Abstract
The current progress in the study of the spatial organization of interphase chromosomes became possible owing to the development of the chromosome conformation capture (3C) protocol. The crucial step of this protocol is the proximity ligation—preferential ligation of DNA fragments assumed to be joined within nuclei by protein bridges and solubilized as a common complex after formaldehyde cross-linking and DNA cleavage. Here, we show that a substantial, and in some cases the major, part of DNA is not solubilized from cross-linked nuclei treated with restriction endonuclease(s) and sodium dodecyl sulphate and that this treatment neither causes lysis of the nucleus nor drastically affects its internal organization. Analysis of the ligation frequencies of the mouse β-globin gene domain DNA fragments demonstrated that the previously reported 3C signals were generated predominantly, if not exclusively, in the insoluble portion of the 3C material. The proximity ligation thus occurs within the cross-linked chromatin cage in non-lysed nuclei. The finding does not compromise the 3C protocol but allows the consideration of an active chromatin hub as a folded chromatin domain or a nuclear compartment rather than a rigid complex of regulatory elements.
INTRODUCTION
Recent advances in the studies of the spatial organization of the eukaryotic genome became possible owing to the development of chromosome conformation capture (3C) and a set of derivative experimental procedures that permit the estimation of the relative spatial proximity of different genomic fragments within the nucleus (1–3). These experimental procedures are commonly known as C-methods. Using the original 3C protocol, de Laat and coworkers have demonstrated that, in mouse erythroblasts, distant regulatory elements of the β-globin gene domain are organized in a common complex to which the promoters of globin genes are recruited in a developmental stage-specific fashion (4). Based on these observations, the active chromatin hub (ACH) model has been proposed, which is presently supported by substantial experimental evidence (4–10). The full-genome variants of the 3C protocol (11–14) were used to study many important biological questions, including the elucidation of general principles of higher order chromatin folding in eukaryotic chromosomes (13,15,16) and analysis of the composition of transcription factories (17). Taking into account the range of applications of the C-methods and significance of conclusions drawn based on the results obtained using these methods, it is important to know how these methods work and what the limitations of their application are. Therefore, it should be noted that the crucial step of all C-methods is the so-called proximity ligation procedure (2). In the original 3C protocol (1,4), cells are treated with formaldehyde to cross-link proteins to nearby proteins and DNA. After the lysis of nuclei by sodium dodecyl sulphate (SDS) and the solubilization of proteins that were not cross-linked, the resulting DNA–protein network is subjected to cleavage by a restriction enzyme(s), which is followed by an appropriate dilution and ligation at a low DNA concentration. It is assumed that after cleavage, the cross-linked and separate DNA fragments are solubilized (1,4). In a diluted solution, ligation between cross-linked DNA fragments should be strongly favored over ligation between individual DNA fragments. This is the primary assumption of the proximity ligation procedure. After ligation, the cross-links are reversed, and ligation products are detected and quantified by polymerase chain reaction (PCR). It is obvious that the solubilization of chromatin fragments is a requirement for the implementation of the proximity ligation procedure, at least in the above-described fashion. Meanwhile, the solubilization of chromatin fragments from formaldehyde-fixed nuclei constitutes a difficult task unless ultrasonication is used for the physical destruction of both chromatin fibers and internal nuclear compounds. Here, we show that in a standard 3C protocol, <15% of the DNA is solubilized from mouse fetal liver cells after digestion with HindIII and treatment with SDS, and ∼40% of the DNA is solubilized if MboI is used instead of HindIII. Most importantly, using the classical model system of the mouse β-globin gene domain in erythroid cells, we demonstrated that it is in the non-solubilized material that the expected 3C signals are generated. Therefore, we conclude that the proximity ligation occurs predominantly within the cross-linked nuclei, wherein the mobility of the ends of DNA fragments is highly restricted by links between adjacent chromatin fibers.
MATERIALS AND METHODS
3C analysis
3C was performed as previously described (4,18). Briefly, fetal livers or brains were isolated from E14.5 mouse embryos, disrupted by pipetting in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (FBS) and, in the case of the brain, 1.25 mg/ml collagenase I and were passed through a 40-µm cell strainer to produce a single-cell suspension. An aliquot containing 107 cells was treated with 2% formaldehyde in phosphate buffered saline (PBS)/10% FBS for 10 min at room temperature, and then the reaction was stopped by the addition of glycine to 0.125 M. After washing with PBS/10% FBS, the fixed cells were incubated for 10 min in an ice-cold lysis solution [10 mM Tris, pH 8.0, 10 mM NaCl, 0.2% Nonidet P40 and a protease inhibitor cocktail (Fermentas)] at a concentration of 2 × 107 cells/ml to release the nuclei. The nuclei were harvested and suspended in 0.5 ml of 1.2× restriction buffer 2 (New England Biolabs) for subsequent HindIII digestion or 0.25 ml of 1.2× restriction buffer 3 (New England Biolabs) for MboI digestion. SDS was added to a final concentration of 0.3%, and the solution was incubated for 1 h at 37°C with shaking followed, in the case of MboI digestion, by the addition of 0.25 ml of 1.2× restriction buffer 3. Triton X-100 was added to a final concentration of 1.8%, and the solution was further incubated for 1 h at 37°C to sequester the SDS. The DNA was digested by overnight incubation with 600 U of HindIII or 800 U of MboI (New England Biolabs) at 37°C with shaking. The restriction endonuclease was inactivated by the addition of SDS to a final concentration of 1.6% and incubation for 20 min at 65°C, which was followed by sonication when desired (VirTis VirSonic 100 sonicator, setting 7). With or without sonication, the material was then processed either accordingly to the standard 3C protocol, or the soluble and the insoluble portions of the 3C material were separated by centrifugation (16 000g, 20 min). After collection of the supernatant, the pellet was resuspended in a buffer that matched the composition of the supernatant.
The solution was diluted by adding 7 ml of 1× ligation buffer (Fermentas). Triton X-100 was added to a final concentration of 1%, and the solution was incubated at 37°C for 1 h while shaking. Next, 100 U of T4 DNA Ligase (Fermentas) was added, and the DNA was ligated for 4.5 h at 16°C and then for 30 min at room temperature with slow agitation. Cross-links were reversed by incubation at 65°C for 16 h in the presence of Proteinase K (40 μg/ml). After cross-link reversion, RNase A was added to a final concentration of 40 μg/ml, and RNA was digested for 45 min at 37°C. DNA was purified by extraction with phenol, phenol-chloroform and chloroform followed by precipitation with ethanol.
The ligation products were analyzed by TaqMan real-time PCR. A random ligation standard was generated using a bacterial artificial chromosome carrying the murine β-globin gene locus along with flanking sequences (BAC clone RP24-79I7, CHORI BACPAC Resources Center), which was digested with HindIII or Sau3A (isoschisomer of MboI insensitive to dam methylation) and then ligated at a high DNA concentration. For data normalization, the frequencies of ligation between fragments of the gene Ercc3 were determined. If 3C material was divided into soluble and insoluble parts, ligation frequencies determined for each part were normalized to the sum of ligation frequencies for Ercc3 gene determined in the soluble and insoluble parts. The sequences of primers and TaqMan probes are presented in Supplementary Table S1.
Immunofluorescence and FISH
Cells or insoluble 3C material were cytospinned and fixed with 1% paraformaldehyde (PFA) for 10 min at room temperature, then treated with 1% Triton X-100 for 10 min and washed three times (5 min each) with PBS. After washing, the samples were pre-incubated in PBS supplemented with 1% bovine serum albumin (BSA) for 30 min and then incubated in the same solution with primary antibodies for 1 h at room temperature. The following primary antibodies were used: mouse anti-Sc35 (Abcam ab11826), rabbit anti-nucleolin (Sigma N2662), rabbit anti-topoisomerase II alpha (Abcam ab12318), rabbit anti-H3K9me3 (Abcam ab8898) and rabbit anti-H3K27me3 (LPBio AR-0171). After incubation with primary antibodies, the samples were washed three times (5 min each) with PBS supplemented with 0.2% BSA and 0.05% Tween 20. Primary antibodies bound to antigens were visualized using Alexa Fluor 488-conjugated secondary antibodies. The following secondary antibodies were used: rabbit anti-mouse AF488 (Invitrogen A11059) and goat anti-rabbit AF 488 (Invitrogen A11008). DNA was counterstained with DAPI (4′,6-diamidino-2-phenylindole).
To perform fluorescence in situ hybridization (FISH), samples were cytospinned and fixed with cold methanol-acetic acid (3:1) for 20 min at 4°C, then treated with RNAse A (100 μg/μl) for 1 h at 37°C and washed with 2× saline-sodium citrate (SSC). After washing, samples were digested with 0.1% pepsin in 0.01 N HCl for 10 min at 37°C. For pepsin inactivation, slides were washed twice with PBS supplemented with 50 mM MgCl2. Post-fixation was performed in 1% PFA for 10 min followed by washing with PBS three times (5 min each). After post-fixation, samples were dehydrated in ethanol series (70%–80%–96%, 5 min each). Chromosome painting using the XCyting Mouse Chromosome Painting Probe for mouse chromosome 7 labeled with a green emitting fluorochrome (comparable with fluorescein isothiocyanate) (MetaSystems) was performed according to the manufacturer’s protocol. The DNA was counterstained with DAPI for 10 min at room temperature. The samples were mounted using Dako Fluorescent mounting medium (Dako/Invitrogen).
Slides were examined and photographed with an Eclipse Ti-E inverted fluorescent microscope (Nikon, Japan) equipped with an ×60/NA = 1.4 lens and iXon cooled EMCCD camera (Andor), under the control of NIS-Elements 4.0 software. Serial optical sections were deconvolved using the AutoQuant blind deconvolution algorithm included in the NIS-Elements package.
Electron microscopy
Samples at different steps of the 3C procedure were fixed with 2.5% neutralized glutaraldehyde in the requisite buffer for 2 h at room temperature, post-fixed with 1% aqueous OsO4 and embedded in Epon. Sections of 100-nm thickness were cut and counterstained with uranyl acetate and lead citrate. Sections were examined and photographed with a JEM 1400 transmission electron microscope (JEOL, Japan) equipped with a QUEMESA bottom-mounted CCD-camera (Olympus SIS, Japan) and operated at 100 kV.
Immunoblotting and coomassie staining
Aliquots of the soluble and the insoluble 3C material were sonicated (VirTis VirSonic 100 sonicator, setting 15, 30-s pulse) to reduce the sizes of DNA fragments. The proteins were then separated by 15% SDS-polyacrylamide gel electrophoresis and either stained with Coomasie Blue R-250 according to the standard protocol (19) or blotted onto polyvinylidene difluoride membranes (Hybond-P, Amersham Biosciences). The membranes were blocked overnight in 5% dry milk in PBS containing 0.1% Tween 20 (PBS-T) and incubated for 1 h with primary antibodies (anti-Histone H3, Abcam ab1791) diluted in PBS containing 0.02% Tween 20 and 5% dry milk. After three washes with PBS-T, the membranes were incubated for 1 h with secondary antibodies (horseradish peroxidase-conjugated anti-rabbit immunoglobulin G) in PBS containing 0.02% Tween 20 and 5% dry milk. The immunoblots were visualized using an Amersham ECL kit. Protein band quantification was carried out using ImageJ software.
RESULTS
Distribution of DNA and histones between soluble and insoluble fractions on treatment of cross-linked nuclei with SDS and restriction enzymes
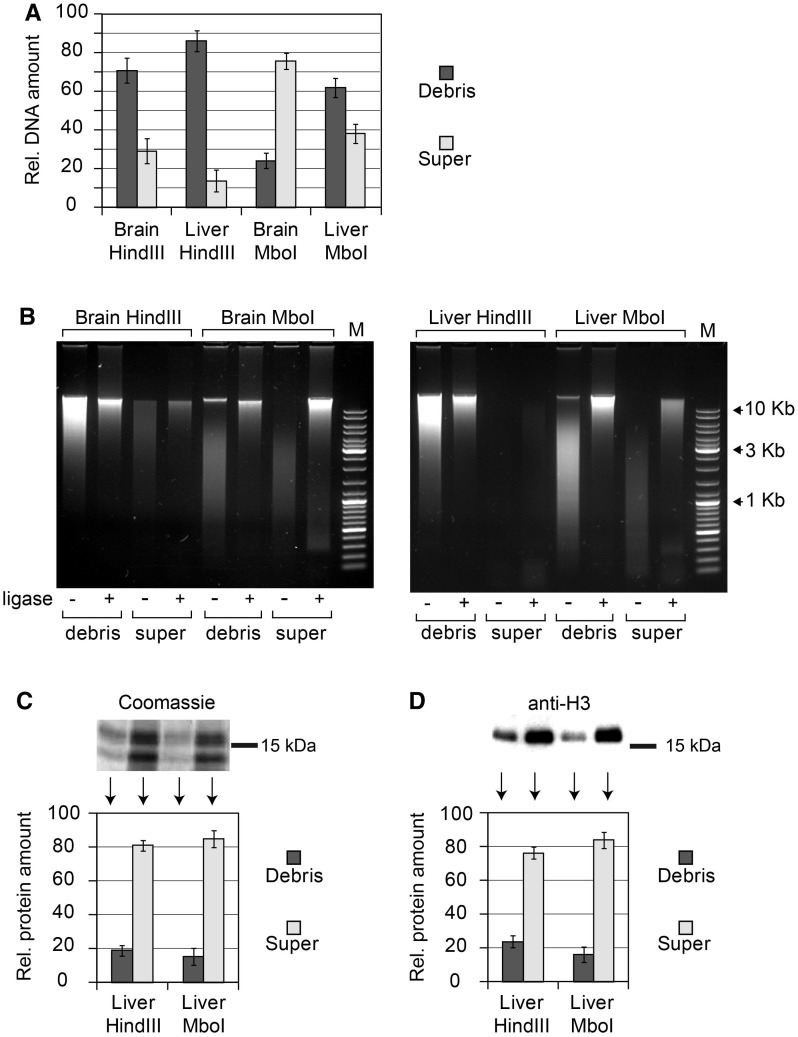
We chose mouse liver and brain cells to study the solubilization of chromatin fragments from cross-linked nuclei. The reason behind this decision was that the frequencies of ligation of the β-globin gene domain fragments can be subsequently analyzed separately in soluble and insoluble material, and the results can be compared with the previously published data (4). In the first set of experiments, the cross-linked nuclei were lysed with SDS solution [exactly as proposed in the original 3C protocol (1,4)] and treated with the HindIII restriction enzyme. The reaction was terminated by the addition of SDS at a final concentration of 1.6% and incubation at 65°C for 20 min. The soluble and insoluble materials were then separated by centrifugation (16 000g, 20 min). After collection of the supernatant, the insoluble material (debris) was resuspended in a buffer that matched the composition of the supernatant. This suspension and the collected supernatant were then diluted by the addition of 1× ligation buffer as in the standard 3C protocol (1,4). Aliquots were taken from both samples for (i) DNA isolation and (ii) ligation followed by DNA isolation. Surprisingly, in both liver and brain cells, the major portion of DNA (85 and 70%, respectively) remained in the insoluble fraction (Figure 1A, ‘Hind’ bars). The size distribution of DNA fragments was similar in the soluble and insoluble fractions, and in all cases, the fragments were ligatable (Figure 1B, ‘Hind’ panels). Next, we repeated the above-described experiments using an MboI restriction enzyme to cut DNA in cross-linked nuclei. This enzyme recognises four base pairs of DNA and cuts DNA into shorter fragments than HindIII, which recognises six base pairs. Indeed, analysis of the size distribution of the DNA fragments isolated from cross-linked nuclei treated with MboI demonstrated that the fragments were much shorter than those observed in the experiment that used the HindIII restriction enzyme (Figure 1B, ‘Mbo’ panels). The degree of solubilization of these short DNA fragments increased (Figure 1A, ‘Mbo’ bars). Nevertheless, a significant portion of DNA (∼25 and ∼60% in brain and liver cells, respectively) remained in the insoluble fraction. It is also of note that the MboI fragments from both soluble and insoluble fractions were ligatable, as were the HindIII fragments (Figure 1B, ‘Mbo’ panels).
Figure 1.
Partitioning of DNA and histones between soluble and insoluble portions of the 3C material and size distribution of DNA fragments. (A) Relative amounts of DNA in soluble (super) and insoluble (debris) portions of the 3C material, as determined by fluorometric assays (Qubit, Invitrogen). In each experiment, the total amount of DNA in two fractions is set as 100. (B) Electrophoretic separation of DNA from soluble and insoluble portions of the 3C material before and after ligation (agarose gel, ethidium bromide staining). M—DNA size marker (Fermentas, SM0331). (C and D) Partitioning of histones between the soluble and the insoluble portions of the 3C material. Proteins present in equal portions of the soluble and the insoluble 3C material were separated by PAAG and visualized by Coomassie staining (C) or by immunoblotting with antibodies against histone H3 (D). The intensity of bands was quantified using ImageJ software. In each experiment, the total amount of histones in two fractions is set as 100. The error bars represent SEM for three independent experiments.
We have next analyzed distribution of histones between soluble and insoluble fractions in the course of preparation of the 3C material. With this aim, the samples were sonicated to decrease the sizes of DNA fragments and incubated overnight at 65°C to reverse cross-links; proteins were then separated in 15% Polyacrylamide gel (PAAG) and either directly stained with Coomassie blue (Figure 1C) or transferred to a membrane followed by visualization of histone H3 by immunostaining (Figure 1D). The results of a representative experiment are shown on the top, and quantification of the data obtained in three independent experiments is presented below. It is evident that distribution of histones does not closely match the distribution of DNA. In the case of fetal liver cell nuclei digested by HindIII, a significant portion of histones (75–80%) is solubilized, while most of the DNA (∼85%) remains in the nuclei (see above). These can be easily explained by incomplete cross-linking of histones.
Preferential ligation between the fragments that are thought to be assembled into an active chromatin hub occurs mainly in the insoluble fraction
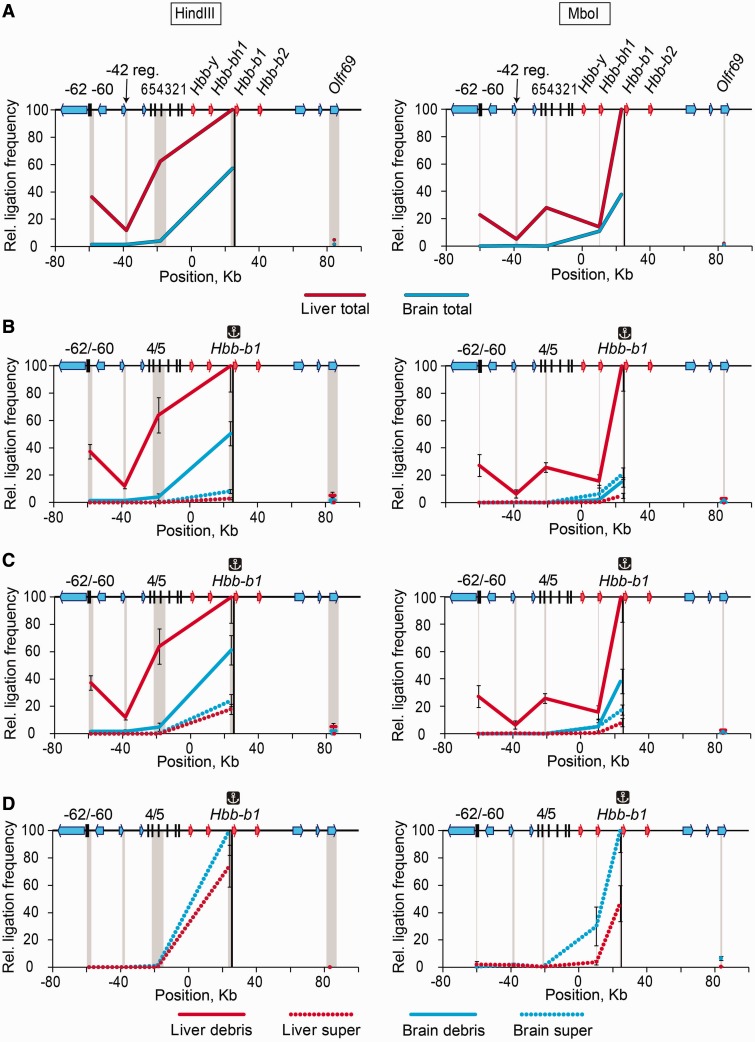
We first reproduced the 3C experiments on the β-globin gene domain reported by Tolhuis et al. (4). In the first set of experiments, the HindIII restriction enzyme was used to cut DNA in cross-linked nuclei that were lysed by SDS. In agreement with the results of the above-cited authors, in fetal liver (erythroid) cells, the anchor placed on the promoter of the major β-globin gene Hbb-b1exhibited elevated ligation frequency with the DNase I hypersensitive sites 4/5 (HS4/5) of the locus control region (LCR) and with the HS-62/-60, while the frequency of ligation with the -42 region was lower (Figure 2A, left graph). In brain cells, the above-mentioned peaks were not observed. The ligation frequencies decreased with an increase in the distance between the anchor and a test fragment, again in agreement with the previously published results (1). The principal results of the HindIII-3C analysis were confirmed in experiments with MboI-digested DNA in cross-linked nuclei (Figure 2A, right graph).
Figure 2.
Frequencies of ligation of the fragment harboring the Hbb-b1 promoter with several selected fragments of the β-globin gene domain in soluble and insoluble portions of the 3C material. (A) Results of standard 3C analysis performed without fractionating the 3C material. (B) Results of 3C analysis performed separately on soluble (super) and insoluble (debris) fractions. (C) The same as (B) after normalization of the ligation frequencies to the amount of DNA in the samples. (D) The same as (C), soluble fraction only. On the top of each graph, a map of the domain is shown, with β-globin genes, olfactory receptor genes and DNase I hypersensitive sites shown by red arrows, blue arrows and black vertical lines, respectively. Plotted on the horizontal axis are the fragment positions. The scale is in kilobases, and according to GenBank entry NT_039433, the ‘0’ point corresponds to the start of the Hbb-y gene. The black rectangle in the background of each graph shows the anchor fragment, and the gray rectangles indicate test fragments. Plotted on the vertical axis are the ligation frequencies; the highest ligation frequency observed is set to 100 [the frequency of ligation between the anchor fragment and the upstream restriction fragment in the total 3C material from fetal liver cells (A) or in the insoluble portion of the 3C material from fetal liver cells (B and C) or the soluble portion of the 3C material from fetal brain cells (D)]. Red and blue lines show the results for liver and brain cells, respectively; solid lines show the results for the total 3C material (A) or the insoluble portion of the 3C material (B and C); dotted lines show the results for the soluble portion of the 3C material. Ligation frequencies of HindIII and MboI fragments are presented on the left and the right graphs, respectively. The error bars represent SEM for three independent experiments.
One can consider multiple ways of generation of the 3C profiles obtained in the above-described experiments. Both soluble and insoluble portions of the initial 3C material may contribute to the 3C profile without any preference. Another possibility is that the soluble or the insoluble portion predominantly or even exclusively determines the 3C profile. To choose between these possibilities, we analyzed the ligation frequencies separately in the soluble and insoluble portions of the 3C material obtained using either HindIII or MboI to digest the DNA in the cross-linked nuclei. We first analyzed the ligation frequencies in equal aliquots of the soluble and insoluble material, disregarding the fact that the amounts of DNA in these aliquots differed significantly. The results of these experiments (Figure 2B) clearly demonstrated that virtually all 3C signals observed in the experiment using non-fractionated 3C material came from the insoluble fraction. Indeed, in the insoluble fraction of the 3C material prepared from erythroid cells, elevated ligation frequencies of the anchor fixed on the Hbb-b1 promoter with the HS4/5 of the LCR and the HS-62/-60, and a very low ligation frequency of the same anchor with the -42 fragment were observed. As in the experiments with the non-fractionated 3C material (see above), the results obtained with the MboI digestion of cross-linked nuclei closely matched those obtained with HindIII digestion. When the soluble portion of the 3C material from erythroid cells was analyzed, no characteristic increase of the ligation frequencies with the HS4/5 of the LCR and the HS-62/-60 was detected. Furthermore, even the frequencies of ligation of the anchor to the adjacent DNA fragments were diminished. In the soluble portion of the material from brain cells, the frequency of ligation of the fragments adjacent to the anchor was also diminished (Figure 2B). The observed characteristics of the insoluble and the soluble portions of the 3C material did not change when the experimentally determined ligation frequencies were normalized to the amount of DNA in different samples (Figure 2C). When the soluble fractions were plotted and normalized separately (to better see weak signals), the characteristic 3C pattern still was not visible (Figure 2D).
Because of the above-described results, the possibility of non-equal solubilization of different DNA fragments from the cross-linked nuclei should be considered. In the extreme case, some fragments may be entirely absent in either the soluble or insoluble fraction. Naturally, this would affect the experimentally determined ligation frequencies. To address this problem, we determined the relative amounts in the soluble and insoluble fractions of all fragments of the β-globin gene domain studied in our experiments. In the case of HindIII digestion, we found that with an increase of fragment length, the degree of fragment solubilization decreased (Supplementary Figure S1A and B). However, there was no correlation between the relative amount of a fragment in the insoluble or soluble portion of the 3C material and the observed ligation frequency. For example, in the case of liver cells, two long HindIII fragments containing the HS4/5 and Olfr59 gene were found almost entirely (∼95%) in the insoluble fraction, and of these two fragments, only the one containing HS4/5 was preferentially ligated to the Hbb-b1 promoter. In the soluble fraction, the highest ligation frequency was observed for the fragment located immediately upstream of the anchor fragment, whereas this fragment was not the fragment represented at the highest level in this fraction (Supplementary Figure S1B). The MboI fragments were partitioned almost randomly between the soluble and the insoluble portions of the 3C material irrespective of their size, and the differences in fragment distribution fell into the 50% range (Supplementary Figure S1C and D). Again, no correlation was observed between the relative amount of a fragment in the insoluble or soluble portion of the 3C material and the observed ligation frequency.
The insoluble fraction of the 3C material is composed of non-lysed nuclei
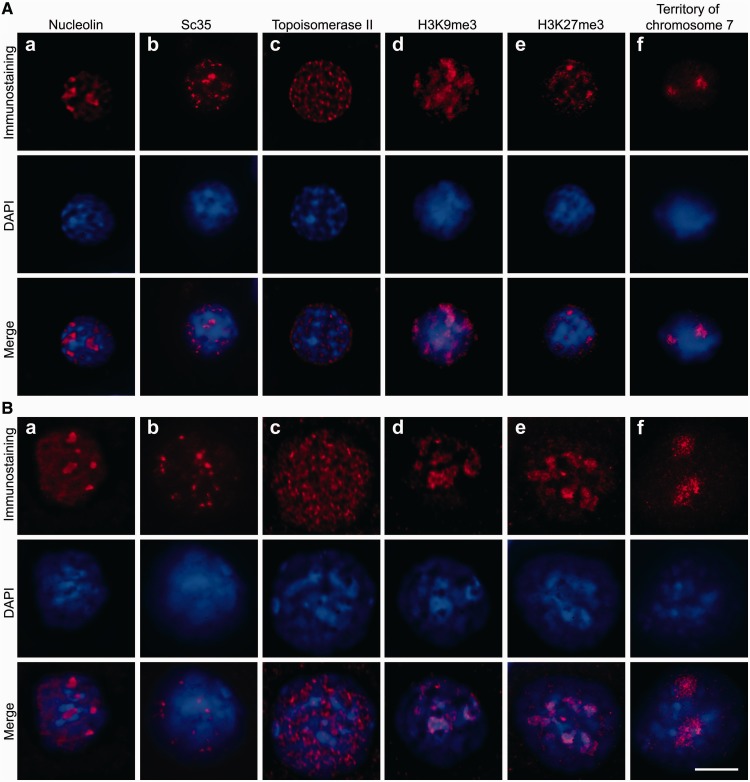
Because the proximity ligation that generates the characteristic 3C signals proceeds predominantly in the insoluble portion of the cross-linked material, it was important to understand the nature of this insoluble fraction. To address this problem, we inspected the insoluble material under a fluorescence microscope after staining with DAPI to visualize DNA and immunostaining with antibodies that recognise characteristic components of different nuclear compartments. The results of these experiments are presented in Figure 3. First, it became evident that the insoluble portion of the 3C material was composed of non-lysed nuclei that retain a spherical shape and some characteristic features of the internal organization after extraction with 0.3% SDS solution and subsequent treatment with a restriction enzyme followed by extraction with 1.6% SDS solution. Indeed, using antibodies against nucleolin, Sc35 and DNA topoisomerase II, we were able to visualize nucleoli, splicing speckles and the nuclear matrix in the non-lysed nuclei that were collected after performing all steps of the 3C protocol that precede DNA ligation (Figure 3B, panels a–c). Hence, all of the above-mentioned compartments survive in cross-linked nuclei that are subjected to treatment with a restriction endonuclease and SDS extraction.
Figure 3.
Visualization of nuclear compartments and chromatin domains in non-treated liver cells (A) and the same cells treated according to the 3C protocol up to the ligation step (B). The insoluble fraction was collected after HindIII digestion and 1.6% SDS extraction. (a–e) Immunostaining with antibodies against nucleolin (a), Sc35 (b), DNA topoisomerase II (c), H3K9me3 (d) and H3K27me3 (e). (f) Visualization of the chromosome 7 territory (FISH with a library of the chromosome 7–specific probes). In both sections of the Figure, the results of immunostaining are shown in the first row (red) and counterstaining of DNA with DAPI is shown in the second row (blue). The superimposition of the immunostaining and counterstaining of DNA is shown in the third row. Scale bar: 5 µm.
Inspection of samples of precipitated 3C material stained with DAPI fully confirmed the results of the quantitative analysis of the DNA distribution between the soluble and the insoluble portions of the 3C material. It is evident that residual nuclei obtained after treatment with a restriction enzyme and SDS extraction still contain a substantial amount of DNA. However, these treatments cause an increase in the apparent size of the residual nuclei. This may be due to the partial decondensation of chromatin after extraction of those histones that have not been cross-linked. It is important to know whether this decondensation provokes redistribution of chromatin within the nucleus. To address this question, we analyzed the fate of heterochromatin domains in the course of treatment of cross-linked nuclei with restriction enzymes and SDS. Toward this aim, the nuclei were immunostained with antibodies recognizing histone modifications typical for constitutive [tri-methylation of histone H3 lysine 9 (H3K9 me3)] and facultative [tri-methylation of histone H3 lysine 27 (H3K27 me3)] heterochromatin (20). In non-treated cells, both antibodies visualized restricted heterochromatic regions (Figure 3A, panels d and e). Although enlarged along with the whole nucleus, these heterochromatic regions were clearly observed after digestion of chromatin with a restriction endonuclease and SDS extraction (Figure 3B, panels d and e). This strongly suggests that harsh treatments performed during preparation of 3C material do not destroy formaldehyde-fixed nuclei and do not cause intermingling of distinct chromatin domains. To verify this conclusion, we stained the whole chromosome 7 using FISH with chromosome-specific probes. The discrete chromosomal territory was clearly visible both before and after treatment of formaldehyde-fixed cells with a restriction enzyme and SDS (Figure 3A and B, panels f). Furthermore, the next step of the 3C procedure, incubation in ligation buffer, affects neither the distribution of heterochromatic regions and chromosome territories nor the visualization of nucleoli, splicing speckles and the nuclear matrix (data not shown). Finally, the degree of chromatin cleavage does not significantly affect the specific distribution of either chromatin (as revealed by staining of chromosome territories and heterochromatic regions) or the nuclear compartments analyzed in our experiments. Indeed, similar results were obtained on analysis of cross-linked nuclei treated with MboI instead of Hind III (Supplementary Figure S2). Thus, formaldehyde cross-linking preserves nuclei from lysis by SDS and strongly interferes with chromatin solubilization. Consequently, it can be concluded that in the course of the preparation of 3C material, the proximity ligation proceeds within the partially decondensed chromatin that is retained within nuclear remnants. This conclusion is valid for both liver and brain cells, as similar results were obtained when the above-described experiments were repeated on brain cells (Supplementary Figure S3).
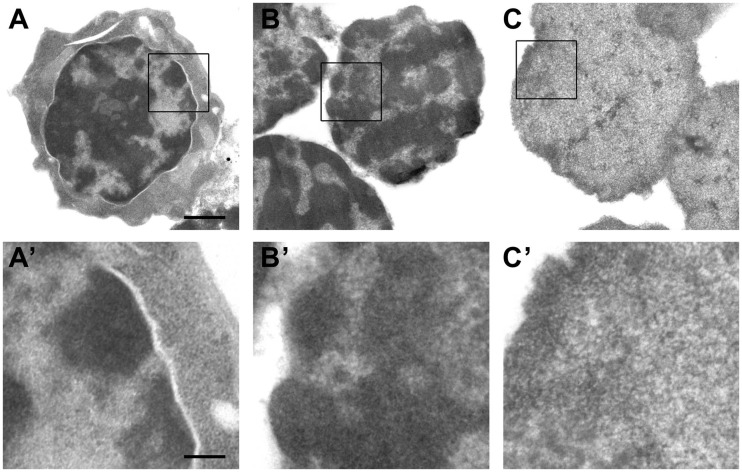
To obtain more information about the structural milieu of the proximity ligation, the nuclear remnants obtained after treatment of fixed nuclei with a restriction enzyme and SDS were inspected under an electron microscope. Figure 4 shows the typical morphology of a mouse fetal liver cell with a considerable amount of highly condensed heterochromatin located at the nuclear periphery (Figure 4A and A’). This characteristic nuclear morphology remains well preserved after cell lysis and extraction with 0.3% SDS/1.8% Triton X-100, which almost completely removes the cytoplasm (Figure 4B and B’). DNA digestion with HindIII restriction endonuclease and subsequent extraction with 1.6% SDS resulted in significant changes in chromatin structure, rendering nuclei less electron dense and with almost the complete disappearance of structurally defined condensed heterochromatin (Figure 4C and C’). The main structural component of these nuclei is represented by a meshwork of thin fibers (10–25 nm in diameter), apparently of chromatin, as suggested by DNA and histone staining (see Figure 3). The fibers are almost homogeneously distributed throughout the nuclear volume, and only a slight increase in fiber density is observed at the nuclear periphery, apparently corresponding to regions of heterochromatin. Interfibrillar distances range from 20–25 nm at the nuclear periphery to 30–50 nm in central regions of the nucleus. Similar results were obtained on electron microscopy analysis of fixed nuclei treated with MboI instead of HindIII (Supplementary Figure S4).
Figure 4.
Electron microscopic analysis of the insoluble 3C material from liver cells at different steps of the 3C procedure. After formaldehyde cross-linking (A and A’), after isolation of nuclei and extraction with 0.3% SDS followed by 1.8% Triton X-100 (B and B’) and after digestion with HindIII restriction endonuclease followed by extraction with 1.6% SDS (C and C’). Panels below show the enlarged framed region of the above images. Scale bars: 1 µm (A–C) and 250 nm (A’–C’).
Disruption of the residual nuclei results in a decrease of the 3C signals
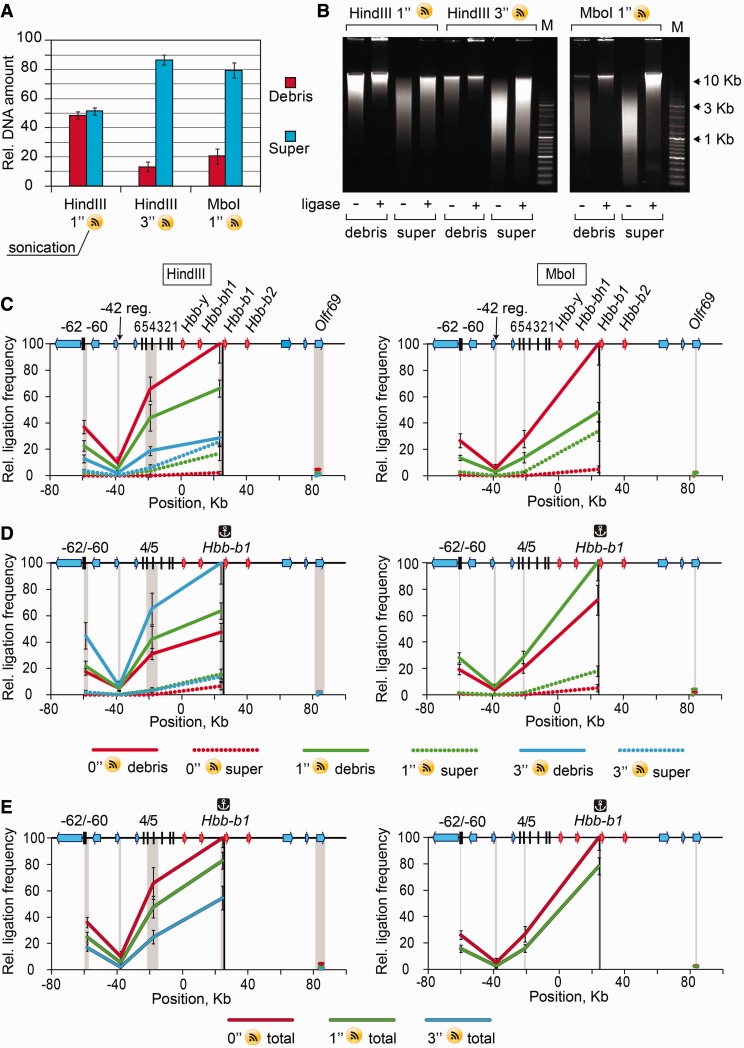
Collectively, the data discussed above may indicate that preservation of the internal nuclear organization is essential for producing specific 3C signals. The other possibility is that the nuclear remnants just interfere with solubilization of the ligated DNA–protein complexes without affecting the specificity and/or efficiency of ligation. To choose between the two possibilities, it is necessary to destroy the residual nuclei under conditions that do not affect the integrity of the putative DNA–protein complexes and then perform the proximity ligation in a solution. We have tested several strong dissociating agents, such as guanidine hydrochloride and guanidine isothiocyanate, but failed to lyse nuclei fixed by formaldehyde. Other researchers reported that sonication increased solubilization of the 3C material (21) and even increased the resolution of the C-protocols (22). We have thus sonicated the 3C material before the ligation step and then looked for the 3C signals in the total 3C material and separately in the soluble and insoluble fractions. The experiment was performed on E14.5 mouse embryo liver cells. Cross-linked chromatin was digested with either HindIII or MboI. As expected, even mild sonication (1-s pulse) caused the release of an important portion (∼50% in the case of Hind III digestion and ∼80% in case of MboI digestion) of cross-linked chromatin fragments into the soluble fraction. More intensive sonication (3-s pulse) ensured a solubilization of ∼85% of DNA from HindIII-treated nuclei (Figure 5A). Under these conditions, the average size of the released fragments decreased a little, but the possibility to ligate the solubilized fragments was not visibly affected (Figure 5B). Solubilization of an important (or even the major) portion of the cross-linked DNA fragments after sonication correlated with the apparition of the detectable 3C signals in the soluble fraction. However, the level of these signals was much lower than that observed in the insoluble fraction (Figure 5C). It was especially evident after normalization to the amount of DNA present in each fraction (Figure 5D). Furthermore, the total 3C signals (i.e. the signals detected in the unfractionated 3C material) decreased with the increase of the percentage of the solubilized DNA fragments. For example, in the experiments with HindIII endonuclease, the relative ligation frequency of the Hbb-b1 promoter to the HS4/5 of the LCR in the total 3C material decreased from ∼65 to ∼25% (Figure 5E). One may suggest that this drop in the 3C signals is because of the breakage of restriction fragments caused by sonication. To find out if this indeed happens under conditions of sonication used, we analyzed the levels of circularization of the anchor HindIII restriction fragment on ligation of a mixture of restriction fragments prepared from the control 3C material and the 3C material subjected to 1-s or 3-s pulses of sonication. The results obtained demonstrated that under our conditions of sonication, the level of breakage of the tested 1-Kb HindIII fragment was <5% (data not shown). Although larger Hind III fragments could be more seriously damaged by sonication, this is not likely to affect the possibility of ligation of their ends to the anchor fragment and thus the formation of bona fide amplicons that were very short (100–200 bp) in all our experiments. Thus, it is likely that the observed decrease of the level of total 3C signals observed in experiments with sonication of cross-linked nuclei is caused by the disruption of the internal nuclear organization rather than by additional fragmentation of DNA.
Figure 5.
Effect of sonication on the DNA partitioning between soluble and insoluble portions of the 3C material and on the frequencies of ligation of the β-globin gene domain fragments. (A) Relative amounts of DNA in the soluble and the insoluble portions of the sonicated 3C material. (B) Electrophoretic separation of DNA from soluble and insoluble portions of the sonicated 3C material before and after ligation. (C) Results of 3C analysis performed separately on soluble (super) and insoluble (debris) portions of the sonicated 3C material. (D) The same as (C) after normalization of the ligation frequencies to the amount of DNA in the samples. (E) Results of standard 3C analysis performed on the sonicated 3C material without fractionating into the soluble and the insoluble portions. The sonication time is indicated in seconds. The error bars represent SEM for two independent experiments. Other designations are as in Figure 1 and 2.
DISCUSSION
The proximity ligation procedure is a crucial step of all C-methods (2). It has been assumed that DNA fragments joined via protein bridges are solubilized from the formaldehyde-fixed nuclei after DNA cleavage by restriction enzymes and extraction with SDS. In a diluted solution, the cross-ligation of joined DNA fragments should be highly favored over the cross-ligation of separate fragments (1,4). The primary observation made in the present study is that the proximity ligation that generates characteristic 3C signals proceeds in the insoluble fraction within the cross-linked nuclei that turned out to be stable enough to resist SDS extraction. First, we demonstrated that a significant portion of DNA cannot be extracted from cross-linked nuclei after fragmentation by restriction endonucleases and SDS extraction. The amount of soluble material depends on the degree of DNA cleavage. A higher portion of DNA is solubilized after the treatment of cross-linked nuclei with MboI than with HindIII, which cut DNA into fragments ranging in sizes from ∼0.02 to ∼1.4 Kb and from ∼0.15 to ∼10 Kb, respectively. The degree of solubilization also depends on the nature of the cells. In our experiments, under similar conditions, more DNA was solubilized from brain cells than from erythroid cells. Most important, the characteristic 3C signals were generated in the insoluble fraction regardless of the portion of solubilized DNA. It is especially evident on comparing the analysis of ligation frequencies in the insoluble and soluble 3C material obtained from fetal liver cells using HindIII (solubilization of ∼15% of total DNA) and MboI (solubilization of ∼40% of total DNA) restriction enzymes. In both cases, the preferential ligation of the fragments bearing regulatory elements of the β-globin gene domain was observed in the insoluble 3C material only. Analysis of the insoluble material obtained in the course of the normal 3C procedure demonstrated that the nuclei cross-linked by formaldehyde are sufficiently resistant to SDS extraction. This treatment causes a significant decompactization of chromatin but does not provoke drastic redistribution of chromatin domains. It has been reported previously that, contrary to what one would expect based on the rationales of the 3C procedure (1,4,23), dilution of the 3C material before ligation is not essential to observe characteristic profiles of the 3C signals (21). We have made similar observations (data not shown). These observations receive an easy explanation after recognition of the fact that the proximity ligation proceeds within non-lysed nuclei. Taking into account this fact, one may ask whether characteristic 3C ligation products can only be generated within non-lysed nuclei. Two possibilities can be considered. First, ‘freezing’ of relatively large folded chromatin domains within the nucleus may be essential for generation of the characteristic 3C ligation products. These folded domains may be stabilized both by stochastic cross-links between neighboring chromatin fibers and by interaction with non-chromatin structures such as nuclear lamina, nuclear skeleton (matrix), Sc35 speckles et cetera. If this scenario is correct, mechanical destruction of cross-linked nuclei by sonication before ligation may be expected to interfere with the possibility to generate specific 3C ligation products. The other possibility is that complexes of regulatory elements linked via interacting proteins are fixed in the nuclei as originally proposed (1,4,23) but simply cannot be solubilized due to mechanical obstacles (such as the surrounding mesh of cross-linked chromatin fibers or the nuclear lamina stabilized by formaldehyde cross-links). In this case, mechanical destruction should not interfere with generation of the specific patterns of the 3C ligation products. Indeed, these products will be generated in a diluted solution exactly as postulated in the original 3C protocol (1,4,23). It has been reported previously that sonication improve solubilization of the 3C material but does not improve the resolution of the 3C procedure (21). Here we have studied the effect of sonication more systematically and made two important observations: (i) although sonication stimulated solubilization of the 3C material, the major portion of the characteristic 3C ligation products was generated in the insoluble fraction even after solubilization of >80% of cross-linked chromatin fragments (Figure 5C); and (ii) solubilization of cross-linked chromatin fragments correlated with the decrease of the total 3C signals (i.e. the signals detected in the unfractionated 3C material). Both observations better agree with a supposition that characteristic 3C ligation products can only be generated within non-lysed nuclei. We suggest that in formaldehyde-fixed nuclei, the mutual positions of regulatory elements located close to each other are maintained owing to cross-links between chromatin fibers, regardless of the nature of non-histone proteins that directly interact with these regulatory elements (Figure 6). Stochastic links between neighboring chromatin fibers will ‘freeze’ the spatial configuration of large chromosomal domains. The relative spatial positions of different genomic elements within these domains will not change after introducing a limited number of double-stranded breaks into DNA as long as each of the fragments is bound to other fragments by formaldehyde links (i.e. remains a part of the cross-linked chromatin mesh). Meanwhile, the DNA ends will possess local mobility sufficient for ligation to other DNA ends located in close proximity. At first approximation, it will be correct to say that chromatin fibers are joined by formaldehyde cross-links randomly. Occasionally, even relatively long fragments can be excluded from a cross-linked network. These fragments will be solubilized by SDS extraction. With the decrease of the average sizes of fragments, the portion of fragments that are not cross-linked to other fragments should increase. This explains the observed higher degree of DNA (chromatin) solubilization after the treatment of cross-linked nuclei with a restriction enzyme that cuts frequently (MboI). However, the mutual positions of different genomic elements will not be preserved in soluble fractions that are composed (at least for the most part) of separated DNA fragments. Thus, we do not see the expected 3C signals in the soluble fraction. Even the ligation frequency of adjacent DNA fragments, which should be cross-linked rather effectively via histones, is notably diminished in this fraction compared with the insoluble fraction. This observation can be easily explained by the lower probability of solubilization of the cross-linked fragments because both fragments will stay with the cross-linked chromatin network even if only one of these fragments is linked to this network.
Figure 6.
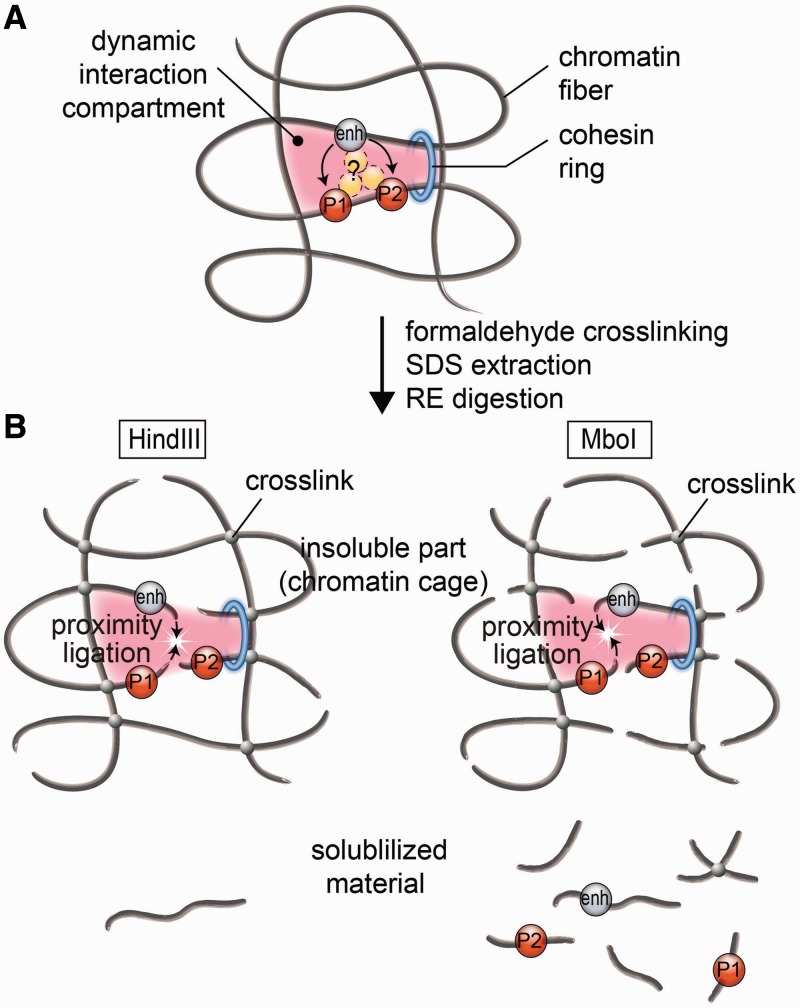
The proposed model of an active chromatin hub/compartment. (A) A schematic showing a putative dynamic interaction compartment containing two promoters (P1 and P2) controlled by a distant enhancer (enh). The enhancer is located in close spatial proximity to P1 and P2 owing to the formation of a chromatin loop stabilized by a cohesin ring (blue ring in the schematic). A complex of regulatory proteins (yellow circles) may or may not directly join the enhancer and promoter(s). After formaldehyde fixation, the mutual positions of the enhancer and the promoters became ‘frozen’ owing to the cross-links between closely located chromatin fibers. For the simplicity of presentation, these fibers are shown crossing each other. (B) After SDS extraction, the putative multicomponent protein complexes joining the enhancer and the promoter(s) are disintegrated (because of non-effective fixation of such complexes by formaldehyde), while the chromatin ‘cage’ stabilized by formaldehyde cross-links survives. Cleavage of DNA with restriction enzymes produces cohesive ends that possess a certain (limited) mobility within a chromatin cage so that the cohesive ends of different chromatin/DNA fibers located in close spatial proximity can be cross-ligated. Linking of chromatin fibers by formaldehyde is a stochastic process. In our model, the interaction compartment is stabilized by four cross-links between chromatin fibers. A failure to produce some of these cross-links will result in the separation of DNA fragments bearing the enhancer or the promoters from a chromatin cage. These fragments will be solubilized. However, in a solution, the fragments bearing the enhancer and the promoters will no longer be held in spatial proximity. Consequently, ligation of solubilized fragments will proceed without special preference and will not result in the generation of characteristic 3C signals.
Mechanical destruction of cross-linked nuclei by sonication is likely to produce a variety of products from separate chromatin fragments to relatively large pieces of the cross-linked chromatin mesh (chromatin aggregates), which will be partitioned between the soluble and the insoluble portions of the 3C material, the heaviest being precipitated under conditions that we have used for the separation. Within large chromatin aggregates, the relative spatial positions of individual genomic elements are likely to be preserved. This should allow for generation of a correct pattern of the 3C products on subsequent ligation. With a decrease in the size of the chromatin aggregates, the positioning information will be lost owing to a separation/breakage of individual fragments and increase of flexibility of the whole structure. Indeed, in our experiments with sonication of cross-linked nuclei, the correct 3C ligation products were generated predominantly in the insoluble portion of the material composed of large chromatin aggregates and nuclear remnants.
It should be stated that recognition of the fact that in the 3C protocol the proximity ligation is mediated within non-lysed nuclei does not question the existence and functional significance of chromatin loops that juxtapose distant regulatory elements. As a matter of fact, the first experimental evidence for such looping was produced using the nuclear ligation assay, an experimental procedure based on the proximity ligation in non-lysed nuclei (24,25). The model of the proximity ligation inside the chromatin cage (Figure 6) allows for a certain degree of flexibility of an ACH. As soon as the rigid fixation of all participating elements via formation of a common complex with interacting proteins is not obligatory, the chromatin hub can be considered to be a folded chromatin domain or a nuclear compartment, where regulatory elements and the promoters of transcribed genes are recruited [an expression hub, as proposed by Kosak and Groudine (26)]. The proximal positions of all of these regulatory elements may be supported by proteins that do not directly interact with any of them, for example, by CTCF and cohesin holding together two insulators (27–29) or by SATB1/2 interacting with MAR elements (30). The proposed model allows for short-lived alternating associations of regulatory elements with different promoters present within the active compartment. These temporal associations may explain the previously observed alternating transcription of β-globin genes (31,32) that appear to be recruited to the common ACH (4). The possibility that enhancers present within the nuclear compartment serve as nucleation centers for attraction of RNA Polymerase II, chromatin remodeling factors, histone acetylases, et cetera, also should not be ruled out.
Most of the 3C-derivative methods (4C, 5C, Hi-C) do not really depend on solubilization of cross-linked chromatin fragments. The observations made using Hi-C protocol (13,16,33) mostly concern the folding/shape of large chromosomal domains. Consequently, identification of multiple interactions between neighboring chromatin fibers should be of primary importance. This can be easily achieved without solubilization of chromatin from the nucleus (see Figure 6).
In contrast to the 4C, 5C and Hi-C, the ChIA-PET (14), ChIP-loop (34) and e4C (17) procedures strictly depend on solubilization of cross-linked chromatin fragments, as a specific portion of these fragments is selected for further analysis using immunoprecipitation from solution. In the ChIA-PET protocol (14), sonication is used to solubilize relatively short cross-linked chromatin fragments. As shown here, sonication causes massive drop in the detectable 3C signals. Under conditions used to prepare ChIA-PET material, only ‘true’ complexes of regulatory elements joined by protein bridges can be identified. It is obvious that such complexes also exist. Otherwise, the ChIA-PET method would not work. However, the percentage of such cross-linked complexes is likely to be rather low. Indeed, ‘very deep’ sequencing is necessary to detect ChIA-PET signals (2). It does not constitute a problem as long as one realizes that interactions identified by ChIA-PET and by most of the other C-methods may be basically different. The 3C protocol as well as the 4C and the 5C are indeed based on the fixation of chromosome conformation (‘freezing’ of the proximal location of distinct regulatory elements within a chromosomal territory) and not on the fixation of interactions between promoters and enhancers via regulatory proteins (protein complexes). Correspondingly, the results of the 3C analysis show the relative proximities of fragments within the nucleus rather than the frequencies of the interaction of these fragments. The ChIP-loop (34) and e4C (17) protocols do not include sonication step. Yet only the soluble portion of the 3C material is analyzed, which is subjected to immunoprecipitation before the proximity ligation. In our cellular model, only 15% of total DNA (DNA solubilized after treatment of nuclei with a 6-bp-cutting restriction enzyme) would be available for the ChIP-loop or e4C analysis. Furthermore, this would not be a representative portion of the total 3C material because of the non-equal solubilization of DNA fragments of different sizes and underrepresentation of interacting fragments that can be identified by the analysis of the proximity ligation products. It may happen that the degree of cross-linked chromatin fragment solubilization differs in different cellular models. Still, it is obvious that one should use the ChIP-loop and e4C procedures with extreme caution. In particular, the degree of solubilization of the cross-linked chromatin fragments should be strictly controlled.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figures 1–4.
FUNDING
Ministry of Science and Education of the Russian Federation [contracts 14.740.12.1344, 16.740.11.0483 and grant 8052]; Russian Foundation for Support of Basic Research [11-04-00361-а, 11-04-91334-NNIO_а, 12-04-00036-а, 12-04-33040 and12-04-93109_CNRS]; Presidium of the Russian Academy of Sciences (grants from the Program on Molecular and Cellular Biology); President of the Russian Federation [MK-3813.2012.4]; Dmitri Zimin’s foundation ‘Dynasty’. Experiments were performed using the equipment of the IGB RAS facilities supported by the Ministry of Science and Education of the Russian Federation [contract 16.552.11.7067]. Funding for open access charge: Dmitri Zimin’s foundation Dynasty.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 2.de Wit E, de Laat W. A decade of 3C technologies: insights into nuclear organization. Genes Dev. 2011;26:11–24. doi: 10.1101/gad.179804.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ethier SD, Miura H, Dostie J. Discovering genome regulation with 3C and 3C-related technologies. Biochim. Biophys. Acta. 2012;1819:401–410. doi: 10.1016/j.bbagrm.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 5.de Laat W, Grosveld F. Spatial organization of gene expression: the active chromatin hub. Chromosome Res. 2003;11:447–459. doi: 10.1023/a:1024922626726. [DOI] [PubMed] [Google Scholar]
- 6.Palstra RJ, de Laat W, Grosveld F. Beta-globin regulation and long-range interactions. Adv. Genet. 2008;61:107–142. doi: 10.1016/S0065-2660(07)00004-1. [DOI] [PubMed] [Google Scholar]
- 7.Palstra RJ. Close encounters of the 3C kind: long-range chromatin interactions and transcriptional regulation. Brief. Funct. Genomic Proteomic. 2009;8:297–309. doi: 10.1093/bfgp/elp016. [DOI] [PubMed] [Google Scholar]
- 8.Vernimmen D, De Gobbi M, Sloane-Stanley JA, Wood WG, Higgs DR. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. EMBO J. 2007;26:2041–2051. doi: 10.1038/sj.emboj.7601654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vernimmen D, Marques-Kranc F, Sharpe JA, Sloane-Stanley JA, Wood WG, Wallace HA, Smith AJ, Higgs DR. Chromosome looping at the human alpha-globin locus is mediated via the major upstream regulatory element (HS -40) Blood. 2009;114:4253–4260. doi: 10.1182/blood-2009-03-213439. [DOI] [PubMed] [Google Scholar]
- 10.Gavrilov AA, Razin SV. Spatial configuration of the chicken alpha-globin gene domain: immature and active chromatin hubs. Nucleic Acids Res. 2008;36:4629–4640. doi: 10.1093/nar/gkn429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat. Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 12.Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat. Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 13.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, Lee C, Shendure J, Fields S, Blau CA, Noble WS. A three-dimensional model of the yeast genome. Nature. 2011;465:363–367. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, McCord RP, Ho YJ, Lajoie BR, Hildebrand DG, Simon AC, Becker MS, Alt FW, Dekker J. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148:908–921. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat. Genet. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagege H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forne T. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nat. Protoc. 2007;2:1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- 19.Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, New York, USA: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 20.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 21.Comet I, Schuettengruber B, Sexton T, Cavalli G. A chromatin insulator driving three-dimensional Polycomb response element (PRE) contacts and Polycomb association with the chromatin fiber. Proc. Natl Acad. Sci. USA. 2011;108:2294–2299. doi: 10.1073/pnas.1002059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fullwood MJ, Ruan Y. ChIP-based methods for the identification of long-range chromatin interactions. J. Cell. Biochem. 2009;107:30–39. doi: 10.1002/jcb.22116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Splinter E, Grosveld F, de Laat W. 3C technology: analyzing the spatial organization of genomic loci in vivo. Methods Enzymol. 2004;375:493–507. doi: 10.1016/s0076-6879(03)75030-7. [DOI] [PubMed] [Google Scholar]
- 24.Cullen KE, Kladde MP, Seyfred MA. Interaction between transcription regulatory regions of prolactin chromatin. Science. 1993;261:203–206. doi: 10.1126/science.8327891. [DOI] [PubMed] [Google Scholar]
- 25.Gothard LQ, Hibbard JC, Seyfred MA. Estrogen-mediated induction of rat prolactin gene transcription requires the formation of a chromatin loop between the distal enhancer and proximal promoter regions. Mol. Endocrinol. 1996;10:185–195. doi: 10.1210/mend.10.2.8825558. [DOI] [PubMed] [Google Scholar]
- 26.Kosak ST, Groudine M. Form follows function: the genomic organization of cellular differentiation. Genes Dev. 2004;18:1371–1384. doi: 10.1101/gad.1209304. [DOI] [PubMed] [Google Scholar]
- 27.Hou C, Dale R, Dean A. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc. Natl Acad. Sci. USA. 2010;107:3651–3656. doi: 10.1073/pnas.0912087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mishiro T, Ishihara K, Hino S, Tsutsumi S, Aburatani H, Shirahige K, Kinoshita Y, Nakao M. Architectural roles of multiple chromatin insulators at the human apolipoprotein gene cluster. EMBO J. 2009;28:1234–1245. doi: 10.1038/emboj.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sofueva S, Hadjur S. Cohesin-mediated chromatin interactions–into the third dimension of gene regulation. Brief. Funct. Genomics. 2012;11:205–216. doi: 10.1093/bfgp/elr048. [DOI] [PubMed] [Google Scholar]
- 30.Zhou LQ, Wu J, Wang WT, Yu W, Zhao GN, Zhang P, Xiong J, Li M, Xue Z, Wang X, et al. The AT-rich DNA binding protein SATB2 promotes expression and physical association of human Ggamma- and Agamma-Globin genes. J. Biol. Chem. 2012;287:30641–30652. doi: 10.1074/jbc.M112.355271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wijgerde M, Grosveld F, Fraser P. Transcription complex stability and chromatin dynamics in vivo. Nature. 1995;377:209–213. doi: 10.1038/377209a0. [DOI] [PubMed] [Google Scholar]
- 32.Gribnau J, de Boer E, Trimborn T, Wijgerde M, Milot E, Grosveld F, Fraser P. Chromatin interaction mechanism of transcriptional control in vivo. EMBO J. 1998;17:6020–6027. doi: 10.1093/emboj/17.20.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat. Genet. 2005;37:31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.