Abstract
An artificial small RNA (afsRNA) scaffold was designed from an Escherichia coli sRNA, SibC. Using the lacZ reporter system, the gene silencing effects of afsRNAs were examined to explore the sRNA-mediated gene-silencing mechanisms in E. coli. Substitution of the original target recognition sequence with a new sequence recognizing lacZ mRNA led to effective reduction of lacZ gene expression. Single-strandedness of the target recognition sequences in the scaffold was essential for effective gene silencing. The target recognition sequence was shortened to 10 nt without significant loss of gene silencing, although this minimal length was limited to a specific target mRNA sequence. In cases where afsRNAs had mismatched (forming internal loops) or unmatched (forming bulges) regions in the middle of the target recognition sequence, internal loop-forming afsRNAs were more effective in gene silencing than those that formed bulges. Unexpectedly, gene silencing by afsRNA was not decreased but increased on hfq disruption in E. coli, particularly when interactions between afsRNA and mRNA were weak, suggesting that Hfq is possibly involved in destabilization of the RNA–RNA duplex, rather than enhancement of base pairing.
INTRODUCTION
Small non-coding RNAs (sRNAs) have a variety of regulatory functions in diverse species, ranging from bacteria to mammals (1–4). To date, ∼100 sRNA species in Escherichia coli and some of their functions have been identified (5,6). Among these, regulation of translation and/or stability of target mRNAs are the most common function (7–10). This regulation proceeds by RNA–RNA interaction through base pairing of sRNA, which could make it possible to discriminate its cognate target from non-cognate targets, even though it is usually difficult to identify actual RNA–RNA interaction because base pairing in the cell is a very sophisticated and, sometimes, unpredictable process (11–14). Interactions between sRNA and its target mRNA usually suppress translation of target mRNA (13,15). The majority of gene silencing by bacterial sRNAs is proposed to be mediated by an Hfq-containing complex (16–19).
Discrimination of cognate targets from non-cognates by sRNA depends on a number of factors, including base pairing, accessibility and involvement of cellular factors such as RNA chaperones (20–22). Although base pairing is believed to be the most effective determinant of target discrimination, accessibility and chaperone proteins may play a role in determining specificity (23). The single strandedness of interacting sequences in both sRNAs and target mRNAs may be the key factor in determining accessibility. Hfq, a highly abundant RNA chaperone protein, is proposed to enhance base pairing (23,24). Owing to this complexity, it is difficult to draw conclusions about the number of base pairings required for target recognition by sRNAs in the cell, although a previous in vitro study with synthetic oligonucleotides showed that a 14 nt antisense RNA was sufficient to suppress translation (25). Furthermore, base-paired regions between natural sRNAs and their target mRNAs are usually not contiguous, and prediction of precise RNA–RNA interactions is difficult (20,26–28). Nevertheless, it is a generally accepted view that the number of base pairings determines sRNA specificity in target discrimination.
The target discrimination mechanism of sRNA through base pairing with target mRNA could be explored using artificial sRNA (afsRNA) loaded with various target-recognition sequences, as afsRNAs can be designed to have defined target-recognition sequences for overcoming the complexity faced by natural sRNA (29). Recently, an afsRNA prototype in E. coli was developed by Man et al. (30). This afsRNA containing the target-recognition sequence at the 5′ region, an Hfq-binding site in the middle and a terminator hairpin at the 3′ region specifically suppressed target gene expression in bacteria, confirming the efficiency of this approach for specific gene silencing in E. coli. However, the afsRNAs differ in gene silencing activity from one another depending on which target recognition sequences are loaded, possibly owing to structural variations as a result of sequence changes. Thus, their afsRNA may not be suitable for examining the target discrimination mechanism of sRNA through base paring with target mRNA, as it would be difficult to distinguish between the sequence and structure effects.
Recently, we identified two target-recognition sequences, TRD1 and TRD2, of cis-acting sRNA, SibC RNA (31,32). Each TRD is ∼20 nt and sufficient to suppress target ibsC translasion, although sRNA and target mRNA contain a 140 nt stretch of complementary sequences. In particular, TRD2 is functional only when embedded in a specialized structure. This scaffold includes the P1 stem at the 5′-end and terminator hairpin of SibC at the 3′-end. RNA structural probing studies suggest that the majority of TRD2 sequences should reside in a single-stranded region within the secondary structure model (32). In this study, we took advantage of the single strandedness of TRD2 in the specialized structure to design afsRNA including target-recognition sequences in a single-stranded accessible region, with little structural variation by the altered sequences. Using this afsRNA scaffold, we examined the base-pairing effects on gene silencing with regard to the position of the target-recognition sequence, minimal base pairing, mismatch of base pairing and Hfq dependency. Our results showed that the single strandedness of the target recognition sequences is crucial for effective gene silencing. The 10 nt target-recognition sequence was sufficient for gene silencing by afsRNA. However, this minimal number of base pairings depends on the location of the target sequence. The presence of mismatched or unmatched regions in the middle of the target-recognition sequences hindered gene silencing by afsRNA. Interestingly, internal loop-forming afsRNA was more effective in gene silencing than bulge-forming afsRNA. Unexpectedly, gene silencing by afsRNA was not reduced by disruption of the hfq gene but in fact increased, particularly when the base pairing between afsRNA and mRNA was weak. Based on these findings, we propose a novel function of Hfq in increasing specificity via removal of sRNAs from their interacting target mRNAs through short base pairing.
MATERIALS AND METHODS
Bacterial strains, plasmids and oligonucleotides
The E. coli K-12 strain, DH5α, was used for plasmid construction. A lysogen containing an ssrS::lacZ transcriptional fusion was constructed using strain DJ480, as described previously (33). Briefly, the promoter region containing the sequence between positions −180 and +10 of ssrS relative to the +1 transcription start site was amplified, and the fragment was cloned between the EcoRI and BamHI sites of pRS1553 vector to generate an ssrS::lacZ transcriptional fusion plasmid. DJ480 was transformed with the fusion plasmid and transfected with λRS468 to construct the ssrS::lacZ lysogen. Single-copy integration was confirmed using polymerase chain reaction (PCR) (34). P1-mediated transduction was used (35) to generate the hfq knockout (hfq−) in a DJ480 ssrS::lacZ lysogen from a Keio strain (hfq::kan) (36). The strain was confirmed by sequence analysis of the amplified knockout region. Plasmid pHM-tac, an isopropyl β-D-1-thiogalactopyranoside (IPTG)-inducible RNA expression vector, was constructed by replacing the AatII site of plasmid pHM1 (32) with EcoRI and XbaI sites. RNA-coding sequences were amplified by PCR and cloned into the EcoRI/XbaI sites of the pHM-tac vector for generating artificial sRNAs. Each artificial sRNA itself carried the sibC terminator of the P7 stem and loop as a termination signal for IPTG-induced RNA transcription. When the target-recognition sequence was grafted onto the sibC terminator, its termination efficiency was drastically reduced, and then the run-through transcripts were terminated at the further downstream rrnB terminator, generating extended transcripts of ∼250 nt. The oligonucleotides used are listed in Table 1.
Table 1.
Oligonucleotide sequences
| Name | Sequences | Use |
|---|---|---|
| BamHIss6F | CGGGATCCATAAATGTGAGCGGATAACATTGACATTGTGAGCGG | pHM-tac |
| HindIIIss6R | CCCAAGCTTGTCGACTCTAGACGCGCGGAATTCATTATATTGTTATCCGCTCACAATGTC | pHM-tac |
| ERIARlacZ1F | CGGAATTCAACAGCAAAGTATGATGACAAGTCGCATCATAGCTGTTTCCTGTGTGAGC | ARlacZ1 |
| ARlacZ1XbaIR | GCTCTAGAGGTAAAGCCCTCACCGAAGCGAGGGCTCACACAGGAAACAGCTATG | ARlacZ1 |
| ARlacZ2F | CGGAATTCAACAGCAAAGTAACTTGACAAGTCGCAAGTTAGCTGTTTCCTGTGTG | ARlacZ2 |
| ERIARlacZ3F | CGGAATTCAAGGGTAAAGTATGATGACAAGTCGCATCATAACCCTTTAGCTGTTTCC | ARlacZ3 |
| ARlacZ3XbaIR | GCTCTAGAGGAAAACCTGTGTGCGATCACACAGGAAACAGCTAAAGGGTTATGATG | ARlacZ3 |
| ERIARlacZ4F | CGGAATTCAAGGGTAAAGTATGATGACAAGTCGCATCATAACCCTTCTCCTTTAGC | ARlacZ4 |
| ARlacZ4XbaIR | GCTCTAGAGGTAAACTGTTTCCTCACACAGGAAACAGCTAAAGGAGAAGGGTTATGATG | ARlacZ4 |
| ARlacZ5F | CATAGCTGTTTCCTGTGTGATTCAAGCCCTCGCTTCGGTGAGGGC | ARlacZ5 |
| ERIARlacZ6F | CGGAATTCTCACACTAAGGGGAAACTCATAGCTGTTTCCTGTGTGACTCCTTCAAGC | ARlacZ6 |
| ARlacZ6XbaIR | GCTCTAGAGGTAAAGCCCTCACCGAAGCGAGGGCTTGAAGGAGTCACACAGG | ARlacZ6 |
| ERIARlacZ7F | CGGAATTCACAGGACAAGAACAGTTCAAGTCATAGCTGTTTCCTGTGTGATAAGAGC | ARlacZ7 |
| ARlacZ7XbaIR | GCTCTAGAGGTAAAGCCCTCACCGAAGCGAGGGCTCTTATCACACAGGAAACAGC | ARlacZ7 |
| ARlacZ8F | GATAAGATAGGAATACCCTCTCCTTCAAGCCCTCGCTTCG | ARlacZ8 |
| ARlacZ9F | CCTGTGTGATAAGATCTCCTTCAAGCCCTCGCTTCG | ARlacZ9 |
| ARlacZ19NF | CGGAATTCAACAGCAAAGTATCTTGACAAGTCGCAAGATAGCTGTTTCCTGTGTGAG | ARlacZ19N |
| ARlacZ17NF | ATCATAGCTGTTTCCTGTGTAGGCCCTCGCTTCGGTGAGGGC | ARlacZ17N |
| ARlacZ15NF | GCATCATAGCTGTTTCCTGTTCAGGCCCTCGCTTCGGTGAGGGC | ARlacZ15N |
| ARlacZ12NF | CGGAATTCAGCTAAAAACCCTTATGTCAAGCATCATAAGGTTAGCTGTTTCCTGCCCTC | ARlacZ12N |
| ARlacZ11NF | CGGAATTCAGCTTAAAACCCTTATGTCAAGCATCATAAGGTAAGCTGTTTCCTGCCCTC | ARlacZ11N |
| ARlacZ10NF | CGGAATTCAGCATAAAACCCTTATGTCAAGCATCTAAGGTATGCTGTTTCCTGCCCTC | ARlacZ10N |
| ARlacZ9NF | CGGAATTCAGGATAAAACCCTTATGTCAAGCATCATAAGGTATCCTGTTTCCTGCCCTC | ARlacZ9N |
| ARlacZ10N − 6F | CGGAATTCAGGGGTAAACCCTTATGTCAAGCATCATAAGGATTCCTGTGTGAGCCCTCGCTTCGGTGAGGGC | ARlacZ10N − 6 |
| ARlacZ10N − 5F | CGGAATTCAAGGTTAAACGCTTATGTCAAGCATCATAAGCAGCCTTCCTGTGTGGCCCTCGCTTCGGTGAGGGC | ARlacZ10N − 5 |
| ARlacZ10N − 4F | CGGAATTCAAAGGCAAACGCTTATGTCAAGCATCATAAGCGCCTTTCCTGTGTGCCCTCGCTTCGGTG AGGGC | ARlacZ10N − 4 |
| ARlacZ10N − 3F | CGGAATTCAACGCAAAACGCTTATGTCAAGCATCATAAGCTGCGTTTCCTGTGGCCCTCGCTTCGGTGAGGGC | ARlacZ10N − 3 |
| ARlacZ10N − 2F | CGGAATTCACACGATTACGGTTATGTCAAGCATCATAACCTCGTGTTTCCTGTGCCCTC | ARlacZ10N − 2 |
| ARlacZ11N − 2F | CGGAATTCACAGGATTACGGTTATGTCAAGCATCATAACCTCCTGTTTCCTGTGCCCTC | ARlacZ11N − 2 |
| ARlacZ12N − 2F | CGGAATTCACAGCAAAAGGGTTATGTCAAGCATCATAACCTGCTGTTTCCTGTGCCCTC | ARlacZ12N − 2 |
| ARlacZ10N − 1F | CGGAATTCATAGAAGAAACCCTTATGTCAAGCATCATAAGGCTTCTGTTTCCTGGCCCTCGCTTCGGTGAGGGC | ARlacZ10N − 1 |
| ARlacZ10N + 1F | CGGAATTCAGTTGACAAACTCTTATGTCAAGCATCATA AGGGTCAGCTGTTTCCGCCCTCGCTTCGGTGAGGGC | ARlacZ10N + 1 |
| ARlacZ10N + 2F | CGGAATTCAGTTAGAAACCCTTATGTCAAGCATCATAAGGCTAGCTGTTTCGCCCTCGCTTCGGTGAGGGC | ARlacZ10N + 2 |
| ARlacZ10N + 3F | CGGAATTCATCTGGAAACCCTTATGTCAAGCATCATAA GGCCAGATAGCTGTTTGCCCTCGCTTCGGTGAGGGC | ARlacZ10N + 3 |
| ARlacZ10N + 4F | CGGAATTCATGCTGAAACCCTTATGTCAAGCATCATAA GGCAGCATAGCTGTTGCCCTCGCTTCGGTGAGGGC | ARlacZ10N + 4 |
| ARlacZ10N + 5F | CGGAATTCATGGGTTAAACCCTTATGTCAAGCATCATA AGGAACTCATAGCTGTGCCCTCGCTTCGGTGAGGGC | ARlacZ10N + 5 |
| ARlacZ14NF | CGGAATTCATAGAGGTTTTACTGGTACAAGTCGACCAGTACTCTGTTTCCTGTGTGGCCCTCGCTTCGGTG | ARlacZ14N |
| ARlacZ12L2F | CGACCAGTACTCTGTCACCTGTGTGGCCCTCG | ARlacZ12L2 |
| ARlacZ14L2F | CGGAATTCATAGCTGTTTTACTGGTACAAGTCGACCAGTAAGCTGTCACCTGTGTGGCCCTCGCTTCGGTG | ARlacZ14L2 |
| ARlacZ14L4F | CGGAATTCAGAGCTCTTTATTTGGTACAAGTCGACCAGATAGCTCTCACCTGTGTGGCCC | ARlacZ14L4 |
| ARlacZ14L8F | CGGAATTCAGCTTACTTTATGACCTACAAGTCGAGGTCATTGAGCTCACCTGTGTGGCCC | ARlacZ14L8 |
| ARlacZ14B2F | CGGAATTCAAGCAGTTTTAGTACTGACAAGTCGCAGTACTCTGTTTCACCTGTGTGGCCCTCGCTTCGGTG | ARlacZ14B2 |
| ARlacZ14B4F | CGGAATTCAGAAACTTTTAGTACTGACAAGTCGCAGTACTGTTTCTCACCTGTGTGGCCC | ARlacZ14B4 |
| ARlacZ14B8F | CGGAATTCAGCTTATTTTAAACAGGACAAGTCGCCTGTTTTGAGCTCACCTGTGTGGCCC | ARlacZ14B8 |
| ARlacZ14L2 − 5F | CGGAATTCATGGAGAAAAGAAAGTCACAAGTCGGACTTTCCTCCGTGATAAGTGCC | ARlacZ14L2 − 5 |
| ARlacZ14B2 − 5F | CGGAATTCATGGACTAAAAGGAATTACAAGTCGGATTCCTGTCCGTGATAAGTGCCCTCGC | ARlacZ14B2 − 5 |
| ARlacZ14L2 − 4F | CGGAATTCAGTGGATAAGAACTTTGACAAGTCGCAAAGTTTCCACTGTGATAAAAGCCCTCGCTTCGGTG | ARlacZ14L2 − 4 |
| ARlacZ14B2 − 4F | CGGAATTCAGTCAGAATCGAATATGACAAGTCGCATATTCCTGACTGTGATAAAAGCCCTCGCTTCGGTG | ARlacZ14B2 − 4 |
| ARlacZ14L2 + 1F | CGGAATTCAGTCAGAAACCTAATTGACAAGTCGCAATTAGCTGACTCCTGTGTAAGCCCTCGCTTCGGTG | ARlacZ14L2 + 1 |
| ARlacZ14B2 + 1F | CGGAATTCAGTAACTATCAGCATATACAAGTCGATATGCTGTTACTCCTGTGTAAGCCCTCGCTTCGGTG | ARlacZ14B2 + 1 |
| ARlacZ14L2 + 3F | CGGAATTCAGTGCTTAAGATGTATGACAAGTCGCATACATAGCACTTTCCTGTAAGCCCTCGCTTCGGTG | ARlacZ14L2 + 3 |
| RlacZ14B2 + 3F | CGGAATTCAGTCAGAAAGCTATATGACAAGTCGCATATAGCTGACTTTCCTGTAAGCCCTCGCTTCGGTG | ARlacZ14B2 + 3 |
| ARlacZ14L2 + 5F | CGGAATTCACGTATAAAAGACGTACACAAGTCGGTACGTCATACGTGTTTCCTGCCC | ARlacZ14L2 + 5 |
| ARlacZ14B2 + 5F | CGGAATTCATGGCTAAAAATGATACACAAGTCGGTATCATAGCCATGTTTCCTGCCCTCGC | ARlacZ14LB + 5 |
| ARlacZ14N + 1F | CGGAATTCAAACAGATTCCATCTTGACAAGTCGCAAGATGCTGTTTCCTGTGTAAGCCCTC | ARlacZ14N + 1 |
| ARlacZ12L2 + 1F | CGGAATTCAGTCAGAAACCATATTGACAAGTCGCAATATGCTGACTCCTGTGTAAGCCCTC | ARlacZ12L2 + 1 |
| ARlacZ14L4 + 1F | CGGAATTCAGTACGAAACCTATGTTACAAGTCGAACATAGCGTACTCCTGTGTAAGCCCTC | ARlacZ14L4 + 1 |
| ARlacZ14L8 + 1F | CGGAATTCAGTGTCAAACGATTGATACAAGTTGGTCAATCGACACTCCTGTGTAAGCCCTC | ARlacZ14L8 + 1 |
| ARlacZ14B4 + 1F | CGGAATTCAGTACATATCACAGTATTGAAGAGTATGCTGTTGTACTCCTGTGTAAGCC CTC | ARlacZ14B4 + 1 |
| ARlacZ14B8 + 1F | CGGAATTCAGTGTCTATCGATAACAACAAGTGCTGTTATCGACACTCCTGTGTAAGCCCTC | ARlacZ14B8 + 1 |
| Rzym_R | CCGATCCGTTTCGTCCTCACGGACTCATCAG | SibC ribozyme |
| T7_Rz_SCsh + 1 | ATTAATACGACTCACTATAGGGATCCGTCAAGGGTAAAGTATGATG | SibC ribozyme |
| T7_Rzym_ARL1_F | ATTAATACGACTCACTATAGGGAGAGGCTGTTCTGATGAGTCCGTGAG | ARlacZ1 ribozyme |
| T7_Rz_ARL1 + 1 | ATTAATACGACTCACTATAGGGATCCGTCAACAGCAAAGTATGATG | ARlacZ1 ribozyme |
| T7_Rz_ARL2 + 1 | ATTAATACGACTCACTATAGGGATCCGTCAACAGCAAAGTAACTTG | ARlacZ2 ribozyme |
| T7_Rzym_ARL3_F | ATTAATACGACTCACTATAGGGAGAGACCCTTCTGATGAGTCCGTGAG | ARlacZ3,4 ribozyme |
| T7_Rz_ARL3 + 1 | ATTAATACGACTCACTATAGGGATCCGTCAAGGGTAAAGTATGATG | ARlacZ3,4 ribozyme |
| AR3R | GGAAAACCTGTGTGCGATCAC | ARlacZ3 ribozyme |
| AR4R | GGTAAATCTGTTTCCTCACAC | ARlacZ4 ribozyme |
| T7_Rzym_ARL5_F | ATTAATACGACTCACTATAGGGAGAGTCCTGTCTGATGAGTCCGTGAG | ARlacZ5,7 ribozyme |
| T7_Rz_ARL5 + 1 | ATTAATACGACTCACTATAGGGATCCGTCACAGGACAAGAACAGTTC | ARlacZ5,7 ribozyme |
| T7_Rzym_ARL6_F | ATTAATACGACTCACTATAGGGAGAGGTGTGACTGATGAGTCCGTGAG | ARlacZ6 ribozyme |
| T7_Rz_ARL6 + 1 | ATTAATACGACTCACTATAGGGATCCGTCTCACACTAAGGGGAAAC | ARlacZ6 ribozyme |
| T7_Rzym_ARL8_F | ATTAATACGACTCACTATAGGGAGAGTAAGATCTGATGAGTCCGTGAG | ARlacZ8 ribozyme |
| T7_Rz_ARL8 + 1 | ATTAATACGACTCACTATAGGGATCCGTCATCTTACGCATTCACATAG | ARlacZ8 ribozyme |
| T7_Rzym_ARL9_F | ATTAATACGACTCACTATAGGGAGAGTACCCTCTGATGAGTCCGTGAG | ARlacZ9 ribozyme |
| T7_Rz_ARL9 + 1 | ATTAATACGACTCACTATAGGGATCCGTCAGGGTAGCTGTTTCCTGAGGGC | ARlacZ9 ribozyme |
| Dhfq 5′ | TCAGAATCGAAAGGTTCAAAGTACAAATAAGCATATAAGGAAAAGAGAGATCAGAAGAACTCGTCAAGAAG | Hfq deletion |
| Dhfq 3′ | CGGGGAACGCAGGATCGCTGGCTCCCCGTGTAAAAAAACAGCCCGAAACCTATGGACAGCAAGCGAACCG | Hfq deletion |
| Hfq aug − 80F | TACAATTGAGACGTATCGTGCGC | Deletion confirm |
| Hfq uaa + 80R | AACAAGCGTATAACCCTCTAAATAG | Deletion confirm |
| 5S + 90R | GAGACCCCACACTACCATCGG | 5S Northern probe/ qRT-PCR |
| ARlacZnp | GGTCACACAGGAAACAGCTA | ARlacZ Northern probe |
| ARlacZR74 | GGTAAAGCCCTCACCGAAGCGAG | ARlacZ Northern probe |
| lacZsdR253 | TCAGGAAGATCGCACTCCAG | qRT-PCR |
| T7MicA + 1F | ATTAATACGACTCACTATAGAAAGACGCGCATTTG | MicA |
| MicA + 78R | AAAAGAAAAAGGCCACTCGTGAGTG | MicA |
| T7OxyS + 1F | ATTAATACGACTCACTATAGAAACGGAGCGGCACC | OxyS |
| OxyS + 110R | AGCGGATCCTGGAGATCCGCAAAAG | OxyS |
| T7ssrSlacZ + 1F | ATTAATACGACTCACTATAGAAGACAAAATTTCTCTG | LacZ370 |
| lacZsdR199 | CAGGCAAAGCGCCATTCGCC | LacZ370/qRT-PCR |
β-galactosidase assay
Three colonies for each strain were pooled and grown overnight in Luria-Bertani media (LB) containing ampicillin (50 μg/ml). After 1:100 dilution of the overnight culture in fresh LB, cells were grown at 37°C for 2 h in the presence of IPTG. In case of hfq−, cells were grown for >2 h because hfq− cells grew with a slower rate than the wild-type (the wild-type cells reached an optical density 600 (OD600) of 0.5 after ∼2 h growth, whereas hfq− cells reached the same OD after ∼3 h). Relative β-galactosidase activities were determined, as described previously (37). At least three independent measurements were performed for each strain.
Northern blot analysis
Total cellular RNA was prepared from the same cells used for β-galactosidase assays using hot phenol extraction, as described previously (38). RNA samples were separated on a 5% polyacrylamide gel containing 7 M urea and electrotransferred to a Hybond N+ membrane (Amersham Biosciences). Oligonucleotides were labelled with [γ−32P]ATP and T4 polynucleotide kinase (Takara). The labelled oligonucleotides, ARlacZnp and ARlacZR74, were used for artificial sRNAs and 5S + 90R for 5S RNA (Table 1). Hybridization was performed according to the manufacturer’s instructions. Membranes were visualized and quantified using Image Analyzer FLA 7000 (Fuji).
In vitro transcription
To prepare SibC(1–8::77–141) and ARlacZ1 RNA (Figure 1), direct in vitro transcription using the T7 promoter or SP6 promoter was inefficient, as the first nucleotide of both RNAs is A, which is unfavourable for transcription initiation by the phage RNA polymerases (39). Instead, precursor RNAs carrying the extra sequence at the 5′-end were transcribed in vitro with T7 RNA polymerase and treated with ribozymes to obtain the correct 5′-containing RNA. The corresponding hammerhead ribozymes were designed and prepared with in vitro transcription using T7 RNA polymerase, as described previously (40). DNA templates for precursor RNAs and ribozyme RNAs were obtained using PCR with the corresponding primer pairs (Table 1). The in vitro transcripts were purified via gel elution (41) and used for the ribozyme reactions. The ratio of the ribozyme RNA to precursor RNA was 1:10. Ribozyme reactions were performed in 50 mM Tris–HCl (pH 7.9), 28 mM MgCl2 at 55°C for 1 h and cleaved RNA was purified via gel elution. To prepare MicA, OxyS and LacZ370 (a transcript consisting of 370 nt from the 5′-end of ssrS-lacZ fusion mRNA), DNA templates were obtained via PCR using primer pairs of T7MicA + 1F/MicA + 78R, T7OxyS + 1F/OxyS + 110R and T7ssrSlacZ + 1F/lacZsdR199, respectively (Table 1). In vitro transcription was carried out using T7 RNA polymerase (Promega). Purified RNAs were 32P-labelled at the 5′-end using [γ−32P] ATP and T4 polynucleotide kinase (Takara).
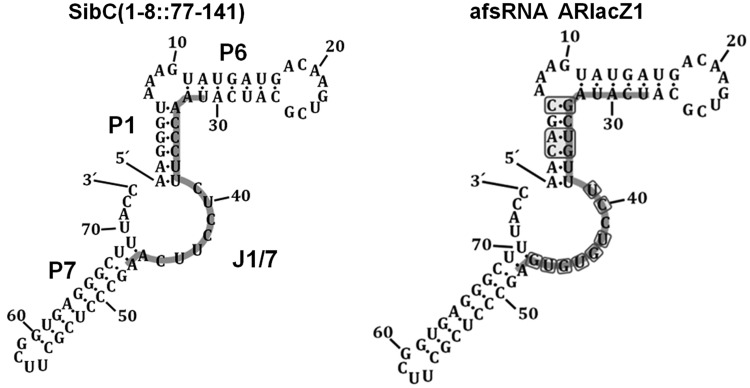
Figure 1.
Secondary structure models of SibC(1–8::77–141) and afsRNA ARlacZ1. The sequences indicated with the light grey line in SibC(1–8::77–141) and ARlacZ1 are TRD2 of SibC and an antisense sequence to lacZ mRNA, respectively. The replaced bases in ARlacZ1 are boxed.
Chemical and enzymatic structure probing
32P-labelled RNAs were renatured by heating for 1 min at 95°C and slowly cooling to 25°C in a 10 μl of RNA structure buffer (Ambion). After pre-incubation of RNAs with 1 μg yeast tRNA for 15 min at 37°C, 1 μl of S1 nuclease (0.4, 2 or 10 U, Promega), RNase V1 (0.0008, 0.004 or 0.02 U, Ambion) and Pb(II) (1 mM, 5 mM or 25 mM Lead(II)-acetate, Sigma Aldrich) were added, followed by incubation at 25°C for 5, 15 and 25 min, respectively. Reactions were stopped by adding 1 μl of 0.5 M ethylenediaminetetraacetic acid and 5 μl of gel loading buffer II (Ambion) and heating at 95°C for 3 min. Cleavage products were analysed with a 8% (v/v) polyacrylamide-9 M urea sequencing gel. RNase T1 and OH ladders were obtained according to the manufacturer’s specifications (Ambion).
Quantitative real-time reverse transcriptase PCR
Total cellular RNAs were treated with Turbo DNase (Ambion) to remove any residual DNA. DNase was heat inactivated, and RNA samples were subjected to reverse transcription with Moloney murine leukemia virus reverse transcriptase (MMLV RT) (Enzynomics) using primers lacZsdR253 for lacZ mRNA and 5S + 90R for 5S rRNA. For quantification standards, total cellular RNAs isolated from DJ480 cells lacking lacZ mRNA were mixed with known amounts of in vitro transcribed LacZ370 consisting of 370 nucleotides from the 5′-end of ssrS-lacZ fusion mRNA. Quantitative real-time reverse transcriptase RT-PCR (qRT-PCR) was done by Exicycler 96 (Bioneer) with AccuPower 2× Greenstar qPCR Master Mix (Bioneer). The abundance of lacZ mRNA was normalized to the amount of 5S RNA. Data were analysed using ExiAnalysis software (Bioneer).
Gel mobility shift assay
His-tagged Hfq was purified from cells containing ASKA-hfq plasmid, as previously reported (42). 32P-labelled RNAs of 20 nM were renatured by heating for 1 min at 95°C and slowly cooling to 25°C in a 10 μl of RNA binding buffer (10 mM Tris–HCl (pH 8.0), 100 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol (DTT), 0.01% NP40, 5% glycerol). The renatured RNAs were incubated with the purified Hfq protein for 20 min at 25°C. The reaction mixtures were then analysed on 5% polyacrylamide gels, as described previously (43).
RESULTS
Design of artificial sRNAs
We selected the ssrS-lacZ mRNA fusion as a target for artificial sRNAs (afsRNAs) and monitored repression by afsRNAs via measurement of β-galactosidase activity. For this purpose, we constructed a lysogen carrying the ssrS-lacZ transcriptional fusion in which the ssrS P1 promoter was fused to the lacZ coding sequence, including its translation initiation region (TIR). Transcription from the ssrS P1 promoter is constitutive during the exponential phase (44). The TIR of ssrS-lacZ mRNA was used as the target sequence for afsRNAs, as this site may be highly accessible to a ribosome. To design afsRNAs, we adopted a RNA scaffold from SibC(1–8::77–141) (32) containing target-recognition domain 2 (TRD2) (Figure 1). SibC(1–8::77–141), a derivative of the cis-acting sRNA SibC, efficiently suppresses expression of the target ibsC toxin gene through TRD2. TRD2 spans positions +99 to +115, which mostly lies within J1/7 and one strand of the P1 stem in the secondary structure model of SibC. Kinetic analysis of interactions between SibC and target ibsC mRNA disclosed that J1/7 is a single-stranded region before the interaction, with the single strand extending to P1 as the interaction proceeds (32). TRD2 or its related sequences were replaced with antisense sequences as new target recognition sequences. As sequence replacement with antisense sequence would disrupt stem P1 or/and stem P6 in afsRNA, complementary alterations were additionally incorporated so that afsRNAs could retain the similar stem structure and stability. Nucleotide bases for the complementary alterations were selected in such a way that the calculated energy of each stem was approximately the same by introducing nucleotide changes for appropriate base pairings in the opposite strand of the antisense sequence. The MFOLD programme was used as a guide to designing afsRNAs that would have the similar stability and share the core RNA scaffold as much as possible, despite the incorporation of different antisense sequences (45). The designed afsRNA sequences were cloned into an RNA expression vector, pHM-tac, in which transcription was inducible with IPTG.
Suppression of ssrS-lacZ expression by artificial sRNAs
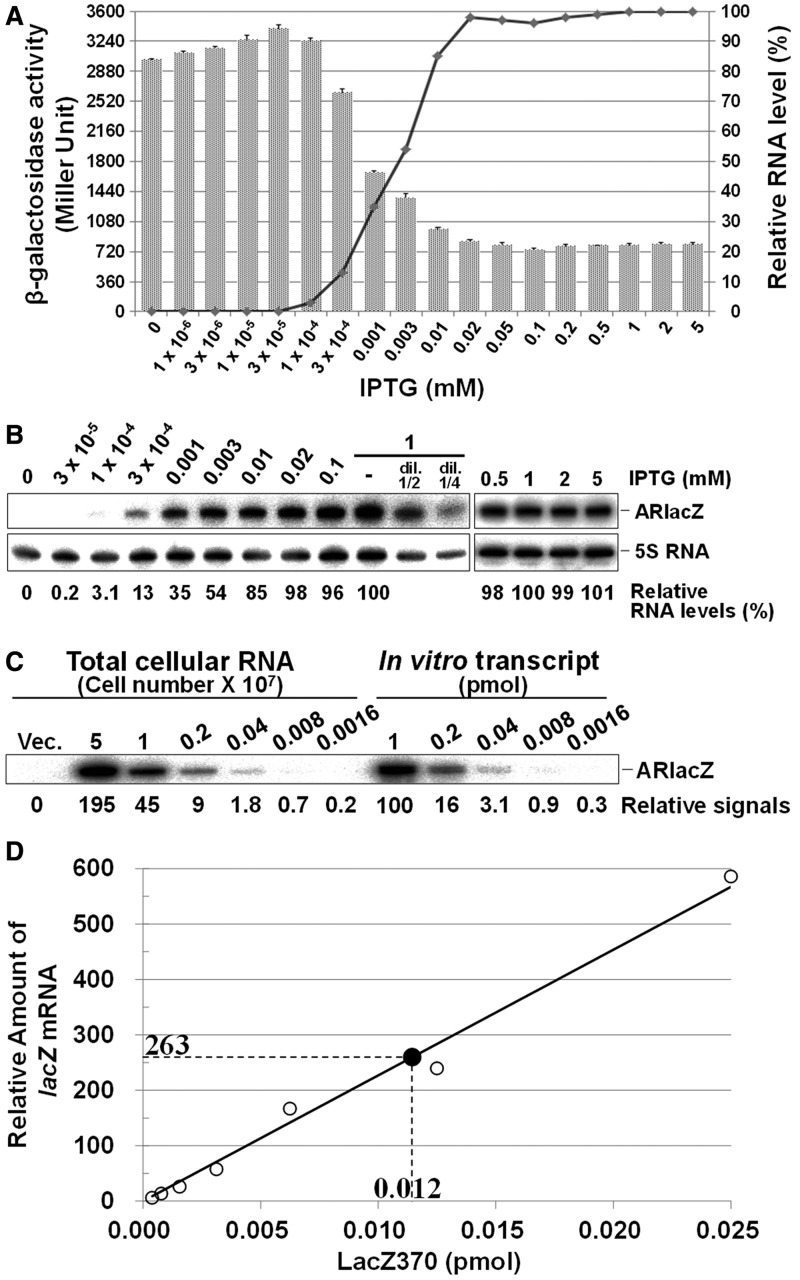
TRD2 (17 nt) in SibC(1–8::77–141) was replaced with the 18 nt complementary sequence from position −17 to +1, relative to the +1 translation initiation site of ssrS-lacZ mRNA, as a new target recognition sequence to generate the afsRNA, ARlacZ1 (Figure 1). Inadvertently, ARlacZ1 contained an extra region able to form a base pair with the +2 to +4 region of ssrS-lacZ mRNA. Therefore, ARlacZ1 could form a base pair with the 21 nt sequence from positions −17 to +4 of ssrS-lacZ mRNA. Following induction of ARlacZ1 expression with 1 mM IPTG, β-galactosidase expression was reduced to 20% (Figuer 2). Steady-state concentrations of ARlacZ1 expressed from the plasmid were analysed with increasing concentrations of IPTG (from 0.0001 to 5 mM) in the cell. ARlacZ1 expression increased with the IPTG concentration and appeared saturated at 0.02 mM. The repression levels of β-galactosidase expression also increased with the IPTG concentration and showed a plateau at 0.02 mM. The level of suppression was inversely proportional to the steady-state level of ARlacZ1 afsRNA when concentrations were above the threshold, corresponding to the amount of ARlacZ1 induced by 0.0001 mM IPTG. The relationship between the cellular concentration of ARlacZ1 and its gene silencing effects may be used to evaluate gene silencing by other afsRNAs, along with their cellular levels. We quantitated the cellular levels of ARlacZ1 and lacZ mRNA using known amounts of each in vitro transcript as a standard. Total cellular RNAs from ARlacZ1-expressing DJ480 ssrS::lacZ cells were used for analysis of ARlacZ1, whereas those from DJ480 ssrS::lacZ cells containing the vector were analysed for lacZ mRNA to examine its cellular level in the absence of repression by afsRNA. ARlacZ1 and lacZ mRNA were quantitated using northern blot (Figure 2C) and qRT-PCR (Figure 2D), respectively. For qRT-PCR, total cellular RNAs isolated from DJ480 cells lacking lacZ mRNA were mixed with known amounts of in vitro transcript consisting of 370 nt from the 5′-end of ssrS-lacZ mRNA and used as quantification standards. We estimated 36 000 molecules of the afsRNA and 2400 molecules of lacZ mRNA in a cell treated with 1 mM IPTG, suggesting that ∼15 times excess afsRNA are required for efficient repression (Figure 2). However, all 2400 molecules of lacZ mRNA might not need to be intact mRNA because any degradation intermediates could be substrates for qRT-PCR as long as they carried the 5′ 370 nucleotides.
Figure 2.
Expression of ARlacZ1 and gene silencing effects. (A) Cells containing the ARlacZ1-expressing plasmid were treated with IPTG at increasing concentrations from 0 to 5 mM. β-galactosidase activities were measured after IPTG induction, and they are represented by bar graph. The indicated values are calculated from at least three independent experiments. The levels of ARlacZ1 analysed in Figure 2B are represented by line graph. (B) Total cellular RNA was prepared from IPTG-treated cells and subjected to northern blot analysis. ARlacZ1 afsRNA and 5S RNAs were probed with the oligonucleotides, ARlacZnp and 5S + 90R, respectively. RNA quantities are expressed relative to ARlacZ1 induced with 1 mM IPTG, using a semi-standard curve with serial dilutions of total cellular RNA from 1 mM IPTG-induced cells after normalization to 5S RNA. (C) The number of ARlacZ1 in a cell was estimated with serial dilutions of total cellular RNA and known amounts of in vitro transcribed ARlacZ1 as standards. Total cellular RNAs from the indicated numbers of ARlacZ1-expressing DJ480 ssrS::lacZ cells, treated with 1 mM IPTG, were subjected to northern blot analysis. Relative northern blot signals are indicated below each lane. About 0.6 pmole of ARlacZ1 was present in 1 × 107 cells. (D). The lacZ mRNA was quantitated by qRT-PCR. Total cellular RNA from 3 × 106 DJ480 ssrS::lacZ cells containing the vector, treated with 1 mM IPTG, was subjected to qRT-PCR (solid circle). For the standard curve, total cellular RNA prepared from the same number of DJ480 cells lacking lacZ mRNA was mixed with known amounts of LacZ370, an in vitro transcript consisting of 370 nucleotides from the 5′-end of ssrS-lacZ fusion mRNA, and also subjected to qRT-PCR (open circle). The abundance of lacZ mRNA was normalized to the amount of 5S RNA and depicted as relative levels. Values represent the average from three independent experiments. About 0.04 pmole of lacZ mRNA was present in 1 × 107 cells.
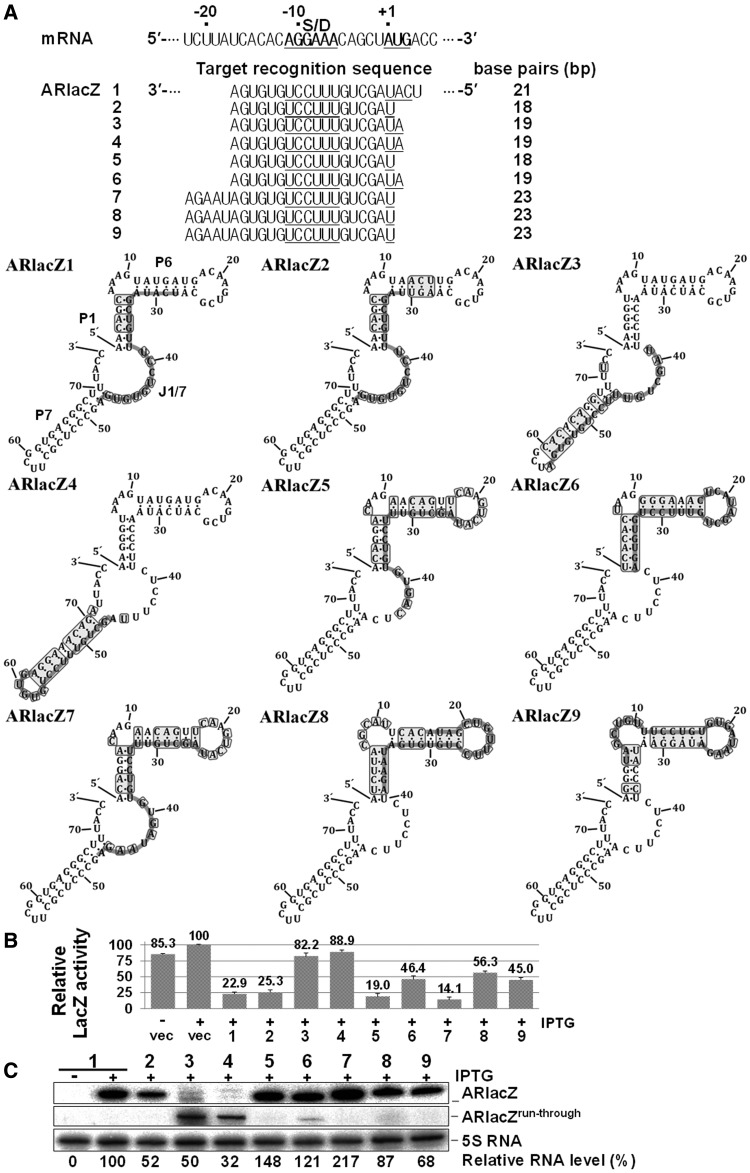
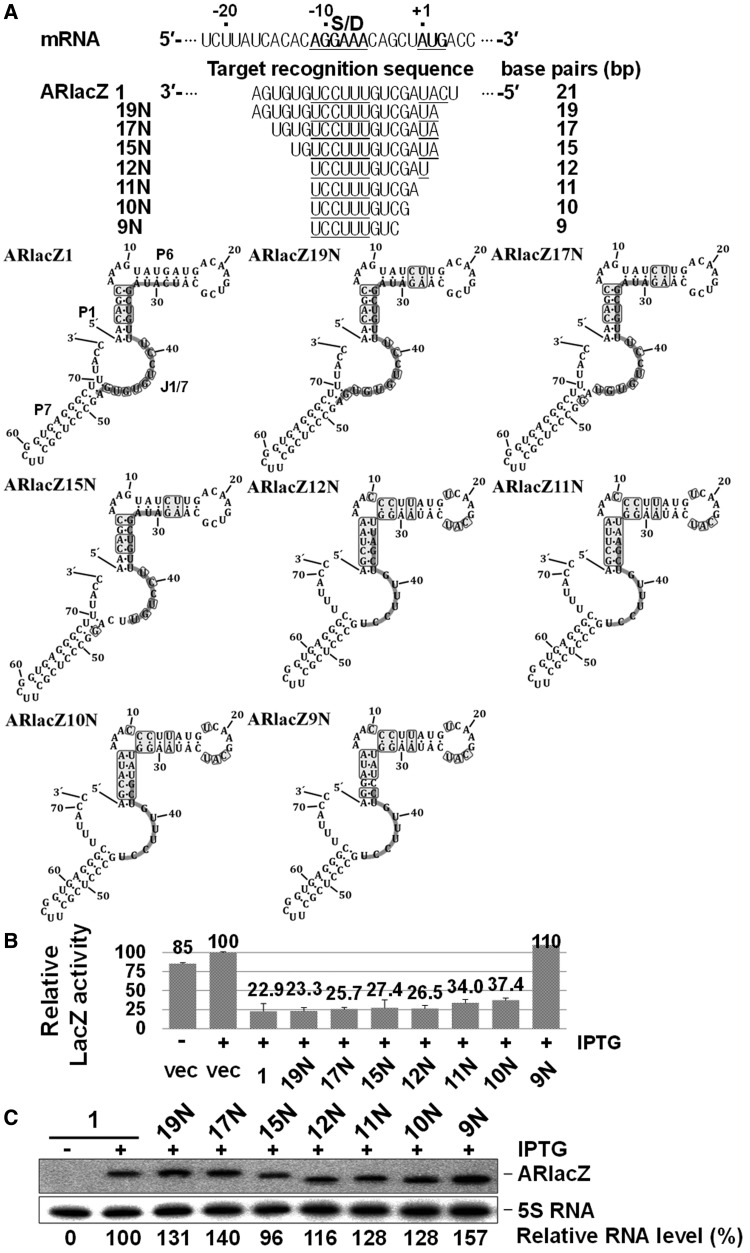
We constructed ARlacZ2, a derivative of ARlacZ1, by replacing the extra base-pairing region at positions +2 to +4 of ssrS-lacZ mRNA, with the complementary bases. ARlacZ2, containing a target-recognition sequence of 18 nt complementary to the −17 to +1 region of ssrS-lacZ mRNA, induced similar lacZ repression as parental ARlacZ1 (Figure 3). The 18 or 19 nt target-recognition sequence was also substituted with sequences containing regions other than TRD2 in SibC(1–8::77–141) to examine the potential impact on the location of the target-recognition sequence within afsRNAs. Both ARlacZ1 and ARlacZ2 had the target-recognition sequence fully embedded with the J1/7 region. When part of the target-recognition sequence was embedded in J1/7 (as in ARlacZ5), a similar level of repression was observed. However, ARlacZ6, which had no target-recognition sequence in J1/7, but had the whole target-recognition sequence embedded in the P1 and P6 regions, induced moderate repression (46.4%). ARlacZ6 was modified to ARlacZ8 and ARlacZ9 containing an extended target-recognition sequence (23 nt) that were more embedded in P6. Both showed moderate gene silencing effects comparable with ARlacZ6. ARlacZ2 was also modified to ARlacZ7 containing the same 23 nt target recognition sequence with additional sequences embedded in P6. ARlacZ7 showed a higher gene silencing effect than ARlacZ2. As ARlacZ7 expression was higher than ARlacZ2, this high expression might have contributed to the high gene silencing effect. When the target-recognition sequence was embedded in the terminator hairpin, as for ARlacZ3 and ARlacZ4, transcription was not terminated at that terminator site but at the further downstream rrnB terminator, generating a longer run-through product of ∼250 nt. The gene silencing effects of these transcripts were considerably smaller in magnitude, even considering the lower expression levels. These results suggest that the presence of the target-recognition sequence in the J1/7 region is essential, at least as a seeding sequence, for the effective function of afsRNAs.
Figure 3.
Effects of the location of target-recognition sequences on gene silencing. (A) The target-recognition sequences are presented below the lacZ mRNA sequence. The embedded region of the target-recognition sequence in each afsRNA scaffold is denoted with a light grey line, and the nucleotides that were changed from the parental scaffold are boxed. The Shine-Dalgarno sequence and translation start codon sequence are underlined. (B) Relative β-galactosidase activities of cells expressing afsRNAs are shown. β-Galactosidase activities were expressed relative to that of cells containing the vector and treated with 1 mM IPTG. Values are presented as an average of at least three independent experiments. (C) Total cellular RNA was prepared from IPTG-treated cells and subjected to northern blot analysis as for Figure 2B. As ARlacZ3 and ARlacZ4 had the target-recognition sequence sembedded in the terminator hairpin, most transcription was not terminated at that terminator, but at the further downstream rrnB terminator, generating a longer run-through transcript of ∼250 nt (ARlacZrun-through). Vec, vector control.
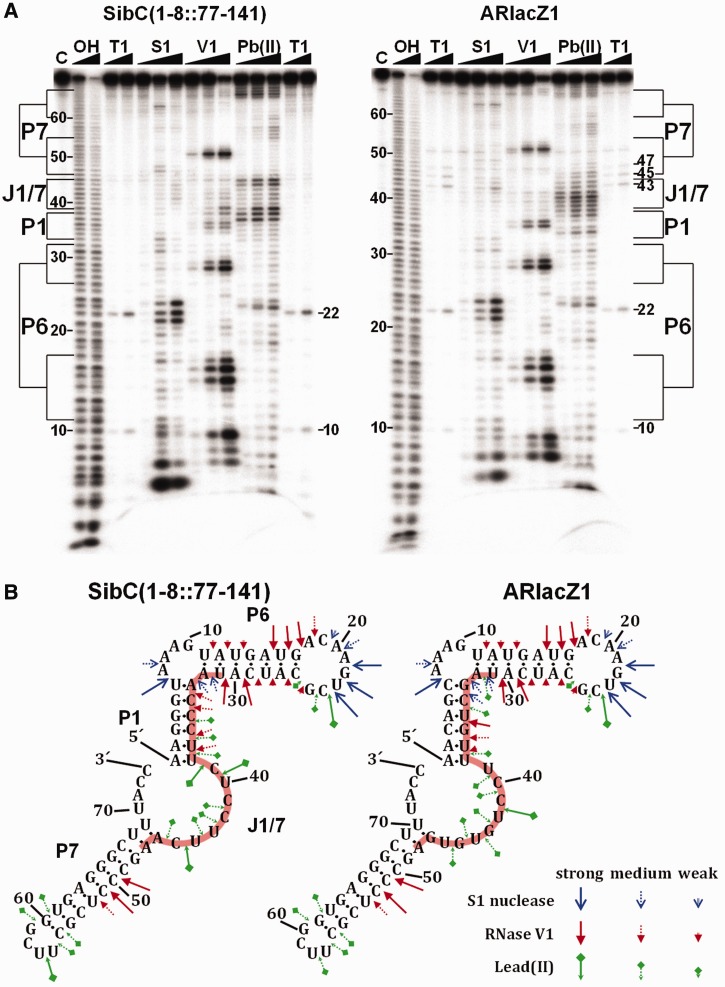
Our secondary structure models of afsRNAs might not represent the real RNA structures. Therefore, we performed enzymatic and chemical mapping analyses with ARlacZ1 and SibC(1–8::77–141) to ascertain whether afsRNAs would not deviate much from the original scaffold. The enzymatic and chemical cleavage patterns of the two RNAs were similar in most of regions, although cleavages by RNase V1 differed a little bit in stem P1 (Figure 4). As grafting of the corresponding antisense sequence alone onto SibC(1–8::77–141) to generate ARlacZ1 could disrupt the P1 stem, additional base alterations were introduced to regenerate the stem. Therefore, the P1 stem of ARlacZ1 had different base pairs from that of SibC(1–8::77–141). This difference of the structural details was probably the main cause of the slightly different cleavage patterns in the P1 stem. We further probed a subset of afsRNAs depicted in Figure 3. Strong RNase V1 cleavages were observed on the P6 and P7 stems, whereas relatively mild cleavages were observed on the P1 stem (Supplementary Figure S1). As expected, the J1/7 region was much less cleavable. Although the actual structure of each afsRNA might differ from its secondary structure model, the probing data suggest common structural features of afsRNAs that the P6 and P7 regions had strong base pairings and that the J1/7 region and maybe the P1 region were rich in single strandedness. These structural features were consistent with the results that gene silencing was more effective with target-recognition sequences incorporated into the J/1 and P1 regions than for those into the P6 or P7 region (Figure 3). It is also noteworthy that the P1 stem is the region melting immediately after the J1/7 sequence recognizes the target sequence (32). In subsequent experiments, therefore, the target-recognition sequences were fixed in the entire J1/7 and part of P1 in afsRNA.
Figure 4.
Structural mapping of SibC(1–8::77–141) and ARlacZ1. (A) 32P-labelled RNA (20 nM) was partially digested with S1 nuclease (0.4, 2 and 10 U), RNase V1 (0.0008, 0.004 and 0.02 U) and lead (II) (1, 5 and 25 mM) in a 10 μl of reaction volume. Untreated RNA and alkaline ladders are shown in lanes C and OH, respectively. Lane T1 corresponds to the RNase T1 ladders of denatured RNA treated with 0.1 and 0.5 U enzyme. The positions of cleaved G residues and some OH-cleaved products are marked. The regions corresponding to possible structural domains of afsRNAs are also marked. (B) S1 nuclease, RNase V1 and lead (II) cleavage sites are shown in the secondary structure predicted using Mfold. The cleavage levels are indicated with different arrows.
Effects of the number of base pairs on gene silencing
To determine the minimal number of base pairs required for target recognition, the 21 nt target-recognition sequence of ARlacZ1 was serially shortened to 9 nt in both the 5′ or 3′ directions, retaining the middle UCCUUU sequence complementary to the Shine–Dalgarno sequence (Figure 5). No significant effects were observed until the length was reduced to 10 nt, although sequences of both 10 and 11 nt led to slightly weakened repression. However, no repression was observed when the length was reduced to 9 nt, suggesting that 10 nt is sufficient to suppress ssrS-lacZ expression. As the dosage-dependent effectiveness of the 10 nt and 11 nt constructs (ARlacZ10N or ARlacZ11N) for gene silencing may be different from that of longer constructs such as ARlacZ1, their gene silencing effects were also examined at lower concentrations than 1 mM IPTG that we normally used for ensuring full expression of afsRNAs. The IPTG dependency of these two afsRNAs for gene silencing appeared to be very similar to that of ARlacZ1 (Supplementary Figure S2), suggesting that the gene silencing effects by ARlacZ11N and ARlacZ10N are proportional to their cellular levels as in case of ARlacZ1.
Figure 5.
Minimal length of target-recognition sequences for gene silencing. (A) The target-recognition sequences. Gene silencing effects (B) and cellular levels of afsRNAs (C) were analysed as for Figure 3.
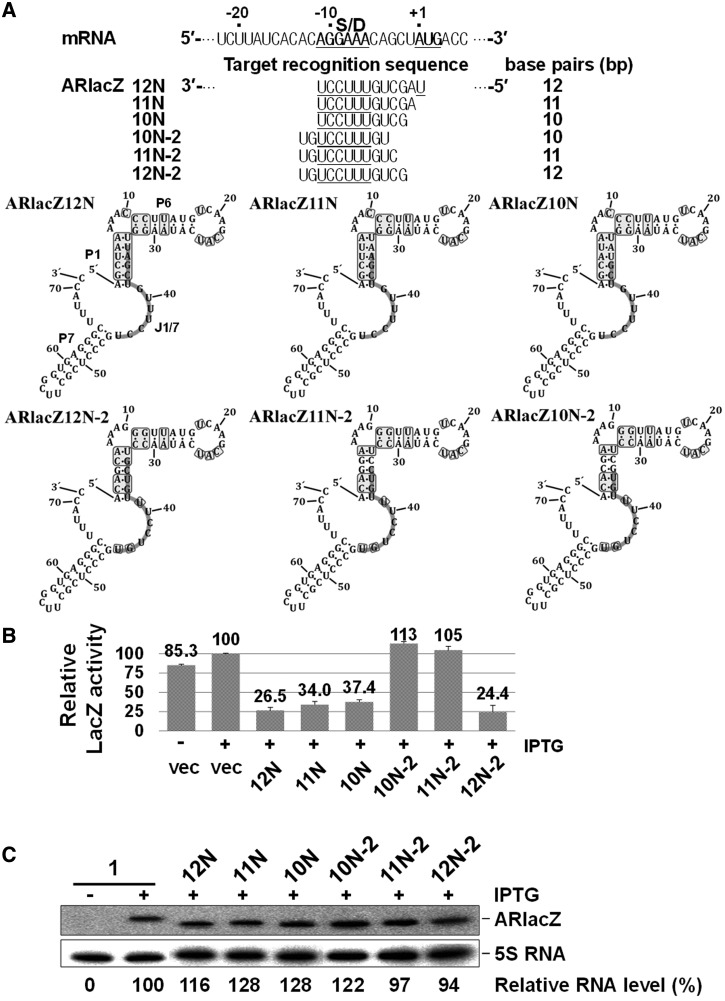
The target sequence of ARlacZ10N was located between positions −11 and −2 of ssrS-lacZ mRNA. To establish whether this minimal number depends on the target-sequence position, we moved the target sequence 2 nt upstream to generate ARlacZ10N−2 (Figure 6). This translocation of the 10 nt window did not induce gene silencing. When energies of base paring between the target-recognition sequence and target mRNA were calculated by using Freiburg RNA Tools (46), the value for ARlacZ10N−2 was −12.7 kcal/mol, which was weaker than that for ARlacZ10N (−13.4 kcal/mol). Considering that the lack of gene silencing by ARlacZ10N−2 may arise from its lower energy of base pairing with target mRNA, we designed a 11 nt construct (ARlacZ11N−2) by adding 1 nt to the 5′-side that could interact with one nucleotide further downstream of the target site. Notably, this extra base pairing did not increase repression, although the base-pairing energy (−15.5 kcal/mol) was higher than that in ARlacZ10N. Therefore, the lack of repression by the 10 nt construct (ARlacZ10N−2) may be attributed to the position of the target sequence, rather than lower energy of base pairing. On extension of the target region to 12 nt (ARlacZ12N−2), repression was observed. ARlacZ10N and ARlacZ12N−2 could recognize the −2C target sequence, but not ARlacZ10N−2 and ARlacZ11N−2, suggesting that this specific region is crucial for repression by sequences with limited base pairing.
Figure 6.
Optimal location of the minimal sequence for gene silencing. (A) The target recognition sequences. Gene silencing effects (B) and cellular levels of afsRNAs (C) were analysed as for Figure 3. ARlacZ1 was used as an internal control for evaluation of RNA expression levels.
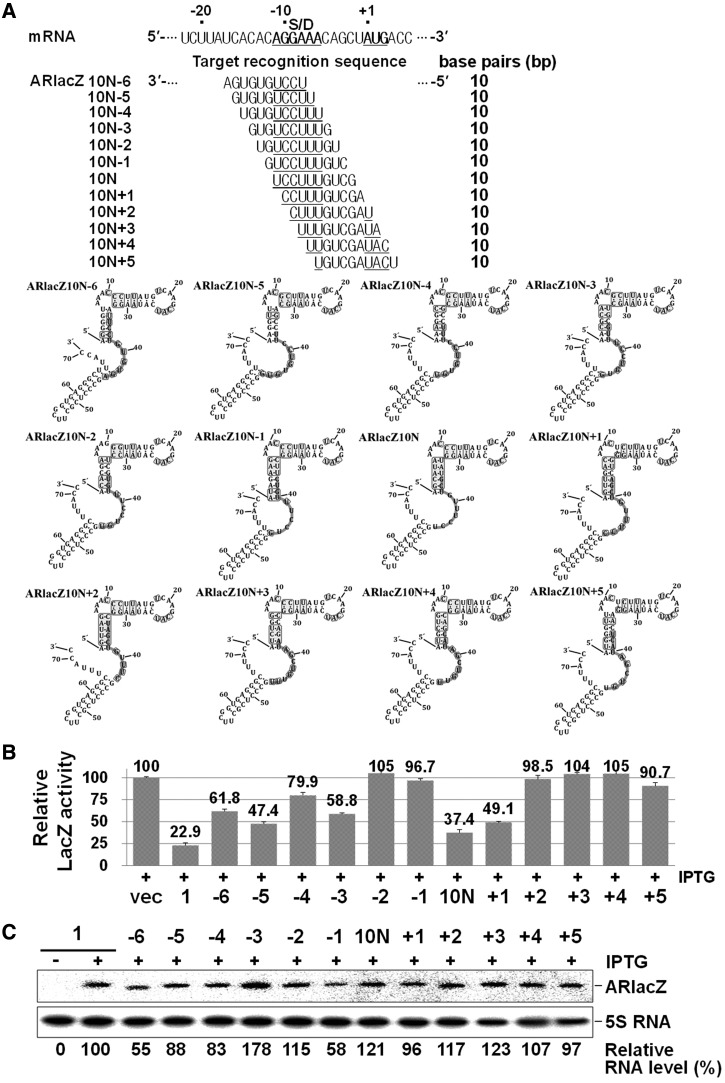
We have analysed more target regions in the TIR of mRNA with the corresponding 10 nt target-recognition sequences (Figure 7). When we scanned the TIR from −17 to +4 of mRNA with a 10 nt window of target-recognition sequence, we found that the parental 10 nt afsRNA, ARlacZ10N, was the most effective one. Some afsRNAs showed slightly weakened repression, and others had no effects. ARlacZ10N+1 and ARlacZ10N−5 were prominent among those showing repression. ARlacZ10N+1 could recognize the sequences from positions −10 to −1, whereas ARlacZ10N−5 could recognize the sequences from positions −16 to −7. These results further support that the 10 nt target-recognition sequence was sufficient for gene silencing by afsRNA and that gene silencing through this minimal length depends on the location of target sequences in mRNA.
Figure 7.
Scanning of TIR of mRNA with a 10 nt window of target-recognition sequence. (A) The target-recognition sequences. Gene silencing effects (B) and cellular levels of afsRNAs (C) were analysed as for Figure 3. ARlacZ1 was used as a control for evaluation of gene silencing effects and RNA expression levels.
Mismatch effects
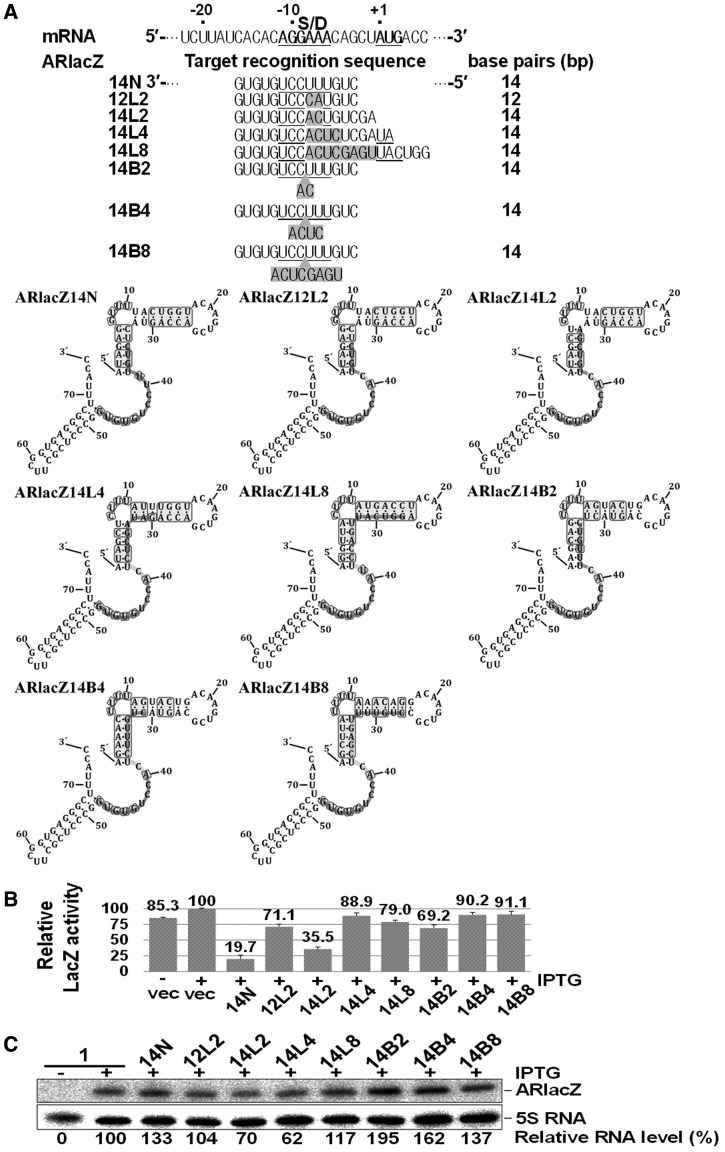
Naturally occurring sRNAs usually contain partially mismatched regions with their target mRNAs, which generate bulges or internal loops in the partial RNA duplexes. We further examined the effects of bulges or internal loops on gene silencing by afsRNAs. Although a variety of different structures can be generated by partially matched duplexes, we simply constructed a series of afsRNAs in which 14 base-pairing regions were disrupted into two helices of 8 and 6 base pairs (8 + 6) by introducing single-strand or double-strand mismatching (Figure 8). In this 8 + 6 base pairing, we referred to the 8 bp region as a major target-recognition sequence and the 6 bp region as an ancillary sequence. Single-strand mismatch mutation allowed afsRNAs to form bulge-containing duplexes with their target mRNAs, whereas double-strand mismatch led to internal loop-containing duplexes. When all 14 base-pairing regions were contiguous (ARlacZ14N), gene expression was reduced to 19.7%, but incorporation of a 2-base double-stranded mismatch (ARlacZ12L2) leading to 8 + 4 base pairing lowered gene silencing by reducing gene expression to only 71.1%. With the same double-stranded mismatch, the target-recognition sequence was extended by two bases at the 5′-end to generate an 8 + 6 construct (ARlacZ14L2) displaying repression to 35.5%, indicating that the 6 base-pair region contributes to gene silencing. Another 8 + 6 construct capable of forming a bulge-containing duplex, ARlacZ14B2, was generated via a two-base single-strand mismatch. ARlacZ14B2 showed a significantly lower gene silencing effect (repression to 69.2%) than ARlacZ14L2. These data suggest that internal loops in the RNA duplex formed between sRNA and mRNA could weaken gene silencing to a lower extent than bulges.
Figure 8.
Effects of mismatched target-recognition sequences on gene silencing. (A) The target-recognition sequences are presented below the lacZ mRNA sequence. Base substitutions within a consecutive sequence generated afsRNAs ARlacZ12L2, ARlacZ14L2, ARlacZ14L4 and ARlacZ14L8 capable of forming internal loops with mRNA, whereas base insertions produced afsRNAs ARlacZ14B2, ARlacZ14B4 and ARlacZ14B8 that formed bulges. The mismatched sequences are shown in shaded boxes. Gene silencing effects (B) and cellular levels of afsRNAs (C) were analysed as for Figure 3. ARlacZ1 was used as a control for evaluation of RNA expression levels.
To further confirm this, we generated more afsRNAs carrying mismatched regions by changing target locations in the TIR of mRNA. Ten additional 8 + 6 constructs capable of forming either a bulge-containing (ARLacZ14B2 − 5, 14B2 − 4, 14B2 + 1, 14B2 + 3 and 14B2 + 5) or internal loop-containing duplex (ARLacZ14L2 − 5, 14L2 − 4, 14L2 + 1, 14L2 + 3 and 14L2 + 5) were generated via a two-base mismatch (Supplementary Figure S3). We found that their gene silencing activities were depending on where the target sequence was located. Only ARlacZ14L2 + 1 besides ARlacZ14L2 induced significant repression, whereas the other afsRNAs showed little repression whether they were internal loop forming or bulge forming. ARlacZ14L2 + 1 capable of forming an internal loop displayed repression to 30%, whereas the cognate bulge-forming afsRNA, ARlacZ14B2 + 1, generated little repression. All together, our results support that loop-forming afsRNAs are more effective in gene silencing than those forming bulges, even though the gene silencing depends on the location of target sequences in mRNA. We also examined gene silencing when the sizes of bulges and internal loops were increased to four or more bases (Figure 8 and Supplementary Figure S4). When the size of mismatch that loop-forming ARlacZ14L2 (Figure 8) and ARlacZ14L2 + 1 (Supplementary Figure S4) had was increased to four bases (ARlacZ14L4 and ARlacZ14L4 + 1) or eight bases (ARlacZ14L8 and ARlacZ14L8 + 1), gene silencing was significantly decreased. The effects of the increase of the mismatch size were also observed with bulge-forming ARlacZ14B2 versus ARlacZ14B4 and ARlacZ14B8 (Figure 8) and ARlacZ14B2 + 1 versus ARlacZ14B4 + 1 and ARlacZ14B8 + 1 (Supplementary Figure S4), although ARlacZ14B2 and ARlacZ14B2 + 1 showed only very weak gene silencing effects. Therefore, it is likely that ancillary target-recognition sequences are effective only when in close proximity to the major contiguous sequence.
Effects of Hfq on gene silencing
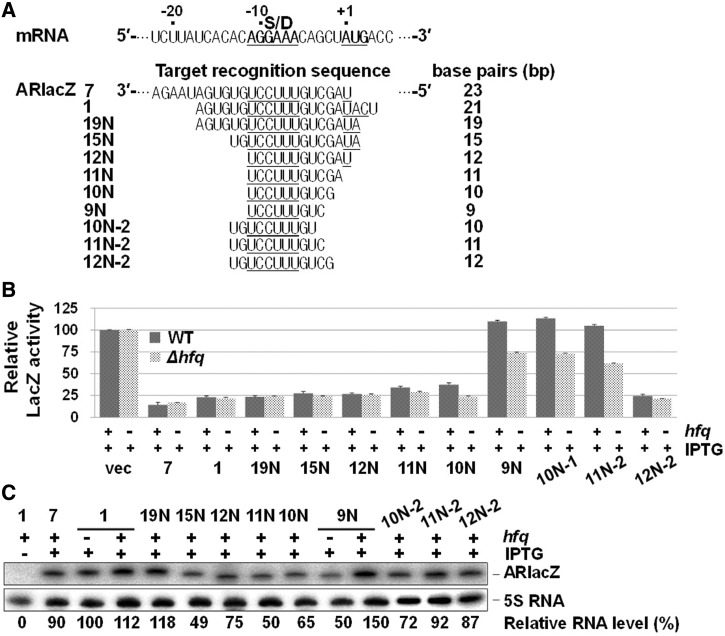
Hfq is known to enhance base pairing between sRNA and its target mRNA (24), but the underlying mechanism is currently unclear. We examined whether gene silencing by afsRNA is Hfq dependent. First, hfq knockout was introduced into the ssrS::lacZ lysogenic strain to generate hfq− cells, and β-galactosidase activity was assayed in hfq− cells expressing afsRNAs (Figure 9). Gene silencing by ARlacZ1 was not affected in the absence of Hfq. Moreover, hfq knockout had no effect on gene silencing by ARlacZ7 containing a more extended target-recognition sequence than ARlacZ1. As SibC RNA was not listed in Hfq-bound sRNAs, it is not expected to respond to Hfq (47). However, its Hfq independency had not been yet experimentally proven. One possible function of Hfq is to enhance gene silencing by sRNAs, and this enhancement of gene silencing can be achieved by stabilizing the duplexes between sRNA and its target mRNA. Therefore, we attempted to see the effects of the hfq knockout on gene silencing by afsRNAs having serially shortened target-recognition sequences (Figure 9). Similar gene silencing effects were observed in both hfq+ and hfq− cells until the length was reduced 12 nt, but the reduction to 11 or 10 nt slightly increased gene silencing. Furthermore, when the base pairing was shortened to 9 nt, hfq− cells showed gene silencing, whereas hfq+ cells did not. These are unexpected results, as Hfq is considered a positive factor for gene silencing. As the hfq− mutant cells grew with a slower rate than the wild-type, hfq+ and hfq− cells grown to OD600 of ∼0.5 were subjected to the aforementioned analysis. However, one may argue that these results might come from difference in cell physiology rather than in base pairing. Therefore, we analysed cells grown for different growth times to see whether this difference in growth conditions would affect the Hfq influence on ARlacZ10N and ARlacZ11N (showing the modest Hfq effects) as well as ARlacZ9N (showing the significant Hfq effect). Mutant hfq− cells grown for 2, 2.5, 3 and 3.5 h were assayed for β-galactosidase and compared with hfq+ cells grown for ∼2 h whose OD600 was the same as that of hfq− cells grown for ∼3 h (Supplementary Figure S5). The Hfq effects on ARlacZ9N were observed, regardless of growth times. We also observed the Hfq effects with ARlacZ10N and ARlacZ11N, but not with ARlacZ12N or ARlacZ1 in cells grown for 3 h or longer, suggesting that the Hfq effects reflect differences in base pairing rather than in cell physiology. In view of these results, we propose a novel function of Hfq as a negative factor in cases where the base-paired recognition region is short. The induction of gene silencing by the hfq knockout was additionally observed with ARlacZ11N−2 and ARlacZ10N−2 with base-paired regions of 11 nt, and 10 nt, respectively, although they induced no gene silencing in wild-type cells. The results collectively support the theory that Hfq is involved in suppressing gene silencing caused by short base pairing. Moreover, this type of suppression mechanism may be physiologically used in the cell to discriminate sRNAs from non-cognate target mRNAs.
Figure 9.
Effects of Hfq on gene silencing by afsRNAs. (A) The target-recognition sequences. (B) The gene silencing effects of various afsRNAs were examined in hfq+ and hfq− cells. ß-Galactosidase activities were expressed relative to that of cells containing the vector and treated with 1 mM IPTG for each strain. Values are presented as an average of at least three independent experiments. (C) Total cellular RNA from was prepared from IPTG-treated hfq− cells and subjected to northern blot analysis as for Figure 2B. Total cellular RNA from hfq+ cells expressing ARlacZ1 or ARlacZ9N was used as an internal control for comparing RNA expression levels between hfq+ and hfq− cells.
Then an interesting question would be how the effects of the hfq− mutation would happen without an Hfq’s preferred binding site. We speculated that Hfq could bind to afsRNAs even in the absence of the preferred binding site, and that this binding may cause duplex destabilization when the duplex stability is marginal for gene silencing. We performed Hfq-binding assays for SibC(1–8::77–141), ARlacZ1 and LacZ370, and their affinities were compared with those of known Hfq-binding sRNAs, MicC and OxyS (48). We found that Hfq was still able to bind to SibC(1–8::77–141), ARlacZ1 and LacZ370, although their binding affinities were lower than those of MicC and OxyS (Supplementary Figure S6), suggesting that Hfq-induced repression of the gene silencing effect could occur through Hfq binding to afsRNAs and/or lacZ mRNA. Yet, it still remains to be determined how this lower affinity could contribute to enhancing destabilization of RNA duplexes with marginal stability and whether the Hfq function in RNA duplex destabilization can be applied to RNAs containing Hfq’s preferred binding sequences.
DISCUSSION
In the current study, we used the specific features of a TRD2 of cis-acting sRNA, SibC, to design afsRNAs (32). A truncated version, SibC(1–8::77–141), including TRD2 within a structure similar to intact SibC, has been reported to suppress IbsC toxin expression (31,32). We designed afsRNAs based on SibC(1–8::77–141) and used these structures to investigate the target-recognition mechanism of sRNA. These afsRNAs contained no sequences complementary to target mRNAs, apart from the specific target-recognition sequence. Previous studies have shown that TRD2 is mainly located in the J1/7 junction, a single-stranded region, in the secondary structure model of SibC (32). In the present investigation, we confirmed that TRD2 lies within a single-stranded region, even in the SibC(1–8::77–141) scaffold. On replacement of TRD2 with a sequence, which is complementary to the TIR of lacZ mRNA as a target, the new sequence was similarly embedded within a single-stranded region and led to repression of lacZ expression. The generation of afsRNAs with various TRD2 sequences at different positions in the scaffold allowed us to examine base-pairing effects on gene silencing with regard to single strandedness of target-recognition sequences, minimal base pairing and mismatch regions and Hfq dependency. We concluded that single strandedness is important for target recognition, based on the finding that gene silencing was reduced when the target-recognition sequence was located within a base-paired region, although the importance of single strandedness has been shown many times in naturally occurring sRNA (49). We additionally determined the minimal base-pairing region required for efficient gene silencing. Our data showed that a length of 10 bp is sufficient to suppress expression of target mRNA, but the base-pairing number is variable, depending on the location of the target sequence. For instance, in the case of lacZ mRNA, the sequence immediately upstream of the start codon is essential for efficient gene silencing by the 10 nt base-pairing region. Although an in vitro approach to determine the minimal base-pairing region for the gene silencing function of natural sRNAs has been reported using defined oligonucleotides (25), determination of this specific region in vivo is difficult, as deletion or mutation of sRNAs can affect RNA structure, thereby influencing target recognition efficacy and in vivo stability. The presence of partial base-pairing regions may further complicate determination of the minimal base-paring region. The afsRNAs may be useful to overcome these problems and thus effectively used for defining the crucial base-pairing region in vivo.
The finding that the 10 nt base-pairing region is sufficient for gene silencing raises the issue of whether increasing the antisense nucleotide number would be helpful for gene silencing. If the target-recognition sequence is 10 nt, target sites occur at a frequency of one in ∼106 (410). As the location of the target sequence in mRNA is also important for gene silencing, the 10 nt region may have little chance of recognizing multiple target sites in prokaryotic mRNAs. However, when the number of base pairings increases, the additional sequences in sRNA may facilitate the recognition of other targets (7,50–52), although this may contribute to increasing the stability of RNA–RNA complexes. Therefore, the increase in base-pairing number may not be beneficial in terms of enhancing sRNA target specificity. This aspect requires consideration while designing antisense-based oligonucleotides, such as ribozymes, as well as afsRNA to inhibit the expression of a specific gene with reduced off-target effects.
As naturally occurring sRNAs can bind to target mRNAs through imperfect complementarities, afsRNAs were used to examine the mechanisms by which mismatched regions affect recognition of target mRNA. The RNA duplexes formed by partially mismatched afsRNAs and target mRNA simply involve the major and ancillary helices linked by the bulge or the internal loop. The disruption of the RNA duplexes generally hindered gene silencing, but some afsRNAs able to form 8 + 6 base pairing with a two-base mismatch displayed repression comparable with that by the parental afsRNA carrying the contiguous 14 nt target-recognition sequence. We found that afsRNAs capable of forming an internal loop-containing helix were more effective than those forming a bulge-containing helix. An ancillary recognition sequence helix that is ≥4 nt away from the major recognition sequence contributes to gene silencing to a lower extent than that 2 nt away, suggesting that the distance between the major and ancillary recognition sequences is important for the effectiveness of gene silencing.
Our experiments showed that gene silencing by afsRNA is not Hfq-dependent in the presence of long target-recognition sequences of ≥12 nt. This may be attributed to the lack of Hfq-binding sites (23,53) within afsRNA or/and near the target site of mRNA. Nonetheless, the silencing effectiveness of afsRNA is comparable with that of an afsRNA prototype developed by Man et al. (30), which is strictly Hfq-dependent. The non-dependency of afsRNA on Hfq suggests that gene silencing by afsRNA may be based solely on RNA–RNA interactions. Alternatively, RNA chaperone proteins other than Hfq may be involved in this gene silencing process. On the other hand, Hfq surprisingly suppressed the gene silencing effects of afsRNA with shorter target-recognition sequences. Although the underlying mechanism of Hfq-induced repression of the gene silencing effect remains to be established, one possibility is that repression is accomplished by melting the base-paired region between sRNA and mRNA, as Hfq has both RNA annealing acceleration and duplex stabilization activities (54–56). This finding is opposite to the known function of Hfq in enhancing gene silencing activity (23,24). Suppression of gene silencing by Hfq may minimize the off-target effects of sRNAs by rewinding relatively short base-paired regions. To our knowledge, this is the first report showing that Hfq decreases the gene silencing effects of sRNA.
afsRNAs provide a useful tool for investigating RNA–RNA interactions in the cell and may thus be used to precisely define the interacting regions and examine structural requirements for the interactions. For example, our studies with lacZ mRNA as a target have disclosed that the sequence between positions −3 and −12 is the most effective target site for gene silencing by sRNA. Naturally derived afsRNAs are expected to physiologically behave like natural sRNAs. Therefore, the target-recognition sequence of sRNA can be validated with afsRNA, if incorporated into the afsRNA scaffold. In addition, the gene silencing systems of lacZ and afsRNA may be effectively used to determine the role of Hfq in this process by incorporating Hfq-binding sites into afsRNA or/and mRNAs. Furthermore, afsRNA can be used as a gene silencing tool for prokaryotes in more elaborate ways. This may applied as a powerful experimental technique, such as gene silencing in eukaryotes, facilitating the effective investigation of gene function in prokaryotes. In particular, our inducible knockdown system can be used for detailed in vivo studies on the functions of bacterial essential genes, which are not possible with knockout analyses. Our experiments confirm that modulation by recognition sequences and their locations in the sRNA structure as well as the location of target sites in mRNA can be used as variables for the fine tuning of gene regulation with enhanced accuracy.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–6.
FUNDING
The National Research Foundation of Korea (NRF) Grant by the Korea government (MEST) [2010-0029167, 2011-0020322]; the Intelligent Synthetic Biology Center of Global Frontier Project funded by MEST [2012M3A6A8054837]. Funding for open access charge: The National Research Foundation of Korea (NRF) Grant by the Korea government (MEST) [2010-0029167].
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Racz Z, Kaucsar T, Hamar P. The huge world of small RNAs: regulating networks of microRNAs (review) Acta. Physiol. Hung. 2011;98:243–251. doi: 10.1556/APhysiol.98.2011.3.1. [DOI] [PubMed] [Google Scholar]
- 2.Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell. 2011;43:880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suh N, Blelloch R. Small RNAs in early mammalian development: from gametes to gastrulation. Development. 2011;138:1653–1661. doi: 10.1242/dev.056234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma CM, Vogel J. Experimental approaches for the discovery and characterization of regulatory small RNA. Curr. Opin. Microbiol. 2009;12:536–546. doi: 10.1016/j.mib.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Brantl S. Bacterial chromosome-encoded small regulatory RNAs. Future Microbiol. 2009;4:85–103. doi: 10.2217/17460913.4.1.85. [DOI] [PubMed] [Google Scholar]
- 7.Prevost K, Salvail H, Desnoyers G, Jacques JF, Phaneuf E, Masse E. The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis. Mol. Microbiol. 2007;64:1260–1273. doi: 10.1111/j.1365-2958.2007.05733.x. [DOI] [PubMed] [Google Scholar]
- 8.Frohlich KS, Vogel J. Activation of gene expression by small RNA. Curr. Opin. Microbiol. 2009;12:674–682. doi: 10.1016/j.mib.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 9.Urban JH, Vogel J. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 2007;35:1018–1037. doi: 10.1093/nar/gkl1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pfeiffer V, Papenfort K, Lucchini S, Hinton JC, Vogel J. Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat. Struct. Mol. Biol. 2009;16:840–846. doi: 10.1038/nsmb.1631. [DOI] [PubMed] [Google Scholar]
- 11.Richter AS, Schleberger C, Backofen R, Steglich C. Seed-based INTARNA prediction combined with GFP-reporter system identifies mRNA targets of the small RNA Yfr1. Bioinformatics. 2010;26:1–5. doi: 10.1093/bioinformatics/btp609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papenfort K, Podkaminski D, Hinton JC, Vogel J. The ancestral SgrS RNA discriminates horizontally acquired Salmonella mRNAs through a single G-U wobble pair. Proc. Natl Acad. Sci. USA. 2012;109:E757–E764. doi: 10.1073/pnas.1119414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawamoto H, Koide Y, Morita T, Aiba H. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol. Microbiol. 2006;61:1013–1022. doi: 10.1111/j.1365-2958.2006.05288.x. [DOI] [PubMed] [Google Scholar]
- 14.Sharma V, Yamamura A, Yokobayashi Y. Engineering artificial small RNAs for conditional gene silencing in Escherichia coli. ACS Synth. Biol. 2011;1:6–13. doi: 10.1021/sb200001q. [DOI] [PubMed] [Google Scholar]
- 15.Chen S, Zhang A, Blyn LB, Storz G. MicC, a second small-RNA regulator of Omp protein expression in Escherichia coli. J. Bacteriol. 2004;186:6689–6697. doi: 10.1128/JB.186.20.6689-6697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geissmann TA, Touati D. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J. 2004;23:396–405. doi: 10.1038/sj.emboj.7600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moon K, Gottesman S. Competition among Hfq-binding small RNAs in Escherichia coli. Mol. Microbiol. 2011;82:1545–1562. doi: 10.1111/j.1365-2958.2011.07907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carpousis AJ. The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu. Rev. Microbiol. 2007;61:71–87. doi: 10.1146/annurev.micro.61.080706.093440. [DOI] [PubMed] [Google Scholar]
- 19.Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eggenhofer F, Tafer H, Stadler PF, Hofacker IL. RNApredator: fast accessibility-based prediction of sRNA targets. Nucleic Acids Res. 2011;39:W149–W154. doi: 10.1093/nar/gkr467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dai X, Zhao PX. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 2011;39:W155–W159. doi: 10.1093/nar/gkr319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peer A, Margalit H. Accessibility and evolutionary conservation mark bacterial small-RNA target-binding regions. J. Bacteriol. 2011;193:1690–1701. doi: 10.1128/JB.01419-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011;9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soper T, Mandin P, Majdalani N, Gottesman S, Woodson SA. Positive regulation by small RNAs and the role of Hfq. Proc. Natl Acad. Sci. USA. 2010;107:9602–9607. doi: 10.1073/pnas.1004435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maki K, Morita T, Otaka H, Aiba H. A minimal base-pairing region of a bacterial small RNA SgrS required for translational repression of ptsG mRNA. Mol. Microbiol. 2010;76:782–792. doi: 10.1111/j.1365-2958.2010.07141.x. [DOI] [PubMed] [Google Scholar]
- 26.Cao Y, Zhao Y, Cha L, Ying X, Wang L, Shao N, Li W. sRNATarget: a web server for prediction of bacterial sRNA targets. Bioinformation. 2009;3:364–366. doi: 10.6026/97320630003364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tjaden B. TargetRNA: a tool for predicting targets of small RNA action in bacteria. Nucleic Acids Res. 2008;36:W109–W113. doi: 10.1093/nar/gkn264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Busch A, Richter AS, Backofen R. IntaRNA: efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics. 2008;24:2849–2856. doi: 10.1093/bioinformatics/btn544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stefan A, Schwarz F, Bressanin D, Hochkoeppler A. Shine-Dalgarno sequence enhances the efficiency of lacZ repression by artificial anti-lac antisense RNAs in Escherichia coli. J. Biosci. Bioeng. 2010;110:523–528. doi: 10.1016/j.jbiosc.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 30.Man S, Cheng R, Miao C, Gong Q, Gu Y, Lu X, Han F, Yu W. Artificial trans-encoded small non-coding RNAs specifically silence the selected gene expression in bacteria. Nucleic Acids Res. 2011;39:e50. doi: 10.1093/nar/gkr034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fozo EM, Kawano M, Fontaine F, Kaya Y, Mendieta KS, Jones KL, Ocampo A, Rudd KE, Storz G. Repression of small toxic protein synthesis by the Sib and OhsC small RNAs. Mol. Microbiol. 2008;70:1076–1093. doi: 10.1111/j.1365-2958.2008.06394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han K, Kim KS, Bak G, Park H, Lee Y. Recognition and discrimination of target mRNAs by Sib RNAs, a cis-encoded sRNA family. Nucleic Acids Res. 2010;38:5851–5866. doi: 10.1093/nar/gkq292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beran RK, Simons RW. Cold-temperature induction of Escherichia coli polynucleotide phosphorylase occurs by reversal of its autoregulation. Mol. Microbiol. 2001;39:112–125. doi: 10.1046/j.1365-2958.2001.02216.x. [DOI] [PubMed] [Google Scholar]
- 34.Powell BS, Rivas MP, Court DL, Nakamura Y, Turnbough CL., Jr Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acids Res. 1994;22:5765–5766. doi: 10.1093/nar/22.25.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl Acad. Sci. USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100050. 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X, Bremer H. Control of the Escherichia coli rrnB P1 promoter strength by ppGpp. J. Biol. Chem. 1995;270:11181–11189. doi: 10.1074/jbc.270.19.11181. [DOI] [PubMed] [Google Scholar]
- 38.Kim S, Kim H, Park I, Lee Y. Mutational analysis of RNA structures and sequences postulated to affect 3′ processing of M1 RNA, the RNA component of Escherichia coli RNase P. J. Biol. Chem. 1996;271:19330–19337. doi: 10.1074/jbc.271.32.19330. [DOI] [PubMed] [Google Scholar]
- 39.Milligan JF, Uhlenbeck OC. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- 40.De la Pena M, Gago S, Flores R. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 2003;22:5561–5570. doi: 10.1093/emboj/cdg530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morl M, Schmelzer C. A simple method for isolation of intact RNA from dried polyacrylamide gels. Nucleic Acids Res. 1993;21:2016. doi: 10.1093/nar/21.8.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 43.Chae H, Han K, Kim KS, Park H, Lee J, Lee Y. Rho-dependent termination of ssrS (6S RNA) transcription in Escherichia coli: implication for 3′ processing of 6S RNA and expression of downstream ygfA (putative 5-formyl-tetrahydrofolate cyclo-ligase) J. Biol. Chem. 2011;286:114–122. doi: 10.1074/jbc.M110.150201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim KS, Lee Y. Regulation of 6S RNA biogenesis by switching utilization of both sigma factors and endoribonucleases. Nucleic Acids Res. 2004;32:6057–6068. doi: 10.1093/nar/gkh939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith C, Heyne S, Richter AS, Will S, Backofen R. Freiburg RNA tools: a web server integrating INTARNA, EXPARNA and LOCARNA. Nucleic Acids Res. 2010;38:W373–W377. doi: 10.1093/nar/gkq316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- 48.Olejniczak M. Despite similar binding to the Hfq protein regulatory RNAs widely differ in their competition performance. Biochemistry. 2010;50:4427–4440. doi: 10.1021/bi102043f. [DOI] [PubMed] [Google Scholar]
- 49.Richter AS, Backofen R. Accessibility and conservation: General features of bacterial small RNA-mRNA interactions? RNA Biol. 2012;9:954–965. doi: 10.4161/rna.20294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beisel CL, Storz G. The base-pairing RNA spot 42 participates in a multioutput feedforward loop to help enact catabolite repression in Escherichia coli. Mol. Cell. 2011;41:286–297. doi: 10.1016/j.molcel.2010.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guillier M, Gottesman S, Storz G. Modulating the outer membrane with small RNAs. Genes Dev. 2006;20:2338–2348. doi: 10.1101/gad.1457506. [DOI] [PubMed] [Google Scholar]
- 52.Pulvermacher SC, Stauffer LT, Stauffer GV. The role of the small regulatory RNA GcvB in GcvB/mRNA posttranscriptional regulation of oppA and dppA in Escherichia coli. FEMS Microbiol. Lett. 2008;281:42–50. doi: 10.1111/j.1574-6968.2008.01068.x. [DOI] [PubMed] [Google Scholar]
- 53.Salim NN, Feig AL. An upstream Hfq binding site in the fhlA mRNA leader region facilitates the OxyS-fhlA interaction. PLoS. One. 2010;5:e13028. doi: 10.1371/journal.pone.0013028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arluison V, Hohng S, Roy R, Pellegrini O, Regnier P, Ha T. Spectroscopic observation of RNA chaperone activities of Hfq in post-transcriptional regulation by a small non-coding RNA. Nucleic Acids Res. 2007;35:999–1006. doi: 10.1093/nar/gkl1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hopkins JF, Panja S, McNeil SA, Woodson SA. Effect of salt and RNA structure on annealing and strand displacement by Hfq. Nucleic Acids Res. 2009;37:6205–6213. doi: 10.1093/nar/gkp646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Doetsch M, Stampfl S, Furtig B, Beich-Frandsen M, Saxena K, Lybecker M, Schroeder R. Study of E. coli Hfq's RNA annealing acceleration and duplex destabilization activities using substrates with different GC-contents. Nucleic Acids Res. 2013;41:487–497. doi: 10.1093/nar/gks942. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.