Abstract
As researchers begin probing deep coverage sequencing data for increasingly rare mutations and subclonal events, the fidelity of next generation sequencing (NGS) laboratory methods will become increasingly critical. Although error rates for sequencing and polymerase chain reaction (PCR) are well documented, the effects that DNA extraction and other library preparation steps could have on downstream sequence integrity have not been thoroughly evaluated. Here, we describe the discovery of novel C > A/G > T transversion artifacts found at low allelic fractions in targeted capture data. Characteristics such as sequencer read orientation and presence in both tumor and normal samples strongly indicated a non-biological mechanism. We identified the source as oxidation of DNA during acoustic shearing in samples containing reactive contaminants from the extraction process. We show generation of 8-oxoguanine (8-oxoG) lesions during DNA shearing, present analysis tools to detect oxidation in sequencing data and suggest methods to reduce DNA oxidation through the introduction of antioxidants. Further, informatics methods are presented to confidently filter these artifacts from sequencing data sets. Though only seen in a low percentage of reads in affected samples, such artifacts could have profoundly deleterious effects on the ability to confidently call rare mutations, and eliminating other possible sources of artifacts should become a priority for the research community.
INTRODUCTION
Recent technical developments (1–4) and decreasing costs have enabled cost effective deep sequencing coverage of the gene-coding regions of the human genome across a large number of samples. This ultra-deep coverage of the human exome enables researchers to push beyond previous biological limitations such as stromal admixture or clonal heterogeneity to robustly detect somatic mutations present in a lower fraction of the cells. Some recently identified low allelic fraction events appear to play important roles in cancer initiation and progression (5–9), indicating that routine characterization of these events will become increasingly critical to the interpretation of cancer genomes.
However, there are many challenges to the robust detection of events present in only a few percentage of cells, including, but not limited to algorithmic limitations, sequencing errors, and sample preparation artifacts. Error rates for the Illumina HiSeq sequencer chemistry are low and well understood (10) as is the sensitivity of commonly used analysis tool suites [Cancer Genome Analysis Toolkit: http://confluence.broadinstitute.org/display/CGATools/Home, (11)]. Additionally, the most commonly used enzymes in next generation sequencing (NGS) sample preparation for both fragment end polishing (12) and PCR (13–15) have been thoroughly characterized for fidelity and error rate. However, it is less well known how other common processes and reagents used during DNA extraction and sample preparation might affect the fidelity of downstream mutation calling. A survey of the literature reveals that no studies have been published to date, specifically examining the effects that DNA extraction methods, storage conditions or DNA fragmentation could have on the fidelity of low frequency mutation calling in NGS data. The possible error rates of these processes are currently unknown.
Here, we describe the discovery and characterization of a previously unreported source of artifactual mutations occurring during the NGS sample preparation process. We detail the analysis methods used to discover the artifact in ultra-deep coverage-targeted capture sequencing data and present novel sequence data metrics that can be used to detect and measure these artifacts in the primary analysis pipeline, before mutation calling. We outline the experimental results, which elucidated the source of the artifact as an oxidative mechanism during high-powered DNA shearing and demonstrate detection of abnormally high levels of the oxidation product 8-oxog in affected samples following shearing. We also describe recommended laboratory process changes that can be readily adopted to reduce opportunities for oxidation in the NGS sample preparation and propose analytical methods for identifying and screening out obvious oxidation artifacts already present in Illumina sequence data. Finally, we discuss how such artifacts could adversely affect the ability to identify true rare somatic mutations and the impact that the discovery of process induced artifacts could have on protocol development in the NGS field.
MATERIALS AND METHODS
Illumina sequencing library preparation and Agilent SureSelect targeted capture process
Automated Illumina DNA library construction was performed as described by Fisher et al. (4) with the following modifications: (i) initial genomic DNA input into shearing was reduced from 3 µg to 100 ng in 50 µl and (ii) for adapter ligation, Illumina paired-send adapters were replaced with palindromic forked adapters with unique eight base index sequences embedded within the adapter to enable library multiplexing before sequencing. DNA shearing too was performed on a Covaris E210 instrument using standard crimp-cap AFA vessels. To achieve the 150-bp fragment size for targeted capture libraries, the Covaris was programed with the following settings: 20% duty cycle, intensity 5 and cycles per burst 200 for 165 s. For 500-bp shearing for whole genome shotgun libraries, the Covaris settings were as follows: 1% duty cycle, intensity 5 and cycles per burst 200 for 300 s. Automated targeted hybridization and capture was performed using the Agilent SureSelect system also as described in Fisher et al. (4).
Illumina cluster amplification, sequencing and data processing
Following sample preparation, libraries were quantified using quantitative PCR with primers specific to the ends of the Illumina adapters (KAPA Biosystems). Based on qPCR quantification, libraries were normalized to 2 nM and then denatured using 0.1 N NaOH. Cluster amplification of denatured templates was performed according to the manufacturer’s protocol (Illumina) using HiSeq 2000 v2, HiSeq v3 or MiSeq cluster chemistry and flowcells. HiSeq flowcells were sequenced on HiSeq 2000 instruments using HiSeq v2 or v3 Sequencing-by-Synthesis Kits, then analysed using RTA v1.10.15 or RTA v.1.12.4.2. MiSeq flowcells were amplified and sequenced on MiSeq instruments running Control Software v1.1.1 and analysed with the Illumina RTA v1.13 pipeline. For pooled libraries prepared using forked, indexed adapters, Illumina’s Multiplexing Sequencing Primer Kit was used, and a third, 8-base sequencing read was performed to read the 8-base molecular indices. Output from Illumina software was processed by the Picard data-processing pipeline to yield Binary Alignment Map (BAM) files containing well-calibrated, aligned reads. The Artifact-Q (ArtQ) script as described in the Results section was then run on the annotated BAM files generated by Picard.
Sample preparation and sequencing on the Ion Torrent Personal Genome Machine (PGM)
For this analysis, Ion Torrent libraries were created by ligating Ion Torrent PGM paired-end adapters onto already made Illumina libraries. Preparative Ion Sphere templating and PGM sequencing was performed according to manufacturer’s recommended protocols. The Illumina adapter sequence was trimmed from the sequencing reads before alignment and artifact detection.
Sequence analysis and mutation calling
A detailed description of the sequence analysis methods for Illumina data used here has been previously described (16). Owing to low purity, genome complexity and subclonal mutations found in cancer samples, this analysis workflow is tuned to detect low allelic fraction variants. Briefly, sequencing reads were aligned to the human genome reference GRCh37 (hg19) using bwa (17), quality scores recalibrated using the Genome Analysis Toolkit (11) and sequencing metrics calculated using the Picard suite of tools (http://picard.sourceforge.net). As additional quality controls, we ensured concordance of the sequence data with 24 single nucleotide polymorphisms (SNPs) independently genotyped by mass spectrometry (Sequenom) and copy number profiles derived from an Affymetrix SNP 6.0 microarray run for each sample. Possible cross-contamination of samples during library construction and sequencing was scored using ConTest (18). Somatic substitutions and indels were detected using muTect and IndelLocator, respectively. All mutation calls were annotated using Oncotator. These and other analysis tools are described on our website, http://www.broadinstitute.org/cancer/cga.
Enzyme-linked immunosorbent assay for 8-oxog
The enzyme-linked immunosorbent assay (ELISA) kit was obtained from Enzo Biosciences and contains a monoclonal antibody specific for 8-oxog in extracted DNA. All ELISA tests were run following the manufacturer’s recommended protocol with slight modifications: the amount of primary anti-8-oxoG antibody used was increased by 15%, and primary antibody incubation time was also increased to 2 h to improve sensitivity. For all ELISA experiments, 200 ng of total DNA from each sample as measured by Pico Green was used as input to the assay. Calculation of 8-oxoG concentration against the provided standard was performed as recommended by the manufacturer.
Bead-based buffer exchange and antioxidant evaluation
Buffer exchange of incoming samples before DNA shearing with the Covaris E210 was achieved by performing a solid phase reversible immobilization (SPRI) magnetic bead clean up by adding 50 ul of Ampure XP beads (Beckman Genomics) to 50 ul of DNA and following the standard manufacturer protocol. Following SPRI clean up genomic DNA samples were eluted in 50 ul of antioxidant buffers or in 10 mM Tris–HCl alone. Antioxidant agents ethylenediaminetetraacetic acid (EDTA), deforoxamine mesylate (DFAM) and butylated hydroxytoluene (BHT) were obtained from Sigma-Aldrich. For stock solutions, EDTA and DFAM were both dissolved in water and BHT in ethanol per manufacturer’s recommendations. Final concentrations of 1 mM EDTA, 100 uM DFAM and 100 uM BHT were added to the standard 10 mM Tris–HCl alone, in pairs, and all three together (see Results).
RESULTS
Discovery and initial characterization of a low allelic fraction artifact
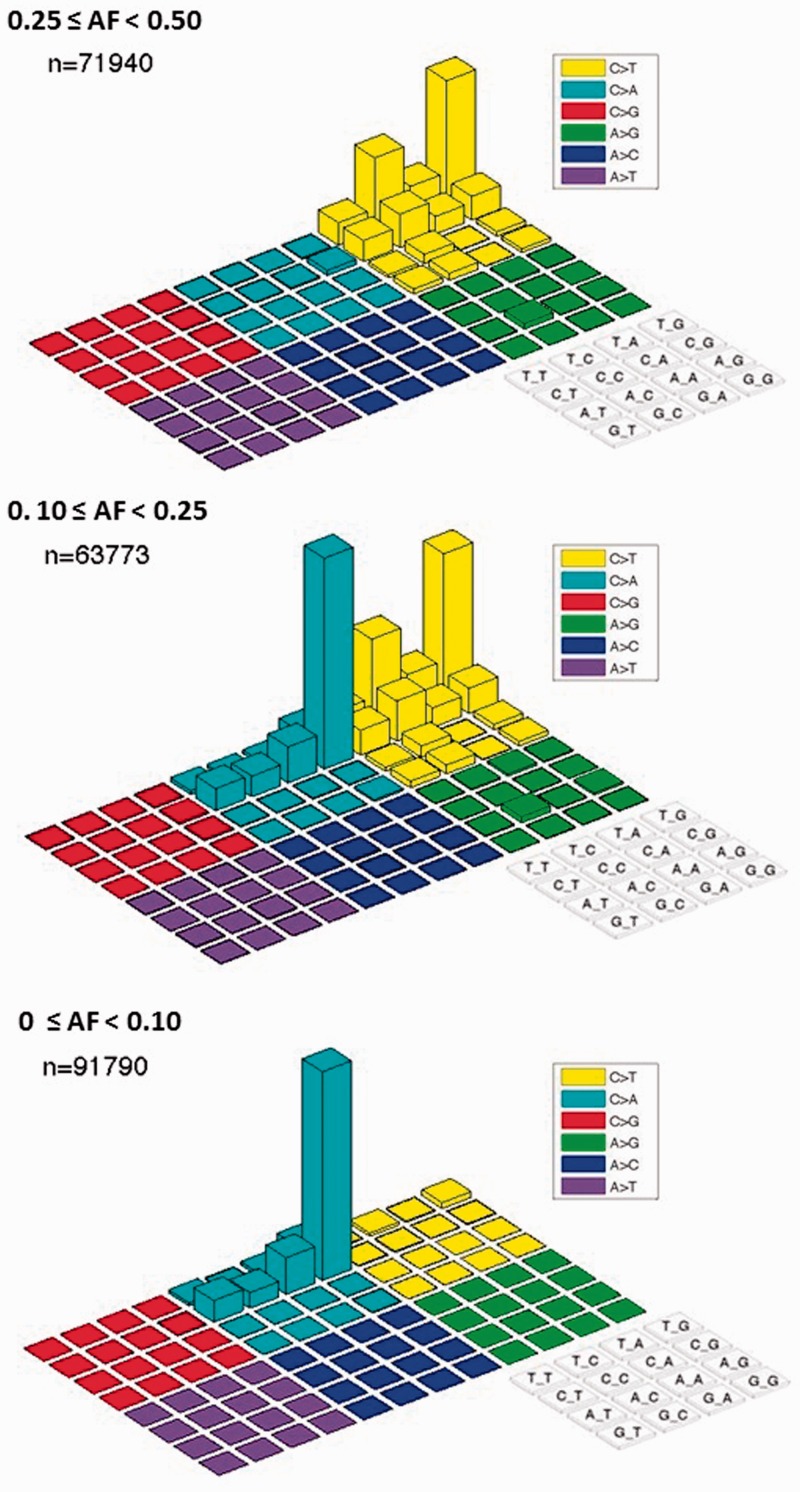
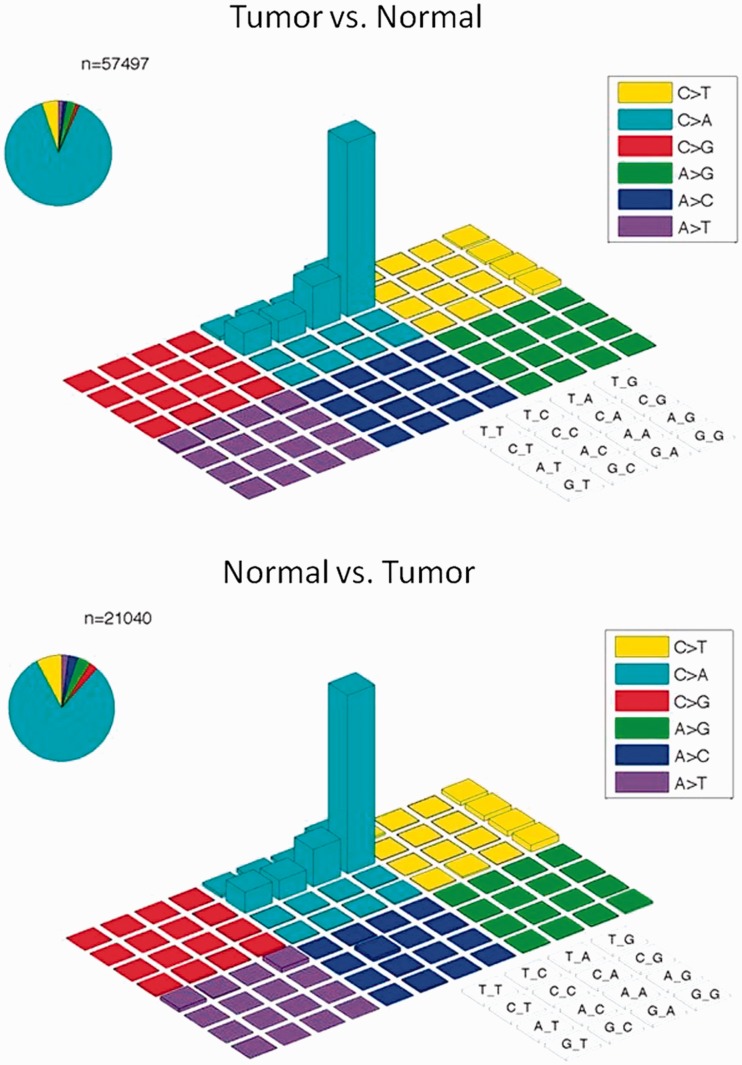
In a deep coverage exome study of 221 tumors and matched normal tissues from melanoma, we observed an unexpectedly high number of variants at allelic fractions <20% (Figure 1). These variants were not consistent with the expected dominance of C > T transitions in melanoma owing to ultraviolet damage as previously reported (19,20). Instead, the analysis uncovered thousands of apparent C > A/G > T variants in these samples with a specific sequence context of CCG > CAG, and most strikingly were not restricted to tumor material. They were also found at a similarly high rate when mutation calling was reversed to call ‘variants’ off the normal using the tumor as the reference, further indicating these base changes were likely not due to a disease mechanism (Figure 2). Further, we noted that the artifacts had a specific strand orientation in that G > T errors always presented in the first Illumina HiSeq instrument read, whereas the C > A errors were always found in the second HiSeq read. Lastly, these variants were not supported by matching RNA sequence data obtained from either the tumor or normal DNA of affected samples (data not shown). Closer inspection of deep coverage exome data from other cancer types also uncovered these same C > A/G > T transversions at low allelic fractions in subsets of samples, including cancers with known lower mutation rates such as neuroblastoma (21,22) and chronic lymphocytic leukemia (23–25) (data not shown). Combined, these characteristics led us to believe that these variants were not biological in nature. Rather, we hypothesized that these base changes were caused by some artifact induced in the sample collection, extraction, library preparation or sequencing processes.
Figure 1.
Distribution of mutation calls across a variety of base motifs and allelic fractions in Melanoma samples. Lego plots consisting of counts of all single base mutations for given bases and contexts at various allele fractions (AF) for 221 melanoma samples. Each colored region is a different mutation type where the reference base ‘C’ or ‘A’ listed includes the reverse complement ‘G’ or ‘T’. Within each colored region are the 4 × 4 combinations of possible preceding (5′) or trailing (3′) bases as labeled in the grid at the lower right. At allele fractions <10%, the CCG > CAG mutation dominates all other variants.
Figure 2.
Presence of CCG > CAG mutations in both tumor and normal samples. CCG > CAG mutations can also be detected at an abnormally high rate in this set of neuroblastoma samples when mutation calling was flipped to call variants off normal samples using the matched tumors as a reference.
To better understand the full scope of the artifact’s presence in exome data, we developed a sensitive and accurate metric using the unique context, strand specificity and read orientation characteristics of the artefact that could quickly process large data sets to measure the rate of C > A/G > T artifacts. We started by looking at all high quality (>Q20) base observations at C/G sites in the genome and classifying them into the following: (i) reference basecalls; (ii) alternative basecalls (A/T) that are consistent with the artifact characteristics; (iii) alternative basecalls (A/T) that are inconsistent with the artifact characteristics; and (iv) all other bases. As in aggregate there should be no correlated bias between C > A and G > T error modes and instrument read order for true biological variation, we assume that the errors from sources other than that of the artifact should be symmetric and arrive at the following definition of ArtQ:
This yields a phred scaled error rate attributable to G > T/A > C transversions with the specific artifact characteristics we had observed in our data. An ArtQ score >30 means that less than 1 in a thousand bases are attributable to artifact error. For the purposes of our analysis, we considered a sample with an ArtQ score >30 to be ‘unaffected’ by this artifact.
Determining the origin of the artifact
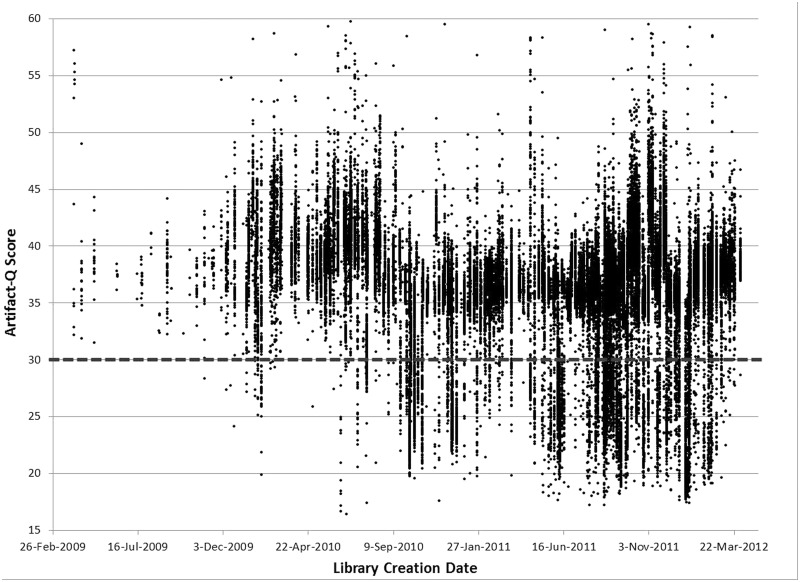
We calculated the ArtQ metric for nearly all human targeted capture and whole genome projects sequenced at our institute since 2009, encompassing >50 000 libraries from hundreds of different initiatives and disease projects (Figure 3). Immediately, it became clear that the prevalence of the artifact, though always there, had increased in frequency during the previous year, and that there was a large amount of project-to-project variation in the artifact prevalence (see Discussion). We began to investigate possible sources of error in the laboratory, and we were first able to rule out that the Illumina HiSeq and cluster amplification chemistry was not inducing these base changes by sequencing the same affected libraries on Illumina HiSeq V2, V3 and MiSeq chemistries, as well as on Life Technologies Ion Torrent PGM chemistry. The Illumina sequencing versions tested generated no significant differences in ArtQ scores for each library (Table 1). Although ArtQ scores could not be calculated in the same fashion for Ion data owing to the lack of pairing information, C > A base changes found in the samples at given base positions in Illumina data were also found in Ion PGM data for each library tested (data not shown). These observations strongly indicated that the base changes had occurred before the sequencing step.
Figure 3.
ArtQ metric over time for Broad’s Targeted Capture pipeline. ArtQ by library creation date.
Table 1.
Artifact-Q scores
| HiSeq V2 | HiSeq V3 | MiSeq | |
|---|---|---|---|
| ‘Affected’ library 1 | 18.9 | 19.0 | 19.2 |
| ‘Affected’ library 2 | 18.1 | 18.2 | 18.0 |
| ‘Unaffected’ library | 30.9 | 30.4 | 30.8 |
Sequencing chemistry does not induce C > A/G > T artifact. The same libraries were sequenced using HiSeq V2, HiSeq V3 and MiSeq flowcells. The ArtQ values for each library varied little between Illumina sequencing chemistries, indicating that sequencing was not the artifact source.
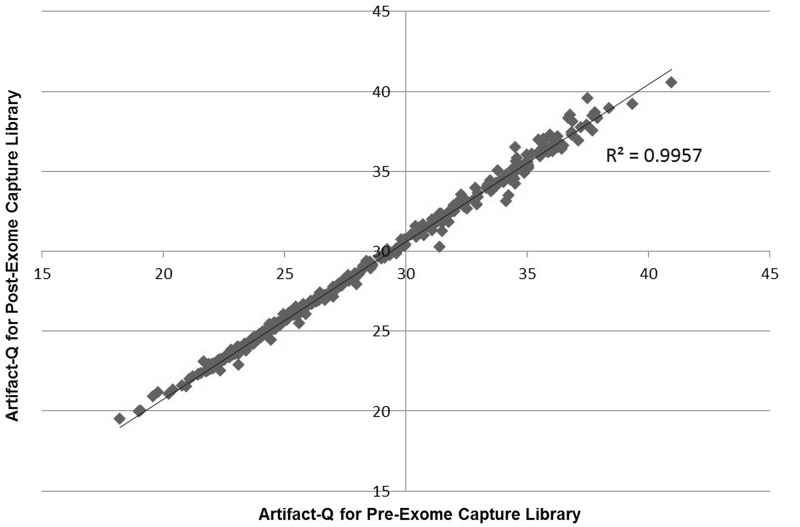
We then observed that whole genome samples contained little to no evidence of significant artifactual C > A/G > T transversions compared with targeted capture samples processed during the same period (Supplementary Figure S1). This finding then led us to investigate our automated SureSelect targeted enrichment process itself. We sequenced a pool of 370 pre-exome enrichment libraries along with their 370 corresponding post-exome enrichment captured libraries. We found that for all samples, the ArtQ scores for both were highly correlated (Figure 4), indicating that the base changes were already been induced upstream of targeted capture. Because we had streamlined our laboratory processes, the production protocols used to create libraries destined for either targeted capture or whole genome processing were nearly identical. The one exception was the acoustic shearing protocol used to fragment the DNA for either the exome process (a high powered 150-bp fragmentation) or whole genome (a lower powered 500-bp fragmentation) (see Methods).
Figure 4.
ArtQ for Pre- versus Post-targeted capture. For a set of 370 samples, both the pre- and post-exome enrichment libraries were sequenced. ArtQ was well correlated (R2 = 0.9957), indicating that the artifactual base changes had already been introduced before exome capture.
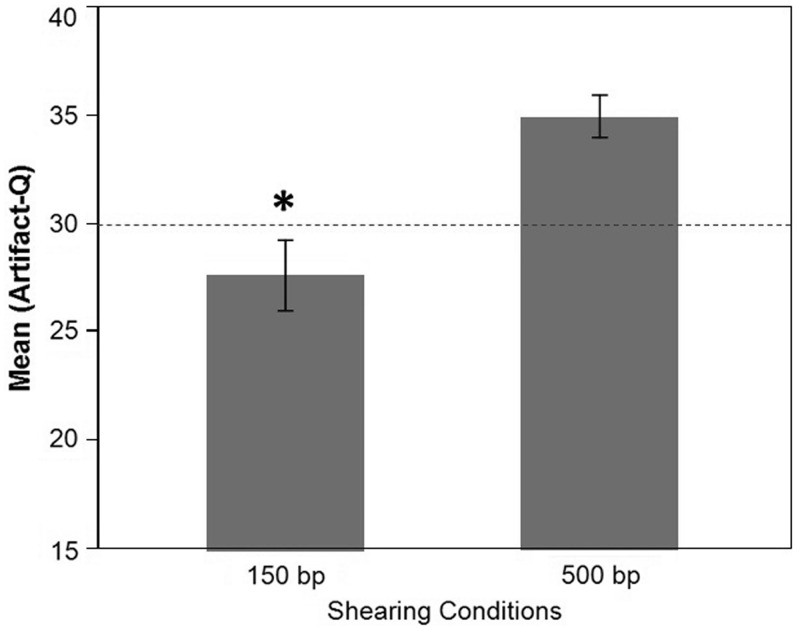
To determine whether the 150-bp shearing protocol could be introducing the C > A/G > T artifacts, we took DNA samples that when sequenced previously had low ArtQ scores and made new 150 bp and 500 bp libraries. All other steps in the library preparation protocol were kept the same, and the libraries were sequenced immediately after PCR without any further processing or size selection steps. The sequencing results show a clear and significant increase in the prevalence of the artifact, as measured by ArtQ, when the same samples were sheared using the 150-bp protocol as compared with the 500-bp protocol (Figure 5). These data provided the first proof that a major contributing source of the artifact was the higher powered 150-bp acoustic shearing protocol used to create these targeted capture libraries.
Figure 5.
Comparison of 150-bp versus 500-bp shearing conditions. Average ArtQ scores post-sequencing for the same set of six samples sheared with using both 150-bp and 500-bp Covaris protocols. The 150-bp shear protocol had significantly lower ArtQ values (P < 0.05) than the 500 bp for all samples tested.
Interaction of shearing protocol with incoming genomic sample quality
Although the 150-bp shearing step was demonstrated to induce these artifacts, we saw that less than a half of all exome samples receiving that 150-bp shearing protocol were significantly affected (Figure 3), and for samples that traveled together on 96-well plates throughout the entire process, often only a subset of samples would be affected. Moreover, as described in the previous section, we consistently observed that new sequencing libraries made from the same source DNA as a previously affected library were also highly affected at similar ArtQ scores. Taken together, these observations strongly suggested that the 150-bp shearing protocol alone was not sufficient to cause the artifact. Rather, some inherent property of the incoming sample made it more or less susceptible to damage during the 150-bp shearing process.
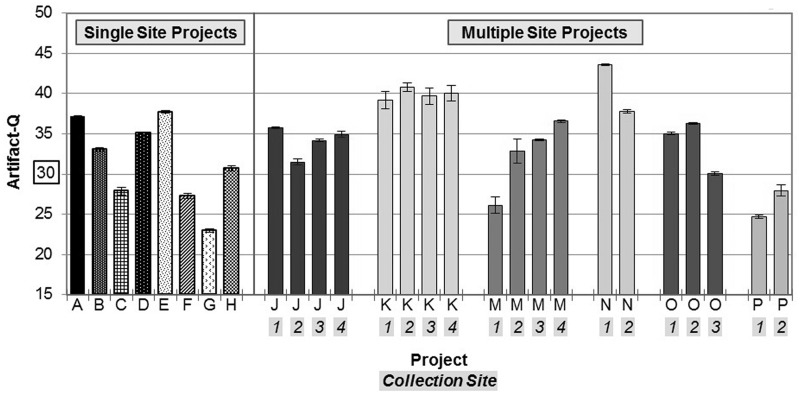
Within and between each disease project, the rate of artifact prevalence varied widely without any immediately obvious patterns or correlations (Figure 6). However, for a given disease project, we often receive pre-extracted DNA samples from multiple collaborators or institutions, which we term as separate ‘collection sites’ within a project. These separate laboratories do not necessarily use the same DNA extraction or handling methods. By further separating disease projects into their constituent collections, we observed that the artifact’s prevalence was clustering by DNA collection site (Figure 6). This finding added credence to our hypothesis that varied upstream extraction methods may be causing a subset of incoming samples to be more susceptible to artifact generation during shearing.
Figure 6.
ArtQ by project and collection site within a project. ArtQ scores vary significantly between disease projects, despite consistent protocols and automation used during the targeted capture preparation process. For a subset of six projects shown here receiving DNA from multiple collection sites using various extraction methods, the prevalence of the artifact was also variable from site to site within the same project. All projects shown consist of >100 samples; all sites listed contributed at least 12 samples to their respective project.
Confirmation of DNA oxidation mediated mutation during DNA shearing
We next set out to determine the molecular basis for these artifacts. The fact that these were C > A/G > T transversions and that they occur most frequently in the context of CCG → CAG led us to hypothesis that the cause was oxidation of DNA, specifically the conversion of guanine to 8-oxoG. 8-oxoG is a common lesion in DNA generated via oxidation, and it is known to pair with both cytosine and adenosine during PCR leading to C > A/G > T transversions (26,27). Oxidation of guanine to 8-oxogG has also been demonstrated to exhibit a distinct sequence context preference (28–30) in which the likelihood of a base being targeted for oxidation is highly dependent on both the 5′ and 3′ bases surrounding it, with the CCG/GGC we observed here being the context with the highest oxidation potential in demonstrated in these previous studies. Oxidation of DNA can come from a variety of commonly encountered sources, including DNA extraction methods, long-term storage of DNA in aqueous buffers, heat, exposure to trace metals and sonication (31–37). During the 150-bp shearing protocol, we observed that the contents of the shearing tube increased in temperature from 10°C to ∼30°C, despite being submerged in a 10°C water bath. Conversely, the temperature did not increase during 500-bp shear protocol. The presence of both powerful acoustic sonication energy and heat accumulation provided further indications that the 150-bp shearing protocol could be oxidizing DNA.
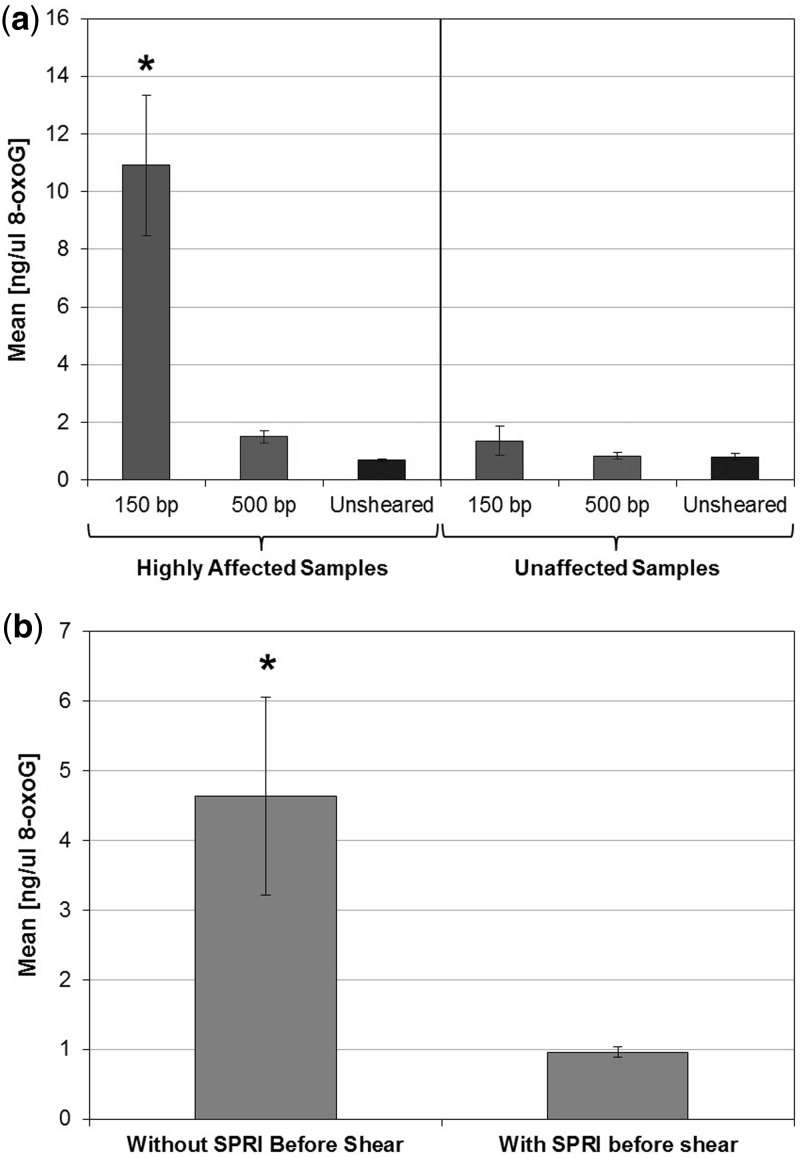
To confirm the presence of 8-oxog in affected samples, we performed an 8-oxoG specific ELISA assay (Enzo Biosciences) on remaining DNA from six melanoma that were highly affected in previous sequencing (ArtQ < 20) and six samples that were relatively unaffected (ArtQ > 30). Each sample was split in three equal aliquots and sheared with the 150-bp protocol, 500-bp protocol or left unsheared and then assayed via ELISA (Figure 7a). The results clearly show significantly elevated levels of 8-oxoG (P < 0.05) were present only in the previously highly affected melanoma samples when sheared with the 150-bp shearing protocol. The levels of 8-oxoG generated following the lower powered 500-bp shear were not significant even in known susceptible samples, which was consistent with previous observations of lower artifact prevalence in 500-bp whole genome samples. The difference in 8-oxoG levels between affected and unaffected samples further confirmed that shearing alone is not enough to induce oxidation, but that there was some contaminant in some samples that leads to increased oxidation activity during shearing. To confirm this, we compared 8-oxoG levels on samples sheared with or without buffer exchange using Ampure XP SPRI beads (Figure 7b). The results show a significant decrease (P < 0.05) in the presence of 8-oxog in the samples that underwent buffer exchange, further confirming that there were contaminants in the DNA buffers that were contributing to oxidation.
Figure 7.
8-oxoG ELISA results. (a) Bar chart of mean ng/ml of 8-oxoG for affected and unaffected samples processed with different shearing conditions. The ‘Highly Affected’ samples with high artifact rate sheared to 150 bp had a significantly higher level of 8-oxoG post shearing as compared with all other samples and conditions (asterisk denotes P < 0.01). (b) Bar chart of mean ng/ml of 8-oxoG, showing the significant decrease in 8-oxoG levels post shearing for samples following buffer exchange (asterisk denotes P < 0.05).
These ELISA results provided confirmation for the shearing-induced oxidation hypothesis and demonstrated that affected DNA samples contain some contaminants in their source buffers that when exposed to the strong acoustic energy and/or heat generated during the 150-bp shear create a highly oxidative environment. These 8-oxoG bases then persist throughout the NGS protocol to the PCR enrichment step, where the C > A and G > T base changes occur owing to Hoogsteen base pairing of 8-oxoG and A (27). In addition, this mechanism fits with the sequencer read specificity we observed in our data: the G > T base change is always observed on the read 1 strand, and the C > A is always on the opposite read 2 strand. If the artifact is being induced in shearing, this is before ligation of the forked Illumina adapters. Because of the nature of these palindromic adapters and subsequent PCR reaction mechanics, the original DNA strands containing the 8-oxoG lesion containing the G > T error will always end up on the read 1 side of the final library fragment, and the strand generated during PCR enrichment (containing the C > A error) will be read during read 2 (Supplementary Figure S2).
Development of methods to reduce DNA oxidation during shearing
We next explored the addition of antioxidants to samples before shearing as an attempt to both rescue susceptible samples and also gain insight into the reaction mechanism. Additives tested included two metal chelators, 1 mM EDTA and 100 uM DFAM, and a phenolic antioxidant and free radical scavenger, 100 uM BHT (38,39). Susceptible melanoma sample material (previous ArtQ = 22) was diluted in triplicate in our standard 10 mM Tris–HCl pH 8 buffer without or with these additives, alone and in combination, before 150-bp shearing (see Materials and Methods). Following standard library construction and sequencing, the ArtQ metrics were used to determine the effectiveness of each additive or combination of additives at preventing the oxidation artifact (Figure 8, bottom pane). The results of this experiment demonstrated a significant reduction in oxidation artifacts as measured by ArtQ for samples when either of the chelators, EDTA or DFAM was included in the shearing buffer. The hydroxyl radical scavenger BHT appeared to have little protective effect. The success of the chelators strongly suggests that some form of metal ions present in samples after DNA extraction may be involved in the oxidation mechanism during shearing. However, as DFAM significantly reduced yields from our library construction process (Figure 8, top), only EDTA showed a clear benefit without risking, reducing the robustness of our library preparation process.
Figure 8.
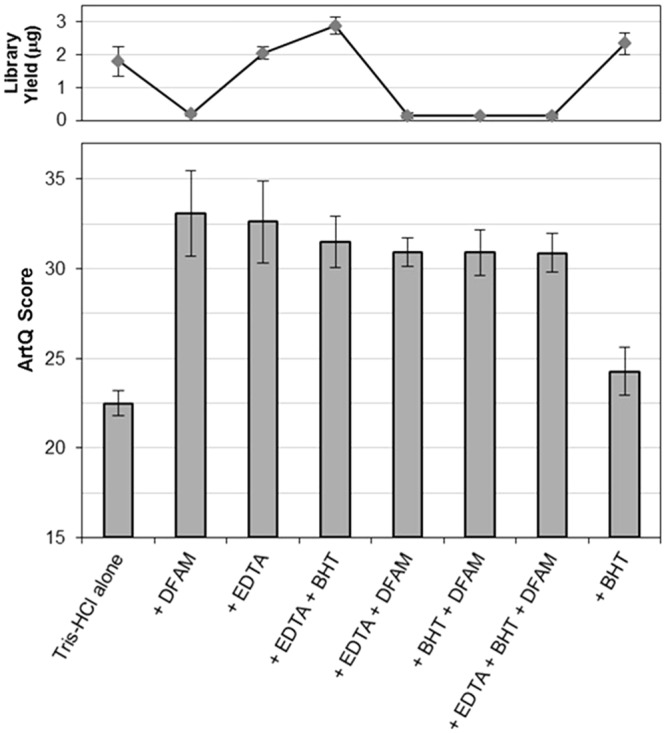
Antioxidant additives in shearing. Bottom pane: Mean ArtQ score for oxidation susceptible melanoma samples (n = 3 sample per condition) sheared with antioxidant conditions compared with Tris–HCl alone, showing significant improvement in ArtQ scores with addition of EDTA and/or DFAM. Top pane: Mean library yield following enrichment PCR in total ug, showing DFAM inhibits the Illumina library preparation process, whereas EDTA does not.
These results in combination with the results from the previously described buffer exchange experiments have allowed us to develop a protocol to protect DNA from oxidation during shearing. All incoming DNA samples are buffer exchanged into Tris-EDTA (TE) buffer (10 mM Tris, 1 mM EDTA) as a preventative measure to remove any possible contaminants present in the source buffer and to add EDTA to protect against shearing induced oxidation. Although metal-catalysed oxidation appears to be the primary mechanism of artifact induction for this particular subset of samples tested, it is highly likely that other oxidation mechanisms could exist. We are continuing to evaluate more samples from a wider variety of sources and extraction methods, as addition of metal chelation with EDTA may not be a cure all for shearing induced oxidation in all samples.
Development of a filter-based method for removing oxidation artifacts from sequencing data during mutation calling
The presence of these artifactual C > A/G > T transversions in sequencing data could lead to obvious issues in somatic mutation calling. Reprocessing the samples with low ArtQ scores through library preparation using the previously detailed laboratory process improvements is preferred, but for affected data sets that cannot be reprocessed in the laboratory, we have developed a post-processing filtering method that can be used to screen out oxidation-induced artifacts in sequencing data with high confidence to improve the fidelity of mutation calling at low allelic fractions. As the artifact was originally found at low allelic fractions, we have discovered that a universal threshold cannot be applied to throw out possible artifacts, as bona fide somatic mutations can be found at similar or even lower allele fractions. In particular, in high mutation rate tumors such as lung adenocarcinomas C > A artifacts are likely to co-mingle with previously characterized smoking-induced C > A mutations (Supplementary Figure S3) (40,41).
Mutation calls from the program used here, MuTect (Cibulskis et al. in preparation, http://confluence.broadinstitute.org/display/CGATools/Home), depend on well-calibrated base qualities to distinguish real mutations from sequencing errors, but in the case of the CCG > CAG artifacts reported here, the base qualities do not reflect the probability that a non-reference base call is the result of a true mutation. As MuTect already applies post-processing filters to remove various types of previously known sequencing artifacts from the final list of somatic point mutations, we designed a new filter to specifically remove CCG > CAG oxidation artifacts based on their unique properties that are inconsistent with real mutations including the base context, sequencing read orientation and characteristic low allelic fraction prevalence. Artifact base changes are seen as G > T only in read 1 and C > A only in read 2 as previously described, which we represent in the filter as the fraction of alternate allele supporting reads comprising G > T changes on read 1 and C > A changes on read (‘FoxoG’). Artifactual base changes should have high FoxoG ratios as compared with real somatic C > A mutations that are not read orientation specific. To determine the FoxoG value for the non- C > A or G > T mutations, we adopted a convention that ‘A’ and ‘G’ alternate alleles on read 2 and ‘C’ and ‘T’ alternate alleles on read 1 are counted in the numerator of the FoxoG value. To capture the characteristic low allele fraction of 8-oxoG artifact for purposes of artifact filtering, we quantified this property in terms of the MuTect ‘tumor_lod’ score, which is the estimated log odds that the observed number of alternate allele reads from the tumor sample could have arisen from a reference allele. The filter therefore takes into account both the FoxoG and tumor_lod measures to determine whether C > A base changes are most likely artifactual in nature and should be excluded.
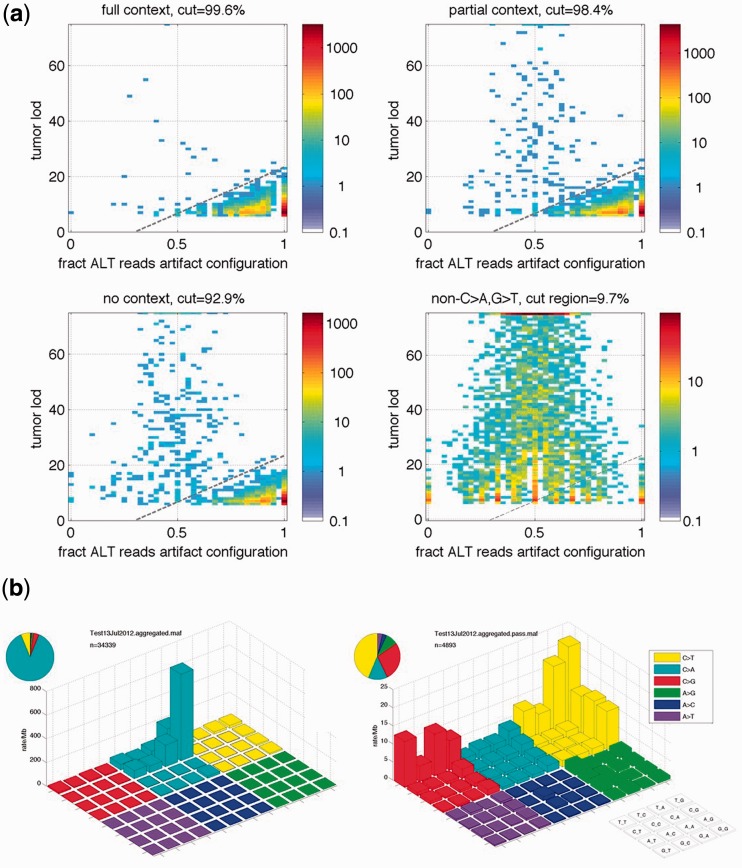
To train the filter, we used a set of 31 samples from seven different tumor types prepared and sequenced at different periods during 2011 and early 2012 (Supplementary Table S1). Here, we defined ‘full’ context mutations as C > A base changes in the context of both a proceeding C and a trailing G base (CCG > CAG). ‘Partial’ context mutations are defined as having either a preceding C or a trailing G, but not both (NCG > NAG or CCN > CAN), and ‘no’ context C > A mutations lack both the preceding C and trailing G bases. All non- C > A or G > T mutations served as a null model for non-oxidation-induced base changes. Two-dimensional histograms of FoxoG versus the tumor_lod for C > A and G > T mutations at each of the three levels of sequence context (Figure 9) showed an excess enrichment for high FoxoG and low tumor_lod mutations (lower right corner area of plots) as compared with the non- C > A or G > T mutation distribution. As expected, the proportion of artifact prevalence increased with sequence context specificity from ‘No’ to ‘Partial’ to ‘Full’ as the oxidation potential of the G base in question increased. Based on the distribution of context-specific low allelic fraction and high FoxoG C > A/G > T mutations seen in the training set, we determined the filter threshold to be applied (represented by the dashed line in the plots) to be the following: tumor_lod > −10 + (100/3) FoxoG. Once applied, the filter was able to successfully eliminate the excess number of CCG > CAG mutations that demonstrated artifactual characteristics (Figure 9b). Using the null model from non- C > A or G > T mutations, we estimated that the fraction of true biological C > A mutations removed by the filter was only 1.4% (1.2–1.6%, 95% confidence interval), whereas the fraction of passed mutations that could be attributed to oxidation artifacts was reduced to 0.1% (0–1.6%, 95% confidence interval) in this test set. Application of this filter to MuTect mutation calls therefore removed nearly all artifactual C > A mutations with high specificity, high confidence and a low chance of false positive mutation calls attributed to oxidative base changes in shearing.
Figure 9.
Training of CCG > CAG artifact mutation call filter. (a) Two dimensional histograms showing the filter criteria as distributions of FoxoG (the fraction of alternate allele reads in the oxoG artifact configuration, horizontal axis) and Tumor_lod (log odds that the mutation could arise from the reference allele, vertical axis) for C > A or G > T mutations in various contexts and for the non- C > A or G > T mutations. Colors correspond to the count of mutations in a bin. The ‘non-C>A,G>T’ data serves as a null model (non-OxoG artifact). The proportion of non-artifact depends on sequence context, but the region dominated by OxoG in the lower right corner below the dotted line is consistent across contexts. Each panel is labeled with the fraction of mutations below the threshold, although in the case of the ‘non-C>A,G>T’ data, all mutations including those under the cut line, are passed by the filter. (b) Before (left) and after (right) application of the OxoG filter in MuTect in the set of 31 samples from a variety of tumor types. The vertical scale is the mutation rate (mutations per Mb of bases covered in the whole exome targeted capture data); note the scales are different owing to the excess of C > A artifacts in the unfiltered data.
DISCUSSION
This discovery of a previously unreported mechanism of DNA damage inflicted during common sample preparation methods that can lead to anomalous base changes has an obvious and considerable impact on downstream data analysis. Although many projects such as medical or population genetics studies may not be looking at highly rare events, an increasing number of recent cancer projects have focused on attempting to detect extremely low frequency variants at the lowest allelic fractions. In the past year, publications have emerged detailing NGS methods for mutation calling at 0.1% allelic fraction (42), detection of circulating tumor cell DNA in human blood extracts (43) and single cell whole exome sequencing of human kidney tumor cells to understand tumor genetic heterogeneity (44). Outside of cancer, other groups have published work detailing sequencing methods for non-invasive analysis of chromosomal abnormalities from free circulating fetal cells in the mother’s blood (45,46). The introduction of artifacts at low allelic fractions like those described here could certainly derail the accuracy and limit of detection of such projects. Although we describe an oxidation-specific artifact induced in our high-powered acoustic shearing protocol, many laboratories use similar DNA fragmentation methods as used in our laboratory, and further, there are likely other mechanisms for both oxidation and non-oxidation-mediated base changes that still need to be discovered. This discovery therefore has broad implications for all NGS laboratories.
Vendors and research institutions alike have been focusing much effort in the past few years on the reduction of DNA input requirements for the library construction process. Yet, as we continue to reduce inputs, the effects of random stochastic damage events such as the oxidation mechanism described here are likely to be amplified. In looking at our data for this particular transversion, we observed that this phenomenon did in fact worsen progressively over time, as we reduced our input into the exome process from 3 ug to 100 ng (Figure 3). Considering our model of shearing-induced base damage, it makes sense that reducing the DNA input into the shearing vessel by 30-fold but maintaining the intensity of the acoustic power applied to each sample led to the effect being amplified. As groups go even lower with input requirements and single cell technologies begin to take off, the way we examine the data produced while developing these protocols needs to be rethought to detect these types of random stochastic molecular changes.
Further, the effects that upstream sample acquisition, nucleic acid extraction and sample storage techniques can have on DNA base composition and fidelity of downstream single nucleotide polymorphisms (SNPs) calling are still not well understood. We are now implementing a best practice approach of performing a buffer exchange of all incoming samples to reduce the risk of oxidative agents damaging DNA during shearing. Although we observed protection from oxidation with the addition metal chelators, we still do not know the exact identity of these contaminants that make some DNA samples more susceptible to oxidation than others. To help provide answers, we are currently devising experiments to identifying these agents by methods such as mass spectroscopy or high pressure (or high performance) liquid chromatography in hopes to design more targeted methods to counteract oxidation and provide valuable information that can be used to develop safer methods of DNA extraction.
Finally, the discovery of this particular oxidation-driven error mechanism has led us to think on a much larger scale about other non-biological base substitutions that may be lurking in sequencing data. The obvious deleterious effects that the existence of such artifacts can have on the field of cancer research could be dramatic. If multiple common processes in the laboratory can significantly alter the physical base sequence of DNA, it begs the question of whether we can truly be confident that the rare mutations we are searching for can actually be attributed to true biological variation. We have invested much time and effort into characterizing this one particular oxidation mediated event, but this is one of the myriad of possible low frequency errors that could be induced during NGS sample preparation. The discovery of this oxidation-induced artifact in NGS sample preparation demonstrated that the development of new protocols for NGS sample preparation can lead to changes in to DNA at a molecular and chemical level that may be subtle and difficult (if not impossible) to see with conventional sequence quality measures. A systematic review of a wide variety of data obtained using different protocols from different laboratories needs to be undertaken by the sequencing community to identify whether there are any types of other artifacts that may be induced during extraction and/or library preparation that could be wrongly attributed to the biology of a given disease.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–3.
FUNDING
Funding for open access charge: National Human Genome Research Institute [HG03067-05].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
From the Broad Institute, the authors thank Sheli Dookran, Laura Lambiase, Emily Wheeler, Andrew Cheney, Kristian Cibulskis, Kristin Ardlie and Andreas Gnirke for technical assistance and advice; Carrie Sougnez, Elizabeth Bevilacqua and Lizz Gottardi for project management support; and Wendy Winckler and Niall Lennon for their helpful feedback on the manuscript. From MIT’s Department of Biological Engineering, the authors thank Professor Peter Dedon for invaluable advice regarding DNA oxidation.
REFERENCES
- 1.Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat. Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, Nayir A, Bakkaloğlu A, Ozen S, Sanjad S, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc. Natl Acad. Sci. USA. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adey A, Morrison HG, Asan, Xun X, Kitzman JO, Turner EH, Stackhouse B, MacKenzie AP, Caruccio NC, Zhang X, et al. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 2010;11:R119. doi: 10.1186/gb-2010-11-12-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fisher S, Barry A, Abreu J, Minie B, Nolan J, Delorey TM, Young G, Fennel TJ, Allen A, Ambrogio L, et al. A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol. 2011;12:R1–R15. doi: 10.1186/gb-2011-12-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K, Werner L, Sivachenko A, DeLuca DS, Zhang L, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N. Engl. J. Med. 2011;365:2497–2506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Notta F, Mullighan CG, Wang JC, Poeppl A, Doulatov S, Phillips LA, Ma J, Minden MD, Downing JR, Dick JE. Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature. 2011;469:362–367. doi: 10.1038/nature09733. [DOI] [PubMed] [Google Scholar]
- 7.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pugh TJ, Weeraratne SD, Archer TC, Pomeranz-Krummel DA, Auclair D, Bochicchio J, Carneiro MO, Carter SL, Cibulskis K, Erlich RL, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106–110. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keats JJ, Chesi M, Egan JB, Garbitt VM, Palmer SE, Braggio E, Van Wier S, Blackburn PR, Baker AS, Dispenzieri A, et al. Clonal competition with alternating dominance in multiple myeloma. Blood. 2012;120:1067–1076. doi: 10.1182/blood-2012-01-405985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mioche A, Dohm JC, Himmelbauer H. Evaluation of genomic high-throughput sequencing data generated on Illumina HiSeq and Genome Analyzer systems. Genome Biol. 2011;12:R112–R126. doi: 10.1186/gb-2011-12-11-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kunkel TA, Loeb LA, Goodman MF. On the fidelity of DNA Replication. J. Biol. Chem. 1984;259:1539–1544. [PubMed] [Google Scholar]
- 13.Cline J, Braman JC, Hogrefe HH. PCR fidelity of PFU polymerase and other thermostable DNA polymerases. Nucleic Acids Res. 1996;24:3546–3551. doi: 10.1093/nar/24.18.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilje B, Heikkilä R, Oltedal S, Tjensvoll K, Nordgård O. High-fidelity DNA polymerase enhances the sensitivity of a peptide nucleic acid clam PCR assay for K-ras mutations. J. Mol. Diagn. 2008;10:325–331. doi: 10.2353/jmoldx.2008.070183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zagordi O, Klein R, Däumer M, Beerenwinkel N. Error correction of next-generation sequencing data and reliable estimation of HIV quasispecies. Nucleic Acids Res. 2010;38:7400–7409. doi: 10.1093/nar/gkq655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet JP, Ahmann GJ, Adli M, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cibulskis K, McKenna A, Fennell T, Banks E, DePristo M, Getz G. ContEst, estimating cross-contamination of human samples in next-generation sequencing data. Bioinformatics. 2011;27:2601–2602. doi: 10.1093/bioinformatics/btr446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, DiGiovanna JJ, Stern JB, Hornyak TJ, Raffeld M, Khan SG, Oh KS, Hollander MC, Dennis PA, Kraemer KH. Evidence of ultraviolet type mutation sin xeroderma pigmentosum melanomas. Proc. Natl Acad. Sci. USA. 2009;106:6279–6284. doi: 10.1073/pnas.0812401106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berger MF, Hodis E, Heffernan TP, Deribe YL, Lawrence MS, Protopopov A, Ivanova E, Watson IR, Nickerson E, Ghosh P, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–506. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hosoi G, Hara J, Okamura T, Osugi Y, Ishihara S, Fukuzawa M, Okada A, Okada S, Tawa A. Low frequency of the p53 gene mutations in neuroblastoma. Cancer. 1994;73:3087–3093. doi: 10.1002/1097-0142(19940615)73:12<3087::aid-cncr2820731230>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 22.Shukla N, Ameur N, Yilmaz I, Nafa K, Lau CY, Marchetti A, Borsu L, Barr FG, Ladanyi M. Oncogene mutation profiling of pediatric solid tumors reveals significant subsets of embryonal rhabdomyosarcoma and neuroblastoma with mutated genes in growth signaling pathways. Clin. Cancer Res. 2012;18:748–757. doi: 10.1158/1078-0432.CCR-11-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown RJ, Levine RL, Thompson C, Basile G, Gilliland DG, Freedman AS. Systematic genomic screen for tyrosine kinase mutations in CLL. Leukemia. 2008;22:1966–1969. doi: 10.1038/leu.2008.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fabbri G, Rasi S, Rossi D, Trifonov V, Khiabanian H, Ma J, Grunn A, Fangazio M, Capello D, Monti S, et al. Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. J. Exp. Med. 2011;208:1389–1401. doi: 10.1084/jem.20110921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puente XS, Pinyol M, Quesada V, Conde L, Ordóñez GR, Villamor N, Escaramis G, Jares P, Beà S, González-Díaz M, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–110. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McAuley-Hecht KE, Leonard GA, Gibson NJ, Thomson JB, Watson WP, Hunter WN, Brown T. Crystal structure of a DNA duplex containing 8-hydroxydeoxyguanine-adenine base pairs. Biochemistry. 1994;33:10266–10270. doi: 10.1021/bi00200a006. [DOI] [PubMed] [Google Scholar]
- 27.Beard WA, Batra VK, Wilson SH. DNA polymerase structure-based insight on the mutagenic properties of 8-oxoguanine. Mutat. Res. 2010;703:18–23. doi: 10.1016/j.mrgentox.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saito I, Nakamura T, Kakatani K, Yoshioka Y, Yamaguchi K, Sugiyama H. Mapping of the hot spots for DNA damage by one-electron oxidation: efficacy of GG doublets and GGG triplets as a trap in long-range hole migration. J. Am. Chem. Soc. 1998;120:12686–12687. [Google Scholar]
- 29.Margolin Y, Cloutier JF, Shafirovich V, Geacintov NE, Dedon PC. Paradoxical hotspots for guanine oxidation by a chemical mediator of inflammation. Nature Chem. Biol. 2006;2:365–366. doi: 10.1038/nchembio796. [DOI] [PubMed] [Google Scholar]
- 30.Margolin Y, Shafirovich V, Geacintov NE, DeMott MS, Dedon PC. DNA sequence context as a determinant of the quantity and chemistry of guanine oxidation produced by hydroxyl radicals and one-election oxidants. J. Biol. Chem. 2008;283:35569–35578. doi: 10.1074/jbc.M806809200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ravanat JL, Douki T, Duez P, Gremaud E, Herbert K, Hofer T, Lasserre L, Saint-Pierre C, Favier A, Cadet J. Cellular background level of 8-oxo-7.,8-dihydro-2′-deoxyguanosine: an isotope based method to evaluate artefactual oxidation of DNA during its extraction and subsequent work-up. Carcinogenesis. 2002;23:1911–1918. doi: 10.1093/carcin/23.11.1911. [DOI] [PubMed] [Google Scholar]
- 32.Finnegan MTV, Herbert KE, Evans MD, Giffiths HR, Lunec K. Evidence for sensitization of DNA to oxidative damage during isolation. Free Rad. Biol. Med. 1996;20:93–98. doi: 10.1016/0891-5849(95)02003-9. [DOI] [PubMed] [Google Scholar]
- 33.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 34.Bruskov VI, Malakhova LV, Maslimov ZK, Cherikov AV. Heat-induced formation of reactive oxygen species and 8-oxoguanine, a biomarker of damage to DNA. Nucleic Acids Res. 2002;30:1354–1363. doi: 10.1093/nar/30.6.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kennedy LJ, Moore K, Caulfield JL, Tannenbaum SR, Dedon PC. Quantitation of 8-oxoguanine and strand breaks produced by four oxidizing agents. Chem. Res. Toxicol. 1997;10:386–392. doi: 10.1021/tx960102w. [DOI] [PubMed] [Google Scholar]
- 36.Fuciarelli AF, Sisk EC, Thomas RM, Miller DL. Induction of base damage in DNA solutions by ultrasonic cavitation. Free Rad. Biol. Med. 1995;18:231–238. doi: 10.1016/0891-5849(94)00119-5. [DOI] [PubMed] [Google Scholar]
- 37.Milowska K, Gabryelak T. Reactive oxygen species and DNA damage after ultrasound exposure. Biomol. Eng. 2007;24:236–237. doi: 10.1016/j.bioeng.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 38.Burton GW, Ingold KU. Autoxidation of biological molecules. 1. Antioxidant activity of vitamin E and related chain-breaking phenolic antioxidants in vitro. J. Am. Chem. Soc. 1981;103:6472–6477. [Google Scholar]
- 39.Taghizadeh K, McFaline JL, Pang B, Sullivan M, Dong M, Plummer E, Dedon PC. Quantification of DNA damage products resulting from deamination, oxidation and reaction with products of lipid peroxidation by liquid chromatography isotope dilution tandem mass spectrometry. Nat. Protoc. 2008;3:1287–1298. doi: 10.1038/nprot.2008.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Husgafvel-Pursiainen K, Boffetta P, Kannio A, Nyberg F, Pershagen G, Mukeria A, Constantinescu V, Fortes C, Benhamou S. p53 mutations and exposure to environmental tobacco smoke in a multicenter study on lung cancer. Cancer Res. 2000;60:2906–2911. [PubMed] [Google Scholar]
- 41.Hainaut P, Pfiefer GP. Patterns of p53 G→T transversions in lung cancers reflect the primary mutatgenic signature of DNA-damage by tobacco smoke. Carcinogenesis. 2001;22:367–374. doi: 10.1093/carcin/22.3.367. [DOI] [PubMed] [Google Scholar]
- 42.Flaherty P, Natsoulis G, Muralidharan O, Winters M, Buenrostro J, Bell J, Brown S, Holodniy M, Zhang N, Ji HP. Ultrasensitive detection of rare mutations using next-generation targeted resequencing. Nucleic Acids Res. 2012;40:e2. doi: 10.1093/nar/gkr861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DW, Kaper F, Dawson SJ, Piskorz AM, Jimenez-Linan M, Bentley D, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci. Transl. Med. 2012;4:136ra68. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- 44.Xu X, Hou Y, Yin X, Bao L, Tang A, Song L, Li F, Tsang S, Wu K, Wu H, et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148:886–895. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kinde I, Papadopoulos N, Kinzler KW, Vogelstein B. FAST-SeqS: a simple and efficient method for the detection of aneuploidy by massively parallel sequencing. PLoS ONE. 2012;7:e41162. doi: 10.1371/journal.pone.0041162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kitzman JO, Snyder MW, Ventura M, Lewis AP, Qiu R, Simmons LE, Gammill HS, Rubens CE, Santillan DA, Murray JC, et al. Noninvasive whole-genome sequencing of a human fetus. Sci. Transl. Med. 2012;4:137ra76. doi: 10.1126/scitranslmed.3004323. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.