Abstract
Alveolar epithelial cell (AEC) trans-differentiation is a process where type II alveolar epithelial cells (AEC II) trans-differentiate into type I alveolar epithelial cells (AEC I) during lung recovery after various injuries, in which AEC I are damaged. This process is critical for lung tissue repair. MicroRNAs are a group of small RNAs that regulate gene expression at the post-transcriptional level. They have the potential to regulate almost every aspect of cell physiology. However, whether AEC trans-differentiation is regulated by microRNAs is completely unknown. In this study, we found that miR-375 was downregulated during AEC trans-differentiation. The overexpression of miR-375 with an adenoviral vector inhibited alveolar epithelial trans-differentiation as indicated by an increase in the AEC II marker, surfactant protein C, and decreases in the AEC I markers, T1α and advanced glycosylation end product-specific receptor. miR-375 also inhibited the Wnt/β-catenin pathway. The constitutively activation of Wnt/β-catenin signaling with a stabilized form of β-catenin blocked the miR-375 effects. Frizzled 8 was identified as a target of miR-375. In summary, our results demonstrate that miR-375 regulates AEC trans-differentiation through the Wnt/β-catenin pathway. This discovery may provide new targets for therapeutic intervention to benefit lung recovery from injuries.
INTRODUCTION
The discovery of microRNAs (miRNAs) has opened up new avenues of research into regulation of gene expression and mechanisms of diseases. miRNAs are a group of endogenous non-coding regulatory RNAs. They are ∼22-nt long and regulate the expression of their target genes at the post-transcriptional level by cleavage of a target mRNA, translational inhibition and mRNA deadenylation (1–4). So far, >1000 miRNAs have been discovered in humans. The known functions of miRNAs in animals have covered almost every aspect of cell physiology, including regulation of development timing, cell proliferation and differentiation, apoptosis, fat and lipid metabolisms, exocytosis, cancers, diabetes and other diseases (5–7). According to the computational analysis, the majority of mammalian mRNAs are under selective pressure to be conserved targets of miRNAs (8).
miR-375 has previously been reported as a pancreatic islet-specific miRNA. It can regulate insulin secretion and pancreatic islet development (9–11). The identified targets of miR-375 include 3′phosphoinositide-dependent protein kinase-1 (PDK1) and Myotrophin (Mtpn). Recently, miR-375 has been shown to be a proliferation inhibitor and a tumor suppressor. The involved targets include yes-associated protein, Janus kinase 2 and PDK1 (12–14). We have previously reported that miR-375 is expressed in the rat lung (15). The function of miR-375 in the lung is of particular interest to us.
The epithelium of the lung is composed of cuboidal type II alveolar epithelial cells (AEC II) and squamous type I alveolar epithelial cells (AEC I). AEC II are multifunctional cells involved in surfactant synthesis and secretion, fluid transport and recovery from lung injury (16). The main functions of AEC I are gas exchange and fluid transport (17). AEC I can also protect lung epithelium from hyperoxic injury (18).
During the saccular phase of lung development, columnar epithelial cells differentiate into AEC II, which contain distinctive lamellar bodies in the cytoplasm. As the air sacs expand, AEC I begin to derive from AEC II and undergo a thinning process. The squamous type I epithelium and the capillary endothelium form a thin air–blood barrier. Under a variety of disease conditions, AEC I are damaged and AEC II proliferate. Some of these AEC II keep their morphologic characteristics, whereas others trans-differentiate into AEC I (19–21). Radioactive tracing experiment after injury reveals that tritiated thymidine is first incorporated in ACE II and is subsequently observed in AEC I, which further confirms the AEC II as progenitor cells of AEC I (22,23). When they are cultured in plastic dishes, AEC II gradually lose their morphologic characteristics and their ability to synthesize and secrete surfactant. On the other hand, these cells obtain the characteristics of AEC I, which include the squamous appearance and expression of all known AEC I markers such as T1α and advanced glycosylation end product-specific receptor (RAGE) (24–27). This is a well-established in vitro model that mimics the AEC trans-differentiation in vivo. We have previously reported that TGF-β regulates the AEC trans-differentiation through the Smad pathway (27). However, the roles of miRNAs in the regulation of this trans-differentiation process are completely unknown.
Wnt/β-catenin signaling pathway plays a critical role in the regulation of lung epithelial fate determination. In the canonical Wnt/β-catenin pathway, Wnt ligands bind to transmembrane receptors, Frizzled (FZD) and the co-receptors low-density lipoprotein receptor-related protein-5/6 (LRP5/6) (28,29). This binding leads to the activation of Dishevelled, which results in the inhibition of β-catenin phosphorylation by glycogen synthase kinase-3β (GSK-3β) and degradation of β-catenin. The stabilized β-catenin then translocates into the nucleus, binds with T-cell factor/lymphoid-enhancing factor (TCF/LEF) transcriptional factors and regulates the expression of downstream β-catenin dependent genes, such as cyclin D1 (CCND1) and c-myc. In respiratory epithelial cells, Wnt/β-catenin signaling is necessary for branching morphogenesis and distal airway cell specification (30–32). Some Wnt ligands, including Wnt5a and Wnt7b, are expressed in the lung epithelium (33–35). FZD8 is also found in the lung epithelium (36). A recent study has demonstrated that Wnt/β-catenin promotes trans-differentiation in the AEC culture model (37).
In this study, we found that miR-375 was significantly downregulated during AEC trans-differentiation. Furthermore, we provided evidence that miR-375 regulates alveolar epithelial trans-differentiation through the Wnt/β-catenin pathway and identified FZD8 as one of the targets of miR-375.
MATERIALS AND METHODS
Isolation of AEC II
AEC II were isolated from male Sprague–Dawley rats (200–250 g) as previously described (27). All the animal experiments in this study followed the protocols approved by the Oklahoma State University Animal Care and Use committee. In brief, the rat lungs were perfused and isolated. They were then lavaged, and digested with elastase. The lung lobes were chopped and incubated with DNase. The mixture was then filtered through a 160-, 37- and 15-μm nylon mesh in sequence. The cells were incubated in rat immunoglobulin G (IgG)-coated plastic dishes twice for 30 min each. Cells were then pelleted and resuspended in complete medium [minimum essential medium (MEM) supplemented with 10% fetal bovine serum (FBS)]. The cell purity was >90% as determined by the modified Papanicolaou staining.
Alveolar epithelial trans-differentiation
Freshly isolated AEC II were seeded into 35-mm tissue culture dishes at a density of 0.8 × 106/dish. Cells were treated with different conditions after seeding and cultured for 1–5 days with medium changed on Day 1 and every other day.
RNA extraction
Total RNA were extracted from freshly isolated AEC II and cultured AEC II using mirVanaTM miRNA isolation kit (Ambion, Austin, TX) following the manufacturer’s instructions.
Fetal lung isolation
Whole lungs were isolated from rat fetuses on gestational day 16, 19, 21 (E16, E19, E21), new born (P0), postnatal day 6 and 14 (P6 and P14) and adult rats. For fetal lungs, pregnant Sprague–Dawley rats were sacrificed with CO2, and the lungs were isolated from the fetuses. For pup and adult lungs, male rats were anesthetized and sacrificed before isolation of the lungs. The lungs were homogenized immediately after isolation, and total RNA was extracted as described earlier in the text.
Quantitative reverse transcriptase-polymerase chain reaction
For mRNA quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR), total RNAs were treated with TURBO DNA-freeTM (Ambion) to eliminate any genomic DNA contaminations. One micrograms of total RNA was reverse transcribed into cDNA using Moloney Murine Leukemia Virus reverse transcriptase. Real-time PCR was performed using SYBR Green master mix from Eurogentec (Seraing, Belgium) on an ABI 7500 fast system (Applied Biosystems, Foster City, CA). The following primers were used in this study: T1α forward, GCCATCGGTGCGCTAGAAGATGATCTT and T1α reverse, GTGATCGTGGTCGGAGGTTCCTGAGGT; RAGE forward, CTACCTATTCCTGCAGCTTC and RAGE reverse, CTGATGTTGACAGGAGGGCTTTCC; β-Catenin forward, CGAGGACTCAATACCATTCC and β-Catenin reverse, AGCCGTTTCTTGTAGTCCTG; FZD8 forward, AAGGGCATCGGTTACAACTACAC and FZD8 reverse, TCTAGGCCCGCCTCATCTT; 18 S forward, TCCCAGTAAGTGCGGGTCATA and 18 S reverse, CGAGGGCCTCACTAAACCATC. miRNA qRT-PCR was performed with as described (38). Total RNAs were treated with DNase and were purified by phenol/chloroform extraction and ethanol precipitation. One micrograms of DNase-treated RNA were poly A-tailed and purified by phenol/chloroform extraction and ethanol precipitation. Poly A-tailed RNA was reverse transcribed into cDNA with polyT adapter as the primer, GCGAGCACAGAATTAATACGACTCACTATAGG TTTTTTTTTTTTVN. Real-time PCR was performed with a universal reverse primer, GCGAGCACAGAATTAATACGACTCAC, and a forward primer with the same sequence as mature miR-375. The real-time PCR thermal conditions were 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 60°C for 30 s and 65°C for 30 s. The endogenous reference gene was U6 snRNA. The comparative computed tomography method was used, and the relative expression of miRNA was calculated with the equation 2−(CTmiRNA−CTU6).
Construction of adenoviral vectors
The pre-miRNA sequence of hsa-miR-375 with flanking sequences of 350 nt on both the 5′ and 3′ ends was PCR amplified from human genomic DNA with the following primers: forward primer cacctcgagGCACAGCCTCTCCCACCCGTA and reverse primer gagaattcCGTGTCAGCCGCAGATGCGT. The sequence was inserted into the pENTR plasmid (Invitrogen), downstream of the cytomegalovirus-green fluorescent protein (GFP) (39,40). The insert was then switched to the adenoviral vector through LR recombination. After digested by Pac I, the vector was transfected into 293A cells to produce adenovirus overexpressing miR-375 (Ad-miR-375). A control virus was also constructed from an empty vector with GFP only. For the β-catenin silencing adenovirus vector (si-β-catenin), we used a new method developed in our laboratory (41). Four short hairpin RNAs were driven by four different promoters: mU6, hU6, H1 and 7SK. The four small RNA sequences for β-catenin silencing were as follows: GGACCAGGTGGTCGTTAATAA, GTGGATTCCGTACTGTTCTAC, GAATGCCGTTCGCCTTCATTA and ACTGTTGGATTGATCCGAAAC. The expression cassette was then cloned into the adenovirus vector as described earlier in the text (41). A vector expressing four non-relevant siRNA sequences was used as a control (si-control). For the Ad-ΔGSK-β-catenin adenoviral vector, the eGFP-ΔGSK-β-catenin expression sequence was transferred from pEGFP-C1 vector into a pENTR vector. This sequence contained mutations at four GSK3β phosphorylation sites (Ser33, Ser37, Thr41 and Ser45) (42). The pEGFP-C1 vector expressing ΔGSK-β-catenin was a gift of Dr. Angela Barth from Stanford University.
3′-Untranslated region luciferase assay
The 3′-untranslated region (3′-UTR) of rat FZD8 was PCR amplified and cloned into pGL3 vector downstream of a firefly luciferase reporter gene. The following primers for PCR were used: forward, CGCGAATTCCTGAACGGAAGCCCAGAAG, and reverse, GACTCTAGAGCTGCTGTTAGTGTAAGTGGC. The forward primer includes an EcoRI restriction site, and the reverse primer contains an XbaI restriction site. One day after plating 293A cells in a 96-well plate, 50 ng of miRNA overexpression plasmid pENTR-miR-375 was co-transfected into 239A cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) together with 5 ng of the reporter construct and 0.4 ng of control pRL-TK, which contains a Renilla luciferase gene (Promega). The cells were harvested, and luciferase activities were detected 48 h post-transfection using Dual Luciferase Reporter Assay System (Promega). In brief, 35 μl of passive lysis buffer was applied to each well. After shaking, 7 μl of cell lysate was taken for the dual-luciferase reporter assay. Firefly luciferase activity and Renilla luciferase activity were measured by the FLUOstar OPTIMA microplate fluorometer (BMG LABTECH, Offenburg, Germany).
Western blotting
The following primary antibodies were used in western blotting: mouse monoclonal anti-T1α (1:2000) from Dr. Mary Williams (Boston U), mouse monoclonal anti-acctive-β-catenin (ABC) (#05-665, 1:500) from Millipore (Billerica, MA), mouse monoclonal anti-β-catenin (#610154, 1:2000) from BD (Franklin Lakes, NJ), rabbit polyclonal anti-casein kinase 2, α 1 (CSNK2A1) (#2656, 1:1000) from Cell signaling (Danvers, MA), goat polyclonal anti-FZD8 (sc-33504, 1:200) from Santa Cruz (Santa Cruz, CA), rabbit polyclonal anti-PDK1 (#3062, 1:1000) from Cell signaling and rabbit polyclonal anti-β-actin from Sigma (A-2066, 1:2000). For western blots, 35 μg of protein was loaded for each sample. After being incubated with primary antibodies, the membranes were washed with Tris-Buffered Saline and Tween 20 and incubated with horseradish peroxidase-conjugated anti-mouse or rabbit secondary antibodies (1:2000) for 1 h. The target proteins were visualized with SuperSignal West Pico Chemiluminescent Substrate (Pierce) and Molecular Imager VersaDoc MP 5000 System (Bio-Rad).
Immunocytochemistry
The cultured cells were fixed in 4% paraformaldehyde, permeabilized with 0.4% Triton X-100 and then blocked in 10% FBS. The cells were incubated with mouse anti-T1α (1:250) or rabbit polyclonal anti-surfactant protein C (SP-C) (Santa Cruz, sc-13979, 1:100) antibodies at 4°C overnight followed by incubation with Alexa 546-conjugated anti-mouse or anti-rabbit secondary antibodies (1:250). After washing, the cells were examined under a Nikon Eclipse E600 fluorescence microscope.
Chromatin immunoprecipitation assay
The Chromatin immunoprecipitation (ChIP) assay was performed with EZ-ChIP™ ChIP kit from Millipore (Billerica, MA). Briefly, freshly isolated AEC II or cultured cells were fixed in 1% formaldehyde. The cross-linked DNA was then sheared into ∼200–1000 base pairs in length with sonication. One percent of the sheared DNA was set aside as an input control. Anti-β-catenin antibody or normal mouse IgG was incubated with sheared DNA at 4°C overnight with rotation. Protein G Agarose was then added. Protein/DNA complexes were then eluted from the agarose, and the cross-linking then was reversed to free DNA. Purified DNA was then analyzed with PCR using the primers flanking LEF-1-binding site in the CCND1 promoter. The glyceraldehyde 3-phosphate dehydrogenase (GAPDH) promoter without LEF-1 biding sites was used as a negative control. The following primers were used: CCND1 forward TTCTCTGCCCGGCTTTGAT, CCND1 reverse CACAGGAGCTGGTGTTCCATG, GAPDH forward GTGCAAAAGACCCTGAACAATG and GAPDH reverse GAAGCTATTCTAGTCTGATAACCTCC.
Statistical analysis
All experiments were performed with at least three independent replicates. For statistical analysis, student t-test was used, and P < 0.05 was considered significant. All values were shown as means ± S.E.
RESULTS
miR-375 expression during alveolar epithelial trans-differentiation
We first determined miR-375 expression levels during AEC trans-differentiation. Total RNAs from freshly isolated AEC II and those cultured in plastic dishes for 1–5 days (D0, D1, D3 and D5) were assayed for their expression of miR-375 using real-time PCR. The expression of miR-375 was decreased significantly during alveolar epithelial trans-differentiation. On Day 3 and Day 5, the expression of miR-375 was only 16 and 11% of that on Day 0 (Figure 1), respectively.
Figure 1.
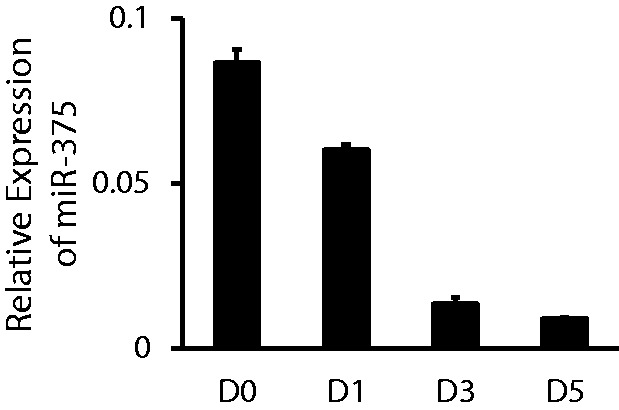
The expression pattern of miR-375 in the AEC trans-differentiation. Freshly isolated AEC II were cultured for 0, 1, 3 or 5 days (D0, D1, D3 or D5) on plastic dishes. Relative expression of miR-375 was determined by qRT-PCR. The expression level of miR-375 at each time point was normalized to U6. Data shown are means ± S.E. from three independent cell preparations, each assayed in duplicate.
miR-375 is enriched in AEC II
The relative expression of miR-375 in freshly isolated AEC II and the whole lung was measured using qRT-PCR. The relative expression level of miR-375 was 2.5-fold higher than that in the whole lung (Figure 2).
Figure 2.
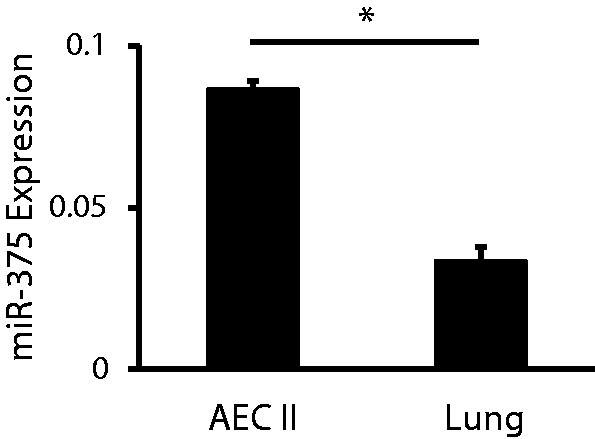
Expression of miR-375 in AEC II and the lung. The expression of miR-375 in freshly isolated AEC II and the whole lung was determined by qRT-PCR and normalized to U6. Data shown are means ± S.E. from three independent replicates. *P < 0.0001.
Expression pattern of miR-375 in lung development
To study the functions of miR-375 in alveolar epithelial trans-differentiation, we also determined the expression of miR-375 during fetal lung development that involves trans-differentiation using qRT-PCR. The development of the fetal lung can be divided into four stages: the embryonic phase, the glandular or pseudoglandular phase, the canalicular phase and the saccular phase. We selected different time points that represent different stages of lung development and determined the expression of miR-375 at these time points. In rat, the canalicular phase starts on Day 18 and ends on Day 20. The saccular phase lasts from Day 20 to full term. AEC II first appear in the transition from the canalicular to the saccular phase. During the saccular stage, the differentiation of AEC II becomes more evident. As determined by qRT-PCR, the expression of miR-375 increased at this late stage (E21 and P0) (Figure 3), which could result from more differentiated AEC II in the lung. The change in miR-375 level during lung development was lower compared with that in the trans-differentiation process. This is likely owing to the fact that miR-375 levels were measured in whole lung tissue, not in a specific cell type. It is also possible that an miRNA expression could be shifted from one type of cells to another type of cells during development. For example, we have previously observed that miR-127 is dominantly localized in mesenchymal cells at embryonic day 19 and shifted toward the epithelial cells at embryonic day 21 (39,40).
Figure 3.
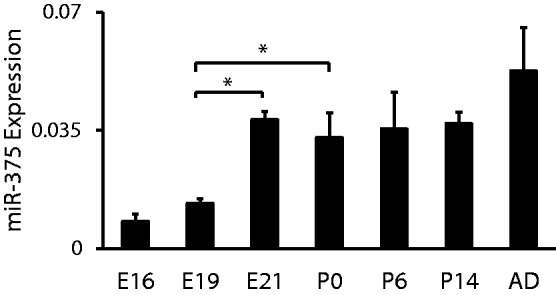
The expression pattern of miR-375 during fetal lung development. Whole lungs were isolated from rat fetuses on gestational day 16, 19, 21 (E16, E19, E21), new born (P0), postnatal day 6 and 14 (P6 and P14) and adult (AD) rats. The expression levels of miR-375 were determined with qRT-PCR. Expression data were normalized to U6. *P < 0.001. Data shown are means ± S.E. from three independent replicates.
miR-375 inhibits AEC trans-differentiation
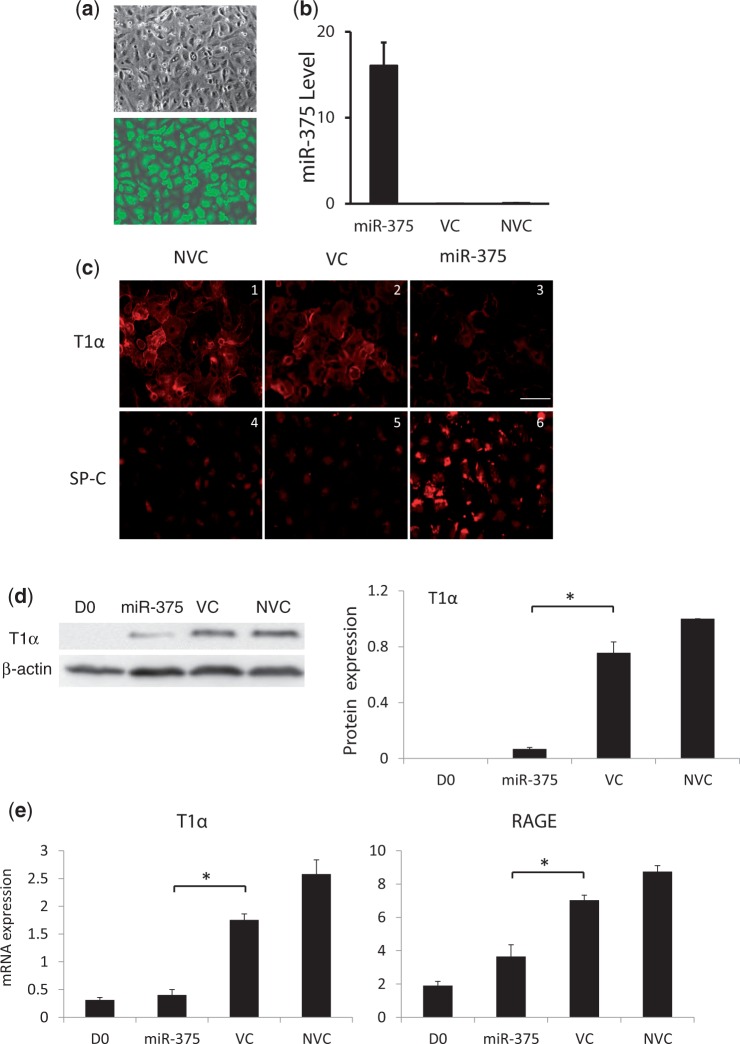
The expression of miR-375 significantly decreased during alveolar epithelial trans-differentiation. It is reasonable to expect that the decrease of endogenous miR-375 is important for this process. To study the function of miR-375 in this process, an adenovirus overexpressing miR-375 was used to transduce AEC II. The overexpression efficiency was assessed by eGFP signals and qRT-PCR for miR-375 on Day 4. Nearly 100% of the cells were expressing eGFP (Figure 4a). The expression of miR-375 was markedly increased compared with that in cells transduced with the control virus (Figure 4b).
Figure 4.
Effect of miR-375 overexpression on the trans-differentiation of AEC II to AEC I. Freshly isolated AEC II were transduced with a miR-375 overexpression virus or a virus control (VC) (MOI = 100) or none (NVC) and cultured for 4 days. (a) Bright-field and GFP fluorescence of miR-375-transduced cells. (b) miR-375 levels revealed by real-time PCR. Data were normalized to U6. (c) Immunofluorescence of T1α and SP-C using anti-T1α and anti-SP-C antibodies. Because of GFP, double-labeling is not practical. (d) The expression of T1α was detected and quantitated with western blots. Data were normalized to β-actin. (e) The mRNA levels of T1α and RAGE were determined by qRT-PCR. Data shown are means ± S.E. from three independent cell preparations. *P < 0.001.
The effect of miR-375 overexpression on AEC trans-differentiation was monitored after culturing for 4 days. T1α, a marker protein for AEC I, and SP-C, a marker protein for AEC II, were detected with immunofluorescence. In non-virus and virus controls, SP-C disappeared and T1α increased significantly, indicating that AEC II trans-differentiated into AEC I (Figure 4c). In cells overexpressing miR-375, T1α was significantly decreased, and SP-C was increased in comparison with controls (Figure 4c). The expression level of T1α was further determined using western blots. The amount of T1α in cells overexpressing miR-375 decreased significantly compared with that in cells transduced with the control virus (P < 0.001). The control virus had no effects on T1α expression (Figure 4d). In addition, the mRNA levels of T1α and RAGE, AEC I markers, were dramatically increased during trans-differentiation and were significantly inhibited by miR-375 (Figure 4e). These results suggest that alveolar epithelial trans-differentiation is inhibited by miR-375.
miR-375 inhibits Wnt/β-catenin pathway
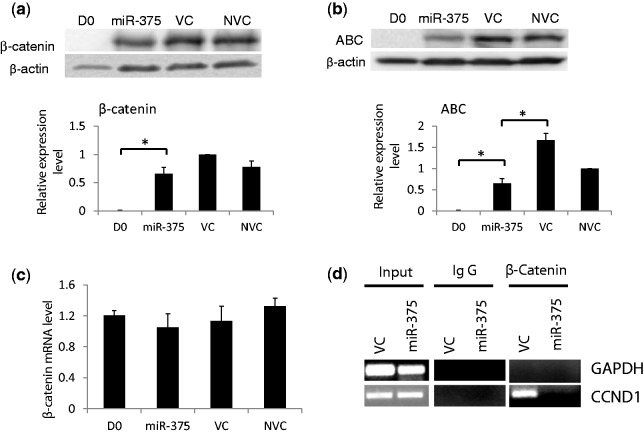
It has been shown that Wnt/β-catenin signaling pathway can promote AEC trans-differentiation in the in vitro model (37). We tested whether this pathway was affected by miR-375. In freshly isolated AEC II, there was almost no β-catenin that could be detected. After culturing for 4 days, there was an abundance of β-catenin (Figure 5a). Interestingly, the mRNA level of β-catenin did not change during culture (Figure 5c). The overexpression of miR-375 slightly decreased the total amount of β-catenin, but did not reach a significant level. The activated β-catenin (ABC) can be detected with the antibody that recognizes β-catenin dephosphorylated on Ser37 or Thr41. ABC was decreased significantly after miR-375 overexpression (Figure 5b).
Figure 5.
Effect of miR-375 on the Wnt/β-catenin pathway. Freshly isolated AEC II (D0) were transduced with Ad-miR-375 (miR-375) or control virus (VC) (MOI = 100) or none (NVC) and cultured for 4 days. The relative amounts of total β-catenin and activated β-catenin (ABC) were detected with western blots (a and b) and qRT-PCR (c) after miR-375 overexpression in the AEC trans-differentiation. Data shown are means ± S.E. from three independent replicates. *P < 0.01. (d) ChIP assays were used to detect the association of β-catenin with the promoter of CCND1 in AEC II transduced with Ad-miR-375 or the virus control and cultured for 4 days. Mouse normal IgG was used as a negative control. The promoter of GAPDH was used as another negative control.
Cyclin D1 (CCND1) is a direct transcriptional target of the β-catenin/LEF-1 pathway. The expression of CCND1 is regulated by this pathway through a LEF-1 binding site in the CCND1 promoter (43). We further used ChIP assay to detect the binding of β-catenin to the promoter of CCND1 during the process of trans-differentiation after transduction of miR-375 or control virus (VC). Fractions of the sheared DNA samples were set aside as input controls. The rest of the DNA was precipitated with the β-catenin antibody or mouse normal IgG. The purified DNAs from the precipitations were amplified using the primers for LEF-1 binding site and the flanking sequences of the CNND1 promoter. As expected, the overexpression of miR-375 eliminated the association between β-catenin and the promoter of CCND1 (Figure 5d). β-catenin did not bind to the GAPDH promoter that did not have the LEF-1 binding sites. The negative control using normal IgG did not yield signals. These data provided evidence that miR-375 can inhibit the canonical Wnt/β-catenin pathway.
Knockdown of β-catenin inhibits AEC trans-differentiation
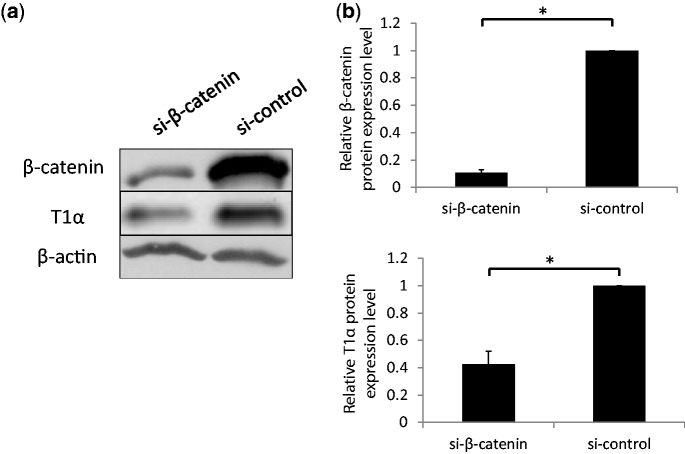
To test whether the inhibition of alveolar epithelial trans-differentiation is caused by inhibition of the Wnt/β-catenin pathway, we decided to knockdown β-catenin to block this pathway. We used a novel method to construct an adenovirus vector that expresses four shRNAs targeting to β-catenin (si-β-catenin) (41). AEC II were treated with si-β-catenin, or control virus, on D0 with a multiplicity of infection (MOI) of 100. The cells were collected on Day 4, and the expression of β-catenin was detected by western blot. As shown in Figure 6, the expression of β-catenin was reduced by >90%. The silencing of β-catenin led to the decrease in T1α, indicating the inhibition of AEC trans-differentiation.
Figure 6.
Effect of silencing β-catenin on trans-differentiation. AEC II were transduced with β-catenin silencing virus (si-β-catenin) or control virus (si-control) and then cultured for 4 days. The expression of β-catenin and T1α was detected with western blots (a). The signals were quantitated, and the expression levels were shown as the fractions of those in the si-controls (b). Data shown are means ± S.E. from three independent replicates. *P < 0.01.
Stabilized β-catenin blocks the effect of miR-375
To demonstrate that the regulation of miR-375 on the AEC trans-differentiation is through downregulation of activated β-catenin, we transduced AEC II with adenovirus Ad-ΔGSK-β-catenin, which expressed a stabilized form of β-catenin without GSK3β phosphorylation sites (ΔGSK-β-catenin) (42). The expression of ΔGSK-β-catenin was confirmed by western blots with ABC antibody (Figure 7a). The expression of T1α was decreased by miR-375 overexpression in the absence of ΔGSK-β-catenin. In the cells treated with Ad-ΔGSK-β-catenin, the expression of T1α was not affected by miR-375 anymore (Figure 7a). The quantitated data are presented in Figure 7b. These results showed that expression of stabilized β-catenin and thus constitutively activation of Wnt/β-catenin pathway blocked the effect of miR-375 on the trans-differentiation.
Figure 7.
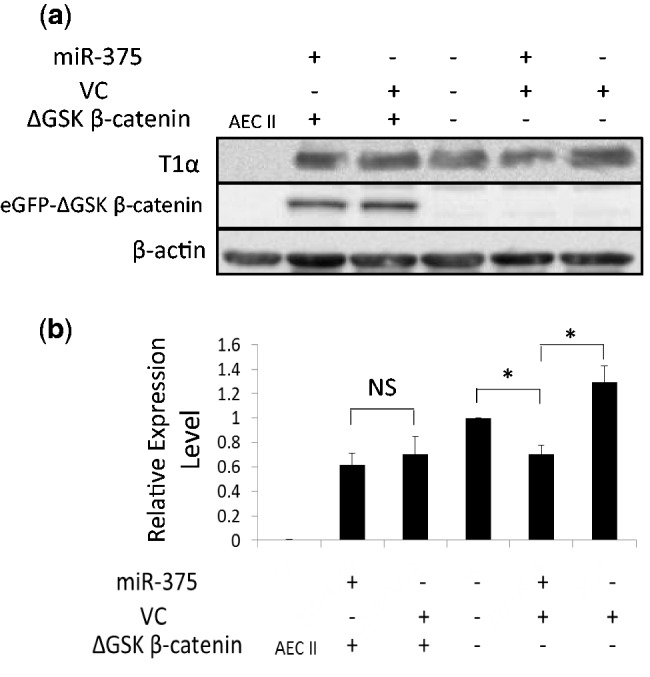
Effect of stabilized β-catenin on the trans-differentiation. Freshly isolated AEC II were transduced with Ad-miR-375 or virus control with or without the expression of eGFP-ΔGSK β-catenin as indicated in the figure (Ad-miR-375 MOI = 50 and Ad-ΔGSK β-catenin MOI = 5). Cells were then cultured for 4 days. (a) The expression of T1α and eGFP-ΔGSK β-catenin was detected by western blots using anti-T1α and anti-ABC antibodies, respectively. (b) The western blots were quantitated. Data shown were means ± S.E. from four independent experiments. *P < 0.001. NS: not significant.
FZD8 is a target of miR-375 in the Wnt/β-catenin pathway
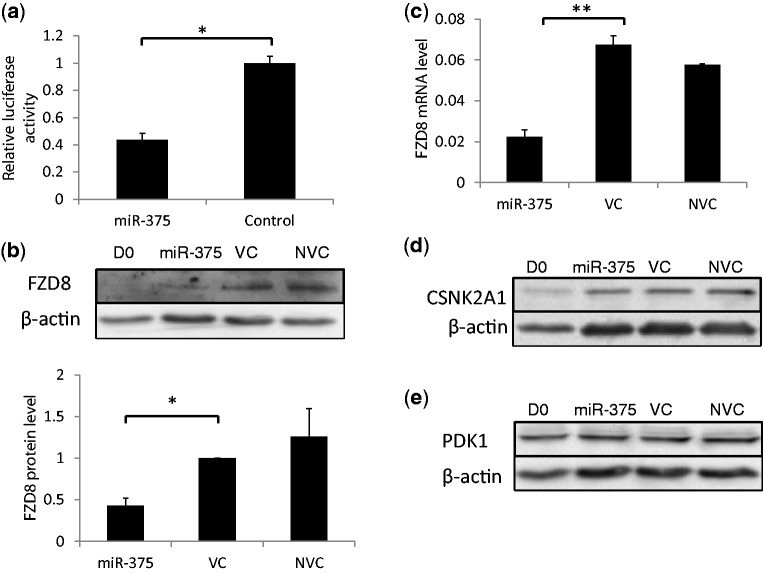
The web-based software TargetScan and PicTar predicted FZD8 as a target of miR-375. To verify the prediction, we produced a reporter construct by fusing the 3′-UTR of FZD8 with a firefly luciferase reporter gene. The miRNA-375 overexpression plasmid and the luciferase reporter construct were transfected into 293A cells together with a control vector pRL-TK, which contains a Renilla luciferase gene. As shown in Figure 8a, the luciferase activity of the reporter construct containing the 3′-UTR of FZD8 was significantly decreased by miR-375.
Figure 8.
Identification of target genes. (a) Luciferase assay. 3′-UTR of FZD8 was fused with the firefly luciferase gene. miR-375 overexpression plasmids were co-transfected with the luciferase-3′-UTR constructs into 293 A cells. Luciferase activity was determined 48 h post-transfection. The firefly luciferase activity was first normalized to the Renilla luciferase activity and then expressed as a ratio of the control plasmid. (b and c) Effect of miR-375 on the expression of endogenous FZD8. AEC II were transduced with Ad-miR-375 or virus control (MOI = 100) and then cultured for 4 days. The protein and mRNA levels of FZD8 were detected with western blots (b) and qRT-PCR (c). The expression levels of CSNK2A1 (d) and PDK1 (e) were detected with western blots. Data represent means ± S.E. from at least three replicates. *P < 0.01, **P < 0.001. The blots shown are representatives of three independent replicates.
To confirm that endogenous FZD8 is regulated by miR-375, we examined the effects of miR-375 on the expression of FZD8 during AEC trans-differentiation. The protein level of FZD8 was decreased significantly by miR-375 as shown in Figure 8b. The mRNA level of FZD8 was also decreased by 60% after miR-375 overexpression compared with cells infected with the control virus (Figure 8c). All together, miR-375 inhibits the expression of FZD8 at both protein and mRNA levels.
Casein kinase II, alpha 1 (Csnk2a1) polypeptide is another predicted target of miR-375 in the canonical Wnt signaling pathway. However, the protein expression of CSNK2A1 was not affected by miR-375 overexpression (Figure 8d). These data ruled out the possibility that miR-375 regulates Wnt/β-catenin signaling through CSNK2A1 during AEC trans-differentiation.
It has been reported that PDK1 is a direct target of miR-375 in gastric carcinomas (14). To test whether it is the same in our case, we also examined the expression of PDK1 during AEC trans-differentiation (Figure 8e). The expression of PDK1 was not changed during the trans-differentiation (from D0 to D4) and was not decreased by miR-375 overexpression. These data indicate that PDK1 is not the target of miR-375 in AEC, probably because of the differences in cell environment.
miR-375 is downregulated in idiopathic pulmonary fibrosis
Idiopathic pulmonary fibrosis (IPF) is a chronic progressive interstitial lung disease, characterized by fibrosis and excessive deposition of collagen in the pulmonary interstitium (44). IPF is also characterized by AEC II hyperplasia (44,45). A role of alveolar epithelial injury, AEC II proliferation and trans-differentiation in IPF pathogenesis and progression has been demonstrated (44). In this study, we determined the expression of miR-375 in the lung samples from IPF patients using qRT-PCR. The expression of miR-375 was decreased in severe IPF [50–80% forced vital capacity (FVC) and <50% FVC] compared with less severe IPF (>80%) (Figure 9). This result is consistent with our discovery that miR-375 is decreased during AEC II trans-differentiation.
Figure 9.
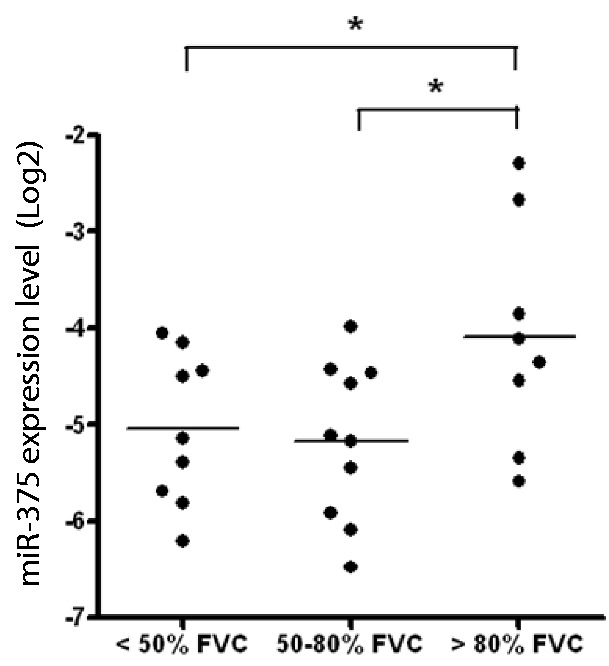
The expression of miR-375 is downregulated in IPF. The expression of miR-375 in the lungs of patients with different stages of IPF was determined with qRT-PCR. Data were normalized to U44. n = 10 for < 50% FVC, n = 10 for 50–80% FVC and n = 8 for > 80% FVC. *P < 0.05.
DISCUSSION
In this study, miR-375 was found to be downregulated during AEC differentiation. Further studies demonstrated that miR-375 inhibited the process of trans-differentiation when it was overexpressed in isolated AEC II. miR-375 also inhibited the Wnt/β-catenin pathway and constitutively activated β-catenin reversed the miR-375 effects, indicating that miR-375 acts at upstream of β-catenin. FZD8 was identified as one of the targets of miR-375. Together, these data indicate that miR-375 inhibits AEC trans-differentiation through the inhibition of Wnt/β-catenin pathway, and effects of miR-375 on FZD8 could play a role in this process (Figure 10).
Figure 10.
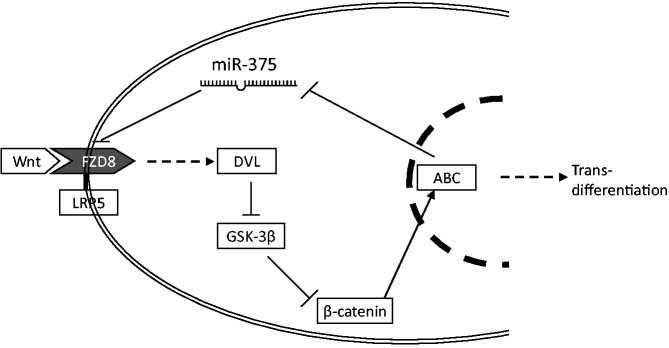
miR-375 inhibits AEC trans-differentiation through the inhibition of Wnt/β-catenin pathway.
To our knowledge, this is the first report studying the function of miR-375 in the lung epithelial cell differentiation. When miR-375 was first identified, it was found to be restricted only to pancreatic islets, but not in other tissues (9). However, we have shown that miR-375 is highly expressed in the rat lung in comparison with heart, liver, kidney, spleen and brain (15). This result was verified by northern blots. In this study, we discovered that miR-375 was enriched in AEC II, and the expression of miR-375 was dramatically downregulated during the trans-differentiation of AEC II to AEC I. In addition, miR-375 expression in fetal lungs was lower than that in the adult lung. All of the expression data suggest a potential role of miR-375 in the lung.
There have been several studies regarding the function of miR-375 in the pancreas. miR-375 suppresses insulin secretion by direct targeting of the mRNA of Mtpn (9). miR-375 not only regulates the function of the pancreatic islet but also affects the development of pancreatic islets. Knockdown of miR-375 results in aberrant formation of the pancreas islet (10). In addition, miR-375 can maintain normal pancreatic α and β cell mass and therefore normal glucose homeostasis (46).
In spite of all these studies in the pancreas, there are few functional studies of miR-375 in other organs. It has been reported that the expression of miR-375 is downregulated in hepatocellular tumors (47,48). An expression ratio of miR-221:miR-375 showed a high sensitivity and specificity for head and neck squamous cell carcinoma, indicating that miR-375 may be used as a diagnostic marker for cancers (47).
We have demonstrated that miR-375 inhibits pulmonary surfactant secretion from AEC II (40). This inhibition is carried out via the reorganization of cytoskeleton, rather than effects on surfactant synthesis or the formation of lamellar bodies (40). The result is reminiscent of the function of miR-375 in the regulation of insulin secretion in pancreas islets (9).
The Wnt signaling pathway has been proven to have widespread roles in tissue differentiation and organogenesis (49), including fetal lung development. This pathway regulates branching morphogenesis and epithelial cell differentiation (30,31). Wnt7b and Wnt5a induce lung branching morphogenesis through epithelial-mesenchymal communications, possibly via N-myc, bone morphogenetic protein 4 (BMP4) and fibroblast growth factor (FGF) signaling. An activated form of β-catenin causes ectopic differentiation of AEC II-like cells in airways, indicating a critical role for the Wnt/β-catenin signaling pathway in the differentiation of the lung epithelial cells (32).
A recent study, which used the same AEC II culture model as we did, showed the pivotal function of β-catenin pathway during alveolar epithelial trans-differentiation (37). Constitutive β-catenin signaling cannot be detected in adult AEC II in vivo or in freshly isolated AEC II. This signaling is activated after the lung is subjected to bleomycin-induced injury or during the culture of AEC II. Activation of Wnt/β-catenin pathway promotes AEC trans-differentiation, whereas forced inhibition of this pathway leads to an increase in cell death (37). In our study, we also found that this pathway was activated during this process of trans-differentiation, and the silencing of β-catenin inhibited the trans-differentiation. Total β-catenin protein could not be detected in freshly isolated AEC II and increased markedly in cultured AEC II. However, the mRNA level of β-catenin did not change in freshly isolated and cultured AEC II. This could result from detachment of β-catenin from cell–cell adhesion complex and degradation by proteasomes during AEC II isolation.
miR-8 negatively regulates Wnt/Wingless pathway at multiple levels by directly targeting wntless and CG32767 and by repressing the protein level of T-cell factor in Drosophila (50). The depression of the Wnt signaling by miR-8 leads to promotion of adipogenesis in mammals (50). Canonical Wnt signaling is important for osteoblast differentiation. miR-29a modulates osteoblast differentiation by directly targeting the negative regulators of Wnt/β-catenin signaling, namely Dkk1, Kremen2 and sFRP2. On the other hand, the miR-29a promoter activity is under the regulation of canonical Wnt pathway (51). The activation of canonical Wnt pathway induces miR-29a transcription, which subsequently downregulates Wnt antagonists and induces the Wnt signaling even further.
Although we are the first to report that the Wnt/β-catenin pathway is regulated by miR-375, we are not the only group that is interested in the relation between miR-375 and this pathway. The downregulation of miR-375 is associated with β-catenin mutations in hepatocellular tumors (48), suggesting that the activation of β-catenin signaling represses miR-375. Our results showed that miR-375 can inhibit the Wnt/β-catenin pathway. During trans-differentiation, miR-375 is downregulated and the β-catenin signaling is activated, which could lead to further depression of miR-375. It is likely that miR-375 regulates AEC trans-differentiation through this feedback loop. In this process, miR-375 may work as a switch of trans-differentiation. Once miR-375 decreases at the beginning of trans-differentiation, the Wnt/β-catenin signaling is activated, which leads to further depression of miR-375 expression, and the process cannot go backward.
To investigate the mechanism of how miR-375 regulates AEC trans-differentiation through the Wnt/β-catenin pathway, we identified FZD8 as a target of miR-375. FZD8 is one of the Wnt receptors. The binding of Wnt ligands and the FZD8 receptor leads to the stabilization of cytoplasmic β-catenin and activate canonical Wnt pathway. Whole-mount RNA in situ hybridizations have shown that FZD8 is highly expressed throughout the epithelium during the early stage of the mouse lung development (36). We demonstrate in this study that miR-375 downregulates the mRNA and protein levels of FZD8 during AEC trans-differentiation, leading to the inhibition of the Wnt/β-catenin pathway.
FZD8 is not likely the only target of miR-375. Targetscan software predicts 141 and 117 targets for human and rat miR-375. However, many of the predictions cannot be verified experimentally. Furthermore, the relationship between miRNAs and targets is also cell content- and developmental stage-dependent. One of the components of the Wnt/β-catenin signaling, CSNK2A1 (casein kinase II), is another predicted target of miR-375; PDK1 has been experimentally verified as a direct target of miR-375 in gastric carcinomas (11). However, both CSNK2A1 and PDK1 protein levels were not changed during the trans-differentiation of AEC II to AEC I and thus are not likely targets of miR-375 in AECs. Other targets identified in literature including yes-associated protein, neuronal RNA-binding protein HuD, Dexamthasone-induced Ras-related protein 1 (RASD1) and Mtpn are not obviously related to Wnt/β-catenin signaling (12,52,53).
AEC trans-differentiation is involved in IPF pathogenesis and progression. It has been reported that Wnt/β-catenin pathway is significantly activated in AEC II from IPF patients (54,55). Increased Wnt/β-catenin signaling induces AEC proliferation and probably trans-differentiation. In another word, a process similar to AEC trans-differentiation is involved in IPF. There is a negative correlation between the expression of miR-375 and activation of Wnt/β-catenin signaling in IPF, which further indicates that miR-375 may modulate AEC trans-differentiation through the regulation of the canonical Wnt signaling pathway in the diseased state.
AEC trans-differentiation is important for the recovery of injured alveolar epithelium resulting from a variety of disease conditions. Thus, the findings from this study may shed light on the recovery of lung injuries. The discovery that downregulation of miR-375 and activation of the Wnt/β-catenin pathway are necessary for this process may provide potential targets for therapeutic intervention. In addition, it is also well known that abnormal activation of Wnt signaling resulting from several different genetic defects causes cancer (56). It is possible that the inhibition of Wnt signaling by miR-375 can be used as a therapeutic method to treat cancer.
In summary, we identified a new function of miR-375 in AEC trans-differentiation. The regulation of the trans-differentiation is through the inhibition of the canonical Wnt pathway likely by direct targeting of FZD8. This discovery may provide potential targets for therapeutic intervention in the recovery from lung injuries.
FUNDING
Funding for open access charge: National Institutes of Health [HL071628, HL087884 and HL095383 to L.L.]; American Heart Association predoctoral fellowships [08100162 to Y.W.], [06101432 to T.W.] and [09PRE2300211 to Y.G.].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Dr. Mary Williams (Boston University) and Dr. Angela Barth (Stanford University) for kindly providing anti-T1α antibodies and ΔGSK-β-catenin construct. This study used biological specimens and data provided by the Lung Tissue Research Consortium (LTRC), supported by the National Heart, Lung, and Blood Institute (NHLBI).
REFERENCES
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y, Stricker HM, Gou D, Liu L. MicroRNA: past and present. Front. Biosci. 2007;12:2316–2329. doi: 10.2741/2234. [DOI] [PubMed] [Google Scholar]
- 4.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 5.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 6.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu. Rev. Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 7.Gangaraju VK, Lin H. MicroRNAs: key regulators of stem cells. Nat. Rev. Mol. Cell Biol. 2009;10:116–125. doi: 10.1038/nrm2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 10.Kloosterman WP, Lagendijk AK, Ketting RF, Moulton JD, Plasterk RH. Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development. PLoS Biol. 2007;5:e203. doi: 10.1371/journal.pbio.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El Ouaamari A, Baroukh N, Martens GA, Lebrun P, Pipeleers D, van Obberghen E. miR-375 targets 3'-phosphoinositide-dependent protein kinase-1 and regulates glucose-induced biological responses in pancreatic beta-cells. Diabetes. 2008;57:2708–2717. doi: 10.2337/db07-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu AM, Poon RT, Luk JM. MicroRNA-375 targets Hippo-signaling effector YAP in liver cancer and inhibits tumor properties. Biochem. Biophys. Res. Commun. 2010;394:623–627. doi: 10.1016/j.bbrc.2010.03.036. [DOI] [PubMed] [Google Scholar]
- 13.Ding L, Xu Y, Zhang W, Deng Y, Si M, Du Y, Yao H, Liu X, Ke Y, Si J, et al. MiR-375 frequently downregulated in gastric cancer inhibits cell proliferation by targeting JAK2. Cell Res. 2010;20:784–793. doi: 10.1038/cr.2010.79. [DOI] [PubMed] [Google Scholar]
- 14.Tsukamoto Y, Nakada C, Noguchi T, Tanigawa M, Nguyen LT, Uchida T, Hijiya N, Matsuura K, Fujioka T, Seto M, et al. MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer Res. 2010;70:2339–2349. doi: 10.1158/0008-5472.CAN-09-2777. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Weng T, Gou D, Chen Z, Chintagari NR, Liu L. Identification of rat lung-specific microRNAs by micoRNA microarray: valuable discoveries for the facilitation of lung research. BMC Genomics. 2007;8:29. doi: 10.1186/1471-2164-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fehrenbach H. Alveolar epithelial type II cell: defender of the alveolus revisited. Respir. Res. 2001;2:33–46. doi: 10.1186/rr36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams MC. Alveolar type I cells: molecular phenotype and development. Annu. Rev. Physiol. 2003;65:669–695. doi: 10.1146/annurev.physiol.65.092101.142446. [DOI] [PubMed] [Google Scholar]
- 18.Chen J, Chen Z, Chintagari NR, Bhaskaran M, Jin N, Narasaraju T, Liu L. Alveolar type I cells protect rat lung epithelium from oxidative injury. J. Physiol. 2006;572:625–638. doi: 10.1113/jphysiol.2005.103465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adamson IY, Bowden DH. The type 2 cell as progenitor of alveolar epithelial regeneration. A cytodynamic study in mice after exposure to oxygen. Lab. Invest. 1974;30:35–42. [PubMed] [Google Scholar]
- 20.Crandall ED, Matthay MA. Alveolar epithelial transport. Basic science to clinical medicine. Am. J. Respir. Crit. Care Med. 2001;163:1021–1029. doi: 10.1164/ajrccm.163.4.2006116. [DOI] [PubMed] [Google Scholar]
- 21.Kasper M, Haroske G. Alterations in the alveolar epithelium after injury leading to pulmonary fibrosis. Histol. Histopathol. 1996;11:463–483. [PubMed] [Google Scholar]
- 22.Evans MJ, Cabral LJ, Stephens RJ, Freeman G. Renewal of alveolar epithelium in the rat following exposure to NO2. Am. J. Pathol. 1973;70:175–198. [PMC free article] [PubMed] [Google Scholar]
- 23.Evans MJ, Cabral LJ, Stephens RJ, Freeman G. Transformation of alveolar type 2 cells to type 1 cells following exposure to NO2. Exp. Mol. Pathol. 1975;22:142–150. doi: 10.1016/0014-4800(75)90059-3. [DOI] [PubMed] [Google Scholar]
- 24.Dobbs LG, Williams MC, Brandt AE. Changes in biochemical characteristics and pattern of lectin binding of alveolar type II cells with time in culture. Biochim. Biophys. Acta. 1985;846:155–166. doi: 10.1016/0167-4889(85)90121-1. [DOI] [PubMed] [Google Scholar]
- 25.Cheek JM, Evans MJ, Crandall ED. Type I cell-like morphology in tight alveolar epithelial monolayers. Exp. Cell Res. 1989;184:375–387. doi: 10.1016/0014-4827(89)90337-6. [DOI] [PubMed] [Google Scholar]
- 26.Dobbs LG. Isolation and culture of alveolar type II cells. Am. J. Physiol. 1990;258:L134–L147. doi: 10.1152/ajplung.1990.258.4.L134. [DOI] [PubMed] [Google Scholar]
- 27.Bhaskaran M, Kolliputi N, Wang Y, Gou D, Chintagari NR, Liu L. Trans-differentiation of alveolar epithelial type II cells to type I cells involves autocrine signaling by transforming growth factor beta1 through the Smad pathway. J. Biol. Chem. 2007;282:3968–3976. doi: 10.1074/jbc.M609060200. [DOI] [PubMed] [Google Scholar]
- 28.Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev. Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mucenski ML, Wert SE, Nation JM, Loudy DE, Huelsken J, Birchmeier W, Morrisey EE, Whitsett JA. beta-Catenin is required for specification of proximal/distal cell fate during lung morphogenesis. J. Biol. Chem. 2003;278:40231–40238. doi: 10.1074/jbc.M305892200. [DOI] [PubMed] [Google Scholar]
- 31.Shu W, Guttentag S, Wang Z, Andl T, Ballard P, Lu MM, Piccolo S, Birchmeier W, Whitsett JA, Millar SE, et al. Wnt/beta-catenin signaling acts upstream of N-myc, BMP4, and FGF signaling to regulate proximal-distal patterning in the lung. Dev. Biol. 2005;283:226–239. doi: 10.1016/j.ydbio.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 32.Mucenski ML, Nation JM, Thitoff AR, Besnard V, Xu Y, Wert SE, Harada N, Taketo MM, Stahlman MT, Whitsett JA. Beta-catenin regulates differentiation of respiratory epithelial cells in vivo. Am. J. Physiol. Lung. Cell. Mol. Physiol. 2005;289:L971–L979. doi: 10.1152/ajplung.00172.2005. [DOI] [PubMed] [Google Scholar]
- 33.Li C, Xiao J, Hormi K, Borok Z, Minoo P. Wnt5a participates in distal lung morphogenesis. Dev. Biol. 2002;248:68–81. doi: 10.1006/dbio.2002.0729. [DOI] [PubMed] [Google Scholar]
- 34.Weidenfeld J, Shu W, Zhang L, Millar SE, Morrisey EE. The WNT7b promoter is regulated by TTF-1, GATA6, and Foxa2 in lung epithelium. J. Biol. Chem. 2002;277:21061–21070. doi: 10.1074/jbc.M111702200. [DOI] [PubMed] [Google Scholar]
- 35.Morrisey EE. Wnt signaling and pulmonary fibrosis. Am. J. Pathol. 2003;162:1393–1397. doi: 10.1016/S0002-9440(10)64271-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Langhe SP, Sala FG, Del Moral PM, Fairbanks TJ, Yamada KM, Warburton D, Burns RC, Bellusci S. Dickkopf-1 (DKK1) reveals that fibronectin is a major target of Wnt signaling in branching morphogenesis of the mouse embryonic lung. Dev. Biol. 2005;277:316–331. doi: 10.1016/j.ydbio.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 37.Flozak AS, Lam AP, Russell S, Jain M, Peled ON, Sheppard KA, Beri R, Mutlu GM, Budinger GR, Gottardi CJ. Beta-catenin/T-cell factor signaling is activated during lung injury and promotes the survival and migration of alveolar epithelial cells. J. Biol. Chem. 2010;285:3157–3167. doi: 10.1074/jbc.M109.070326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi R, Chiang VL. Facile means for quantifying microRNA expression by real-time PCR. Biotechniques. 2005;39:519–525. doi: 10.2144/000112010. [DOI] [PubMed] [Google Scholar]
- 39.Bhaskaran M, Wang Y, Zhang H, Weng T, Baviskar P, Guo Y, Gou D, Liu L. MicroRNA-127 modulates fetal lung development. Physiol. Genomics. 2009;37:268–278. doi: 10.1152/physiolgenomics.90268.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang H, Mishra A, Chintagari NR, Gou D, Liu L. Micro-RNA-375 inhibits lung surfactant secretion by altering cytoskeleton reorganization. IUBMB Life. 2009;62:78–83. doi: 10.1002/iub.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gou D, Weng T, Wang Y, Wang Z, Zhang H, Gao L, Chen Z, Wang P, Liu L. A novel approach for the construction of multiple shRNA expression vectors. J. Gene Med. 2007;9:751–763. doi: 10.1002/jgm.1080. [DOI] [PubMed] [Google Scholar]
- 42.Barth AI, Stewart DB, Nelson WJ. T cell factor-activated transcription is not sufficient to induce anchorage-independent growth of epithelial cells expressing mutant beta-catenin. Proc. Natl. Acad. Sci. USA. 1999;96:4947–4952. doi: 10.1073/pnas.96.9.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shtutman M, Zhurinsky J, Simcha I, Albanese C, D'Amico M, Pestell R, Ben-Ze'ev A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc. Natl. Acad. Sci. USA. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meltzer EB, Noble PW. Idiopathic pulmonary fibrosis. Orphanet J. Rare Dis. 2008;3:8. doi: 10.1186/1750-1172-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Willis BC, Liebler JM, Luby-Phelps K, Nicholson AG, Crandall ED, du Bois RM, Borok Z. Induction of epithelial-mesenchymal transition in alveolar epithelial cells by transforming growth factor-beta1: potential role in idiopathic pulmonary fibrosis. Am. J. Pathol. 2005;166:1321–1332. doi: 10.1016/s0002-9440(10)62351-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poy MN, Hausser J, Trajkovski M, Braun M, Collins S, Rorsman P, Zavolan M, Stoffel M. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc. Natl. Acad. Sci. USA. 2009;106:5813–5818. doi: 10.1073/pnas.0810550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Avissar M, Christensen BC, Kelsey KT, Marsit CJ. MicroRNA expression ratio is predictive of head and neck squamous cell carcinoma. Clin. Cancer Res. 2009;15:2850–2855. doi: 10.1158/1078-0432.CCR-08-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ladeiro Y, Couchy G, Balabaud C, Bioulac-Sage P, Pelletier L, Rebouissou S, Zucman-Rossi J. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47:1955–1963. doi: 10.1002/hep.22256. [DOI] [PubMed] [Google Scholar]
- 49.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 50.Kennell JA, Gerin I, MacDougald OA, Cadigan KM. The microRNA miR-8 is a conserved negative regulator of Wnt signaling. Proc. Natl. Acad. Sci. USA. 2008;105:15417–15422. doi: 10.1073/pnas.0807763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kapinas K, Kessler C, Ricks T, Gronowicz G, Delany AM. miR-29 modulates Wnt signaling in human osteoblasts through a positive feedback loop. J. Biol. Chem. 2010;285:25221–25231. doi: 10.1074/jbc.M110.116137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abdelmohsen K, Hutchison ER, Lee EK, Kuwano Y, Kim MM, Masuda K, Srikantan S, Subaran SS, Marasa BS, Mattson MP, et al. miR-375 inhibits differentiation of neurites by lowering HuD levels. Mol. Cell Biol. 2010;30:4197–4210. doi: 10.1128/MCB.00316-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Souza Rocha Simonini P, Breiling A, Gupta N, Malekpour M, Youns M, Omranipour R, Malekpour F, Volinia S, Croce CM, Najmabadi H, et al. Epigenetically deregulated microRNA-375 is involved in a positive feedback loop with estrogen receptor alpha in breast cancer cells. Cancer Res. 2010;70:9175–9184. doi: 10.1158/0008-5472.CAN-10-1318. [DOI] [PubMed] [Google Scholar]
- 54.Chilosi M, Poletti V, Zamo A, Lestani M, Montagna L, Piccoli P, Pedron S, Bertaso M, Scarpa A, Murer B, et al. Aberrant Wnt/beta-catenin pathway activation in idiopathic pulmonary fibrosis. Am. J. Pathol. 2003;162:1495–1502. doi: 10.1016/s0002-9440(10)64282-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Konigshoff M, Balsara N, Pfaff EM, Kramer M, Chrobak I, Seeger W, Eickelberg O. Functional Wnt signaling is increased in idiopathic pulmonary fibrosis. PLoS One. 2008;3:e2142. doi: 10.1371/journal.pone.0002142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]