60 | 145 | 55 (64.7) | 90 (55.5) | 0.165 | 55 | 26 (50.9) | 29 (56.8) | 0.551 |
| ≥60 | 102 | 30 (35.3) | 72 (45.5) | 47 | 25 (49.1) | 22 (43.2) | ||
| Gender | ||||||||
| Male | 160 | 51 (60.0) | 109 (67.3) | 0.255 | 59 | 26 (50.9) | 33 (64.7) | 0.160 |
| Female | 87 | 34 (40.0) | 53 (42.7) | 43 | 25 (49.1) | 18 (35.3) | ||
| Tumor size | ||||||||
| <5 cm | 81 | 36 (42.3) | 45 (27.8) | 0.020* | 34 | 10 (19.6) | 24 (47.1) | 0.003* |
| ≥5 cm | 166 | 49 (57.7) | 117 (72.2) | 68 | 41 (80.4) | 27 (52.9) | ||
| Tumor depth | ||||||||
| m/sm/mp | 101 | 40 (47.0) | 61 (37.7) | 0.153 | 46 | 16 (31.4) | 30 (58.8) | 0.005* |
| ss/se/si | 146 | 45 (53.0) | 101 (62.3) | 56 | 35 (68.6) | 21 (41.2) | ||
| Differentiation status | ||||||||
| Well | 55 | 25 (29.4) | 30 (18.5) | 0.023* | 22 | 7 (13.7) | 15 (29.4) | 0.139 |
| Moderate | 67 | 27 (31.7) | 40 (24.6) | 29 | 17 (33.3) | 12 (23.5) | ||
| Poor and others | 125 | 33 (38.8) | 92 (56.9) | 51 | 27 (53.0) | 24 (47.1) | ||
| Lymph node invasion | ||||||||
| Absent | 77 | 37 (43.5) | 40 (24.1) | 0.002* | 31 | 8 (15.7) | 23 (45.1) | 0.001* |
| Present | 170 | 48 (56.5) | 122 (75.9) | 71 | 43 (84.3) | 28 (54.9) | ||
| Venous invasion | ||||||||
| Absent | 166 | 60 (70.6) | 106 (65.4) | 0.412 | 76 | 35 (68.6) | 41 (80.3) | 0.173 |
| Present | 81 | 25 (29.4) | 56 (34.6) | 26 | 16 (31.4) | 10 (19.7) | ||
| Peritoneal dissemination | ||||||||
| Absent | 215 | 77 (90.6) | 138 (85.2) | 0.230 | 90 | 44 (86.2) | 46 (90.1) | 0.539 |
| Present | 32 | 8 (9.4) | 24 (14.8) | 12 | 7 (13.8) | 5 (9.9) | ||
| Liver metastasis | ||||||||
| Absent | 202 | 75 (88.2) | 127 (78.3) | 0.057 | 78 | 35 (68.6) | 43 (84.3) | 0.062 |
| Present | 45 | 10 (11.8) | 35 (11.7) | 24 | 16 (31.4) | 8 (15.7) | ||
| TNM stage | ||||||||
| I–II | 80 | 39 (45.8) | 41 (25.3) | 0.001* | 34 | 11 (21.6) | 23 (45.1) | 0.012* |
| III–IV | 167 | 46 (44.2) | 121 (74.7) | 68 | 40 (78.4) | 28 (54.9) | ||
| PXN mRNA | ||||||||
| Low | 51 | 24 (80.0) | 27 (37.5) | <0.001* | 51 | 20 (39.2) | 31 (60.8) | 0.029* |
| High | 51 | 6 (20.0) | 45 (62.5) | 51 | 31 (60.8) | 20 (39.2) | ||
| miR-137 | ||||||||
| Low | 51 | 12 (40.0) | 39 (54.2) | 0.009* | ||||
| High | 51 | 18 (60.0) | 33 (45.8) | |||||
m: tumor invasion of mucosa; sm: submocosa; mp: muscularis propria; ss: subserosa; se: serosa penetration; si: invasion to adjacent structures.
*P < 0.05 (chi-square test).
PXN and miR-137 are associated with clinicopathological factors and prognosis in CRC
We hypothesized that overexpression of PXN by suppression of miR-137 might contribute to tumor progression and metastasis and thus lead to poor prognosis. To analyze the clinicopathological significance of PXN, the patients were divided into a PXN high expression group (n = 162) and a PXN low expression group (n = 85) based on IHC scores. The correlations between clinicopathological factors and PXN expression were analyzed (Table I). High PXN expression was positively associated with larger tumor size (P = 0.020), adverse differentiation status (P = 0.023), more extensive lymph node invasion (P = 0.002) and higher TNM stage (P = 0.001). Kaplan–Meier analysis and log-rank test was used to evaluate the prognostic value of PXN for CRC patients. A significant difference of overall survival was found between patients with high PXN expression and patients with low PXN expression. Kaplan–Meier survival curves showed that high PXN expression was associated with poor overall survival (P < 0.001, Figure 1C). Univariate analysis demonstrated patients with high PXN expression tended to have a higher risk of death (hazard ratio = 4.91, 95% CI, 2.82–8.55; P < 0.001, Table II). To assess the clinical significance of miR-137, patients were divided into two groups, the miR-137 high-level group (n = 51) and the miR-137 low-level group (n = 51) according to the median value of miR-137 expression. As expected, high miR-137 level was negatively correlated with tumor size (P = 0.003), tumor depth (P = 0.005), lymph node invasion (P = 0.001) and TNM stage (P = 0.012) (Table I). Kaplan–Meier survival curve showed patients with high miR-137 level tended to have more favorable prognosis (P < 0.001, Figure 1C). In addition, tumor size, lymph node invasion and TNM stage proved to be associated with overall survival as indicated by univariate analysis (Table II). However, age, gender, differentiation status and tumor depth had no prognostic significance in this studied population. Multivariate analysis showed that PXN expression and TNM stage were independent prognostic factors for CRC patients (P = 0.001 and 0.033, respectively, Table II).
Table II.
Univariate and multivariate analyses of various potential prognostic factors in CRC patients
| Factors | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| Case no. | HR (95% CI) | P | HR (95% CI) | P | |
| Age (<60/≥60) | 102/145 | 0.86 (0.57–1.30) | 0.496 | — | — |
| Gender (male/female) | 160/87 | 0.71 (0.48–1.04) | 0.083 | — | — |
| Differentiation (well, moderate/poor) | 122/125 | 1.43 (0.93–2.21) | 0.099 | — | — |
| Tumor depth (m, sm, mp/ss, se, si) | 101/146 | 1.64 (1.03–2.59) | 0.084 | — | — |
| Tumor size (≥5cm/<5cm) | 166/81 | 1.74 (1.05–2.88) | 0.039* | 1.53 (0.77–3.04) | 0.219 |
| Lymph node invasion (present/absent) | 170/77 | 1.92 (1.51–2.43) | 0.008* | 1.26 (0.78–2.03) | 0.337 |
| TNM stage (I–II/III–IV) | 80/167 | 1.78 (1.16–2.74) | 0.001* | 1.55 (0.88–2.75) | 0.033* |
| miR-137 (high/low) | 51/51 | 0.67 (0.35–0.89) | 0.002* | 0.70 (0.51–0.97) | 0.126 |
| PXN protein (high/low) | 162/85 | 4.91 (2.82–8.55) | <0.001* | 4.81 (1.95–11.88) | 0.001* |
HR: hazard ratio; CI: confidence interval; m: tumor invasion of mucosa; sm: submocosa; mp: muscularis propria; ss: subserosa; se: serosa penetration; si: invasion to adjacent structures.
*P < 0.05.
PXN is modulated by miR-137 in CRC cell lines
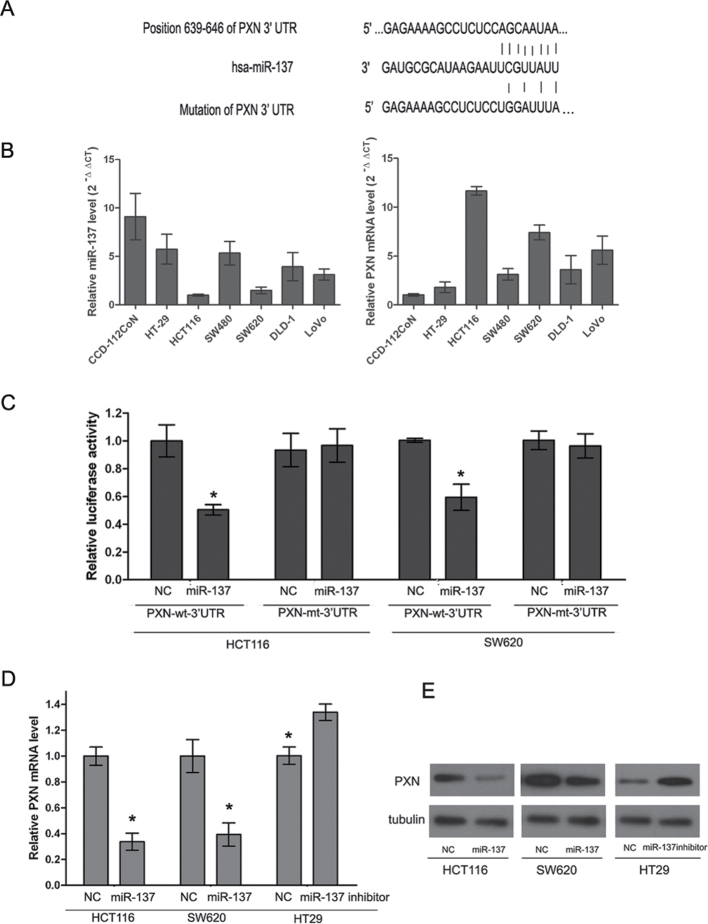
It is generally acknowledged that miRNAs exert their effects by regulation of target genes. In this study, the putative miRNAs that might target PXN were predicted by using bioinformatic algorithms, such as TargetScan and miRanda (26–28). The results showed that PXN possesses potential miR-137 binding sites in the 3′UTR (Figure 2A). To explore whether the expression of PXN is negatively associated with miR-137 in CRC cell lines, six CRC cell lines and a normal colonic cell line were used to measure the PXN and miR-137 expression by RT-qPCR. The expression level of PXN mRNA in all the cell lines was inversely associated with miR-137 expression (Figure 2B). To verify a direct binding and repression effect, a luciferase assay was performed. The PXN-wt-3′UTR and its mutant type (PXN-mt-3′UTR) were amplified and cloned downstream of a luciferase reporter gene in the pGL3-basic vector. Cotransfection of HCT116 and SW620 cells with pcDNAmiR-137 and wild-type reporter vector markedly reduced luciferase activity compared with the NC (both P < 0.05, Figure 2C). However, no difference in luciferase activity was observed in cells cotransfected with PXN-mt-3′UTR and pcDNAmiR-137. To further investigate whether miR-137 can regulate endogenous PXN expression, HCT116 and SW620 cells, which have high levels of PXN and low level of miR-137, were transfected with miR-137 mimics, whereas HT-29 cells that possess a low level of PXN and high level of miR-137 were transfected with the miR-137 inhibitor. Based on real-time PCR, we found that the mRNA levels of PXN were dramatically reduced in miR-137 overexpressing HCT116 and SW620 cell lines compared with that of the NC cells (P < 0.05, Figure 2D). Furthermore, the protein levels of PXN were also markedly decreased in the HCT116 and SW620 cell lines after ectopic overexpression of miR-137 as detected by western blotting (Figure 2E). However, both the mRNA and protein levels were significantly increased in the HT-29 cells after infection with miR-137 inhibitor (P < 0.05, Figure 2D and E).
Fig. 2.
PXN is modulated by miR-137 in CRC cell lines. (A) The binding sites of miR-137 in the PXN 3′UTR. (B) Relative PXN mRNA (normalized to β-actin) and miR-137 (normalized to U6) expression levels were detected by real-time reverse transcription PCR in six CRC cell lines and a normal colonic cell line CCD-112-CoN. Data are presented as means ± SD. (C) The pcDNAmiR-137 expression plasmid or the NC, a pGL3 luciferase vector containing the wild or mutant type of PXN 3′UTR, were cotransfected into HCT116 and SW620 cells and relative firefly luciferase activity was measured. Data are presented as means ± SD of three independent experiments (*P < 0.05). (D) mRNA levels of PXN in CRC cell lines treated with miR-137 mimics or miR-137 inhibitor. HCT116 and SW620 cells were infected with miR-137 mimics and HT-29 cells were infected with miR-137 inhibitor. (E) Protein levels of PXN in CRC cell lines treated with miR-137 mimics or miR-137 inhibitor. HCT116 and SW620 cells were infected with miR-137 mimics and HT-29 cells were infected with miR-137 inhibitor.
Overexpression of PXN following miR-137 repression promotes cell proliferation, migration and invasion abilities in vitro
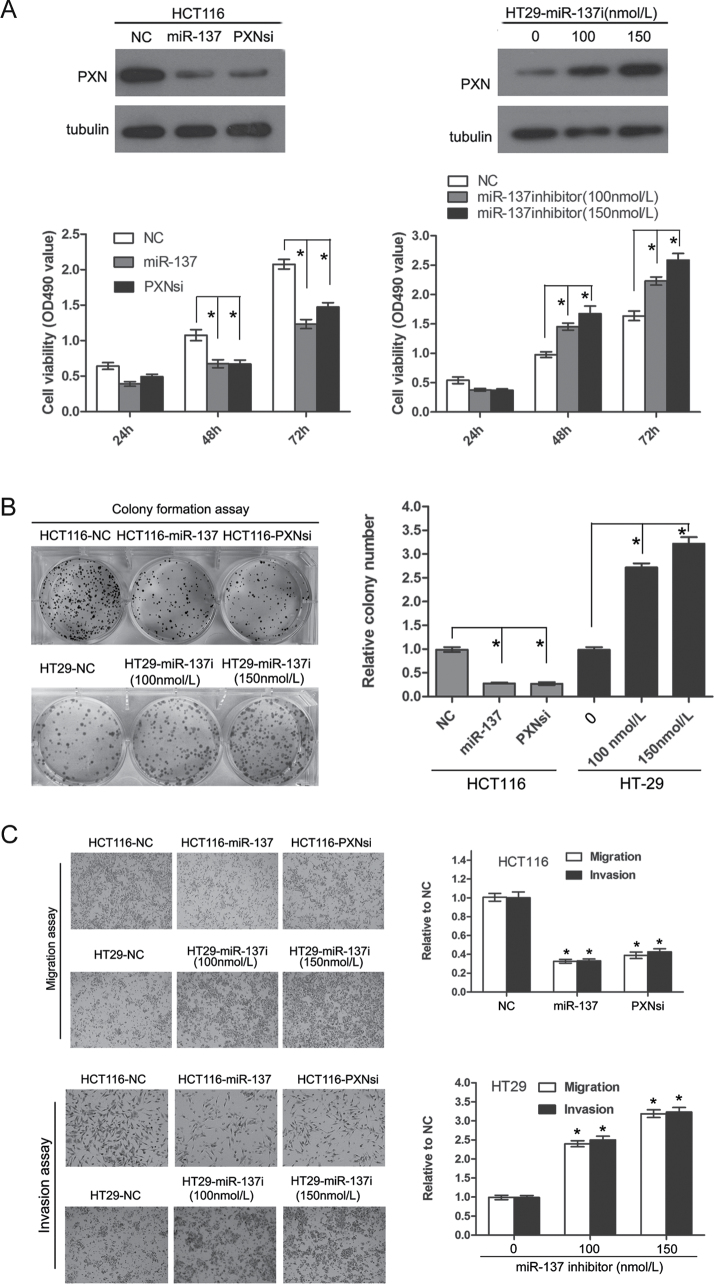
Based on the results listed above, we hypothesized that overexpression of PXN by a reduced miR-137 expression may affect the proliferation, migration and invasion ability of CRC cell lines. The HCT116 cell line, which had high levels of PXN and low levels of miR-137, was transfected with PXN siRNA or miR-137 mimics. Another CRC cell line HT-29 that possessed relative low levels of PXN and high level of miR-137 was treated with the miR-137 inhibitor. The MTT assay showed that knockdown of PXN or ectopic expression of miR-137 could significantly inhibit the proliferative ability in HCT116 cells (P < 0.05, Figure 3A). Similarly, colony formation capacity was repressed after downregulation of PXN or overexpression of miR-137 (P < 0.05, Figure 3B). Conversely, upon transfection of the miR-137 inhibitor, the growth capacity of the HT-29 cells was stimulated as indicated by the MTT and colony formation assays (P < 0.05, Figure 3A and B). Cell motility was measured by the migration and invasion assay. Compared with that of NC cells, the migration and invasion ability were markedly impaired in HCT116 cells, which were transfected with the PXN siRNA and miR-137 mimics (P < 0.05, Figure 3C). Likewise, repression of miR-137 could apparently promote the migration and invasion of HT-29 cells (P < 0.05, Figure 3C). To determine whether overexpression of PXN could reverse the effects of miR-137, HCT116 cells stably expressing miR-137 (HCT116-LV-miR-137), or with a knockdown of PXN (HCT116-sh-PXN), were transfected with a pcDNA3.1PXN plasmid, which contains the coding sequence of PXN, but lacks the 3′UTR. As expected, overexpression of PXN could at least partially rescue the proliferation, migration and invasion abilities repressed by miR-137 (P < 0.05, Supplementary Figure S2, available at Carcinogenesis Online).
Fig. 3.
Upregulation of PXN induced by miR-137 reduction promotes cell proliferation, migration and invasion. (A) Proliferation rate of HCT116 cells with or without miR-137 mimics and PXN knockdown plasmid treatment and of HT-29 cells with or without miR-137 inhibitor treatment on the basis of MTT assay. The change in PXN protein was confirmed by western blotting (upper), and the absorbance at OD490 is shown at the bottom (*P < 0.05). (B) Representative colony formation assay of HCT116 cells with and without miR-137 mimics and PXN knockdown plasmid treatment and of HT-29 cells with or without miR-137 inhibitor treatment (*P < 0.05). (C) Representative transwell migration and invasion assay of HCT116 cells with and without miR-137 mimics and PXN knockdown plasmid treatment and of HT-29 cells with and without miR-137 inhibitor treatment (*P < 0.05, compared with NC).
Knockdown of PXN and (or) ectopic expression of miR-137 inhibits the proliferative and metastatic abilities of CRC cells in vivo
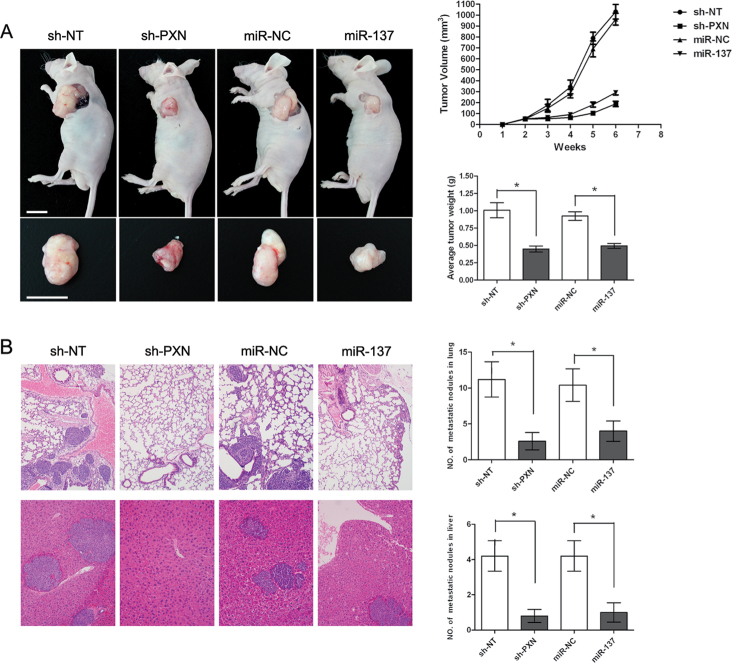
To evaluate the in vivo effects of PXN and miR-137 on CRC cells, we constructed four stable cell lines by using the lentivirus vector to mediate the knockdown of PXN and expression of miR-137; the resulting cells were termed HCT116-sh-PXN, HCT116-sh-NT, HCT116-LV-miR-137 and HCT116-LV-miR-NC. These four cell lines were injected into the left flanks of nude mice. Tumor size was measured over time; at 6 weeks, mice were killed and tumors were dissected out. The results showed that the volume and weight of the tumors from the mice injected with HCT116-sh-PXN cells were significantly less than those of HCT116-sh-NT cells (P < 0.05, Figure 4A). Similarly, the volume and weight of tumors from HCT116-LV-miR-137 were markedly reduced compared with those of HCT116-LV-miR-NC cells (P < 0.05, Figure 4A).
Fig. 4.
Knockdown of PXN or ectopic expression of miR-137 inhibit tumor growth and metastasis of CRC cells in vivo. (A) Xenograft tumor growth in nude mice. The graph shows the representative tumor growth 6 weeks after injection. The tumor sizes were measured every week after injection and tumor volume was estimated. At the end, the mice were killed and tumors were removed and weighted; all data are presented as mean ± SD (*P < 0.05) (scale bar, 1cm). (B) Lung and liver metastases in nude mice. The graph shows the representative hematoxylin- and eosin-stained sections of lung metastasis (upper) and liver metastasis (under) (×100); all the data are shown as means ± SD (*P < 0.05).
To explore the inhibition of in vivo metastasis, the four cell lines were injected into the tail vein of nude mice. Six weeks later, mice were killed and lung and liver metastases were examined. Consistent with the in vitro results, the numbers of micrometastases in the lungs and livers were significantly less in mice injected with HCT116-sh-PXN cells than those of HCT116-sh-NT cells (P < 0.05, Figure 4B). Likewise, the numbers of metastatic nodules in the lungs and livers were apparently reduced in the HCT116-LV-miR-137 group in relation to those in the HCT116-LV-miR-NC group (P < 0.05, Figure 4B).
Discussion
Although abnormal expression of PXN has been observed in CRC (11), this study is the first report on the clinicopathological and prognostic significance of PXN in CRC. Our study revealed that upregulation of PXN is significantly associated with aggressive and invasive phenotype (tumor size, tumor depth, lymph node invasion, tumor stage and distant metastasis) and with adverse prognosis of CRC patients as well. These results show overexpression of PXN might be a common incident in CRC and could serve as an independent prognostic marker to identify patients with poor outcome. Similar to our findings, upregulation of PXN was observed in prostate cancer (29) and non-small cell lung cancer (8). Expression of PXN was reported in correlation with clinicopathological characteristics in lung cancer (30), hepatocellular carcinoma (31) and salivary adenoid cystic carcinoma (32). However, further investigation is needed to confirm whether PXN could be used as an effective biomarker and prognostic indicator for neoplasm.
Many studies have characterized the expression profiles of miRNAs in tumors and have revealed that miRNAs are frequently downregulated in tumor tissues compared with normal tissues (33,34). Furthermore, studies have found that miRNAs could function as oncogenes or tumor suppressors, based on the genes they regulate (35). In regard to CRC, a series of miRNAs have been found to be involved in tumorigenesis, tumor progression and invasion, chemotherapy sensitivity and prognosis; these effects have been reviewed by Jia et al. (36). Downregulation of miR-137 has been frequently observed in cancer cell lines. For example, miR-137 was reported to be epigenetically silenced and that it might act as a tumor suppressor in oral cancer cells (37). Similarly, low expression of miR-137 leads to stimulated cell proliferation and invasion in glioblastoma cell lines (38). However, miR-137 was found to be upregulated in squamous cell carcinoma of the tongue (39). This implies that miR-137 may play different roles depending on the tumor type. In this study, we found that miR-137 levels were significantly lower in CRC cell lines than in normal colonic cells. This result is in line with previous studies (23,40). Furthermore, our study revealed the clinicopathological significance of miR-137 in CRC.
In the present study, using bioinformatic tools, we found 34 putative miRNAs that might target PXN. Their functions in tumors were summarized in Supplementary Table II, available at Carcinogenesis Online. Among these miRNAs, miR-137, miR-142 and miR-30a have been reported to be associated with CRC progression and metastasis. However, only miR-137 was consistently downregulated in CRC tissues and cell lines based on our preliminary experiments. Therefore, we focused on PXN and miR-137 in the present study. A luciferase activity assay confirmed the interaction between miR-137 and PXN. Ectopic expression of miR-137 markedly reduced endogenous PXN mRNA and protein levels. Moreover, in clinical tissue samples and CRC cell lines, an inverse correlation between miR-137 level and PXN expression level was observed. Taken together, these data provided sound evidence that PXN is negatively regulated by miR-137 in CRC cell lines.
PXN is a focal adhesion protein that may enhance the adhesion between tumor cells and the surrounding cells and molecules, thus promote the migration and invasion capacities of tumor cells. Previous reports have indicated that PXN plays a critical role in cell adhesion and invasion (41). The effect of PXN on cell motility and invasion is mainly mediated by tyrosine/serine phosphorylation (42). In breast cancer cells, Chen et al. (43) found that cell migration and invasion stimulated by breast tumor kinase was mediated by phosphorylation of PXN. In colon cancer, PXN phosphorylation at tyrosines 31 and 118 together is necessary for pressure-induced cell adhesion and metastasis (44). More recently, Jun et al. (12) demonstrated overexpression of wild-type PXN plasmids promoted cell proliferation and enhanced migration, invasive capacity and metastasis of HCT-8 cells, whereas inhibition of PXN by siRNA repressed these abilities. In the present study, we found that HT-29 and SW480 cells were characterized with relatively lower PXN mRNA and higher miR-137, whereas HCT116 and SW620 cells with relatively higher PXN mRNA and lower miR-137. To our interest, in a nude mouse model, we found that HT-29 and SW480 cells were less proliferative and less invasive than HCT116 and SW620 cells (data not shown). In fact, SW480 and SW620 cells are known to derive from different stages of colon cancer in the same patients (45); SW480 was isolated from the primary tumor, and SW620 was isolated from a lymph node metastasis. Moreover, it has been reported that SW480 was less invasive than SW620 (46). Based on these results, we evaluated the effect of PXN and/or miR-137 on CRC cell lines. We showed knockdown of PXN inhibited cell proliferation, migration and invasion in vitro. In addition, in vivo xenograft tumor growth and metastasis assays demonstrated that stable knockdown of PXN in CRC cell line HCT116 could also suppress tumor growth and metastasis to lung and liver. Importantly, ectopic expression of miR-137 presented a similar tumor-repressive effect of PXN knockdown in CRC cell lines. This is in line with past reports that indicated miR-137 could inhibit cell proliferation and invasion in breast cancer cells, glioblastoma cells and CRC cell lines (22,38,40). Moreover, overexpression of PXN, in the HCT116-LV-miR-137 cell line stably expressing miR-137 from a pcDNA3.1PXN plasmid, could at least partially nullify the antiproliferation and antimetastasis effect of miR-137. These results further support the hypothesis that miR-137 exert its effect by regulating PXN. However, there may be other genes regulated by miR-137 as PXN overexpression could not completely counteract the effect of miR-137.
There remain some limitations in the present study. We could not determine an absolute value to distinguish patients with high PXN expression from those with low PXN expression; PXN might also be regulated by other miRNAs and molecules. Further studies are needed to address these issues.
In conclusion, our study demonstrated that upregulation of PXN in response to miR-137 reduction may promote tumor growth and metastasis and that PXN and miR-137 may be indicators of prognosis in CRC. Based on these results, we think PXN could be used as a potential therapeutic target in CRC patients.
Supplementary material
Supplementary Tables I and II and Figures S1 and S2 can be found at http://carcin.oxfordjournals.org/
Funding
National Natural Science Foundation of China (30672408); Guangzhou Bureau of Science and Technology (2006Z3-E0041); Sun Yat-sen University 985 Program Initiation Fund (China).
Conflict of Interest Statement: None declared.
Supplementary Material
Acknowledgement
We thank Zhizhong Pan and Weihua Jia for allowing access to fresh CRC tissue samples.
Glossary
Abbreviations:
- CRC
colorectal cancer
- IHC
immunohistochemistry
- miRNA
microRNA
- mRNA
messenger RNA
- MTT
3-(4,5-dimethylthiazole-2-yl)-2,5-biphenyl tetrazolium bromide
- NC
negative control
- PXN
paxillin
- siRNA
small interfering RNA
- 3′UTR
3′-untranslated region
- TNM
tumor, lymph nodes, distant metastasis.
References
- 1. Jemal A, et al. (2010). Cancer statistics, 2010. CA Cancer J. Clin., 5, 277–300 [DOI] [PubMed] [Google Scholar]
- 2. Wood L.D, et al. (2007). The genomic landscapes of human breast and colorectal cancers. Science, 5853, 1108–1113 [DOI] [PubMed] [Google Scholar]
- 3. Kudo Y, et al. (2004). Invasion and metastasis of oral cancer cells require methylation of E-cadherin and/or degradation of membranous beta-catenin. Clin. Cancer Res., 16, 5455–5463 [DOI] [PubMed] [Google Scholar]
- 4. Kumar R, et al. (2006). p21-activated kinases in cancer. Nat. Rev. Cancer, 6, 459–471 [DOI] [PubMed] [Google Scholar]
- 5. Schaller M.D. (2001). Paxillin: a focal adhesion-associated adaptor protein. Oncogene, 44, 6459–6472 [DOI] [PubMed] [Google Scholar]
- 6. Turner C.E. (2000). Paxillin and focal adhesion signalling. Nat. Cell Biol., 12, E231–E236 [DOI] [PubMed] [Google Scholar]
- 7. Brown M.C, et al. (2004). Paxillin: adapting to change. Physiol. Rev., 4, 1315–1339 [DOI] [PubMed] [Google Scholar]
- 8. Wu D.W, et al. (2010). Paxillin predicts survival and relapse in non-small cell lung cancer by microRNA-218 targeting. Cancer Res., 24, 10392–10401 [DOI] [PubMed] [Google Scholar]
- 9. Kasai M, et al. (2003). The Group 3 LIM domain protein paxillin potentiates androgen receptor transactivation in prostate cancer cell lines. Cancer Res., 16, 4927–4935 [PubMed] [Google Scholar]
- 10. Li B.Z, et al. (2008). Increased expression of paxillin is found in human oesophageal squamous cell carcinoma: a tissue microarray study. J. Int. Med. Res., 2, 273–278 [DOI] [PubMed] [Google Scholar]
- 11. Ayaki M, et al. (2001). Reduced expression of focal adhesion kinase in liver metastases compared with matched primary human colorectal adenocarcinomas. Clin. Cancer Res., 7, 3106–3112 [PubMed] [Google Scholar]
- 12. Jun Q, et al. (2011). Effects of paxillin on HCT-8 human colorectal cancer cells. Hepatogastroenterology, 112, 1951–1955 [DOI] [PubMed] [Google Scholar]
- 13. Pillai R.S. (2005). MicroRNA function: multiple mechanisms for a tiny RNA? RNA, 12, 1753–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zamore P.D, et al. (2005). Ribo-gnome: the big world of small RNAs. Science, 5740, 1519–1524 [DOI] [PubMed] [Google Scholar]
- 15. Bartel D.P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell, 2, 281–297 [DOI] [PubMed] [Google Scholar]
- 16. Esquela-Kerscher A, et al. (2006). Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer, 4, 259–269 [DOI] [PubMed] [Google Scholar]
- 17. Calin G.A, et al. (2006). MicroRNA signatures in human cancers. Nat. Rev. Cancer, 11, 857–866 [DOI] [PubMed] [Google Scholar]
- 18. Bueno M.J, et al. (2008). Control of cell proliferation pathways by microRNAs. Cell Cycle, 20, 3143–3148 [DOI] [PubMed] [Google Scholar]
- 19. Nicoloso M.S, et al. (2009). MicroRNAs—the micro steering wheel of tumour metastases. Nat. Rev. Cancer, 4, 293–302 [DOI] [PubMed] [Google Scholar]
- 20. Bemis L.T, et al. (2008). MicroRNA-137 targets microphthalmia-associated transcription factor in melanoma cell lines. Cancer Res., 5, 1362–1368 [DOI] [PubMed] [Google Scholar]
- 21. Langevin S.M, et al. (2010). MicroRNA-137 promoter methylation in oral rinses from patients with squamous cell carcinoma of the head and neck is associated with gender and body mass index. Carcinogenesis, 5, 864–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhao Y, et al. (2012). MiR-137 targets estrogen-related receptor alpha and impairs the proliferative and migratory capacity of breast cancer cells. PLoS ONE, 6, e39102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Balaguer F, et al. (2010). Epigenetic silencing of miR-137 is an early event in colorectal carcinogenesis. Cancer Res., 16, 6609–6618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Teng K.Y, et al. (2010). DNA polymerase eta protein expression predicts treatment response and survival of metastatic gastric adenocarcinoma patients treated with oxaliplatin-based chemotherapy. J. Transl. Med., 8, 126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang S, et al. (1998). Loss of AP-2 results in downregulation of c-KIT and enhancement of melanoma tumorigenicity and metastasis. EMBO J., 15, 4358–4369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. John B, et al. (2004). Human microRNA targets. PLoS Biol., 11, e363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krek A, et al. (2005). Combinatorial microRNA target predictions. Nat. Genet., 5, 495–500 [DOI] [PubMed] [Google Scholar]
- 28. Lewis B.P, et al. (2003). Prediction of mammalian microRNA targets. Cell, 7, 787–798 [DOI] [PubMed] [Google Scholar]
- 29. Sen A, et al. (2010). Paxillin regulates androgen- and epidermal growth factor-induced MAPK signaling and cell proliferation in prostate cancer cells. J. Biol. Chem., 37, 28787–28795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mackinnon A.C, et al. (2011). Paxillin expression and amplification in early lung lesions of high-risk patients, lung adenocarcinoma and metastatic disease. J. Clin. Pathol., 1, 16–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li H.G, et al. (2005). Clinicopathological significance of expression of paxillin, syndecan-1 and EMMPRIN in hepatocellular carcinoma. World J. Gastroenterol., 10, 1445–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shi J, et al. (2010). Paxillin expression levels are correlated with clinical stage and metastasis in salivary adenoid cystic carcinoma. J. Oral Pathol. Med., 7, 548–551 [DOI] [PubMed] [Google Scholar]
- 33. Volinia S, et al. (2006). A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl Acad. Sci. U.S.A., 7, 2257–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lu J, et al. (2005). MicroRNA expression profiles classify human cancers. Nature, 7043, 834–838 [DOI] [PubMed] [Google Scholar]
- 35. Zhang B, et al. (2007). MicroRNAs as oncogenes and tumor suppressors. Dev. Biol., 1, 1–12 [DOI] [PubMed] [Google Scholar]
- 36. Jia Y, et al. (2013). Epigenetic changes in colorectal cancer. Chin. J. Cancer 32 21–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kozaki K, et al. (2008). Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res., 7, 2094–2105 [DOI] [PubMed] [Google Scholar]
- 38. Chen L, et al. (2012). miR-137 is frequently down-regulated in glioblastoma and is a negative regulator of Cox-2. Eur. J. Cancer 48 3104–11 [DOI] [PubMed] [Google Scholar]
- 39. Wong T.S, et al. (2008). Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin. Cancer Res., 9, 2588–2592 [DOI] [PubMed] [Google Scholar]
- 40. Liu M, et al. (2011). miR-137 targets Cdc42 expression, induces cell cycle G1 arrest and inhibits invasion in colorectal cancer cells. Int. J. Cancer, 6, 1269–1279 [DOI] [PubMed] [Google Scholar]
- 41. Panetti T.S. (2002). Tyrosine phosphorylation of paxillin, FAK, and p130CAS: effects on cell spreading and migration. Front. Biosci., 7, d143–d150 [DOI] [PubMed] [Google Scholar]
- 42. Schaller M.D, et al. (2001). Multiple stimuli induce tyrosine phosphorylation of the Crk-binding sites of paxillin. Biochem. J., 360 (Pt 1)57–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen H.Y, et al. (2004). Brk activates rac1 and promotes cell migration and invasion by phosphorylating paxillin. Mol. Cell. Biol., 24, 10558–10572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Downey C, et al. (2008). Pressure activates colon cancer cell adhesion via paxillin phosphorylation, Crk, Cas, and Rac1. Cell. Mol. Life Sci., 9, 1446–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Leibovitz A, et al. (1976). Classification of human colorectal adenocarcinoma cell lines. Cancer Res., 12, 4562–4569 [PubMed] [Google Scholar]
- 46. Ding Q, et al. (2011). APOBEC3G promotes liver metastasis in an orthotopic mouse model of colorectal cancer and predicts human hepatic metastasis. J. Clin. Invest., 11, 4526–4536 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.