Abstract
MicroRNAs (miRNAs) constitute an important class of small regulatory RNAs that are derived from distinct hairpin precursors (pre-miRNAs). In contrast to mature miRNAs, which have been characterized in numerous genome-wide studies of different organisms, research on global profiling of pre-miRNAs is limited. Here, using massive parallel sequencing, we have performed global characterization of both mouse mature and precursor miRNAs. In total, 87 369 704 and 252 003 sequencing reads derived from 887 mature and 281 precursor miRNAs were obtained, respectively. Our analysis revealed new aspects of miRNA/pre-miRNA processing and modification, including eight Ago2-cleaved pre-miRNAs, eight new instances of miRNA editing and exclusively 5′ tailed mirtrons. Furthermore, based on the sequences of both mature and precursor miRNAs, we developed a miRNA discovery pipeline, miRGrep, which does not rely on the availability of genome reference sequences. In addition to 239 known mouse pre-miRNAs, miRGrep predicted 41 novel ones with high confidence. Similar as known ones, the mature miRNAs derived from most of these novel loci showed both reduced abundance following Dicer knockdown and the binding with Argonaute2. Evaluation on data sets obtained from Caenorhabditis elegans and Caenorhabditis sp.11 demonstrated that miRGrep could be widely used for miRNA discovery in metazoans, especially in those without genome reference sequences.
INTRODUCTION
miRNAs constitute an important class of small non-coding RNAs that regulate gene expression at the post-transcriptional level through sequence-specific base pairing (1). Most miRNAs are transcribed by the RNA polymerase II to generate primary miRNA (pri-miRNA) transcripts. For canonical miRNAs, the pri-miRNAs bearing one or more imperfect inverted repeats are cleaved by the RNase III enzyme Drosha to yield pre-miRNAs hairpins (2,3). Alternatively, pre-miRNAs can be generated from debranched short introns with hairpin-forming potential (mirtron) by the spliceosome complex (4–9), or can be derived from other small non-coding RNAs such as snoRNAs (9–16). After being transported into the cytoplasm by the exportin-5 complex (17,18), pre-miRNAs are further processed by another RNase III enzyme Dicer (19) into double-stranded miRNA:miRNA* duplexes, of which one strand is incorporated into the RNA-induced silencing complex (RISC) (20–22) and guides the effector complex to the target mRNA (1). In mammals, at least one-third of protein-coding genes are thought to be under miRNA regulation (23,24), and accumulating evidence has implicated miRNAs in an ever-increasing list of biological processes.
The early discovery of miRNA genes was achieved by Sanger sequencing of cDNAs cloned from small RNAs (25–27). Due to the limit of affordable sequencing depth, only abundant miRNAs could be identified while those with low expression or present only in a certain development stage or specific cell populations were difficult to detect. With the recent introduction of massive parallel sequencing technology, which can sequence DNAs orders of magnitude faster and at much lower cost, the detection sensitivity has been dramatically improved (28,29). For instance, using Illumina technology, modENCODE project has sequenced mature miRNAs from Drosophila melanogaster in unprecedented depth (14). In addition to profiling the expression of known miRNAs in different fly tissues, Berezikov et al. identified dozens of novel miRNA loci and elucidated several new features of miRNA biogenesis and post-transcriptional modifications (14).
In contrast to the analysis of mature miRNAs, attempts to profile pre-miRNAs are rather limited. To date, the expression patterns of known pre-miRNAs have been analysed by using northern blot, in situ hybridization and qPCR. Due to the relatively laborious procedure, such experiments have seldom been performed at the global level. The precise sequences of most, if not all, pre-miRNAs were not directly determined by sequencing experiments. Instead, they were often inferred from the sequences of the corresponding miRNA and miRNA*, therefore ambiguity could arise when the miRNA* was not identified. Most recently, Hammond’s lab has developed a high throughput sequencing method to profile pre-miRNAs, but only in a gene-specific manner (30). Burroughs et al. profiled pre-miRNAs in Hela cells through the application of locked nucleic acids and revealed 5′/3′arm variation including concomitant cleavage and polyuridylation patterns (31).
In this study, in order to gain a deeper understanding of mammalian miRNAs, we sequenced in parallel miRNAs and pre-miRNAs derived from 10 different tissues of adult mice. With the sequence information of both, we revealed several new aspects of processing and modification of known mouse miRNAs, including Ago2-cleaved pre-miRNAs, new instances of miRNA editing events, as well as exclusively 5′ tailed mirtrons. Furthermore, we developed a computational pipeline, miRGrep (miRNA Genome Reference free Prediction), to search for genuine miRNA genes solely based on our sequencing data set, without using genome sequences. Using miRGrep, 239 known mouse pre-miRNAs could be recovered and 41 novel ones were predicted with high confidence. Similar as known ones, the mature miRNAs derived from most of these novel loci showed reduced abundance following Dicer knockdown. Moreover, Argonaute2 immunoprecipitation (Ago2 IP) experiment confirmed that novel miRNAs could bind to Ago2/RISC complex. Evaluation on data sets obtained from Caenorhabditis elegans and Caenorhabditis sp.11 demonstrated that miRGrep could be widely used for miRNA discovery in different metazoans, especially in the absence of a reference genome.
MATERIALS AND METHODS
Cell culture and siRNA knockdown
Mouse Neuroblastoma (N2a) cells were cultured in DMEM supplemented with 10% FBS. siRNA duplex targeting the Dicer-1 ORF (Ambion s101208) was transfected into mouse N2a cells using Lipofectamine™ RNAiMAX (Invitrogen) as described in the manufacturer’s instructions. Cells were harvested 3 days after transfection, then total RNA was extracted and used for quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) or deep-sequencing.
To validate the efficiency of Dicer knockdown, cDNA was synthesized from 1 μg total RNA from unperturbed and Dicer silenced cells using SuperScript II according to the manufacturer’s instructions (Invitrogen). Quantitative PCR (qPCR) was carried out with SYBR Green assay (Applied Biosystems). mGAPDH (NM_008084) was used as endogenous control and amplified with the following primer pair: mGAPDH_F: 5′-AACTTTGGCATTGTGGAAGG-3′, mGAPDH_R: 5′-GGATGCAGGGATGATGTTCT-3′. Mouse Dicer1 mRNA (NM_148948) was detected with: mDICER1_F: 5′-ACCAAGTGATCCGTTTACGC-3′, mDICER1_R: 5′-CAACCGTACACTGTCCATCG-3′. The expression of Dicer was normalized to endogenous GAPDH mRNA level using the ΔΔCT method (32).
Argonaute2 immunoprecipitation
Mouse N2a cells were lysed in the buffer containing 20 mM Tris–HCl (pH 7.5), 150 mM NaCl, 0.25% NP-40 and 1.5 mM MgCl2.
For immunoprecipitation, anti-mouse Ago2 monoclonal antibody (Wako) was used. IP was performed as described before (33) except that Protein-G Dynabeads instead of Protein-G-sepharose beads were used. In brief, 100 μl Protein-G Dynabeads (Invitrogen) was washed with CPBT (citrate phosphate buffer pH 5.0 with 0.01% Tween 20) and incubated with anti-mouse Ago2 monoclonal antibody at room temperature with gentle agitation for 40 min. After washes with CPBT, beads were incubated with N2a cell lysate at 4°C with gentle agitation overnight. Then the antibody-coated beads were extensively washed with washing buffer (300 mM NaCl, 50 mM Tris-glycine pH 7.5, 5 mM MgCl2, 0.05% NP40) followed by a wash with PBS.
RNA extraction
Total RNA was isolated from adult mouse tissues (cerebellum, cortex, heart, kidney, liver, lung, ovary, skeletal muscle, spleen and testes), unsynchronous C. elegans as well as C. sp.11 sample containing worms of all stages and mouse N2a cells with/without siDicer treatment using TRIZOL reagent (Invitrogen) following the manufacturer’s protocol. RNA from Ago2 IP samples was isolated with 100 µg Proteinase K in 150 µl of Proteinase K buffer (300 mM NaCl, 200 mM Tris-glycine pH 7.5, 25 mM EDTA, 2% SDS) followed by Phenol/Chloroform extraction and Ethanol precipitation. After treating with DNase I (Ambion), we measured the RNA concentration by NanoDrop ND-1000 (Thermo). The total RNAs from 10 mouse tissues were pooled in equal amount for pre-miRNA sequencing (see below).
Small RNA sequencing libraries preparation
Small RNA sequencing libraries were prepared using Illumina small RNA library preparation kits. In brief, first, small RNA fraction with a size range of 10–40 nucleotide (nt) from 10 mouse tissues, unsynchronous C. elegans, unsynchronous C. sp.11 and mouse N2a cells as well as small RNAs of a size range of 50–100 nt from the mixture of 10 mouse tissues, unsynchronous C. elegans and unsynchronous C. sp.11 were separated using flashPAGE Fractionator (Ambion) according to the manufacturer’s instructions. Then, the small RNA fractions were ligated sequentially at the 3′ and 5′ end with synthetic RNA adapter, reverse transcribed and amplified using Illumina sequencing primers. Specifically, for 50–100 nt small RNA, the first strand cDNA synthesis was performed at 65°C for 50 min using SuperScript III reverse transcriptase (Invitrogen). Twelve cycles (98°C for 10 s, 60°C for 30 s and 72°C for 15 s) and 15 cycles (95°C for 10 s, 60°C for 30 s and 72°C for 30 s) of PCR amplification were performed on cDNAs derived from 10–40 nt RNA and 50–100 nt RNAs, respectively. The amplified libraries were subsequently purified by polyacrylamide gel electrophoresis (PAGE) according to the expected product size. Using the same procedure, the small RNA sequencing library was also constructed from Ago2 IP RNA.
Normalization of small RNA (50–100 nt fraction) sequencing library
The small RNA (50–100 nt fraction) sequencing library was normalized by using Duplex-specific Nuclease (DSN, Evrogen) according to the manufacturer’s instructions. Briefly, an aliquot (100 ng, 4 µl) of amplified small RNA (50–100 nt fraction) library was mixed with 1 µl of 4× hybridization buffer (200 mM HEPES pH 7.5, 2 M NaCl), overlaid with mineral oil, denatured at 98°C for 3 min and allowed to renature at 68°C for 5 h. After 5 h of incubation, 5 µl of 2× DSN master buffer (100 mM Tris–HCl pH 8.0, 10 mM MgCl2 and 2 mM dithiothreitol) preheated to 68°C was added to the reaction mixture and then incubated for 10 min. Next, 0.5 units of DSN enzyme were added to the reaction, and the incubation was continued for 25 min. DSN was subsequently inactivated by the addition of 10 µl of DSN stop solution (5 mM EDTA). After DSN inactivation, DNA was purified using the SPRI beads (Agencourt AMPure) and eluted in a final volume of 20 µl. An aliquot (5 µl) was used for PCR (95°C for 10 s, 60°C for 30 s and 72°C for 30 s, 12 cycles) with Illumina primers, followed by purification using the SPRI beads.
To validate the efficiency of DSN normalization, qPCR was carried out with SYBR Green Assays (Applied Biosystems). The primer sequences were listed in Supplementary Table S1. Five microlitre/ten nanograms of non-normalized or normalized sequencing libraries were used as PCR templates.
Small RNA sequencing
Small RNA (10–40 nt fraction) libraries for mouse and C. elegans were sequenced for 36 cycles, each on a separate lane, using Illumina GAIIx. Ago2 IP RNA library was sequenced for 50 cycles using Illumina HiSeq 2000. Normalized small RNA (50–100 nt) libraries were sequenced for 100 cycles, each on one lane, using Illumina HiSeq 2000. Both libraries (10–40 and 50–100 fraction) for C. sp.11 were sequenced for 100 cycles using Illumina HiSeq 2000.
Small RNA sequence reads mapping
First, 3′ adapter sequences were removed from the sequencing reads using an in-house Perl script. The reads of length between 17 and 30 nt from small RNA 10–40 nt fraction were retained. The reads from mouse and C. elegans samples were mapped to genome reference sequences (UCSC genome browser mm9 and ce6) and known pre-miRNA sequences deposited in miRBase (mouse and C. elegans, v16.0) (http://www.mirbase.org/) (34) without allowing any mismatch using soap1 and soap.short (35), respectively. To be considered as a known miRNA, the 5′ and 3′ ends of a sequencing read should be within 1 and 3 nt from the 5′ and 3′ ends of the miRNA annotated in miRBase v16.0, respectively. The 5′ or 3′ ends of 13 mouse miRNAs in miRBase v16.0 were manually corrected because: (i) 5′ ends of >90% of our sequencing reads mapped to the miRNA loci were at least 2 nt away from the annotated 5′ end. (ii) The corrected annotation of 5′ or 3′ ends could better fit with characteristics of miRNA biogenesis (Supplementary Table S2). For the small RNA 50–100 nt fraction, the sequencing reads of length between 40 and 94 nt were retained. After removing the last 5 nt at the 3′ end, which often contain the sequences with low quality, we aligned them to genome reference sequences (UCSC genome browser mm9 and ce6) allowing two mismatches using soap2 (36). To determine the mouse reads derived from full-length pre-miRNAs, we mapped the first 40 nt to the mouse pre-miRNA sequences deposited in miRBase v16.0 (34) allowing two mismatches using soap2 and then further extended the alignment to the 3′ end. The 5′ and 3′ ends of mouse pre-miRNAs in miRBase were manually annotated based on the secondary structure if miRNA* has not been identified (Supplementary Table S3). Reads to be considered as full-length pre-miRNAs should satisfy the following criteria: (i) the 5′ and 3′ end of the alignment were within 2 and 5 nt from 5′ and 3′ end of the pre-miRNA, respectively. (ii) No more than five mismatches were found in the alignment.
To predict miRNAs based on the sequencing reads obtained from the two small RNA fractions corresponding to potential miRNAs and pre-miRNAs, we mapped the sequencing reads of length between 17 and 30 nt on the sequencing reads of length between 40 and 94 nt using soap.short without allowing any mismatch.
The genome annotation of mouse and C. elegans non-miRNA/pre-miRNA sequencing reads was based on Ensembl Genes 59 for mouse and Ensembl Genes 52 for C. elegans (www.biomart.org) (37).
Identification of Ago2-cleaved pre-miRNA (ac-pre-miRNA)
After aligning the sequencing reads of length between 40 and 94 nt on known mouse pre-miRNA sequences as above, we applied the following filters to extract the reads derived from potential ac-pre-miRNAs: (i) Compared with annotated ends of pre-miRNAs, while one end of the alignment was within a distance of 2 nt, the other was truncated by 9–12 nt. (ii) The truncated part consisted of 8–10 nt that could form base pairs with the nucleotides on the other arm. (iii) No bulge located within 4 nt from the potential cleavage site. Finally, ac-pre-miRNA candidates should be supported by at least two reads.
Identification of miRNA-editing events
To examine the mouse miRNA-editing events, we mapped the non–genome-mapping reads from mouse small RNA (10–40 nt) libraries to mouse reference miRNA sequences, allowing one mismatch. The uniquely mapped reads with one mismatch at least 1 nt away from the 3′ or 5′ end of known miRNAs were retained. For each of the mismatches identified in these reads, we calculated the fraction of certain mismatch at one position as the number of reads bearing that mismatch divided by the number of all reads containing mismatches at the same position. We obtained a set of highly confident A-I editing sites by searching for A-G changes that could pass the following filters: (i) The fraction was higher than 90%. (ii) The change was found in at least 10 reads. (iii) The same change was found in at least one pre-miRNA read, and the Illumina sequencing quality score of that base was higher than 30. (iv) The same change was not annotated as an SNP in dbSNP (build 128). Editing frequency was calculated as the number of reads containing the edited A-G change divided by the total number of reads mapped to the same miRNA.
Extraction of potential pre-miRNA sequences
We selected the 40–94 nt (long) reads as potential pre-miRNAs on which the mapping pattern of 17–30 nt (short) reads was compatible with Dicer processing in the following four steps.
(i) On one long read, a cluster of mapped short reads was defined as all short reads with overlapping mapping positions, and the maximal distance between the start position of any two reads within one cluster did not exceed 14 nt. If the long and short reads were originated from genuine precursor and mature miRNAs, the short reads should form at most three clusters at the 5′ end, 3′ end and the middle of the long read, corresponding to the miRNA/miRNA* and the loop, respectively. Also, we would expect that the 5′ and 3′ end clusters contained many more short reads than the middle cluster. Furthermore, given the length distribution of canonical mature miRNAs, the majority of short reads from 5′ and 3′ end clusters should be of length between 17 and 25 nt. Therefore, based on these rules, we discarded the long reads if they had any of the following patterns:
Number of clusters exceeded 3.
Minimal distance between any two reads in different clusters was within 5 nt.
Number of reads in the middle cluster exceeded that in the 5′ end and 3′ end cluster.
Less than 66% of distinct/non-redundant short reads or <90% of all short reads from 5′ and 3′ end cluster were of length between 17 and 25 nt.
(ii) After filtering out the obvious non-Dicer compatible reads, we further selected the potential pre-miRNA reads. For each remaining long read, we first identified the most abundant distinct/non-redundant short reads from the 5′ and 3′ end clusters. The long reads were retained only if the most abundant short reads start or end <5 nt away from the 5′ or 3′ end of the long read, respectively. We then counted the number of short reads that start at most 1 nt away from the 5′ end of the most abundant reads in the 5′ and 3′ end clusters. The term ‘Sharpness’ denoted the percentage of these reads out of all short reads mapped on the same long read. Because most short reads that mapped to a genuine pre-miRNA should origin from miRNA/miRNA*, we selected long reads with a sharpness value above the threshold of 0.75. The selected reads were then clustered if (a) The most abundant distinct/non-redundant short reads mapped on their 5′ and 3′ clusters were identical, (b) They differed <5 nt in length and (c) Their sequence similarities were >90%. One representative read with the highest abundance from each cluster was selected.
(iii) We predicted the secondary structures of the selected long reads using RNAfold (parameters: −p −d 2 −noLP) (38) and randfold (parameter: −d 199) (39), respectively. Only the long reads that could fold into unbifurcated hairpin structures were retained.
(iv) The remaining long reads satisfying the following criteria were selected as potential pre-miRNA candidates. The rest were used as ‘background’ in the probabilistic scoring of potential pre-miRNA candidates.
(a) The randfold P-value was smaller than 0.2
(b) More than 60% of the nucleotides in the ‘mature’ part (the most abundant distinct/non-redundant short reads from 5′ or 3′ end clusters) were base paired.
Probabilistic scoring of potential pre-miRNA candidates
We scored the potential pre-miRNA candidates using a Naïve Bayesian classifier with six features:
-
f1: Minimal folding free energy calculated by RNAfold divided by the sequence length
f2: Randfold P-value
f3: Number of unpaired nucleotides at 5′ end
f4: Length of 3′ overhang (number of unpaired nucleotides at 3′ end minus that at 5′ end)
f5: Average length of the most abundant distinct/non-redundant short reads from the 5′ and 3′ end cluster that corresponded to potential miRNA/miRNA*
f6: Length of candidate pre-miRNA
The ‘positive training data set’ was pre-miRNAs from miRBase v16.0. We calculated the probability of a given potential pre-miRNA candidate to be a genuine pre-miRNA using the following formula:
-
Pr(pre|data) = P(data|pre) × P(pre)/[P(data|pre) × P(pre) + P(data|non) × P(non)]
where P(data|pre) = P(f1|pre) × P(f2|pre) × P(f3|pre) × P(f4|pre) × P(f5|pre) × P(f6|pre) and P(data|non) = P(f1|non) × P(f2|non) × P(f3|non) × P(f4|non) × P(f5|non) × P(f6|non)
P(pre) was the prior probability that a long read was a genuine miRNA precursor.
P(non) was the prior probability that a long read was non-miRNA background stem-loop and was equal to 1–P(pre). Both P(pre) and P(non) were set to 0.5 by default, but could be changed based on the expected pre-miRNA sequences in the deep sequencing samples.
P(f1|pre) to P(f6|pre) was the probability that a miRBase pre-miRNA had the value of f1–f6.
P(f1|non) to P(f6|non) was the probability that a background stem-loop sequence had the value of f1–f6.
Estimation of small RNA expression change upon Dicer knockdown
After removing 3′ adapter sequences using an in-house Perl script, we mapped the small RNA sequencing reads from the Dicer-silenced and the unperturbed N2a cells to the following sequences: tRNAs, rRNAs, known pre-miRNAs and our predicted novel pre-miRNAs using soap1 with the following options: −r 2 −v 0 −s 6 −n 1. The sequences of tRNA and rRNA were obtained from Ensembl Genes 59 (http://biomart.org/) (37). Six hundred seventy-two known mouse pre-miRNA sequences were downloaded from miRBase v16.0 (http://www.mirbase.org/) (34). The expression level of each annotated RNA was estimated as the number of reads mapped on the respective sequences. After adding a pseudo count of one, we calculated the log2 fold-change of expression level between Dicer-silenced and unperturbed samples for each RNA gene.
Estimation of small RNA abundance in Ago2 IP sample and input sample
The abundance of each annotated RNA in Ago2 IP and input sample, including tRNAs, rRNAs, known pre-miRNAs and our predicted novel pre-miRNAs, was estimated in the same way as described above.
In vitro transcription
Two novel pre-miRNA candidates (CandidateMMU32 and CandidateMMU33), with the same sequences as their respective pre-miRNA sequencing reads, were obtained by in vitro transcription, in which the templates were amplified from mouse genomic DNA using specific primer pairs linked with T7 promoter sequences (Supplementary Table S4) and purified from a 2.5% agarose gel using the QIAquick Gel Extraction kit (Qiagen). One microgram of each template was transcribed by T7 RNA polymerase (Promega) at 37°C for 2 h in the presence of UTP or [α-32P] UTP (25 µCi at concentration of 10 µCi/µl). In vitro transcribed RNAs were purified by phenol/chloroform extraction and precipitated with 100% isopropanol in the presence of 0.3 M ammonium acetate. Pellets were dissolved in RNase free water at the concentration of 20 000 cpm/µl.
In vitro recombinant Dicer (rDicer) processing assay
RNA was incubated with rDicer (Invitrogen) (0.1 U/reaction) in 75 mM NaCl, 20 mM Tris–HCl, pH 7.5, 3 mM MgCl2 and 0.1 U/µl RNase inhibitor at 37°C for 15, 30, 60 and 120 min. Processing products were resolved at 10% denaturing PAGE gel, which were then exposed for 1 h to a phosphorimaging screen and visualized on FLA 7000 imager (GE healthcare).
Northern Blot
Ten microgram of RNA with size of 10–200 nt from mouse liver and unsynchronized C. sp. 11 sample was separated on 15% denaturing PAGE gel and transferred to a nylon membrane (GE Healthcare) by semidry electro blot. After ultraviolet cross-linking, the membranes were pre-hybridized for 1 h, and hybridized overnight at 50°C with [γ-32P] ATP end-labelled probes. After hybridization, membranes were washed twice 10 min with 5× SSC and once 10 min with 1× SSC. All radioisotopic images were recorded using phosphorimaging screen on FLA 7000 imager (GE healthcare). The probe sequences were listed in Supplementary Table S20.
Taqman assay
To validate the expression of the novel miRNA, CandidateMMU33, TaqMan® MicroRNA Assay from Applied Biosystems was custom designed (Target Sequence: 5′-GGAGGAUUAUGUGUGACAGACA-3′) and used according to the manufacturer’s instructions. TaqMan® MicroRNA Assays from Applied Biosystems for a known miRNA, miR-137, as well as control assay for snoRNA-202, were also applied. In brief, the reverse transcription was carried with TaqMan® MicroRNA Reverse Transcription Kit (Applied Biosystems). The PCR mix for each reaction contained 1.33 µl of product from RT reaction, 10 μl of TaqMan 2 × Universal PCR Master Mix, No AmpErase UNG (Applied Biosystems), and 1 µl of 20 × TaqMan small RNA assay in a total volume of 20 μl. Standard reactions were performed using the following cycle parameters: AmpliTaq activation 95°C for 10 min; PCR: denaturation 95°C for 15 s and annealing/extension 60°C for 1 min (repeated 40 times). All experiments were carried out in triplicate.
Luciferase assay
The synthesized oligos containing sequences complementary to CandiMMU32 or CandiMMU33 (Supplementary Table S19) were annealed and inserted into the 3′UTR of the Renilla luciferase gene in psiCHECK-2 plasmid (Promega), using the XhoI and NotI sites. As a control, the oligos with scrambled sequences (Supplementary Table S19) were cloned into the same site. HEK293 cells were plated at 3 × 104/well in 96-well plates one day prior to transfection. The cells were then transfected with the psiCHECK2 reporter vector (10 ng), either without or together with in vitro transcripts of CandiMMU32 or CandiMMU33 (10 ng) using Lipofectamine 2000 (Invitrogen). After 24 h transfection, Firefly and Renilla luciferase activity were measured consecutively using the Dual-Glo Luciferase Assay System (Promega). Each plasmid was tested in three independent experiments. Renilla luciferase activity was normalized to that of firefly luciferase.
RESULTS AND DISCUSSIONS
miRNA and pre-miRNA sequencing
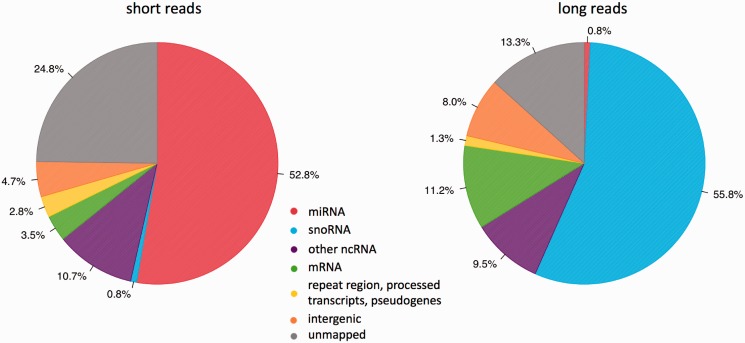
To profile mouse mature miRNAs, we sequenced small RNA (10–40 nt) libraries from 10 different mouse tissues (see methods) and obtained 167 million reads between 17 and 30 nt in length (‘short reads’) (Supplementary Table S5). Of these, 75.2% aligned to the mouse genome without mismatch and 52.8% derived from known mouse miRNA loci (Figure 1) (see methods).
Figure 1.
Genome annotation of sequencing reads from 10 mouse tissues. The pie charts depicted the percentage of 10–40 nt short reads (A) and 50–100 nt long reads (B) that were mapped to the indicated genome features based on miRBase v16.0 (34) and Ensembl Genes 59 (www.biomart.org) (37). ‘Other ncRNA’ included rRNA, tRNA, scRNA, snRNA and srpRNA.
In parallel, we also sequenced pre-miRNAs. To characterize as many pre-miRNAs as possible, we pooled the total RNA from the 10 mouse tissues equally and extracted small RNA between 50 and 100 nt in length. It is known that this RNA fraction contains a variety of other small RNAs such as highly abundant C/D box snoRNAs that have both 5′ (monophosphate) and 3′ (hydroxyl) ends (40) compatible with our small RNA cloning procedure. To reduce the representation of the extremely abundant non-pre-miRNA transcripts, we normalized the cDNA library before sequencing (see Supplemental Information and Supplementary Figure S1 for normalization efficiency) (41). With one lane of Illumina HiSeq 2000 sequencing run, we obtained >57 million reads between 40 and 94 nt in length (long reads), of which 86.7% could be mapped to the mouse genome (see methods). In contrast to mature miRNA sequencing, only 0.80% of the long reads were derived from known pre-miRNA loci whereas the vast majority was from snoRNAs (Figure 1).
In a recent attempt of unbiased pre-miRNA sequencing, Burroughs et al. obtained an even lower percentage (0.2%) of pre-miRNA sequencing reads after locked nucleic acids were used to exclude highly expressed classes of other noncoding RNAs before sequencing (31). In comparison, the PCR-based approach developed in Hammond’s lab has a much higher efficiency in increasing the fraction of pre-miRNA sequencing reads. With a primer pool directed at 219 miRNA precursors, >50% of their sequencing reads were mapped on the target pre-miRNA loci (30). However, this method can only be used to study known miRNAs in gene-specific manner.
Profiling of known miRNAs and pre-miRNAs
In total, 87 369 704 short reads matched 887 known mouse miRNAs and miRNAs* derived from 568 pre-miRNAs. Six hundred eighty-seven miRNA/miRNA* from 481 pre-miRNAs were expressed (defined as RPM >1; RPM: Reads Per Million total miRNA reads) in at least one tissue (Supplementary Table S5) (see methods). In comparison, only 252 003 long reads generated from the tissue mixture sample matched 281 full length known pre-miRNAs (Supplementary Table S5) (see methods). The distribution of length and RNA secondary structure of these 281 pre-miRNAs did not differ from that of all mouse pre-miRNAs deposited in miRBase, indicating that our detection of pre-miRNAs was not biased towards particular subsets of pre-miRNAs (Supplementary Figure S5). However, out of >50 million reads we obtained, pre-miRNAs constituted only <1% even after cDNA normalization. Furthermore, most of the 281 pre-miRNAs that we identified had only a few mapped reads. This might be explained by the combination of two following possibilities, i.e. (i) the abundance of most pre-miRNAs is indeed low, for they serve as an intermediate during miRNA maturation and most of them might be rapidly processed by Dicer, (ii) their stable hairpin structures make the cloning extremely inefficient. Very likely, the cloning efficiency would be different for different pre-miRNAs, depending on their sequences and/or structures. Therefore, it is worthy to note that the number of pre-miRNA sequencing reads should not be used to infer their cellular abundance.
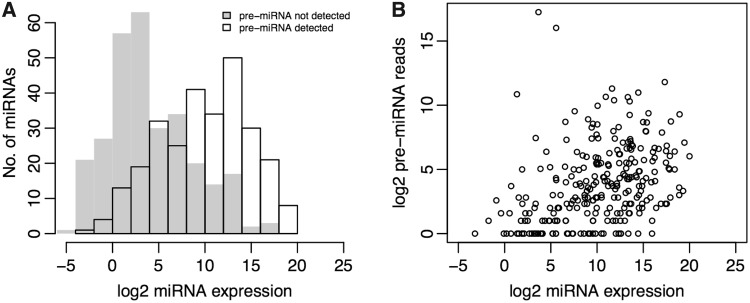
Two hundred seventy-eight out of 281 detected pre-miRNAs had the corresponding miRNA/miRNA* present in at least one tissue. As shown in Figure 2, miRNAs with their precursors detected were expressed at a significant higher level than those without (two-sided Wilcox rank-sum test, P < 2.2e-16), whereas the correlation between the abundance of miRNA and that of its precursor was low (R2 = 0.1501). Such low correlation was most likely due to the fact that the majority of the detected pre-miRNAs have only few mapped reads (Supplementary Figure S6), the different biases in cloning miRNAs versus the pre-miRNAs, and the variations in miRNA/pre-miRNA stability, although it might also be explained by the regulation of pre-miRNA processing via Dicer and its cofactors. The numbers of sequence reads were listed for each pre-miRNA and miRNA in Supplementary Table S6.
Figure 2.
Distribution of the miRNA and pre-miRNA abundance. (A) Histogram of the log2 transformed expression level (sum of RPMs from 10 tissues, RPM: Reads Per Million total miRNA reads) was shown for the known miRNAs. The miRNAs with precursor detected (black) expressed at a higher level than those without (grey). (B) Comparison of the miRNA and pre-miRNA abundance. Each circle represented one pre-miRNA locus. X-axis denoted the log2 transformed sum of miRNA/miRNA* RPM derived from the pre-miRNA locus in 10 tissues, Y-axis denoted the log2 transformed number of pre-miRNA sequencing reads.
Almost half of the long reads mapped to known miRNA loci did not cover full-length pre-miRNAs. Whereas most of these reads possibly represented degradation product from pre-miRNAs or pri-miRNAs, some were derived from miRNA-processing intermediates. For example, the long reads that missed one arm of the hairpin but contained the other arm and full loop sequences might have been abnormally processed by Dicer (31). Another kind of long reads matching pre-miRNAs at one end but truncated from the other end resembled an endogenous intermediate resulted from Ago2-mediated endonucleolytic cleavage within one arm of the hairpin precursor, which has been identified before in human cells and termed as Ago2-cleaved pre-miRNAs (ac-pre-miRNA) (42). In total, we found eight potential ac-pre-miRNAs in mouse (Table 1), out of which seven were from let-7 families. The cleavage sites were found always at the 3′ arms that also represented passenger strands (Supplementary Table S7). This finding was consistent with the proposed function of Ago2 cleavage at hairpin precursor, i.e. to facilitate removal of the nicked passenger strand from RISC after maturation. Interestingly, for all the ac-pre-miRNAs, we observed uridylation at the 3′ end, an additional evidence that our ac-pre-miRNA candidates represented more likely true processing intermediates than degradation products (Table 1). Moreover, we could demonstrate that the cleavage of pre-let-7 was indeed mediated by Ago2 (Supplementary Information).
Table 1.
Number of sequencing reads derived from full-length pre-miRNA, ac-pre-miRNAs, (poly-)uridylated ac-pre-miRNAs and the mature miRNAs at 5′ and 3′ arm
| miRNA | Full-length pre-miRNA | Ac-pre- miRNA | Uridylated ac-pre-miRNA | 5′ arm miRNA | 3′ arm miRNA |
|---|---|---|---|---|---|
| mmu-let-7a-1 | 46 | 25 | 10 | 1 682 015 | 420 |
| mmu-let-7b | 34 | 178 | 100 | 2 222 105 | 299 |
| mmu-let-7c-2 | 84 | 22 | 8 | 3 347 997 | 419 |
| mmu-let-7d | 6 | 2 | 2 | 389 631 | 7748 |
| mmu-let-7f-1 | 29 | 297 | 62 | 1 940 949 | 72 |
| mmu-let-7i | 93 | 49 | 2 | 111 553 | 1533 |
| mmu-mir-98 | 24 | 4 | 3 | 18 033 | 73 |
| mmu-mir-30b | 144 | 2 | 2 | 88 407 | 3040 |
Identification of miRNA-editing sites with high confidence
A-to-I editing has been reported in mammalian pre-miRNAs, and such events can affect miRNA biogenesis as well as target selection (43). To identify potential editing sites at mature miRNA sequences, we first searched for non–genome-mapping reads that can be uniquely mapped to miRBase precursor sequences with one mismatch. Given the prominent expression of adenosine deaminases (ADARs) in brain, we focused our analysis on two libraries generated from cerebellum and cortex. After excluding reads with mismatches at the 3′ end, the possible untemplated 3′ terminal modification, we obtained 292 573 (cortex) and 324 408 (cerebellum) reads, corresponding to 2.18% (cortex) and 2.38% (cerebellum) of the reads that were mapped to known miRNAs without mismatch. Of these reads, the percentage of those containing A-to-G change was 51% (cortex) and 54% (cerebellum). Most of these changes could be background noise caused by errors introduced during sequencing library preparation or sequencing process. To distinguish the true editing events from noise, we first calculated the fraction of a nucleotide change as the number of reads bearing that change divided by the number of all reads containing a mismatch at the same position. We then considered a nucleotide change as a good editing candidate if its fraction exceeded 90% and was found in at least 10 reads (see methods). Of the 165 274 (cortex) and 202 227 (cerebellum) candidate changes, A-to-G changes made up of 82% (cortex) and 82% (cerebellum). Finally, we took advantage of our pre-miRNA sequencing data and retained only the changes that were also found in at least one pre-miRNA sequencing reads with sufficient sequencing quality. After this final filtering, 36 280 (cortex) and 64 173 (cerebellum) A-to-G changes were found on 13 (cortex) and 11 (cerebellum) pre-miRNAs, accounting for 99.4% (cortex) and 99.8% (cerebellum) of all retained nucleotide changes (Supplementary Table S8). As listed in Table 2, nine editing sites were found both in cortex and cerebellum, but their editing frequencies were still different in these two different brain regions. Eleven of 15 sites located in the seed regions of miRNAs, which might affect selection of mRNA targets, as previously described for the miR-376 cluster (44). Of the remaining four sites outside of seed regions, the A-to-I editing at pre-mir-497 could affect its secondary structure by forming ‘I-C’ base pair with the cytosine on the opposite arm, thereby impacting the processing by Dicer and even loading into RISC.
Table 2.
Inferred A-to-I editing sites in miRNAs
| miRNA | Position | No. of pre-miRNA reads | Fraction edited (cortex) | Fraction edited (cerebellum) |
|---|---|---|---|---|
| mmu-mir-137-3p | 11 | 2 | 0.0053 | 0 |
| mmu-mir-151-3p | 3 | 5 | 0.1179 | 0.0266 |
| mmu-mir-186-5p | 3 | 1 | 0.0006 | 0 |
| mmu-mir-27a-5p | 7 | 14 | 0.1229 | 0 |
| mmu-mir-376a-5p | 4 | 2 | 0.0831 | 0.1115 |
| mmu-mir-376b-5p | 6 | 2 | 0 | 0.3781 |
| mmu-mir-376b-3p | 6 | 7 | 0.4612 | 0.4390 |
| mmu-mir-376c-3p | 6 | 3 | 0.2342 | 0.2603 |
| mmu-mir-378-3p | 16 | 1 | 0.0606 | 0.1814 |
| mmu-mir-381-3p | 4 | 1 | 0.3940 | 0.2278 |
| mmu-mir-384-5p | 4 | 1 | 0.0115 | 0.0096 |
| mmu-mir-497-3p | 20 | 104 | 0.8846 | 0.9769 |
| mmu-mir-540-5p | 3 | 1 | 0.2661 | 0.4810 |
| mmu-mir-770-5p | 4 | 1 | 0 | 0.0206 |
| mmu-mir-770-3p | 11 | 2 | 0.0023 | 0 |
Recently, by deep sequencing miRNAs from brain samples, Chiang et al. discovered 16 editing sites with editing frequency >5% (45). Seven of those sites were also identified in our study. Among the remaining nine sites, we observed A-to-I editing of eight sites (except mmu-mir-219-2-3 p) at the mature miRNA sequencing reads, but not at the respective pre-miRNA reads. Compared with those seven known editing sites, the eight new sites that we discovered in this study were generally edited with lower frequency (Supplementary Figure S7), indicating the capability of our approach in the identification of such editing events. In a complex tissue with extremely heterogeneous cell types such as brain, the miRNAs detected with low editing frequency in an only anatomically distinct region could well be much more significantly edited in a specific cell type. Also, depending on the miRNA expression level, even at low editing frequency, the absolute number of edited molecules can still be high enough to be functionally relevant. Therefore the new editing events that we have identified, although of low frequency, might well have significant effects.
miRGrep predicted 239 known and 41 novel mouse pre-miRNAs
To identify potential miRNAs based on both short reads (corresponding to miRNA/miRNA*) and long reads (corresponding to pre-miRNAs), we developed a computational pipeline, miRGrep. In miRGrep, after mapping the short reads to the long reads, potential pre-miRNA sequence candidates were extracted by selecting the long reads that could form a stable hairpin and had a pattern of mapped short reads compatible with Dicer processing. We then scored these candidates and finally reported a list of highly confident pre-miRNAs/miRNAs.
In more detail, we first investigated where short reads were mapped on long reads. If a long read and its mapped short reads represented a genuine pre-miRNA and its Dicer-processing products, the short reads should be clustered to a maximum of three positions, i.e. at 5′ end, 3′ end and the middle of the long read, corresponding to the miRNA/miRNA* and the loop, respectively. Given the rapid degradation of loop fragments, we would also expect that short reads from the 5′ and 3′ end clusters should be much more abundant than that from the middle cluster. After discarding the long reads that did not fit with such criteria and most likely arose from other small non-coding RNAs or degradation products from longer RNA transcripts, we predicted the secondary structure of the remaining long reads and kept only those that could form stable hairpin structures as potential candidates. These candidates were finally scored based on the characteristics of known miRNAs and pre-miRNAs, including pre-miRNA thermodynamic stability, 5′ and 3′ end duplex overhang as well as pre-miRNA and mature miRNA length (see methods, Supplementary Figures S8 and S9).
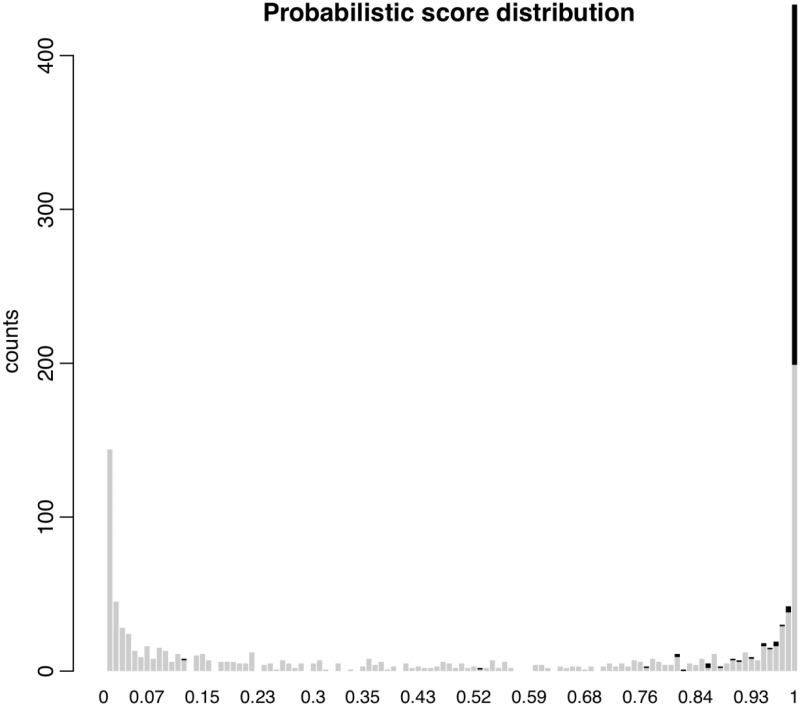
We applied miRGrep to predict mouse miRNAs based on our sequencing data set. In total, 155 760 811 short reads were perfectly mapped to 5 789 406 distinct long reads. From these, 5 524 656 long reads were discarded because the mapping position of short reads did not fit with the model of Dicer processing. The remaining 264 750 long reads were then merged into 131 207 clusters, and one representative read from each cluster was selected to predict the secondary structure (see methods). Out of these, 1277 long reads that could form stable hairpin structures entered the probabilistic scoring process. As a result, 538 long reads were scored >0.95 (Figure 3), of which 324 with at least five mapped short reads were retained as pre-miRNA candidates.
Figure 3.
Probabilistic score distribution of pre-miRNA candidates. Histogram showed the probabilistic score distribution of the potential pre-miRNA candidates using a Naïve Bayesian classifier with six features of known miRNA, including pre-miRNA minimal folding free energy, thermodynamic stability, 5′ and 3′ end duplex overhang as well as pre-miRNA and mature miRNA length. Known pre-miRNAs were shown in black, the remaining in grey.
Among these 324 pre-miRNA candidates, 245 corresponded to 239 known mouse pre-miRNAs (Supplementary Table S9). Forty-seven pre-miRNAs, which were detected but not reported by miRGrep, were filtered out at different stages of our prediction pipeline (Supplementary Table S10). Whereas most of these unreported known pre-miRNAs might represent false negatives, some could as well be falsely annotated in miRBase. For instance, miR-1944 and miR-715, which were not reported by miRGrep, have been recently removed from miRBase v18.0.
After mapping 79 novel candidates on the mouse genome reference sequence, 11 were found to derive from chimerical reads representing ligation artifacts (Supplementary Table S11). Another 27 were considered unlikely to be genuine pre-miRNA after further manual inspection by applying the stringent filtering criteria: (i) mature miRNAs with a relatively uniform 5′ terminus; (ii) absence of loop reads protruding into stems; (iii) number of unpaired nucleotides at 3′ and 5′ end compatible with Dicer processing; and (iv) absence of proximal reads suggesting the candidate is derived from degradation product (Supplementary Table S12).
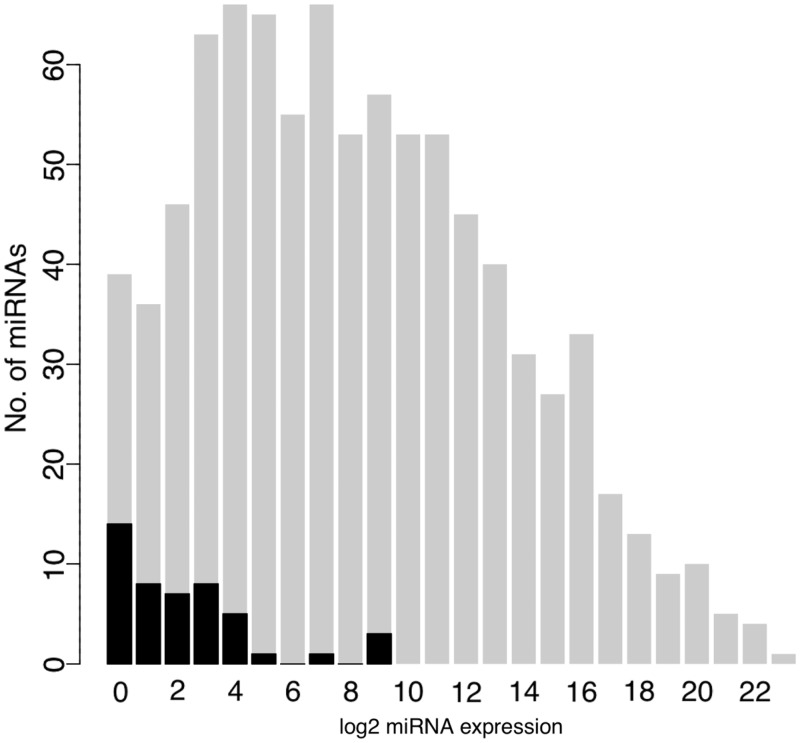
Of the 41 novel pre-miRNA candidates retained with high confidence, three were recently registered in miRBase v18.0. Two could be mapped to LINE, SINE or LTR repetitive loci, consistent with previous finding that miRNA genes could originate from transcribed transposons (46,47). Four located to known snoRNA loci. The similar finding has been reported in the study of human cells (10). The remaining 32 predicted novel pre-miRNAs mapped to either intergenic (3), exonic (1), exon–intron boundary (1) and intronic (27) regions of protein-coding genes. From the 41 novel pre-miRNAs, 66 miRNA/miRNA* were identified with short sequencing reads. Compared with known miRNAs, these novel miRNAs were expressed at a much lower level (Figure 4). Detailed information about these novel candidates can be found in Supplementary Table S13, and two examples were also shown in Figure 5.
Figure 4.
Abundance of known and novel mouse miRNAs. Histogram of the log2 transformed expression level (sum of sequencing reads in 10 mouse tissues) of known miRNAs (grey) and novel miRNAs (black) in 10 mouse tissues. Compared with known miRNAs, novel miRNAs expressed at a much lower level.
Figure 5.
Examples of novel pre-miRNAs discovered in this study. (A) A novel mouse pre-miRNA within the intron of the protein-coding gene Camk2a. (B) A novel mouse 5′ tailed mirtron derived from the protein-coding gene Ankrd23. (C) A novel C. elegans pre-miRNA from the 5′UTR of the protein-coding gene F35G12.2.
In total, 32 novel pre-miRNAs predicted in this study located to introns of protein-coding genes. Among these, 11 had both 5′ and 3′ end at least 10 nt away from the corresponding end of the introns. Of the remaining 21 intron-containing pre-miRNAs, whereas four had the ‘nearly’ exact boundary as the hosting introns and thus resembled canonical mirtrons, 17 had only one end generated by spliceosome while the other end likely matured through Drosha-independent trimming (6,7,48). Interestingly, all the pre-miRNAs from the latter category were 5′ tailed mirtrons, which shared only their 3′ ends with the hosting introns. Indeed, we also found the long reads possibly derived from the intermediate tailing products for several tailed mirtrons (Supplementary Figure S10). To check whether the tailed mirtrons in mouse were exclusively 5′ tailed, we analysed the boundary of known mouse pre-miRNAs located in introns and found that all 21 known tailed mirtrons are tailed from 5′ end. In contrast to our findings in mouse, the tailed mirtrons identified so far in Drosophila are all from 3′ end (8,9). It awaits further investigation whether the inconsistence between the two organisms is due to the difference in underlying processing mechanisms such as more efficient usage of 5′–3′ (mouse) versus 3′–5′ (fly) exoribonuclease after splicing.
Experimental validation of novel mouse miRNAs
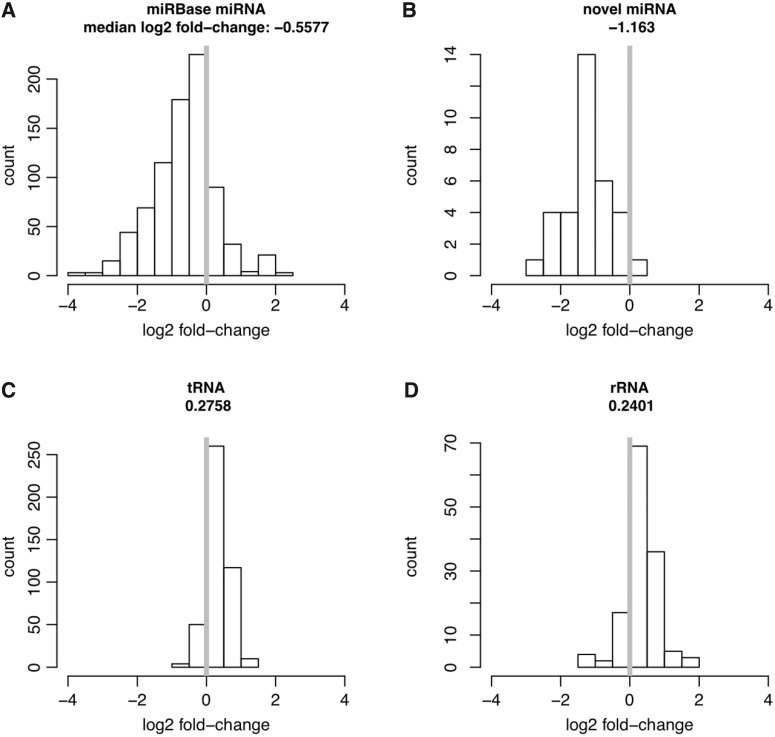
To investigate whether the novel miRNAs we identified were indeed dependent on Dicer for expression, we used RNA interference to knockdown Dicer in a mouse N2a cell line (see methods). RT-qPCR showed that in cells treated with siRNA, the level of Dicer mRNA transcripts was reduced to 15% of that in unperturbed cells (Supplementary Figure S11). After sequencing the small RNAs (10–40 nt) from unperturbed and treated cells, we compared the abundance of different non-coding RNA-derived transcripts between the two samples (see methods). More specifically, we counted the sequencing reads mapped to tRNAs and rRNAs, transcripts that are believed not to be processed by Dicer, or to known miRNA loci deposited in miRBase, as well as to the novel pre-miRNAs that we have identified. As the results, 45 novel miRNAs derived from 35 novel pre-miRNAs were expressed in N2a cells (Supplementary Table S14). As shown in Figure 6 (A–D) and Supplementary Figure S12, whereas both rRNAs and tRNAs showed a median increase of 21% and 19% after silencing Dicer, both known and novel miRNAs decreased in abundance with a median reduction of 32% and 55%, respectively. Using TaqMan assay, we confirmed that the expression level of CandidateMMU33 decreased after Dicer knockdown, similar as a known miRNA miR-137 (Supplementary Figure S13). These results demonstrated that the novel miRNAs predicted in this study were enriched in Dicer-dependent small RNAs.
Figure 6.
Novel miRNAs are dependent on Dicer for expression. RNA interference was used to silence Dicer in a mouse N2a cell line. Log2 fold-changes of small RNA expression after Dicer knockdown were noted for known miRNAs (A), novel miRNAs (B), tRNAs (C) and rRNAs (D).
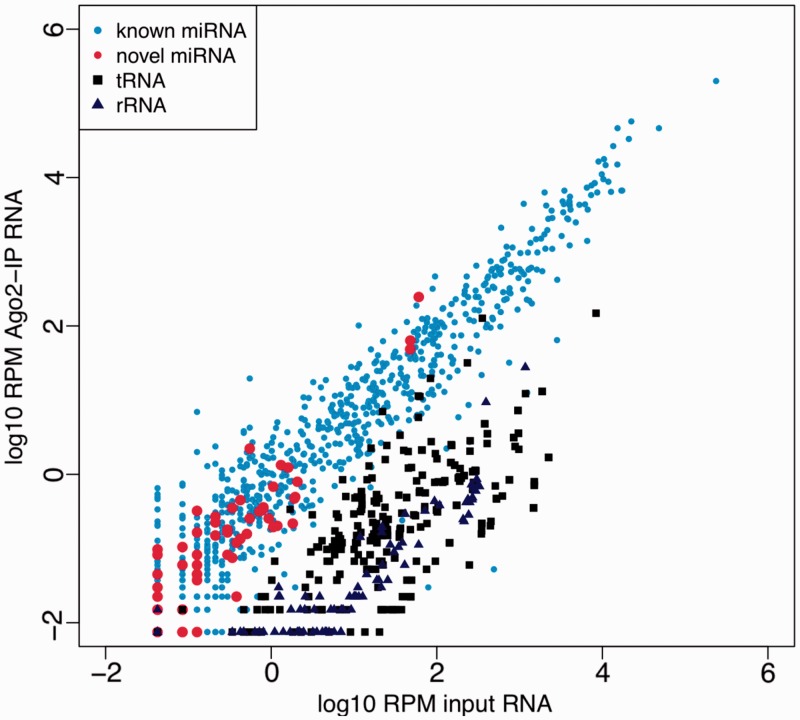
Mature miRNAs directly bind with Argonaute proteins to mediate target mRNA silencing. To confirm the functionality of our miRNA candidates, we isolated and sequenced Ago2-associated RNA in N2a cells by Ago2 IP (see methods). The specificity of Ago2 IP was shown by the depletion of sequencing reads derived from highly abundant rRNAs (0.01%) and tRNAs (0.1%). A total of 49 novel miRNAs derived from 37 candidate pre-miRNAs could be detected in IP sample. We then compared the abundance of known miRNAs and novel miRNAs in the IP sample with that in the input sample. As shown in Figure 7, the novel miRNAs showed a similar Ago2 binding profile as that of known miRNAs, whereas both tRNAs and rRNAs showed significant depletion. The enrichment of one known miRNA, miR-137, and one novel miRNA, CandidateMMU33, were also validated by Taqman assay (Supplementary Figure S14). These results showed that our novel miRNAs were indeed incorporated into Ago2/RISC complex, indicating their potential functionality.
Figure 7.
Novel miRNAs show similar Ago2-binding profile as known miRNAs. X-axis denoted the abundance (log10 transformed RPM value) of known miRNAs (light blue dots), novel miRNAs (red dots), tRNAs (black solid squares) and rRNAs (dark blue solid triangles) in the input sample. y-axis denoted their abundance in Ago2 IP sample. Novel miRNAs showed similar Ago2-binding profile as known miRNAs, whereas both tRNAs and rRNAs were significantly depleted in Ago2 IP sample.
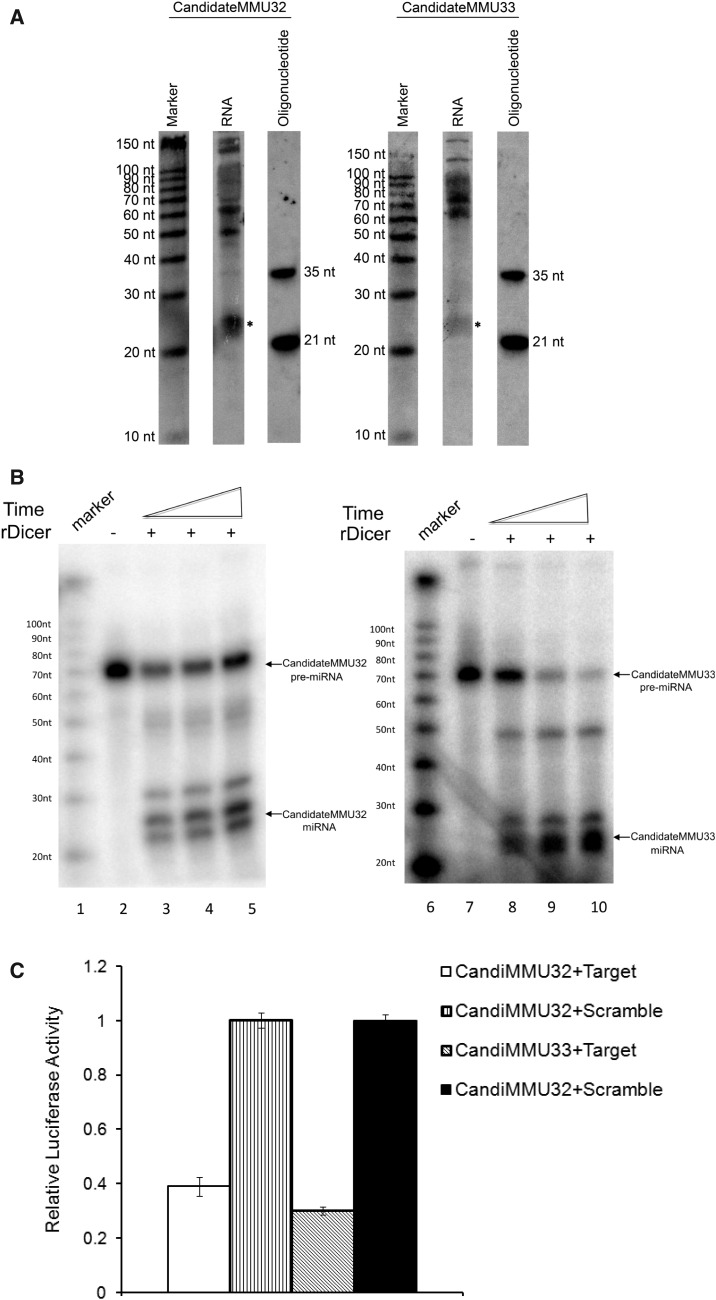
Furthermore, two most abundant novel pre-miRNA loci (CandidateMMU32 and CandidateMMU33) were chosen for further experimental investigation. As shown in Figure 8A, using northern blot, with a probe complementary to the potential mature miRNA, we could detect the band of the size corresponding to the mature miRNAs that we predicted. To investigate whether the processing of the two pre-miRNA candidates was dependent on Dicer, we incubated the in vitro transcribed and 32P-labelled pre-miRNAs with recombinant Dicer. As shown in Figure 8B, both candidates could be efficiently processed. To further test their functions in vivo, we performed the luciferase assay (Material and Methods). As shown in Figure 8C, compared to the scrambled control, presence of target sequences of the candidate miRNAs in 3′ UTR resulted in a 61% (CandidateMMU32) and 70% (CandidateMMU33) reduction of protein translation.
Figure 8.
Experimental validation of the two novel miRNA candidates. (A) The RNA with size of 10–200 nt was separated using flashPAGE Fractionator (Ambion) and loaded onto the denaturing PAGE gel for northern blot. Northern blot of two novel miRNAs (CandidateMMU32 and CandidateMMU33) revealed bands corresponding to the mature miRNA (marked as *). (B) In vitro rDicer processing. 32P-labelled CandidateMMU32 pre-miRNA (lane 3–5) and CandidateMMU33 pre-miRNA (lane 8–10) were incubated with rDicer for an increasing time. Lane 1 and 6 showed the size marker. Lane 2 and 7 were templates without adding rDicer. Both candidates could be efficiently processed by rDicer. (C) Luciferase assay. The sequences complementary to candidate miRNAs (Target) or scrambled control sequences (Scramble) were inserted into the 3′UTR of the Renilla luciferase gene in psiCHECK-2 plasmid. HEK293 cells were transfected with the plasmid alone or together with candidate pre-miRNA in vitro transcripts. All of the experiments were preformed in triplicates. Compared with scrambled controls, presence of target sequences of the candidate miRNAs in 3′UTR resulted in significant reduction of protein translation.
Further evaluation of the performance of miRGrep
To our knowledge, most miRNA discovery tools based on analysing small RNA sequencing data sets rely on the alignment of sequencing reads to reference genome sequences (49–51). Obviously, these tools have limited usage in the study of organisms whose genome has not been sequenced. Some other tools that do not depend on genome sequences make use of evolutionary conservation to identify the miRNAs with orthologs already found in other organisms (52), but will miss the miRNAs specific to certain lineages. In contrast to these available tools, miRGrep takes advantage of parallel sequencing of potential mature and precursor miRNAs. miRGrep successfully identified 239 pre-miRNAs detected in our sample, corresponding to 35% of all mouse pre-miRNAs deposited in miRBase. Of 438 known mature miRNAs recovered by miRGrep, 48 are not conserved in other species and could not be identified only by homology search. It suggests that our miRGrep can discover not only the well-conserved miRNAs, but also lineage specific ones.
In probabilistic scoring of pre-miRNA candidates, we used the known mouse miRNAs to estimate the model parameters. In this case, it could be argued that our model was over-trained for predicting mouse miRNAs. If the performance is dependent on a particular training data set, it will be questionable that our approach could work in an organism in which nothing is known about its miRNAs. To investigate the potential bias, we trained our model again using known human, fruit fly and C. elegans miRNAs, respectively. As illustrated in Supplementary Figure S15, the predictions based on known miRNAs from different organisms were nearly identical, indicating that the features included in our model represent the miRNA characteristics common to all metazoans.
Furthermore, to assess whether miRGrep could be applied in other metazoans, we sequenced mature and precursor miRNAs from C. elegans (see Supplementary Table S15) and applied the pipeline to predict C. elegans miRNAs. In total, 91% (16 270 159) of short reads could be mapped to 97% (49 702 713) of long reads without mismatch. After discarding the long reads with mapped short reads incompatible with Dicer processing, the remaining long reads could be merged into 47 052 clusters (see methods). Out of the representative reads, one from each cluster, 6007 could form stable hairpin structures. Finally, based on probabilistic scoring, 126 with a score higher than 0.95 and with at least five mapped short reads were considered as pre-miRNA candidates. Ninety-eight out of these candidates corresponded to 88 known C. elegans pre-miRNAs. Among the 28 novel candidates, one was derived from E. coli transcript, representing potential contamination. After mapping the remaining candidates to C. elegans genome, 10 were found to derive from chimerical reads and another seven were considered unlikely as genuine pre-miRNAs after manual inspection by applying the same criteria as described before (Supplementary Tables S11 and S16). Of the remaining 10 pre-miRNA candidates, one could be mapped to DNA transposable element, nine to either intergenic (6), exonic (1) or intronic (2) regions of protein-coding genes. Detailed information about the novel candidates can be found in Supplementary Table S17, and one example was also shown in Figure 5.
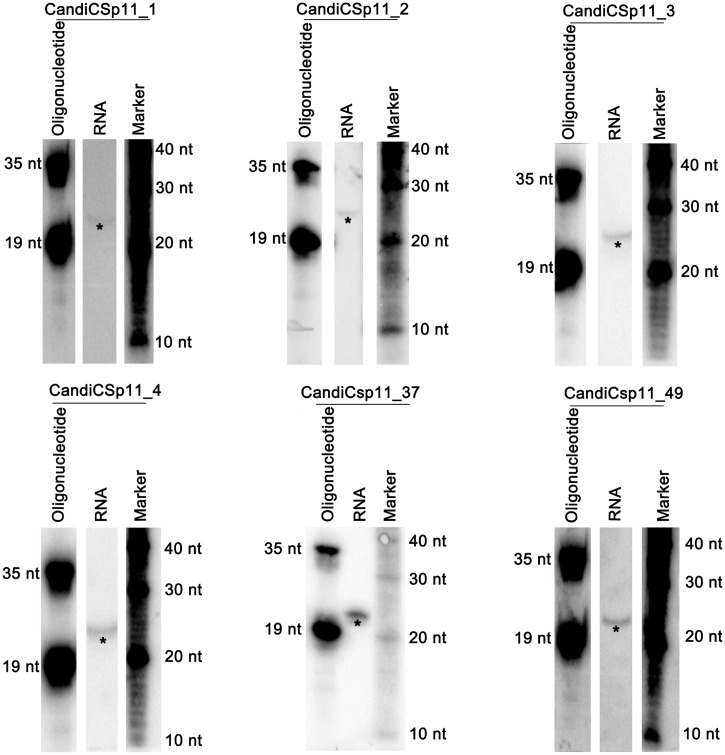
Finally, we predicted miRNAs in C. sp.11, a recently described hermaphroditic worm species closely related to C. elegans, whose genome has not yet been sequenced (53). In total, 79.45% (90 458 802) of short reads could be mapped to 99.66% (141 326 437) of long reads without mismatch (see Supplementary Table S18). We went through the miRGrep pipeline as described for C. elegans except that we required at least 40 supporting short reads, due to the increased number of short reads. Ninety-seven long reads were considered as pre-miRNA candidates after manual inspection (see Supplementary Tables S11 and S21). Sixty-one of them have homologues in C. elegans that can derive 74 mature miRNAs. The remaining 36 pre-miRNA candidates yielded 58 novel mature miRNAs with supporting short reads. Four most abundant novel miRNAs and two miRNAs with homologues in C. elegans were further validated by northern blot (Figure 9). Based on one run of mature and pre-miRNA sequencing, miRGrep alone can identify 132 miRNAs. Our method, if combined with homology-based approach, could offer an even more comprehensive miRNA annotation in this species (see Supplementary Figure S16).
Figure 9.
Northern Blot validation of six novel miRNAs in C. sp. 11. *Signals denoted the putative mature miRNAs. Note that CandiCsp11_37 and CandiCsp11_49 have corresponding homologues in C. elegans as Cel-mir-58 and Cel-let-7, respectively.
CONCLUSION
In summary, we have performed unbiased genome-wide parallel profiling of mature and precursor miRNAs. Comparing with mature miRNA sequencing, our pre-miRNA sequencing has rather limited efficiency and awaits further technical improvement. However, even with the current data set, we could improve the understanding of the miRNA processing and modification. More importantly, we developed a novel miRNA discovery pipeline, miRGrep, which did not rely on the available genome reference sequences. We believe miRGrep could be widely used in the study of miRNAs not only in the metazoans whose genome has not yet been sequenced, but also in samples where the genome differs significantly from the reference sequences, such as cancer.
AVAILABILITY
Illumina sequencing data have been submitted to the Short Read Archive at NCBI (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?) and are accessible through accession no. SRA046016.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–21 and Supplementary Figures 1–16.
FUNDING
Federal Ministry for Education and Research (BMBF), Germany and Senate of Berlin, Berlin, Germany (BIMSB 0315362A, 0315362C); China Scholarship Council (CSC) and Illumina (N.L.); CSC (T.C. and H.D.); EMBO long-term fellowship [ALTF 225-2011] (to M.R.F.); MDC Systems Biology Network (MSBN) as a participator of the Helmholz-Alliance on Systems Biology (S.D.M.). Funding for open access charge: Max-Delbruck-Centrum für Molekulare Medizin (MDC).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Mirjam Feldkamp and Claudia Langnick for their excellent technical assistance.
REFERENCES
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 3.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol. Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flynt AS, Greimann JC, Chung WJ, Lima CD, Lai EC. MicroRNA biogenesis via splicing and exosome-mediated trimming in Drosophila. Mol. Cell. 2010;38:900–907. doi: 10.1016/j.molcel.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang JS, Lai EC. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol. Cell. 2011;43:892–903. doi: 10.1016/j.molcel.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol. Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 11.Saraiya AA, Wang CC. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000224. e1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scott MS, Avolio F, Ono M, Lamond AI, Barton GJ. Human miRNA precursors with box H/ACA snoRNA features. PLoS Comput. Biol. 2009;5 doi: 10.1371/journal.pcbi.1000507. e1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS. Small RNAs derived from snoRNAs. RNA. 2009;15:1233–1240. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berezikov E, Robine N, Samsonova A, Westholm JO, Naqvi A, Hung JH, Okamura K, Dai Q, Bortolamiol-Becet D, Martin R, et al. Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome Res. 2011;21:203–215. doi: 10.1101/gr.116657.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brameier M, Herwig A, Reinhardt R, Walter L, Gruber J. Human box C/D snoRNAs with miRNA like functions: expanding the range of regulatory RNAs. Nucleic Acids Res. 2011;39:675–686. doi: 10.1093/nar/gkq776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ono M, Scott MS, Yamada K, Avolio F, Barton GJ, Lamond AI. Identification of human miRNA precursors that resemble box C/D snoRNAs. Nucleic Acids Res. 2011;39:3879–3891. doi: 10.1093/nar/gkq1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 18.Okada C, Yamashita E, Lee SJ, Shibata S, Katahira J, Nakagawa A, Yoneda Y, Tsukihara T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science. 2009;326:1275–1279. doi: 10.1126/science.1178705. [DOI] [PubMed] [Google Scholar]
- 19.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 20.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 21.Leuschner PJ, Ameres SL, Kueng S, Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 23.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 24.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 25.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 26.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 27.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 28.Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 29.Shendure JA, Porreca GJ, Church GM. Overview of DNA sequencing strategies. Curr. Protoc. Mol. Biol. 2008 doi: 10.1002/0471142727.mb0701s81. Chapter 7, Unit 7 1. [DOI] [PubMed] [Google Scholar]
- 30.Newman MA, Mani V, Hammond SM. Deep sequencing of microRNA precursors reveals extensive 3′ end modification. RNA. 2011;17:1795–1803. doi: 10.1261/rna.2713611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burroughs AM, Kawano M, Ando Y, Daub CO, Hayashizaki Y. pre-miRNA profiles obtained through application of locked nucleic acids and deep sequencing reveals complex 5′/3′ arm variation including concomitant cleavage and polyuridylation patterns. Nucleic Acids Res. 2011;40:1424–1437. doi: 10.1093/nar/gkr903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Beitzinger M, Meister G. Experimental identification of microRNA targets by immunoprecipitation of Argonaute protein complexes. Methods Mol. Biol. 2011;732:153–167. doi: 10.1007/978-1-61779-083-6_12. [DOI] [PubMed] [Google Scholar]
- 34.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 36.Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 37.Kinsella RJ, Kahari A, Haider S, Zamora J, Proctor G, Spudich G, Almeida-King J, Staines D, Derwent P, Kerhornou A, et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database (Oxford) 2011;2011 doi: 10.1093/database/bar030. bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hofacker IL, Stadler PF. Memory efficient folding algorithms for circular RNA secondary structures. Bioinformatics. 2006;22:1172–1176. doi: 10.1093/bioinformatics/btl023. [DOI] [PubMed] [Google Scholar]
- 39.Bonnet E, Wuyts J, Rouze P, Van de Peer Y. Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics. 2004;20:2911–2917. doi: 10.1093/bioinformatics/bth374. [DOI] [PubMed] [Google Scholar]
- 40.Underwood JG, Uzilov AV, Katzman S, Onodera CS, Mainzer JE, Mathews DH, Lowe TM, Salama SR, Haussler D. FragSeq: transcriptome-wide RNA structure probing using high-throughput sequencing. Nat. Methods. 2010;7:995–1001. doi: 10.1038/nmeth.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adamidi C, Wang Y, Gruen D, Mastrobuoni G, You X, Tolle D, Dodt M, Mackowiak SD, Gogol-Doering A, Oenal P, et al. De novo assembly and validation of planaria transcriptome by massive parallel sequencing and shotgun proteomics. Genome Res. 2011;21:1193–1200. doi: 10.1101/gr.113779.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 43.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawahara Y, Zinshteyn B, Sethupathy P, Iizasa H, Hatzigeorgiou AG, Nishikura K. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smalheiser NR, Torvik VI. Mammalian microRNAs derived from genomic repeats. Trends Genet. 2005;21:322–326. doi: 10.1016/j.tig.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 47.Piriyapongsa J, Jordan IK. A family of human microRNA genes from miniature inverted-repeat transposable elements. PLoS One. 2007;2:e203. doi: 10.1371/journal.pone.0000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 49.Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 50.Hackenberg M, Sturm M, Langenberger D, Falcon-Perez JM, Aransay AM. miRanalyzer: a microRNA detection and analysis tool for next-generation sequencing experiments. Nucleic Acids Res. 2009;37:W68–W76. doi: 10.1093/nar/gkp347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hendrix D, Levine M, Shi W. miRTRAP, a computational method for the systematic identification of miRNAs from high throughput sequencing data. Genome Biol. 2010;11:R39. doi: 10.1186/gb-2010-11-4-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gerlach D, Kriventseva EV, Rahman N, Vejnar CE, Zdobnov EM. miROrtho: computational survey of microRNA genes. Nucleic Acids Res. 2009;37:D111–D117. doi: 10.1093/nar/gkn707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kiontke KC, Félix MA, Ailion M, Rockman MV, Braendle C, Pénigault JB, Fitch DH. A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. BMC Evol. Biol. 2011;11:339. doi: 10.1186/1471-2148-11-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.