Abstract
Zinc-finger recombinases (ZFRs) represent a potentially powerful class of tools for targeted genetic engineering. These chimeric enzymes are composed of an activated catalytic domain derived from the resolvase/invertase family of serine recombinases and a custom-designed zinc-finger DNA-binding domain. The use of ZFRs, however, has been restricted by sequence requirements imposed by the recombinase catalytic domain. Here, we combine substrate specificity analysis and directed evolution to develop a diverse collection of Gin recombinase catalytic domains capable of recognizing an estimated 3.77 × 107 unique DNA sequences. We show that ZFRs assembled from these engineered catalytic domains recombine user-defined DNA targets with high specificity, and that designed ZFRs integrate DNA into targeted endogenous loci in human cells. This study demonstrates the feasibility of generating customized ZFRs and the potential of ZFR technology for a diverse range of applications, including genome engineering, synthetic biology and gene therapy.
INTRODUCTION
Site-specific DNA recombination systems, such as Cre-loxP, FLP-FRT and ϕC31-att have emerged as powerful tools for genetic engineering (1,2). The enzymes that promote these conservative DNA rearrangements—known as site-specific recombinases—recognize short (30–40 bp) sequences and coordinate DNA cleavage, strand exchange and re-ligation by a mechanism that does not require DNA synthesis or a high-energy cofactor (3). This simplicity has allowed researchers to study gene function with extraordinary spatial and temporal sensitivity. However, the strict sequence requirements imposed by site-specific recombinases have limited their application to cells and organisms that contain artificially introduced recombination sites or pre-existing pseudo-recognition sites. To address this limitation, directed evolution has been used to alter the sequence specificity of several site-specific recombinases towards naturally occurring DNA sequences (4–8). Yet, despite advances (7,8), the widespread adoption of this technology has been hindered by the need for complex mutagenesis and selection strategies (4,7) coupled with the finding that re-engineered recombinase variants routinely demonstrate relaxed substrate specificity (4,6–8).
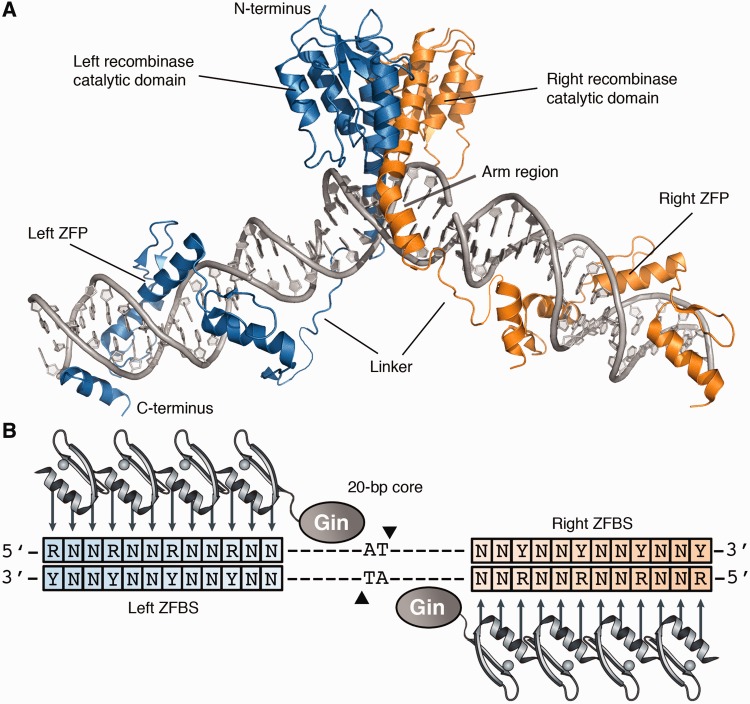
Zinc-finger recombinases (ZFRs) represent a versatile alternative to conventional site-specific recombination systems (9,10). These chimeric enzymes are composed of an activated catalytic domain derived from the resolvase/invertase family of serine recombinases and a zinc-finger DNA-binding domain, which can be custom-designed to recognize almost any DNA sequence (11–16) (Figure 1A). ZFRs catalyse recombination between specific ZFR target sites (17) that consist of two inverted zinc-finger–binding sites (ZFBS) flanking a central 20-bp core sequence recognized by the recombinase catalytic domain (18) (Figure 1B). In contrast to zinc-finger (19–21) and transcription activator-like (TAL) effector nucleases (22,23), ZFRs function autonomously and can excise and integrate transgenes in human and mouse cells without activating the cellular DNA damage response pathway (9,24–26). However, as with conventional site-specific recombinases, applications of ZFRs have been restricted by sequence requirements imposed by the recombinase catalytic domain, which dictate that ZFR target sites contain a 20-bp core derived from a native serine resolvase/invertase recombination site.
Figure 1.
Structure of the zinc-finger recombinase dimer bound to DNA. (A) Each ZFR monomer (blue or orange) consists of an activated serine recombinase catalytic domain linked to a custom-designed zinc-finger DNA-binding domain. Model was generated from crystal structures of the γδ resolvase and Aart zinc-finger protein (PDB IDs: 1GDT and 2I13, respectively). (B) Cartoon of the ZFR dimer bound to DNA. ZFR target sites consist of two-inverted ZFBS flanking a central 20-bp core sequence recognized by the ZFR catalytic domain. ZFPs can be designed to recognize distinct ‘left’ or ‘right’ half-sites (blue and orange boxes, respectively). Abbreviations are as follows: N indicates A, T, C or G; R indicates G or A; and Y indicates C or T.
To address this problem, we previously described a knowledge-based approach for re-engineering serine recombinase catalytic specificity (27). This strategy, which was based on the saturation mutagenesis of specificity-determining DNA-binding residues, was used to generate recombinase variants that showed >10 000-fold shift in specificity. Significantly, this strategy focused exclusively on amino acid residues located outside the recombinase dimer interface (Supplementary Figure S1). As a result, we found that catalytic domains re-engineered by this method could associate to form ZFR heterodimers, and that designed ZFR pairs could recombine pre-determined DNA sequences with exceptional specificity. Taken together, these results led us to hypothesize that an expanded catalogue of specialized catalytic domains developed by this method could be used for the design of ZFRs with custom specificity. Here, we expand on our previous work by combining substrate specificity analysis and directed evolution to develop a diverse collection of Gin recombinase catalytic domains capable of recognizing an estimated 3.77 × 107 unique 20-bp core sequences. We show that ZFRs assembled from these re-engineered catalytic domains recombine user-defined sequences with high specificity, and that designed ZFRs integrate DNA into targeted endogenous loci in human cells. To our knowledge, this report describes the first generalized approach for the design of customizable site-specific recombinases and also provides the first demonstration of targeted integration into endogenous human loci by custom-designed site-specific recombinases.
MATERIALS AND METHODS
Plasmids
The split gene reassembly vector (pBLA) was derived from pBluescriptII SK (−) (Stratagene) and modified to contain a chloramphenicol resistance gene and an interrupted TEM-1 β lactamase gene under the control of a lac promoter. ZFR target sites were introduced as previously described (8). Briefly, GFPuv (Clontech) was polymerase chain reaction (PCR) amplified with the primers GFP–ZFR–XbaI–Fwd and GFP–ZFR–HindIII–Rev and cloned into the SpeI and HindIII restriction sites of pBLA to generate pBLA–ZFR substrates. All primer sequences are provided in Supplementary Table S1.
To generate luciferase reporter plasmids, the Simian vacuolating virus 40 (SV40) promoter was PCR amplified from pGL3-Prm (Promega) with the primers SV40–ZFR–BglIII–Fwd and SV40–ZFR–HindIII–Rev. PCR products were digested with BglII and HindIII and ligated into the same restriction sites of pGL3-Prm to generate pGL3–ZFR-1, 2, 3 … 18. The pBPS–ZFR donor plasmids were constructed as previously described (24,27) with the following exception: the ZFR-1, 2 and 3 recombination sites were encoded by primers 3′ CMV (Cytomegalovirus)–PstI–ZFR-1, 2 or 3–Rev. Correct construction of each plasmid was verified by sequence analysis.
Recombination assays
ZFRs were assembled by PCR as previously described (9,27). PCR products were digested with SacI and XbaI and ligated into the same restrictions sites of pBLA. Ligations were transformed by electroporation into Escherichia coli TOP10F′ (Invitrogen). After 1-h recovery in Super Optimal Broth with Catabolite suppression (SOC) medium, cells were incubated with 5 ml of Super broth (SB) medium with 30 µg ml−1 of chloramphenicol and cultured at 37°C. At 16 h, cells were harvested; plasmid DNA was isolated by Mini-prep (Invitrogen); and 200 ng of pBLA was used to transform E. coli TOP10F′. After 1-h recovery in SOC, cells were plated on solid Lysogeny broth (LB) media with 30 µg ml−1 of chloramphenicol or 30 µg ml−1 of chloramphenicol and 100 µg ml−1 of carbenicillin, an ampicillin analogue. Recombination was determined as the number of colonies on LB media containing carbenicillin and chloramphenicol divided by the number of colonies on LB media containing only chloramphenicol. Colony number was determined by automated counting using the GelDoc XR Imaging System (Bio-Rad).
Selections
The ZFR library was constructed by overlap extension PCR as previously described (27). Mutations were introduced into the Gin catalytic domain at positions 120, 123, 127, 136 and 137 with the degenerate codon NNK (N: A, T, C or G and K: G or T), which encodes all 20 amino acids. PCR products were digested with SacI and XbaI and ligated into the same restriction sites of pBLA. Ligations were ethanol precipitated and used to transform E. coli TOP10F′. Library size was routinely determined to be ∼5 × 107. After 1-h recovery in SOC medium, cells were incubated in 100 ml of SB medium with 30 µg ml−1 of chloramphenicol at 37°C. At 16 h, 30 ml of cells were harvested; plasmid DNA was isolated by Mini-prep; and 3 µg plasmid DNA was used to transform E. coli TOP10F′. After 1-h recovery in SOC, cells were incubated with 100 ml of SB medium with 30 µg ml−1 of chloramphenicol and 100 µg ml−1 of carbenicillin at 37°C. At 16 h, cells were harvested, and plasmid DNA was isolated by Maxi-prep (Invitrogen). Enriched ZFRs were isolated by SacI and XbaI digestion and ligated into fresh pBLA for further selection. After four rounds of selection, sequence analysis was performed on individual carbenicillin-resistant clones. Recombination assays were performed as described earlier in the text.
ZFR construction
Recombinase catalytic domains were PCR amplified from their respective pBLA selection vector with the primers 5′ Gin–HBS–Koz and 3′ Gin–AgeI–Rev. PCR products were digested with HindIII and AgeI and ligated into the same restriction sites of pBH (9) to generate the SuperZiF-compitable subcloning plasmids: pBH-Gin-α, β, γ, δ, ε or ζ. Zinc-fingers were assembled by SuperZiF (28) and ligated into the AgeI and SpeI restriction sites of pBH-Gin-α, β, γ, δ, ε or ζ to generate pBH–ZFR-L/R-1, 2, 3 … 18 (L: left ZFR; R: right ZFR) (Supplementary Table S2). ZFR genes were released from pBH by SfiI digestion and ligated into pcDNA 3.1 (Invitrogen) to generate pcDNA–ZFR-L/R-1, 2, 3 … 18. Correct construction of each ZFR was verified by sequence analysis (Supplementary Table S3).
Luciferase assays
Human embryonic kidney (HEK) 293 and 293 T cells (ATCC) were maintained in Dulbecco’s modified Eagle’s medium containing 10% (vol/vol) Fetal Bovine Serum (FBS) and 1% (vol/vol) Antibiotic-Antimycotic (Anti-Anti; Gibco). HEK293T cells were seeded onto 96-well plates at a density of 4 × 104 cells per well and established in a humidified 5% CO2 atmosphere at 37°C. At 24 h after seeding, cells were transfected with 150 ng of pcDNA–ZFR-L 1–18, 150 ng of pcDNA–ZFR-R 1–18, 2.5 ng of pGL3–ZFR-1, 2, 3 … or 18 and 1 ng of pRL–CMV using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. At 48 h after transfection, cells were lysed with Passive Lysis Buffer (Promega), and luciferase expression was determined with the Dual-Luciferase Reporter Assay System (Promega) using a Veritas Microplate Luminometer (Turner Biosystems).
Integration assays
HEK293 cells were seeded onto 6-well plates at a density of 5 × 105 cells per well and maintained in serum-containing media in a humidified 5% CO2 atmosphere at 37°C. At 24 h after seeding, cells were transfected with 1 µg of pcDNA–ZFR-L-1, 2 or 3 and 1 µg of pcDNA–ZFR-R-1, 2 or 3 and 200 ng of pBPS–ZFR-1, 2 or 3 using Lipofectamine 2000 according to the manufacturer’s instructions. At 48 h after transfection, cells were split onto 6-well plates at a density of 5 × 104 cells per well and maintained in serum-containing media with 2 µg ml−1 of puromycin. Cells were harvested on reaching 100% confluence, and genomic DNA was isolated with the Quick Extract DNA Extraction Solution (Epicentre). ZFR targets were PCR amplified with the following primer combinations: ZFR–Target-1, 2 or 3–Fwd and ZFR–Target-1, 2 or 3–Rev (Unmodified target); ZFR–Target-1, 2 or 3–Fwd and CMV–Mid–Prim-1 (Forward integration); and CMV–Mid–Prim-1 and ZFR–Target-1, 2 or 3–Rev (Reverse integration) using the Expand High Fidelity Taq System (Roche). For clonal analysis, at 2 days post-transfection, 1 × 105 cells were split onto a 100-mm dish and maintained in serum-containing media with 2 µg ml−1 of puromycin. Individual colonies were isolated with 10- × 10-mm open-ended cloning cylinders with sterile silicone grease (Millipore) and expanded in culture. Cells were harvested on reaching 100% confluence, and genomic DNA was isolated and used as template for PCR, as described earlier in the text. For colony counting assays, at 2 days post-transfection, cells were split into 6-well plates at a density of 1 × 104 cells per well and maintained in serum-containing media with or without 2 µg ml−1 of puromycin. At 16 days, cells were stained with a 0.2% crystal violet solution, and genome-wide integration rates were determined by counting the number of colonies formed in puromycin-containing media divided by the number of colonies formed in the absence of puromycin. Colony number was determined by automated counting using the GelDoc XR Imaging System (Bio-Rad).
RESULTS
Specificity profile of the Gin recombinase
To effectively re-engineer serine recombinase catalytic specificity, we first sought to develop a detailed understanding of the factors underlying substrate recognition by this family of enzymes. To accomplish this, we evaluated the ability of an activated mutant of the catalytic domain of the DNA invertase Gin (29) to recombine an extensive set of symmetrically substituted target sites. In nature, the Gin catalytic domain recombines a pseudo-symmetric 20-bp core that consists of two 10-bp half-site regions. Our collection of mutant recombination sites, therefore, contained each possible single-base substitution at positions 10, 9, 8, 7, 6, 5 and 4 and each possible two-base combination at positions 3 and 2 and the dinucleotide core. We determined recombination by split gene reassembly (8), a previously described method that links recombinase activity to antibiotic resistance.
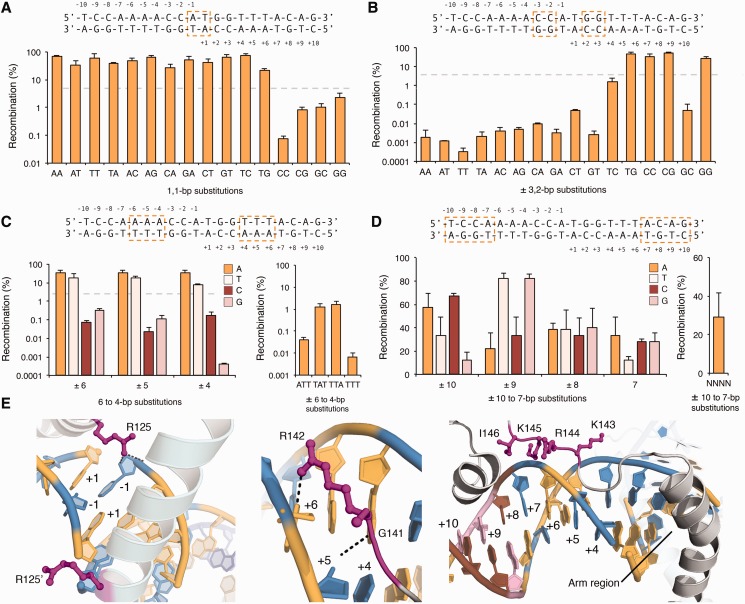
In general, we found that Gin tolerates: (i) 12 of the 16 possible two-base combinations at the dinucleotide core (AA, AT, AC, AG, TA, TT, TC, TG, CA, CT, GA and GT); (ii) 4 of the 16 possible two-base combinations at positions 3 and 2 (CC, CG, GG and TG); (iii) a single A to T substitution within positions 6, 5, or 4; and (iv) all 16 possible single-base combinations at positions 10, 9, 8, and 7 (Figure 2A–D). Furthermore, we found that Gin recombined a target site library containing >106 (of a possible 4.29 × 109) unique base combinations at positions 10, 9, 8 and 7 within each 20- bp target (Figure 2D). These findings are consistent with observations made from crystal structures of the γδ resolvase (30,31), which indicate that (i) the interactions made by the recombinase dimer across the dinucleotide core are asymmetric and predominately non-specific; (ii) the interactions between an evolutionarily conserved Gly–Arg motif in the recombinase arm region and the DNA minor groove impose a requirement for adenine or thymine at positions 6, 5 and 4; and (iii) there are no sequence-specific interactions between the arm region and the minor groove at positions 10, 9, 8 or 7 (Figure 2E). These results are also consistent with studies that focused on determining the DNA-binding properties of the closely related Hin recombinase (32–34).
Figure 2.
Specificity of the Gin recombinase catalytic domain. (A–D) Recombination was measured on DNA targets that contained (A) each possible two-base combination at the dinucleotide core, (B) each possible two-base combination at positions 3 and 2, (C) each possible single-base substitution at positions 6, 5 and 4 and (D) each possible single-base substitution at positions 10, 9, 8 and 7. Substituted bases are boxed above each panel. Recombination was evaluated by split gene reassembly and measured as the ratio of carbenicillin-resistant to chloramphenicol-resistant transformants (‘Materials and Methods’ section). Dotted lines indicate threshold for which sequences were considered non-functional. Error bars indicate standard deviation (n = 3). (E) Interactions between the γδ resolvase dimer and DNA at (left) the dinucleotide core, (middle) positions 6, 5 and 4 and (right) positions 10, 9, 8 and 7 (PDB ID: 1GDT). Interacting residues are shown as magenta sticks. Bases are coloured as follows: A, yellow; T, blue; C, brown; and G, pink.
Re-engineering Gin recombinase catalytic specificity
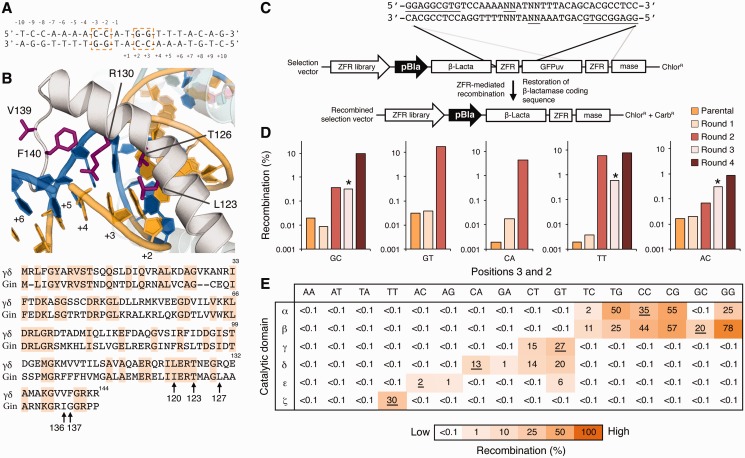
Based on the finding that Gin tolerates conservative substitutions at positions 3 and 2 (i.e. CC, CG, GG and TG), we next investigated whether Gin catalytic specificity could be re-engineered to recognize core sequences containing each of the 12 base combinations not tolerated by the native enzyme (Figure 3A). To identify the specific amino acid residues involved in DNA recognition by Gin, we examined the crystal structures of two related serine recombinases, the γδ resolvase (30) and Sin recombinase (35), in complex with their respective DNA targets. Based on these models, we identified five residues that contact DNA at positions 3 and 2: Leu 123, Thr 126, Arg 130, Val 139 and Phe 140 (numbered according to the γδ resolvase) (Figure 3B). We randomly mutagenized the equivalent residues in the Gin catalytic domain (Ile 120, Thr 123, Leu 127, Ile 136 and Gly 137) by overlap extension PCR and constructed a library of ZFR mutants by fusing these catalytic domain variants to an unmodified copy of the ‘H1’ zinc-finger protein (ZFP) (9), which recognizes the sequence 5′-GGAGGCGTG-3. The theoretical size of this library was 3.3 × 107 variants.
Figure 3.
Re-engineering Gin recombinase catalytic specificity. (A) The canonical 20-bp core recognized by the Gin catalytic domain. Positions 3 and 2 are boxed. (B) (Top) Structure of the γδ resolvase in complex with DNA (PDB ID: 1GDT). Arm region residues selected for mutagenesis are shown as magenta sticks. (Bottom) Sequence alignment of the γδ resolvase and Gin recombinase catalytic domains. Conserved residues are shaded orange. Black arrows indicate arm region positions selected for mutagenesis. (C) Schematic representation of the split gene reassembly selection system. Expression of active ZFR variants leads to restoration of the β-lactamase reading frame and host-cell resistance to ampicillin. Solid lines indicate the locations and identity of the ZFR target sites. Positions 3 and 2 are underlined. (D) Selection of Gin mutants that recombine core sites containing GC, GT, CA, TT and AC base combinations at positions 3 and 2. Asterisks indicate selection steps in which incubation time was decreased from 16 h to 6 h (‘Materials and Methods’ section). (E) Recombination specificity of the selected catalytic domains (β, γ, δ, ε and ζ, wild-type Gin indicated by α) for each possible two-base combination at positions 3 and 2. Intended DNA targets are underlined. Recombination was determined by split gene reassembly and performed in triplicate.
We cloned the ZFR library into substrate plasmids containing one of five base combinations not tolerated by the native enzyme (GC, GT, CA, AC or TT) and enriched for active ZFRs by split gene reassembly (8) (Figure 3C). After four rounds of selection, we found that the activity of each ZFR population increased >1000-fold on DNA targets containing GC, GT, CA and TT substitutions and >100-fold on a DNA target containing AC substitutions (Figure 3D). We sequenced individual recombinase variants from each population and found that a high level of amino acid diversity was present at positions 120, 123 and 127, and that >80% of selected clones contained Arg at position 136 and Trp or Phe at position 137 (Supplementary Figure S2). These results suggest that positions 120, 123 and 127 play critical roles in the specific recognition of unnatural core sequences, and that positions 136 and 137 are important structural determinants for DNA-binding. We evaluated the ability of each selected enzyme to recombine its target DNA and found that nearly all recombinases showed high activity (>10% recombination) and displayed a >1000-fold shift in specificity towards their intended core sequence (Supplementary Figure S3). As with the parental Gin, we found that several recombinases tolerated conservative substitutions at positions 3 and 2 (i.e. cross-reactivity against GT and CT or AC and AG), indicating that a single re-engineered catalytic domain could be used to target multiple core sites (Supplementary Figure S3).
To further investigate recombinase specificity, we determined the recombination profiles of five Gin variants (hereafter designated Gin β, γ, δ, ε and ζ) shown to recognize 9 of the 12 possible two-base combinations at positions 3 and 2 not tolerated by the parental enzyme (GC, TC, GT, CT, GA, CA, AG, AC and TT) (Table 1). We found that Gin β, γ and ζ recombined their intended core sequences with activity and specificity near that of the parental enzyme (hereafter referred to as Gin α), and that Gin γ, δ and ζ were able to recombine their intended core sequences with specificity exceeding that of Gin α (Figure 3E). Each recombinase displayed a >1000-fold preference for adenine or thymine at positions 6, 5 and 4 and showed no base preference at positions 10, 9, 8 and 7 (Supplementary Figure S4). These results indicate that mutagenesis of the DNA-binding arm allows for reprogramming of recombinase specificity at positions 3 and 2 without compromising recognition elsewhere. We were unable to select for Gin variants capable of tolerating AA, AT or TA substitutions at positions 3 and 2. One possibility for this result is that DNA targets containing >4 consecutive A–T base pairs might exhibit bent DNA conformations that interfere with recombinase binding and/or catalysis.
Table 1.
Catalytic domain substitutions and intended DNA targets
| Catalytic domain | Target | Positions |
||||
|---|---|---|---|---|---|---|
| 120 | 123 | 127 | 136 | 137 | ||
| α | CCa | Ile | Thr | Leu | Ile | Gly |
| β | GC | Ile | Thr | Leu | Arg | Phe |
| γ | GT | Leu | Val | Ile | Arg | Trp |
| δ | CA | Ile | Val | Leu | Arg | Phe |
| εb | AC | Leu | Pro | His | Arg | Phe |
| ζc | TT | Ile | Thr | Arg | Ile | Phe |
aWild-type DNA target.
bThe ε catalytic domain also contains the substitutions E117L and L118S.
cThe ζ catalytic domain also contains the substitutions M124S, R131I and P141R.
Engineering ZFRs to recombine user-defined sequences
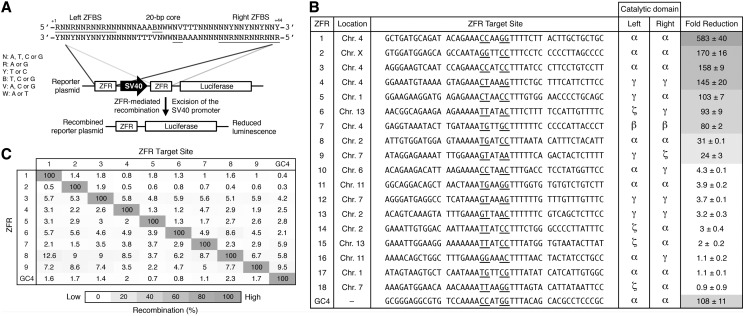
We next investigated whether ZFRs composed of the re-engineered catalytic domains could recombine pre-determined sequences. To test this possibility, we searched the human genome (GRCh37 primary reference assembly) for potential ZFR target sites using a 44-bp consensus recombination site predicted to occur approximately once every 7.44 × 106 bp of random DNA (Figure 4A). This ZFR consensus target site, which was derived from the core sequence profiles of the selected Gin variants, includes ∼3.77 × 107 (of a possible 1.0955 × 1012) unique 20-bp core combinations predicted to be tolerated by the 21 possible catalytic domain combinations and conservatively excludes low-affinity or unavailable 5′-CNN-3′ and 5′-TNN-3′ triplets within each ZFBS. Using ZFP specificity as the primary determinant for selection (36), we identified 18 possible ZFR target sites across eight human chromosomes (Chromosome 1, 2, 4, 6, 7, 11, 13 and X) at non-protein coding loci. On average, each 20-bp core showed ∼46% sequence identity to the core sequence recognized by the native Gin catalytic domain (Figure 4B). We constructed each corresponding ZFR by modular assembly (28) (‘Materials and Methods’ section).
Figure 4.
ZFRs recombine user-defined sequences in mammalian cells. (A) Schematic representation of the luciferase reporter system used to evaluate ZFR activity in mammalian cells. ZFR target sites flank an SV40 promoter that drives luciferase expression. Solid lines denote the 44-bp consensus target sequence used to identify potential ZFR target sites. The consensus ZFR target site consists of two-inverted 12-bp ZFBS flanking a central 20-bp core sequence recognized by the ZFR catalytic domain. Underlined bases indicate zinc-finger targets and positions 3 and 2. (B) Fold-reduction of luciferase expression in HEK293T cells co-transfected with designed ZFR pairs and their cognate reporter plasmid. Fold-reduction was normalized to transfection with empty vector and reporter plasmid. Renilla luciferase expression was used to normalize for transfection efficiency and cell number. The sequence identity and chromosomal location of each ZFR target site and the catalytic domain composition of each ZFR pair are shown. Underlined bases indicate positions 3 and 2. Standard errors were calculated from three independent experiments. ZFR amino acid sequences are provided in Supplementary Table S3. (C) Specificity of ZFR pairs. Fold-reduction of luciferase expression was measured for ZFR pairs 1 through 9 and GinC4 for each non-cognate reporter plasmid. Recombination was normalized to the fold-reduction of each ZFR pair with its cognate reporter plasmid. Assays were performed in triplicate.
To determine whether each ZFR pair could recombine its intended DNA target, we developed a transient reporter assay that correlates ZFR-mediated recombination to reduced luciferase expression (Figure 4A and Supplementary Figure S5). To accomplish this, we introduced ZFR target sites upstream and downstream an SV40 promoter that drives expression of a luciferase reporter gene. HEK293T cells were co-transfected with expression vectors for each ZFR pair and the corresponding reporter plasmid. Luciferase expression was measured 48 h after transfection. Of the 18 ZFR pairs analysed, 38% (7 of 18) reduced luciferase expression by >75-fold and 22% (4 of 18) decreased luciferase expression by >140-fold (Figure 4B). In comparison, GinC4, a positive ZFR control designed to target the core sequence recognized by the native Gin catalytic domain, reduced luciferase expression by 107-fold. Overall, we found that 50% (9 of 18) of the evaluated ZFR pairs decreased luciferase expression by >20-fold. The remaining ZFR pairs, however, had a negligible affect on luciferase expression. Importantly, virtually every catalytic domain that displayed significant activity in bacterial cells (>20% recombination) was successfully used to recombine at least one naturally occurring sequence in mammalian cells.
To evaluate ZFR specificity, we separately co-transfected HEK293T cells with expression plasmids for the nine most active ZFRs with each non-cognate reporter plasmid. Every ZFR pair demonstrated high specificity for its intended DNA target, and 77% (7 of 9) of the evaluated ZFRs showed an overall recombination specificity nearly identical to that of the positive control, GinC4 (Figure 4C). To establish that reduced luciferase expression was the product of the intended ZFR heterodimer and not the byproduct of recombination-competent ZFR homodimers, we measured the contribution of each ZFR monomer to recombination. Co-transfection of the ZFR 1 ‘left’ monomer with its corresponding reporter plasmid led to nearly a 130-fold reduction in luciferase expression (total contribution to recombination: ∼22%), but the vast majority of individual ZFR monomers (16 of 18) did not significantly contribute to recombination (<10% recombination), and many (7 of 18) showed no activity (Supplementary Figure S6). Taken together, these studies indicate that ZFRs can be engineered to recombine user-defined sequences with high specificity.
Engineered ZFRs target integration into the human genome
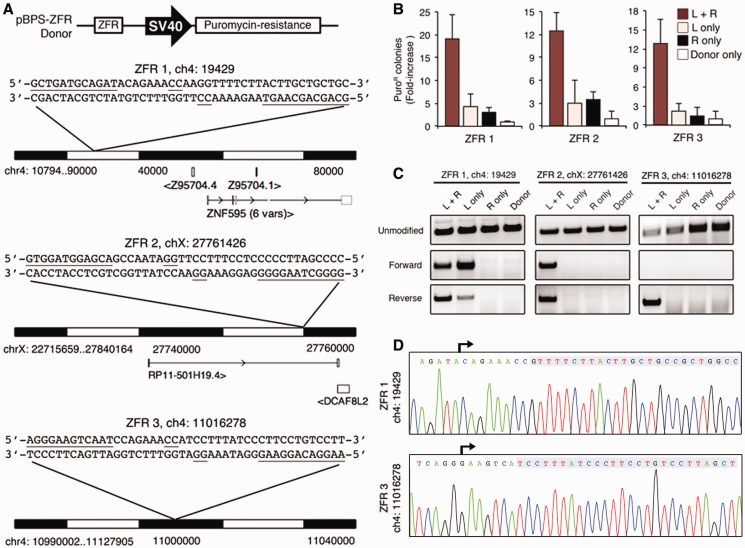
We next evaluated whether ZFRs could integrate DNA into endogenous loci in human cells. To accomplish this, we co-transfected HEK293 cells with ZFR expression vectors and a corresponding DNA donor plasmid that contained a specific ZFR target site and a puromycin-resistance gene under the control of an SV40 promoter (24) (Figure 5A). For this analysis, we used ZFR pairs 1, 2 and 3, which were designed to target non-protein coding loci on human chromosomes 4, X and 4, respectively (Figure 5A). At 2 days post-transfection, we incubated cells with puromycin-containing media and measured genome-wide integration rates by determining the number of puromycin-resistant (puroR) colonies. We found that (i) co-transfection of the donor plasmid and the corresponding ZFR pair led to a >12-fold increase in puroR colonies in comparison with transfection with donor plasmid only, and (ii) co-transfection with both ZFRs led to a 6- to 9-fold increase in puroR colonies in comparison with transfection with individual ZFR monomers (Figure 5B). The overall integration rates for ZFR pairs 1, 2 and 3 were determined to be 0.14 ± 0.06%, 0.24 ± 0.02% and 0.31 ± 0.1%, respectively. By comparison, the genome-wide integration rate of our internal ZFR positive control, GinC4, towards a pre-introduced target site (24,25) was previously determined to be ∼1%. To evaluate whether each ZFR pair correctly targeted integration, we isolated genomic DNA from puroR populations and amplified the targeted loci by PCR. The PCR products corresponding to integration in the forward and reverse orientation were observed at the loci targeted by ZFR pairs 1 and 2 (Figure 5C). ZFR pair 3 was found to target integration only in the reverse orientation. The reason for this bias remains unclear, but it could be explained by preferential formation of a particular synaptic complex topology (37). To determine the overall specificity of ZFR-mediated integration, we isolated genomic DNA from clonal cell populations and evaluated plasmid insertion by PCR. This analysis revealed targeting specificities of 14.2% (5 of 35 clones), 8.3% (1 of 12 clones) and 9.1% (1 of 11 clones) for ZFR pairs 1, 2 and 3, respectively (Supplementary Figure S7). Sequence analysis of each PCR product confirmed ZFR-mediated integration (Figure 5D); however, we observed mutations within the donor plasmid nearby the anticipated junctions for each ZFR pair. The mechanism underlying how these mutations were introduced remains unknown. Taken together, these results indicate that ZFRs can be designed to integrate DNA into endogenous loci. Finally, we note that the ZFR-1 ‘left’ monomer was found to target integration into the ZFR-1 locus in the absence of the corresponding ‘right’ ZFR monomer (Figure 5C). This result is consistent with the luciferase reporter studies described earlier in the text (Supplementary Figure S6) and indicates that recombination-competent ZFR homodimers have the capacity to mediate off-target integration. The comprehensive evaluation of off-target integration events and the development of optimized obligate heterodimeric ZFR architectures should lead to the design of ZFRs that show greater targeting efficiency and specificity.
Figure 5.
ZFRs target integration into the human genome. (A) Schematic representation of the donor plasmid (top) and the genomic loci targeted by ZFRs 1, 2 and 3. Open boxes indicate neighbouring exons. Arrows indicate transcript direction. The sequence and location of each ZFR target is shown. Underlined bases indicate zinc-finger targets and positions 3 and 2. (B) Genome-wide ZFR-mediated integration rates. Data were normalized to data from cells transfected with donor plasmid only. Error bars indicate standard deviation (n = 3). (C) PCR analysis of ZFR-mediated integration. PCR primer combinations amplified (top) unmodified locus or integrated plasmid in (middle) the forward or (bottom) the reverse orientation. (D) Representative chromatograms of PCR-amplified integrated donor for ZFRs 1 and 3. Arrows indicate sequencing primer orientation. Shaded boxes denote genomic target sequences.
DISCUSSION
Targeted genome engineering is driving progress in new areas of research in gene therapy, synthetic biology and basic science. Although improvements in the design and assembly of zinc-finger and TAL effector nucleases have been central to this revolution, the development of new methods that do not rely on DNA double-strand breaks and thus, do not carry the risk of non-homologous end joining-mediated mutagenesis, are necessary to improve the safety of genome engineering. ZFRs capable of autonomously catalysing recombination between DNA targets represent one such alternative. Yet, despite their promise, the use of ZFRs has been limited by the strict sequence requirements imposed by the ZFR catalytic domain. In the present study, we have addressed this problem by combining substrate specificity analysis and directed evolution to establish a user-friendly toolbox of modified serine recombinase catalytic domains suitable for the design of ZFRs with custom specificity. Guided by an extensive evaluation of serine recombinase catalytic specificity, we have developed a collection of re-engineered Gin recombinase catalytic domains that recognize an estimated 3.77 × 107 unique 20-bp core sequences. We have shown that ZFRs assembled from these re-engineered catalytic domains recombine user-defined sequences with high specificity and that designed ZFRs integrate DNA into pre-determined endogenous loci in human cells. Although previous studies have shown that site-specific recombinases, such as the ϕC31 integrase, can mediate integration into the human (38) and mouse genomes (39), these efforts were based on the presence of pseudo-recognition sites tolerated by the native enzyme (40), did not require catalytic reprogramming, and thus did not allow for targeting of user-defined sequences. To our knowledge, this report describes the first general approach for the design of site-specific recombinases with customizable specificity and also provides the first demonstration of targeted integration into endogenous human loci by customized site-specific recombinases.
Based on our current archive of >45 pre-selected zinc-finger modules, we estimate that ZFRs can now be designed to recognize between 5000 and 20 000 unique 44-bp DNA sequences in the human genome (Supplementary Note). This corresponds to approximately one potential ZFR target site for every 160 000–620 000 bp of random sequence and represents a substantial improvement in targeting capacity compared with conventional site-specific recombinases, which typically require complex evolutionary methods for reprogramming (4,7). Currently, the requirement for adenine by the Gin recombinase within positions 6, 5 and 4 represents the only major sequence restriction with the strategy described. To alleviate this constraint, structurally and functionally related serine recombinase variants (18) with broad or complementary sequence requirements at these positions could be subjected to the types of directed evolution described in this study. This approach may effectively expand the targeting repertoire of this custom-designed site-specific recombinase family. Additional improvements in the targeting capacity of this technology could be envisioned with the incorporation of alternate DNA-binding domains; in particular, we anticipate that the re-engineered catalytic domains described herein should be compatible with recently described TAL effector recombinases (41). Application of more sophisticated and high-throughput methods for specificity profiling (42) should lead to more effective use of the evolved catalytic domains and may also improve ZFR activity. Finally, although the efficiency of ZFR-mediated integration is lower than that achieved by zinc-finger (43,44) or TAL effector (22) nuclease-based approaches, we anticipate that optimization of the ZFR architecture will lead to reduced off-target integration events and higher targeting efficiency. Additional studies aimed at evaluating whether ZFR activity is cell type (25) or chromatin structure dependent (45) may also help establish limitations and clarify opportunities for ZFR targeting. In conclusion, we have developed a diverse collection of re-engineered Gin recombinase catalytic domains suitable for the design of ZFRs with custom specificity. We have shown that ZFRs can be assembled to recombine user-defined DNA targets, and that designed ZFRs integrate DNA into endogenous genomic loci. This work illustrates the potential of ZFRs for a wide range of applications, including genome engineering, synthetic biology and gene therapy.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–7 and Supplementary Note.
FUNDING
National Institutes for Health (NIH) [DP1CA174426]; National Institute of General Medicine Sciences fellowship [T32GM080209 to T.G.]. Funding for open access charge: NIH [CA174426].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank R.M. Gordley for contributing to preliminary studies and the Barbas laboratory for discussion of the manuscript. T.G. and C.F.B. designed research; T.G., A.C.M., S.J.S., R.M.G. and H.L.S. performed experiments; T.G., A.C.M., S.J.S., R.M.G. and C.F.B. analysed data; and T.G., S.J.S. and C.F.B wrote the manuscript.
REFERENCES
- 1.Sorrell DA, Kolb AF. Targeted modification of mammalian genomes. Biotechnol. Adv. 2005;23:431–469. doi: 10.1016/j.biotechadv.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 2.Branda CS, Dymecki SM. Talking about a revolution: the impact of site-specific recombinases on genetic analyses in mice. Dev. Cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- 3.Grindley ND, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 4.Buchholz F, Stewart AF. Alteration of Cre recombinase site specificity by substrate-linked protein evolution. Nat. Biotechnol. 2001;19:1047–1052. doi: 10.1038/nbt1101-1047. [DOI] [PubMed] [Google Scholar]
- 5.Sclimenti CR, Thyagarajan B, Calos MP. Directed evolution of a recombinase for improved genomic integration at a native human sequence. Nucleic Acids Res. 2001;29:5044–5051. doi: 10.1093/nar/29.24.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolusani S, Ma CH, Paek A, Konieczka JH, Jayaram M, Voziyanov Y. Evolution of variants of yeast site-specific recombinase Flp that utilize native genomic sequences as recombination target sites. Nucleic Acids Res. 2006;34:5259–5269. doi: 10.1093/nar/gkl548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sarkar I, Hauber I, Hauber J, Buchholz F. HIV-1 proviral DNA excision using an evolved recombinase. Science. 2007;316:1912–1915. doi: 10.1126/science.1141453. [DOI] [PubMed] [Google Scholar]
- 8.Gersbach CA, Gaj T, Gordley RM, Barbas CF., 3rd Directed evolution of recombinase specificity by split gene reassembly. Nucleic Acids Res. 2010;38:4198–4206. doi: 10.1093/nar/gkq125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gordley RM, Smith JD, Graslund T, Barbas CF., 3rd Evolution of programmable zinc finger-recombinases with activity in human cells. J. Mol. Biol. 2007;367:802–813. doi: 10.1016/j.jmb.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 10.Akopian A, He J, Boocock MR, Stark WM. Chimeric recombinases with designed DNA sequence recognition. Proc. Natl Acad. Sci. USA. 2003;100:8688–8691. doi: 10.1073/pnas.1533177100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Segal DJ, Dreier B, Beerli RR, Barbas CF., 3rd Toward controlling gene expression at will: selection and design of zinc finger domains recognizing each of the 5′-GNN-3′ DNA target sequences. Proc. Natl Acad. Sci. USA. 1999;96:2758–2763. doi: 10.1073/pnas.96.6.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beerli RR, Segal DJ, Dreier B, Barbas CF., 3rd Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc. Natl Acad. Sci. USA. 1998;95:14628–14633. doi: 10.1073/pnas.95.25.14628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dreier B, Beerli RR, Segal DJ, Flippin JD, Barbas CF., 3rd Development of zinc finger domains for recognition of the 5′-ANN-3′ family of DNA sequences and their use in the construction of artificial transcription factors. J. Biol. Chem. 2001;276:29466–29478. doi: 10.1074/jbc.M102604200. [DOI] [PubMed] [Google Scholar]
- 14.Dreier B, Fuller RP, Segal DJ, Lund CV, Blancafort P, Huber A, Koksch B, Barbas CF., 3rd Development of zinc finger domains for recognition of the 5′-CNN-3′ family DNA sequences and their use in the construction of artificial transcription factors. J. Biol. Chem. 2005;280:35588–35597. doi: 10.1074/jbc.M506654200. [DOI] [PubMed] [Google Scholar]
- 15.Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol. Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Curtin SJ, Blackburn JS, Thibodeau-Beganny S, Qi Y, et al. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA) Nat. Methods. 2011;8:67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prorocic MM, Wenlong D, Olorunniji FJ, Akopian A, Schloetel JG, Hannigan A, McPherson AL, Stark WM. Zinc-finger recombinase activities in vitro. Nucleic Acids Res. 2011;39:9316–9328. doi: 10.1093/nar/gkr652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith MC, Thorpe HM. Diversity in the serine recombinases. Mol. Microbiol. 2002;44:299–307. doi: 10.1046/j.1365-2958.2002.02891.x. [DOI] [PubMed] [Google Scholar]
- 19.Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011;188:773–782. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 21.Gaj T, Guo J, Kato Y, Sirk SJ, Barbas CF., 3rd Targeted gene knockout by direct delivery of zinc-finger nuclease proteins. Nat. Methods. 2012;9:805–807. doi: 10.1038/nmeth.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 23.Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordley RM, Gersbach CA, Barbas CF., 3rd Synthesis of programmable integrases. Proc. Natl Acad. Sci. USA. 2009;106:5053–5058. doi: 10.1073/pnas.0812502106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gersbach CA, Gaj T, Gordley RM, Mercer AC, Barbas CF., 3rd Targeted plasmid integration into the human genome by an engineered zinc-finger recombinase. Nucleic Acids Res. 2011;39:7868–7878. doi: 10.1093/nar/gkr421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nomura W, Masuda A, Ohba K, Urabe A, Ito N, Ryo A, Yamamoto N, Tamamura H. Effects of DNA binding of the zinc finger and linkers for domain fusion on the catalytic activity of sequence-specific chimeric recombinases determined by a facile fluorescent system. Biochemistry. 2012;51:1510–1517. doi: 10.1021/bi201878x. [DOI] [PubMed] [Google Scholar]
- 27.Gaj T, Mercer AC, Gersbach CA, Gordley RM, Barbas CF., 3rd Structure-guided reprogramming of serine recombinase DNA sequence specificity. Proc. Natl Acad. Sci. USA. 2011;108:498–503. doi: 10.1073/pnas.1014214108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonzalez B, Schwimmer LJ, Fuller RP, Ye Y, Asawapornmongkol L, Barbas CF., 3rd Modular system for the construction of zinc-finger libraries and proteins. Nat. Protoc. 2010;5:791–810. doi: 10.1038/nprot.2010.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klippel A, Cloppenborg K, Kahmann R. Isolation and characterization of unusual gin mutants. EMBO J. 1988;7:3983–3989. doi: 10.1002/j.1460-2075.1988.tb03286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang W, Steitz TA. Crystal structure of the site-specific recombinase gamma delta resolvase complexed with a 34 bp cleavage site. Cell. 1995;82:193–207. doi: 10.1016/0092-8674(95)90307-0. [DOI] [PubMed] [Google Scholar]
- 31.Li W, Kamtekar S, Xiong Y, Sarkis GJ, Grindley ND, Steitz TA. Structure of a synaptic gammadelta resolvase tetramer covalently linked to two cleaved DNAs. Science. 2005;309:1210–1215. doi: 10.1126/science.1112064. [DOI] [PubMed] [Google Scholar]
- 32.Hughes KT, Youderian P, Simon MI. Phase variation in Salmonella: analysis of Hin recombinase and hix recombination site interaction in vivo. Genes Dev. 1988;2:937–948. doi: 10.1101/gad.2.8.937. [DOI] [PubMed] [Google Scholar]
- 33.Glasgow AC, Bruist MF, Simon MI. DNA-binding properties of the Hin recombinase. J. Biol. Chem. 1989;264:10072–10082. [PubMed] [Google Scholar]
- 34.Hughes KT, Gaines PC, Karlinsey JE, Vinayak R, Simon MI. Sequence-specific interaction of the Salmonella Hin recombinase in both major and minor grooves of DNA. EMBO J. 1992;11:2695–2705. doi: 10.1002/j.1460-2075.1992.tb05335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mouw KW, Rowland SJ, Gajjar MM, Boocock MR, Stark WM, Rice PA. Architecture of a serine recombinase-DNA regulatory complex. Mol. Cell. 2008;30:145–155. doi: 10.1016/j.molcel.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mandell JG, Barbas CF., 3rd Zinc finger tools: custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res. 2006;34:W516–W523. doi: 10.1093/nar/gkl209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bai H, Sun M, Ghosh P, Hatfull GF, Grindley ND, Marko JF. Single-molecule analysis reveals the molecular bearing mechanism of DNA strand exchange by a serine recombinase. Proc. Natl Acad. Sci. USA. 2011;108:7419–7424. doi: 10.1073/pnas.1018436108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thyagarajan B, Olivares EC, Hollis RP, Ginsburg DS, Calos MP. Site-specific genomic integration in mammalian cells mediated by phage phiC31 integrase. Mol. Cell Biol. 2001;21:3926–3934. doi: 10.1128/MCB.21.12.3926-3934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olivares EC, Hollis RP, Chalberg TW, Meuse L, Kay MA, Calos MP. Site-specific genomic integration produces therapeutic Factor IX levels in mice. Nat. Biotechnol. 2002;20:1124–1128. doi: 10.1038/nbt753. [DOI] [PubMed] [Google Scholar]
- 40.Chalberg TW, Portlock JL, Olivares EC, Thyagarajan B, Kirby PJ, Hillman RT, Hoelters J, Calos MP. Integration specificity of phage phiC31 integrase in the human genome. J. Mol. Biol. 2006;357:28–48. doi: 10.1016/j.jmb.2005.11.098. [DOI] [PubMed] [Google Scholar]
- 41.Mercer AC, Gaj T, Fuller RP, Barbas CF., 3rd Chimeric TALE recombinases with programmable DNA sequence specificity. Nucleic Acids Res. 2012;40:11163–11172. doi: 10.1093/nar/gks875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jarjour J, West-Foyle H, Certo MT, Hubert CG, Doyle L, Getz MM, Stoddard BL, Scharenberg AM. High-resolution profiling of homing endonuclease binding and catalytic specificity using yeast surface display. Nucleic Acids Res. 2009;37:6871–6880. doi: 10.1093/nar/gkp726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 44.Moehle EA, Rock JM, Lee YL, Jouvenot Y, DeKelver RC, Gregory PD, Urnov FD, Holmes MC. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc. Natl Acad. Sci. USA. 2007;104:3055–3060. doi: 10.1073/pnas.0611478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Rensburg R, Beyer I, Yao XY, Wang H, Denisenko O, Li ZY, Russell DW, Miller DG, Gregory P, Holmes M, et al. Chromatin structure of two genomic sites for targeted transgene integration in induced pluripotent stem cells and hematopoietic stem cells. Gene Ther. 2013;20:201–214. doi: 10.1038/gt.2012.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.