Abstract
Short hairpin RNAs (shRNAs) are widely used to induce RNA interference (RNAi). We tested a variety of shRNAs that differed in stem length and terminal loop size and revealed strikingly different RNAi activities and shRNA-processing patterns. Interestingly, we identified a specific shRNA design that uses an alternative Dicer-independent processing pathway. Detailed analyses indicated that a short shRNA stem length is critical for avoiding Dicer processing and activation of the alternative processing route, in which the shRNA is incorporated into RISC and processed by the AGO2-mediated slicer activity. Such alternatively processed shRNAs (AgoshRNAs) yield only a single RNA strand that effectively induces RNAi, whereas conventional shRNA processing results in an siRNA duplex of which both strands can trigger RNAi. Both the processing and subsequent RNAi activity of these AgoshRNAs are thus mediated by the RISC-component AGO2. These results have important implications for the future design of more specific RNAi therapeutics.
INTRODUCTION
RNA interference (RNAi) is an evolutionarily conserved post-transcriptional gene silencing mechanism in eukaryotes that is triggered by double-stranded RNA (dsRNA) (1,2). The key players in the RNAi mechanism are small non-coding dsRNAs of 20–30 basepairs (bp) with a 3′ dinucleotide overhang, a 5′ monophosphate and 3′ hydroxyl group (3,4). Several classes of small non-coding RNAs exist, including microRNAs (miRNAs) and small interfering RNAs (siRNAs). MiRNAs are transcribed in the nucleus as primary miRNAs (pri-miRNAs) by RNA polymerase (Pol II, occasionally Pol III) and are subsequently processed by the Microprocessor complex, comprising ribonuclease (RNase) III Drosha and subunit DGCR8, into ∼70 nucleotide (nt) pre-miRNAs (5,6). The pre-miRNA associates with Exportin 5 that recognizes the 2-nt 3′ overhang and facilitates RNA export to the cytoplasm (7–9). The cytoplasmic complex that contains RNase III Dicer, TAR RNA-binding protein (TRBP) and protein kinase R (PKR) activator (PACT) processes pre-miRNAs into mature miRNA duplexes of 20–24 bp. The miRNA duplex associates with a member of the Argonaute protein family (AGO) within the precursor RNAi-induced silencing complex (pre-RISC) (10,11). Either miRNA strand can mediate gene silencing, but many miRNAs show asymmetry with preferential loading of one of the strand (guide) into RISC. Thermodynamic properties largely determine which strand of the duplex will be incorporated as guide into RISC (12,13). The remaining strand of the duplex (passenger) is cleaved and degraded. The mature RISC contains the single-stranded guide RNA annealed to a complementary target mRNA for post-transcriptional gene silencing (PTGS). The RNAi pathway can also be induced by artificial substrates, transiently by siRNAs that directly feed into RISC or stably by intracellularly expressed shRNA precursors that are processed by Dicer into siRNAs (14–16).
A key component of the RNAi pathway is the AGO protein. Human cells have four Ago genes (Ago1–4), and the siRNAs and miRNA templates are assorted among these AGO proteins (17,18). In worms and flies, the sorting is influenced by prior Dicer processing, the structure of the small RNA duplex (thermodynamic properties) and its terminal nucleotides (19–26), whereas in mammalian cells a strict system for small RNA sorting is lacking (17,27,28). Only the AGO2 protein is known to have ‘slicer’ activity, executed by a catalytically active RNaseH-like domain that cleaves the target mRNA (29). Endonucleolytic cleavage of the target mRNA occurs opposite nucleotide position 10 and 11 of the annealed siRNA guide strand. The mRNA cleavage products are released from RISC and interact with the 5′ → 3′ exonuclease XRN1, leading to decapping and subsequent mRNA degradation (30).
Most research on the description of the RNAi machinery and improvement of the RNAi knockdown efficiency has focused on synthetic siRNAs or cellular miRNAs, but much less on shRNAs (31–36). The sequence and structure of shRNAs have been shown to influence the RNAi activity (16,37–44). However, specific sequences or elements within the shRNAs that determine the processing efficiency remain poorly defined. Recently, evidence has accumulated for non-canonical processing of miRNAs and shRNAs (45–50). Processing of the pre-miRNA-451 has been shown to occur by AGO2 instead of Dicer cleavage (45–47). The short 17 bp stem and 4 nt loop of miR-451 are the major determinants for this alternative processing route (45–47). Two groups previously described chemically synthesized shRNAs of minimal size that potently induced RNAi via mRNA cleavage, yet were not processed by Dicer in vitro and in vivo (34,51). The Siolas report suggested that the shRNA loop could be cleaved by a cellular endonuclease of unknown origin. The Dallas group suggested that AGO2 may be involved in the processing of shRNAs with short stem lengths of ≤19 bp. Indirect evidence for AGO2 involvement was obtained by chemical modification and introduction of mismatches at and around the cleavage site, which resulted in decreased processing and knockdown activities (49).
In this study, we generated shRNA variants with different duplex lengths and loop sequences to test the effect on shRNA processing and RNAi activity. Reporter constructs that score guide and passenger strand activity revealed a nearly complete switch from the regular guide to the passenger strand for a certain shRNAs design. Further analyses confirmed an alternative shRNA-processing mechanism that is independent of Dicer, yet carries all hallmarks of AGO2-mediated processing. AGO2 co-immunoprecipitation studies revealed that these alternatively processed shRNAs are indeed incorporated into RISC and processed by AGO2. These findings indicate that the shRNA stem length is a critical factor for Dicer-independent processing. Importantly, the alternatively processed shRNAs induce an exquisite strand-specific RNAi-mediated gene knockdown compared with conventionally processed shRNAs.
MATERIALS AND METHODS
Plasmids
ShRNA expression plasmids targeting HIV-1 sequences pol47, pol9 and R/T5 were constructed as previously described (16,39,52). The RNA secondary structure of the shRNA transcripts was predicted by the Mfold program (53). The firefly luciferase reporter plasmids were constructed by insertion in the 3′ untranslated region (3′UTR) of a 50–70-basepair fragment with the actual 19-nucleotide target in the centre. For this, we used the EcoRI and PstI sites of the pGL-3 plasmid (54). The luciferase reporters with the sense target sequences were described previously (52), whereas the reporters harbouring the antisense target sequences were newly constructed as described above. The wild-type AGO2 protein, the catalytic mutant (D597A) and the N domain mutant (F181A) were expressed from the pIRESneo-Flag-haemagglutinin-AGO2 vector and were kindly provided by Y. Tomari (55).
Cell culture
Human embryonic kidney 293T adherent cells and HCT-116 cells were grown as monolayer in Dulbecco’s modified Eagle’s medium (Life Technologies, Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal calf serum (FCS), penicillin (100 U/ml) and streptomycin (100 μg/ml) in a humidified chamber at 37°C and 5% CO2.
DNA transfection
Co-transfection of pGL-3 (Firefly luciferase reporter) with the shRNA vector was performed in 96-well format. Per well, 2 × 104 293T cells were seeded in 100 µl DMEM with 10% FCS without antibiotics. The next day, 25 ng pGL-3, 10 ng shRNA vector and 0.5 ng pRL (Renilla luciferase reporter) were transfected using 0.5 µl Lipofectamine 2000 according to the manufacturer’s instructions (Life Technologies). Cells were lysed 48 h post transfection to measure firefly and renilla luciferase activities using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) according to the manufacturer’s instructions. The ratio between firefly and renilla luciferase activity was used for normalization of experimental variations such as differences in transfection efficiencies. Transfection experiments were corrected for between-session variations as described previously (56).
siRNA detection by Northern blotting
Northern blots were performed as previously described (57). Briefly, 1.5 × 106 HCT-116 cells were transfected with equimolar quantities (5 μg) of shRNA constructs using Lipofectamine 2000. Total cellular RNA was extracted 2 days post-transfection with the mirVana miRNA isolation kit (Life Technologies, Ambion, Austin, TX, USA) according to the manufacturer’s protocol. The RNA concentration was measured using the Nanodrop 1000 (Thermo Fisher Scientific). For Northern blot analysis, 15 μg total RNA or 1 µg of AGO2-immunoprecipitated RNA samples were electrophoresed in a 15% denaturing polyacrylamide gel (precast Novex TBU gel, Life Technologies). RNA molecular weight marker (Life Technologies) was prepared according to the manufacturer’s protocol and run alongside the cellular RNA. To check for equal sample loading, ribosomal RNA was stained with 2 µg/ml ethidium bromide and visualized under UV light. The RNA in the gel was electro-transferred to a positively charged nylon membrane (Boehringer Mannheim, GmbH, Mannheim, Germany) and cross-linked to the membrane using UV light at a wavelength of 254 nm (1200 µJ × 100). Hybridizations were performed at 42°C with radiolabelled locked nucleic acid (LNA) oligonucleotides in 10 ml ULTRAhyb hybridization buffer (Life Technologies, Austin, TX, USA) according to the manufacturer’s instructions. LNA oligonucleotide probes were 5′-end labelled with the kinaseMax kit (Life Technologies) in the presence of 1 µl [γ-32 P] ATP (0.37 MBq/µl, Perkin Elmer). To remove unincorporated nucleotides, the probes were purified on Sephadex G-25 spin columns (Amersham Biosciences) according to the manufacturer’s protocol. We used the following oligonucleotides to detect the antisense strand of the siRNA (LNA positions underlined): 5′-ATGGCAGGAAGAAGCGGAG-3′ (R/T5), 5′-GTGAAGGGGCAGTAGTAAT-3′ (pol47) and 5′-TAGCAGGAAGATGGCCAGT-3′ (pol9). To detect the sense strand of the siRNA, the following oligonucleotides were used (LNA positions underlined): 5′-CTCCGCTTCTTCCTGCCAT-3′ (R/T5), 5′-ATTACTACTGCCCCTTCAC-3′ (pol47) and 5′-ACTGGCCATCTTCCTGCTA-3′ (pol9). The signal was detected by autoradiography and quantified using a phosphorimager (Amersham Biosciences).
AGO2 co-immunoprecipitation of small RNAs
HCT-116 cells (5 × 106 cells) were co-transfected with 5 µg Flag-tagged AGO2 plasmid and 20 µg of the various shRNA vectors. At 36 h post-transfection, cytoplasmic cell extracts were prepared by the treatment of cells on ice for 20 min with IsoB-NP-40 [10 mM Tris-HCl (pH 7.9), 150 mM NaCl, 1.5 mM MgCl2, 1% NP-40] followed by a centrifugation at 12 000g for 10 min at 4°C. The supernatant was incubated with 75 µl of anti-FLAG M2 agarose beads (Sigma) with constant rotation overnight at 4°C. The beads were washed three times in NET-1 buffer [50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 2.5% Tween 20]. Small RNAs associated with AGO2 were isolated by phenol chloroform extraction followed by DNAse treatment using the TURBO DNA-free kit (Life Technologies). The RNAs were subsequently analysed on Northern blots (as described above) or cloned and sequenced.
Cloning of small RNAs
The purification and cloning strategy of small RNAs was previously described (58). Briefly, small RNAs were polyadenylated using the A-Plus™ Poly(A) Polymerase Tailing Kit (EPICENTRE Biotechnologies) according to the manufacturer’s protocol, and a 5′ adapter was ligated to the polyadenylated RNAs. Next, the 5′ adaptor-small RNA-poly(A)n molecule was reverse transcribed into cDNA followed by amplification of the cDNA by PCR. PCR products were gel-purified and subsequently used for TOPO-TA cloning (TOPO-TA cloning kit, Life Technologies). Positive clones were sequenced with the T7 or M13rp primer using the BigDye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems).
Results
Reversal of strand selection for a particular shRNA design
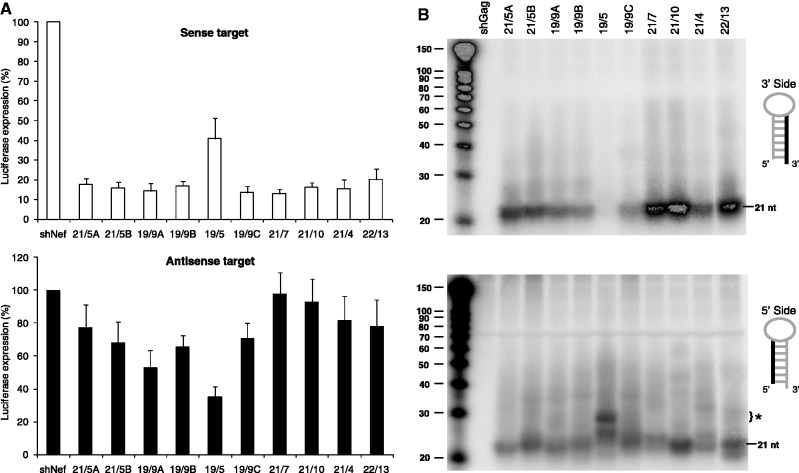
To study whether we could improve the shRNA design, we tested several variants of shPol47 (Figure 1) with the anti-HIV guide sequence encoded on the 3′ side of the hairpin duplex that targets a conserved sequence in the HIV-1 Pol gene. These shPol47 variants differ in length of the basepaired stem and terminal loop sequence. Several shRNA variants have been described in a previous study where we studied the impact of the terminal loop, varying in size and structure, on shRNA-induced gene silencing (39). We renamed the shRNAs according to the stem length and loop size, e.g. shPol47 with a stem length of 21 bp and a loop sequence of 5 nt was named 21/5. The 21/5 A design that was originally proposed by Brummelkamp and co-workers is commonly used in many laboratories (16,52,59–62). We tested both the knockdown activity mediated by the 5′ and 3′ side of the shRNA duplex by co-transfection with luciferase reporter constructs encoding the respective targets. An irrelevant shRNA (shNef) served as negative control that resulted in maximal luciferase expression (set at 100%). Most shPol47 variants efficiently inhibited gene expression of the luciferase reporter with the sense HIV-target and exercised little activity on the antisense reporter, indicating that the 3′ strand of the shRNA is indeed selected as guide (Figure 2A). However, the 19/5 design showed a significant reduction in sense luciferase knockdown with a concomitant increase in antisense luciferase knockdown. Moderate activity on the antisense reporter was also observed for three other variants with a shortened 19 bp duplex (19/9 A, B and C), but not for shRNAs with 21 bp. Thus, the 19 bp duplex seems to result in a shift in guide strand selection from the 3′ to the 5′ side of the shRNA.
Figure 1.
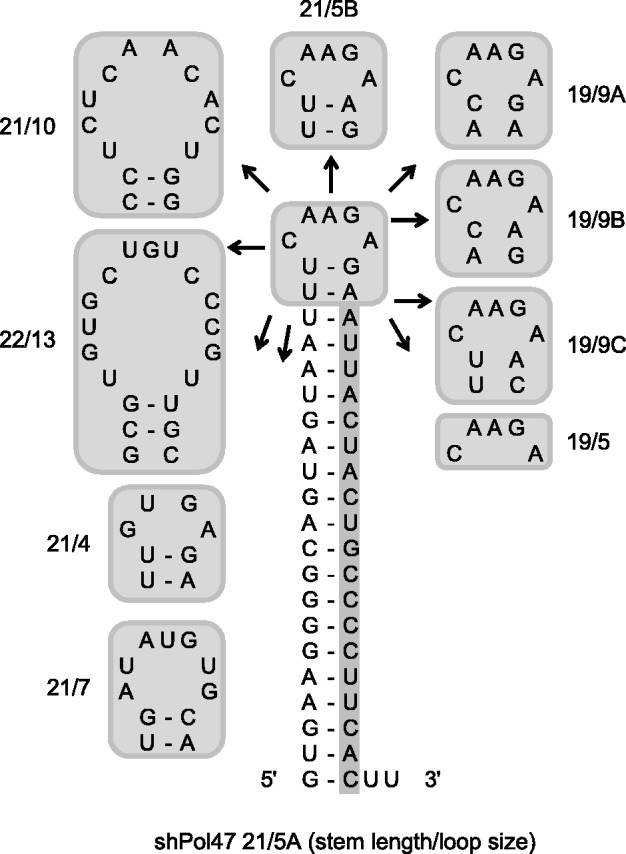
Design of a set of shPol47 variants. The encoded siRNA sequence targeting the Pol region of HIV-1 is boxed. The name of each shRNA indicates the stem length and loop size, e.g. a stem length of 19 bp and a loop sequence of 5 nt was named 19/5.
Figure 2.
Knockdown activity of the 3′/5′ strands of the shPol47 variants. (A) The knockdown activity of the 3′ and the 5′ strands of the different shRNAs was determined by co-transfection of a luciferase reporter encoding either the sense- or antisense-target sequence, respectively. 293T cells were co-transfected with 25 ng of the respective firefly luciferase reporter plasmid, 0.5 ng of renilla luciferase plasmid and 5 ng of the corresponding shRNA constructs. An irrelevant shRNA (shNef) served as negative control, which was set at 100% luciferase expression. (B) Processing of the 3′ strand (upper panel) and 5′ strand (lower panel) of shPol47 was analysed by Northern blot analysis. HCT-116 cells were transfected with 5 μg of the shRNA constructs. An irrelevant shRNA (shGag) was used as negative control. Northern blot analysis was performed on total cellular RNA. The RNA size marker (nt) is shown on the left. The regular 21 nt siRNA products and the new ∼30 nt product are indicated (marked as *).
To investigate this phenomenon in more detail, Northern blot analyses were performed to analyse the processing of the shPol47 variants. We used LNA oligonucleotides to detect shRNA products derived from the 3′ and 5′ side (Figure 2B, upper and lower panel, respectively). An irrelevant shRNA (shGag) served as negative control. We observed 3′ side-derived siRNAs (∼21 nt) for most shRNA variants, except for the 19/5 variant and to a lesser extent for the other 19/9 designs. This nearly complete loss of the 3′ signal (Figure 2B, top) correlated for construct 19/5 with reduced knockdown activity on the sense luciferase reporter (Figure 2A, top). A low level of regular ∼21 nt fragments was detected for all constructs with the 5′ side probe, reflecting the passenger strand. Interestingly, a new and abundant RNA fragment of ∼30 nt was observed for the 19/5 variant (marked * in Figure 2B, bottom), which coincides with the knockdown activity gained on the antisense luciferase reporter (Figure 2A, bottom). The size of this RNA product is not consistent with regular Dicer processing.
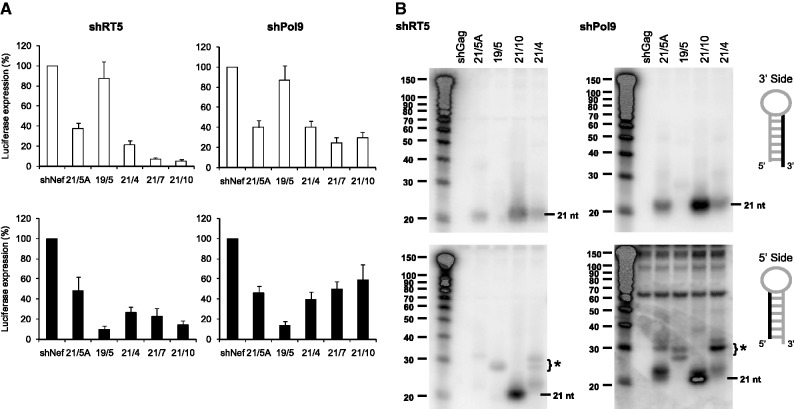
To study whether this phenomenon also applies to unrelated shRNAs with a completely different stem sequence, we tested several stem/loop variants of the validated shRT5 and shPol9 HIV-1 inhibitors for knockdown activity on the two reporters, including the special 19/5 design (Figure 3A). As expected, for most shRNAs, we observed more potent inhibition on the sense reporter (normal guide or 3′ side shRNA activity) than the antisense reporter (passenger or 5′ side shRNA activity). However, the activity of the 19/5 shRNA variants was shifted from 3′ towards the 5′ side, indicating a reversal of strand selection. Moreover, we again observed the loss of the regular 3′ side RNA fragment and appearance of a new ∼30 nt 5′ side RNA fragment for the 19/5 shRNAs (Figure 3B). These ∼30 nt RNA fragments do not resemble regular Dicer-processed RNA products. They seem to consist of 19 nt from the 5′ side of the shRNA, 5-nt loop and ∼6 nt from the upper 3′ side.
Figure 3.
Knockdown activity of the 3′/5′ strands of several shRT5 and shPol9 variants. (A) The knockdown activity of the 3′ and the 5′ strands of the indicated shRNAs was determined by co-transfection of a luciferase reporter encoding the sense (white bars) or antisense target sequence (black bars), respectively, in 293T cells. See Figure 2A for details. (B) Processing of the 3′/5′ strands of the indicated shRNAs was analysed by Northern blot analysis. We used LNA oligonucleotides to detect the 3′ (upper panel) and 5′ strand (lower panel) of the siRNA. An irrelevant shRNA (shGag) was used as negative control. The regular 21 nt products are marked and the * indicates the ∼30 nt RNA products.
The hairpin stem length is critical for alternative shRNA processing
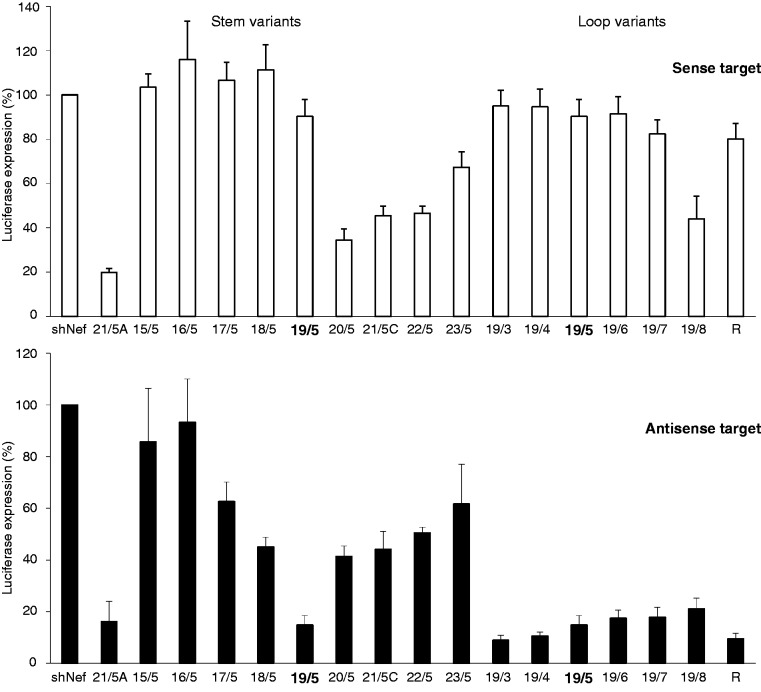
The shRNA mutant data indicate that variation in the stem and/or loop can induce alternative shRNA processing. An additional set of shRNAs was designed to critically test this idea. The stem of shRT5 was shortened or extended at the bottom of the hairpin (Figure 4, stem variants 15/5–23/5). The hairpin loop was varied between 3 and 8 nt (Figure 4, loop variants 19/3–19/8), and in addition, we reversed the 5-nt loop sequence (19/5R). We tested the knockdown activity of the 5′ and 3′ strands of these shRNAs on the respective luciferase reporter targets. The irrelevant shNef served as negative control for which luciferase expression was set at 100%. The original 21/5 A hairpin design was included as a positive control. The guide strand switch from 3′ to 5′ side, resulting in a switch from knockdown on the sense to antisense reporter, was apparent for all constructs with a 19 bp duplex (Figure 5, loop variants 19/3–19/8). A partial reversal of strand activity was seen for the shRNAs with a duplex of 17 or 18 bp, but all RNAi activity was lost for shorter shRNAs (15/5 and 16/5). The shRNAs of at least 20 bp do show the regular 3′ side activity on the sense reporter (20/5–23/5). The loop size does clearly modulate the 5′ side activity on the antisense reporter, with optimal activity for 19/3 and a gradual decrease towards larger loops. In fact, 19/7 and 19/8 seem to regain some regular 3′ side activity at the expense of 5′ side activity.
Figure 4.
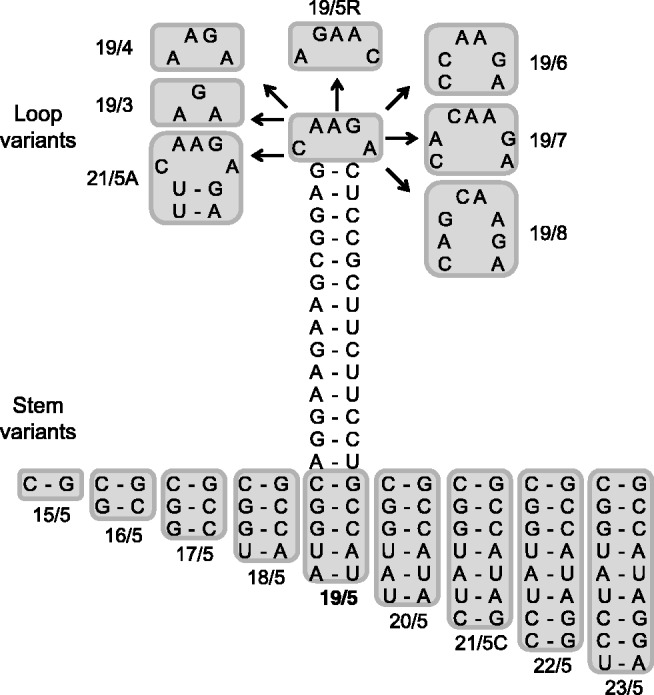
Design of additional shRT5 mutants varying in loop size and stem length. The shRT5 with a 19 bp stem and a 5-nt loop (19/5) was used as backbone for this design. The shRNA stem length was reduced/extended from the bottom of the hairpin, resulting in shRT5 variants 15/5–23/5. In addition, shRNA terminal loops ranging in size from 3 to 8 nt were designed (19/3–19/8) and the loop sequence was reversed (19/5R). The original shRT5 21/5A variant was also included.
Figure 5.
Knockdown activity of the shRT5 variants. RNAi activity of the 3′ side (lower panel) and 5′ side (upper panel) of the shRT5 was determined by co-transfection of a luciferase reporter encoding the sense or antisense target sequence, respectively, in 293T cells. See Figure 2A for details.
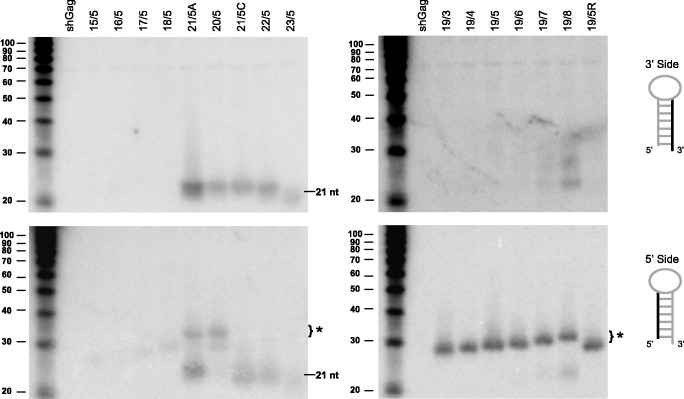
We next investigated the processing of these shRT5 variants by Northern blot analyses. We observed regular 3′ strand siRNA production (∼21 nt) for hairpins with a stem length of 20–23 bp, with the original 21/5 A design being the most optimal (Figure 6, upper, left panel). These shRNAs also showed some passenger 5′ strand ∼21 nt siRNA production (Figure 6, lower, left panel). Consistent with the luciferase knockdown data, we observed no 3′ strand siRNA production for the 19/3–19/6 loop mutants, but a modest regain of regular siRNA production (∼21 nt) was apparent for 19/7 and especially 19/8 (Figure 6, upper, right panel). Loss of regular 3′ side product coincides with the appearance of the new ∼30 nt product from the 5′ side (Figure 6, lower, right panel). Note that the size of this new product gradually increases for the loop variants (19/3–19/8), demonstrating that the loop nucleotides are an integral part of this new processing product. The 17/5 and 18/5 variants also produced some 5′ strand-derived RNAs of ∼30 nt. The 20/5 and 21/5 A variants showed weak expression of the ∼30 nt RNA fragment (Figure 6, lower, left panel). These shRNAs are therefore likely processed in different ways by the conventional and a novel processing route. The shRT5 variants of 15 and 16 bp did not show any 3′ or 5′ siRNA expression, consistent with the lack of knockdown activity.
Figure 6.
The processing pattern and production of the 3′ and 5′ strand of the shRT5 variants. Processing of the 3′ strand (upper panel) and 5′ strand (lower panel) of shRT5 variants was analysed by Northern blot analysis. See Figure 2B for details.
These combined results indicate that an shRNA with minimal duplex length (19 > 18 > 17 bp) in combination with a small loop (3 > 4 > 5 > 6 > 7 nt) favours an alternative shRNA-processing route. The alternative processing route produces an RNA fragment that does not resemble conventional Dicer-processed shRNA products and most likely consists of 19 nt 5′ side, the terminal loop and ∼6–8 nt from the upper 3′ side of the shRNA.
AGO2-mediated processing of 17–19 basepair shRNAs
The results described above indicate that the new shRNA-processing products of ∼30 nt are active in the RNAi mechanism and are causing efficient knockdown of reporter gene expression, which implies that they end up in the mature RISC complex and associate with AGO2. Recent miRNA and shRNA studies implicated AGO2 in the processing step (45–50). To test this for our 19/5 shRNA design and to determine the exact nature of the processed RNA fragments, we performed immunoprecipitation and sequencing of the AGO2-associated small RNAs. HCT-116 cells were co-transfected with FLAG-tagged AGO2 plasmid and a vector encoding the shRT5 variants 19/5 or 21/5 A. At 36 h post transfection, cytoplasmic cell extracts were prepared and the AGO2 complexes were captured with an anti-FLAG M2 agarose resin. The RNA content of purified AGO2 complexes was extracted, cloned and subsequently sequenced to determine the identity. Many of the small RNA sequences found in AGO2 complexes were of miRNA origin that served as positive controls. The identified small RNA sequences that originate from either shRT5 19/5 or 21/5 A are listed in Table 1. Perhaps surprisingly, no regular 21 nt siRNAs were detected for these shRNAs. However, shRNA 19/5 yielded seven identical sequences that represent the new siRNA of 33 nt including the complete 5′ strand, terminal loop and part of the 3′ strand of the shRNA. Thus, the shRNA is cleaved half-way the 3′ side of the duplex. This finding implicates the involvement of AGO2, as its slicer activity will cleave duplexes between nucleotide 10 and 11 from the 5′ end of the guide strand, which coincides exactly with the identified cleavage site. A similar fragment was observed for shRT5 21/5 A (1×), albeit 37 nt in length due to the extra 2 bp. Both the 33 and 37 nt RNA fragments were observed on Northern blot (Figure 6, lower right and left panel).
Table 1.
AGO2-associated small RNAs from transfected HCT-116 cells
| AGO2 product | shRT5 19/5 | shRT5 21/5A |
|---|---|---|
| n = 48a | n = 48a | |
| 5′ 21 nt siRNA | 0× | 0× |
| 3′ 21 nt siRNA | 0× | 0× |
| ∼30 nt siRNA | 7× (33 nt) | 1× (37 nt) |
| miRNA | 4× | 16× |
| tRNA | 1× | – |
| Other RNAs (rRNA, mitRNA) | 4× | 1× |
aA total of 48 clones were sequenced.
To demonstrate that the catalytic slicer activity of AGO2 is indeed required for this alternative shRNA-processing path, we compared the small RNA products processed from the shRT5 19/5 and 21/5A hairpins in cells transfected with the wild-type AGO2 (wt-AGO2) or the catalytically dead AGO2 mutant (cat-AGO2) (55). Additionally, we included the F181A AGO2 protein with a mutation in the N domain (N-AGO2) that is defective in siRNA unwinding (55). Cytoplasmic extracts were prepared from cells expressing the different AGO2 constructs and shRT5 variant 19/5 or 21/5A. The AGO2 complexes were immunoprecipitated, and the small RNAs were isolated for Northern blot analysis. We first probed for the regular 3′ side-derived shRNA products. Moderate ∼21 nt 3′ side siRNA and precursor shRNA signals were detected for 21/5A in association with the wt and mutant AGO2 proteins (Figure 7, upper panel). The highest amount of ∼21 nt RNAs was detected in association with the catalytically deficient AGO2 mutant. For the 19/5 hairpin, neither precursor nor 3′ side-processed siRNAs could be detected in association with the wt-, N- or cat-AGO2 protein. These results are consistent with the processing pattern previously observed for 19/5 shRNAs (Figures 2B, 3B and 6).
Figure 7.
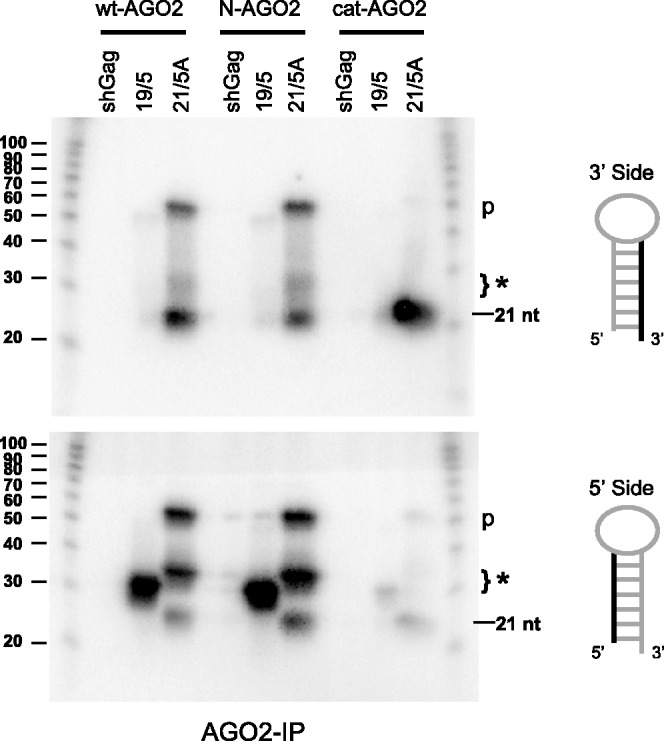
The short RNAs derived from the 3′ and 5′ strand of the shRT5 19/5 and 21/5A that associate with wild-type and mutant AGO2 proteins. AGO2-IP experiments were performed on cell lysates from HCT-116 cells co-transfected with shRT5 19/5 or 21/5A together with the wild-type (wt-AGO2), or mutant AGO2 proteins (N-AGO2 or cat-AGO2). The small RNAs associated with the AGO2 proteins were extracted and used for Northern blot analyses to detect the 3′ and 5′ strand of the shRNAs. The RNA size marker (nt) is shown on the left. The precursor shRNAs are indicated with a p and the new 33 or 37 nt product are indicated with *.
Using a probe to detect the 5′ side of the shRNAs, we exclusively detected alternatively processed 33 nt RNA fragments for the 19/5 shRNA in association with wt- and N-AGO2 proteins (Figure 7, lower panel), whereas nearly no 33 nt RNA was found in association with cat-AGO2. Notably, this product was present in whole cell extracts of cat-AGO2-transfected cells (data not shown). This observation indicates that AGO2 slicer activity is required for generation of the alternatively processed 33 nt RNA fragment. The weak 33 nt siRNA signal may represent processing by the endogenous wt AGO2 protein and subsequent uptake by the FLAG-tagged cat-AGO2 mutant. For the 21/5A construct, we detected the precursor shRNA, the alternatively processed 37 nt RNA fragment and the regular ∼21 nt 5′ side siRNAs in association with wt- and N-AGO2. This mixed profile of regular and alternative shRNA processing was previously observed for different 21/5A constructs (Figures 3B, 6 and 7). The catalytically dead AGO2 mutant contained little precursor 21/5A and 5′ siRNAs of ∼21 nt, whereas no 37 nt siRNAs were detected. This result confirms that AGO2 slicer activity is required for alternative shRNA processing. For unknown reasons, the catalytic AGO2 seems to favour the loading of ∼21 nt 3′ siRNAs (Figure 7, upper panel) derived from the 21/5A over 5′ siRNAs. Notably, mutation of the unwinding domain did not hinder alternative processing for both 19/5 and 21/5A shRNAs (Figure 7, lower panel).
DISCUSSION
The sequence and structure of natural miRNAs are important features that influence their processing and RNAi activity. Exact features in man-made shRNAs that determine the processing efficiency have not been examined in detail. The shRNA loop sequence is likely to have an effect on Dicer-mediated recognition and processing (37,39,63,64), but also the stem length is an important determinant for RNAi activity (51,65,66). We tested a variety of shRNAs and identified a specific shRNA design with a minimized stem length and a small loop that triggers an alternative processing route to form a new characteristic RNA fragment of 33 nt. The new cleavage event half-way the 3′ side of the shRNA duplex suggested a role for AGO2, which cleaves target mRNA or the passenger strand of an siRNA duplex between nt 10 and 11 from the 5′ end of the guide strand (10,67). Sequencing of AGO2-associated small RNAs confirmed this exact cleavage event. Such non-canonical processing has recently been reported for miR-451 and has been suggested for several other miRNAs and shRNAs (45–50). It would be interesting to compare the processing kinetics of AgoshRNAs versus conventional shRNAs and the underlying binding activities for Dicer and AGO2.
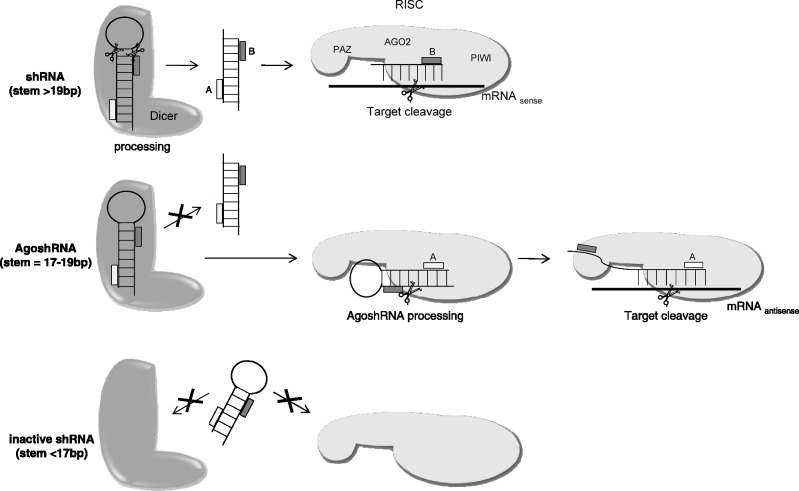
Production of the 33 nt RNA fragments was abolished in the presence of a catalytic AGO2 mutant, demonstrating that processing is mediated by AGO2 slicer activity. The minimized shRNAs of 17–19 bp are likely too small to be recognized by Dicer (34) and end up in AGO2 for alternative processing and subsequent RNAi silencing of an appropriate mRNA (Figure 8, middle panel). The new shRNA design that elicits this non-canonical processing pathway is termed AgoshRNA. We confirmed this specific AgoshRNA design for other shRNAs, demonstrating the universal value of the new shRNA design (19 bp stem and a small loop). Notably, this AgoshRNA processing yields only a single RNAi-active RNA fragment, whereas the conventional shRNA design potentially yields two active siRNA strands of ∼21 nt (Figure 8). This AgoshRNA feature is important to restrict RNAi-induced off target effects via the passenger strand of the siRNA.
Figure 8.
Alternative shRNA processing mechanisms. Processing of shRNAs by the RNAi machinery can use two competing pathways, depending largely on the shRNA stem length. For shRNAs >19 bp, processing occurs via the conventional pathway in which the shRNA is cleaved by Dicer into an siRNA (19–21 nt) followed by RISC incorporation and AGO2-mediated cleavage of the target mRNA. In principle, each siRNA strand can instruct RISC for cleavage, but there is usually a strand preference (e.g. the 3′ side of the shRNA 21/5A). For shRNAs of 17–19 bp, processing occurs independent of Dicer. We refer to this design as AgoshRNA. Consistent with several literature reports, such minimal shRNA templates are not efficient substrates for Dicer (34,51). We propose that the AgoshRNA is loaded into RISC and cleaved by AGO2 on the 3′ side, resulting in the new 33 nt RNA product that, upon unfolding, can instruct target mRNA cleavage. RNAi is mediated by AGO2 cleavage of the antisense target mRNA. Thus, AGO2 is instrumental twice, for the shRNA processing (new function) and mRNA cleavage (old function). If the shRNA is <17 bp, no processing by the RNAi machinery occurs.
The frequently used shRNA 21/5A design (16) exhibited a mixed processing profile according to the conventional Dicer and the alternative AGO2 mechanism. This may relate to the fact that the upper 2 bp of this shRNA design are relatively weak (U-G and U-A). Thus, this hairpin may exist in solution as a mixture of structures with 19, 20 and 21 bp. The conformers with 19 bp stem will be alternatively processed, whereas conformers with an extended stem of 20 or 21 bp will be processed conventionally. A recent report by Yang et al. (48) showed that a conventional hairpin of 21 bp with similar design was alternatively processed in a Dicer-independent manner. However, this study only addressed the required specifics to induce alternative processing for pre-miRNAs and not for shRNAs.
We described the specific AgoshRNA design that bypasses the Dicer endonuclease and instead uses AGO2 for processing. During processing of the AgoshRNA, AGO2 cleaves in the 3′ strand of the hairpin and the short 3′ end product is likely degraded. The remaining 33 nt RNA is used as a guide to target the complementary mRNA for cleavage. The determinants for AGO2 loading of the unprocessed AgoshRNA hairpin are the stem lengths and loop sizes. Our results further support the notion that RISC loading does not require prior Dicer-mediated processing of the RNAi inducer (17,68).
The loop size of the AgoshRNA molecules has a modest influence on its activity with higher knockdown efficiencies observed for smaller loops. Increasing the loop size from 3 to 7 or 8 nt caused a partial return to regular Dicer processing as detected by Northern blot analysis, suggesting that the two processing routes are in competition. We hypothesize that a large loop (e.g. >7 nt) sterically hinders the efficient uptake of AgoshRNA by AGO2 (Figure 8, middle panel). Consistent with our findings that increasing the loop size of the 19 bp shRNAs can cause a shift from alternative to conventional processing, pre-miRNAs appear to be more efficiently processed by Dicer when they contain larger loops (69).
It seems that alternatively processed AgoshRNAs can be more active in RNAi knockdown experiments than conventional shRNAs. More potent target knockdown was observed in the luciferase reporter assays with the 17–19 bp minimized AgoshRNA compared with the 21 bp regular shRNAs, but a direct comparison remains difficult, as different guide strands are generated that are probed on different reporter constructs. The design of such optimized AgoshRNA therapeutics may allow one to reduce the RNA dosage, thus reducing the chance of adverse effects, e.g. due to off targeting (70). Increased activity may be related to the extended length of the 33 nt AgoshRNA products compared with ∼21 nt shRNA products, although this does not extend the basepairing potential with the targeted mRNA reporter. It remains possible that a further increase of the basepairing complementarity of the ∼30 nt AgoshRNA fragment with the mRNA target, e.g. by loop sequence adaptation, will further increase the RNAi efficiency. Increased activity may also be related to the production of only a single guide strand, which will also reduce the chance of off-target effects.
AgoshRNAs may have additional theoretical benefits as AgoshRNAs form smaller hairpin duplexes than regular shRNAs. The former may exhibit a better safety profile concerning activation of the dsRNA-induced protein kinase R (PKR)/interferon pathways (71). Because AgoshRNAs do not mature via Dicer, they also do not compete with this aspect of miRNA biogenesis. AGO2-mediated processing may also yield more distinct RNA molecules as Dicer creates imprecise ends (72). Finally, AgoshRNAs are attractive molecules to silence target genes in Dicer-compromised cells, and they represent the only option in Dicer-deficient cells, which relates to a growing number of human diseases and cancers (73–75) and monocytes that lack Dicer expression (76).
In conclusion, we describe a specific shRNA design that yields only a single active strand (guide), as the other strand (passenger) is destroyed by AGO2 cleavage. This feature is an important property to restrict off-target effects induced by the passenger strand. Experiments are ongoing to test whether these AgoshRNAs are safer than conventional shRNAs and whether conversion of the regular loop sequences into anti-HIV sequences will generate more potent and possibly escape-proof antivirals.
FUNDING
RNAi research in the Berkhout laboratory is sponsored by the Nederlandse Organisatie voor Wetenschappelijk Onderzoek Chemische Wetenschappen (NWO-CW) (Top grant) and ZonMW (Translational Gene Therapy program). Funding for open access charge: NWO-CW (Top grant).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank the members of the RNAi group for stimulating discussions and useful suggestions. Y.P.L. performed and designed experiments, analysed data and wrote the manuscript. N.C.T.S. performed and designed experiments, analysed data and wrote the manuscript. B.B. designed experiments, provided funding and wrote the manuscript.
REFERENCES
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Napoli C, Lemieux C, Jorgensen R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell. 1990;2:279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 4.Nykanen A, Haley B, Zamore PD. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 5.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 6.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 7.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 9.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 11.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 12.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 13.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 14.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 15.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 16.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 17.Yoda M, Kawamata T, Paroo Z, Ye X, Iwasaki S, Liu Q, Tomari Y. ATP-dependent human RISC assembly pathways. Nat. Struct. Mol. Biol. 2010;17:17–23. doi: 10.1038/nsmb.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burroughs AM, Ando Y, de Hoon MJ, Tomaru Y, Suzuki H, Hayashizaki Y, Daub CO. Deep-sequencing of human Argonaute-associated small RNAs provides insight into miRNA sorting and reveals Argonaute association with RNA fragments of diverse origin. RNA Biol. 2011;8:158–177. doi: 10.4161/rna.8.1.14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel DJ. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frank F, Sonenberg N, Nagar B. Structural basis for 5'-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010;465:818–822. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- 21.Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5'-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA. 2010;16:43–56. doi: 10.1261/rna.1972910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 24.Hu HY, Yan Z, Xu Y, Hu H, Menzel C, Zhou YH, Chen W, Khaitovich P. Sequence features associated with microRNA strand selection in humans and flies. BMC. Genomics. 2009;10:413. doi: 10.1186/1471-2164-10-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okamura K, Liu N, Lai EC. Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol. Cell. 2009;36:431–444. doi: 10.1016/j.molcel.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 29.Ender C, Meister G. Argonaute proteins at a glance. J. Cell Sci. 2010;123:1819–1823. doi: 10.1242/jcs.055210. [DOI] [PubMed] [Google Scholar]
- 30.Braun JE, Truffault V, Boland A, Huntzinger E, Chang CT, Haas G, Weichenrieder O, Coles M, Izaurralde E. A direct interaction between DCP1 and XRN1 couples mRNA decapping to 5' exonucleolytic degradation. Nat. Struct. Mol. Biol. 2012;19:1324–1331. doi: 10.1038/nsmb.2413. [DOI] [PubMed] [Google Scholar]
- 31.Zeng Y, Cullen BR. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 2004;32:4776–4785. doi: 10.1093/nar/gkh824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li L, Lin X, Khvorova A, Fesik SW, Shen Y. Defining the optimal parameters for hairpin-based knockdown constructs. RNA. 2007;13:1765–1774. doi: 10.1261/rna.599107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boudreau RL, Martins I, Davidson BL. Artificial microRNAs as siRNA shuttles: improved safety as compared to shRNAs in vitro and in vivo. Mol. Ther. 2009;17:169–175. doi: 10.1038/mt.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siolas D, Lerner C, Burchard J, Ge W, Linsley PS, Paddison PJ, Hannon GJ, Cleary MA. Synthetic shRNAs as potent RNAi triggers. Nat. Biotechnol. 2005;23:227–231. doi: 10.1038/nbt1052. [DOI] [PubMed] [Google Scholar]
- 35.Sun X, Rogoff HA, Li CJ. Asymmetric RNA duplexes mediate RNA interference in mammalian cells. Nat. Biotechnol. 2008;26:1379–1382. doi: 10.1038/nbt.1512. [DOI] [PubMed] [Google Scholar]
- 36.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McManus MT, Petersen CP, Haines BB, Chen J, Sharp PA. Gene silencing using micro-RNA designed hairpins. RNA. 2002;8:842–850. doi: 10.1017/s1355838202024032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hinton TM, Wise TG, Cottee PA, Doran TJ. Native microRNA loop sequences can improve short hairpin RNA processing for virus gene silencing in animal cells. J. RNAi Gene Silencing. 2008;4:295–301. [PMC free article] [PubMed] [Google Scholar]
- 39.Schopman NC, Liu YP, Konstantinova P, Ter Brake O, Berkhout B. Optimization of shRNA inhibitors by variation of the terminal loop sequence. Antiviral Res. 2010;86:204–211. doi: 10.1016/j.antiviral.2010.02.320. [DOI] [PubMed] [Google Scholar]
- 40.Miyagishi M, Sumimoto H, Miyoshi H, Kawakami Y, Taira K. Optimization of an siRNA-expression system with an improved hairpin and its significant suppressive effects in mammalian cells. J.Gene Med. 2004;6:715–723. doi: 10.1002/jgm.556. [DOI] [PubMed] [Google Scholar]
- 41.Li L, Lin X, Khvorova A, Fesik SW, Shen Y. Defining the optimal parameters for hairpin-based knockdown constructs. RNA. 2007;13:1765–1774. doi: 10.1261/rna.599107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawasaki H, Taira K. Short hairpin type of dsRNAs that are controlled by tRNA(Val) promoter significantly induce RNAi-mediated gene silencing in the cytoplasm of human cells. Nucleic Acids Res. 2003;31:700–707. doi: 10.1093/nar/gkg158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei JX, Yang J, Sun JF, Jia LT, Zhang Y, Zhang HZ, Li X, Meng YL, Yao LB, Yang AG. Both strands of siRNA have potential to guide posttranscriptional gene silencing in mammalian cells. PLoS One. 2009;4:e5382. doi: 10.1371/journal.pone.0005382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vlassov AV, Korba B, Farrar K, Mukerjee S, Seyhan AA, Ilves H, Kaspar RL, Leake D, Kazakov SA, Johnston BH. shRNAs targeting hepatitis C: effects of sequence and structural features, and comparision with siRNA. Oligonucleotides. 2007;17:223–236. doi: 10.1089/oli.2006.0069. [DOI] [PubMed] [Google Scholar]
- 45.Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, Papapetrou EP, Sadelain M, O'Carroll D, Lai EC. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc. Natl Acad. Sci. USA. 2010;107:15163–15168. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang JS, Maurin T, Lai EC. Functional parameters of Dicer-independent microRNA biogenesis. RNA. 2012;18:945–957. doi: 10.1261/rna.032938.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dallas A, Ilves H, Ge Q, Kumar P, Shorenstein J, Kazakov SA, Cuellar TL, McManus MT, Behlke MA, Johnston BH. Right- and left-loop short shRNAs have distinct and unusual mechanisms of gene silencing. Nucleic Acids Res. 2012;40:9255–9271. doi: 10.1093/nar/gks662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dueck A, Ziegler C, Eichner A, Berezikov E, Meister G. microRNAs associated with the different human Argonaute proteins. Nucleic Acids Res. 2012;40:9850–9862. doi: 10.1093/nar/gks705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ge Q, Ilves H, Dallas A, Kumar P, Shorenstein J, Kazakov SA, Johnston BH. Minimal-length short hairpin RNAs: the relationship of structure and RNAi activity. RNA. 2010;16:106–117. doi: 10.1261/rna.1894510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ter Brake O, Konstantinova P, Ceylan M, Berkhout B. Silencing of HIV-1 with RNA interference: a multiple shRNA approach. Mol. Ther. 2006;14:883–892. doi: 10.1016/j.ymthe.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 53.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Westerhout EM, Ooms M, Vink M, Das AT, Berkhout B. HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucleic Acids Res. 2005;33:796–804. doi: 10.1093/nar/gki220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kwak PB, Tomari Y. The N domain of Argonaute drives duplex unwinding during RISC assembly. Nat. Struct. Mol. Biol. 2012;19:145–151. doi: 10.1038/nsmb.2232. [DOI] [PubMed] [Google Scholar]
- 56.Ruijter JM, Thygesen HH, Schoneveld OJ, Das AT, Berkhout B, Lamers WH. Factor correction as a tool to eliminate between-session variation in replicate experiments: application to molecular biology and retrovirology. Retrovirology. 2006;3:1–8. doi: 10.1186/1742-4690-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu YP, Haasnoot J, Ter Brake O, Berkhout B, Konstantinova P. Inhibition of HIV-1 by multiple siRNAs expressed from a single microRNA polycistron. Nucleic Acids Res. 2008;36:2811–2824. doi: 10.1093/nar/gkn109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun G, Li H, Rossi JJ. Cloning and detecting signature microRNAs from mammalian cells. Methods Enzymol. 2007;427:123–138. doi: 10.1016/S0076-6879(07)27007-7. [DOI] [PubMed] [Google Scholar]
- 59.Ter Brake O, 't Hooft K, Liu YP, Centlivre M, von Eije KJ, Berkhout B. Lentiviral vector design for multiple shRNA expression and durable HIV-1 Inhibition. Mol. Ther. 2008;16:557–564. doi: 10.1038/sj.mt.6300382. [DOI] [PubMed] [Google Scholar]
- 60.Huang M, Chan DA, Jia F, Xie X, Li Z, Hoyt G, Robbins RC, Chen X, Giaccia AJ, Wu JC. Short hairpin RNA interference therapy for ischemic heart disease. Circulation. 2008;118:S226–S233. doi: 10.1161/CIRCULATIONAHA.107.760785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Asparuhova MB, Barde I, Trono D, Schranz K, Schumperli D. Development and characterization of a triple combination gene therapy vector inhibiting HIV-1 multiplication. J. Gene Med. 2008;10:1059–1070. doi: 10.1002/jgm.1238. [DOI] [PubMed] [Google Scholar]
- 62.Bernards R, Brummelkamp TR, Beijersbergen RL. shRNA libraries and their use in cancer genetics. Nat. Methods. 2006;3:701–706. doi: 10.1038/nmeth921. [DOI] [PubMed] [Google Scholar]
- 63.Vermeulen A, Behlen L, Reynolds A, Wolfson A, Marshall WS, Karpilow J, Khvorova A. The contributions of dsRNA structure to Dicer specificity and efficiency. RNA. 2005;11:674–682. doi: 10.1261/rna.7272305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu YP, von Eije KJ, Schopman NC, Westerink JT, Ter Brake O, Haasnoot J, Berkhout B. Combinatorial RNAi against HIV-1 using extended short hairpin RNAs. Mol. Ther. 2009;17:1712–1723. doi: 10.1038/mt.2009.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McIntyre GJ, Yu YH, Lomas M, Fanning GC. The effects of stem length and core placement on shRNA activity. BMC. Mol. Biol. 2011;12:34. doi: 10.1186/1471-2199-12-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 68.Ye X, Huang N, Liu Y, Paroo Z, Huerta C, Li P, Chen S, Liu Q, Zhang H. Structure of C3PO and mechanism of human RISC activation. Nat. Struct. Mol. Biol. 2011;18:650–657. doi: 10.1038/nsmb.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Feng Y, Zhang X, Graves P, Zeng Y. A comprehensive analysis of precursor microRNA cleavage by human Dicer. RNA. 2012;18:2083–2092. doi: 10.1261/rna.033688.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 71.Bridge AJ, Pebernard S, Ducraux A, Nicoulaz AL, Iggo R. Induction of an interferon response by RNAi vectors in mammalian cells. Nat. Genet. 2003;34:263–264. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- 72.Gu S, Jin L, Zhang Y, Huang Y, Zhang F, Valdmanis PN, Kay MA. The loop position of shRNAs and Pre-miRNAs is critical for the accuracy of dicer processing in vivo. Cell. 2012;151:900–911. doi: 10.1016/j.cell.2012.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pampalakis G, Diamandis EP, Katsaros D, Sotiropoulou G. Down-regulation of dicer expression in ovarian cancer tissues. Clin. Biochem. 2010;43:324–327. doi: 10.1016/j.clinbiochem.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 74.Karube Y, Tanaka H, Osada H, Tomida S, Tatematsu Y, Yanagisawa K, Yatabe Y, Takamizawa J, Miyoshi S, Mitsudomi T, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hill DA, Ivanovich J, Priest JR, Gurnett CA, Dehner LP, Desruisseau D, Jarzembowski JA, Wikenheiser-Brokamp KA, Suarez BK, Whelan AJ, et al. DICER1 mutations in familial pleuropulmonary blastoma. Science. 2009;325:965. doi: 10.1126/science.1174334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Coley W, Van DR, Carpio L, Guendel I, Kehn-Hall K, Chevalier S, Narayanan A, Luu T, Lee N, Klase Z, et al. Absence of DICER in monocytes and its regulation by HIV-1. J. Biol. Chem. 2010;285:31930–31943. doi: 10.1074/jbc.M110.101709. [DOI] [PMC free article] [PubMed] [Google Scholar]