Abstract
MicroRNAs (miRNAs) are small non-coding RNAs that can posttranscriptionally regulate gene expression by targeting messenger RNAs. During miRNA biogenesis, the star strand (miRNA*) is generally degraded to a low level in the cells. However, certain miRNA* express abundantly and can be recruited into the silencing complex to regulate gene expression. Most miRNAs function as suppressive regulators on gene expression. Group B coxsackieviruses (CVB) are the major pathogens of human viral myocarditis and dilated cardiomyopathy. CVB genome is a positive-sense, single-stranded RNA. Our previous study shows that miR-342-5p can suppress CVB biogenesis by targeting its 2C-coding sequence. In this study, we found that the miR-10a duplex could significantly up-regulate the biosynthesis of CVB type 3 (CVB3). Further study showed that it was the miR-10a star strand (miR-10a*) that augmented CVB3 biosynthesis. Site-directed mutagenesis showed that the miR-10a* target was located in the nt6818–nt6941 sequence of the viral 3D-coding region. MiR-10a* was detectable in the cardiac tissues of suckling Balb/c mice, suggesting that miR-10a* may impact CVB3 replication during its cardiac infection. Taken together, these data for the first time show that miRNA* can positively modulate gene expression. MiR-10a* might be involved in the CVB3 cardiac pathogenesis.
INTRODUCTION
MicroRNAs (miRNAs) are small non-coding RNAs that can posttranscriptionally regulate gene expression by interacting with the target messenger RNAs (mRNAs) (1). MiRNAs play a critical role in modulating cell differentiation, proliferation, apoptosis and various pathological processes including virus infection (1–9). Accumulated evidence shows that viral biosynthesis and replication can be regulated by cellular miRNAs (10–12). On the other hand, miRNAs encoded by certain viruses can also modulate the expression of their own as well as cellular mRNAs (13–15).
During the miRNA biogenesis, a RNA duplex of ∼22–24 nucleotides (nt) is generated in the cytoplasm from the double-stranded pre-miRNA by the cleavage of RNase III enzyme Dicer (16). A strand from the RNA duplex, termed the guide strand or the mature miRNA, is recruited into the Argonaute (AGO) complex and guided to complementary transcripts for regulation. The other strand, known as the star strand (miRNA*) or passenger strand, is degraded and maintained at a lower level in the cells (17–19). Therefore, it is generally believed that the guide strand regulates gene translation. However, studies revealed that certain miRNA* is expressed abundantly in the cells, and the miRNA/miRNA* ratio varies dramatically among developmental stages (7,20–22). Moreover, the miRNA* strand can also be recruited into the silencing complex and exert regulatory effect on gene expression (23).
While most miRNAs serve as suppressive regulators on gene expression, there are a few miRNAs, e.g. miR-10a (24) and miR-122 (25), with positive impact on the translation of their targets. MiR-10a targets the 5′-untranslated region (5′-UTR) of ribosomal protein mRNAs and reduces the translational suppression of the ribosomal protein mRNAs when amino acid starvation occurs (24). MiR-122 can up-regulate hepatitis C virus (HCV) replication by targeting the 5′-UTR of HCV genome (25). MiR-122 is the most abundant miRNA in the liver, and thus, it is widely accepted that miR-122 is one of the tissue tropism determinants of HCV infection (25). It is possible that the different species of miRNAs exert different influences on the translation of their targets. In addition, the involvement of either guide or star strand in the RNA silencing complex brings more complexities to the functions of miRNAs.
Group B coxsackieviruses (CVB), including six serotypes (CVB1–CVB6), are the human enterovirus B species of the Picornaviridae family (26). CVBs are the major pathogens of human viral myocarditis that can lead to dilated cardiomyopathy and cardiac failure (27–30). CVB genome is an ∼7.4-kb positive-sense single-stranded RNA (+ssRNA). CVB genome is composed of three parts: the 5′-UTR, the single open reading frame (ORF) and the 3′-UTR (31). The 5′-UTR plays a critical role in guiding the processes of virus translation and replication (32). The ORF encodes a polyprotein that is processed into the capsid proteins and non-structural proteins via a series of cleavages by the viral proteases 2A and 3C (32). Because of its positive polarity nature, theoretically, CVB genome can be a direct target of cellular miRNAs. Indeed, our previous study showed that miR-342-5p could suppress the biogenesis and replication of CVBs by targeting its 2C-coding sequence (33).
In the present study, we initially found a surprising effect of the miR-10a duplex that could significantly up-regulate the biosynthesis of CVB3 when we screened the miRNAs expressed in mouse cardiac tissues. Further study showed that, unlike miR-122 and miR-10a, it was the star strand of miR-10a (miR-10a*) that augmented the CVB3 biosynthesis. The target sequence of miR-10a* was located in the 3D-coding region of CVB3 genome. These findings for the first time show that the miRNA* can also positively modulate gene expression. MiR-10a* might be involved in the pathogenesis of CVB3 cardiac infection.
MATERIALS AND METHODS
Cells and mice
HeLa cells were cultured in Dulbecco Modified Eagle Medium (DMEM) (Invitrogen, Carlsbad, CA, USA) supplemented with 10% (growth medium) or 5% (maintaining medium) fetal bovine serum (FBS) (Biologica Industries, Israel), 50 U/ml penicillin and 50 mg/ml streptomycin at 37°C with 5% CO2. Pathogen-free 1–3-week-old Balb/c mice were obtained from the Harbin Medical University Experimental Animal Center. Mice were maintained and sacrificed in accordance with the Regulations on the Usage of Experimental Animals of Harbin Medical University.
Viruses
Two CVB3 variants, EGFP-CVB3 and RLuc-CVB3, which expressed enhanced green fluorescence protein (EGFP) and Renilla luciferase (RLuc), respectively, were constructed previously based on pMKS1, a plasmid containing the full-length CVB3 genomic complementary DNA (cDNA) (34). CVB3 H3 strain was also used in this study. The viruses were titered by plaque assay as described previously (35,36).
RNA transfection
The miRNAs, including miR-10a (5′-UACCCUGUAGAUCCGAAUUUGUG-3′), miR-10a* (5′-CAAAUUCGUAUCUAGGGGAAUA-3′), miR-10a duplex, mock miRNA (miR-mock) and so forth, were synthesized by GenePharma (Shanghai, China). The anti-miR-10a oligonucleotide (AMO-10a), a complementary sequence against the miR-10a guide strand (5′-CACAAAUUCGGAUCUACAGGGUA-3′) with a 2′-O-methyl group, was also synthesized by GenePharma. HeLa cells were transfected with miRNAs as described previously (33,37). For example, in a 12-well plate, HeLa cells were seeded at the density of 1 × 105 cells/well in DMEM with 5% FBS, 50 U/ml penicillin and 50 mg/ml streptomycin and cultured at 37°C with 5% CO2 for 18 h. Before transfection, the culture medium was replaced by antibiotics-free medium. Equal volume (2 μl) of Lipofectamine 2000 (Invitrogen) and miRNA were mixed in 100 μl of Opti MEM (Invitrogen) at room temperature for 15 min, and then pipetted to the cells. The transfected cells were cultured at 37°C with 5% CO2 untill the preset time points.
Luciferase assay
RLuc and Firefly luciferase (Luc) activities were measured by the Luc assay reagents (Promega, Madison, WI, USA) following the manufacturer’s protocol. To measure the RLuc expression in the cells infected with RLuc-CVB3, briefly, HeLa cells in 48-well plates (n = 6) were transfected with 20 pmol/well of miRNA using Lipofectamine 2000. The transfected cells were infected with 0.01 multiplicity of infection (MOI) of RLuc-CVB3. Before testing the RLuc expression, the cells were washed with phosphate-buffered solution, and vigorously mixed with 1× RLuc Lysis Buffer (80 μl/well) for 15 min at room temperature. The cell lysate (20 μl) was thoroughly mixed with 100 μl of RLuc Assay Reagent for ∼1–2 s. The mixture was applied to RLuc detection with a bioluminometer 20/20 n (Turner BioSystems, Sunnyvale, CA, USA). All detections were conducted at least three times.
Three Luc-expressing plasmids with the target sequences of miR-10a, miR-10a* and miR-mock in the 3′-UTR of Luc gene were generated and designated as pLuc-10a, pLuc-10a* and pLuc-mock, respectively, in this study. We first inserted a cytomegalovirus promoter upstream to the multiple cloning region of pGL4.17 (Promega). Then the target sequences of miR-10a (CACAAATTCGGATCTACAGGGTA), miR-10a* (TATTCCCCTAGATACGAATTTG) and miR-mock (TTGTACTACACAAAAGTACTG) were inserted to the Dra III–BamH I site in the 3′-UTR of Luc gene, respectively, by overlapping polymerase chain reaction (PCR). The modified plasmids were confirmed by sequencing (Supplementary Figure S1). The effect of miR-10a, miR-10a* and miR-mock on the pLuc-10a, pLuc-10a* and pLuc-mock was validated by determing the Luc expression in the cells with the plasmids and corresponding miRNAs. To determine the potential interference of the endogenous miR-10a and miR-10a* in HeLa cells, the HeLa cells in 48-well plates were co-transfected with pLuc-10a + pGL4.75 (Promega), pLuc-10a* + pGL4.75 and pLuc-mock + pGL4.75, respectively. Dual-Luc reporter reagents (Promega) were used to determine the RLuc and Luc expression. The RLuc expression was used as the transfection control.
EGFP quantitation
EGFP expression was quantitated by counting the EGFP-positive cell under a fluorescence microscope Axiovert 200 (Carl Zeiss, Gottingen, Germany) and by detecting the fluorescence intensity with a fluorescence spectrometer NanoDrop 3300 (Thermo, Rockford, IL, USA) as described previously (33,37).
Reverse transcription-quantitative polymerase chain reaction
Total RNA was extracted from HeLa cells or mouse tissues by TRIzol reagent (Invitrogen). The abundance of miRNAs and CVB3 genomic RNA was measured by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) assay as described previously (33). Briefly, 1 µg of total RNA was applied for reverse transcription with PrimeScript RT Enzyme Mix I (TaKaRa, Otsu, Shiga, Japan) and a reverse transcription primer. About 1 µl of the cDNA product was mixed with SYBR PrimeScript Ex Taq II (TaKaRa) and the primers in the final volume of 20 μl and detected with a LightCycler 2.0 (Roche, Basel, Switzerland). The 2−ΔΔCt method was used to calculate the relative abundance of miRNAs and viral genomic RNA (38). U6 small nuclear RNA (snRNA) was used as loading control for quantifying miRNA expression. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was used as loading control for measuring CVB3 RNA. The primers for RT-qPCR detection are listed in Supplementary Table S1.
miRNA target prediction
The potential targets of miR-10a* in the CVB3 genome were predicted by RNAHybrid 2.2 (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid) and Miranda 3.2a (http://www.microrna.org/microrna) according to the complementary sequence and minimum free energy (mfe) (39).
Site-directed mutagenesis
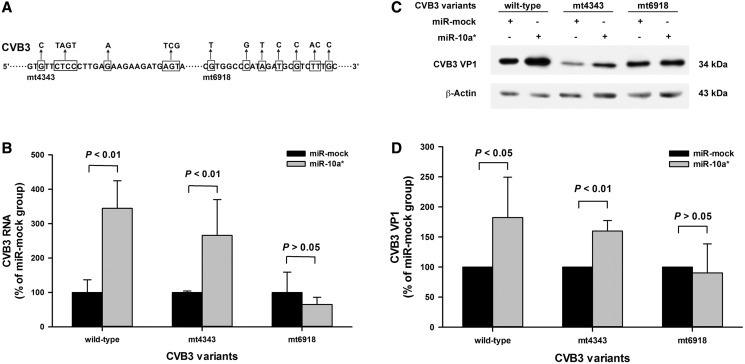
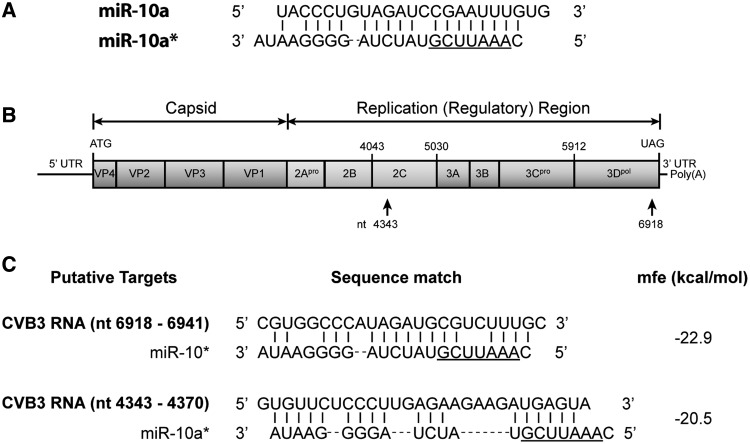
To verify the putative miR-10a* targets in the CVB3 genome, two CVB3 mutants were constructed with mutations at the putative target sites (Figure 5A). The nucleotides were replaced according to the codon degeneracy so that the mutated codons were synonymous with the wild-type. The site-directed mutagenesis assay was conducted by overlapping PCR as described previously (33). The primers were listed in Supplementary Table S2. The constructed mutants were designated as mt4343 and mt6918, respectively (Figure 5A). The sequences of the mutants were confirmed by sequencing. The mutants were recovered and purified by plaque assay in HeLa cells.
Figure 5.
The verification of miR-10a* targets in the CVB3 genome. (A) The putative miR-10a* target sequences in the CVB3 genome were mutated by site-directed mutagenesis. Two CVB3 mutants mt4343 and mt6918 were generated. The framed nucleotides were substituted to minimize the complementation between the putative targets and miR-10a*. (B) The effect of miR-10a* on the viral RNA expression of the mutants. HeLa cells were treated as described above. Total RNA of the cells was extracted and applied to RT-qPCR to detect the 5′-UTR of the CVB3 genome. The viral RNA level in the miR-10a*-treated cells was normalized to that in the miR-mock-treated cells. Error bars represent the SEs (n = 3). (C) The effect of miR-10a* on the capsid protein VP1 expression of the mutants. HeLa cells cultured in 6-well plates were transfected separately with miR-10a* and miR-mock, and infected with the mutants (MOI = 0.01) 24 h posttransfection. The cells were harvested for protein extraction and applied to western blot analysis with an anti-enteroviral VP1 antibody (clone 5-D8/1). (D) The VP1 expression was quantified according to the gray-scale intensity of VP1 blots in panel C. The VP1 levels in the miR-10a*-treated cells was normalized to that in the miR-mock-treated cells. Error bars represent the SDs (n = 3).
Western blotting
The western blotting for CVB3 capsid protein VP1 was performed as described previously (33). Briefly, HeLa cells were seeded in 6-well plates at the density of 2 × 105 cells/well and cultured in DMEM with 5% FBS for 24 h. The cells were transfected with 0.64 μg of miRNAs mixed with equal volume of Lipofectamine 2000, and infected with CVB3 variants (MOI = 0.01) at 24 h posttransfection. The infected cells were harvested at various time points and applied to protein extraction. About 10 μg of the protein lysates was subjected to sodium dodecylsulfate polyacrylamide gel electrophoresis. The separated proteins were transferred to a polyvinylidene fluoride membrane (Millipore, Billerica, MA, USA). A monoclonal mouse anti-enteroviral VP1 antibody (clone 5-D8/1) (Dako, Glostrup, Denmark) was used to detect CVB3 VP1. A horseradish peroxidase–conjugated goat anti-mouse IgG antibody (Millipore) was used as the secondary antibody. β-Actin was detected as loading control with a polyclonal antibody (sc-130301) (Santa Cruz Biotechnology, Santa Cruz, CA, USA). The blots were imaged with a charge-couple camera LAS4000 (Fujifilm, Tokyo, Japan). The experiments were repeated three times.
In vivo infection
Three-day-old Balb/c mice were peritoneally inoculated with 2 × 106 50% tissue culture infection dose (TCID50) of wild-type CVB3 (Wt) or mt6918. The myocardial tissues were collected at day 0, 1, 2 and 3 postinfection (p.i.). Total RNA and protein were extracted and detected by RT-qPCR and western blotting for CVB3 genomic RNA and VP1 capsid protein (n = 3).
Statistical analysis
Data are presented as mean ± standard deviation (SD) or mean ± standard error (SE). Statistical significance (P < 0.05) was determined by Student’s t-test using SigmapStat 3.1 (Systat Software, Richmond, CA, USA).
RESULTS
Endogenous miR-10a/miR-10a* expression is extremely low in HeLa cells
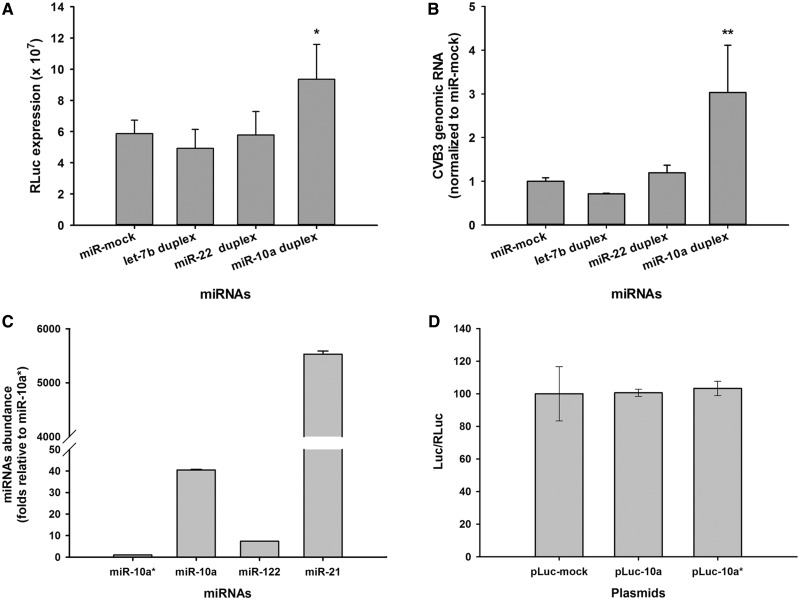
The abundance of miR-10a/miR-10a* in HeLa cells was quantitated to ensure that there was no interference from endogenous miR-10a/miR-10a*. The miR-122 and miR-21 were also detected as references. RT-qPCR detection showed that the abundance of miR-10a and miR-10a* was extremely low in HeLa cells (Figure 1C), while the quantitation of miR-122 and miR-21 in HeLa cells was consistent with previous studies (25,40–42). To confirm this detection, HeLa cells were co-transfected with pGL4.75 and pLuc-10a, pLuc-10a* and pLuc-mock, respectively. Dual-Luc assay showed that in the HeLa cells transfected with these plasmids, the Luc expression was basically identical to each other at 24 h posttransfection (Figure 1D), suggesting that there was no endogenous miR-10a/miR-10a* intervention on the Luc expression. Therefore, we concluded that the endogenous interference of miR-10a/miR-10a* could be excluded in HeLa cells.
Figure 1.
The effect of miR-10a duplex on CVB3 biosynthesis in HeLa cells. (A) HeLa cells in 96-well plates were transfected with various miRNA duplexes. The transfected cells were infected with RLuc-CVB3 (MOI = 0.01) 24 h posttransfection. The RLuc expression in the infected cells was detected 24 h p.i. with a bioluminometer. Error bars represent the SEs (n = 6). (B) HeLa cells were transfected with various miRNA duplexes and infected with RLuc-CVB3 as described above. The total RNA of the infected cells was extracted and detected for CVB3 genomic RNA with RT-qPCR assay. The RNA level of each sample was normalized to that of miR-mock-treated cells. Error bars represent the SDs (n = 4). (C) MiR-10a/miR-10a* abundance in HeLa cells. The total RNA of HeLa cells was extracted by TRIzol reagents. The miRNA abundance was detected by RT-qPCR assay with primers listed in Table S1 (Supplementary data). U6 snRNA was used as loading control. The miRNA abundance was normalized to that of miR-10a*. Error bars represent SEs (n = 4). (D) Quantitation of Luc expression in the HeLa cells transfected with pLuc-10a, pLuc-10a* by dual-Luc assay. The pLuc-10a, pLuc-10a* and pLuc-mock carried a Luc gene with the target sequences of miR-10a, miR-10a* and miR-mock in its 3′-UTR, respectively (Supplementary Figure S1). HeLa cells were co-transfected with pLuc-mock + pGL4.75, pLuc-10a + pGL4.75 and pLuc-10a* + pGL4.75, respectively. Dual-Luc assay was used to determine the Luc and RLuc expression at 24 h posttranfection. The relative Luc expression (Luc/RLuc) in the pLuc-10a- and pLuc-10a*-treated cells was normalized to that in the pLuc-mock-treated cells. Error bars represent SDs (n = 4). * and ** represent P < 0.05 and P < 0.01, respectively, compared with miR-mock group.
MiR-10a duplex significantly up-regulates the biosynthesis of RLuc-CVB3
HeLa cells cultured in 96-well plates were transfected with various miRNA mimics. The miRNA mimics were synthesized and delivered as duplex form for the sake of stability. The transfected cells were infected with RLuc-CVB3 (MOI = 0.01) at 24 h posttransfection. Luc assay showed that the RLuc expression was up-regulated ∼1.6 times in the infected cells with miR-10a duplex transfection at 24 h p.i. (P < 0.05, n = 6) (Figure 1A), while the RLuc expression was not significantly changed in the cells transfected with miR-mock, let-7b and miR-22.
Consistently, RT-qPCR showed that the CVB3 genomic RNA in the infected cells with miR-10a duplex was elevated about three times compared with that in the infected cells with miR-mock (P < 0.01, n = 4) (Figure 1B). These data show that miR-10a duplex could up-regulate the biosynthesis of RLuc-CVB3.
It is not miR-10a but rather miR-10a* that affects the biosynthesis of RLuc-CVB3
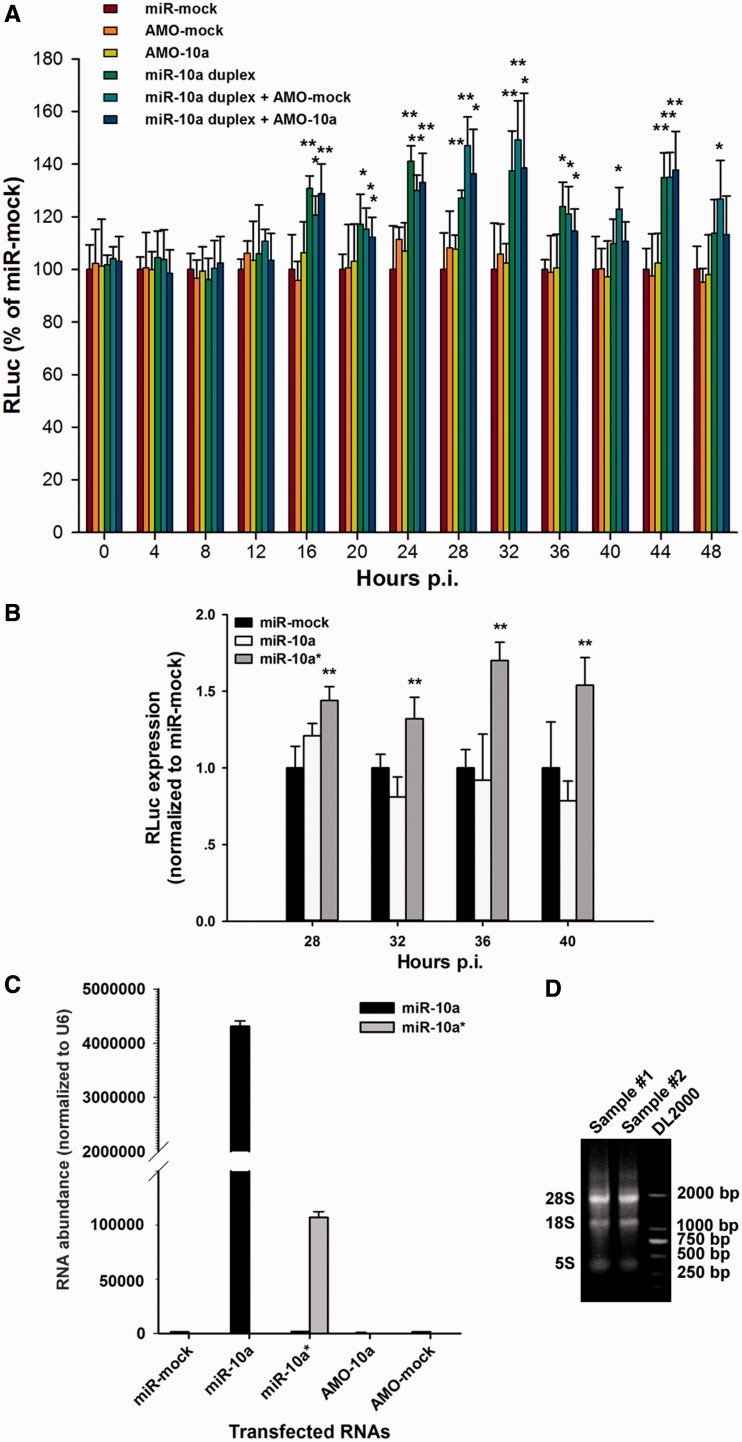
To verify the effect of miR-10a duplex on RLuc-CVB3, AMO-10a was used to suppress the effect of miR-10a. HeLa cells were transfected with miR-mock, AMO-mock, AMO-10a, miR-10a duplex, miR-10a duplex + AMO-mock, miR-10a duplex + AMO-10a. At 24 h posttransfection, the total RNA of these cells was extracted to detect the transfectant abundance. The ratio of 28 S/18S RNA was nearly 1.8 in our extractions (Figure 2D). High miR-10a and miR-10a* abundance was found in the cells transfected with miR-10a and miR-10a*, respectively (Figure 2C). The transfected cells were infected with RLuc-CVB3 (MOI = 0.01) at 24 h posttransfection and harvested to determine RLuc expression at 4-h intervals from 0 to 48 h p.i. (n = 6). At the time points from 16 h p.i., the RLuc levels in the cells transfected with miR-10a duplex were significantly elevated compared with that in the cells transfected with miR-mock (P < 0.01) (Figure 2A). However, the RLuc level in the cells transfected with miR-10a duplex + AMO-10a was also significantly increased at these time points compared with the mock group (Figure 2A). The fact that AMO-10a could not suppress the effect of miR-10a duplex in the CVB3-RLuc-infected cells suggested that it was not the miR-10a but rather the miR-10a* that regulated the RLuc-CVB3 biosynthesis.
Figure 2.
MiR-10a* up-regulates the biosynthesis of CVB3 in HeLa cells. (A) HeLa cells were transfected with miR-mock, AMO-mock, AMO-10a, miR-10a duplex, miR-10a duplex + AMO-mock and miR-10a duplex + AMO-10a. The cells were then infected with RLuc-CVB3 (MOI = 0.01), and harvested for RLuc detection at 4-h intervals from 0 to 40 h p.i. The RLuc level was normalized to that of the miR-mock-treated cells. Error bars represent SDs (n = 6). (B) HeLa cells were transfected with single-stranded miR-10a* or single-stranded miR-10a as described above. The cells were then infected with RLuc-CVB3 (MOI = 0.01), and harvested for RLuc detection at 4-h intervals from 28 to 40 h p.i. The RLuc level was normalized to that of the miR-mock-treated cells. Error bars represent SDs (n = 6). (C) The transfected RNA abundance in the treated HeLa cells. HeLa cells were transfected with miR-mock, single-stranded miR-10a, single-stranded miR-10a*, AMO-10a and AMO-mock. The miR-10a and miR-10a* abundance was detected by RT-qPCR at 24 h posttransfection and normalized to the U6 snRNA abundance in the cells. Error bars represent the SDs (n = 3). (D) The integrity of RNA extraction of HeLa cells. The total RNA was extracted from the transfected HeLa cells with TRIzol reagents. About 2.5 μg of RNA (sample #1 and #2) and 1 μg of DNA ladder DL2000 were loaded to 0.7% agarose gel for electrophoresis. The ratio of 28S/18S RNA was ∼1.8. * and ** represent P < 0.05 and P < 0.01, respectively, compared with the miR-mock-treated group by Student’s t-test.
To validate our speculation, the single-stranded miR-10a and miR-10a* were synthesized and applied to transfect HeLa cells. The transfected cells were infected with RLuc-CVB3 as described above. Interestingly, we found that the RLuc expression was elevated in the cells transfected with miR-10a* at 28, 32, 36 and 40 h p.i. (P < 0.01, n = 6), but did not in the cells transfected with miR-10 a and miR-mock (P > 0.05, n = 6) (Figure 2B). This observation confirmed that it was the miR-10 a* that affected the biosynthesis of RLuc-CVB3.
MiR-10 a* up-regulates the biosynthesis and replication of EGFP-CVB3
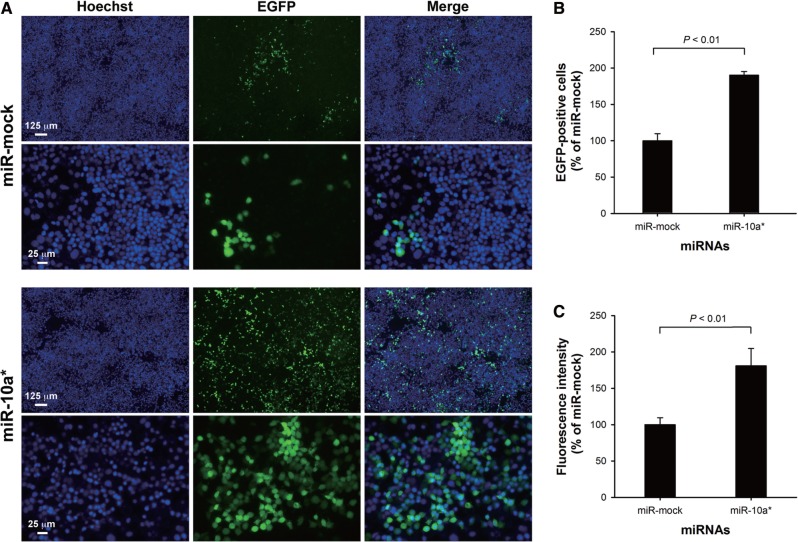
Although miR-10 a* could up-regulate the RLuc-CVB3 biosynthesis, there was a possibility that the up-regulation might be a consequence of the interaction between miR-10 a* and the reporter gene. To exclude the potential bias caused by the RLuc-coding sequence, EGFP-CVB3 variant (MOI = 0.01) was used to infect HeLa cells after the miR-10 a* transfection. The EGFP expression was observed at 32 h p.i. by fluorescence microscopy and spectrometry. Microscopic observation showed that more progeny viruses produced in the infected cells transfected with miR-10a*. The number of the EGFP-expressing cells in the miR-10a*-transfected group was ∼2.0-fold higher than that in the cells transfected with miR-mock (P < 0.01, n = 6) (Figure 3A and B). The EGFP fluorescence intensity in the cells transfected with miR-10a* was also ∼1.8-fold higher than that in the cells transfected with miR-mock (P < 0.01, n = 6) (Figure 3C). A further test using EGFP-expressing plasmid pEGFP-C1 and various miRNAs demonstrated that miR-10a* could not apparently affect the EGFP expression (Supplementary Figure S2). These results indicate that miR-10a* was the functional regulator on both the biosynthesis and replication of CVB3.
Figure 3.
The effect of miR-10a* on the biosynthesis of EGFP-CVB3. (A) MiR-10a* and miR-mock were separately transfected into HeLa cells. The transfected cells were infected with EGFP-CVB3 (MOI = 0.01) 24 h posttransfection. Hoechst 33342 was added to the culture medium to stain the nuclei at 12 h p.i. The EGFP expression in these cells was analyzed at 24 h p.i. (B) The EGFP-positive cell counts were first normalized to the number of nuclei in each sample. The relative counts of EGFP-positive cells of miR-10a*-treated groups were calculated by normalizing to the miR-mock-treated groups. Error bars represent the SDs (n = 6). (C) The EGFP intensity was first normalized to the Hoechst 33342 intensity of the same sample. The normalized EGFP intensities in the miR-10a*-treated cells were further normalized to that in the miR-mock-treated cells. Error bars represent the SDs (n = 6).
MiR-10a* target locates in the 3D-coding region of the CVB3 genome
To search the miR-10a* target in the CVB3 genome, two algorithms for miRNA target prediction, Miranda 3.2a and RNAhybrid 2.2, were used in this study. Based on the mfe scores, two potential miR-10a* targets were identified in the CVB3 genome: nt6918–nt6941 (mfe = −22.9 kcal/mol) and nt4343–nt4370 (mfe = −20.5 kcal/mol) (Figure 4). Both putative targets were located in the viral ORF. No potential target sequence was found in the untranslated regions. The nt6918–nt6941 sequence was located in the 3D (RNA polymerase)-coding region, while the nt4343–nt4370 sequence was located in the 2C-coding region. The modulation of a miRNA on its target mRNA is highly dependent on the seed sequence, which is usually composed of 7–8 nucleotides at the 5′-end of the miRNAs (23). Although the predicted targets have consecutive matching nucleotides against miR-10a* with low mfe values, they do not match well with the putative seed sequence of miR-10a* (Figure 4C).
Figure 4.
Prediction of the miR-10a* targets in the CVB3 genome. The genome of CVB3 H3 strain (Genbank accession: U57056) was scanned by Miranda 3.2a and RNAHybrid 2.2. The potential targets were ranked by two criteria: mfe and seed sequence matching. (A) The sequences of the miR-10a guide and star strands. The underlined region is the putative miR-10a* seed sequence. (B) The CVB3 genome and the locations of the putative miR-10a* targets. (C) The putative miR-10a* target sequences. The number of the nucleotide is the distance to the first nucleotide at the 5′-end of the CVB3 genome.
To verify the predicted targets of miR-10a*, two CVB3 mutants, designated as mt4343 and mt6918, were constructed with mutations at each of the putative targets (Figure 5A). The mutated codons were synonymous with the wild-type. Therefore, these mutations did not lead to the alteration of the encoded protein. Sequencing showed that the mutants carried the mutations exactly as we expected (Supplementary Figure S3).
Because these CVB3 mutants did not express a reporter, RT-qPCR and western blot were used to determine the viral genomic RNA and the capsid protein VP1 to evaluate the impact of miR-10a*. HeLa cells were transfected with miR-10a* and then infected with CVB3 mutants (MOI = 0.01) at 24 h posttransfection. RT-qPCR detection showed that, compared with the miR-mock-transfected cells, the viral RNA levels in the miR-10a*-transfected cells infected with the Wt and mt4343 were significantly increased (P < 0.05, n = 3), while the viral RNA level in the miR-10a*-transfected cells infected with mt6918 remained unchanged (P > 0.05, n = 3) (Figure 5B). Western blotting demonstrated that miR-10a* significantly up-regulated the VP1 expression in the cells infected with the Wt and mt4343 (P < 0.05 and P < 0.01, respectively, n = 3). However, the VP1 expression in the miR-10a*-transfected cells infected mt6918 was not elevated (P > 0.05, n = 3) (Figure 5B). These results suggest that the miR-10a* target located in the nt6918–nt6941 region of the CVB3 genome.
MiR-10a* is detectable in suckling mouse cardiac tissues
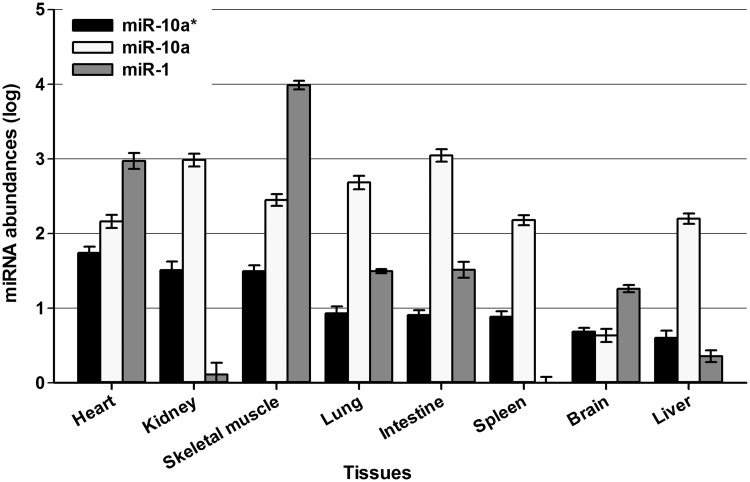
To estimate the impact of miR-10a* on CVB3 infection under in vivo scenario, the miR-10a* expression profile in mouse tissues was detected by RT-qPCR assay. Healthy 3-week-old Balb/c mice were anesthetized with phenobarbital sodium (100 mg/kg intraperitoneally). Organs including brain, heart, lung, liver, spleen, kidney, bone and intestine were quickly removed for total RNA extraction. The miR-1 expression was also detected as reference. The miR-1 and miR-10a expression detected in these tissues was in accordance with previous reports (43–45). Our detection showed that miR-10a* was detectable in the cardiac tissues of 3-week-old Balb/c mice, although its abundance was much lower than that of miR-1 (Figure 6). We also observed that the miR-10a* expression decreased in the cardiac tissues of adult Balb/c mice (8-week-old) compared with that of the suckling mice (1-week-old) (Supplementary Figure S4).
Figure 6.
The abundance of miR-10a* in various tissues of 3-week-old Balb/c mouse. The brain, heart, lung, liver, spleen, kidney, bone and intestine tissues were quickly harvested from phenobarbital sodium-anesthetized 3-week-old healthy Balb/c mice. The abundance of miR-10a*, miR-10a and miR-1 in these tissues was detected by RT-qPCR assay. The error bars represent the SEs (n = 3).
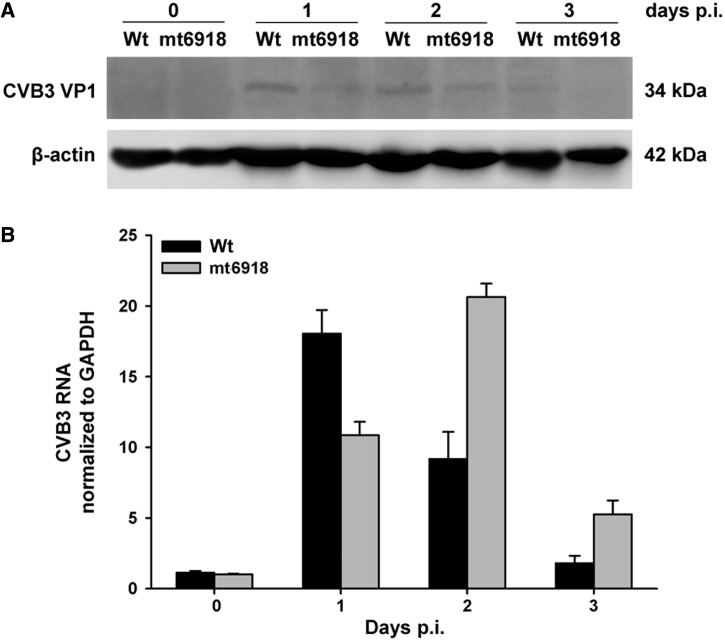
Furthermore, we tried to peritoneally infect 3-day-old Balb/c mice with 2 × 106 TCID50 of Wt and mt6918. In the myocardial tissues, western blotting showed that the VP1 protein expression in the mt6918-infected mice was continuously lower than that in the Wt-infected mice (Figure 7A). RT-qPCR showed that the viral RNA level in the mt6918-infected mice was lower at day 1 p.i. but higher at day 2 and 3 p.i. than that in the wilt-type CVB3-infected mice (Figure 7B). These data imply that although the viral RNA of mt6918 was more abundant, its protein biosynthesis was not as efficient as the Wt. It seems that there is a posttranscriptional regulation that promotes the protein synthesis of Wt under in vivo condition.
Figure 7.
The expression of CVB3 RNA and VP1 protein in the suckling mice infected with Wt and mt6918. Three-day-old Balb/c mice were peritoneally inoculated with 2 × 106 TCID50 of Wt and mt-6918. The myocardial tissues were collected at day 0, 1, 2 and 3 p.i. (A) CVB3 VP1 protein expression detected by western blotting. (B) RT-qPCR detection of CVB3 RNA. The error bars represent the SDs (n = 3).
DISCUSSION
MiRNA* is a by-product of the miRNA biogenesis. During miRNA maturation, the miRNA precursor is generated as a miRNA/miRNA* duplex (16). Usually, only one strand in the duplex is matured into the functional miRNA, while the star strand miRNA* is degraded afterwards (16). The mature miRNA is directed into the AGO complex, which enables the miRNA to interact with its target mRNA. However, in some cases, a substantial fraction of miRNA* species can also be actively sorted into AGO complex and repress the target mRNA (7,20–22). Although the star strand abundance of most miRNAs is less than that of the guide strands, in certain stages of development or in certain organs of adult organisms, the star strands of a few miRNAs were even more abundant than the guide strands (23).
Accumulated data indicate that miRNAs play important roles in the pathogenesis of virus infections (5,8,10). As a +ssRNA, the CVB genome is supposed to be the target of miRNAs. Our previous study demonstrated that miR-342-5p could significantly repress the biosynthesis and replication of CVB by targeting its 2C-coding sequence (33). In the present study, we surprisingly found that the miR-10a duplex could significantly up-regulate the CVB3 biosynthesis. We further clarified that it was the miR-10a* that modulated CVB3 by targeting its 3D-coding sequence. Our data suggest that miRNA* can also positively modulate gene expression.
The regulatory effect of miR-10a* was initially speculated because AMO-10a could not block the effect of miR-10a duplex on CVB3 biosynthesis (Figure 2). Therefore, we assumed that miR-10a* was the functional regulator for CVB3 biosynthesis and replication. Further study with the cells transfected with either single-stranded miR-10a or single-stranded miR-10a* demonstrated that miR-10a* could significantly promote CVB3 biosynthesis while miR-10a could not (Figures 2 and 3). Interestingly, although the AMO-10a sequence was similar to miR-10a*, AMO-10a could not modulate CVB3 biosynthesis (Figure 2A). It seems that the mismatched nucleotides between AMO-10a and miR-10a* are necessary for the interaction with CVB3 RNA.
To search the target of miR-10a* in CVB3 genome, we identified two stretches of sequences located in the 2C-coding region (nt4343–nt4370) and the 3D-coding region (nt6918–nt6941) of the CVB3 genome as the potential targets of miR-10a* (Figure 4). The 3D-coding region, which encodes viral RNA polymerase, is adjacent to the 3′-UTR (29). Site-directed mutagenesis demonstrated that the effect of miR-10a* on the CVB3 biosynthesis depended on the sequence of nt6918–nt6941 (Figure 5).
Generally, miRNAs function to repress the gene expression by interacting with the target mRNA (16). However, evidence shows that certain miRNAs are capable of positively regulating the expression of their target gene (24,25,46). In addition, the target sequences of miRNAs do not always reside in the 3′-UTR of mRNAs. For example, miR-122 can significantly promote HCV replication (25,46). The target of miR-122 is located in the 5′ UTR of HCV genome (47). MiR-10a can also interact with the 5′-UTR of ribosomal protein mRNAs and alleviate the translational repression of the ribosomal protein mRNAs during amino acid starvation (24). To our knowledge, there is no report about the up-regulating function of miRNA* on gene expression so far.
The role of miR-10a* in CVB3 infection in vivo is unclear yet. A recent study showed that miR-10a* expression was significantly up-regulated in the temozolomide (TMZ)-resistant glioblastoma multiforme cells (48). Suppression of miR-10a* did not affect the cell growth, but caused modest cell killing effect in the presence of TMZ (48). This finding indicates that miR-10a* is not merely a passenger RNA molecule but rather a functional RNA molecule in the cells.
MiRNA expression is tissue-dependent and the abundance of a particular miRNA may present a clue whether it functions in the tissue or not (20). The gut, heart and brain are the main organs that CVB3 tends to infect (30). We found that miR-10a* expression was detectable in the myocardial tissue of 3-week-old Balb/c mice (Figure 6), but its level decreased in the adult mouse heart (Supplementary Figure S4). The data of miR-10a* expressing in the neonatal mouse heart is consistent with the fact that the neonatal mouse is more sensitive to the CVB3 cardiac infection (30). However, it is hard to speculate the consequence of the miR-10a* impact on CVB3 cardiac infection. MiR-342-5p, which expresses in cardiac tissue, can interact with CVB3 and suppress its biosynthesis and replication (33). A recent study showed that viral infection could deregulate the cardiac miRNA expression (49). Among the up-regulated miRNAs, miR-155 in the infiltrating macrophages and T lymphocytes was recognized as an adverse mediator of cardiac inflammation. MiR-155 overexpression caused by viral infection could reduce myocardial damage during acute myocarditis (49). Therefore, the outcome of a viral infection is the synergistic and antagonistic consequence of various cellular and viral factors presented in the infected tissue.
To estimate the role of miR-10a* in the CVB3 in vivo infection, the wild-type and mt6918 CVB3 strains were peritoneally inoculated to 3-day-old Balb/c mice. In the myocardial tissues, the VP1 protein levels in the mt6918-infected mice were continuously lower than that in the Wt-infected mice (Figure 7A), while the viral RNA levels in the mt6918-infected mice were higher than that in the Wt-infected mice except the time point of day 1 p.i. (Figure 7B). It is unclear why the mt6918 RNA level was not consistent with its protein expression. Nonetheless, the inconsistency between the viral translation and transcription suggests a posttranscriptional regulation involving in the infection. This is a preliminary observation. Further in vivo investigation is needed to elucidate the role of miR-10a* in the myocardial pathogenesis of CVB3.
To monitor the viral biosynthesis, RLuc and EGFP were integrated into the genome of CVB3 in this study. A fundamental question is whether there is target sequence of miR-10a* in these reporter genes. Based on bioinformatics analysis, we could not find miR-10a* target sequence in these reporter genes. In vitro tests also confirmed that miR-10a* could not significantly affect the expression of RLuc and EGFP genes (Supplementary Figure S2). Hence we concluded that there was no miR-10a* target in the RLuc and EGFP genes.
Taken together, in this study we demonstrated that miR-10a* could up-regulate CVB3 biosynthesis by targeting its 3D-coding sequence. MiR-10a* may play a role in the pathogenesis of CVB3 infection. This finding suggests a notion that both miRNAs and their star strands may play important roles in the diseases caused by viral infections.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1, Supplementary Figures 1–4 and 2.
FUNDING
Natural Science Foundation of China (NSFC) [30872231 and 81271825 to Z.Z.], [81101234 to T.L., 31270198 to W.Z. and 81101235 to Y.W.]. Funding for open access charge: NSFC [30872231 and 81101234].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Heilongjiang Provincial Key Laboratory of Pathogens and Immunity, Harbin 150081, China, for laboratorial support.
REFERENCES
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 3.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 4.Barnes D, Kunitomi M, Vignuzzi M, Saksela K, Andino R. Harnessing endogenous miRNAs to control virus tissue tropism as a strategy for developing attenuated virus vaccines. Cell Host Microbe. 2008;4:239–248. doi: 10.1016/j.chom.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 6.Nilsen TW. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007;23:243–249. doi: 10.1016/j.tig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 7.Okamura K, Phillips MD, Tyler DM, Duan H, Chou Y, Lai EC. The regulatory activity of microRNA star species has substantial influence on microRNA and 3' UTR evolution. Nat. Struct. Mol. Biol. 2008;15:354–363. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ. Res. 2007;100:416. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- 9.Yeung ML, Bennasser Y, Jeang KT. miRNAs in the biology of cancers and viral infections. Curr. Med. Chem. 2007;14:191–197. doi: 10.2174/092986707779313417. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh Z, Mallick B, Chakrabarti J. Cellular versus viral microRNAs in host–virus interaction. Nucleic Acids Res. 2009;37:1035. doi: 10.1093/nar/gkn1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hariharan M, Scaria V, Pillai B, Brahmachari SK. Targets for human encoded microRNAs in HIV genes. Biochem. Biophys. Res. Commun. 2005;337:1214–1218. doi: 10.1016/j.bbrc.2005.09.183. [DOI] [PubMed] [Google Scholar]
- 12.Umbach JL, Cullen BR. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 2009;23:1151. doi: 10.1101/gad.1793309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, Chi JTA, Braich R, Manoharan M, Soutschek J, Ohler U. A viral microRNA functions as an orthologue of cellular miR-155. Nature. 2007;450:1096–1099. doi: 10.1038/nature05992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gottwein E, Cullen BR. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe. 2008;3:375–387. doi: 10.1016/j.chom.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfeffer S, Zavolan M, Grässer FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C. Identification of virus-encoded microRNAs. Science. 2004;304:734. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 16.Liu N, Okamura K, Tyler DM, Phillips MD, Chung WJ, Lai EC. The evolution and functional diversification of animal microRNA genes. Cell Res. 2008;18:985–996. doi: 10.1038/cr.2008.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 18.Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 19.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 20.Ro S, Park C, Young D, Sanders KM, Yan W. Tissue-dependent paired expression of miRNAs. Nucleic Acids Res. 2007;35:5944. doi: 10.1093/nar/gkm641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jagadeeswaran G, Zheng Y, Sumathipala N, Jiang H, Arrese E, Soulages J, Zhang W, Sunkar R. Deep sequencing of small RNA libraries reveals dynamic regulation of conserved and novel microRNAs and microRNA-stars during silkworm development. BMC Genomics. 2010;11:52. doi: 10.1186/1471-2164-11-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA. 2010;16:43. doi: 10.1261/rna.1972910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT, Lai EC. The regulatory activity of microRNA* species has substantial influence on microRNA and 3' UTR evolution. Nat. Struct. Mol. Biol. 2008;15:354–363. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5'UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 25.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 26.Kemball CC, Alirezaei M, Whitton JL. Type B coxsackieviruses and their interactions with the innate and adaptive immune systems. Future Microb. 2010;5:1329–1347. doi: 10.2217/fmb.10.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knowlton K. CVB infection and mechanisms of viral cardiomyopathy. Curr. Top. Microbiol. Immunol. 2008;329:315–335. doi: 10.1007/978-3-540-75546-3_15. [DOI] [PubMed] [Google Scholar]
- 28.Whitton JL. Immunopathology during coxsackievirus infection. Springer Semin. Immunopathol. 2002;24:201–213. doi: 10.1007/s00281-002-0100-4. [DOI] [PubMed] [Google Scholar]
- 29.Whitton JL, Cornell CT, Feuer R. Host and virus determinants of picornavirus pathogenesis and tropism. Nat. Rev. Microbiol. 2005;3:765–776. doi: 10.1038/nrmicro1284. [DOI] [PubMed] [Google Scholar]
- 30.Yajima T, Knowlton KU. Viral myocarditis: from the perspective of the virus. Circulation. 2009;119:2615–2624. doi: 10.1161/CIRCULATIONAHA.108.766022. [DOI] [PubMed] [Google Scholar]
- 31.Bedard KM, Semler BL. Regulation of picornavirus gene expression. Microbes Infect. 2004;6:702–713. doi: 10.1016/j.micinf.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 32.Sean P, Semler BL. Coxsackievirus B RNA replication: lessons from poliovirus. Curr. Top. Microbiol. Immunol. 2008;323:89–121. doi: 10.1007/978-3-540-75546-3_5. [DOI] [PubMed] [Google Scholar]
- 33.Wang L, Qin Y, Tong L, Wu S, Wang Q, Jiao Q, Guo Z, Lin L, Wang R, Zhao W, et al. MiR-342-5p suppresses coxsackievirus B3 biosynthesis by targeting the 2C-coding region. Antiviral Res. 2012;93:270–279. doi: 10.1016/j.antiviral.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 34.Tong L, Lin L, Zhao W, Wang B, Wu S, Liu H, Zhong X, Cui Y, Gu H, Zhang F, et al. Destabilization of Coxsackievirus B3 Genome Integrated with Enhanced Green Fluorescent Protein Gene. Intervirology. 2011;54:268–275. doi: 10.1159/000321351. [DOI] [PubMed] [Google Scholar]
- 35.Yuan J, Cheung PKM, Zhang H, Chau D, Yanagawa B, Cheung C, Luo H, Wang Y, Suarez A, McManus BM. A phosphorothioate antisense oligodeoxynucleotide specifically inhibits coxsackievirus B3 replication in cardiomyocytes and mouse hearts. Lab. Invest. 2004;84:703–714. doi: 10.1038/labinvest.3700083. [DOI] [PubMed] [Google Scholar]
- 36.Zhong Z, Li X, Zhao W, Tong L, Liu J, Wu S, Lin L, Zhang Z, Tian Y, Zhang F. Mutations at nucleotides 573 and 579 within 5'-untranslated region augment the virulence of coxsackievirus B1. Virus Res. 2008;135:255–259. doi: 10.1016/j.virusres.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 37.Wu X, Wu S, Tong L, Luan T, Lin L, Lu S, Zhao W, Ma Q, Liu H, Zhong Z. miR-122 affects the viability and apoptosis of hepatocellular carcinoma cells. Scand. J. Gastroenterol. 2009;44:1332–1339. doi: 10.3109/00365520903215305. [DOI] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2[-Delta Delta C(T)] method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Rajewsky N. microRNA target predictions in animals. Nat. Genet. 2006;38(Suppl.):S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 40.Li S, Zhu J, Fu H, Wan J, Hu Z, Liu S, Li J, Tie Y, Xing R, Sun Z, et al. Hepato-specific microRNA-122 facilitates accumulation of newly synthesized miRNA through regulating PRKRA. Nucleic Acids Res. 2012;40:884–891. doi: 10.1093/nar/gkr715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma L, Liu J, Shen J, Liu L, Wu J, Li W, Luo J, Chen Q, Qian C. Expression of miR-122 mediated by adenoviral vector induces apoptosis and cell cycle arrest of cancer cells. Cancer Biol. Ther. 2010;9:554–561. doi: 10.4161/cbt.9.7.11267. [DOI] [PubMed] [Google Scholar]
- 42.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beuvink I, Kolb FA, Budach W, Garnier A, Lange J, Natt F, Dengler U, Hall J, Filipowicz W, Weiler J. A novel microarray approach reveals new tissue-specific signatures of known and predicted mammalian microRNAs. Nucleic Acids Res. 2007;35:e52. doi: 10.1093/nar/gkl1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang B, Lin H, Xiao J, Lu Y, Luo X, Li B, Zhang Y, Xu C, Bai Y, Wang H, et al. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat. Med. 2007;13:486–491. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]
- 46.Roberts AP, Lewis AP, Jopling CL. miR-122 activates hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucleic Acids Res. 2011;39:7716–7729. doi: 10.1093/nar/gkr426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jopling CL, Yi MK, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 48.Ujifuku K, Mitsutake N, Takakura S, Matsuse M, Saenko V, Suzuki K, Hayashi K, Matsuo T, Kamada K, Nagata I, et al. miR-195, miR-455-3p and miR-10a* are implicated in acquired temozolomide resistance in glioblastoma multiforme cells. Cancer Lett. 2010;296:241–248. doi: 10.1016/j.canlet.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 49.Corsten MF, Papageorgiou A, Verhesen W, Carai P, Lindow M, Obad S, Summer G, Coort SL, Hazebroek M, van Leeuwen R, et al. MicroRNA profiling identifies microRNA-155 as an adverse mediator of cardiac injury and dysfunction during acute viral myocarditis. Circ. Res. 2012;111:415–425. doi: 10.1161/CIRCRESAHA.112.267443. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.