Abstract
Degenerate splice site sequences mark the intron boundaries of pre-mRNA transcripts in multicellular eukaryotes. The essential pre-mRNA splicing factor U2AF65 is faced with the paradoxical tasks of accurately targeting polypyrimidine (Py) tracts preceding 3′ splice sites while adapting to both cytidine and uridine nucleotides with nearly equivalent frequencies. To understand how U2AF65 recognizes degenerate Py tracts, we determined six crystal structures of human U2AF65 bound to cytidine-containing Py tracts. As deoxy-ribose backbones were required for co-crystallization with these Py tracts, we also determined two baseline structures of U2AF65 bound to the deoxy-uridine counterparts and compared the original, RNA-bound structure. Local structural changes suggest that the N-terminal RNA recognition motif 1 (RRM1) is more promiscuous for cytosine-containing Py tracts than the C-terminal RRM2. These structural differences between the RRMs were reinforced by the specificities of wild-type and site-directed mutant U2AF65 for region-dependent cytosine- and uracil-containing RNA sites. Small-angle X-ray scattering analyses further demonstrated that Py tract variations select distinct inter-RRM spacings from a pre-existing ensemble of U2AF65 conformations. Our results highlight both local and global conformational selection as a means for universal 3′ splice site recognition by U2AF65.
INTRODUCTION
Pre-mRNA splicing removes non-coding introns and regulates most human transcripts (1); however, the mechanisms by which the splice sites are identified and regulated are not well understood. Splicing fidelity is assisted by an ATP-dependent series of checkpoints during the assembly of >100 proteins and five small nuclear (sn)RNAs into the active spliceosome [reviewed in (2)]. Consensus sequences mark the 5′ and 3′ splice sites at the intron-exon boundaries of the pre-mRNA. Nevertheless, to be distinguished and regulated in a specific manner, the ‘weak’ regulated splice sites of multicellular organisms often deviate from the optimum consensus of ‘strong’ constitutive splice sites (3). The spliceosome is directed to the 3′ splice site of the pre-mRNA by a polypyrimidine (Py) tract located between the branchpoint sequence and an AG dinucleotide (Figure 1A). The Py tract exemplifies the variability of human splice site signals. Cytidines precede the 3′ splice site at frequencies approaching those of uridine (∼35 and 45%, respectively) (4,5), yet cytidine and uridine confer different Py tract activities. Increasing the number of uridines in a Py tract generally increases use of an adjacent 3′ splice site (6–8). Isolated cytidine nucleotides in a uridine-rich Py tract can support or even enhance the use of an adjacent 3′ splice site (6,9), yet several consecutive cytidines can abolish detectable splicing (7,9). Mutations that compromise recognition of splice site signals can be lethal or lead to genetic diseases and cancer [reviewed in (10,11)], such as shortening of a cftr Py tract that is responsible for some cases of cystic fibrosis (12,13). Consequently, the spliceosomal proteins and snRNAs must overcome the challenge of recognizing degenerate signal sequences embedded within pre-mRNAs that are thousands of nucleotides long.
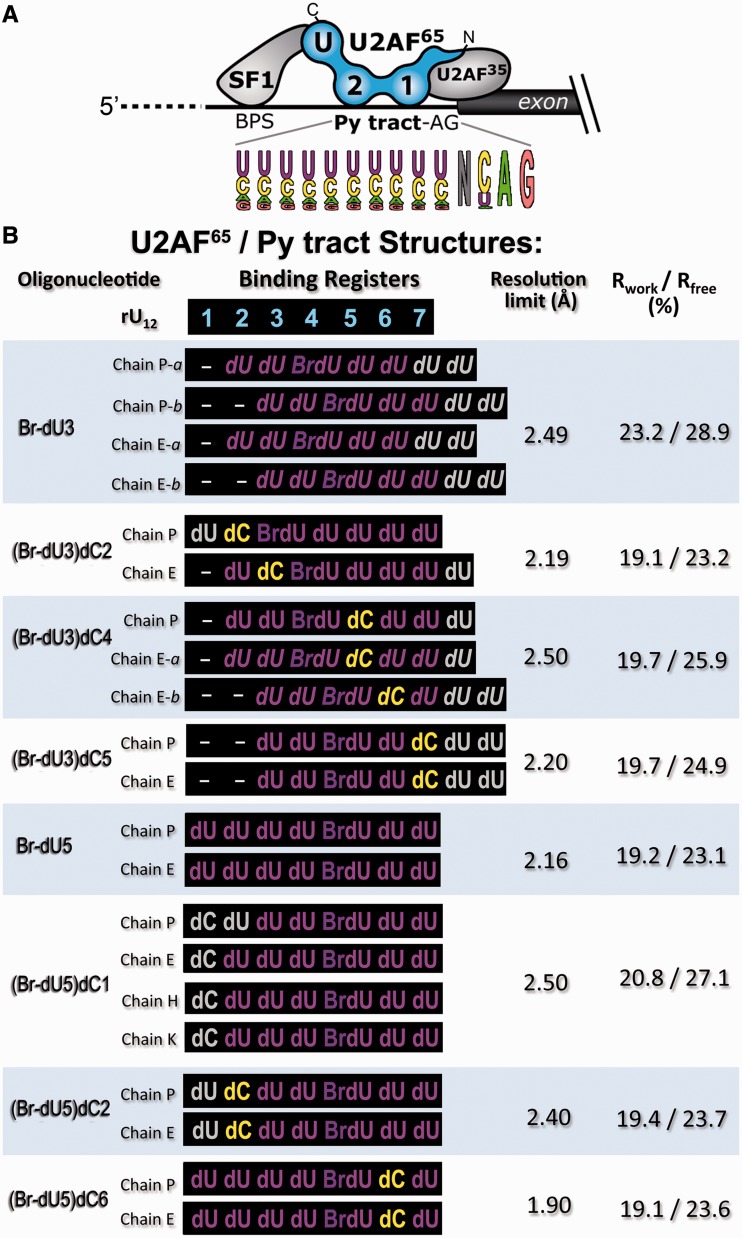
Figure 1.
Crystal structures of U2AF65 bound to diverse Py tracts. (A) Splicing factors recognizing the 3′ splice site. A pictogram of the primate Py tract consensus derived from (4) is inset below. The C- to N-terminal orientation of the U2AF65 domains recognize the 5′–3′- direction of the pre-mRNA. BPS, branchpoint sequence; Py tract, polypyrimidine tract; AG dinucleotide. The U2AF65 RRMs are numbered in sequence and ‘U’ indicates the C-terminal UHM. The U2AF65 domains and inter-RRM linker sequences are given in Supplementary Figure S1. (B) The nomenclature, Py tract sequences, resolution limits and crystallographic Rfactors are listed for the dU2AF651,2 crystal structures. The binding registers of the Py tracts are aligned relative to the seven nucleotide binding sites of the original dU2AF651,2/rU12 structure (PDB ID 2G4B). Electron density maps such as those shown in Supplementary Figure S2 were used to assign the Br-dU positions. Py tracts of distinct copies in the crystallographic asymmetric unit are listed separately (oligonucleotides ‘P’ and ‘E’; (Br-dU5)dC1 also includes oligonucleotides ‘H’ and ‘K’). Alternative binding registers marked ‘a’ and ‘b’ are italicized. Nucleotides marked with grey font lack interpretable electron density and were not included in the final structures. Supplementary Table S1 details the crystallographic data and refinement statistics.
The U2 small nuclear ribonucleoprotein auxiliary factor 65 kDa (U2AF65) universally recognizes degenerate Py tracts preceding 3′ splice sites during the early stages of pre-mRNA splicing (14–17) (Figure 1A), and there facilitates ATP-dependent association of the U2 snRNP (14). U2AF65 is essential for vertebrate development (18), and specific U2AF65 deficiencies are associated with cystic fibrosis (19), myotonic dystrophy (20) and several cancers (21–23). U2AF65 recognizes the Py tract via two central RNA recognition motifs (RRM1 and RRM2) (17,24) (Figure 1A, Supplementary Figure S1A). Our structure of the core RRM1 and RRM2-containing domain of U2AF65 bound to an optimal poly-uridine (poly-rU) RNA (25) reveals that U2AF65 forms specific hydrogen bonds with the edges of the uracil bases, including the N3 and O4 atoms where cytosine differs from uracil. Subsequent NMR characterizations confirmed the interactions of the individual U2AF65 RRMs with uridines in solution (26), although the inter-RRM arrangements differ from the solution configuration owing to an internal deletion of the inter-RRM linker that was required for crystallization (27). The observation of apparently specific hydrogen bonds between U2AF65 and the uracil bases served to highlight the question: how can U2AF65 universally target the diverse Py tracts of multicellular eukaryotes?
To address this question, here we report six crystal structures of human U2AF65 bound to cytosine-containing Py tracts. We also determined and compare the original RNA-bound structure with two crystal structures of the deoxy-uridine (dU) containing counterparts, as deoxy-ribose backbones were required for co-crystallization of these Py tracts. Structural differences, coupled with the region-dependent Py tract affinities of U2AF65 and site-directed mutants and small-angle X-ray scattering (SAXS) data of U2AF65 complexes with various Py tracts, suggest that U2AF65 adapts to degenerate splice site signals through conformational selection of a promiscuous N-terminal RRM1 and a stringent C-terminal RRM2.
MATERIALS AND METHODS
Crystallographic analyses
Given that 7 of 12 co-crystallized uridines were observed in the original structure (27), we obtained new crystal forms of a U2AF65 variant (dU2AF651,2, including residues 148–237 and 258–336) bound to minimal, seven-nucleotide sites and marked by 5-bromo-uridines (Br-dU) to define the oligonucleotide-binding registers (Figure 1B, Supplementary Table S1, Supplementary Figure S2). A deoxy-ribose backbone for the uracil bases was required to co-crystallize the 7-mer oligonucleotides; the U2AF65 protein binds deoxy-ribonucleotides with reasonable affinity (25,28) and contacts only a single 2′ hydroxyl group at the 5′ terminus of the poly-rU RNA (25). We previously demonstrated that the U2AF65 variant (dU2AF651,2, including residues 148–237 and 258–336) used for crystallization exhibits similar RNA affinities, splicing efficiencies and protein/RNA contacts as the unmodified U2AF65 counterpart (25,27,29). The interdomain linker is poorly conserved (Supplementary Figure S1B), and residues 238–257 can be replaced with unrelated sequences without penalizing the affinity or ability of U2AF65 to support pre-mRNA splicing (29).
Human U2AF65 constructs were expressed and purified as described (27). Structures were determined by molecular replacement using the RRMs of PDB ID 2G4B as search models and refined using REFMAC5 (30). Purified DNA oligonucleotides were purchased from Integrated DNA Technologies, Inc. Protein and DNA concentrations were estimated using the respective absorbance at 280 nm or 260 nm and calculated molar extinction coefficients (31,32). Protein (20 mg/mL) and DNA were mixed in 1:1.2 molar ratio with 4 mM [N,N'-Bis(3-D-gluconamidopropyl) deoxycholamide]. Crystals were grown by the hanging drop method against a reservoir solution of 1.5–1.6 M ammonium sulfate, 10% dioxane and 0.1 M 2-(N-morpholino)ethanesulfonic acid (pH 6.0) at 4°C and flash cooled following stepwise transfer to 21% glycerol as the cryoprotectant. Data collection and refinement statistics are reported in Supplementary Table S1.
Surface plasmon resonance experiments
Surface plasmon resonance (SPR) experiments were completed as described previously (25) with minor modifications. To reduce steric effects, a 14-atom linker separated the 5′ biotin used for immobilization from the 5′ phosphate of the oligonucleotide. As the on/off rates approach the limit for reliable measurement, the average equilibrium response at each concentration was fit to a steady-state binding model. Repeated washes with the running buffer were sufficient to regenerate the surface following each injection. At least two independent experiments were repeated for each oligonucleotide/protein combination, and the average KDs are reported in Table 1. Representative sensorgrams and binding curves are shown in Supplementary Figure S6. Isothermal titration calorimetry (ITC) experiments described in Supplementary Figure S7 independently confirmed the preference of U2AF651,2 to bind 5′-4rU over 3′-4rU RNAs.
Table 1.
U2AF65 affinities for degenerate Py tracts
| U2AF65 proteina | Py tract name | Py tract sequenceb | <KD> (µM)c | Fold penaltyd |
|---|---|---|---|---|
| U2AF651,2 | rAdML13 | C.C.C.U.U.U.U.U.U.U.U.C.C | 12.5 ± 1.6 | 1.0 |
| U2AF651,2 | rU13 | U.U.U.U.U.U.U.U.U.U.U.U.U | 13.1 ± 0.9 | 1.0 |
| Pyrimidine mutations: | ||||
| U2AF651,2 | rC(5) | C.C.C.U.C.U.U.U.U.U.U.C.C | 13.9 ± 1.0 | 1.1 |
| U2AF651,2 | rC(7) | C.C.C.U.U.U.C.U.U.U.U.C.C | 9.3 ± 0.4 | 0.8 |
| U2AF651,2 | rC(9) | C.C.C.U.U.U.U.U.C.U.U.C.C | 13.7 ± 1.3 | 1.1 |
| U2AF651,2 | rC(10) | C.C.C.U.U.U.U.U.U.C.U.C.C | 11.9 ± 0.8 | 0.9 |
| U2AF651,2 | rC(7,9) | C.C.C.U.U.U.C.U.C.U.U.C.C | 11.0 ± 0.6 | 0.9 |
| U2AF651,2 | rC(7,10) | C.C.C.U.U.U.C.U.U.C.U.C.C | 14.5 ± 0.9 | 1.2 |
| U2AF651,2 | rC(7,9,10) | C.C.C.U.U.U.C.U.C.C.U.C.C | 11.6 ± 0.8 | 0.9 |
| U2AF651,2 | rC(4,5,8,10) | C.C.C.C.C.U.U.C.U.C.U.C.C | 27.9 ± 1.3 | 2.2 |
| U2AF651,2 | rU(3)C(4,6,9,10,11) | C.C.U.C.U.C.U.U.C.C.C.C.C | 14.4 ± 1.1 | 1.2 |
| U2AF651,2 | rC(4–11) | C.C.C.C.C.C.C.C.C.C.C.C.C | 125 ± 10 | 10.0 |
| Uridine/Cytidine tracts: | ||||
| U2AF651,2 | 5′-3rU | C.C.U.U.U.C.C.C.C.C.C.C.C | 22.0 ± 0.9 | 1.8 |
| U2AF651,2 | 3′-3rU | C.C.C.C.C.C.C.C.U.U.U.C.C | 89.5 ± 3.9 | 7.2 |
| U2AF651,2 | 5′-4rU | C.C.U.U.U.U.C.C.C.C.C.C.C | 14.3 ± 0.6 (3.5 ± 0.3) | 1.1 |
| U2AF651,2 | 3′-4rU | C.C.C.C.C.C.C.U.U.U.U.C.C | 69.4 ± 3.1 (13.5 ± 0.9) | 5.6 |
| U2AF651,2 | 5′-5rU | C.C.U.U.U.U.U.C.C.C.C.C.C | 14.4 ± 0.8 | 1.1 |
| U2AF651,2 | 3′-5rU | C.C.C.C.C.C.U.U.U.U.U.C.C | 45.2 ± 3.4 | 3.6 |
| U2AF65 mutations: | ||||
| U2AF651,2MUT | 5′-4rU | C.C.U.U.U.U.C.C.C.C.C.C.C | 14.0 ± 1.6 | 1.1 |
| U2AF651,2MUT | 3′-4rU | C.C.C.C.C.C.C.U.U.U.U.C.C | 12.5 ± 1.3 | 1.0 |
aThe U2AF651,2 protein comprises the core RRM1 and RRM2 domains connected by the wild-type linker, and the U2AF651,2MUT has four mutations in RRM2 (D293N/K329Q/L331K/Q333E).
bThe nucleotide substitutions to be compared are emphasized in bold font.
cApparent equilibrium dissociation constants for the indicated U2AF651,2 protein/Py tract combination are averages of at least two experiments, and a representative sensorgram is shown in Supplementary Figure S6. Affinities determined by ITC are given in parentheses, and representative isotherms are shown in Supplementary Figure S7. Splicing activities of the rC(10), rC(7,10) and rC(4–11) substrates were reported previously (9).
d‘Fold Penalty’ is the fold decrease in affinity relative to wild-type U2AF651,2 binding the rAdML13 Py tract. >3-fold penalties are highlighted in bold.
SAXS analyses
Following size exclusion chromatography to prepare monodisperse samples, the appropriate protein:RNA stoichiometries were verified by the absorbance ratios of the final SAXS samples at 280 nm and 260 nm. SAXS samples were free of interparticle effects based on the agreement of scattering and Guinier plots over a range of concentrations (Supplementary Figure S8). Ensembles of 20 different PDB structure files were fit using the program EOM (33) and significantly improved the χ2 values over single models (Supplementary Table S2). As the 13-nucleotide Py tracts contribute only 11% of the total scattering mass, the SAXS data primarily reflect the protein conformations. The rigid body models are composed of the RRM1 and RRM2 structures from PDB ID 2G4B and U2AF Homology Motif (UHM) structure from PDB ID 1OPI connected by ab initio linkers.
RESULTS
Structures of human U2AF65 bound to cytidine-containing sites
To understand how U2AF65 adapts to cytidines, we determined eight structures of dU2AF651,2 bound to oligonucleotides, six of which contained single cytidines (Figure 1B, Supplementary Table S1). We previously determined the structure of a U2AF65 variant (dU2AF651,2 lacking residues 238–257 of the inter-RRM linker) bound to a 12-mer poly-rU RNA (rU12) (25). The dU2AF651,2 protein is required for crystallization (27) and exhibits similar RNA affinities, splicing efficiencies and protein/RNA contacts as the wild-type U2AF651,2 counterpart (25–27,29). To distinguish cytidine from uridine, we co-crystallized dU2AF651,2 with the core deoxy-uridine (dU) 7-mer with the sequence register marked by 5-Br-dU (Supplementary Figure S2). A DNA backbone for the uracil bases was required for co-crystallization of these shorter oligonucleotides with U2AF65. The U2AF65 protein binds a 20-mer dU oligonucleotide with a 4–7-fold apparent decrease in affinity relative to the RNA counterpart, consistent with a single 2′ hydroxyl contact with the 5′ terminal nucleotide in the crystal structure of dU2AF651,2 bound to poly-rU RNA (25).
The structures of two baseline Br-dU-containing complexes (Br-dU3 and Br-dU5) were determined at 2.5 and 2.2 Å resolution, respectively, for comparison with the cytidine-containing counterparts. Based on the omit electron density maps, the Br-dU3 strands exhibited alternative conformations, i.e. a mixed population of the two binding registers with the Br-dU in both the fourth and fifth rather than the third binding sites (Figure 1B, Supplementary Figure S2A). The slipped binding register enables the halogen substituent to participate in favourable aromatic stacking interactions with U2AF65 side chains (34) (Figure S2I) and at the fifth position, is near positively charged K225 and R227 residues (Figure 4A). The introduction of cytidines altered the apparent preference for U2AF65 to bind Br-dU at the fourth or fifth sites, in most cases by stabilizing one of the two pre-existing alternative conformations (Figure 1B). The altered binding registers of the cytidine-containing oligonucleotides, coupled with local changes at the individual sites described later in the text, support the hypothesis that certain U2AF65 sites can accommodate cytidines more readily than others. Based on these observations, we subsequently focused on oligonucleotides containing Br-dU at the fifth position of the sequence (Br-dU5). For all complexes of dU2AF651,2 with Br-dU5-containing oligonucleotides, the bromine appeared at the expected fifth nucleotide binding site (Supplementary Figure S2E).
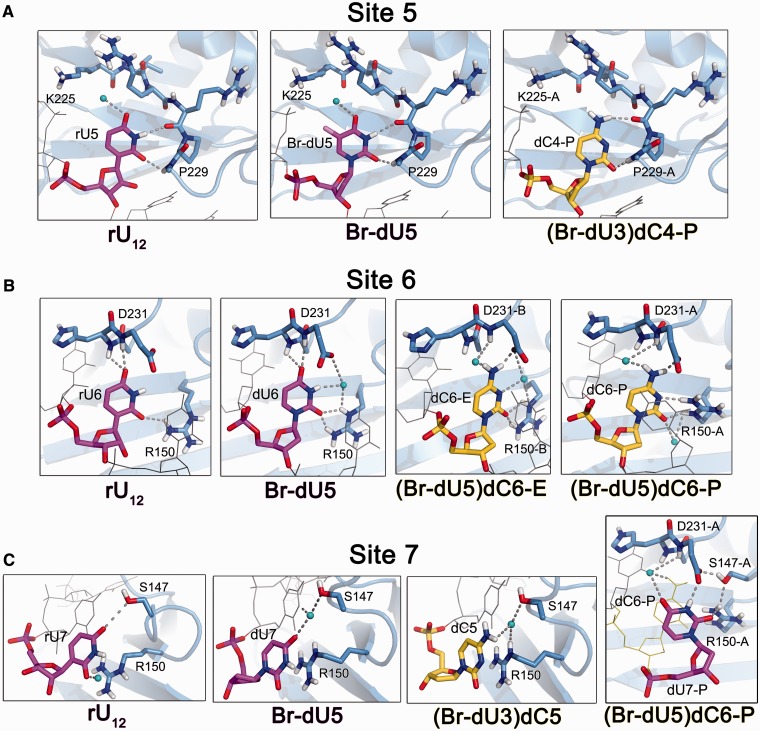
Figure 4.
Structural comparison of U2AF65 RRM1 recognizing uracil versus cytosine. (A) Fifth nucleotide binding sites. The U2AF65 interactions are nearly identical among the Br-dU5 nucleotides from oligonucleotides ‘E’ and ‘P’ of the Br-dU5 structure and the corresponding dU4 nucleotides of the Br-dU3 structure. One example of cytidine in the fifth binding site is available from oligonucleotide ‘P’ of the (Br-dU3)dC4 structure. Supplementary Movie S2 illustrates the cytidine-induced changes at the fifth binding site. (B) Sixth nucleotide binding sites. The U2AF65 interactions are nearly identical among the dU6 nucleotides from oligonucleotides ‘E’ and ‘P’ of the Br-dU5 structure and the corresponding dU5 nucleotides of the Br-dU3 structure. The dC6 interactions of the (Br-dU5)dC6 oligonucleotides ‘E’ and ‘P’ differ slightly in the rotation of the R150 side chain and are shown in separate panels. As shown in C, the movement of R150-A in turn alters dU7-P in the seventh binding site of the (Br-dU5)dC6 oligonucleotide ‘P’. Supplementary Movie S3 illustrates the cytidine-induced changes at the sixth binding site of (Br-dU5)dC6 oligonucleotide ‘P’. (C) Seventh nucleotide binding sites. Cytidine is bound in the seventh binding sites of both oligonucleotides in the (Br-dU3)dC5 structure. The final panel compares the cytidine-induced shift in the dU7 binding site of the (Br-dU5)dC6 oligonucleotide ‘P’.
Each structure offers two crystallographically independent views of the protein/oligonucleotide complex (Figure 2A), with the exceptions of four copies for the dU2AF651,2/(Br-dU5)dC1 structure and one copy for the original dU2AF651,2/rU12 structure. The backbone conformations of the individual RRM1 or RRM2 domains were nearly identical among the structures (rmsd 0.25–0.35 Å between Cα atoms of the RRMs), whereas the relative RRM positions differ (rmsd 6–7 Å between Cα atoms of the overall polypeptide chains) (Supplementary Figure S3A). The overall domain arrangements are likely to be influenced by the internal deletion within inter-RRM linker that is required for crystallization and hence also differ from the NMR-based model for the wild-type U2AF651,2 bound to a 9-mer poly(rU) RNA (26). Nevertheless, the nucleotide interactions by the individual RRMs in the crystal structures agree with the solution data (26) and as such offer the means to illustrate the local U2AF65 interactions with cytidine-substituted Py tracts at high resolution.
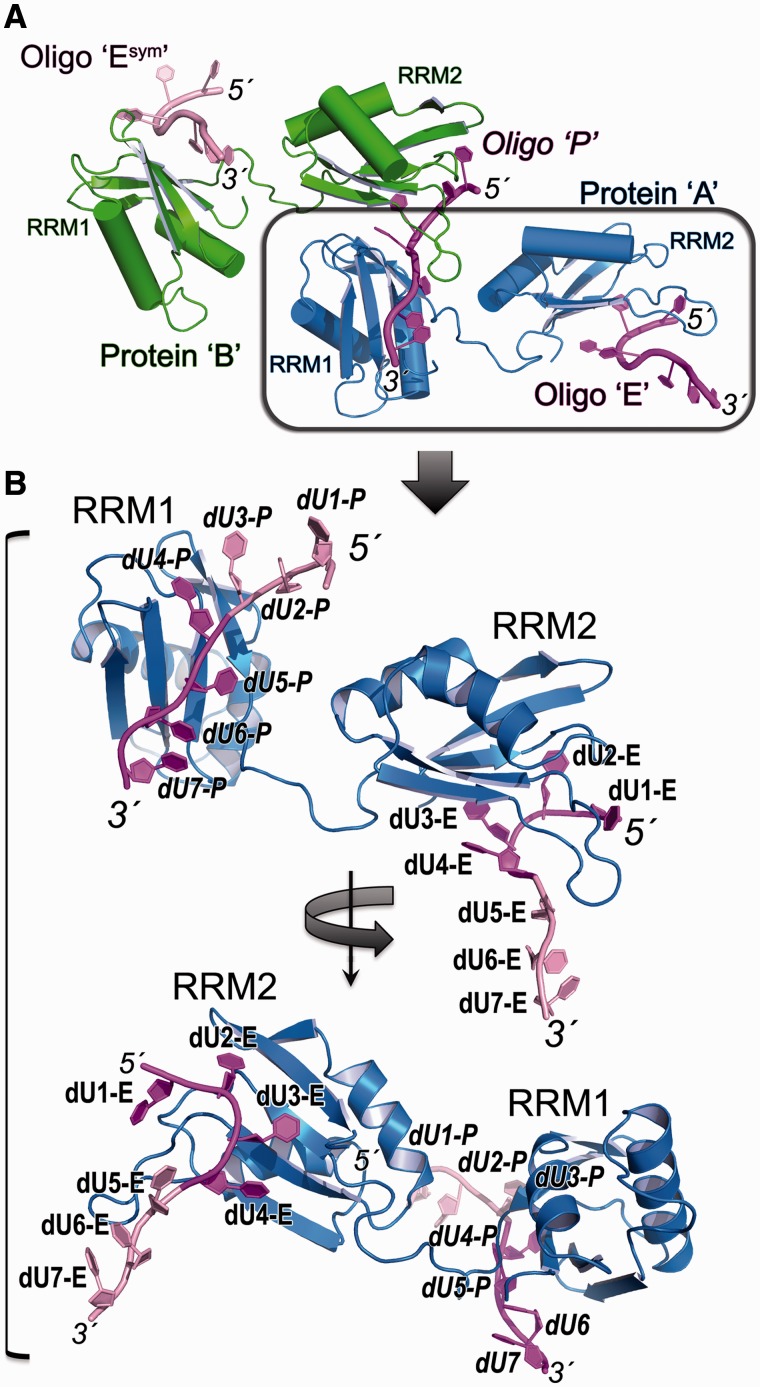
Figure 2.
Overall structures of U2AF65 recognizing Py tracts. (A) Overall packing of the two U2AF65/Py tract complexes in the crystallographic asymmetric units. Protein ‘A’ (blue) respectively binds the 5′ and 3′ regions of oligonucleotides ‘E’ and ‘P’ (magenta); Protein ‘B’ (green) respectively binds the 5′ and 3′ regions of oligonucleotides ‘P’ and symmetry-related ‘Esym’ (pink). The boxed complex is expanded in B. (B) Representative structure of dU2AF651,2 (chain ‘A’) bound to the Br-dU5 oligonucleotide. The four nucleotides contacting each RRM are coloured magenta and numbered as discussed in the text. The central, fourth nucleotide contacts both RRM1 and RRM2 by crystallographic symmetry as shown in A. Top, view into the face of RRM1 bound to the 3′ region in an identical orientation as the boxed polypeptide in A; Bottom, view into the face of RRM2 bound to the 5′ region following 2-fold rotation about the y-axis.
The dU4-dU7 of both complexes and dU1-dU4 in one of the two complexes (oligonucleotide ‘E’) in the crystallographic asymmetric unit correspond to the dU2AF651,2-bound rU12-conformation (rmsd of matching O4, N3, O2 atoms is 0.48 Å for dU4-E–dU7-E/rU4–rU7, 0.50 Å for dU4-P–dU7-P/rU4–rU7 and 0.73 Å rmsd for dU1-E–dU4-E/rU4–rU7) (Supplementary Figure S3B and D). The dU1 and dU2 positions of the other crystallographically independent complex (oligonucleotide ‘P’ in Figure 2) differ from oligonucleotide ‘E’ and the RNA counterpart (rmsd is 6.4 Å for dU1-P – dU4-P/rU1 – rU4 compared with 0.42 Å for dU3-P – dU4-P/rU3 – rU4) (Supplementary Figure S3C). The altered positions of the dU1-P and consequently dU2-P nucleotides likely arise from weakened interactions in the absence of the rU1 5′ terminal phosphate and hydroxyl group (Supplementary Figure S4). Apart from these differences in the first and second nucleotides of oligonucleotide ‘P’, U2AF65 recognizes the dUs in a similar manner as the uridines of the rU12 RNA (Figures 3–4, Supplementary Figure S5).
Figure 3.
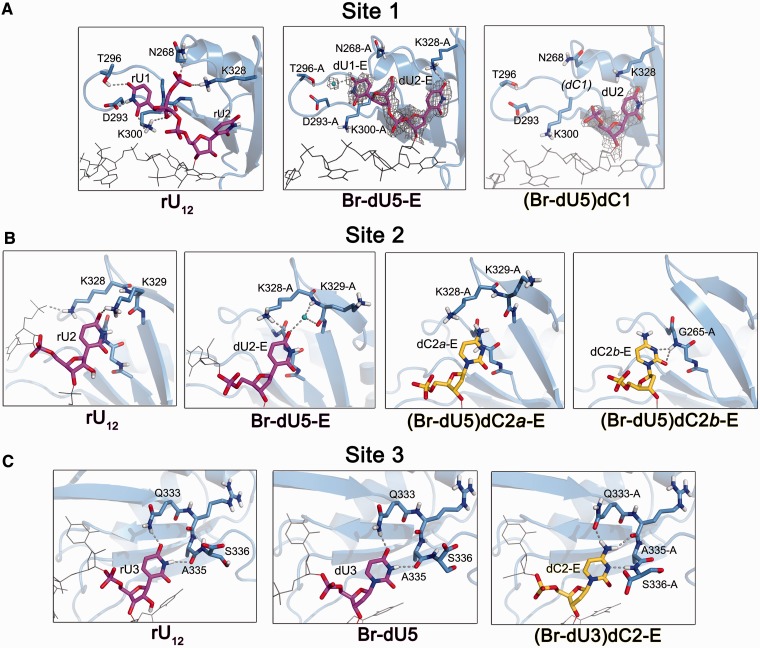
Structural comparison of U2AF65 RRM2 recognizing uracil versus cytosine. (A) First nucleotide binding sites. The dU1 and dU2 of Br-dU5 oligonucleotides ‘E’ correspond to the binding sites of the RNA counterpart and are shown in A and B. The distinct binding sites of dU1 and dU2 from the Br-dU5 ‘P’ oligonucleotides are shown in Supplementary Figure S4A and B. SigmaA-weighted 2|Fo|-|Fc| electron density maps are contoured at 1 σ. The dC1 is disordered and lacks apparent electron density in all four oligonucleotides (‘E’, ‘H’, ‘K’ and ‘P’) of the (Br-dU5)dC1 structure. The neighbouring dU2 of (Br-dU5)dC1 oligonucleotide ‘P’ also is disordered. (B) Second nucleotide binding sites. One example of cytidine in the second binding site of oligonucleotide ‘E’ is available from the (Br-dU5)dC2 structure. The two alternative conformations of dC2-E are shown in separate panels for clarity. The distinct cytidine binding site of (Br-dU5)dC2 oligonucleotide ‘P’ is shown in Supplementary Figure S4C and is nearly identical for (Br-dU3)dC2-P. (C) Third nucleotide binding sites. The U2AF65 interactions are nearly identical with the dU3 nucleotide of the Br-dU5 structure and corresponding dU2 nucleotide of Br-dU3 structure in both oligonucleotides ‘E’ and ‘P’. One example of cytidine in the third binding site is available from oligonucleotide ‘E’ of the (Br-dU3)dC2 structure. Supplementary Movie S1 illustrates the cytidine-induced changes at the third binding site. In Figures 3 and 4, the uridines are coloured magenta, cytidines are yellow, and grey lines show the nucleotides adjoining the site of interest. Polar hydrogens are included for clarity. Dashed lines indicate hydrogen bonds (<3.5 Å between non-hydrogen atoms).
The C-terminal RRM2 of each polypeptide recognizes four nucleotides in the 5′ region of one oligonucleotide (dU1–dU4), and the N-terminal RRM1 recognizes four nucleotides in the 3′ region of a separate oligonucleotide (dU4–dU7) (Figure 2). Consequently, the central dU4 nucleotide is enclosed by RRM1 and RRM2 contributed by distinct polypeptides (Supplementary Figure S5). Despite differences in the relative rotation of the two RRMs among the crystallographically independent complexes, the central dU4 base continues to engage in similar U2AF65 contacts among the structures. The K260, N289 and F304 residues that interact with rU4 (here dU4) contribute significantly to the U2AF651,2 affinity for poly-rU RNA (by 36-fold for K260A/N289A double mutation and 73-fold for F304A mutation) (25). Given the importance of these interactions and the similar interactions, despite crystallographically independent environments, we suggest that the rU4/dU4 interactions are the coalescence of two separable binding sites for nucleotides on U2AF65 RRM1 and RRM2 such that the crystal structure in effect represents eight distinct nucleotide binding sites on the U2AF65 protein. Accordingly, in-depth analyses of the U2AF651,2 affinities for uridine tracts of various lengths suggest a binding site size of 8–9 nucleotides (26). Efforts to substitute a cytidine at the fourth binding site of the crystal structures were unsuccessful, possibly owing to the dual sets of interactions at this site. Instead, we have determined structures that seek to place cytidines at six of the U2AF65/nucleotide binding sites.
The U2AF65 RRM2 structure has difficulty accommodating cytidines in the 5′ region of the Py tract
The structures of dU2AF651,2 bound to either (Br-dU5)dC1, (Br-dU5)dC2 or (Br-dU3)dC2 oligonucleotides suggest that the U2AF65 RRM2 has difficulty forming stable interactions with cytidine substitutions in the 5′ half of the bound oligonucleotide (Figure 3). Based on the 2.2 Å resolution Br-dU5 structure, the dU1 of the oligonucleotide ‘E’ binds a similar site as rU1 of the original 2.5 Å resolution structure of the dU2AF651,2/rU12 structure (Figure 3A). A bound water molecule that mediates a hydrogen bond between dU1-E and T296 of the Br-dU5 structures is likely to be present but unresolved at the lower, 2.5 Å resolution of the dU2AF651,2/rU12 complex, given that the ‘hydrogen bond’ between T296 and rU1 of the RNA-bound complex is relatively long (3.4 Å). As described in more depth for the second site, a K328 side chain that engages the 5′ phosphate of the rU1 nucleotide has shifted in the Br-dU5 structure to interact with the uracil-O4 of the neighbouring dU2. With the intention of placing a cytidine at the first binding site, we determined the structure of dU2AF651,2 bound to a Br-dU5 variant with a single cytidine substituted at the first nucleotide [(Br-dU5)dC1)] at 2.5 Å resolution (Figure 3A, Supplementary Table S1). The four complexes in the crystallographic asymmetric unit of the (Br-dU5)dC1 structure exhibited the Br-dU in the expected fifth binding site (Figure 2B, Supplementary Figure S2F). Rather than attempting to adapt to the differences between uracil and cytosine (N3-H and O4 compared with N3 and N4-H atoms, respectively), the first cytidine (dC1) became disordered and lacked apparent electron density in any of the four complexes (Figure 3A). In addition, electron density for the adjacent dU2 nucleotide could not be interpreted in one of the dU2AF651,2/(Br-dU5)dC1 copies (oligonucleotide ‘P’). Although U2AF65 interactions with the 5′ terminal deoxy-ribose nucleotides are weakened by the absence of the 2′ hydroxyl group and terminal phosphate, comparision of the disordered dC1 and evident dU1 suggests that U2AF65 has more difficulty forming stable contacts with a cytosine than uracil at the first position of the Py tract. One possible explanation is that the negative charge of the D293 side chain is less favourable adjacent a cytosine-N3 than a uracil-N3-H.
To capture a cytidine in the second binding site, we determined the 2.4 and 2.2 Å resolution structures of dU2AF651,2 bound to the (Br-dU5)dC2 and (Br-dU3)dC2 oligonucleotides, respectively (Figure 3B, Supplementary Table S1). Both copies of the Br-dU5)dC2 oligonucleotides in the crystallographic asymmetric unit bound the Br-dU in the fifth and dC2 in the second binding sites of U2AF65 (Supplementary Figure S2G), whereas only the oligonucleotide ‘P’ of the (Br-dU3)dC2 structure placed the dC2 in the second binding site. The rU2 of the dU2AF651,2/rU12 structure chiefly interacts by base stacking with glycines G264 and G265 and a single hydrogen bond between the uracil-O4 and the K329 side chain. The U2AF65 interactions with the dU2 counterpart of Br-dU5 oligonucleotide ‘E’ remained in a similar location as the rU2, except that in the absence of prior engagement by the 5′ phosphate, the K328 side chain replaces the hydrogen bond of K329 with the uracil-O4. The movement of K329 appears to facilitate formation of a water-mediated hydrogen bond between the polypeptide backbone and the uracil-O4 in the higher resolution Br-dU5 structure. In the presence of cytosine, the K328 side chain of the (Br-dU5)dC2 oligonucleotide ‘E’ shifted slightly to avoid the major conformation of the cytosine exocyclic amine. In the minor alternative conformation of the (BrU5)C2 oligonucleotide ‘E’, the cytosine moved to accept a hydrogen bond from the backbone N-H of the glycine residue (G265) (Figure 3B, far right). The shifts in the dC2 positions in turn disrupt the U2AF65 interactions with the adjacent dU1, which is disordered and lacks interpretable electron density in any of the structures with cytidine bound at the second U2AF65 site. The dC2 of the (Br-dU5)dC2 and (Br-dU3)dC2 ‘P’ oligonucleotides rotated too stack against the L330 side chains yet otherwise maintained similar interactions as the major conformation of oligonucleotides ‘E’ (Supplementary Figure S4C). Although these structural changes at the 5′ terminus of the oligonucleotide cannot be assumed to parallel the RNA counterparts, the observed loss of stable U2AF65 interactions suggests that the proximity of a positively charged K328 or K329 lysine could favour a uracil-O4 over a cytosine-N4H3 at the second position of the Py tract.
As aforementioned, the binding register of one of the (Br-dU3)dC2 complexes (oligonucleotide ‘P’) places a cytosine in the second site, where it engages in similar U2AF65 interactions as the (Br-dU5)dC2 oligonucleotide ‘P’. The binding register for the other (Br-dU3)dC2 complex (oligonucleotide ‘E’) places the cytidine in the third and the Br-dU in the fourth of the U2AF65 binding sites (Figure 1B, Supplementary Figure S2B), apparently stabilizing one of the two alternative binding registers of the parental Br-dU3 structure. Among the structures, only the dU2AF651,2/(Br-dU3)dC2 oligonucleotide ‘E’ bound the Br-dU in the third rather than fourth or fifth sites, suggesting that the energetic penalty for fitting a cytidine into the third site is on par with shifting a Br-dU to a less preferable site.
The cytidine at the third site induces relatively large, well-ordered changes in the U2AF65 structure (Figure 3C, Supplementary Movie S1). The U2AF65 interactions are indistinguishable between the rU3 of the original, RNA-bound structure and the dU3 from either the ‘E’ or ‘P’ oligonucleotides of the Br-dU5 structure. Namely, the Q333 side chain and A335 carbonyl respectively engage the uracil-O4 and N3-H in hydrogen bonds. Three structural changes in the U2AF65 protein are observed in response to the uracil-to-cytosine substitution: (i) the Q333 side chain undergoes a torsional rotation to accept a hydrogen bond from the cytosine exocyclic amine; (ii) the backbone carbonyl of R334 moves to accept a second hydrogen bond from this cytosine amine; and (iii) the relatively rigid backbone of S336 undergoes a large ϕ torsional rotation from −142° to +164°, which enables the peptide bond with A335 to donate a hydrogen bond to the cytosine-N3. As the A335 and S336 residues are located at the C-terminus of the crystallized construct of U2AF65, the energetic penalty for adjusting to the third cytidine could be greater and possibly influence downstream residues in the context of the full-length U2AF65 protein.
The U2AF65 RRM1 structure adapts to cytidines in the 3′ region of the Py tract
The structures of dU2AF651,2 bound to either (Br-dU3)dC4, (Br-dU5)dC6 or (Br-dU3)dC5 oligonucleotides suggest that following relatively subtle structural changes, U2AF65 RRM1 can accommodate cytidine substitutions in the 3′ half of the bound oligonucleotide (Figure 4). One of the 2.5 Å resolution dU2AF651,2/(Br-dU3)dC4 complexes (oligonucleotide ‘P’) offered a well-defined view of a cytidine in the fifth binding site (Figure 4A, Supplementary Figure S2C). In the second dU2AF651,2/(Br-dU3)dC4 complex of the crystallographic asymmetric unit (oligonucleotide ‘E’), alternative binding registers place the cytidine with partial occupancy in the fifth and sixth binding sites as observed for the Br-dU3 parent (Figure 1B, Supplementary Figure S2C). This stabilized binding register for one of the complexes suggests a slight preference for U2AF65 to accommodate a cytosine at the fifth over sixth sites. Despite the introduction of a 5-bromo group, the U2AF65 interactions with the fifth dU of both ‘P’ and ‘E’ oligonucleotides in the baseline Br-dU5 structure remain similar to the RNA-bound structure. Namely, the backbone carbonyl of U2AF65 R228 and the amide of H230 respectively form hydrogen bonds with the uracil N3-H and O2 groups. Despite these nucleotide interactions with a relative rigid protein backbone, U2AF65 readily accommodates the switch to the N3 hydrogen bond acceptor of the cytosine at the fifth site. A slight movement of the deoxy-cytidine relative to the uridine enables the U2AF65 R228 carbonyl to accept an analogous hydrogen bond from the cytosine exocyclic amine, and the H230-NH – cytosine-O2 interaction remains unperturbed (Figure 4A, Supplementary Movie S2). An ordered water molecule bound to the uracil-O4 also appears to have been lost, possibly owing to the switch from a hydrogen bond acceptor to a donor for the cytosine exocyclic amine in proximity to the U2AF65 K225 and R227 side chains.
U2AF65 undergoes a number of structural changes to adapt to a cytosine at the sixth binding site (Figure 4B, Supplementary Movie S3). The Br-dU occupies the fifth and cytidine the sixth binding sites in both complexes of the dU2AF651,2/(Br-dU5)dC6 structure (Supplementary Figure S2H), which is the highest resolution among the dU2AF651,2 structures (1.9 Å). The dU6 in both ‘P’ and ‘E’ oligonucleotides of the Br-dU5 structure both exhibit similar U2AF65 interactions as the RNA counterpart. The uracil-O4 accepts hydrogen bonds from the backbone N-H of U2AF65 D231 and H230, the latter of which is bifurcated by the hydrogen bond donated to the preceding dU6-O2. The slightly higher resolution of the Br-dU5 compared with the rU12 structure (2.2 Å and 2.5 Å resolution, respectively) reveals an ordered water molecule that mediates hydrogen bonds among R150, D231 and the uracil-N3-H of both ‘P’ and ‘E’ oligonucleotides. In addition, the R150 side chains of both the ‘P’ and ‘E’ oligonucleotides have rotamers that can donate two hydrogen bonds to the uracil-O2 of dU6.
Following substitution of the cytosine at the sixth site, the D231 side chain rotates to accept a direct hydrogen bond from the cytosine exocyclic amine (Figure 4B). The introduction of an ordered water molecule enables the U2AF65 D231 peptide N-H to donate a water-mediated hydrogen bond to the cytosine exocyclic amine. Remarkably, RNA binding selects from two alternative conformations of the R150 side chain that are apparent in the high resolution structure of the apo-U2AF65 RRM1 (35). The sixth uracil base is only observed to interact with one of two alternative R150 conformations, whereas the hydrogen bond pattern of a cytosine is compatible with either. Accordingly, the sixth cytosine selects a different alternative conformation in each of the two crystallographically distinct dU2AF651,2/(Br-dU5)dC6 copies. In one of the two (Br-dU5)dC6 complexes (oligonucleotide ‘E’), the R150 remains in a similar location as when bound to uracil, where the side chain contributes two direct hydrogen bonds to the cytosine-O2 and a water-mediated hydrogen bond to the cytosine-N3. The R150 side chain of the other dU2AF651,2/(Br-dU5)dC6 copy (oligonucleotide ‘P’) has rotated to replace the water molecule bound to the cytosine-N3. This movement of the R150 side chain alters stacking with the adjacent dU7 base, which in turn shifts to form a distinct set of apparently favourable interactions with U2AF65, including a direct hydrogen bond from the uracil N-H to the available oxygen of the D231 side chain and an indirect hydrogen bond via the water molecule bound to the preceding cytosine amine (Figure 4C, far right panel). Together, these structural changes indicate that cytidine is compatible with a tight network of U2AF65 interactions at the sixth binding site.
Lastly, the dU2AF65/(Br-dU3)dC5 structure provides 2.2 Å resolution views of a cytidine in the seventh binding site (Supplementary Table S1). The binding registers of both dU2AF651,2/(Br-dU3)dC5 complexes shifted to place the Br-dU5 in the fourth and dC5 in the final seventh site (Figure 1B, Supplementary Figure S2D). The stabilization of this binding register compared with the mixture of Br-dU3 at the fourth and fifth sites of the parent suggests a slight preference for cytosine over uracil at the seventh binding site. By comparison with other binding sites, the interactions of U2AF65 with nucleotides in the seventh site are minimal. The seventh uracils of all the poly-rU-containing structures primarily interact by aromatic stacking on the R150 side chain of U2AF65. At the higher resolutions of the Br-dU5 and (Br-dU3)dC5 structures compared with the rU12 counterpart (2.2 Å versus 2.5 Å resolutions), a water-mediated hydrogen bond between the S147 side chain and the uracil-O4 or cytidine-N4H2 becomes apparent. Following a slight shift in the position of an intermediary water molecule, the cytidine at the seventh site of the dU2AF651,2/(Br-dU3)dC5 structure otherwise continues to stack with the R150 in a comparable manner as the uracil.
U2AF65 prefers uridines in the 5′ region of the Py tract and tolerates cytidines in the 3′ region
The different abilities of the U2AF65 RRM structures to adapt to cytidines predicted that U2AF65 would display region-dependent preferences for cytidines and uridines in the Py tract. To facilitate interpretation of the crystal structures, we focused on determining the affinities of the minimal U2AF65 RRM1-RRM2 domain (U2AF651,2) for core Py tracts derived from the prototypical adenovirus major late promoter transcript (rAdML13). Our primary method of steady-state SPR has the advantage of concurrently monitoring the apparent binding stoichiometry (Supplementary Figure S6, see ‘Materials and Methods’ section). ITC experiments independently corroborated the specificities determined by SPR (Supplementary Figure S7).
AdML variants with a few cytidine substitutions in the Py tract previously were shown to support in vitro splicing and spliceosome complex formation at levels comparable with those of the unmodified pre-mRNA parent, whereas a polycytidine tract abolishes detectable use of the adjacent 3′ splice site (9). We first investigated whether these splicing activities correlated with the abilities of U2AF651,2 to bind the Py tract RNAs (Table 1). In agreement with the previously reported splicing activities, the affinities of U2AF651,2 for Py tracts with one, two and even three cytidine mutations at the fifth, seventh, ninth or tenth positions remained similar as for the rAdML13 oligonucleotide. We also found that U2AF651,2 exhibited little or no detectable binding to a polyC tract of equivalent length [rC(4–11)].
We next compared tracts of three, four or five uridines located in either the 5′ or 3′ regions of a Py tract otherwise composed of cytidines (Table 1). In all cases, U2AF65 preferred uridines to cytidines in the 5′ region of the Py tract, whereas cytidines could effectively substitute for uridines in the 3′ region. Consistent with the observation that four nucleotides bind to each U2AF65 RRM and considering the shared fourth nucleotide at the RRM1/RRM2 interface, it is not surprising that the U2AF651,2 affinity for a tract of three uridines in the 5′ region of the Py tract decreased slightly relative to four or five uridines. The specificity of U2AF651,2 protein was greatest for the four-uridine tract (5-fold preference for four consecutive uridines in the 5′ as opposed to 3′ regions of an otherwise cytidine-rich Py tract, respectively named 5′-4rU and 3′-4rU RNAs, Table 1). Given that the U2AF65 RRM1 and RRM2 respectively binds near the 3′ and 5′ termini of the Py tract, the region-dependent uridine/cytidine preferences agreed with the relative ease for the U2AF65 RRM1 structure to accommodate cytidines in comparison with the RRM2.
Residues in U2AF65 RRM2 contribute to region-dependent pyrimidine specificity
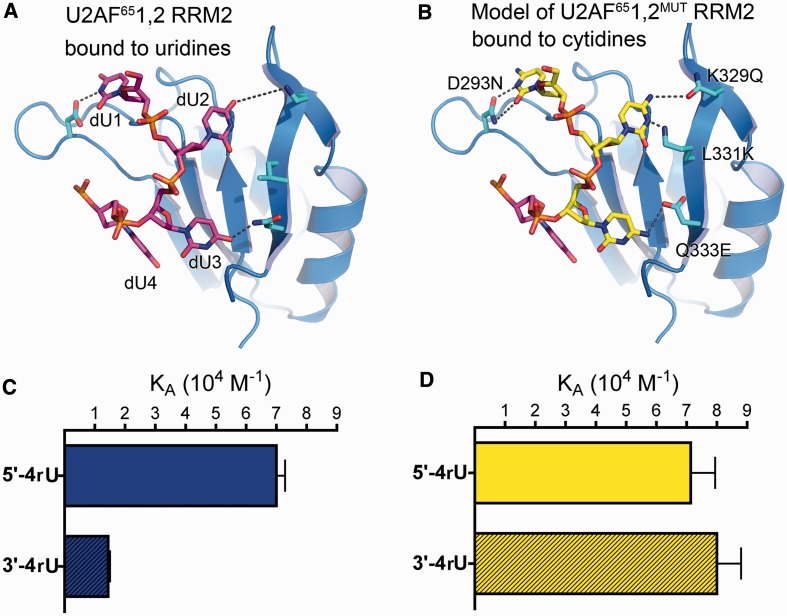
We proceeded to test the direct involvement of the U2AF65 RRM2 in determining the specificity of U2AF65 for uridines in the 5′ region of the Py tract. Based on the structures, we introduced four mutations within the RRM2 of U2AF651,2 (U2AF651,2MUT) that were intended to favour cytidine over uridine binding (Figure 5A and B): (i) D293N, replacing an aspartate that would disfavour a cytosine-N3 at the first site with an asparagine side chain, which is expected to form similar hydrogen bonds with cytosine or uracil; (ii) K329Q, likewise substituting a ‘neutral’ side chain in place of a lysine that contributes an unfavourable electrostatic environment for a cytidine-N4H2 at the second site; (iii) L331K, changing a leucine to a hydrogen bond donor that could potential interact with a cytidine-N3 at the second site; and (iv) Q333E, replacing a glutamine with a stringent hydrogen bond acceptor for interactions with a cytidine-N4H2 at the third site. The U2AF651,2MUT variant continued to bind the 5′-4rU Py tract RNA with indistinguishable affinity as the wild-type protein, indicating that the RRM2 mutations did not detectably penalize recognition of uridines in the 5′ region of the Py tract. As expected based on the crystal structures, the U2AF651,2MUT affinity for the 3′-4rU RNA increased by 5.5-fold to a level comparable with the 5′-4rU counterpart, indicating that the mutated RRM2 had lost the ability to discriminate against cytidines in the 5′ region of the Py tract (Figure 5C and D). These results directly implicated RRM2 in determining the preference of U2AF65 for uridines in the 5′ region of the Py tract.
Figure 5.
U2AF65 RRM2 determines region-dependent uridine/cytidine preferences in Py tracts. (A) Structure of dU2AF651,2 RRM2 bound to dU1-dU4 (from Br-dU5). The unmodified counterparts of the mutated residues are shown as ball-and-stick representations and coloured by atom: carbons, cyan; oxygens, red; nitrogens, blue. (B) Model of U2AF651,2MUT RRM2 showing the predicted interactions of the four mutated D293N/K329Q/L331K/Q333E residues with cytidines (yellow) (C and D) Bar graphs of the 5′-4rU and 3′-4rU (shaded) RNAs affinities for the (C) wild-type U2AF651,2 protein (blue) compared with the (D) D293N/K329Q/L331K/Q333E U2AF651,2MUT protein (yellow).
Given the difficulties of predicting RRM–RNA interactions (36), we did not attempt to enhance the specificity of a promiscuous U2AF65 RRM1. However, several observations indirectly implicated RRM1 in recognizing the 3′ region of the Py tract, including the following: (i) the U2AF651,2 affinity for these Py tracts is higher than expected for RRM2 alone, whereas the inter-RRM linker is not directly involved in RNA binding (26,29), leaving only RRM1; (ii) the U2AF651,2 affinities for Py tracts with identical pyrimidine contents depended on the 5′ versus 3′ locations of the four uridine tract; and (iii) the mutations in RRM2 only affect association with the 3′-4rU RNA but have no effect on binding the 5′-4rU RNA. These strong region-dependent effects call for respective contributions by U2AF65 RRM2 in binding the 5′ nucleotides and RRM1 in binding the 3′ nucleotides of the 13-mer Py tracts.
Py tract variants select different U2AF65 conformations
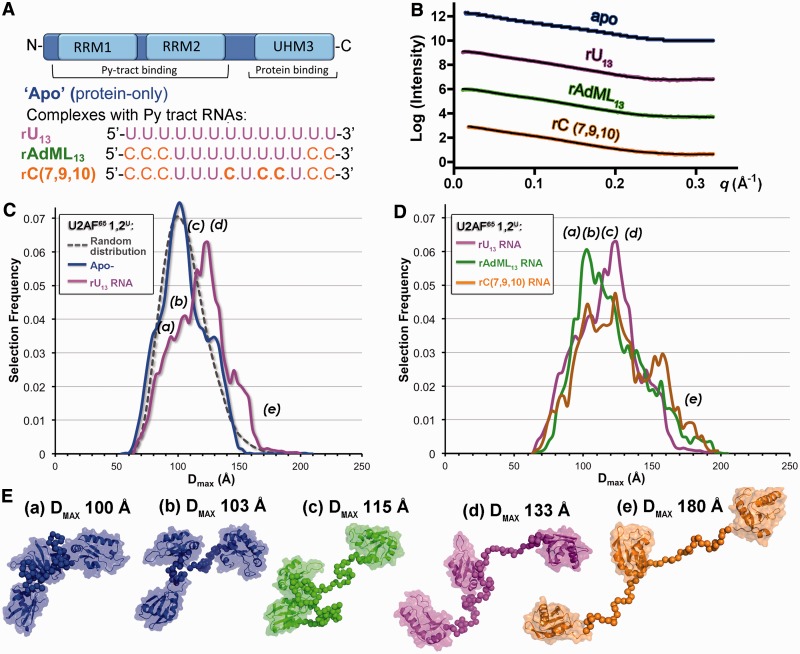
The apparent difference in the specificities for the two U2AF65 RRMs increased the possibility that U2AF65 adapts to degenerate Py tracts by modulating the proximities of a promiscuous RRM1 and stringent RRM2 domains. The prior NMR methods indicated that a 9-mer poly-rU RNA selects an ‘open’ U2AF651,2 conformation with a side-by-side arrangement of the RRMs. In the absence of RNA, the ‘open’ U2AF651,2 conformation was suggested to be minor relative to a major ‘closed’ conformation, in which the RNA binding surface of RRM1 is masked by RRM2 (26). Although broader than the distribution detected by NMR methods, we also observe a range of solution conformations for the apo-U2AF651,2 protein by SAXS (37). Here, we further investigated the influence of binding Py tracts that have distinct cytidine compositions on the distribution of U2AF65 conformations (Figure 6). To ensure that the scattering data primarily reflect the protein rather than the RNA conformations, we used a larger U2AF65 construct (U2AF651,2U) that included the C-terminal UHM in addition to the RRM1 and RRM2 (Figure 6A). The U2AF65 UHM lacks detectable RNA affinity in isolation (39), and NMR spectra suggest that the U2AF651,2 contacts the Py tract in a similar manner in the presence of the UHM domain (26). We also have shown that additional residues surrounding the core U2AF651,2 fragment have little detectable influence on the conformational ensemble (37). As such, we use the U2AF651,2U protein as a means to study the influence of different Py tract RNA sequences on the U2AF65 inter-domain proximities in the solution pool. We compared the effects of three representative Py tracts on the distribution of U2AF651,2U conformations. These Py tracts include a homogeneous 13-mer uridine tract (rU13), the unmodified AdML Py tract composed of an eight-uridine core embedded within five cytidines (rAdML13) and the rC(7,9,10) variant with three internal cytidine substitutions. All three RNAs share a 13-nucleotide length and comparable affinities for U2AF65, despite the different number of cytidines (Figure 6A).
Figure 6.
Py tract sequences redistribute the U2AF65 conformational ensemble. (A) Schematic representation of the U2AF6512U domain and Py tract RNA sequences used for SAXS experiments. (B) Experimental SAXS profiles (coloured circles) compared with profiles calculated from the best fitting ensembles (black lines). Intensity data are offset for clarity. Colours are consistent throughout: rU13, magenta; rAdML13, green; rC(7,9,10), orange. The SAXS profile of apo-U2AF6512U (‘apo’, blue) from reference (38) is shown for comparison. (C) The DMAX distributions of ensembles calculated using EOM (33) (solid coloured lines) for U2AF6512U alone or bound to the rU13 RNA compared with the randomized starting pool (dashed grey line). Lower case letters mark the DMAX values of the models shown in E. (D) The DMAX distributions of U2AF6512U ensembles bound to rU13, rAdML13 or rC(7,9,10) Py tract RNAs. (E) Representative models from the EOM analyses and corresponding DMAX values.
The SAXS data sets for U2AF651,2U with and without RNA appear monodisperse and extend beyond q = 0.30 Å−1 (Figure 6B, Supplementary Figure S8). In the absence of RNA, the average solution shape of the apo-U2AF651,2U protein comprises three lobes corresponding to the relatively separated RRM1, RRM2 and UHM domains (38). We re-analysed the apo-U2AF651,2U SAXS data using an ensemble approach (33), in which a subset of 20 structures that best fit the data was selected from a starting pool of 10 000 conformations comprising the RRM1, RRM2 and UHM structures tethered in randomized orientations and proximities by ab initio linkers. As observed for the apo-U2AF651,2 (37), the distribution of selected apo-U2AF651,2U conformations closely matches that of the randomized starting pool (Figure 6C). Although no change in the average molecular dimensions was apparent following addition of the Py tract RNAs (38) (Supplementary Table S2), the ensemble analyses revealed distinct changes in the distribution of the solution conformations (Figure 6C–E). The distribution of the U2AF651,2U conformations following association with the homogeneous rU13 tract remained broad, but the molecular dimensions of the most prevalent conformations were slightly larger than for the apo-protein (∼120 Å compared with 100 Å). The conformational ensemble of the rAdML13-bound U2AF651,2U also remained broad, but the most prevalent selected conformations shifted to back to a compact arrangement of the U2AF65 RRMs (∼100 Å). The cytidine-interrupted rC(7,9,10) tract increased the prevalence of more extended U2AF651,2U conformations (∼150–180 Å) but lacked clear preference for a conformation of any given size. In summary, the rAdML13, rU13 and rC(7,9,10) Py tracts select subsets of U2AF65 conformations with distinct inter-RRM proximities from the pre-existing solution ensemble of the apo-protein.
DISCUSSION
Here, a series of high resolution U2AF65 structures suggest that a subset of binding sites in the N-terminal RRM1 can tolerate cytidine substitutions of uridine-rich Py tracts more readily than others in the C-terminal RRM2 (Figures 3 and 4). We note that the two deoxy-cytidines in the 5′ terminal positions of the RRM2-bound oligonucleotide are poorly ordered and could be influenced by the minimal backbone. However, the increased disorder of the cytosines relative to the uracils of the dU counterparts is likely to reflect weakened interactions that can be extended to U2AF65 recognition of Py tract RNAs. Accordingly, line broadening of the NMR signals for U2AF65 bound to poly(rU) suggest some flexibility in the RNA interface (26). In light of the C-to-N-terminal orientation of the U2AF65 RRMs bound to the 5′–3′ orientation of the bound RNA strand, the discrimination of U2AF65 against cytidines in the 5′ region of Py tracts and permissiveness towards cytidines in the 3′ region supports the structural conclusion that RRM2 has more difficulty adapting to cytidines than does RRM1 (Table 1). Site-directed mutagenesis confirms a role for the U2AF65 RRM2 in specifying uridines near the 5′ region of the Py tract and indirectly implicates RRM1 in adapting to the 3′ region (Figure 5). Complementary SAXS studies further demonstrate that Py tracts with different cytidine contents select U2AF65 conformations with different inter-RRM spacings from the solution ensemble (Figures 6 and 7).
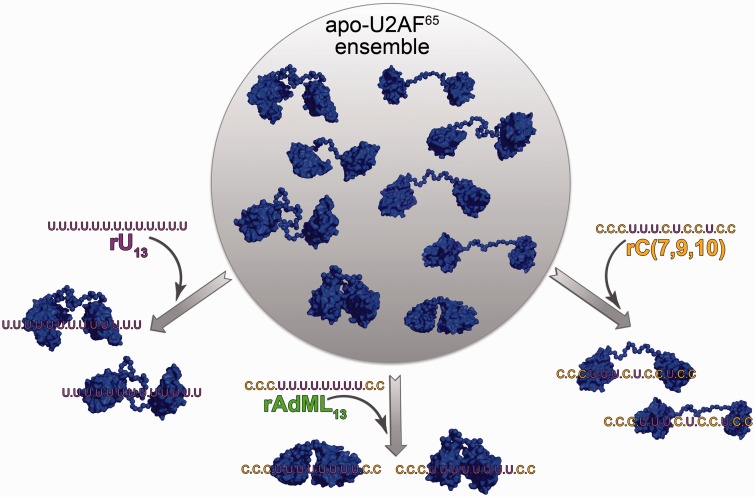
Figure 7.
Model for conformational selection of U2AF65 RRM1-RRM2 by diverse Py tracts. Selection of apo-U2AF651,2 conformations by binding the rU13, rAdML13 and rC(7,9,10) Py tracts used in this study. Representative conformations of tandem RRMs from the selected ensembles are shown with space-filling graphics. Although linear RNA sequences are shown for clarity, the 3′ splice site is likely to exhibit curvature in three-dimensions (38).
The prevalent conformations detected by SAXS of the apo-U2AF651,2U protein are likely to correspond to the ‘closed’ NMR model, with the distinction that the SAXS analyses provides evidence that elongated conformations beyond the radius of the paramagnetic relaxation enhancement labels contribute to the solution ensemble (37). Given that the molecular dimensions of the ‘open’ and ‘closed’ U2AF65 models are comparable at the resolution of the SAXS analyses, the most prevalent conformations of the U2AF651,2U/AdML13 complex are likely to correspond to ‘open’, side-by-side conformations binding the central eight uridines of the AdML13 RNA comparable with the NMR model of the U2AF651,2/rU9 complex (26). We suggest that the increased molecular dimensions of the dominant U2AF651,2U/rU13 conformations arises from the availability of distal RRM binding sites along the longer uridine tract. The U2AF651,2U/rC(7,9,10) ensemble lacks a dominant conformation, which is consistent with a model in which the U2AF65 RRM2 specifies the remaining three contiguous uridines, whereas RRM1 is compatible with a number of cytidine-containing binding sites near the 3′ end of the Py tract sequence (e.g. two redundant CUCC motifs at nucleotides 7–10 or 10–13).
Together, the U2AF65 structures and binding preferences reported here support and refine a recent model for U2AF65 multi-domain conformational selection of pre-mRNA splice sites, which is based on elegant NMR data and biochemical experiments (26). A main feature of this model is that a minor ‘open’ conformation of apo-U2AF65 is selected by Py tract binding, whereas a major, ‘closed’ conformation, in which the RRM1 RNA binding surface is occluded by interactions with the α-helical ‘back’ of RRM2 and hence unavailable for RNA binding. Based on the structural and affinity data that we present here and in reference (37), we propose three key revisions of this model to better explain the ability of U2AF65 to recognize degenerate pre-mRNA splice sites. First, we note that the RRM masked by the ‘closed’ conformation of apo-U2AF651,2 is promiscuous for RNA sequences. This observation suggests that the ‘closed’ conformation could protect U2AF65 against non-specific RNA interactions and hence inappropriate splice site activation. Second, as shown here for the case of R150 at the seventh nucleotide binding site, local conformational selection of pre-existing alternative side chain conformers can contribute to the ability of U2AF65 to adapt to cytidine substitutions. Third, we suggest that the two-state model could be an oversimplification of the U2AF65 conformations available for selection by degenerate Py tracts. Conformations similar to the ‘open’ and ‘closed’ NMR-based U2AF65 conformations contribute to the solution ensembles detected by X-ray scattering. However, both of these NMR structures are characterized by close contacts between the two U2AF65 RRMs, which are insufficient to describe the X-ray scattering data of the apo-U2AF651,2 protein (37). Instead, the apo-U2AF651,2 and U2AF651,2U scattering data are better fit by broad conformational ensembles closely resembling the randomized starting pools of RRM (and UHM) domains connected by ab initio linkers. This subtle discrepancy over the relative populations of compact ‘closed’/‘open’ and more extended conformations suggests that didactic studies are needed to better understand the outcomes of PRE and SAXS techniques when applied to multi-domain proteins. Regardless, distinct U2AF65 conformations with increased molecular dimensions are enriched in the ensembles bound to the homogeneous rU13 or three-uridine rC(7,9,10) Py tracts. This finding emphasizes that the U2AF65 RRM1 and RRM2 can participate independently in identifying 3′ splice site sequences, which increases the diversity of Py tracts that can be recognized by U2AF65.
In summary, we propose that degenerate Py tracts select distinct proximities of a promiscuous RRM1 and stringent RRM2 from the apo-U2AF65 conformational ensemble, which would shift interspersed cytidines to permissive U2AF65 sites without penalizing RNA affinity (Figure 7). Although this result emphasizes conformational selection over an induced fit mechanism of U2AF65/RNA recognition, the apparently weak association between the RRMs could facilitate ‘fine-tuning’ of initial U2AF65 interactions with the RNA. This revised model for U2AF65 recognition of diverse metazoan splice sites clarifies prior site-specific cross-linking analyses that demonstrated broad, overlapping binding sites for U2AF65 RRM1 and RRM2 along the Py tracts of pre-mRNAs (40). In light of the crystal and SAXS structures, the range of cross-linking patterns arises from the independent adjustment of the U2AF65 RRM1 or RRM2 binding registers along a degenerate Py tract. We note that U2AF65 can accommodate up to five consecutive cytidines in a Py tract without altering RNA affinity or pre-mRNA splicing (Table 1) (6,7,9). By contrast, continuous stretches of cytidines saturate the cytidine-compatible binding sites of U2AF65. As such, these cytidines are forced to engage the stringent U2AF65 sites, which strongly inhibits U2AF65-dependent RNA binding and pre-mRNA splicing.
Our results highlight the ability of a promiscuous RRM1 and specific RRM2 to independently seek compatible binding sites as key factors for human U2AF65 to recognize degenerate splice site signals. As these studies are focused on the human U2AF65 homologue, it remains possible that a ‘closed’ U2AF65 conformation plays a greater role in organisms with short, uridine-rich, consensus Py tracts at the 3′ splice sites, such as Caenorhabditis elegans (41) or Saccharomyces cerevisiae (42). In humans, this model for U2AF65 – pre-mRNA splice site recognition is likely to have important implications for disease-associated mutations. For example, shortening in the length of the Py tract preceding a splice acceptor site of the cystic fibrosis transmembrane conductance regulator (cftr) gene is the most common defect in men with cystic fibrosis and infertility owing to congenital bilateral absence of the vas deferens (13). Normal phenotypes are produced by Py tracts that have stretches of 9 Us (UUUUUUUUUAACAG) and 7 Us (UGUUUUUUUAACAG), whereas splicing of the associated 3′ splice site is nearly abolished by shortening of the Py tract to 5 Us (UGUGUUUUUAACAG) (12). Based on the results presented here, the TG expansion in the 5' region of the cftr Py tract is likely to interfere with the sequence-specific association of the U2AF65 RRM2. Future studies will illuminate further roles for conformational selection in spliceosome assembly at regulated pre-mRNA splice sites and its consequences for human genetic disease.
ACCESSION NUMBERS
Coordinates and the structure factors have been deposited in the Protein Data Bank with accession codes 3VAF (BrU3), 3VAG (BrU3C2), 3VAH [(BrU3)C4], 3VAI [(BrU3)C5], 3VAK [BrU5], 3VAL [(BrU5)C1], 3VAM [(BrU5)C2], 3VAJ [(BrU5)C6] (on hold for publication).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1–8, Supplementary Movies 1–3 and Supplementary References [25,28,43].
FUNDING
National Institutes of Health (NIH) [R01 GM070503 to C.L.K. and R01 GM035490 to M.R.G.]; Minority supplement to NIH Grant [R01 GM070503 to J.L.J.]; The University of Rochester Medical Center Structural Biology & Biophysics Facility is supported by NIH NCRR grants [1S10 RR026501 and 1S10 RR027241], NIH NIAID [P30 AI078498] and the School of Medicine and Dentistry; CHESS and the MacCHESS resource are supported by NSF Grant [DMR-0936384] and NIH Grant [GM103485]; SSRL is supported by the DOE and by NIH [Grant P41RR001209]. Funding for open access charge: NIH Grants [R01 GM070503 to C.L.K.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank E. Sickmier and K. Frato for preliminary experiments; T. Blumenthal, J. Wedekind, D. Turner, and S. Kennedy for insightful discussions.
REFERENCES
- 1.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith DJ, Query CC, Konarska MM. “Nought may endure but mutability”: spliceosome dynamics and the regulation of splicing. Mol. Cell. 2008;30:657–666. doi: 10.1016/j.molcel.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Itoh H, Washio T, Tomita M. Computational comparative analyses of alternative splicing regulation using full-length cDNA of various eukaryotes. RNA. 2004;10:1005–1018. doi: 10.1261/rna.5221604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Senapathy P, Shapiro MB, Harris NL. Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol. 1990;183:252–278. doi: 10.1016/0076-6879(90)83018-5. [DOI] [PubMed] [Google Scholar]
- 5.Irimia M, Roy SW. Evolutionary convergence on highly-conserved 3' intron structures in intron-poor eukaryotes and insights into the ancestral eukaryotic genome. PLoS Genet. 2008;4:e1000148. doi: 10.1371/journal.pgen.1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouck J, Litwin S, Skalka AM, Katz RA. In vivo selection for intronic splicing signals from a randomized pool. Nucleic Acids Res. 1998;26:4516–4523. doi: 10.1093/nar/26.19.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coolidge CJ, Seely RJ, Patton JG. Functional analysis of the polypyrimidine tract in pre-mRNA splicing. Nucleic Acids Res. 1997;25:888–896. doi: 10.1093/nar/25.4.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruskin B, Green MR. Role of the 3' splice site consensus sequence in mammalian pre-mRNA splicing. Nature. 1985;317:732–734. doi: 10.1038/317732a0. [DOI] [PubMed] [Google Scholar]
- 9.Roscigno RF, Weiner M, Garcia-Blanco MA. A mutational analysis of the polypyrimidine tract of introns. Effects of sequence differences in pyrimidine tracts on splicing. J. Biol. Chem. 1993;268:11222–11229. [PubMed] [Google Scholar]
- 10.Baralle D, Lucassen A, Buratti E. Missed threads. The impact of pre-mRNA splicing defects on clinical practice. EMBO Rep. 2009;10:810–816. doi: 10.1038/embor.2009.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 2010;24:2343–2364. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu CS, Trapnell BC, Curristin S, Cutting GR, Crystal RG. Genetic basis of variable exon 9 skipping in cystic fibrosis transmembrane conductance regulator mRNA. Nat. Genet. 1993;3:151–156. doi: 10.1038/ng0293-151. [DOI] [PubMed] [Google Scholar]
- 13.Chillon M, Casals T, Mercier B, Bassas L, Lissens W, Silber S, Romey MC, Ruiz-Romero J, Verlingue C, Claustres M, et al. Mutations in the cystic fibrosis gene in patients with congenital absence of the vas deferens. N. Engl. J. Med. 1995;332:1475–1480. doi: 10.1056/NEJM199506013322204. [DOI] [PubMed] [Google Scholar]
- 14.Ruskin B, Zamore PD, Green MR. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell. 1988;52:207–219. doi: 10.1016/0092-8674(88)90509-0. [DOI] [PubMed] [Google Scholar]
- 15.Zamore PD, Green MR. Identification, purification, and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc. Natl Acad. Sci. USA. 1989;86:9243–9247. doi: 10.1073/pnas.86.23.9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zamore PD, Green MR. Biochemical characterization of U2 snRNP auxiliary factor: an essential pre-mRNA splicing factor with a novel intranuclear distribution. EMBO J. 1991;10:207–214. doi: 10.1002/j.1460-2075.1991.tb07937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zamore PD, Patton JG, Green MR. Cloning and domain structure of the mammalian splicing factor U2AF. Nature. 1992;355:609–614. doi: 10.1038/355609a0. [DOI] [PubMed] [Google Scholar]
- 18.Golling G, Amsterdam A, Sun Z, Antonelli M, Maldonado E, Chen W, Burgess S, Haldi M, Artzt K, Farrington S, et al. Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat. Genet. 2002;31:135–140. doi: 10.1038/ng896. [DOI] [PubMed] [Google Scholar]
- 19.Zuccato E, Buratti E, Stuani C, Baralle FE, Pagani F. An intronic polypyrimidine-rich element downstream of the donor site modulates cystic fibrosis transmembrane conductance regulator exon 9 alternative splicing. J. Biol. Chem. 2004;279:16980–16988. doi: 10.1074/jbc.M313439200. [DOI] [PubMed] [Google Scholar]
- 20.Tiscornia G, Mahadevan MS. Myotonic dystrophy: the role of the CUG triplet repeats in splicing of a novel DMPK exon and altered cytoplasmic DMPK mRNA isoform ratios. Mol. Cell. 2000;5:959–967. doi: 10.1016/s1097-2765(00)80261-0. [DOI] [PubMed] [Google Scholar]
- 21.Davies RC, Calvio C, Bratt E, Larsson SH, Lamond AI, Hastie ND. WT1 interacts with the splicing factor U2AF65 in an isoform-dependent manner and can be incorporated into spliceosomes. Genes Dev. 1998;12:3217–3225. doi: 10.1101/gad.12.20.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fay J, Kelehan P, Lambkin H, Schwartz S. Increased expression of cellular RNA-binding proteins in HPV-induced neoplasia and cervical cancer. J. Med. Virol. 2009;81:897–907. doi: 10.1002/jmv.21406. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y, Zhang L, Jones KA. SKIP counteracts p53-mediated apoptosis via selective regulation of p21Cip1 mRNA splicing. Genes Dev. 2011;25:701–716. doi: 10.1101/gad.2002611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banerjee H, Rahn A, Gawande B, Guth S, Valcarcel J, Singh R. The conserved RNA recognition motif 3 of U2 snRNA auxiliary factor (U2AF65) is essential in vivo but dispensable for activity in vitro. RNA. 2004;10:240–253. doi: 10.1261/rna.5153204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sickmier EA, Frato KE, Shen H, Paranawithana SR, Green MR, Kielkopf CL. Structural basis of polypyrimidine tract recognition by the essential pre-mRNA splicing factor, U2AF65. Mol. Cell. 2006;23:49–59. doi: 10.1016/j.molcel.2006.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mackereth CD, Madl T, Bonnal S, Simon B, Zanier K, Gasch A, Rybin V, Valcarcel J, Sattler M. Multi-domain conformational selection underlies pre-mRNA splicing regulation by U2AF. Nature. 2011;475:408–411. doi: 10.1038/nature10171. [DOI] [PubMed] [Google Scholar]
- 27.Sickmier EA, Frato KE, Kielkopf CL. Crystallization and preliminary X-ray analysis of U2AF65 variant in complex with a polypyrimidine tract analogue by use of protein engineering. Acta Crystallogr. 2006;F62:457–459. doi: 10.1107/S1744309106012504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh R, Banerjee H, Green MR. Differential recognition of the polypyrimidine-tract by the general splicing factor U2AF65 and the splicing repressor Sex-lethal. RNA. 2000;6:901–911. doi: 10.1017/s1355838200000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jenkins JL, Shen H, Green MR, Kielkopf CL. Solution conformation and thermodynamic characteristics of RNA binding by the splicing factor U2AF65. J. Biol. Chem. 2008;283:33641–33649. doi: 10.1074/jbc.M806297200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. 2004;D60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 31.Myers JK, Pace CN, Scholtz JM. Denaturant m values and heat capacity changes: relation to changes in accessible surface areas of protein unfolding. Protein Sci. 1995;4:2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cavaluzzi MJ, Borer PN. Revised UV extinction coefficients for nucleoside-5'-monophosphates and unpaired DNA and RNA. Nucleic Acids Res. 2004;32:e13. doi: 10.1093/nar/gnh015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernado P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI. Structural characterization of flexible proteins using small-angle X-ray scattering. J. Am. Chem. Soc. 2007;129:5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- 34.Sternglanz H, Bugg CE. Relationship between the mutagenic and base-stacking properties of halogenated uracil derivatives. The crystal structures of 5-chloro- and 5-bromouracil. Biochim. Biophys. Acta. 1975;378:1–11. doi: 10.1016/0005-2787(75)90130-6. [DOI] [PubMed] [Google Scholar]
- 35.Thickman KR, Sickmier EA, Kielkopf CL. Alternative conformations at the RNA-binding surface of the N-terminal U2AF65 RNA recognition motif. J. Mol. Biol. 2007;366:703–710. doi: 10.1016/j.jmb.2006.11.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clery A, Blatter M, Allain FH. RNA recognition motifs: boring? Not quite. Curr. Opin. Struct. Biol. 2008;18:290–298. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 37.Jenkins JL, Laird KM, Kielkopf CL. A broad range of conformations contribute to the solution ensemble of the essential splicing factor U2AF65. Biochemistry. 2012;51:5223–5225. doi: 10.1021/bi300277t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gupta A, Jenkins JL, Kielkopf CL. RNA induces conformational changes in the SF1-U2AF65 splicing factor complex. J. Mol. Biol. 2010;405:1128–1138. doi: 10.1016/j.jmb.2010.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rain JC, Rafi Z, Rhani Z, Legrain P, Kramer A. Conservation of functional domains involved in RNA binding and protein-protein interactions in human and Saccharomyces cerevisiae pre-mRNA splicing factor SF1. RNA. 1998;4:551–565. doi: 10.1017/s1355838298980335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Banerjee H, Rahn A, Davis W, Singh R. Sex lethal and U2 small nuclear ribonucleoprotein auxiliary factor (U2AF65) recognize polypyrimidine tracts using multiple modes of binding. RNA. 2003;9:88–99. doi: 10.1261/rna.2131603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hollins C, Zorio DA, MacMorris M, Blumenthal T. U2AF binding selects for the high conservation of the C. elegans 3' splice site. RNA. 2005;11:248–253. doi: 10.1261/rna.7221605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patterson B, Guthrie C. A U-rich tract enhances usage of an alternative 3' splice site in yeast. Cell. 1991;64:181–187. doi: 10.1016/0092-8674(91)90219-o. [DOI] [PubMed] [Google Scholar]
- 43.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.