Abstract
In animal ribosomes, two stalk proteins P1 and P2 form a heterodimer, and the two dimers, with the anchor protein P0, constitute a pentameric complex crucial for recruitment of translational GTPase factors to the ribosome. To investigate the functional contribution of each copy of the stalk proteins, we constructed P0 mutants, in which one of the two C-terminal helices, namely helix I (N-terminal side) or helix II (C-terminal side) were unable to bind the P1–P2 dimer. We also constructed ‘one-C-terminal domain (CTD) stalk dimers’, P1–P2ΔC and P1ΔC–P2, composed of intact P1/P2 monomer and a CTD-truncated partner. Through combinations of P0 and P1–P2 variants, various complexes were reconstituted and their function tested in eEF-2-dependent GTPase and eEF-1α/eEF-2-dependent polyphenylalanine synthesis assays in vitro. Double/single-CTD dimers bound to helix I showed higher activity than that bound to helix II. Despite low polypeptide synthetic activity by a single one-CTD dimer, its binding to both helices considerably increased activity, suggesting that two stalk dimers cooperate, particularly in polypeptide synthesis. This promotion of activity by two stalk dimers was lost upon mutation of the conserved YPT sequence connecting the two helices of P0, suggesting a role for this sequence in cooperativity of two stalk dimers.
INTRODUCTION
The dynamic process of mRNA translation is facilitated by interactions of the ribosome with several translational GTPase factors (1–3). The ribosomal large subunits contain a characteristic protein complex in all organisms, termed the stalk complex, responsible for the recruitment of GTPases and GTP hydrolysis (4–9). The stalk complex is composed of multiple copies of a flexible acidic protein, or so-called stalk protein, together with the anchor protein or stalk base. The stalk protein, a key component of this functional unit, is composed of an N-terminal domain (NTD) that participates in dimerization (6,10–14), a C-terminal domain (CTD) responsible for direct binding to GTPases (9,15–18) and a flexible hinge region connecting the N- and C-domains (19). Despite sharing a common functionality, structures and assembly modes of stalk complexes are markedly different between the three domains of life.
In bacteria, the stalk protein L12 forms a homodimer by hydrophobic interaction between the N-terminal 30-residue region, which forms antiparallel V-shaped α1–α2 hairpins. Subsequently, two or three L12–L12 homodimers bind via their NTDs to the C-terminal spine helix of the anchor protein L10 (6). The CTD of bacterial L12 is a globular structure that is composed of 70 amino acids (20). Binding of the CTD to the GTPase factors EF-G, EF-Tu and IF-2 has been demonstrated by analyses of NMR chemical shifts (15), cross-linking (16), cryo-EM (17) and X-ray crystallography (18). In contrast, the eukaryotic stalk proteins P1/P2 and the archaeal counterpart aP1 have a large N-terminal dimerization domain composed of ∼70 amino acids, which form a globular α1–α4 structure (12,14), and a small CTD composed of ∼20 amino acids, which is responsible for binding to GTPases (9). Archaeal aP1 forms a homodimer and three dimers bind to the C-terminal spine helices of the anchor protein aP0 (12), while eukaryotic P1 and P2 form a heterodimer (13,14,21–23) and two P1–P2 heterodimers bind to the C-terminal helices of P0 (24–27). Eukaryotic P1/P2 and archaeal aP1 are related closely to each other, but not to bacterial L12, with respect to amino acid sequence (28,29). It has been therefore suggested that eukaryotic/archaeal stalk protein might not be linked evolutionally to bacterial L12.
Given the essential role of the stalk protein complex in ribosome function, the distinct variations of structure and organization of the complexes in eukaryotes, archaea and bacteria appear to reflect their characteristic functional modes within the different domains of life. The eukaryotic stalk complex is of particular interest. It is composed of three components P0, P1 and P2, which share a common CTD (30). In lower eukaryotes (e.g., Saccharomyces cerevisiae), P1 and P2 have evolved into four different components: P1α, P1β, P2α and P2β (31,32). The most prominent characteristic of eukaryotic stalks including those from S. cerevisiae is that they form a P1–P2 heterodimer, whose structure is asymmetrical (13,14), unlike the archaeal aP1–aP1 homodimer (12). Starting from this asymmetric feature of the P1–P2 structure, Lee et al. (14) recently proposed that two heterodimers assemble on the C-terminal spine helices of P0 in a P2–P1:P1–P2 mode, in which P1 faces itself via hydrophobic interactions. In this model, the two neighbouring P1 components form a cavity that accommodates a loop region containing the conserved YPT motif between the two helices. The conserved YPT sequence, therefore, might be relevant to cooperativity in binding between P0 and P1–P2. In addition, it has been shown by cross-linking experiments that the locations of the CTDs of P1 and P2 in the S. cerevisiae ribosome are different, suggesting distinct roles for P1 and P2 on the ribosome (33). These characteristics of eukaryotic stalk complex are likely to be related to the sophisticated regulation of the stalk assembly and translation mechanism, although there remain issues to be clarified.
The present study focuses on the functional contribution of each copy of the stalk proteins, P1 and P2. P0 mutants lacking the ability to bind to either one of the two P1–P2 dimers were constructed. The stalk complexes were reconstituted in vitro with the P0 mutants, together with one-CTD stalk dimer lacking the CTD of either P1 or P2, and assayed for their activity in GTPase assays and in polypeptide synthesis dependent on eukaryotic elongation factors by using a hybrid ribosome system (34). We could show not only the activity of individual copies of the stalks, but also functional cooperativity between the two stalk dimers. We also detected a novel functional contribution of the conserved YPT motif between two spine helices of P0, to which P1–P2 dimers bind individually, suggesting that the YPT region is involved in the functional cooperativity of the two stalk dimers.
MATERIALS AND METHODS
Plasmid construction, protein expression and purification
The cDNAs for Bombyx mori ribosomal protein full-length P0 and the truncation mutant lacking the C-terminal 55 amino acids (P0ΔC) were amplified by PCR and inserted into the NcoI and BamHI sites of pET15b (Novagen). Double mutations F215Q/V222Q in helix I (P0mutI) and I241Q/L248Q in helix II (P0mutII) were introduced by a PCR-based site-directed mutagenesis method (35) (see Supplementary Table S1 for primers used). The same methodology was used for replacement of the 231-YPTI-234 sequence in the loop region between helices I and II with 231-GGGG-234 (P0mutM), as well as for deletion of this sequence (P0ΔM). The coding sequences for B. mori ribosomal proteins P1, P2 and eL12 (a eukaryotic homologue of prokaryotic L11) were cloned into pET28c or pET3a (Novagen), as described previously (25). The truncation mutants lacking the C-terminal 52 amino acids of P1 (P1ΔC) and lacking 50 amino acids of P2 (P2ΔC) were constructed as described previously (36). The nucleotide sequences of the constructed plasmids were confirmed by DNA sequencing analyses. Expression and purification of each ribosomal protein was performed as described previously (25). The purity of all proteins was checked by SDS-PAGE.
In vitro reconstitution of the stalk complex
The stalk complexes were reconstituted by mixing isolated P0, P1 and P2, or alternative individual mutants, in the presence of 7 M urea at a molar ratio of 1:3:3, as described previously (22). An rRNA fragment containing residues 1841–1939 of rat 28 S rRNA (corresponding to residues 1043–1112 of Escherichia coli 23 S rRNA) was synthesized in vitro using template DNA and SP6 RNA polymerase (25). The P0–P1–P2–rRNA complex was formed by adding 3-fold amounts of the rRNA fragment to 100 pmol of the P0–P1–P2 complex. Individual complexes were analysed by 6% polyacrylamide gel electrophoresis (22). The gel was stained with Coomassie Brilliant Blue.
Ribosomal 50S core and hybrid 50S particles
Ribosomal 50S subunits were prepared from the L11-deficient E. coli strain AM68 (37), as previously described (34). The 50S core particles deficient both in L10 and L7/L12 were prepared by extraction of the L11-deficient 50S subunits, as previously described (34). The B. mori–E. coli hybrid 50S particle was formed by mixing the E. coli 50S core with the reconstituted stalk complex P0–P1–P2 and eL12 (25).
Quantitative analysis of P1–P2 heterodimer incorporated into the ribosome
Bombyx mori ribosomal protein P2 was radiolabelled by phosphorylation using [γ-32P]ATP and casein kinase II (New England Biolabs), as described previously (25). The stalk complex and its variants were reconstituted with the labelled [32P]P2, as described above. The labelled stalk complexes (90 pmol) were incubated with 50S core particles (30 pmol) supplemented with eL12 (90 pmol) in a solution (20 μl) that contained 20 mM Tris–HCl (pH 7.6), 5 mM MgCl2, 50 mM NH4Cl and 5 mM 2-mercaptoethanol at 37°C for 10 min. The mixture was layered onto a 10–28% sucrose gradient and centrifuged at 40 000rpm at 4°C for 3 h in a Hitachi S52ST rotor, as described previously (25). The fraction that contained the 50S particles was collected, and the amount of associated P1–P2 dimer was estimated on the basis of the amount of radioactivity, considering the specific radioactivity of [32P]P2 to be 840 cpm/pmol.
Ribosome functional assays
Eukaryotic eEF-1α and eEF-2 were isolated from pig liver, as previously described (38). The activity of the hybrid ribosome with respect to eEF-2-dependent GTPase activity and eEF-1α/eEF-2-dependent polyphenylalanine synthesis was assayed as described previously (25,34).
RESULTS
Binding sites for P1–P2 at the C-terminal helices of P0
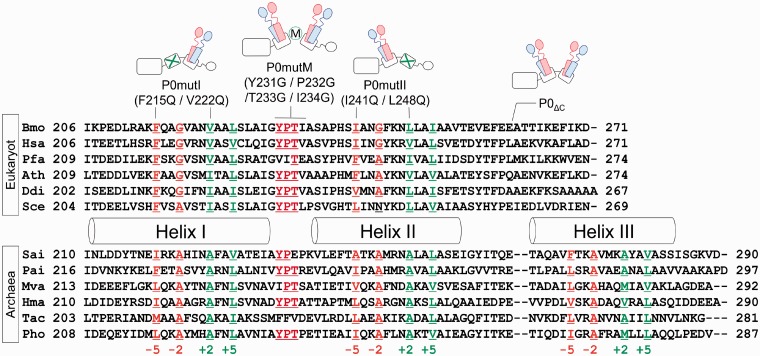
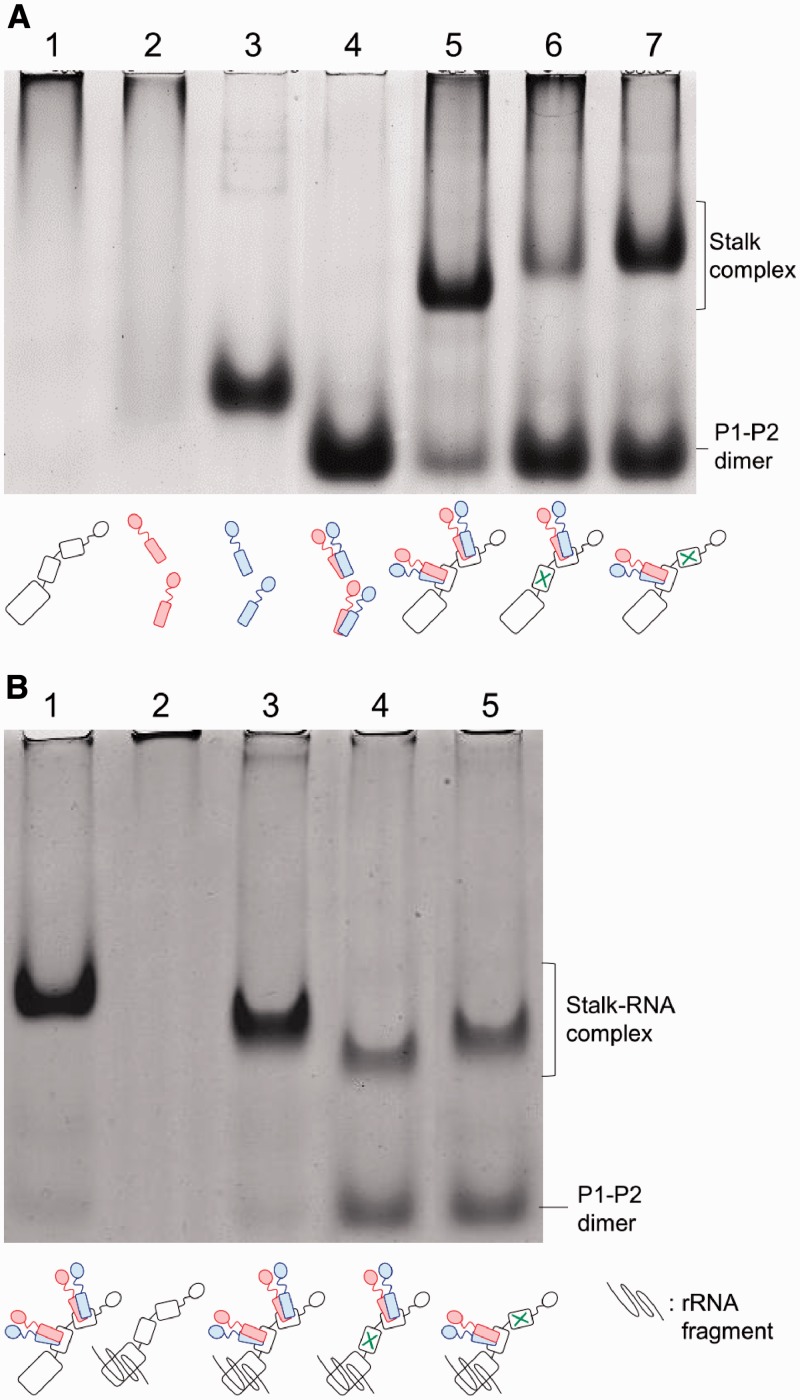
Based on previous data on the archaeal stalk complex, we assumed that the hydrophobic residues F215, G218, V222 and L225 in helix I, and I241, G244, L248 and I251 in helix II of B. moli P0, which are located at positions −5, −2, +2 and +5 in the conserved 2-fold symmetric distribution (Figure 1, Supplementary Figure S1A, also see Supplementary methods), are involved in binding of P1–P2 heterodimer in each of the helicies I and II of P0. We constructed the P0 mutants P0mutI and P0mutII, which contained double mutations at positions −5 and +2, i.e., F215Q/V222Q and I241Q/L248Q, respectively. These P0 mutants were expressed in E. coli cells and purified, as described in ‘MATERIALS AND METHODS’ (Supplementary Figure S2). Each of the P0 mutants was mixed with excess amounts of P1–P2 heterodimer in vitro, and complex formation was analysed by native gel electrophoresis (Figure 2A). In contrast to the failure of isolated P0 to run into the gel (Figure 2A, lane 1), and to the smearing pattern of isolated P1 obtained because of its tendency to aggregate (Figure 2A, lane 2) (39), the P0–P1–P2 stalk complex produced a distinct band (lane 5), whose mobility was clearly lower than that of isolated P2 (lane 3) and P1–P2 heterodimer (lane 4), as described previously (22). The complexes were also formed with P0mutI (lane 6) and P0mutII (lane 7) mutants. The mobility of the complexes with the mutant P0 was slightly lower than that of the wild-type P0 (lane 5). It is noteworthy that, compared with the gel pattern of the wild-type complex sample, there remained larger amounts of free P1–P2 dimer with high gel mobility in samples containing the mutant P0 (lanes 6 and 7), and that the band corresponding to the P0mutI–P1–P2 complex (lane 6) was thinner than that for the P0mutII–P1–P2 complex (lane 7). It has been reported that binding of P0 to an rRNA fragment covering the P0-binding site stabilizes a variant of the P0–P1–P2 complex (25). We therefore performed the same gel electrophoresis after adding the rRNA fragment. As shown in Figure 2B, the electrophoretic mobility of the wild-type P0–P1–P2 complex (Figure 2B, lane 1) became slightly higher upon addition of the RNA (lane 3). The complexes containing the P0 mutants also migrated faster after addition of the RNA (Figure 2B, lanes 4 and 5) when compared with the same samples without the RNA (Figure 2A, lanes 6 and 7, respectively), suggesting that the RNA fragment bound to the P0mutI–P1–P2 and P0mutII–P1–P2 complexes. The thin band produced by the P0mutI–P1–P2 complex observed in Figure 2A (lane 6) became distinct upon RNA addition (Figure 2B, lane 4), suggesting that the RNA binding to P0mutI stabilizes the complex structure of P0mutI–P1–P2. It is again noticeable that even after addition of the RNA, larger amounts of free P1–P2 were observed in samples including the P0 mutants (lanes 4 and 5), compared with the wild-type P0 sample (lane 3). These results suggest that the P0mutI and P0mutII mutants may bind only a single P1–P2 heterodimer.
Figure 1.
Sequence comparison of the stalk-binding sites on the C-terminal helices of eukaryotic P0/archaeal aP0. Amino acid sequences around the C-terminal helices I and II of eukaryotic P0 (Bmo, Bombix mori; Hsa, Homo sapiens; Pfa, Plasmodium falciparum; Ath, Arabidopsis thaliana; Ddi, Dictyostelium discoideum; and Sce, Saccharomyces cerevisiae) and the helices I, II and III of archaeal aP0 (Sac, Sulfolobus acidocaldarius; Pae, Pyrobaculum aerophilum; Mva, Methanococcus vannielii; Hma, Haloarcula marismortui; Tac, Thermoplasma acidophilum; and Pho, Pyrococcus horikoshii) were compared. Helical regions defined by X-ray crystallography of PhoP0 (12) are represented as cylinders. Hydrophobic residues involved in binding of PhoP1 homodimers are located in 2-fold symmetric distribution at ±2 and ±5. These residues are coloured orange for residues at −5 and −2 and green for residues at +2 and +5. Mutations introduced are indicated above the sequences. The conserved YPT sequence located between helices I and II is coloured red. Mutations introduced in this study are indicated above the sequences and in the schematic models of the eukaryotic stalk complex. In this model, P0 is represented as three tandemly linked boxes and a circle (corresponding to the N-terminal domain, helix I, helix II and the CTD of P0, respectively), and P1 and P2 are represented in red and blue, respectively. The mutated helices are marked with a green X. Introduction of the mutations in the YPT sequence is represented by M surrounded by a green circle in these models.
Figure 2.
Formation of stalk complexes with P0 carrying mutations in helices I and II. (A) The complexes were reconstituted by incubation of P0 (lane 5), P0mutI (lane 6) and P0mutII (lane 7) with excess amounts of P1–P2 dimer. Each complex sample including 100 pmol of P0 (or its mutants) was subjected to native gel electrophoresis, as described in ‘Materials and Methods’ section. On the same gel, isolated P0 (100 pmol, lane 1), P1 (300 pmol, lane 2), P2 (300 pmol, lane 3) and P1–P2 dimer (300 pmol, lane 4) were also loaded. The gel was stained with Coomassie Brilliant Blue. (B) The complexes were reconstituted by incubation of P0 (lanes 1, 3), P0mutI (lane 4) and P0mutII (lane 5) with P1–P2 dimer under the same conditions as (A). Each complex including 100 pmol of P0 (or its mutant) was incubated in the presence of the rRNA fragment (300 pmol) for the P0-binding site (lanes 3–5) or in its absence (lane 1), and subjected to native gel electrophoresis under the same conditions as (A). On the same gel, isolated P0 (100 pmol) mixed with the rRNA fragment was loaded (lane 2). The gel was stained with Coomassie Brilliant Blue. Schematic models of the stalk complexes are shown below individual lanes.
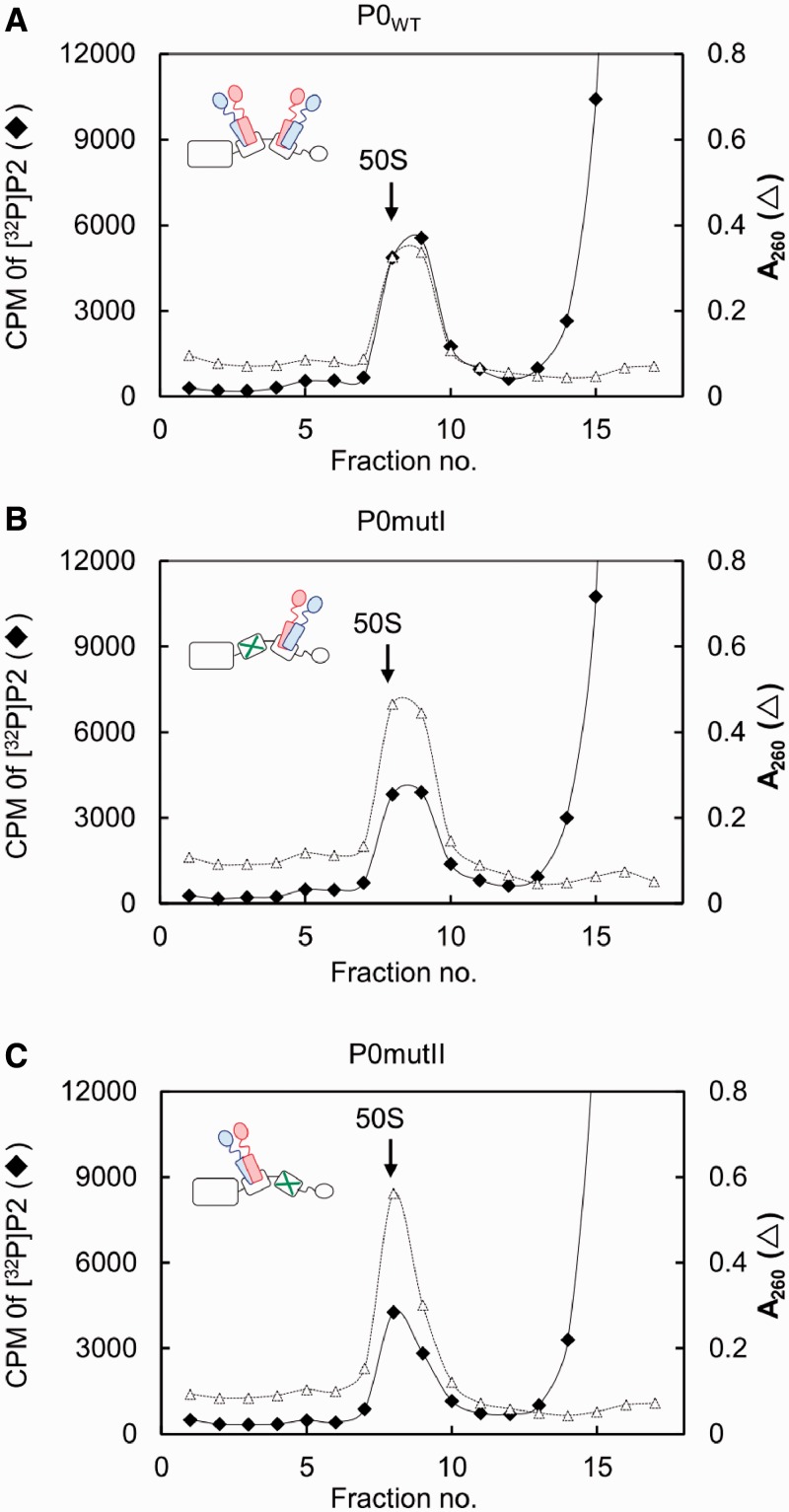
To determine stoichiometry of the complexes P0mutI–P1–P2 and P0mutII–P1–P2, we used the reconstituted radiolabelled P1–[32P]P2 heterodimer in the in vitro assembly of the stalk complexes, as described previously (25). The labelled stalk complex P0–P1–[32P]P2 formed by mixing of P0 and excess amounts of P1–[32P]P2 was substituted for the E. coli stalk complex L10–L12 on the 50S subunit. This eukaryote–bacterium hybrid form, a so-called the ‘hybrid ribosome’ has been successfully used for studies on assembly and function of eukaryotic and archaeal stalk complexes (34,40). In the present study, the P1–[32P]P2 heterodimer bound to P0 or its mutants and assembled onto the E. coli 50S core were separated by sucrose gradient centrifugation (Figure 3). Radioactivity in the 50S fraction was measured, and copy numbers of P1–P2 heterodimer were estimated on the basis of the amounts of 50S particles and bound P1–P2 that were estimated from the A260 and radioactivity values, respectively. The results, summarized in Table 1, clearly indicate that both P0mutI and P0mutII mutants have the ability to bind only a single copy of [32P]P2, implying a single P1–P2 heterodimer binds to individual P0 mutants. Therefore, the amino acid substitutions introduced into helix I and helix II (Figure 1) appear to disrupt P1–P2 binding to individual helices.
Figure 3.
Quantitative analysis of stalk dimers bound to P0 carrying mutations in helices I and II in the 50S particle. The 50S core particles (30 pmol) were mixed with excess amounts of the P0–P1–P2 complexes (90 pmol), which are composed of P1–[32P]P2 dimers (840 cpm/pmol) and P0, and eL12 (90 pmol) (A). The hybrid 50S particles were also reconstituted using P0mutI (B), and P0mutII (C), instead of P0. The hybrid 50S samples were separated by sucrose gradient centrifugation, as described in ‘Materials and Methods’ section. The amount of P2 incorporated to the 50S core was estimated by radioactivity of P2 and the absorbance at 260 nm of the 50S core.
Table 1.
Copy numbers for P2 bound to the 50S core particle in the presence of different P0 variants
| P0 type | Copies of P2 |
|---|---|
| WT | 2.22 ± 0.14 |
| mutI | 1.04 ± 0.14 |
| mutII | 1.14 ± 0.09 |
The copy number of labelled [32P]P2 (840 cpm/pmol) per 50S particle was calculated from absorbance measurements at 260 nm and the radioactivity of the 50S fractions separated by sucrose gradient centrifugation shown in Figure 3, as described in ‘Materials and Methods’ section.
Contributions of individual copies of P1 and P2 to ribosome function
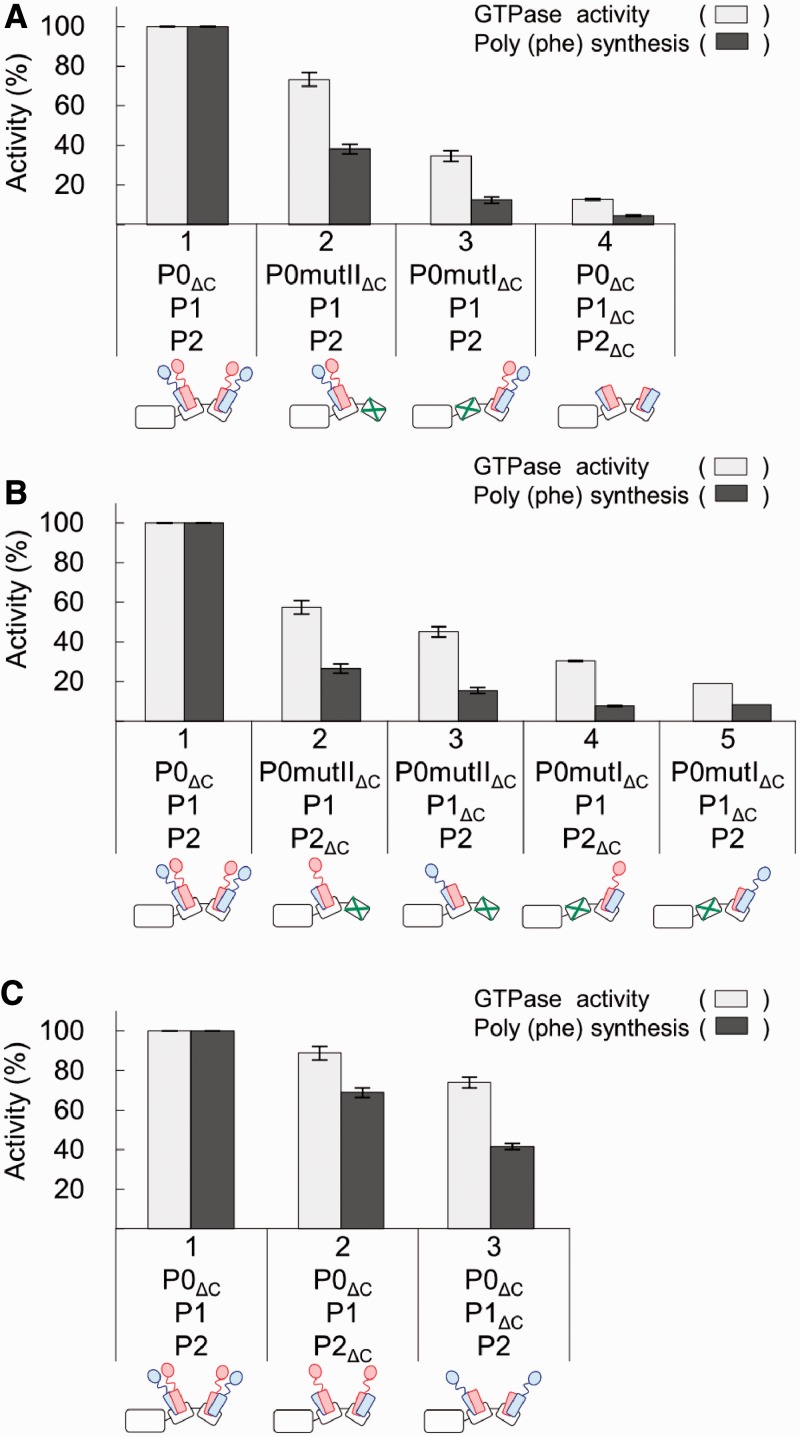
It has long been a question as to why eukaryotic ribosomes have two sets of heterodimers composed of P1 and P2, which share a common CTD, in particular with regards to their functional contribution. In the present study, we used P0mutI and P0mutII mutants, which allow us to investigate the functional contributions of individual P1–P2 dimers at the different locations. To focus on the roles of P1 and P2, we used a P0ΔC mutant lacking the CTD of P0, a sequence shared by P1 and P2. Moreover, we constructed and used the one-CTD stalk dimers P1–P2ΔC and P1ΔC–P2, in which the CTD of either P1 or P2 is truncated (see ‘Materials and Methods’ section). The stalk complexes were reconstituted by various combinations of P0 and P1–P2 variants (Supplementary Figure S3), and incorporated onto the E. coli 50S core. The resultant hybrid ribosomes were assayed for their activity in eEF-2-dependent GTPase and eEF-1α/eEF-2-dependent polyphenylalanine synthesis assays (Figure 4).
Figure 4.
Contributions of individual copies of P1 and P2 to eEF-2-dependent GTPase activity and polyphenylalanine synthesis. The stalk complexes used, as represented in the schematic diagrams for the stalk complexes below the bars, were reconstituted in various combinations with the P0 and P1–P2 variants: (A) P0ΔC, P0mutIΔC or P0mutIIΔC, together with P1–P2 or P1ΔC–P2ΔC; (B) P0ΔC, P0mutIΔC or P0mutIIΔC, together with P1–P2, P1–P2ΔC or P1ΔC–P2; (C) P0ΔC, together with P1–P2, P1–P2ΔC or P1ΔC–P2. For the GTPase acivity, E. coli 50S core particles (2.5 pmol) were preincubated with each reconstituted P0–P1–P2 variant (10 pmol) and 10 pmol eL12, and then assayed for eukaryotic eEF-2-dependent GTPase activity (white bar). The 100% activity corresponds to 80.5 pmol of [32P]GTP hydrolysis per min. For the polypeptide synthetic activity, the 50S core particles (10 pmol) were preincubated with 20 pmol of the same P0–P1–P2 samples and eL12, and then assayed for polyphenylalanine synthesis dependent on eukaryotic eEF-1α and eEF-2 (black bar). The 100% activity corresponds to 16.6 pmol of [14C] phenylalanine polymerized for 10 min.
The hybrid ribosomes containing P0mutIIΔC–P1–P2 (carrying the single P1–P2 dimer bound to helix I) have low, but significant, levels of activity in GTPase assays (73% of P0ΔC–P1–P2) and polyphenylalanine synthesis (38% of P0ΔC–P1–P2), whereas these activities of the ribosomes containing P0mutIΔC–P1–P2 (carrying a single P1–P2 dimer bound to helix II) were only 35% and 12% that of the P0ΔC stalk complex carrying double P1–P2 dimers, respectively (Figure 4A). The results indicate that P1–P2 dimer bound to helix I makes a larger contribution to the factor-dependent activities than the dimer bound to helix II. The same characteristics were also observed when one-CTD stalk dimers P1–P2ΔC and P1ΔC–P2 were used, instead of the P1–P2 dimer, although the activities of one-head stalk dimers were lower than those of P1–P2 dimer (Figure 4B). It is noticeable that the P1–CTD stalk dimer (P1–P2ΔC) has slightly higher activities than P2–CTD dimer (P1ΔC–P2).
The activities of the hybrid ribosomes containing P0ΔC–P1–P2ΔC and P0ΔC–P1ΔC–P2 (carrying double one-CTD dimers bound to both helicies I and II) were also assayed (Figure 4C). The level of GTPase activity of each double one-CTD stalk complex was approximately the sum of the activities of single one-CTD stalk dimers bound to helices I and II (compare Figure 4C with B). In contrast, the level of polypeptide synthesis of each double one-CTD stalk complex was obviously higher than the sum of activities of individual ones (Figure 4C and B). The results suggest that the stalk bound to helix I cooperates with that bound to helix II, particularly in polypeptide synthesis.
It should be noted that the CTD of P0, which is shared by P1 and P2, also contributes to ribosome function (41). To examine its contribution to eEF-2-dependent GTPase and eEF-1α/eEF-2-dependent polyphenylalanine synthesis, we also performed functional assays with intact P0 carrying the CTD, instead of the P0ΔC mutant, and compared the data (Supplementary Figure S4) with that in Figure 4A. Although all of P0–P1–P2, P0mutI–P1–P2 and P0mutII–P1–P2 showed slightly higher activity (Supplementary Figure S4) than individual variants lacking the CTD of P0 (Figure 4A), the effect of mutations at helicies I (P0mutI) and II (P0mutII) was roughly comparable with the effects observed with P0mutIΔC and P0mutIIΔC. It is therefore likely that the CTD of P0 exerts its basal functionality, but both P1–P2 dimers do not depend on the presence of the CTD of P0.
Role of the conserved YPT motif between the two spine helices I and II
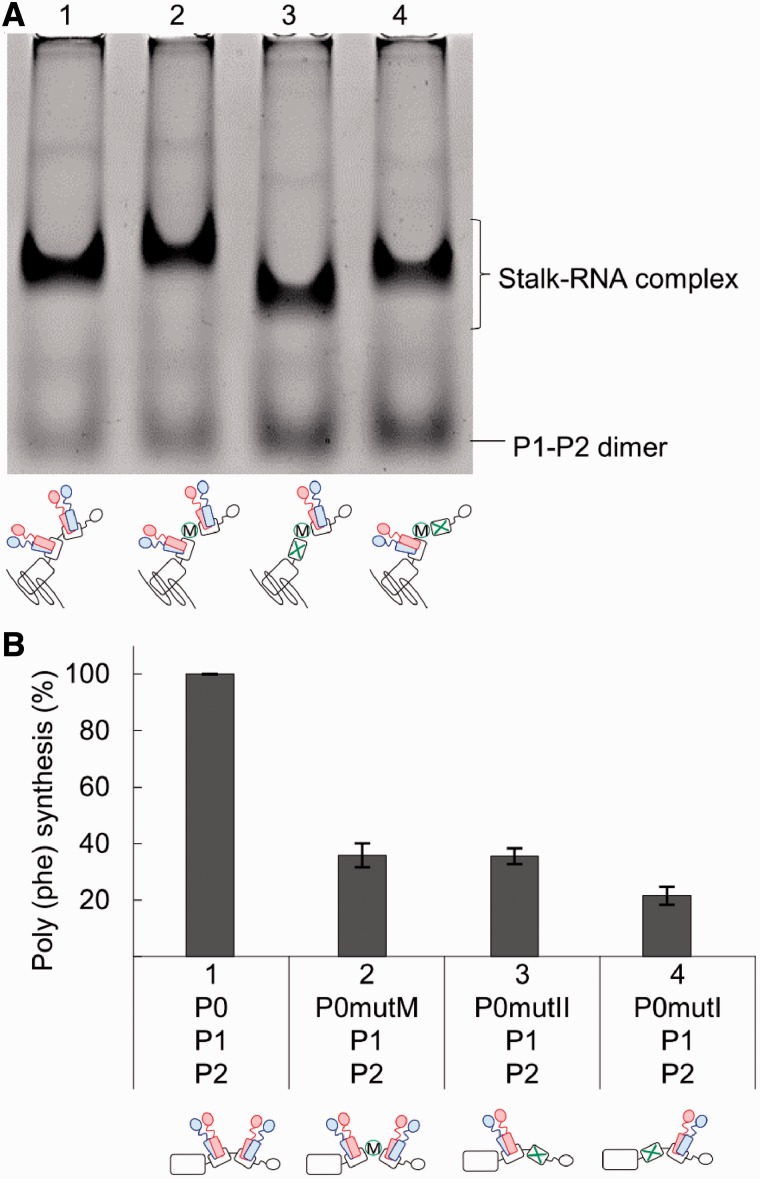
There is a conserved sequence motif, Thy-Pro-Thr, within a loop region between helices I and II of eukaryotic P0 and archaeal aP0. To examine the functional role of this region, we constructed the variants P0mutM, in which the 231-YPTI-234 sequence was changed to 231-GGGG-234 (Figure 5), and P0ΔM, in which the same sequence was deleted (Supplementary Figure S5). These P0 variants were expressed in E. coli cells and purified (Supplementary Figure S2). The ability of P0mutM to bind P1–P2 heterodimer was analysed by native gel electrophoresis in the presence of the rRNA fragment covering the P0-binding site (Figure 5A). A distinct band was observed for the stalk-rRNA complex containing P0mutM (Figure 5A, lane 2), and the intensity of the band was comparable with that of the wild-type complex (lane 1). The result suggests that the YPT motif does not have an essential role in P1–P2 binding. To confirm this suggestion, we introduced additional mutations into P0mutM, i.e., the P1–P2-binding site of either helix I (lane 3) or helix II (lane 4) was disrupted in P0mutM. The results clearly indicate that even after introduction of these mutations, individual P0 mutants still retain the ability to bind P1–P2 dimer (lanes 3 and 4), suggesting that the mutation of the 231-YPTI-234 sequence produced no detectable effect on individual binding of P1–P2 to helices I and II. To examine the functional effect of mutation of the 231-YPTI-234 sequence, the stalk complexes containing P0mutM were assembled into the 50S core and assayed for polyphenylalanine synthesis activity (Figure 5B). Unexpectedly, the activity of the complex containing P0mutM was significantly reduced (33% of P0–P1–P2), although both helices I and II of this mutant retain their P1–P2-binding ability (Figure 5A). A similar result was also obtained with the truncation mutant P0ΔM (Supplementary Figure S5B). It is noteworthy that the levels of activity of the complexes containing P0mutM and P0ΔM were comparable with the complex containing P0mutII (36% of P0–P1–P2), which can bind a single P1–P2 dimer to helix I.
Figure 5.
Role of the conserved YPT motif between the two spine helices I and II. (A) The stalk complexes, as represented in the schematic diagrams of the stalk complexes below the bars, were reconstituted in combinations of P1–P2 dimers with P0 (lane 1), P0mutM (lane 2), P0mutI/mutM (lane 3) and P0mutII/mutM (lane 4). Each complex sample containing 100 pmol of P0 (or its variant) with 300 pmol of the rRNA fragment for the P0-binding region was subjected to native gel electrophoresis, as described in ‘Materials and Methods’ section. The gel was stained with Coomassie Brilliant Blue. (B) The 50S core particles (10 pmol) were preincubated with the stalk complex sample P0–P1–P2, P0mutM–P1–P2, P0mutII–P1–P2 or P0mutI–P1–P2 (20 pmol each) and eL12 (20 pmol), and then assayed for polyphenylalanine synthesis dependent on eukaryotic eEF-1α and eEF-2. The 100% activity corresponds to 21.0 pmol of [14C] phenylalanine polymerized for 10 min.
DISCUSSION
The animal ribosomal stalk is composed of P0 and two copies of P1–P2 heterodimers, which individually bind to two tandem helices (I and II) of P0. In the present study, we identified hydrophobic amino acid residues F215/V222 in helix I and I241/L248 in helix II, as being crucial for individual P1–P2 binding to the helices. The hydrophobicity of these residues is conserved among eukaryotes and archaea. In the crystal structure of the archaeal aP0–aP1–aP1 stalk complex, it has been shown that four hydrophobic residues at positions −5, −2, +2 and +5 in a 2-fold symmetric distribution within each helix interact with α1/α1′ of the aP1–aP1 homodimer. The residues at −5 and −2 make contact with α1 in one aP1 molecule of the homodimer and the residues at +2 and +5 make contact with α1′ of the other aP1 molecule. We infer that the mode of binding between eukaryotic P0 and P1–P2 is similar to that of archaeal aP0 and aP1 homodimer on the basis of the following facts: (i) the 2-fold symmetric distribution of the hydrophobic residues is conserved from archaea to eukaryotes (Figure 1); (ii) the residues F215/V222 in helix I and I241/L248 in helix II at positions corresponding to −5 and +2 are crucially important for P1–P2 binding as revealed in the present study; (iii) it has been reported that the α1/α1′ of eukaryotic P1 and P2 are important for their binding to P0 as well as for dimerization (36,42). It is therefore likely that the four conserved hydrophobic residues in each the helix of eukaryotic P0 participate in binding of the P1–P2 heterodimer. To explore this possibility, we attempted to fit the crystal structural data for helix I of archaeal aP0 (12) to the NMR structural data for human P1–P2 dimer (α1 to α3) (14). In one such model (Supplementary Figure S1B), the four conserved hydrophobic residues of aP0 are able to bind to hydrophobic residues of the α1/α1′ helices of P1 and P2 (Supplementary Figure S1B), i.e., the residues at positions −5 and −2 of P0 can make contact with α1 of P2. Meanwhile, the residues at positions +2 and +5 can make contact with α1′ of P1, assuming that the two P1–P2 dimers assemble on the spine helices of P0 in a P2–P1:P1–P2 mode, as proposed by Lee et al. (14). This putative model, however, contrasts with models proposed previously, in which P1 plays an anchoring role for P2 (43,44).
The resultant mutants P0mutI (F215Q/V222Q) and P0mutII (I241Q/L248Q) actually bind only a single P1–P2 heterodimer (Figure 3). Using the P0mutI and P0mutII mutants, together with the one-CTD stalk heterodimers lacking the CTD of either P1 or P2, we investigated the functional contributions of individual copies of the P1/P2 stalk. Lower, but considerable, levels of eEF-2-dependent GTPase activity were shown by the P1–P2 dimer (Figure 4A) or one-CTD dimers (Figure 4B) bound only to helix I, compared with the activity in the control stalk complex carrying double P1–P2 dimers. Further reduced levels of GTPase activity were also detected in the same dimer samples bound only to helix II. In contrast, further lowering of activity in eEF-1α/eEF-2-dependent polyphenylalanine synthesis assays was observed in the dimer samples bound only to helix I, and a background level of activity was present in dimer samples bound only to helix II. The results suggest that the stalk dimer bound to helix II might make little contribution to polypeptide synthesis. However, when the P1–P2 dimer or one-CTD dimers bind to both helices I and II, high levels of polypeptide synthetic activity were observed (Figure 4A and C). It is therefore more likely that the P1–P2 stalk dimer bound to helix II works cooperatively with the stalk dimer bound to helix I, particularly in polypeptide synthesis. This may relate to the fact that two elongation factors eEF-1α and eEF-2 are required for the peptide polymerization reaction, and that collaboration of two stalk dimers may be required for alternate action of the two elongation factors for efficient polypeptide synthesis. The present experimental system provides straightforward evidence for functional collaboration of the two stalk dimers bound to helices I and II.
The different nature of the CTDs of P1 and P2 within the yeast ribosome has been revealed by previous cross-linking experiments (33); the CTD of P2 shows much higher reactivity towards several other ribosomal components than the CTD of P1. The present study shows that the P1-head dimer, composed of P1 and P2ΔC lacking the CTD, shows slightly, but reproducibly, higher activity than the P2-head dimer, composed of P2 and P1ΔC, with respect to polypeptide synthesis as well as eEF-2-dependent GTPase activity (Figure 4B and C). Our results indicate that both P1 and P2 contribute to ribosomal activity dependent on eEF-1α and eEF-2, and that the degree or quality of contributions seems to be slightly different between P1 and P2. Although it is hard to explain a correlation between the present functional data and previous cross-linking data at present, further detailed investigations with both approaches should provide evidence for differences in the functional nature of P1 and P2.
There is a loop region containing a conserved sequence motif, Thy-Pro-Thr, between helices I and II in eukaryotic P0 and archaeal aP0. Little information is available on the functional role of this region although Lee et al. (14) proposed an interesting hypothesis where they suggest that when two P1–P2 dimers bind individually to helices I and II, the α3 regions of the P1 molecules face each other by hydrophobic interaction and make a hydrophobic cavity, into which the YPT loop can fit. The present study demonstrated that a change of amino acid sequence or deletion of the YPT loop gave no detectable effect on binding of P1–P2 to both the helices of P0 (Figure 5A, Supplementary Figure S5A), but caused a marked effect on polypeptide synthesis (Figure 5B, Supplementary Figure S5B). The present experimental data indicate that the YPT loop is involved in function of the stalk complex, rather than the assembly of the stalk complex. It is, however, unlikely that the YPT loop plays a role in direct contact with translation factors, because of its probable location within a hydrophobic cavity (12,14). Considering the fact that the level of reduced activity of the YPT loop-defective mutants is comparable with that of P0mutII lacking the P1–P2 dimer bound to helix II, it seems likely that the disruption of the YPT loop causes loss in function of the stalk dimer bound to helix II by changing the angle or orientation of helix II with respect to helix I. Because the stalk dimer bound to helix II apparently collaborates with the dimer bound to helix I as discussed above, the YPT loop would seem to play a role in the cooperation of the two stalk dimers.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1–5, Supplementary Methods and Supplementary Reference [45].
FUNDING
Funding for open access charge: Grants-in-Aid for Scientific Research [No. 24370073] from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Kosuke Ito and Dr. Tomohiro Miyoshi of Niigata University for their helpful assistance with the research.
REFERENCES
- 1.Kaziro Y. The role of guanosine 5′-triphosphate in polypeptide chain elongation. Biochim. Biophys. Acta. 1978;505:95–127. doi: 10.1016/0304-4173(78)90009-5. [DOI] [PubMed] [Google Scholar]
- 2.Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 3.Liljas A. Structural aspects of protein synthesis. Singapore: World Scientific Publishing Co.; 2004. pp. 99–158. [Google Scholar]
- 4.Wahl MC, Möller W. Structure and function of the acidic ribosomal stalk proteins. Curr. Protein Pept. Sci. 2002;3:93–106. doi: 10.2174/1389203023380756. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalo P, Reboud JP. The puzzling lateral flexible stalk of the ribosome [Review] Biol. Cell. 2003;95:179–193. doi: 10.1016/s0248-4900(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 6.Diaconu M, Kothe U, Schlünzen F, Fischer N, Harms JM, Tonevitsky AG, Stark H, Rodnina MV, Wahl MC. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell. 2005;121:991–1004. doi: 10.1016/j.cell.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 7.Savelsbergh A, Mohr D, Kothe U, Wintermeyer W, Rodnina MV. Control of phosphate release from elongation factor G by ribosomal protein L7/12. EMBO J. 2005;24:4316–4323. doi: 10.1038/sj.emboj.7600884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mandava CS, Peisker K, Ederth J, Kumar R, Ge X, Szaflarski W, Sanyal S. Bacterial ribosome requires multiple L12 dimers for efficient initiation and elongation of protein synthesis involving IF2 and EF-G. Nucleic Acids Res. 2012;40:2054–2064. doi: 10.1093/nar/gkr1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nomura N, Honda T, Baba K, Naganuma T, Tanzawa T, Arisaka F, Noda M, Uchiyama S, Tanaka I, Yao M, et al. Archaeal ribosomal stalk protein interacts with translation factors in a nucleotide-independent manner via its conserved C terminus. Proc. Natl Acad. Sci. USA. 2012;109:3748–3753. doi: 10.1073/pnas.1112934109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wahl MC, Bourenkov GP, Bartunik HD, Huber R. Flexibility, conformational diversity and two dimerization modes in complexes of ribosomal protein L12. EMBO J. 2000;19:174–186. doi: 10.1093/emboj/19.2.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Briceño V, Camargo H, Remacha M, Santos C, Ballesta JP. Structural and functional characterization of the amino terminal domain of the yeast ribosomal stalk P1 and P2 proteins. Int. J. Biochem. Cell Biol. 2009;41:1315–1322. doi: 10.1016/j.biocel.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 12.Naganuma T, Nomura N, Yao M, Mochizuki M, Uchiumi T, Tanaka I. Structural basis for translation factor recruitment to the eukaryotic/archaeal ribosomes. J. Biol. Chem. 2010;285:4747–4756. doi: 10.1074/jbc.M109.068098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tchórzewski M, Boldyreff B, Issinger OG, Grankowski N. Analysis of the protein-protein interactions between the human acidic ribosomal P-proteins: evaluation by the two hybrid system. Int. J. Biochem. Cell Biol. 2000;32:737–746. doi: 10.1016/s1357-2725(00)00017-0. [DOI] [PubMed] [Google Scholar]
- 14.Lee KM, Yu CW, Chiu TY, Sze KH, Shaw PC, Wong KB. Solution structure of the dimerization domain of the eukaryotic stalk P1/P2 complex reveals the structural organization of eukaryotic stalk complex. Nucleic Acids Res. 2012;40:3172–3182. doi: 10.1093/nar/gkr1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helgstrand M, Mandava CS, Mulder FA, Liljas A, Sanyal S, Akke M. The ribosomal stalk binds to translation factors IF2, EF-Tu, EF-G and RF3 via a conserved region of the L12 C-terminal domain. J. Mol. Biol. 2007;365:468–479. doi: 10.1016/j.jmb.2006.10.025. [DOI] [PubMed] [Google Scholar]
- 16.Nechifor R, Wilson KS. Crosslinking of translation factor EF-G to proteins of the bacterial ribosome before and after translocation. J. Mol. Biol. 2007;368:1412–1425. doi: 10.1016/j.jmb.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 17.Harms JM, Wilson DN, Schluenzen F, Connell SR, Stachelhaus T, Zaborowska Z, Spahn CM, Fucini P. Translational regulation via L11: molecular switches on the ribosome turned on and off by thiostrepton and micrococcin. Mol. Cell. 2008;30:26–38. doi: 10.1016/j.molcel.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 18.Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dey D, Oleinikov AV, Traut RR. The hinge region of Escherichia coli ribosomal protein L7/L12 is required for factor binding and GTP hydrolysis. Biochimie. 1995;77:925–930. doi: 10.1016/0300-9084(95)80003-4. [DOI] [PubMed] [Google Scholar]
- 20.Leijonmarck M, Liljas A. Structure of the C-terminal domain of the ribosomal protein L7/L12 from Escherichia coli at 1.7 A. J. Mol. Biol. 1987;195:555–579. doi: 10.1016/0022-2836(87)90183-5. [DOI] [PubMed] [Google Scholar]
- 21.Zurdo J, González C, Sanz JM, Rico M, Remacha M, Ballesta JP. Structural differences between Saccharomyces cerevisiae ribosomal stalk proteins P1 and P2 support their functional diversity. Biochemistry. 2000;39:8935–8943. doi: 10.1021/bi000363b. [DOI] [PubMed] [Google Scholar]
- 22.Shimizu T, Nakagaki M, Nishi Y, Kobayashi Y, Hachimori A, Uchiumi T. Interaction among silkworm ribosomal proteins P1, P2 and P0 required for functional protein binding to the GTPase-associated domain of 28S rRNA. Nucleic Acids Res. 2002;30:2620–2627. doi: 10.1093/nar/gkf379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tchórzewski M, Krokowski D, Boguszewska A, Liljas A, Grankowski N. Structural characterization of yeast acidic ribosomal P proteins forming the P1A-P2B heterocomplex. Biochemistry. 2003;142:3399–3408. doi: 10.1021/bi0206006. [DOI] [PubMed] [Google Scholar]
- 24.Pérez-Fernández J, Remacha M, Ballesta JP. The acidic protein binding site is partially hidden in the free Saccharomyces cerevisiae ribosomal stalk protein P0. Biochemistry. 2005;44:5532–5540. doi: 10.1021/bi047332r. [DOI] [PubMed] [Google Scholar]
- 25.Hagiya A, Naganuma T, Maki Y, Ohta J, Tohkairin Y, Shimizu T, Nomura T, Hachimori A, Uchiumi T. A mode of assembly of P0, P1, and P2 proteins at the GTPase-associated center in animal ribosome: in vitro analyses with P0 truncation mutants. J. Biol. Chem. 2005;280:39193–39199. doi: 10.1074/jbc.M506050200. [DOI] [PubMed] [Google Scholar]
- 26.Krokowski D, Boguszewska A, Abramczyk D, Liljas A, Tchórzewski M, Grankowski N. Yeast ribosomal P0 protein has two separate binding sites for P1/P2 proteins. Mol. Microbiol. 2006;60:386–400. doi: 10.1111/j.1365-2958.2006.05117.x. [DOI] [PubMed] [Google Scholar]
- 27.Smulski CR, Longhi SA, Ayub MJ, Edreira MM, Simonetti L, Gómez KA, Basile JN, Chaloin O, Hoebeke J, Levin MJ. Interaction map of the Trypanosoma cruzi ribosomal P protein complex (stalk) and the elongation factor 2. J. Mol. Recognit. 2011;24:359–370. doi: 10.1002/jmr.1089. [DOI] [PubMed] [Google Scholar]
- 28.Shimmin LC, Ramirez C, Matheson AT, Dennis PP. Sequence alignment and evolutionary comparison of the L10 equivalent and L12 equivalent ribosomal proteins from archaebacteria, eubacteria, and eucaryotes. J. Mol. Evol. 1989;29:448–462. doi: 10.1007/BF02602915. [DOI] [PubMed] [Google Scholar]
- 29.Grela P, Bernadó P, Svergun D, Kwiatowski J, Abramczyk D, Grankowski N, Tchórzewski M. Structural relationships among the ribosomal stalk proteins from the three domains of life. J. Mol. Evol. 2008;67:154–167. doi: 10.1007/s00239-008-9132-2. [DOI] [PubMed] [Google Scholar]
- 30.Rich BE, Steitz JA. Human acidic ribosomal phosphoproteins P0, P1, and P2: analysis of cDNA clones, in vitro synthesis, and assembly. Mol. Cell Biol. 1987;7:4065–4074. doi: 10.1128/mcb.7.11.4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guarinos E, Remacha M, Ballesta JP. Asymmetric interactions between the acidic P1 and P2 proteins in the Saccharomyces cerevisiae ribosomal stalk. J. Biol. Chem. 2001;276:32474–32479. doi: 10.1074/jbc.M103229200. [DOI] [PubMed] [Google Scholar]
- 32.Tchórzewski M. The acidic ribosomal P proteins [Review] Int. J. Biochem. Cell Biol. 2002;34:911–915. doi: 10.1016/s1357-2725(02)00012-2. [DOI] [PubMed] [Google Scholar]
- 33.Qiu D, Parada P, Marcos AG, Cárdenas D, Remacha M, Ballesta JP. Different roles of P1 and P2 Saccharomyces cerevisiae ribosomal stalk proteins revealed by cross-linking. Mol. Microbiol. 2006;62:1191–1202. doi: 10.1111/j.1365-2958.2006.05445.x. [DOI] [PubMed] [Google Scholar]
- 34.Uchiumi T, Honma S, Nomura T, Dabbs ER, Hachimori A. Translation elongation by a hybrid ribosome in which proteins at the GTPase center of the Escherichia coli ribosome are replaced with rat counterparts. J. Biol. Chem. 2002;277:3857–3862. doi: 10.1074/jbc.M107730200. [DOI] [PubMed] [Google Scholar]
- 35.Kunkel TA, Bebenek K, McClary J. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- 36.Naganuma T, Shiogama K, Uchiumi T. The N-terminal regions of eukaryotic acidic phosphoproteins P1 and P2 are crucial for heterodimerization and assembly into the ribosomal GTPase-associated center. Genes Cells. 2007;12:501–510. doi: 10.1111/j.1365-2443.2007.01067.x. [DOI] [PubMed] [Google Scholar]
- 37.Dabbs ER. Selection for Escherichia coli mutants with proteins missing from the ribosome. J. Bacteriol. 1979;140:734–737. doi: 10.1128/jb.140.2.734-737.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iwasaki K, Kaziro Y. Polypeptide chain elongation factors from pig liver. Methods Enzymol. 1979;60:657–676. doi: 10.1016/s0076-6879(79)60062-9. [DOI] [PubMed] [Google Scholar]
- 39.Grela P, Sawa-Makarska J, Gordiyenko Y, Robinson CV, Grankowski N, Tchórzewski M. Structural properties of the human acidic ribosomal P proteins forming the P1-P2 heterocomplex. J. Biochem. 2008;143:169–177. doi: 10.1093/jb/mvm207. [DOI] [PubMed] [Google Scholar]
- 40.Nomura T, Nakao K, Maki Y, Naganuma T, Takashima T, Tanaka I, Kimura M, Hachimori A, Uchiumi T. In vitro reconstitution of the GTPase-associated centre of the archaebacterial ribosome: the functional features observed in a hybrid form with Escherichia coli 50S subunits. Biochem. J. 2006;396:565–571. doi: 10.1042/BJ20060038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Remacha M, Jimenez-Diaz A, Bermejo B, Rodriguez-Gabriel MA, Guarinos E, Ballesta JP. Ribosomal acidic phosphoproteins P1 and P2 are not required for cell viability but regulate the pattern of protein expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:4754–4762. doi: 10.1128/mcb.15.9.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Camargo H, Nusspaumer G, Abia D, Briceño V, Remacha M, Ballesta JP. The amino terminal end determines the stability and assembling capacity of eukaryotic ribosomal stalk proteins P1 and P2. Nucleic Acids Res. 2011;39:3735–3743. doi: 10.1093/nar/gkq1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zurdo J, Parada P, van den Berg A, Nusspaumer G, Jimenez-Diaz A, Remacha M, Ballesta JP. Assembly of Saccharomyces cerevisiae ribosomal stalk: binding of P1 proteins is required for the interaction of P2 proteins. Biochemistry. 2000;39:8929–8934. doi: 10.1021/bi000362j. [DOI] [PubMed] [Google Scholar]
- 44.Gonzalo P, Lavergne JP, Reboud JP. Pivotal role of the P1 N-terminal domain in the assembly of the mammalian ribosomal stalk and in\ the proteosynthetic activity. J. Biol. Chem. 2001;276:19762–19769. doi: 10.1074/jbc.M101398200. [DOI] [PubMed] [Google Scholar]
- 45.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.