Abstract
Genome-wide association studies (GWASs) have mainly focused on top significant single nucleotide polymorphisms (SNPs), most of which did not have clear biological functions but were just surrogates for unknown causal variants. Studying SNPs with modest association and putative functions in biologically plausible pathways has become one complementary approach to GWASs. To unravel the key roles of mitogen-activated protein kinase (MAPK) pathways in cutaneous melanoma (CM) risk, we re-evaluated the associations between 47 818 SNPs in 280 MAPK genes and CM risk using our published GWAS dataset with 1804 CM cases and 1026 controls. We initially found 105 SNPs with P ≤ 0.001, more than expected by chance, 26 of which were predicted to be putatively functional SNPs. The risk associations with 16 SNPs around DUSP14 (rs1051849) and a previous reported melanoma locus MAFF/PLA2G6 (proxy SNP rs4608623) were replicated in the GenoMEL dataset (P < 0.01) but failed in the Australian dataset. Meta-analysis showed that rs1051849 in the 3ʹ untranslated regions of DUSP14 was associated with a reduced risk of melanoma (odds ratio = 0.89, 95% confidence interval: 0.82–0.96, P = 0.003, false discovery rate = 0.056). Further genotype–phenotype correlation analysis using the 90 HapMap lymphoblastoid cell lines from Caucasians showed significant correlations between two SNPs (rs1051849 and rs4608623) and messenger RNA expression levels of DUSP14 and MAFF (P = 0.025 and P = 0.010, respectively). Gene-based tests also revealed significant SNPs were over-represented in MAFF, PLA2G6, DUSP14 and other 16 genes. Our results suggest that functional SNPs in MAPK pathways may contribute to CM risk. Further studies are warranted to validate our findings.
Introduction
Cutaneous melanoma (CM) is the most aggressive form of skin cancers, and its incidence is increasing annually in Caucasian populations (1). Previous linkage studies have identified some high-penetrance genes that influence CM risk, including cyclin-dependent kinase inhibitor 2A and cyclin-dependent kinase 4 (2). Recent genome-wide association studies (GWASs) on CM have successfully expanded our understanding on low-penetrance loci in CM susceptibility, such as those of MC1R, TYR, HERC2, ATM, MX2, CASP8 and CCND1 (3–9). However, most GWASs have focused on a small number of single nucleotide polymorphisms (SNPs) with the required genome-wide significance level (e.g. P < 10–7), and, as a result, only a small fraction of heritability could be explained by these SNPs. Other SNPs with a moderate significance are largely neglected. In addition, most of the reported significant SNPs do not have clear biological functions and may just be surrogates for the unknown causal SNPs located elsewhere in the genome, with limited application as reliable biomarkers for susceptibility in personalized cancer prevention and cancer therapy (10).
To overcome these limitations in GWASs, several complementary approaches have been proposed recently (11), such as pathway-based analysis and integrating analysis of association results with gene expression (12,13). These applications have successfully revealed several new cancer susceptibility genes and pathways (14,15). Thus, studying SNPs with a moderate significance and putative functions in a biologically plausible pathway may help identify SNPs with a relatively small effect size and provide additional insights into molecular mechanisms of cancer.
Mitogen-activated protein kinase (MAPK) pathways can transduce a large variety of external signals to the nucleus, involving diverse cellular processes, such as cell proliferation, differentiation and apoptosis (16). In mammals, three main MAPK pathways have been characterized. In general, the extracellular signal-regulated kinase (ERK) pathway is preferentially activated by mitogens and plays an important role in cell growth and proliferation, whereas the c-jun N-terminal kinase (JNK) and p38 pathways are mainly responsive to cellular stress and inflammatory signals, often implicated in cellular apoptosis (17). The dynamic balance between ERK and JNK–p38 pathways has been proposed to determine cell survival or apoptosis (18), and their functions in cancer development are also different. Deregulation of the ERK pathway has been involved in oncogenic transformation and tumorigenesis. For example, mutations of NRAS or BRAF, which could lead to constitutive activation of the ERK pathway, have been detected in a series of cancers including melanoma (19,20). JNK and p38 pathways have been generally linked to tumor suppression, and inactivating mutations of MEK4 in these pathways have been observed in several kinds of cancer cells (21).
It is known that MAPK pathways play key roles in CM development and progression (22,23). However, until now, only limited number of candidate genes (such as BRAF and EGF genes) in MAPK pathways had been investigated for their association with CM risk (24–27). Previous GWASs on nevi and CM have identified strong association between genetic variants in one MAPK gene PLA2G6 and CM risk (8,28,29). Considering the broad effects of MAPK pathways on cellular process and cancer development, we hypothesized that other common [with a minor allele frequency (MAF) ≥ 0.05] SNPs in MAPK pathways at a moderate significance level but with putative functions may also contribute to CM risk.
In this study, we first investigated the association of SNPs in three MAPK pathways with CM risk using our published GWAS dataset (4) and performed functional prediction for SNPs with a significance level of ≤10–3. Then, we validated SNPs with putative functions in two other GWAS datasets (5,9). Finally, we evaluated the effects of SNPs that remained significant in the meta-analysis on their corresponding gene expression using the published expression data of the HapMap lymphoblastoid cell lines (30).
Materials and methods
Study population of the discovery dataset at MD Anderson Cancer Center
This study population has been described in the published GWAS study (4). Briefly, the discovery population consisted of 1804 non-Hispanic patients with newly diagnosed CM and 1026 controls, who were recruited from The University of Texas MD Anderson Cancer Center between March 1998 and August 2008. Of these subjects, 931 CM patients and 1026 age- and sex-matched cancer-free controls had completed a lifestyle questionnaire to provide information about their demographic and the known risk factors for CM. A summary table of those factors is presented in Supplementary Table S1, available at Carcinogenesis Online. The remaining 873 CM patients were recruited regardless of treatment and without lifestyle questionnaire data. The study protocol was approved by the institutional review board, and a written informed consent was obtained from all the participants.
Genotyping and imputation in the discovery dataset
Genotyping was performed as described previously (4). Briefly, genomic DNA samples extracted from the whole blood were genotyped with the Illumina HumanOmni1-Quad_v1-0_B array, and the genotypes were called using the BeadStudio algorithm at the John Hopkins University Center for Inherited Disease Research. SNPs with MAF ≤ 0.01, call rate ≤ 95% or Hardy-Weinberg Equilibrium in controls with P ≤ 10–5 were excluded. Finally, 818 237 genotyped autosomal or X chromosome SNPs and 740 pseudo-autosomal SNPs were available for the genome-wide association analysis. Genome-wide imputation had been applied using MACH based on 1000 Genome phase I V2 CEU data (2010–11 data freeze, 2012-02-14 haplotypes) (31). About 7 774 230 SNPs were imputed with r2 ≥ 0.8.
Selection of genes and SNPs from the MAPK pathways
Based on the databases of KEGG (http://www.genome.jp/kegg/) and Biocarta (http://www.biocarta.com/), we selected 280 genes located on autosomal chromosomes from three main MAPK signaling pathways. Genotyped or imputed common SNPs (MAF ≥ 0.05) within these genes or their ±20kb flanking regions were selected for association analysis. As a result, 9076 genotyped SNPs and 38 742 imputed SNPs in MAPK pathways had been extracted from our CM GWAS dataset and used for further analysis. The gene symbols and number of SNPs on each gene were listed in Supplementary Table S2, available at Carcinogenesis Online.
SNP functional prediction
We used one online prediction tool SNPinfo (32), which integrated a variety of in silico tools, to predict the potential functions of SNPs with P ≤ 0.001. SNPs that were predicted to affect protein structure, gene regulation, splicing and micro RNA (miRNA) binding were selected for replication.
In silico replication studies
In silico replication studies were performed using the GWAS data from the GenoMEL consortium and the Australian consortium. Briefly, the GenoMEL participants were recruited from multiple centers across Europe and Israel in two phases. Phase 1 of the original GenoMEL GWAS consisted of samples collected from eight centers across six different European countries. These were supplemented with controls from the Wellcome Trust Case Control Consortium. Standard quality control (QC) measures were applied to both samples and SNPs, given a total of 1353 cases and 3571 controls. Phase 2 of the GenoMEL GWAS samples were collected across 10 centers (four not in phase 1) in eight different European countries and Israel, supplemented again by samples from the Wellcome Trust Case Control Consortium. After QC, 1450 cases and 4047 controls remained. Detailed information about the study population and QC could be found in the published GWAS paper (5). Most of the phase 1 samples were genotyped on the Illumina HumanHap300 BeadChip version 2 duo array (317K SNPs), with the exception of 1905 French controls, which were genotyped on the Illumina Humancnv370k array. The GenoMEL phase 2 samples were genotyped on the Illumina Human610 quad array (610K SNPs). Sample and genotype QC had been applied separately for each platform. Imputation was conducted using IMPUTE v2 based on CEU data from 1000 Genomes Pilot data (March 2012). SNPs from phase 1 and 2 datasets with an imputation accuracy score (equivalent to MACH’s r2) ≥ 0.8 and the meta-analysis results of the two phases data were used in the replication study.
The replication study from the Australian consortium, as described in the published GWAS paper (9), included 2166 cases and 4219 controls, together with 553 controls from the International Barrett’s and Esophageal Adenocarcinoma Consortium. The case samples were recruited from the Queensland Study of Melanoma, Environment and Genetic Associations (Q-MEGA study) and Australian Melanoma Family Study (AMFS) (33,34). The control samples were selected from four sources: Brisbane Adolescent Twin Study, the Queensland Institute of Medical Research, AMFS study and International Barrett’s and Esophageal Adenocarcinoma Consortium. Genomic DNA was extracted from peripheral blood or saliva samples. Controls were genotyped on three Illumina SNP arrays: Omni1-Quad array (20.6%), HumanHap610 or HumanHap670 array (79.4%); cases were genotyped on Illumina Omni1-Quad array (57.2%) or HumanHap610 array (42.8%). Sample and genotype QC had been detailed in the GWAS publication (9). Imputation was performed using MACH with 1000 Genomes Project data obtained from people with ancestry from northern and western Europe. SNPs from imputation with r2 > 0.8 were used in this study. In the replication study, we used the meta-analysis results of the AMFS dataset, Q-MEGA dataset from 610k/670k array, Q-MEGA/International Barrett’s and Esophageal Adenocarcinoma Consortium data from Omni1-Quad array.
Statistical methods
Association between SNPs and CM risk in the discovery dataset was primarily assessed by PLINK v1.07 in an additive model adjusting for the first three principle components. Replication results from GenoMEL and Australian consortia were adjusted for the geographic regions and the first six principle components, respectively. For the meta-analysis, the betas and standard errors from the two validation studies or of all the three studies were combined with the inverse variance-based method as implemented in PLINK (35). Cochran’s Q statistics and I2 were used to assess the heterogeneity of datasets. Fixed effect models were applied when there was no heterogeneity among the datasets (P > 0.10 and I2 < 25); otherwise, random effect models were applied. Benjamini and Hochberg’s false discovery rate (FDR) method was also used to control for the multiple comparisons. Linear regression analysis was used to test the correlations between SNPs and corresponding gene expression obtained from the 90 HapMap CEU lymphoblastoid cell lines (NCBI GEO database, accession GSE6536) (30).
In the stratification analysis, we applied a variety of genetic models (codominant, additive, dominant and recessive). Adjusted odds ratios (ORs) and 95% confidence intervals (CIs) were calculated with adjustment for age, sex, moles, dysplastic nevi, pigmentation score and family history in first-degree relatives with any cancers. As described earlier (36), pigmentation score was a multivariate confounder score summarizing the pigmentation-related variable, including hair color, eye color, skin color, tanning ability, freckling in the sun as a child and history of sunburn. Beta values for each of these variables were calculated using multivariate logistic regression model with risk of CM as the outcome. A summation score for each subject was then calculated using these beta values. Based on the median pigmentation score in controls, participants were dichotomized as low or high levels of pigmentation.
Gene-based test was performed using three approaches: PLINK set-based test (35), hypergeometric method (37) and Versatile Gene-Based Test for Genome-wide Association method (VEGAS) (38). For PLINK set-based test, we first selected N-independent SNPs (r2 < 0.8) within each gene and calculated the average association test statistic across these SNPs as set-statistic; then, 100 000 permutations were performed by randomly shuffling the phenotypes among individuals, and an empirical P value was obtained by calculating the proportion of times that the permuted statistic exceeds the original statistic. VEGAS method also calculates an empirical P value for each gene by using the similar process with the PLINK set-based test. There are two main differences between these methods. The VEGAS method uses the sum association test statistic rather than the average, and the empirical P values are calculated by performing Monte Carlo simulation from a specified multivariate normal distribution rather than phenotype permutation. The hypergeometric method is equivalent to the right-tailed Fisher’s exact test. The P value of this method is the cumulative probability of finding k or more SNPs in one given gene when the probability of sampling k significant SNPs is calculated based on hypergeometric distribution.
LocusZoom was used to produce regional association plots (39). Unless specified otherwise, statistical analyses were performed using the SAS software (version 9.2; SAS Institute, Cary, NC).
Results
In the discovery dataset, a total of 9076 genotyped SNPs and 38 742 imputed SNPs had been extracted from 280 MAPK genes (see Supplementary Figure S1, available at Carcinogenesis Online, of the Manhattan plot). Association of these SNPs with CM risk was tested using trend tests. As shown in Supplementary Table S2, available at Carcinogenesis Online, there were 105 SNPs that reached the significance level of P ≤ 0.001. Based on the in silico prediction results, we found 26 SNPs around seven gene regions (PPP2CA: rs389755 and rs10463914; LTB/LST1: rs9267502; SPON1: rs11369, rs11238, rs16913795 and rs1043237; RRAS2: rs8570; DUSP14: rs1051849; STK4: rs4810446; PLA2G6/MAFF: rs3761444, rs13056506, rs5750558, rs2413507, rs2899297, rs5756968, rs5750561, rs3761445, rs3761447, rs3761449, rs3890451, rs9607517, rs4374456, rs4608623, rs2267372 and rs9610915) with putative functions (Table I).
Table I.
Association between potentially functional SNPs in MAPK pathways and melanoma risk in the MD Anderson dataset (P ≤ 0.001)
| SNP | Chr | Position | Minor/major allele | Nearby gene | Predicted functions | MAF_cases | MAF_controls | OR (95% CI)a | Pa |
|---|---|---|---|---|---|---|---|---|---|
| rs3895755 | 5 | 133561339 | G/A | PPP2CA | TFBS | 0.05 | 0.07 | 0.69 (0.55–0.86) | 9.96E-04 |
| rs10463914 | 5 | 133566760 | A/G | PPP2CA||CDKL3 | TFBS | 0.05 | 0.07 | 0.69 (0.55–0.86) | 8.47E-04 |
| rs9267502 | 6 | 31553194 | A/G | LTB||LST1 | TFBS | 0.08 | 0.06 | 1.45 (1.16–1.81) | 1.00E-03 |
| rs11369 | 11 | 14288096 | G/A | SPON1 | miRNA binding | 0.33 | 0.29 | 1.24 (1.10–1.39) | 4.13E-04 |
| rs11238 | 11 | 14288128 | A/C | SPON1 | miRNA binding | 0.33 | 0.29 | 1.24 (1.10–1.39) | 4.43E-04 |
| rs16913795 | 11 | 14288993 | A/G | SPON1 | miRNA binding | 0.33 | 0.29 | 1.24 (1.10–1.39) | 4.44E-04 |
| rs1043237 | 11 | 14289053 | T/A | SPON1 | miRNA binding | 0.33 | 0.29 | 1.23 (1.10–1.39) | 4.97E-04 |
| rs8570 | 11 | 14300759 | G/C | RRAS2 | miRNA binding | 0.33 | 0.29 | 1.22 (1.09–1.37) | 0.001 |
| rs1051849 | 17 | 35873324 | G/A | DUSP14 | miRNA binding | 0.11 | 0.13 | 0.76 (0.64–0.89) | 0.001 |
| rs4810446 | 20 | 43595868 | A/T | STK4 | TFBS | 0.07 | 0.10 | 0.71 (0.59–0.87) | 7.36E-04 |
| rs3761444 | 22 | 38580371 | G/A | PLA2G6||MAFF | TFBS | 0.40 | 0.45 | 0.82 (0.73–0.91) | 3.65E-04 |
| rs13056506 | 22 | 38580917 | G/T | PLA2G6||MAFF | TFBS | 0.37 | 0.42 | 0.81 (0.72–0.91) | 2.12E-04 |
| rs5750558 | 22 | 38582497 | G/A | PLA2G6||MAFF | TFBS | 0.40 | 0.45 | 0.82 (0.73–0.91) | 3.65E-04 |
| rs2413507 | 22 | 38593428 | A/G | PLA2G6||MAFF | TFBS | 0.40 | 0.45 | 0.82 (0.73–0.92) | 4.89E-04 |
| rs2899297 | 22 | 38594668 | G/A | PLA2G6||MAFF | TFBS | 0.37 | 0.42 | 0.81 (0.73–0.91) | 2.80E-04 |
| rs5756968 | 22 | 38595240 | C/T | PLA2G6||MAFF | TFBS | 0.37 | 0.42 | 0.81 (0.73–0.91) | 2.95E-04 |
| rs5750561 | 22 | 38595260 | A/T | PLA2G6||MAFF | TFBS | 0.37 | 0.42 | 0.81 (0.73–0.91) | 2.95E-04 |
| rs3761445 | 22 | 38595411 | G/A | PLA2G6||MAFF | TFBS | 0.37 | 0.42 | 0.81 (0.73–0.91) | 2.95E-04 |
| rs3761447 | 22 | 38595539 | G/A | PLA2G6||MAFF | TFBS | 0.40 | 0.45 | 0.82 (0.73–0.92) | 4.89E-04 |
| rs3761449 | 22 | 38595615 | C/T | PLA2G6||MAFF | TFBS | 0.38 | 0.43 | 0.82 (0.73–0.91) | 4.21E-04 |
| rs3890451 | 22 | 38595820 | T/G | PLA2G6||MAFF | TFBS | 0.37 | 0.42 | 0.81 (0.73–0.91) | 3.23E-04 |
| rs9607517 | 22 | 38596100 | G/A | PLA2G6||MAFF | TFBS | 0.40 | 0.45 | 0.82 (0.73–0.92) | 4.89E-04 |
| rs4374456 | 22 | 38597377 | G/C | PLA2G6||MAFF | TFBS | 0.40 | 0.45 | 0.82 (0.73–0.91) | 3.57E-04 |
| rs4608623 | 22 | 38597378 | G/T | PLA2G6||MAFF | TFBS | 0.40 | 0.45 | 0.82 (0.73–0.91) | 3.57E-04 |
| rs2267372 | 22 | 38598234 | A/G | MAFF | TFBS | 0.38 | 0.42 | 0.82 (0.73–0.92) | 5.34E-04 |
| rs9610915 | 22 | 38611080 | C/G | MAFF | miRNA binding | 0.44 | 0.49 | 0.83 (0.74–0.92) | 7.14E-04 |
aAdjusted for the first three principle components.
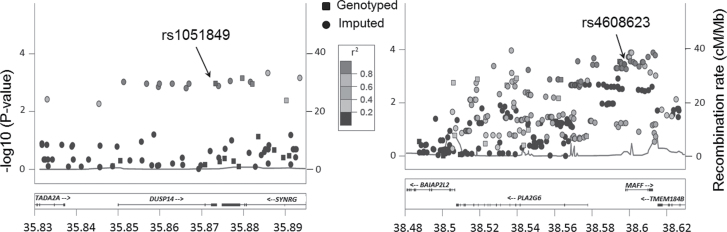
Replication results are presented in Table II, in which 16 SNPs (rs1051849 in DUSP14 and other 15 SNPs in MAFF/PLA2G6) were shown to be significantly associated with CM risk in the GenoMEL study (P < 0.01); however, none of them was significant in the Australian dataset. A meta-analysis combining these two validation datasets was performed to estimate the effect sizes of these SNPs. One functional SNP (rs1051849 in the 3ʹ untranslated region of DUSP14) was found to have the same direction in the effects as in the discovery dataset (OR = 0.89, 95% CI: 0.82–0.96, P = 0.003, FDR = 0.056). Other functional SNPs in MAFF/PLA2G6 were not significant in the meta-analysis because of large heterogeneity between the two validation datasets (I2 > 25). The regional association results from the discovery dataset were plotted for the two gene regions (20kb neighborhood of DUSP14 and PLA2G6/MAFF) (Figure 1). We also listed the replication results for all of the 105 SNPs in Supplementary Table S2, available at Carcinogenesis Online. In addition to the 16 functional SNPs, other 34 non-functional SNPs were replicated in the GenoMEL dataset but not in the Australian dataset: two SNPs around CACNB2; other SNPs were in moderate-to-high linkage disequilibrium (LD) with the functional SNPs in DUSP14 or MAFF/PLA2G6 (eight SNPs were in high LD with rs1051849; 24 SNPs in moderate-to-high LD with rs4608623 in MAFF/PLA2G6) (Supplementary Figure S2, available at Carcinogenesis Online). We used the FDR method to correct for multiple comparisons and found six SNPs in DUSP14 (rs1051849 and five non-functional SNPs: rs4795205, rs117494398, rs147535415, rs4794755 and rs79356259) with a FDR of <0.2. Although no SNPs in MAFF/PLA2G6 were significant in the meta-analysis of the two replication datasets, we still selected rs4608623 as the proxy SNP of this region based on pair-wise LD and used it in further analyses. The types of SNPs (imputed or genotyped) in each validation dataset have been listed in Supplementary Table S3, available at Carcinogenesis Online.
Table II.
Validation results of 26 potentially functional SNPs in GenoMEL and Australian melanoma GWAS datasets
| SNP | GenoMEL dataset | Australian dataset | Meta-analysis of the two studies | I | FDRd | ||||
|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI)a | Pa | OR (95% CI)b | Pb | OR (95% CI) | Pc | Q | |||
| rs3895755 | 0.93 (0.63–1.39) | 0.739 | 0.97 (0.81–1.15) | 0.704 | 0.96 (0.82–1.13) | 0.655 | 0.867 | 0 | 0.696 |
| rs10463914 | 0.94 (0.63–1.39) | 0.745 | 0.97 (0.81–1.15) | 0.707 | 0.96 (0.82–1.13) | 0.657 | 0.873 | 0 | 0.696 |
| rs9267502 | 1.08 (0.96–1.22) | 0.201 | 1.03 (0.89–1.19) | 0.864 | 1.06 (0.97–1.17) | 0.216 | 0.681 | 0 | 0.574 |
| rs11369 | 0.94 (0.88–1.01) | 0.089 | 0.99 (0.91–1.08) | 0.728 | 0.96 (0.91–1.01) | 0.146 | 0.361 | 0 | 0.574 |
| rs11238 | 0.94 (0.88–1.01) | 0.088 | 0.99 (0.91–1.08) | 0.817 | 0.96 (0.91–1.01) | 0.145 | 0.358 | 0 | 0.574 |
| rs16913795 | 0.94 (0.87–1.01) | 0.070 | 1.01 (0.93–1.10) | 0.835 | 0.97 (0.90–1.04) | 0.405 | 0.178 | 44.9 | 0.574 |
| rs1043237 | 0.94 (0.88–1.01) | 0.097 | 0.99 (0.91–1.08) | 0.845 | 0.96 (0.91–1.01) | 0.155 | 0.374 | 0 | 0.574 |
| rs8570 | 0.95 (0.88–1.02) | 0.134 | 1.00 (0.92–1.09) | 0.949 | 0.97 (0.91–1.02) | 0.251 | 0.335 | 0 | 0.574 |
| rs1051849 | 0.86 (0.77–0.95) | 0.003 | 0.93 (0.82–1.05) | 0.264 | 0.89 (0.82–0.96) | 0.003 | 0.303 | 5.7 | 0.056 |
| rs4810446 | 0.94 (0.83–1.06) | 0.310 | 1.01 (0.87–1.17) | 0.929 | 0.97 (0.88–1.06) | 0.491 | 0.449 | 0 | 0.624 |
| rs3761444 | 0.87 (0.81–0.93) | 4.51E-05 | 1.02 (0.86–1.21) | 0.778 | 0.93 (0.80–1.09) | 0.360 | 0.083 | 66.6 | 0.574 |
| rs13056506 | 0.87 (0.82–0.94) | 1.01E-04 | 1.02 (0.86–1.21) | 0.771 | 0.93 (0.80–1.08) | 0.351 | 0.103 | 62.5 | 0.574 |
| rs5750558 | 0.87 (0.81–0.93) | 4.57E-05 | 1.02 (0.86–1.21) | 0.775 | 0.93 (0.80–1.09) | 0.360 | 0.084 | 66.6 | 0.574 |
| rs2413507 | 0.88 (0.82–0.94) | 1.67E-04 | 1.02 (0.87–1.20) | 0.755 | 0.94 (0.81–1.08) | 0.368 | 0.092 | 64.8 | 0.574 |
| rs2899297 | 0.88 (0.82–0.94) | 2.97E-04 | 1.02 (0.87–1.20) | 0.793 | 0.94 (0.81–1.08) | 0.366 | 0.103 | 62.5 | 0.574 |
| rs5756968 | 0.88 (0.82–0.94) | 3.11E-04 | 1.02 (0.87–1.20) | 0.763 | 0.94 (0.82–1.08) | 0.363 | 0.106 | 61.6 | 0.574 |
| rs5750561 | 0.88 (0.83–0.95) | 3.98E-04 | 1.03 (0.87–1.22) | 0.726 | 0.94 (0.81–1.09) | 0.421 | 0.107 | 61.5 | 0.574 |
| rs3761445 | 0.88 (0.82–0.94) | 2.50E-04 | 1.02 (0.87–1.20) | 0.763 | 0.94 (0.81–1.08) | 0.365 | 0.101 | 62.9 | 0.574 |
| rs3761447 | 0.88 (0.82–0.94) | 1.34E-04 | 1.02 (0.87–1.20) | 0.722 | 0.94 (0.81–1.08) | 0.369 | 0.087 | 65.8 | 0.574 |
| rs3761449 | 0.88 (0.83–0.95) | 4.98E-04 | 1.02 (0.84–1.25) | 0.796 | 0.93 (0.81–1.06) | 0.289 | 0.187 | 42.4 | 0.574 |
| rs3890451 | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| rs9607517 | 0.88 (0.82–0.94) | 1.43E-04 | 1.02 (0.87–1.20) | 0.745 | 0.94 (0.81–1.08) | 0.365 | 0.093 | 64.5 | 0.574 |
| rs4374456 | 0.89 (0.83–0.95) | 0.001 | 1.02 (0.85–1.23) | 0.769 | 0.94 (0.82–1.06) | 0.312 | 0.177 | 45.1 | 0.574 |
| rs4608623 | 0.89 (0.83–0.95) | 0.001 | 1.02 (0.85–1.23) | 0.769 | 0.94 (0.82–1.06) | 0.312 | 0.177 | 45.1 | 0.574 |
| rs2267372 | 0.87 (0.81–0.93) | 6.10E-05 | 1.03 (0.87–1.23) | 0.709 | 0.93 (0.79–1.10) | 0.420 | 0.074 | 68.8 | 0.574 |
| rs9610915 | 0.87 (0.82–0.94) | 1.37E-04 | 1.05 (0.86–1.28) | 0.548 | 0.94 (0.79–1.13) | 0.515 | 0.088 | 65.7 | 0.646 |
Significant result in the meta-analysis is in bold.
aAdjusted for geographic regions.
bAdjusted for the first six principle components.
cFixed effect models were used when no heterogeneity was found between studies (P het > 0.10 and I2 < 25); otherwise, random effect models were used.
dFalse discovery rate using the method of Benjamini and Hochberg.
Fig. 1.
Regional association plots in the 20kb neighborhood of DUSP14 (chr 17) and PLA2G6/MAFF (chr 22). The left-hand y-axis shows the association P value of individual SNPs in the discovery dataset, which is plotted as −log10 (P) against chromosomal basepair position. The right-hand y-axis shows the recombination rate estimated from the HapMap CEU population.
Because comprehensive questionnaire data had been obtained from 931 cases and 1026 controls (Supplementary Table S1, available at Carcinogenesis Online), we thus re-examined the observed association between genotypes of these two SNPs (rs1051849 and rs4608623) and CM risk with adjustment for the known major risk factors overall as well as in stratification analyses. For rs1051849, in a dominant model, the G allele was associated with reduced CM risk (P = 0.006) in 1957 samples after adjustment for age, sex, mole status, pigmentation score, dysplastic nevi and family cancer history (Table III). Specifically, compared with the AA genotype, the variant allele carriers (GG + AG genotypes) had a lower CM risk (OR = 0.71, 95% CI: 0.58–0.89). Stratification analysis showed that the associations remained significant among males, younger age (≤52), high pigmentation score, presence of moles and no family cancer history. For SNP rs4608623, the protective effect was more evident in a recessive model (P = 2.00×10–4). Specifically, compared with the common allele carriers (TT + TG genotypes), the GG homozygotes had a lower CM risk (OR = 0.63, 95% CI: 0.49–0.80), and stratification analyses showed that such effects existed only among subjects having moles and a younger age. However, there was no significant heterogeneity in ORs between different strata for both SNPs by the Breslow-Day test (P > 0.1), which indicated that the lack of significance in some strata might be due to small sample sizes.
Table III.
Stratification analysis of the two significant SNPs (rs1051849 and rs4608623) by risk factors in 931 CM cases and 1026 cancer-free controls
| Variable | rs1051849 (cases/controls) | rs4608623 (cases/controls) | ||||||
|---|---|---|---|---|---|---|---|---|
| AA | AG + GG | OR (95%CI)a | P valuea | TT + GT | GG | OR (95% CI)a | P valuea | |
| Overall | 751/768 | 180/258 | 0.73 (0.58–0.91) | 0.006 | 789/793 | 142/233 | 0.63 (0.49–0.80) | 2.00E-04 |
| Gender | ||||||||
| Male | 452/459 | 106/154 | 0.74 (0.55–1.00) | 0.048 | 474/481 | 84/132 | 0.60 (0.41–0.88) | 0.009 |
| Female | 299/309 | 74/104 | 0.70 (0.49–1.00) | 0.049 | 315/312 | 58/101 | 0.65 (0.47–0.89) | 0.008 |
| Age | ||||||||
| ≤52 years old | 385/372 | 84/118 | 0.70 (0.50–0.98) | 0.04 | 405/372 | 64/118 | 0.50 (0.35–0.71) | 1.00E-04 |
| >52 years old | 366/396 | 96/140 | 0.75 (0.55–1.02) | 0.07 | 384/421 | 78/115 | 0.77 (0.55–1.08) | 0.13 |
| Pigmentation scoreb | ||||||||
| Low | 237/384 | 56/126 | 0.74 (0.51–1.07) | 0.112 | 255/397 | 38/113 | 0.55 (0.36–0.84) | 0.005 |
| High | 514/384 | 124/132 | 0.72 (0.54–0.96) | 0.027 | 534/396 | 104/120 | 0.67 (0.49–0.90) | 0.009 |
| Mole | ||||||||
| With | 583/370 | 135/123 | 0.70 (0.53–0.93) | 0.014 | 615/384 | 103/109 | 0.57 (0.42–0.78) | 3.00E-04 |
| Without | 168/398 | 45/135 | 0.74 (0.50–1.10) | 0.138 | 174/409 | 39/124 | 0.74 (0.49–1.11) | 0.141 |
| Cancer family history | ||||||||
| With | 496/465 | 162/119 | 0.79 (0.59–1.05) | 0.100 | 489/509 | 95/149 | 0.65 (0.48–0.89) | 0.006 |
| Without | 286/270 | 61/96 | 0.63 (0.43–0.93) | 0.019 | 300/282 | 47/84 | 0.57 (0.38–0.87) | 0.008 |
aResults were adjusted for five of the six covariates (gender, age, pigmentation score, mole status, dysplastic nevi and family history of cancer) except the stratification variable.
bPigmentation score was dichotomized based on the score median in controls. The score was calculated using the logistic regression coefficients from a multivariate model including skin color, eye color, hair color, tanning ability, sunburns with blistering and freckling in the sun as a child.
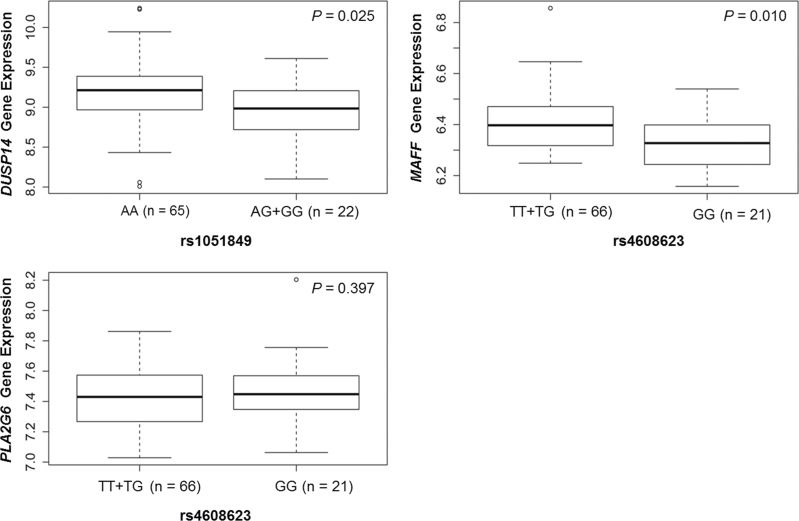
Based on the in silico functional prediction, rs1051849 was located at the 3ʹ untranslated region of DUSP14 and might influence the binding affinity of miRNAs; and rs4608623 was located at the 5ʹ flanking of MAFF and PLA2G6 and might influence the binding affinity of the transcription factors. To provide functional evidence for the association, we further evaluated the correlations between these two functional SNPs and their corresponding messenger RNA (mRNA) expression levels using the published expression data of the 90 HapMap lymphoblastoid cell lines derived from Caucasians (30). Consistent with their association results, the GG + AG genotypes of rs1051849 were shown to be associated with relatively lower levels of mRNA expression of DUSP14, compared with the AA genotype (P = 0.025), whereas for SNP rs4608623, the GG genotype carriers had a relatively lower MAFF expression than those with the TT + TG genotypes (P = 0.010) (Figure 2). However, no significant correlation was found between rs4608623 genotypes and PLA2G6 mRNA expression level (P = 0.397).
Fig. 2.
Analysis of DUSP14 and MAFF expression levels by genotypes of rs1051849 and rs4608623 in 90 HapMap lymphoblastoid cell lines from Caucasians (three with missing data). Consistent with their association results in Table III, genotypes AG + GG of rs1051849 were associated with low mRNA expression levels of DUSP14, compared with that of the AA genotype (P = 0.025); for SNP rs4608623, GG genotype carriers had lower MAFF expression levels than those with TT + TG genotypes (P = 0.010). The y-axis is the normalized gene expression levels. The box represents the central 50% of the data or the interquartile range. The lower edge of the box plot is the first quartile or 25th percentile. The upper edge of the box plot is the third quartile or 75th percentile. The line in the box is the median value. The ends of the vertical lines extend to minimum and maximum unless these values exceed 1.5 × interquartile range.
Because gene-based tests are supposed to be more powerful than the single-locus analysis by combining multiple independent SNPs signals within the same gene, we applied three gene-based methods in this study: PLINK set-based test, hypergeometric method and VEGAS method. By using the PLINK set-based test, we found that significant SNPs were over-represented in 19 MAPK genes (P < 0.05), which included MAFF, PLA2G6, DUSP14, RASGRP3, TNFRSF1A, TNF/LIST1, RRAS2, ARRB1, DUSP2, DUSP8, CASP3, SOS2, MAPKAPK5, PRKCA, TGFB2, STK4, CACNA1G, DUSP4 and PLCB1. These genes were also overlapped with that identified by either the hypergeometric method or the VEGAS method. Table IV lists the association results of these eight genes as well as SNP numbers and top SNPs within each gene. The overall gene-based results are presented in Supplementary Table S4, available at Carcinogenesis Online.
Table IV.
Significant results of gene-based tests using three methods of PLINK set-based test, hypergeometrix and VEGAS
| Gene | Chr: Position (hg18) | # | Most significant SNP | P for gene-based test | |||||
|---|---|---|---|---|---|---|---|---|---|
| Total SNPs | Tag SNPsa | Sig_SNPsb | SNP ID | P value | PLINK methodc | Hypergeometric methodd | VEGAS methodc | ||
| MAFF | 22:36902497..36957012 | 108 | 14 | 11 | rs2267373 | 1.29E-04 | 5.20E-04 | 2.09E-10 | 7.50E-04 |
| PLA2G6 | 22: 36837447..36907707 | 325 | 38 | 13 | rs2012725 | 1.07E-04 | 0.006 | 1.27E-06 | 0.003 |
| DUSP14 | 17: 32924063..32947701 | 81 | 20 | 5 | rs145885981 | 4.64E-04 | 0.011 | 0.062 | 0.007 |
| RASGRP3 | 2: 33514919..33643162 | 397 | 121 | 16 | rs13391694 | 8.56E-04 | 0.011 | 0.012 | 0.013 |
| TNFRSF1A | 12: 6308184..6321544 | 37 | 13 | 5 | rs1468603 | 0.006 | 0.013 | 0.012 | 0.011 |
| TNF/LIST1 | 6: 31651328..31654091 | 54 | 26 | 5 | rs3087617 | 9.39E-05 | 0.015 | 0.143 | 0.011 |
| RRAS2 | 11: 14256041..14337305 | 88 | 17 | 4 | rs4757245 | 2.28E-04 | 0.016 | 0.143 | 0.007 |
| ARRB1 | 11: 74654129..74740521 | 177 | 50 | 10 | rs555875 | 1.87E-04 | 0.019 | 0.009 | 0.017 |
| DUSP2 | 2: 96172634..96174906 | 26 | 5 | 2 | rs1168969 | 0.013 | 0.023 | 0.298 | 0.023 |
| DUSP8 | 11: 1531856..1549726 | 158 | 13 | 2 | rs80025267 | 0.002 | 0.028 | 0.866 | 0.023 |
| CASP3 | 4: 185785843..185807623 | 17 | 12 | 3 | rs4862396 | 0.007 | 0.030 | 0.276 | 0.022 |
| SOS2 | 14: 49653595..49767849 | 328 | 24 | 3 | rs8010248 | 0.003 | 0.035 | 0.792 | 0.031 |
| MAPKAPK5 | 12: 110764661..110815611 | 144 | 6 | 2 | rs77211491 | 0.008 | 0.035 | 0.392 | 0.013 |
| PRKCA | 17: 61729387..62237324 | 1374 | 242 | 22 | rs1003425 | 5.51E-04 | 0.037 | 0.068 | 0.035 |
| TGFB2 | 1: 216586490..216681593 | 40 | 25 | 5 | rs1417488 | 0.015 | 0.038 | 0.142 | 0.039 |
| STK4 | 20: 43028533..43142007 | 220 | 22 | 3 | rs2284271 | 2.30E-04 | 0.039 | 0.726 | 0.011 |
| CACNA1G | 17: 45993447..46059541 | 195 | 69 | 10 | rs2214566 | 0.002 | 0.040 | 0.056 | 0.037 |
| DUSP4 | 8: 29249536..29264104 | 95 | 20 | 5 | rs11780602 | 0.005 | 0.045 | 0.062 | 0.034 |
| PLCB1 | 20: 8061295..8813547 | 1883 | 331 | 29 | rs8123323 | 0.001 | 0.048 | 0.045 | 0.057 |
Sig_SNPs, significant SNPs.
aTagSNPs were selected based on pair-wise linkage disequilibrium r2 ≥ 0.8.
bThe number of tagSNPs with significance level ≤0.05.
cThe empirical P values were calculated 100 000 permutations/simulation using PLINK or VEGAS.
dHypergeometric test was performed using the number of tagSNPs and significant SNPs.
Supplementary Table S5, available at Carcinogenesis Online, summarizes the associations of SNPs in MAPK pathways that were previously identified to be associated with risk of CM or other cancers in the candidate gene-based studies or GWASs. Among the 12 SNPs, four in PLA2G6 were in moderate-to-high LD with the identified functional SNP rs4608623 and showed moderately significant association with CM risk (P = 0.002 for rs132985 and rs738322; P = 0.004 for rs2284063 and P = 0.003 for rs6001027). However, no significant association was found for other selected SNPs.
Finally, we investigated the association between the two functional SNPs (i.e. DUSP14 rs1051849 and PLA2G6/MAFF rs4608623) and pigmentation variables (hair color, eye color, skin color, moles status, tanning ability, freckling and sunburns with blistering), but we found no associations (Supplementary Table S6, available at Carcinogenesis Online).
Discussion
Deregulations of MAPK pathways have been implicated in many cancers (16). Recently, two pathway-based GWASs had reported that genetic variants in MAPK pathways might contribute to the risk of colorectal and breast cancers at the pathway level (14,40), but there is no similar report for CM. In this study, we comprehensively evaluated the association between 47 818 SNPs in 280 MAPK genes and CM risk using our published GWAS dataset, and we found that 105 SNPs were statistically significant (P ≤ 10–3), which is higher than the number expected by chance (n = 48 without considering LD between SNPs). Of the 26 putative functional SNPs, 16 SNPs located in two regions (rs1051849 in DUSP14; rs4608623 and other14 high LD SNPs in MAFF/PLA2G6) were shown to be significantly associated with CM risk in the GenoMEL datasets but failed validation in the Australian dataset. Additional meta-analysis of the validation studies showed that functional SNP rs1051849 located at the 3ʹ untranslated regions of DUSP14 were associated with CM risk in the same direction as in the discovery study. Other functional SNPs including rs4608623 in MAFF/PLA2G6 were not significant in the meta-analysis. Genotype–phenotype correlation analysis using the mRNA expression data from the 90 HapMap CEU lymphoblastoid cell revealed that these two SNPs might influence the mRNA expression levels of DUSP14 and MAFF. The results of gene-based tests provided additional support for the association between MAPK genes and CM risk.
DUSP14 is one of dual-specificity phosphatases (DUSPs) that negatively regulate the MAPK signaling and play critical roles in the development and carcinogenesis (41,42). There is increasing evidence that DUSPs may be abnormally regulated in a number of cancers, but varied effects of DUSPs had been observed in different cancer types and progression stages (41). To date, there is no report about the association between SNPs in DUSP genes and CM risk. In this study, we investigated association between genetic variants in 11 DUSP genes and CM risk through re-analysis of our published CM GWAS dataset. One important finding after the replications by another two GWAS datasets was that carriers of the rs1051849 G variant, which was associated with a relatively lower expression levels of DUSP14, had reduced CM risk compared with those with the AA homozygous genotype. Studies have shown that the inactivation of DUSP14 could cause hyperphosphorylation of the ERK and JNK pathways and enhance interleukin-2 secretion (43), and the latter was known to suppress tumor growth and metastases (44). Furthermore, suppression of the DUSP14 activity would increase JNK phosphorylation and in turn promote apoptosis (45). These studies provide some biological evidence for the molecular mechanisms underlying our observed associations.
PLA2G6 is an A2 phospholipase that has been shown to participate in several signal transduction pathways, including epidermal growth factor receptors, MAPK and MDM2 (46). Previous studies had reported that polymorphisms in or near the PLA2G6 region were associated with mole number and CM risk (7,8,28,47). In this study, we found 15 functional SNPs in MAFF/PLAG6 were associated with reduced melanoma risk in the MD Anderson discovery dataset and GenoMEL replication dataset. The proxy SNP (rs4608623) of them was in a moderate-to-high LD with the four previously identified SNPs (r2= 0.88 for rs132985 and rs738322; r2 = 0.51 for rs2284063 and rs6001027). These four SNPs are located in the intron region of PLA2G6 and do not have any putative functions, whereas SNP rs4608623 is located at the 5ʹ upstream of PLA2G6 and MAFF and has been predicted to influence the binding activity of transcription factors. Additional mRNA expression analysis revealed that this SNP (rs4608623) was correlated only with the gene expression of MAFF, which suggested that functional SNPs in this region might contribute to melanoma risk possibly by regulating MAFF mRNA expression. MAFF is one of the Maf transcription factors that belong to the AP1 superfamily. Several members of the MAFF transcription factors had been reported to be involved in cancer development (48). Based on the currently limited number of studies on the MAFF function, this gene may be involved in oncogenesis by participating in antioxidant responses (48,49). Further functional studies of this gene in CM are warranted to provide biological support for this association.
Previous candidate gene-based studies had identified the associations between SNPs in BRAF (27,50,51), H-RAS (52–54), EGF (55), MAP3K1 (56) and MAP2K4 (57–59) and risk of various cancers. In this study, we also evaluated the association between these SNPs and CM risk, but we did not find any statistical evidence for such association in our study population. These might be due to the heterogeneity in cancer etiology, difference in exposure by geographic areas or different ancestral backgrounds of the study populations.
Although results from in silico replication studies, mRNA expression analysis and gene-based tests have provided evidence for the association between functional SNPs (i.e. DUSP14 rs1051849 and MAFF/PLA2G6 rs4608623 and other 14 high LD SNPs) and CM risk, there were several limitations in this study. First, the association between SNPs in DUSP14 and MAFF/PLA2G6 were only replicated in the GenoMEL study but failed in validation using the Australian GWAS dataset. Although the failure might due to different patterns of exposure to sun (60) or population stratification related to age of disease onset in different studies (the proportions of cases with early-onset age <40 were 20, 23, 26 and 47% for samples in MD Anderson dataset, GenoMEL phase 1 dataset, phase 2 dataset and Australian dataset, respectively (4,5,8,9)), further replication in other population is warranted. Second, because little is known about the functions of DUSP14 and MAFF genes involved in CM development, the biological significance of our findings needs to be investigated by additional functional studies of these two genes. Third, because we have mainly focused on SNPs with a significant level of ≤0.001 and putative functions, the causative SNPs that could not be predicted by currently available in silico tools might have been missed in this study. Further integration of other biological information and prediction tools may be needed to mine the available GWAS datasets more comprehensively to identify additional CM-associated SNPs or genes in the future.
In conclusion, our re-analysis of published CM GWAS datasets identified multiple functional SNPs in DUSP14 (rs1051849) and MAFF/PLA2G6 (proxy SNP rs4608623), which were associated with reduced CM risk in a non-Hispanic population, possibly by a mechanism of altering corresponding mRNA expression. Additional gene-based tests also supported the association between the related genes and CM risk. Further functional studies and replication in other populations are warranted to confirm our findings. This study indicates that re-analyzing SNPs with a moderate significance level in functional pathways might be a useful approach complementary to published GWAS studies.
Supplementary material
Supplementary Figures S1 and S2, Tables S1–S6 and full author list can be found at http://carcin.oxfordjournals.org/.
Funding
Melanoma Research Alliance, the National Institutes of Health/National Cancer Institute (CA88363, CA83115, CA122838, CA87969, CA055075, CA100264, CA133996 and CA49449); the National Health and Medical Research Council of Australia (107359, 200071, 241944, 339462, 380385, 389927, 389875, 389891, 389892, 389938, 402761, 443036, 442915, 442981, 496610, 496675, 496739, 552485 and 552498); the Cancer Councils NSW, Victoria and Queensland, the Cancer Institute New South Wales, the Cooperative Research Centre for Discovery of Genes for Common Human Diseases, Cerylid Biosciences (Melbourne), the Australian Cancer Research Foundation, the Wellcome Trust (WT084766/Z/08/Z) and donations from Neville and Shirley Hawkins. National Health and Medical Research Council of Australia Fellowships scheme (N.K.H. and G.W.M.). S.M. is the recipient of a Career Development Award from the National Health and Medical Research Council of Australia (496674 and 613705). AEC is supported by fellowships from the Cancer Institute NSW and the NHMRC. Cancer Australia grant (1011143 to M.H.L.). GenoMEL has received substantial funding from the European Commission under the Sixth Framework Programme (Contract no: LSHC-CT-2006–018702) from the National Cancer Institute of the US National Institutes of Health (CA83115) and from Cancer Research UK (C588/A4994, C588/A10589 and C8216/A6129). National Institutes of Health (grants R01 CA100264, R01ES011740 and R01CA131274 to Q.W.; P50 CA093459 to E.G.; P30 CA016672 to MD Anderson Cancer Center and HHSN268200782096C to C.A.).
Supplementary Material
Acknowledgements
We thank Yawei Qiao, Min Zhao, Jianzhong He, Kejing Xu and Hongxia Ma for their laboratory assistance; Hongping Yu, Yujing Huang, Ming Yin and Ziyuan Zhou for their help in data analysis and Dakai Zhu for his technical support. The AMFS, Q-MEGA, Queensland Institute of Medical Research and endometriosis studies gratefully acknowledge its participants and the hard work of all its research interviewers, research assistants, technicians, project managers, data as well as sample managers and examiners. We thank Endometriosis Associations for supporting study recruitment and Sullivan Nicolaides and Queensland Medical Laboratory for pro bono collection and delivery of blood samples and other pathology services for assistance with blood collection.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- AMFS
Australian Melanoma Family Study
- CI
confidence interval
- CM
cutaneous melanoma
- DUSP
Dual-specificity phosphatase
- ERK
extracellular signal-regulated kinase
- FDR
false discovery rate
- GWAS
genome-wide association study
- JNK
c-jun N-terminal kinase
- LD
linkage disequilibrium
- MAF
minor allele frequency
- MAPK
mitogen-activated protein kinase
- miRNA
micro RNA
- mRNA
messenger RNA
- OR
odds ratio
- QC
quality control
- Q-MEGA
Queensland Study of Melanoma, Environment and Genetic Associations
- SNP
single nucleotide polymorphism
- VEGAS
Versatile Gene-Based Test for Genome-wide Association.
References
- 1. Howlader N, et al. SEER Cancer Statistics Review, 1975–2008, National Cancer Institute. Bethesda, MD, http://seer.cancer.gov/csr/1975_2008/, based on November 2010 SEER data submission, posted to the SEER web site, 2011 [Google Scholar]
- 2. Hayward N.K. (2003). Genetics of melanoma predisposition. Oncogene, 22, 3053–3062 [DOI] [PubMed] [Google Scholar]
- 3. Nan H, et al. (2011). Genome-wide association study identifies nidogen 1 (NID1) as a susceptibility locus to cutaneous nevi and melanoma risk. Hum. Mol. Genet., 20, 2673–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Amos C.I, et al. ; GenoMEL Investigators; Q-Mega Investigators; AMFS Investigators (2011). Genome-wide association study identifies novel loci predisposing to cutaneous melanoma. Hum. Mol. Genet., 20, 5012–5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barrett J.H, et al. ; GenoMEL Consortium (2011). Genome-wide association study identifies three new melanoma susceptibility loci. Nat. Genet., 43, 1108–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yang X.R, et al. (2010). Associations of 9p21 variants with cutaneous malignant melanoma, nevi, and pigmentation phenotypes in melanoma-prone families with and without CDKN2A mutations. Fam. Cancer, 9, 625–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Duffy D.L, et al. ; GenoMEL (2010). IRF4 variants have age-specific effects on nevus count and predispose to melanoma. Am. J. Hum. Genet., 87, 6–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bishop D.T, et al. (2009). Genome-wide association study identifies three loci associated with melanoma risk. Nat. Genet., 41, 920–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Macgregor S, et al. (2011). Genome-wide association study identifies a new melanoma susceptibility locus at 1q21.3. Nat. Genet., 43, 1114–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Witte J.S. (2010). Genome-wide association studies and beyond. Annu. Rev. Public Health, 31, 9–20 4 p following 20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Freedman M.L, et al. (2011). Principles for the post-GWAS functional characterization of cancer risk loci. Nat. Genet., 43, 513–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang K, et al. (2010). Analysing biological pathways in genome-wide association studies. Nat. Rev. Genet., 11, 843–854 [DOI] [PubMed] [Google Scholar]
- 13. Zhong H, et al. (2010). Integrating pathway analysis and genetics of gene expression for genome-wide association studies. Am. J. Hum. Genet., 86, 581–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Menashe I, et al. (2010). Pathway analysis of breast cancer genome-wide association study highlights three pathways and one canonical signaling cascade. Cancer Res., 70, 4453–4459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tomlinson I.P, et al. ; COGENT Consortium; CORGI Collaborators; EPICOLON Consortium (2011). Multiple common susceptibility variants near BMP pathway loci GREM1, BMP4, and BMP2 explain part of the missing heritability of colorectal cancer. PLoS Genet., 7, e1002105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dhillon A.S, et al. (2007). MAP kinase signalling pathways in cancer. Oncogene, 26, 3279–3290 [DOI] [PubMed] [Google Scholar]
- 17. Ding X.Z, et al. (2001). MEK/ERK-mediated proliferation is negatively regulated by P38 map kinase in the human pancreatic cancer cell line, PANC-1. Biochem. Biophys. Res. Commun., 282, 447–453 [DOI] [PubMed] [Google Scholar]
- 18. Xia Z, et al. (1995). Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis. Science, 270, 1326–1331 [DOI] [PubMed] [Google Scholar]
- 19. Davies H, et al. (2002). Mutations of the BRAF gene in human cancer. Nature, 417, 949–954 [DOI] [PubMed] [Google Scholar]
- 20. Goel V.K, et al. (2006). Examination of mutations in BRAF, NRAS, and PTEN in primary cutaneous melanoma. J. Invest. Dermatol., 126, 154–160 [DOI] [PubMed] [Google Scholar]
- 21. Cunningham S.C, et al. (2006). Targeted deletion of MKK4 in cancer cells: a detrimental phenotype manifests as decreased experimental metastasis and suggests a counterweight to the evolution of tumor-suppressor loss. Cancer Res., 66, 5560–5564 [DOI] [PubMed] [Google Scholar]
- 22. Inamdar G.S, et al. (2010). Targeting the MAPK pathway in melanoma: why some approaches succeed and other fail. Biochem. Pharmacol., 80, 624–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Russo A.E, et al. (2009). Melanoma: molecular pathogenesis and emerging target therapies (Review). Int. J. Oncol., 34, 1481–1489 [DOI] [PubMed] [Google Scholar]
- 24. Jackson S, et al. (2005). No evidence for BRAF as a melanoma/nevus susceptibility gene. Cancer Epidemiol. Biomarkers Prev., 14, 913–918 [DOI] [PubMed] [Google Scholar]
- 25. Casula M, et al. ; Italian Melanoma Intergroup Study (2004). BRAF gene is somatically mutated but does not make a major contribution to malignant melanoma susceptibility: the Italian Melanoma Intergroup Study. J. Clin. Oncol., 22, 286–292 [DOI] [PubMed] [Google Scholar]
- 26. James M.R, et al. (2004). Epidermal growth factor gene (EGF) polymorphism and risk of melanocytic neoplasia. J. Invest. Dermatol., 123, 760–762 [DOI] [PubMed] [Google Scholar]
- 27. James M.R, et al. (2005). BRAF polymorphisms and risk of melanocytic neoplasia. J. Invest. Dermatol., 125, 1252–1258 [DOI] [PubMed] [Google Scholar]
- 28. Falchi M, et al. (2009). Genome-wide association study identifies variants at 9p21 and 22q13 associated with development of cutaneous nevi. Nat. Genet., 41, 915–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Newton-Bishop J.A, et al. (2010). Melanocytic nevi, nevus genes, and melanoma risk in a large case-control study in the United Kingdom. Cancer Epidemiol. Biomarkers Prev., 19, 2043–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stranger B.E, et al. (2007). Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science, 315, 848–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li Y, et al. (2010). MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol., 34, 816–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xu Z, et al. (2009). SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res., 37, W600–W605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cust A.E., et al. (2009) Population-based, case-control-family design to investigate genetic and environmental influences on melanoma risk: Australian Melanoma Family Study. Am. J. Epidemiol., 170, 1541–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Baxter A.J, et al. (2008) The Queensland Study of Melanoma: Environmental and Genetic Associations (Q-MEGA); Study Design, Baseline Characteristics, and Repeatability of Phenotype and Sun Exposure Measures. Twin. Res. Hum. Genet., 11, 183–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Purcell S, et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Miller K.L, et al. (2006). XPA, haplotypes, and risk of basal and squamous cell carcinoma. Carcinogenesis, 27, 1670–1675 [DOI] [PubMed] [Google Scholar]
- 37. Peng G, et al. (2010). Gene and pathway-based second-wave analysis of genome-wide association studies. Eur. J. Hum. Genet., 18, 111–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu J.Z, et al. ; AMFS Investigators (2010). A versatile gene-based test for genome-wide association studies. Am. J. Hum. Genet., 87, 139–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pruim R.J, et al. (2010). LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics, 26, 2336–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lascorz J, et al. (2010). Genome-wide association study for colorectal cancer identifies risk polymorphisms in German familial cases and implicates MAPK signalling pathways in disease susceptibility. Carcinogenesis, 31, 1612–1619 [DOI] [PubMed] [Google Scholar]
- 41. Bermudez O, et al. (2010). The dual-specificity MAP kinase phosphatases: critical roles in development and cancer. Am. J. Physiol., Cell Physiol., 299, C189–C202 [DOI] [PubMed] [Google Scholar]
- 42. Keyse S.M. (2008). Dual-specificity MAP kinase phosphatases (MKPs) and cancer. Cancer Metastasis Rev., 27, 253–261 [DOI] [PubMed] [Google Scholar]
- 43. Marti F, et al. (2001). Negative-feedback regulation of CD28 costimulation by a novel mitogen-activated protein kinase phosphatase, MKP6. J. Immunol., 166, 197–206 [DOI] [PubMed] [Google Scholar]
- 44. Shada A.L, et al. (2010). Interface of signal transduction inhibition and immunotherapy in melanoma. Cancer J., 16, 360–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bai L, et al. (2004). ZBP-89-induced apoptosis is p53-independent and requires JNK. Cell Death Differ., 11, 663–673 [DOI] [PubMed] [Google Scholar]
- 46. Hooks S.B, et al. (2008). Role of Ca2+-independent phospholipase A2 in cell growth and signaling. Biochem. Pharmacol., 76, 1059–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Höiom V, et al. (2011). Genome-wide linkage analysis of Swedish families to identify putative susceptibility loci for cutaneous malignant melanoma. Genes. Chromosomes Cancer, 50, 1076–1084 [DOI] [PubMed] [Google Scholar]
- 48. Motohashi H, et al. (2007). Carcinogenesis and transcriptional regulation through Maf recognition elements. Cancer Sci., 98, 135–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Blank V. (2008). Small Maf proteins in mammalian gene control: mere dimerization partners or dynamic transcriptional regulators? J. Mol. Biol., 376, 913–925 [DOI] [PubMed] [Google Scholar]
- 50. Kelemen L, et al. (2005). BRAF polymorphisms and the risk of ovarian cancer of low malignant potential. Gynecol. Oncol., 97, 807–812 [DOI] [PubMed] [Google Scholar]
- 51. Meyer P, et al. (2003). Polymorphisms of the BRAF gene predispose males to malignant melanoma. J. Carcinog., 2, 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Catela Ivkovic T, et al. (2009). Association of H-ras polymorphisms and susceptibility to sporadic colon cancer. Int. J. Oncol., 35, 1169–1173 [DOI] [PubMed] [Google Scholar]
- 53. Sathyan K.M, et al. (2006). Influence of single nucleotide polymorphisms in H-Ras and cyclin D1 genes on oral cancer susceptibility. Oral Oncol., 42, 607–613 [DOI] [PubMed] [Google Scholar]
- 54. Zhang Y, et al. (2008). Association between H-RAS T81C genetic polymorphism and gastrointestinal cancer risk: a population based case-control study in China. BMC Cancer, 8, 256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Xu W, et al. (2010). Association between EGF promoter polymorphisms and cancer risk: a meta-analysis. Med. Oncol., 27, 1389–1397 [DOI] [PubMed] [Google Scholar]
- 56. Lu P.H, et al. (2011). Association between mitogen-activated protein kinase kinase kinase 1 rs889312 polymorphism and breast cancer risk: evidence from 59,977 subjects. Breast Cancer Res. Treat., 126, 663–670 [DOI] [PubMed] [Google Scholar]
- 57. Wei Y, et al. (2009). The association between -1304T>G polymorphism in the promoter of MKK4 gene and the risk of sporadic colorectal cancer in southern Chinese population. Int. J. Cancer, 125, 1876–1883 [DOI] [PubMed] [Google Scholar]
- 58. Liu B, et al. (2010). A functional variant (-1304T>G) in the MKK4 promoter contributes to a decreased risk of lung cancer by increasing the promoter activity. Carcinogenesis, 31, 1405–1411 [DOI] [PubMed] [Google Scholar]
- 59. Zheng J, et al. (2012). The protective role of polymorphism MKK4 -1304 T>G in nasopharyngeal carcinoma is modulated by Epstein-Barr virus’ infection status. Int. J. Cancer, 130, 1981–1990 [DOI] [PubMed] [Google Scholar]
- 60. Tucker M.A, et al. (2003). Melanoma etiology: where are we? Oncogene, 22, 3042–3052 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.