Abstract
Although artificial RNA motifs that can functionally replace the GNRA/receptor interaction, a class of RNA–RNA interacting motifs, were isolated from RNA libraries and used to generate designer RNA structures, receptors for non-GNRA tetraloops have not been found in nature or selected from RNA libraries. In this study, we report successful isolation of a receptor motif interacting with GAAC, a non-GNRA tetraloop, from randomized sequences embedded in a catalytic RNA. Biochemical characterization of the GAAC/receptor interacting motif within three structural contexts showed its binding affinity, selectivity and structural autonomy. The motif has binding affinity comparable with that of a GNRA/receptor, selectivity orthogonal to GNRA/receptors and structural autonomy even in a large RNA context. These features would be advantageous for usage of the motif as a building block for designer RNAs. The isolated motif can also be used as a query sequence to search for unidentified naturally occurring GANC receptor motifs.
INTRODUCTION
Similar to proteins, several classes of RNAs exhibit their functions depending on their complex and defined 3D structures. Phylogenetic studies of these RNAs, including catalytic RNAs (ribozymes) and gene-controlling RNAs (riboswitches), have revealed that they are composed of recurrently occurring local sequences, so-called RNA motifs. X-ray crystallographic and NMR studies of RNAs have further elucidated that the RNA motifs are conserved at the level of tertiary rather than primary structure (1–5). The finding and elucidation of RNA motifs yield insights into the principles of RNA folding and the design of RNA architectures in a bottom–up manner.
RNA motifs are frequently involved in RNA–RNA tertiary interactions, which consist of ordered and stacked arrays of non-Watson–Crick base pairs (3). Among these interactions, GNRA tetraloops (where N stands for any nucleotide, and R for A or G) and their receptor motifs are most abundantly used in naturally occurring RNA structures (6–9). The most primitive and common receptor motif for a GUAA tetraloop is a tandem of two G:C pairs in the RNA duplex in which recognition is mediated by a lock-and-key mechanism based on the shape complementarity between the two rigid elements (GUAA loop and GG:CC base pairs) (10). The most sophisticated receptor motif is the 11 nucleotide receptor, or R(11 nt), that strongly recognizes a GAAA loop in a highly specific manner (7,9,11–13). In molecular recognition between R(11 nt) and GAAA, an induced-fit mechanism was observed in which the binding of the rigid GAAA loop induces structural rearrangement in the R(11 nt) motif (14).
In addition to the naturally occurring examples, several artificial receptor motifs for GNRA loops, such as GGAA and GUAA, have been generated by in vitro selection, or SELEX (15–17). To expand the repertory of modular RNA–RNA interacting motifs beyond GNRA loops, a receptor motif for a naturally occurring internal loop (C-loop) was isolated using a selection system with the cisDSL ribozyme—a class of RNA–RNA ligases—the catalytic activity of which depends on the tertiary interaction (18). The interacting motif of the C-loop and its artificial receptor is the first and sole example of an artificial interaction that can functionally replace the GNRA/receptor interactions. Naturally occurring RNA uses several classes of tetraloops as modular structural elements (19), but there have been no previous reports of modular receptor motifs for non-GNRA tetraloops. Thus, identification of modular receptors specific to non-GNRA tetraloops from naturally occurring RNA or artificial RNA sequence libraries would expand the variety of designer RNAs.
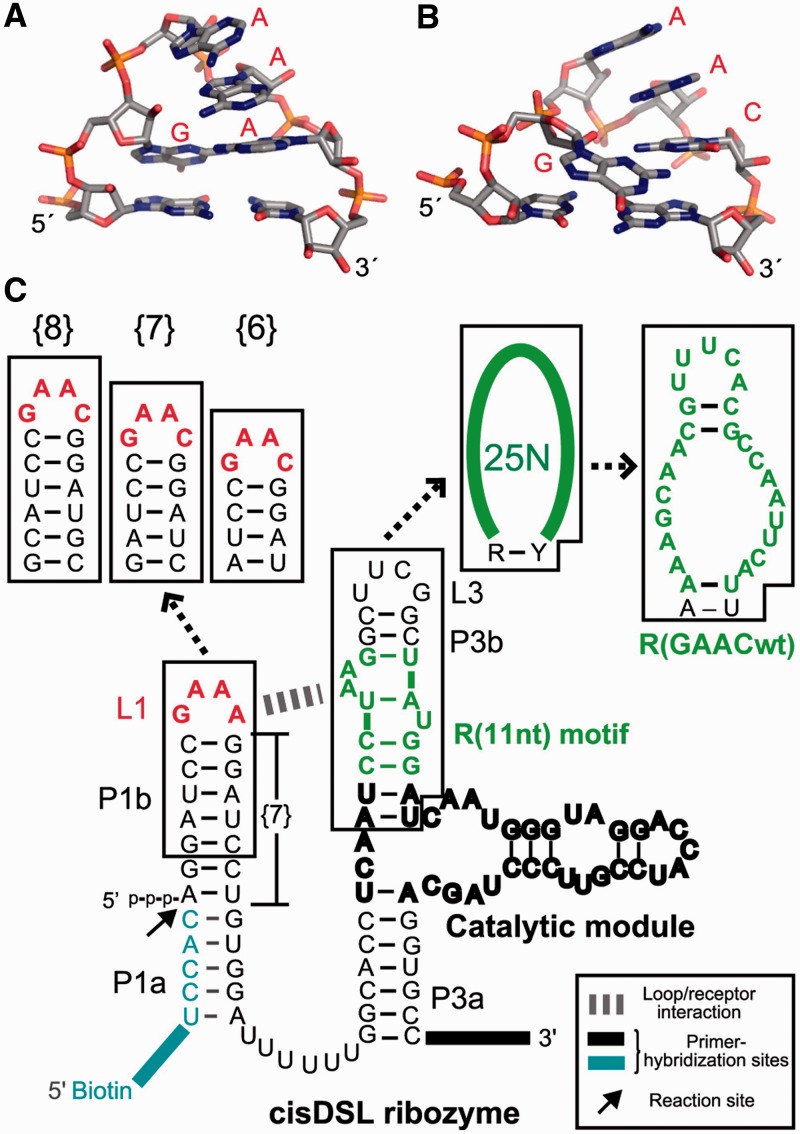
In this study, we identified a motif recognizing a GAAC tetraloop from artificial RNA sequence libraries. The GANC tetraloop has been reported as a novel tetraloop class in group II introns because the GANC loop folds in a manner close to, but distinct from, the GNRA-fold (Figure 1A and B) (20,21). In the crystal structure of a group II intron from Oceanobacillus iheyensis, the GAAC tetraloop forms a stacking interaction between the first A of GAAC and a distant part of the intron. The single base-stacking interaction seemingly contributes to establishment of the 3D structure of the group IIC intron as part of multiple tertiary interactions. This stacking interaction, however, seems too weak to apply as a modular part to construct artificial RNA 3D structures and assemblies. This observation prompted us to carry out in vitro selection of GAAC receptors. The isolated receptor motif is the first modular receptor for the non-GNRA tetraloop. Binding affinity, selectivity and modularity of the selected receptor were characterized with catalytic RNAs and a self-assembling RNA.
Figure 1.
Design of RNA libraries. Comparison of 3D structures of the GAAA (A) (PDB:2ADT), and the GAAC loops (B) (PDB:3EOH) drawn by using the PyMOL Molecular Graphics System (Version 1.3, Schrödinger, LLC.). (C) The secondary structures of the original cisDSL ribozyme, RNA libraries used for selection and the selected receptor. In the libraries, the GAAA tetraloop and its 11 nt receptor in the original DSL were replaced with GAAC and randomized nucleotides, respectively. {n} stands for the number of base pairs in P1b.
MATERIALS AND METHODS
Library design and RNA preparation
RNA libraries were designed based on the cisDSL ribozyme essentially as described previously with several modifications (18). In this study, GAAA loop and R(11 nt) in L1 and P3b region of the DSL ribozyme were replaced by GAAC and 27 random nucleotides, respectively (Figure 1C). The RNA library was synthesized by in vitro run-off transcription of DNA templates containing target GAAC sequence and the randomized region. The resulting RNA was purified on 10% denaturing gel. TectoRNAs, cisDSL and Tetrahymena intron ribozymes were also prepared by run-off transcription using PCR-amplified DNA templates. 5′-end labelled RNAs were purchased from JBios and IDT. 3′-end labelled RNAs with Alexa Fluor 488 hydrazide derivative (Invitrogen) were prepared as previously reported (22). Sequences of oligonucleotides used are listed in Supplementary Table S1, S3 and S6.
In vitro selection
In vitro selection was performed as described previously (18) (Supplementary Methods). Ligation reaction with libraries and biotinylated substrate RNA was performed under 50 mM MgCl2, 25 mM KCl and 30 mM Tris-Cl (pH 7.5) at 37°C. Reaction time was gradually shortened with each round. After five rounds of selection, RNA libraries were subjected to ligation reaction with 5′-FAM-labelled substrate RNA in the selection buffer for 10 h, and then the mixtures were separated on 9% denaturing PAGE. The ligated products visualized by fluorescence from the attached FAM were recovered and used as a template for reverse transcription. The resulting cDNA was amplified by PCR and cloned into pGEM T-Vector (Promega) for blue-white screening. Forty-nine clones were sequenced. One dominant and two minor sequences were subjected to secondary structure prediction using mFold (23). Then, the doped libraries were synthesized by mutation at a rate of 45% or 15% per position into the receptor sequence. Preparation of the RNA libraries was performed as described above. After two rounds of selection, the doped libraries were cloned and sequenced as described above.
Catalytic assay of the ribozymes
The ribozyme dissolved in distilled water was denatured at 80°C for 3 min, and then snap-cooled on ice for 2 min. Tenfold reaction buffer was added prior to incubation at the appropriate temperature and time indicated in the figure legend. Then, reactions were started by adding 5′-FAM-labelled substrate, and stopped by mixing equal amounts of aliquots of reaction mixture and stop solution containing 75% formamide, 0.10 M EDTA and 0.01% bromophenol blue at each time point. The mixtures were separated on denaturing gels (8% acrylamide for cisDSL, and 15% for intron ribozyme). Fluorescence intensities of the bands of products and substrates were quantified by Pharos FX FluoroImager (BioRad). The data were fitted to the following equation:
where t is time, k is the observed rate constant for formation of the product (kobs) and Fa is the calculated final yield. All experiments were repeated at least twice. The mean values are shown in the figures, and error bars indicate the minimal and maximal values.
Gel mobility shift assay of tectoRNA
All of the gel mobility shift assays were analysed on 13 cm × 13 cm × 2 mm thick, non-denaturing polyacrylamide gels (10% acrylamide) containing 89 mM Tris-Borate (pH 8.3) and 15 mM Mg(OAc)2 or 0.10 mM EDTA. The RNA samples containing the 3′-Alexa Fluor 488 (Invitrogen)-labelled (12.5 nM) and partner non-labelled (0–2500 nM) tectoRNAs were denatured at 80°C for 3 min and immediately snap-cooled on ice for 2 min. Folding buffer [89 mM Tris-borate (pH 8.3) and 15 mM Mg(OAc)2 or 0.10 mM EDTA at final concentration] was added, and the solutions were incubated at 30°C for 30 min, and then 4°C for 10 min. A 6-fold loading buffer [folding buffer with 0.01% bromophenol blue, 0.01% xylene cyanol, 50% glycerol] was added prior to load the samples on native gels. Gels were run for 4 h at 40 mA, and scanned using Pharos FX FluoroImager (BioRad). Discrete bands of monomer and dimer were quantified using Quantity One (BioRad). Kds were determined as the concentration at which one-half the RNA molecules are dimerized by using non-linear fitting with Kaleida Graph (ver. 4.1.0) (Supplementary Methods). Kd values represent the average of two independent experiments.
Chemical probing experiments with DMS and CMCT
Chemical modification experiments with dimethyl sulphate (DMS) and 1-cyclohexyl-3-(2-morpholinoethyl)carbodiimide metho-p-toluenesulfonate (CMCT) were performed essentially as described previously (24,25). Chemically modified nucleotides were detected as stops of reverse transcription (26). The resulting cDNA fragments were separated by electrophoresis using DNA sequencer (4300 DNA Analyzer; Li-COR). Semi-Automated Footprinting Analysis software (SAFA) was used to quantify band intensities at single-nucleotide resolution (27). Each band was assigned by comparing the modified sample’s lane to sequencing ladders run on the same gel. Modification intensities of each nucleotide were normalized by dividing it by sum of intensity of the corresponding lane, and normalized values were averaged from two independent experiments.
RESULTS
Selection of GAAC receptor motif
To isolate modular receptor motifs for non-GNRA tetraloops from an RNA library, we chose the GAAC tetraloop, a frequent sequence in the GANC family, as a target and applied a selection system developed by Ohuchi and coworkers (18). This system is based on the cisDSL ribozyme that ligates its 5′ terminus to a short RNA fragment aligned as part of the substrate P1 helix (Figure 1C) (28). Bond-forming reaction in the P1 helix is promoted by the catalytic module in the P3 element, and the association between the P1 helix and catalytic module is established by the GAAA/R(11 nt) interaction between L1 and P3b. Therefore, recognition of the L1 tetraloop by the receptor in P3b is tightly coupled with the catalytic activity of the ribozyme.
In the designed RNA libraries, the L1 GAAA loop and R(11 nt) module in cisDSL were replaced with the GAAC loop and 27 random nucleotides, respectively (Figure 1C). Among the 27 nucleotides, first and last nucleotides were assigned to be purine (R) and pyrimidine (Y) bases, because the R–Y pair is essential to establish the active catalytic module (28,29). Because the docking of the substrate P1 helix to the catalytic module is directly governed by the interaction between L1 and P3b, ligation-dependent selection of the library will enrich receptors for the L1 GAAC loop. Active RNA molecules, which were covalently joined with a biotinylated RNA substrate, were recovered by selective capture with streptavidin-magnetic beads. In addition to the library with a GAAC loop in place of the parent L1 GAAA, we constructed another two libraries with the L1 GAAC loop and a single-base pair insertion or deletion in the P1b region (+1 or −1 bp) because the relative position and orientation between the GAAC loop and the putative receptor are unpredictable.
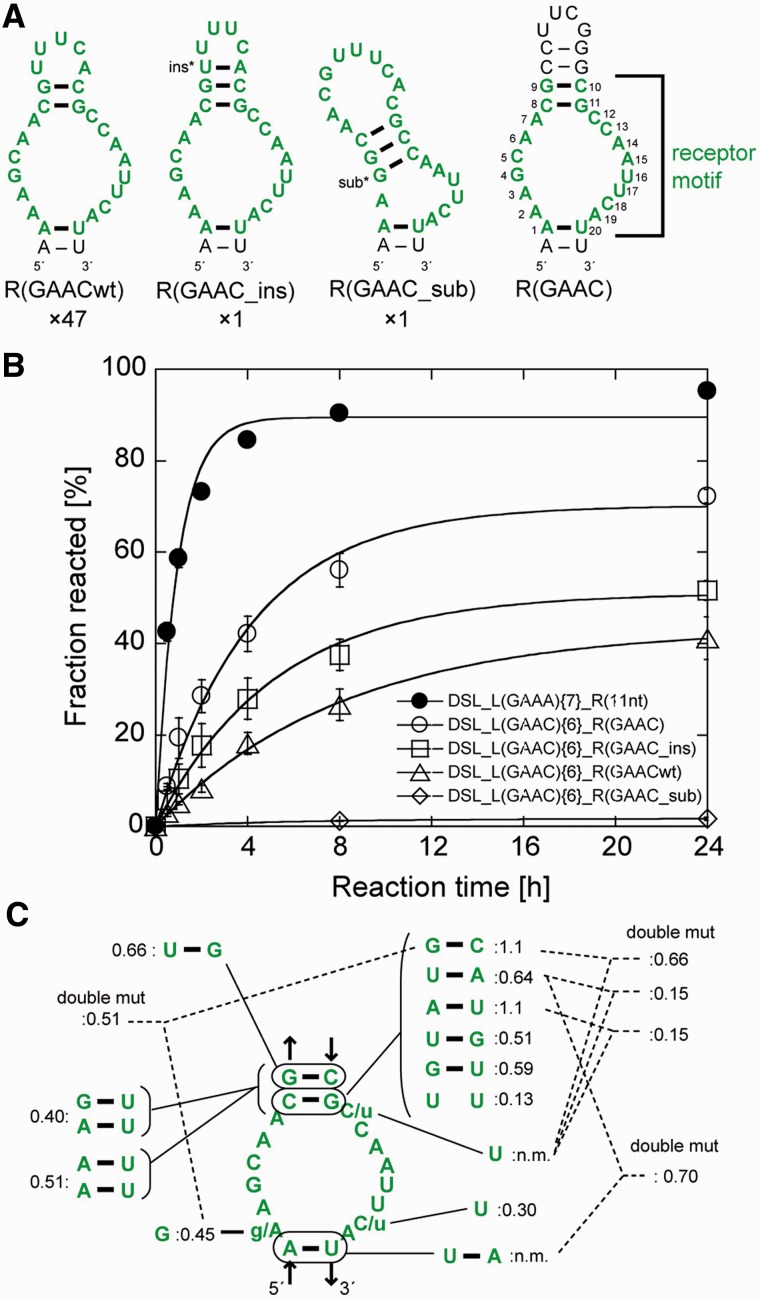
Ligation reactions of the three libraries (designated as libraries {7}, {8} and {6} with 7, 8 and 6 bp in the P1b region, respectively) with the biotinylated substrate RNA were performed individually (Supplementary Table S2). After five rounds of selection, catalytic activity was observed in the pool from library {6}. The ligated products, which were purified by denaturing PAGE, were cloned and sequenced. Of 49 clones picked randomly from the product pool, 47 had an identical sequence, termed R(GAACwt), in the randomized region (Figure 2A). The other two clones also had R(GAACwt)-like sequences with a single-base insertion or substitution, and were designated as R(GAAC_ins) or R(GAAC_sub), respectively.
Figure 2.
Selected receptors for the GAAC loop. (A) Predicted secondary structures of three selected sequences (the left three) and the UUCG-capped mutant (right). The numbers shown below the structures indicate frequency of occurrence of each receptor among 49 sequenced clones. Selected sequences are shown in green, and consensus nucleotides of the receptor are numbered. The inserted or substituted nucleotides are denoted by asterisks. (B) Time courses of ligation reaction by the cisDSLs possessing the L1 GAAC loop and selected receptors. Reaction mixtures include 1.0 µM ribozymes, 0.50 µM 5′-FAM labelled substrate RNA and the buffer used in the selection experiment. (C) Mutations obtained from doped selection. Relative activity is indicated by product yield after 6 h reaction for each mutant with DSL_L(GAAC){6}_R(GAAC) as the standard (1.0). (n.m.) Not-measured because of the absence of the mutants.
Secondary structures of the three receptors were predicted using mFold under the constraint that the first and last nucleotides form a base pair corresponding to the R–Y pair (23). R(GAACwt) can form an asymmetric internal loop with 6 and 8 nucleotides capped with a pentaloop hairpin (Figure 2A). The ligation activity of each clone of the three cisDSL ribozymes was evaluated with the buffer used for selection (Figure 2B). In this study, we will refer to the cisDSL ribozyme consisting of one L1 tetraloop (L1) and one receptor (R) as DSL_L1{n}_R where ‘n’ denotes the number of base pairs (bp) in the P1b region of the ribozyme (Figure 1C). The ligation product of DSL_L(GAAC){6}_R(GAACwt) was observed in 27% yield after 8 h. The activity of DSL_L(GAAC){6}_R(GAAC_ins) (38% after 8 h) was higher than that of DSL_L(GAAC){6}_R(GAACwt). DSL_L(GAAC){6}_R(GAAC_sub) was poorly active presumably due to deformation of the receptor structure induced by base substitution. We therefore prepared a variant [DSL_L(GAAC){6}_R(GAAC)] in which the internal loop was closed with four consecutive base pairs capped by a UUCG loop (Figure 2A right). We used the UUCG loop to cap P3b because the secondary structures of R(GAACwt) and R(GAAC_ins) would be further stabilized by the extraordinary stability of the UNCG hairpin structures (30). The activity of DSL_L(GAAC){6}_R(GAAC) (60% after 8 h) was higher than those of any of the selected clones, indicating the predicted structure involving the asymmetric internal loop with 6 and 8 nucleotides would be active form. Therefore, we defined the 20 nucleotides in the selected sequence as a consensus of the GAAC receptor motif (numbered nucleotides in Figure 2A right).
We then performed the second selection experiment with the doped libraries to optimize the receptor motif and identify the nucleotides important to folding and/or interaction with the GAAC loop. Starting from 45%-doped library based on DSL_L(GAAC){6}_R(GAAC), two rounds of selection enriched sequences maintaining the predicted secondary structure of the parent motif (Figure 2C and Supplementary Figure S1). The high degree of nucleotide conservation of the internal loop region of the receptor indicated that most of the original nucleotides were functionally important. Activity assay indicated that selected variants had activity comparable with or lower than the original clone (Figure 2C). Continuation of the selection round and another selection with the mixed library of 15%- and 45%-doped sub-libraries only resulted in enrichment of the parent R(GAAC) motif (Supplementary Tables S4 and S5).
Evaluation of binding affinity, specificity and modularity of the GAAC/R(GAAC) interaction in three structural contexts
Structural context of the DSL ribozyme where the GAAC/R(GAAC) motif evolved
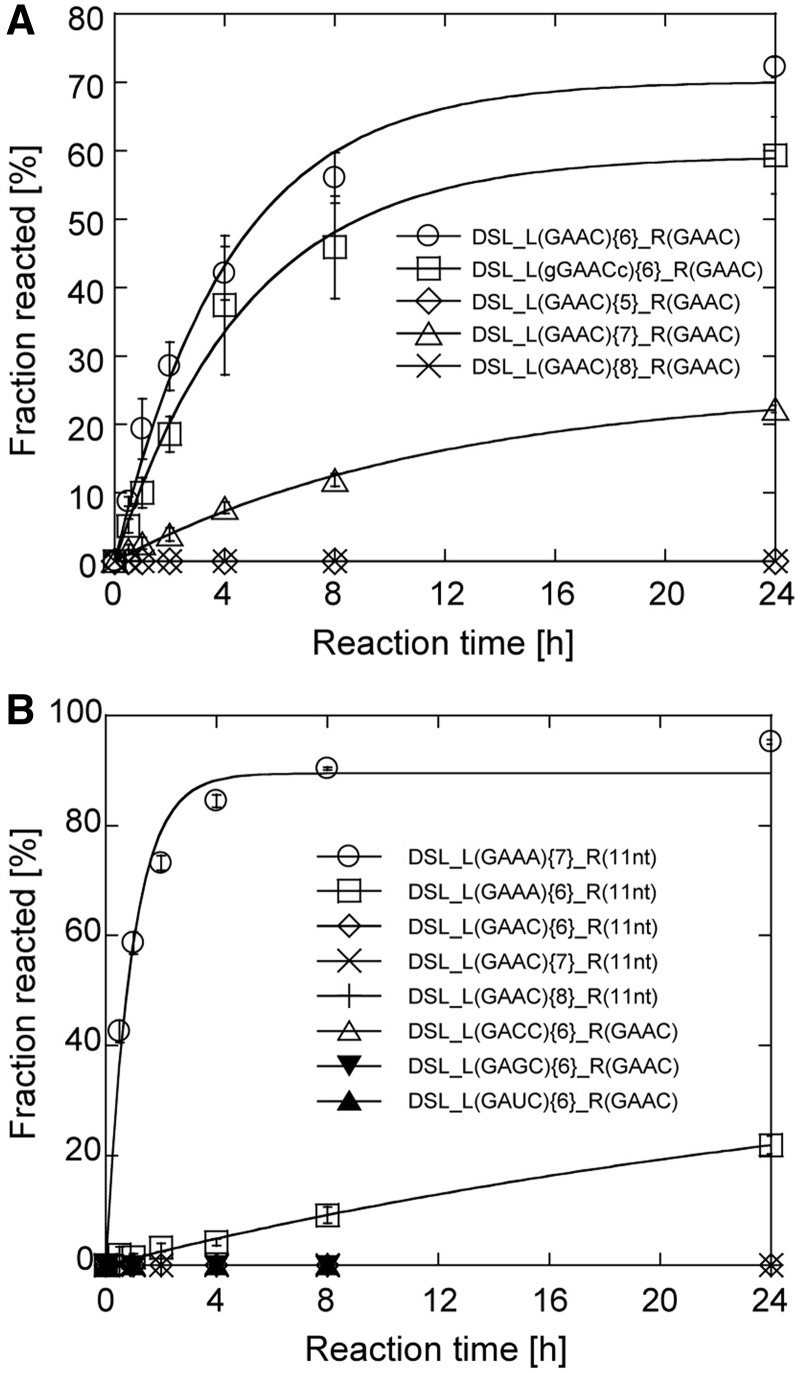
To gain insight into binding properties of the R(GAAC) motif, a series of combinations of tetraloops and receptors were embedded in the L1 and P3b regions of the cisDSL ribozyme scaffold. DSL_L(GAAC){6}_R(GAAC) gave the ligation product in 60% yield in 8 h reaction. The effects of the length of P1b were examined using DSL_L(GAAC){5}_R(GAAC), DSL_L(GAAC){7}_R(GAAC) and DSL_L(GAAC){8}_R(GAAC); these variants were either hardly active or inactive (Figure 3A). These observations indicated that the GAAC/R(GAAC) interaction is critically dependent on the relative position between the loop and the receptor, as observed in the naturally occurring GAAA/R(11 nt) interaction (31). To determine whether recognition by the R(GAAC) requires only the GAAC tetraloop region, we altered the base pair (C–G) closing the L1 loop with a G–C pair. The resulting variant was designated as DSL_L(gGAACc){6}_R(GAAC). This mutation caused no severe reduction of the ribozyme activity, suggesting that the receptor recognizes the L1 GAAC loop in a hairpin form of the P1b element (Figure 3A). We examined whether the receptor generally recognizes the GANC loop family or is highly specific to GAAC. No ligation ability was detected in variants with GAGC, GACC and GAUC L1 loops, indicating that the selected receptor was highly specific to the GAAC loop (Figure 3B). Furthermore, the R(11 nt) motif with L1 GAAC loop also showed no activity, suggesting that GAAC/R(GAAC) and GAAA/R(11 nt) interactions are orthogonal to each other (Figure 3B).
Figure 3.
Selectivity of the GAAC/R(GAAC) interaction. (A and B) Time courses of ligation reaction by the DSL mutants. Conditions were as described in the legend of Figure 2.
Simple structural context of the tectoRNA assembling via loop/receptor interactions
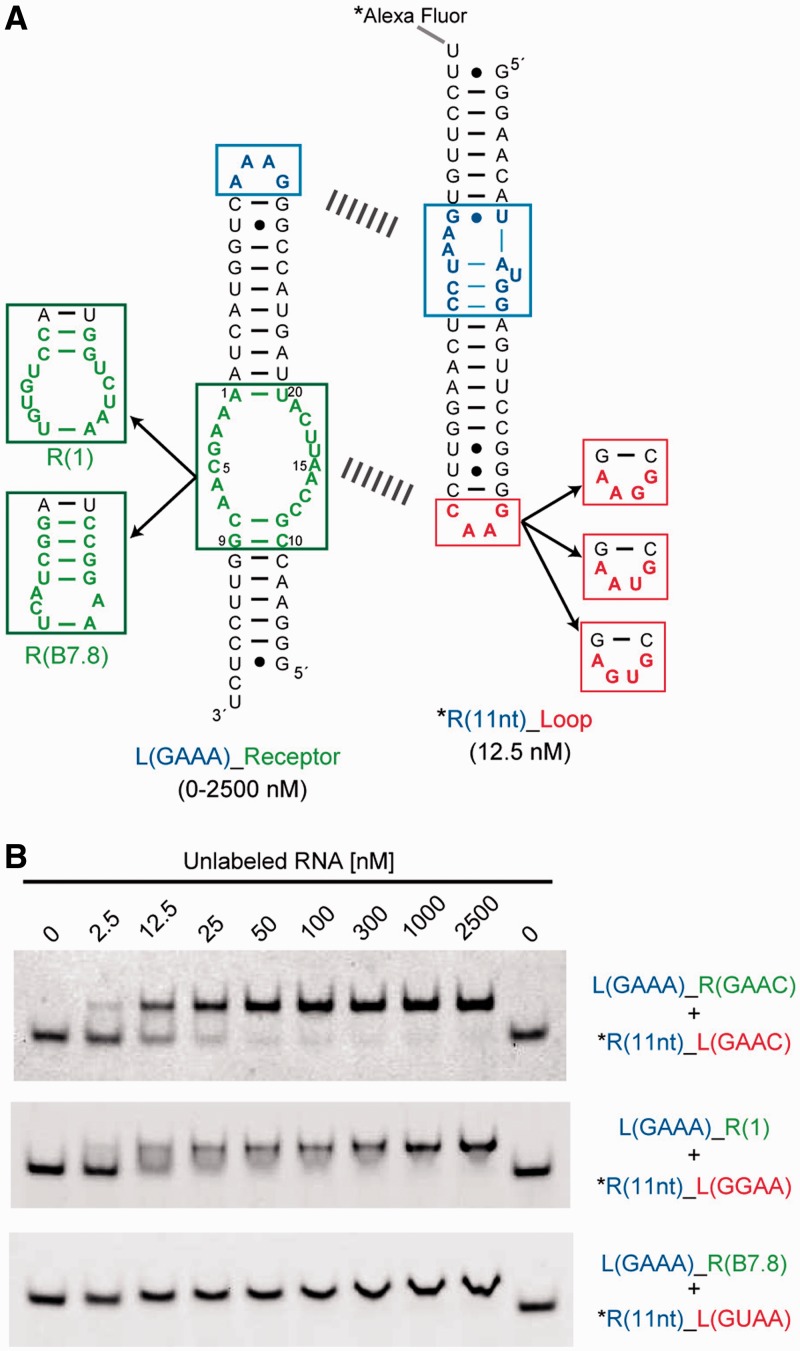
As the GAAC receptor was evolved in the context of the cisDSL ribozyme, it was important to determine whether the motif works as a module in different structural contexts. As an alternative structural context that is simpler than that of the cisDSL, we chose a self-assembling tectoRNA, which assembles via two sets of loop/receptor interacting motifs separated by RNA duplexes with appropriate length (Figure 4A) (32). To compare the binding properties of the selected motif with those of the GNRA receptors, we prepared a series of heterodimeric tectoRNAs bearing one GAAA/R(11 nt) module as a common clamp and another interacting module as an analyte. In this structural context, the loop/receptor interaction of analytes can be readily and semiquantitatively analysed by gel mobility shift assay (17,32). In the presence of 15 mM Mg2+ ions, a heterodimer formed through the GAAC/R(GAAC) interaction showed a dissociation constant (Kd) of 2.4 ± 0.36 nM, which was comparable with Kd (8.3 ± 0.97 nM) of a heterodimer formed through the GGAA/R(1) interaction that has the highest affinity among artificial GNRA loop/receptor interactions (Figure 4B) (17). On the other hand, no complex formation was observed in the case of tectoRNAs having mismatched combinations of loop/receptors, such as GNRA/R(GAAC) and GAAC/R(GNRA) (Supplementary Figure S2). These results were fully consistent with those of the cisDSL ribozyme activity assay and again indicated that the interaction of GAAC and its receptor is highly orthogonal to a class of GNRA loop/receptor modules.
Figure 4.
Self-assembly experiments using tectoRNA. (A) The secondary structure of tectoRNA, a self-assembling RNA consisting of fluorescently labelled R(11 nt)_Loop (right) and unlabelled L(GAAA)_Receptor (left) RNAs. The two RNAs assembled via GAAA/R(11 nt) as a common clamp (upper dashed line) and other tetraloop/receptor interactions as analyte modules (lower dashed line). (B) Representatives of PAGE analysis of assembling tectoRNAs. Upper and lower bands represent the hetero dimer and dye-labelled monomer, respectively. The mobility of the dimer band of R(11 nt)_L(GUAA) + L(GAAA)_R(B7.8) varied continuously with concentration of unlabelled RNA, indicating a fast exchange kinetics between monomer and dimer forms. Assembling was performed as described in the Methods section.
Complex structural context of a naturally occurring group I intron ribozyme
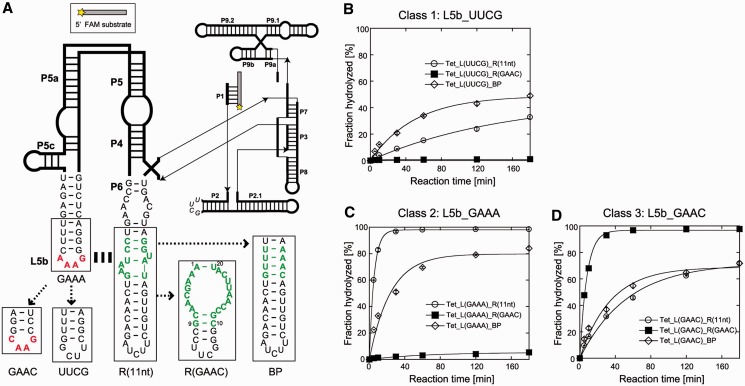
To further demonstrate the structural modularity of the GAAC receptor motif, we introduced the GAAC/R(GAAC) into a third structural context that is more complex than cisDSL. We used a naturally occurring group I ribozyme from the Tetrahymena large ribosomal subunit RNA using the GAAA/R(11 nt) module in its P4–P6 domain. In the cisDSL ribozyme, the substrate recognition was fully dependent on the L1_loop/P3b_receptor interaction. On the other hand, the large and complex Tetrahymena ribozyme structure is supported by multiple redundant tertiary interactions, in which disruption of one interaction [GAAA/R(11 nt) module] reduces but does not abolish its catalytic activity. Multiple redundant tertiary interactions involving the GAAA/R(11 nt) module also determine the multistep folding process of the Tetrahymena ribozyme (33–37). Characterization of the GAAC/R(GAAC) module in a complex RNA structure was carried out with a derivative of the Tetrahymena ribozyme lacking long-range L2–L5c base pairs (Figure 5A). The hydrolytic endonuclease reaction was used because this reaction sensitively reflects the effects of the loop/receptor interaction. We prepared three classes of variant with different L5b tetraloops (UUCG, GAAA or GAAC). Each class had three ribozymes with different P6 regions [BP, R(11 nt) or R(GAAC)]. We used a UUCG loop because the folded UNCG loops not only stabilize the hairpin structure (30) but also are incapable of forming long-range tertiary interactions (38).
Figure 5.
Catalytic activities of the large naturally occurring RNA possessing the loop/receptor derivatives. (A) Secondary structure of the L-21 ScaI Tetrahymena thermophila intron ribozyme lacking long-range L2–L5c interaction. (B–D) Time courses of endonuclease reaction by the ribozymes. Reactions were performed with 1.0 µM ribozymes, 0.50 µM 5′-FAM-labelled substrate RNA, 2 mM Mg2+ and 30 mM Tris-Cl (pH 8.0) at 50°C.
The first class variants shared the L5b UUCG loop disrupting L5b–P6 interaction regardless of the receptor motif in the P6 region. Tet_L(UUCG)_BP and Tet_L(UUCG)_R(11 nt) were moderately active because the core elements could be folded correctly due to other tertiary interaction (Figure 5B). On the other hand, Tet_L(UUCG)_R(GAAC) was almost completely inactive, possibly because the R(GAAC) motif disturbs the tertiary folding of the intron and/or deforms its local structures, such as the neighbouring P6 element involved in the catalytic core.
The second class of variants had the GAAA loop in L5b. The wild-type [Tet_L(GAAA)_R(11 nt)] exhibited the highest activity, indicating establishment of the L5b–P6 interaction (Figure 5C). The variant with L(GAAA)/R(GAAC) motif was still nearly inactive. In the case of P6_BP mutants, replacement of the L5b UUCG loop with the GAAA loop gave modest improvement of the activity (Figure 5B and C), probably due to a gain of A-minor interactions between GNRA-type tetraloops and a minor grove of the RNA duplex (17,39,40).
The third class of variants sharing the L5b GAAC loop were then tested (Figure 5D). The activity of the mutant with R(GAAC) motif was as high as that of the wild-type, indicating docking of the GAAC loop into the R(GAAC) motif in the P6 region. The resulting GAAC/R(GAAC) interaction could fully replace the parent GAAA/R(11 nt) interaction. Interestingly, substitution of the L5b UUCG loop with the GAAC loop modestly improved the activities of the ribozymes with the R(11 nt) motif and the P6_BP element (Figure 5B and D), although the extent of improvement was lower than in the case of substitution with GAAA loop (Figure 5B and C). This observation suggested the presence of weak interactions between the L5b GAAC loop and the R(11 nt) and duplex in P6, which were difficult to detect in simpler structural contexts.
Chemical probing of GAAC/R(GAAC) interaction in the three structural contexts
To obtain structural information regarding the GAAC/R(GAAC) interacting motif in the different contexts, chemical modification was performed with the simple (tectoRNA), standard (cisDSL ribozyme) and complex (Tetrahymena ribozyme) structural contexts.
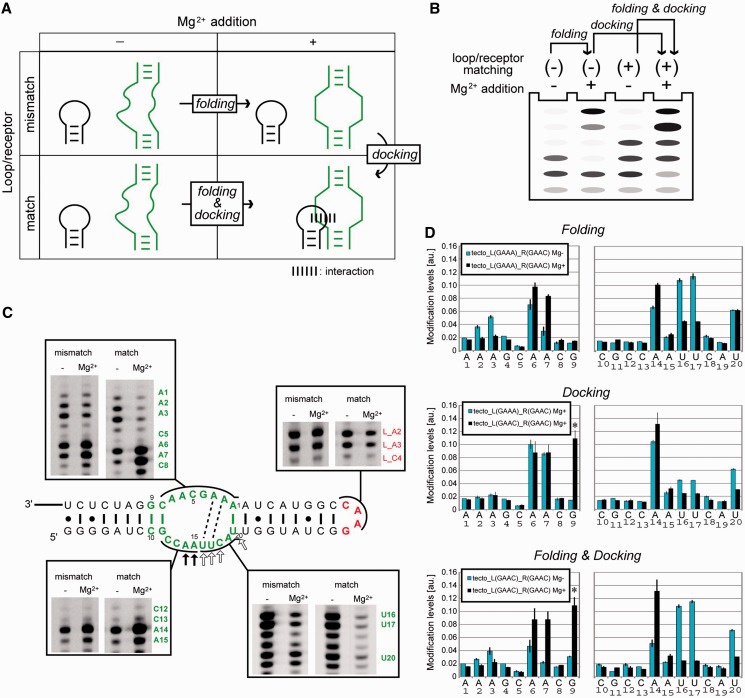
According to the previous structural analysis of the GAAA/R(11 nt) interaction, the structural changes of the tetraloop/receptor interaction can be classified into two steps—Mg2+-dependent folding of the isolated loop and receptor (abbreviated as folding) and docking of the two components (abbreviated as docking) (Figure 6A and B). Structural changes occurring in the respective steps can be evaluated by comparing the chemical modification profiles of matched and mismatched combinations of loop/receptor motifs in the presence or absence of Mg2+ ions.
Figure 6.
Chemical modification of the tectoRNA. (A and B) Scheme (A) and experimental conditions (B) used to show the effects of docking, folding and both. Loop/receptor matching (+) or (–) indicate whether the combination of loop and receptor is appropriate (+) or not (–). (C) Representatives of PAGE gel images of reverse-transcription of modified RNA. The box on the bottom right shows gel for CMCT experiments, and the others for DMS. Dashed lines indicate possible WC-base pairs in the Mg2+-dependent pre-organized structure. In the presence of Mg2+ ions, positions indicated with open and filled arrows exhibited stronger protection and modification respectively in the matched pair than in the mismatched pair. (D) Bar graph representation of relative modification intensity quantified and calculated from raw data (C). Asterisks denote Mg2+-dependent but CMCT-independent stops at G9. The intensities at A and C nucleotides were obtained from DMS modification experiment, and G and U from CMCT modification. Upper, middle and bottom graphs represent the effect of docking, folding and both, against chemical modification in the receptor embedded in tectoRNA.
Mg2+-dependent folding of isolated GAAC loop and R(GAAC)
It is well known that tertiary folding of RNA structures as well as their local elements involving RNA motifs requires metal ions (41,42). Therefore, we evaluated the Mg2+-dependent folding of isolated GAAC loop and R(GAAC) using homodimerization-deficient mutants of tectoRNA (Figure 6C). Chemical probing was carried out using DMS and CMCT, which have been used to probe the solvent accessibility of the Watson-Crick (WC) edge of nucleobases. Modification-dependent termination of reverse transcription allows us to detect DMS modifications at N1 of adenines and N3 of cytosines and CMCT modifications at N1 of guanines and N3 of uridines (26). The structural changes of the isolated GAAC loop were characterized using a homodimerization-deficient RNA lacking R(GAAC) [tecto_L(GAAC)_R(11 nt)]. Modification of L_A2 and L_A3 in the GAAC loop was unchanged upon addition of Mg2+, suggesting that no large structural changes were induced in the GAAC loop by Mg2+ (Figure 6C).
Mg2+-dependent structural changes of the isolated R(GAAC) motif was also evaluated using another homodimerization-deficient mutant lacking the GAAC loop [tecto_L(GAAA)_R(GAAC)]. In the presence of 15 mM Mg2+, three adenines in the receptor (A6, A7 and A14) were strongly modified, whereas four nucleotides in the receptor (A2, A3, U16 and U17) were protected from modification (Figure 6D top), indicating that Mg2+ induced folding of the R(GAAC) in the isolated state. These chemical probing data suggest a possible 2D structure of the R(GAAC) motif that has two canonical AU base pairs in the internal loop (Figure 6C).
Structural change upon docking between GAAC loop and receptor
We next investigated the homodimeric RNA [tecto_L(GAAC)_R(GAAC)] with 15 mM Mg2+ to determine the effects of loop/receptor docking. In the dimerized state, two adenines in the GAAC loop were slightly protected (Figure 6C). In R(GAAC) forming the tectoRNA homodimer, although the modification pattern was similar to that of the dimerization-deficient mutant,U16, U17 and U20 showed further protection upon dimerization (Figure 6D, middle). Considering the fact that U16, U17 and C18 were conserved in the active sequences recovered from doped selection (Supplementary Figure S1), the UUC trinucleotides may directly participate in the recognition of the GAAC tetraloop.
The modification data obtained by probing the folding and docking processes indicated that R(GAAC) was structured in a Mg2+-dependent manner and docking of the loop/receptor motif rigidified the structure of the RNA receptor motif without large structural rearrangement. In the GAAC loop, two adenines were slightly protected upon docking (Figure 6C). This observation, taken together with the observation that the third nucleotide of GAAC loop critically determines the catalytic activity of the cisDSL (Figure 3B), suggested that the WC edges of the two adenines in the GAAC loop would be involved in the interaction with the receptor motif.
Metal ion specificity of the GAAC/R(GAAC) interaction
The roles of metal ions in the functional RNAs are major and important issues in RNA biochemistry (41,42). However, the analysis of metal ion dependency of ribozymes is usually complicated because the effects of metal ions must be classified according to structural and catalytic roles. Therefore, metal ion dependency on the GAAC/receptor interaction was investigated using the tectoRNA, non-catalytic RNA.
DMS modification of the homodimeric construct [tecto_L(GAAC)_R(GAAC)] was performed in the presence of either MgCl2, MnCl2, Co(NH3)6Cl3, CaCl2 or KCl. Among these metals, Ca2+ and Mg2+ afforded similar modification signatures, as clearly seen in the loop and receptor regions (Supplementary Figure S3). These observations strongly suggested that Ca2+ can induce docking of L(GAAC)/R(GAAC). In fact, gel mobility shift assay with Ca2+ ions experimentally confirmed the tectoRNA dimerization induced by Ca2+ (Supplementary Figure S4). In contrast, addition of an excess of K+ did not alter the modification pattern, suggesting that K+ does not support the interaction (Supplementary Figure S3). Modification patterns with Mn2+ or  were closer to that without metal ions than that with Mg2+. These observations were consistent with the fact that Mn2+ ions, which support catalysis by the original DSL ribozyme, did not support the reaction of DSL_L(GAAC){6}_R(GAAC) (Supplementary Figure S5).
were closer to that without metal ions than that with Mg2+. These observations were consistent with the fact that Mn2+ ions, which support catalysis by the original DSL ribozyme, did not support the reaction of DSL_L(GAAC){6}_R(GAAC) (Supplementary Figure S5).
Module independency and mutational effects analysed in the cisDSL context
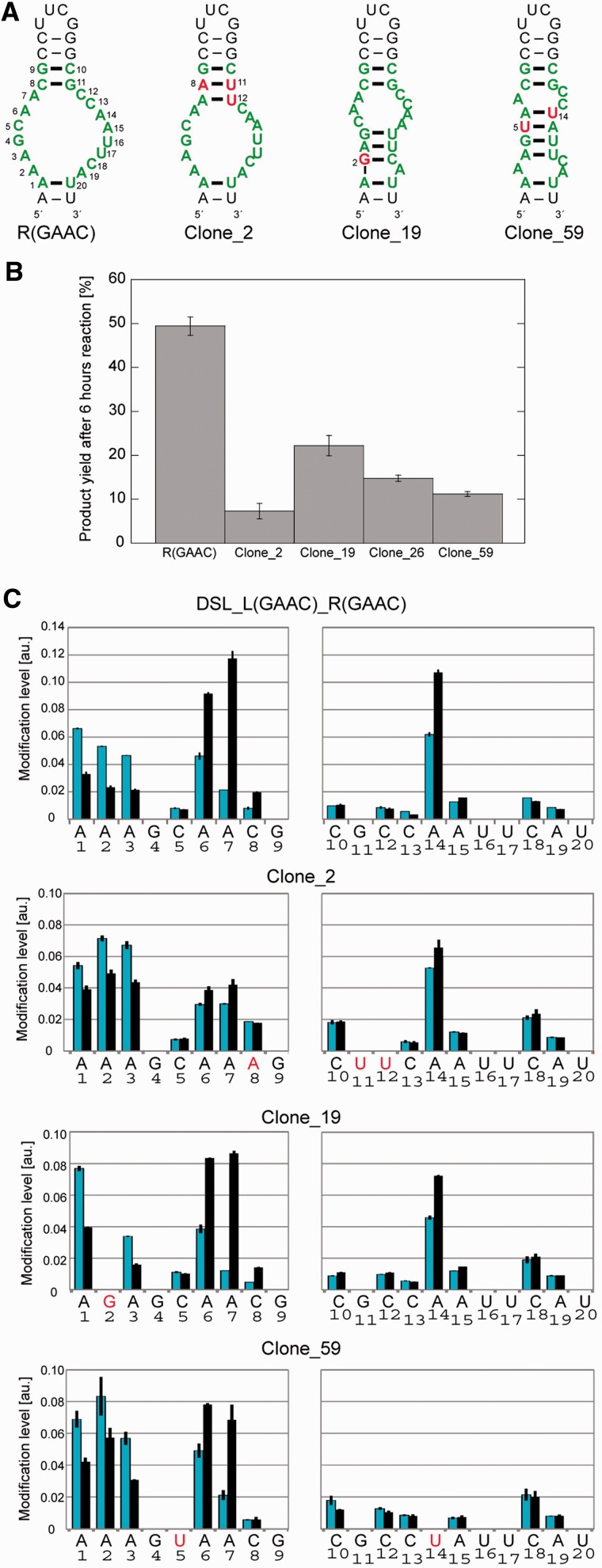
In the context of the cisDSL ribozyme, the P3b receptor motif neighbours the catalytic module. Thus, folding interdependency between the two modules, which may be an issue from the viewpoint of RNA tectonics, was analysed by DMS. Comparative DMS modification analysis of the parent [DSL_L(GAAC){6}_R(GAAC)] and L1 UUCG mutant [DSL_L(UUCG){6}_R(GAAC)] indicated that folding of the catalytic module is dependent on Mg2+ ions but independent of the loop/receptor interaction (Supplementary Figure S6). Structural independency between the catalytic module and R(GAAC) in cisDSL was also confirmed from the observation that DMS modification patterns of the R(GAAC) motif folded by Mg2+ and docked with L1 GAAC loop in the cisDSL ribozyme were closely similar to those in the tectoRNA and the intron ribozyme (Supplementary Figure S7). Thus, the GAAC receptor can be regarded as an independent module, although it should be noted that in the absence of Mg2+, the R(GAAC) motif in cisDSL would be less structured or flexible than the motif in tectoRNA because the motif in the former context is more accessible to DMS (Figure S7). The structure–function relationship of the R(GAAC) motif was analysed using less active cisDSL mutants that survived after doped selection. DMS modification was applied to four variants (clone_2, _19, _26 and _59) (Figure 7A). Modification levels in receptor regions differed among the variants. For example, DSL_L(GAAC){6}_R(GAAC) showed higher modification in A6-7 than A1-3 (Figure 7C). In contrast, these five adenines showed similar extents of modification in the less active clone_2, which had a single-nucleotide substitution that can provide one additional base pair. As the original receptor does not have a canonical base pair in this position, the new base pair may disturb or reduce the correct folding of the receptor motif. DMS modification of the four mutants suggested a correlation between the activity of each mutant and its DMS modification profile because the mutant receptors, the modification patterns of which were similar to that of the parent motif sequence, showed similar catalytic activity to the parent ribozyme (Figure 7B and C).
Figure 7.
Mutant R(GAAC) motifs selected from doped selection. (A) Predicted secondary structures of original (left) doped mutant motifs by mFold. The mutated nucleotides are represented in red and by numbering. (B) Comparison of catalytic activities of cisDSL possessing GAAC loop and original or mutated receptors. Product yields after 6 h reaction are shown. (C) DMS modification levels of the mutated receptors embedded in the cisDSL context with (black bar) or without (blue bar) Mg2+.
Structural probing of the GAAC/R(GAAC) interaction in the complex structural context
DMS modification of the GAAC/R(GAAC) interaction in the context of the Tetrahymena ribozyme provided the relationship between the secondary structure and the ribozyme function that relies on the complex and precise formation of its active 3D structure. For this purpose, modification patterns of fully active Tet_L(GAAC)_R(GAAC) and virtually inactive Tet_L(UUCG)_R(GAAC) were compared. The two intron ribozymes showed similar modification patterns in their R(GAAC) and neighbouring catalytic core (Supplementary Figure S8), suggesting that disruption of GAAC/R(GAAC) interaction does not cause large deformation of secondary structure of the catalytic core and confirmed the modular independency of the receptor motif. The large difference in the catalytic activity between the two ribozymes (Figure 5) may originate from small structural differences at the 3D structure level, such as local misfolding seen in the late folding step of this ribozyme (43).
DISCUSSION
The first non-GNRA tetraloop/receptor motif
The R(GAAC) motif selected in this study is the first receptor motif for non-GNRA tetraloops. All tetraloop receptors artificially generated to date have targeted GNRA-type tetraloops (15,16,17). As the GAAC loop/receptor motif reported here showed modularity between three different contexts and orthogonality to GNRA/receptor motifs, this novel RNA–RNA interacting motif would expand the modular tools for constructing a variety of designer RNAs.
Natural evolution or artificial generation of tetraloop receptors may depend on accessibility of base edges of target tetraloops. The structures of GNRA tetraloops elucidated to date by NMR spectroscopy and X-ray crystallography have the canonical form shown in Figure 1A, in which the last three nucleotides expose their WC edges and upper surface of continuous base stacking. These geometric features of the GNRA tetraloops can act as a handle for interacting with receptors. The structure of the GAAC loop in the group IIC intron is similar to that of the GAAA loop, and biochemical data using DMS modification indicated that base edges of the GAAC tetraloop in the isolated state are accessible by solvents. Therefore, other tetraloops with exposed base-edges, such as UNAC (44) and AUCG (45) tetraloops, could have receptors in nature or selected from RNA libraries. On the other hand, considering the limited number of solvent-accessible base-edges of UNCG tetraloops (38), we estimate that receptors for UNCG tetraloops would be difficult to isolate from RNA libraries or to evolve in natural RNA.
Biochemical properties of the GAAC receptor and its interaction with the GAAC loop
Chemical modification experiments in the three different structural contexts commonly suggested no large structural rearrangement in the receptor motif upon complexation with the GAAC loop. On the other hand, folding of the receptor motif is highly dependent on Mg2+ ions, which can be substituted by Ca2+ ions.
The GAAC receptor motif seems to preorganize its structure in the presence of Mg2+ in the isolated state. Docking of the preorganized GAAC receptor with the GAAC loop would rigidify the structure of the receptor, resulting in a similar, but enhanced, chemical modification pattern (Figure 6D docking). Therefore, in contrast to the GAAA/R(11 nt) that interacts in an induced-fit mechanism (14), the GAAC/R(GAAC) motif may interact in a lock-and-key mechanism. These observations suggest that the R(GAAC) motif forms a particular 3D structure in the presence of Mg2+ ions. At present, prediction of the tertiary structure of the R(GAAC) motif is difficult mainly due to the restricted phylogenetic variation. Direct structural determination with X-ray crystal structural analysis and NMR spectroscopy would be best approaches to elucidate its 3D structure because the motif functions in the structural contexts useful for X-ray crystal structural analysis (Tetrahymena intron) and NMR study (tectoRNA).
Biochemical analysis indicated that the GAAC/R(GAAC) interaction generated in the presence of Mg2+ was supported by Ca2+ but not by Mn2+. In the case of the GAAA/R(11 nt) interaction, which can also be supported by Ca2+ and Mn2+, metal ions contribute mainly to organization of the receptor motif because the GAAA loop does not coordinate divalent metal ions. In the case of the GAAC/R(GAAC) interaction, however, the role of metal ions may be more complex because GAAC loop specifically coordinates a Mg2+ ion in the crystal structure (20,21).
The GAAC/R(GAAC) motif in the three different contexts
We installed the GAAC/R(GAAC) motif in three RNA structures with different complexities by substituting their GAAA/R(11 nt) motifs. For comparison between the GAAC/R(GAAC) motif and the GAAA/R(11 nt) motif, we consider three factors affecting the basic properties of RNA–RNA tertiary interaction that are: (i) the binding affinity between the tetraloop and its receptor; (ii) relative orientation between a tetraloop and its receptor; (iii) structural autonomy by which the structural modularity is preserved in the complex structural context. Analysis using tectoRNA showed that the affinity of the GAAC/R(GAAC) pair was comparable with that of the GGAA/R(1) pair (Figure 4) (17). The relative orientation between the tetraloop and its receptor is crucially important in the case that the interaction directly affects chemical transformations. In the family of GNRA loop and its receptor interactions, the relative orientation between the tetraloop and receptors is highly preserved because the A-minor interaction between conserved G–C pair and the last adenine in the loop is used as a common mode of interaction that determines the relative position and orientation (10,17). On the other hand, the activity assay of the cisDSL ribozyme indicated that the ribozyme with the GAAC/R(GAAC) motif is less active than the parent ribozyme bearing GAAA/R(11 nt). In the context of the class DSL ribozyme, it has been shown that GGAA/R(1) can replace GAAA/R(11 nt) without reduction of activity (46). Thus, these observations suggest that relative orientation and position between the GAAC loop and R(GAAC) motif are slightly deviated from those between the GNRA loops and their receptors. This deviation may sensitively affect the catalytic ability of the DSL ribozyme but be compensated by flexibility of RNA duplex in the contexts of tectoRNA and the Tetrahymena intron ribozyme.
The structural autonomy of the receptor motif is important to ensure the formation of a defined structure within the context with long and complex sequence, such as ribozymes and riboswitches within mRNAs. Although this issue is not important in the context of cisDSL and tectoRNA because their folding process must be simple, it becomes crucial in the context of the Tetrahymena intron. In the Tetrahymena intron with mismatched loop/receptor pair, the mutants with R(GAAC) motif was hardly active while the mutants with R(11 nt) motif retained considerable activity (Figure 5). These observations suggest that the R(GAAC) motif is prone to induce misfolding and/or inactive structure presumably because of its structure lacking WC-base pairs. This observation is consistent with the fact that the order of the activity of mutants having L5b UUCG loop lacking the loop/receptor interaction is BP variant > R(11 nt) > R(GAAC) because stable base pairs in BP mutant were most resistant to form alternative structures. In the context of Tetrahymena intron RNA in this study, the relatively weak structural autonomy of R(GAAC) was completely complemented upon the docking with L5b GAAC loop (Figure 5D). This structural fragility of R(GAAC), however, does not complement in the more complex structural and environmental context because in vivo splicing assay in Escherichia coli, in which the production of LacZ reporter gene is governed by the self-splicing of the ribozyme, showed that the intron with GAAC/R(GAAC) pair had no observable splicing, while the UUCG/BP pair and UUCG/R(11 nt) pair were less efficient than the parent construct with GAAA/R(11 nt) but still produced detectable amounts of the reporter protein (Supplementary Figure S9). This result suggested that R(GAAC) has a negative effect on the Tetrahymena ribozyme in the context of LacZ mRNA expressed in E.coli.
Evolutionary aspect of R(GAAC) motif
Modular receptor motifs for GAAC loop have not been found in naturally occurring RNAs. Therefore, the isolation of the R(GAAC) motif in this study not only suggests the possibility of the existence of naturally occurring modular receptors recognizing GANC loop, but provides a possible probe motif for use in searching for as yet unidentified naturally occurring GANC receptor motifs. On the other hand, natural evolution of the RNA motifs involves selection pressures more complex than those in the in vitro selection/evolution technique. Therefore, further evolution of the R(GAAC) to adapt to in vivo conditions, or artificial generation of the motif family to recognize GANC loops other than GAAC are important issues as the next steps prior to searching for unidentified naturally occurring GANC receptor motifs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–6, Supplementary Figures 1–9, Supplementary Methods and Supplementary References [47,48].
FUNDING
Grants-in-Aid for Scientific Research (B) [23310161 to Y.I.]; JSPS Fellows [11J00683 to J.I.]; and on Innovative Areas ‘Emergence in Chemistry’ [23111717 to Y.I.] Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan. Funding for open access charge: Grant-in-Aid for Scientific Research (B) [23310161] from the MEXT, Japan.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors deeply thank Dr. Shoji Ohuchi for his kind advice on in vitro selection experiments.
REFERENCES
- 1.Hermann T, Patel D. Stitching together RNA tertiary architectures. J. Mol. Biol. 1999;294:829–849. doi: 10.1006/jmbi.1999.3312. [DOI] [PubMed] [Google Scholar]
- 2.Moore PB. Structural motifs in RNA. Annu. Rev. Biochem. 1999;68:287–300. doi: 10.1146/annurev.biochem.68.1.287. [DOI] [PubMed] [Google Scholar]
- 3.Leontis N, Westhof E. Analysis of RNA motifs. Curr. Opin. Struct. Biol. 2003;13:300–308. doi: 10.1016/s0959-440x(03)00076-9. [DOI] [PubMed] [Google Scholar]
- 4.Leontis NB, Lescoute A, Westhof E. The building blocks and motifs of RNA architecture. Curr. Opin. Struct. Biol. 2006;16:279–287. doi: 10.1016/j.sbi.2006.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holbrook SR. Structural principles from large RNAs. Ann. Rev. Biophys. 2008;37:445–464. doi: 10.1146/annurev.biophys.36.040306.132755. [DOI] [PubMed] [Google Scholar]
- 6.Pley HW, Flaherty KM, McKay DB. Model for an RNA tertiary interaction from the structure of an intermolecular complex between a GAAA tetraloop and an RNA helix. Nature. 1994;372:111–113. doi: 10.1038/372111a0. [DOI] [PubMed] [Google Scholar]
- 7.Murphy FL, Cech TR. GAAA tetraloop and conserved bulge stabilize tertiary structure of a group I intron domain. J. Mol. Biol. 1994;236:49–63. doi: 10.1006/jmbi.1994.1117. [DOI] [PubMed] [Google Scholar]
- 8.Jaeger L, Michel F, Westhof E. Involvement of a GNRA tetraloop in long-range RNA tertiary interactions. J. Mol. Biol. 1994;236:1271–1276. doi: 10.1016/0022-2836(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 9.Costa M, Michel F. Frequent use of the same tertiary motif by self-folding RNAs. EMBO J. 1995;14:1276–1285. doi: 10.1002/j.1460-2075.1995.tb07111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Correll CC, Swinger K. Common and distinctive features of GNRA tetraloops based on a GUAA tetraloop structure at 1.4 A resolution. RNA. 2003;9:355–363. doi: 10.1261/rna.2147803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA. Crystal structure of a group I ribozyme domain: Principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 12.Krasilnikov AS, Yang X, Pan T, Mondragon A. Crystal structure of the specificity domain of ribonuclease P. Nature. 2003;421:760–764. doi: 10.1038/nature01386. [DOI] [PubMed] [Google Scholar]
- 13.Davis JH, Tonelli M, Scott LG, Jaeger L, Williamson JR, Butcher SE. RNA helical packing in solution: NMR structure of a 30 kDa GAAA tetraloop-receptor complex. J. Mol. Biol. 2005;351:371–382. doi: 10.1016/j.jmb.2005.05.069. [DOI] [PubMed] [Google Scholar]
- 14.Butcher SE, Dieckmann T, Feigon J. Solution structure of a GAAA tetraloop receptor RNA. EMBO J. 1997;16:7490–7499. doi: 10.1093/emboj/16.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costa M, Michel F. Rules for RNA recognition of GNRA tetraloops deduced by vitro selection: Comparison with in vivo evolution. EMBO J. 1997;16:3289–3302. doi: 10.1093/emboj/16.11.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Juneau K, Cech TR. In vitro selection of RNAs with increased tertiary structure stability. RNA. 1999;5:1119–1129. doi: 10.1017/s135583829999074x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geary C, Baudrey S, Jaeger L. Comprehensive features of natural and in vitro selected GNRA tetraloop-binding receptors. Nucleic Acids Res. 2008;36:1138–1152. doi: 10.1093/nar/gkm1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohuchi SP, Ikawa Y, Nakamura Y. Selection of a novel class of RNA–RNA interaction motifs based on the ligase ribozyme with defined modular architecture. Nucleic Acids Res. 2008;36:3600–3607. doi: 10.1093/nar/gkn206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woese CR, Winker S, Gutell RR. Architecture of ribosomal RNA: Constraints on the sequence of “tetra-loops”. Proc. Natl Acad. Sci. USA. 1990;87:8467–8471. doi: 10.1073/pnas.87.21.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keating KS, Toor N, Pyle AM. The GANC tetraloop: A novel motif in the group IIC intron structure. J. Mol. Biol. 2008;383:475–481. doi: 10.1016/j.jmb.2008.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toor N, Keating KS, Taylor SD, Pyle AM. Crystal structure of a self-spliced group II intron. Science. 2008;320:77–82. doi: 10.1126/science.1153803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Proudnikov D, Mirzabekov A. Chemical methods of DNA and RNA fluorescent labeling. Nucleic Acids Res. 1996;24:4535–4542. doi: 10.1093/nar/24.22.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kladwang W, Cordero P, Das R. A mutate-and-map strategy accurately infers the base pairs of a 35-nucleotide model RNA. RNA. 2011;17:522–534. doi: 10.1261/rna.2516311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tijerina P, Mohr S, Russell R. DMS footprinting of structured RNAs and RNA-protein complexes. Nat. Protoc. 2007;2:2608–2623. doi: 10.1038/nprot.2007.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inoue T, Cech TR. Secondary structure of the circular form of the tetrahymena rRNA intervening sequence: a technique for RNA structure analysis using chemical probes and reverse transcriptase. Proc. Natl Acad. Sci. USA. 1985;82:648–652. doi: 10.1073/pnas.82.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. SAFA: Semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA. 2005;11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ikawa Y, Tsuda K, Matsumura S, Inoue T. De novo synthesis and development of an RNA enzyme. Proc. Natl Acad. Sci. USA. 2004;101:13750–13755. doi: 10.1073/pnas.0405886101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horie S, Ikawa Y, Inoue T. Structural and biochemical characterization of DSL ribozyme. Biochem. Biophys. Res. Commun. 2006;339:115–121. doi: 10.1016/j.bbrc.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 30.Molinaro M, Tinoco I., Jr Use of ultra stable UNCG tetraloop hairpins to fold RNA structures: Thermodynamic and spectroscopic applications. Nucleic Acids Res. 1995;23:3056–3063. doi: 10.1093/nar/23.15.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ikawa Y, Matsumoto J, Horie S, Inoue T. Redesign of an artificial ligase ribozyme based on the analysis of its structural elements. RNA Biol. 2005;2:137–142. doi: 10.4161/rna.2.4.2302. [DOI] [PubMed] [Google Scholar]
- 32.Jaeger L, Westhof E, Leontis NB. TectoRNA: Modular assembly units for the construction of RNA nano-objects. Nucleic Acids Res. 2001;29:455–463. doi: 10.1093/nar/29.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van der Horst G, Christian A, Inoue T. Reconstitution of a group I intron self-splicing reaction with an activator RNA. Proc. Natl Acad. Sci. 1991;88:184–188. doi: 10.1073/pnas.88.1.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laggerbauer B, Murphy FL, Cech TR. Two major tertiary folding transitions of the tetrahymena catalytic RNA. EMBO J. 1994;13:2669. doi: 10.1002/j.1460-2075.1994.tb06557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Treiber DK, Rook MS, Zarrinkar PP, Williamson JR. Kinetic intermediates trapped by native interactions in RNA folding. Science. 1998;279:1943–1946. doi: 10.1126/science.279.5358.1943. [DOI] [PubMed] [Google Scholar]
- 36.Pan J, Woodson SA. The effect of long-range loop-loop interactions on folding of the tetrahymena self-splicing RNA. J. Mol. Biol. 1999;294:955–965. doi: 10.1006/jmbi.1999.3298. [DOI] [PubMed] [Google Scholar]
- 37.Deras ML, Brenowitz M, Ralston CY, Chance MR, Woodson SA. Folding mechanism of the tetrahymena ribozyme P4-P6 domain. Biochemistry. 2000;39:10975–10985. doi: 10.1021/bi0010118. [DOI] [PubMed] [Google Scholar]
- 38.Ennifar E, Nikulin A, Tishchenko S, Serganov A, Nevskaya N, Garber M, Ehresmann B, Ehresmann C, Nikonov S, Dumas P. The crystal structure of UUCG tetraloop. J. Mol. Biol. 2000;304:35–42. doi: 10.1006/jmbi.2000.4204. [DOI] [PubMed] [Google Scholar]
- 39.Doherty EA, Batey RT, Masquida B, Doudna JA. A universal mode of helix packing in RNA. Nat. Struct. Biol. 2001;8:339–343. doi: 10.1038/86221. [DOI] [PubMed] [Google Scholar]
- 40.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc. Natl Acad. Sci. USA. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feig AL, Uhlenbeck OC. In: the RNA World. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY. 1999. The role of metal ions in RNA biochemistry; pp. 287–319. [Google Scholar]
- 42.Pyle AM. Metal ions in the structure and function of RNA. J. Biol. Inorg. Chem. 2002;7:679–690. doi: 10.1007/s00775-002-0387-6. [DOI] [PubMed] [Google Scholar]
- 43.Russell R, Das R, Suh H, Travers KJ, Laederach A, Engelhardt MA, Herschlag D. The paradoxical behavior of a highly structured misfolded intermediate in RNA folding. J. Mol. Biol. 2006;363:531–544. doi: 10.1016/j.jmb.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 44.Zhao Q, Huang HC, Nagaswamy U, Xia Y, Gao X, Fox GE. UNAC tetraloops: to what extent do they mimic GNRA tetraloops? Biopolymers. 2012;97:617–628. doi: 10.1002/bip.22049. [DOI] [PubMed] [Google Scholar]
- 45.Duszczyk MM, Wutz A, Rybin V, Sattler M. The xist RNA a-repeat comprises a novel AUCG tetraloop fold and a platform for multimerization. RNA. 2011;17:1973–1982. doi: 10.1261/rna.2747411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ishikawa J, Matsumura S, Jaeger L, Inoue T, Furuta H, Ikawa Y. Rational optimization of the DSL ligase ribozyme with GNRA/receptor interacting modules. Arch. Biochem. Biophys. 2009;490:163–170. doi: 10.1016/j.abb.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Novikova IV, Hassan BH, Mirzoyan MG, Leontis NB. Engineering cooperative tecto-RNA complexes having programmable stoichiometries. Nucleic Acids Res. 2011;39:2903–2917. doi: 10.1093/nar/gkq1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Williamson CL, Desai NM, Burke JM. Compensatory mutations demonstrate that P8 and P6 are RNA secondary structure elements important for processing of a group I intron. Nucleic Acids Res. 1989;17:675–689. doi: 10.1093/nar/17.2.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.