Abstract
Objective
Retroviral vector proviruses can lead to aberrant expression of nearby genes in hematopoietic repopulating cells, leading to an over-representation of clones with dysregulated genes that affect hematopoiesis. Common integration sites (CISs) identified using the vector provirus as a molecular tag can be used to identify these genes. Here we characterized a retroviral CIS observed at high frequency in baboon hematopoietic repopulating cells that has not been described previously.
Materials and Methods
Gammaretroviral vector integration sites in baboon repopulating cells identified by polymerase chain reaction amplification were localized to the human genome to identify a CIS. The presence of each clone was tracked over time using allele-specific polymerase chain reaction.
Results
In three different animals that received gammaretrovirally transduced CD34-enriched bone marrow cells, vector proviruses were identified at three distinct sites within a window of 664 base pairs between leupaxin and zinc finger protein 91 (ZFP91). All three integrants of the CIS occurred within a CpG island between leupaxin and zinc finger protein 91 (ZFP91).
Conclusions
We describe a novel CIS between leupaxin and ZFP91 in hematopoietic repopulating cells. Our data suggest that leupaxin and/or ZFP91 may play a role in hematopoietic repopulating cells.
When a vector provirus integrates near a gene controlling growth and alters its expression, the host cell may gain a selective advantage. There are several mechanisms whereby retroviral vector proviruses can lead to aberrant expression of nearby genes, including enhancer activation from the vector long terminal repeats (LTRs). After deregulation, expansion of cells with the vector provirus can lead to the formation of a clonal tumor in which each cell contains a vector provirus integrated at the same site. Independent clonal tumors can be analyzed for the presence of common integration sites (CISs) using a vector or virus as a molecular tag. Nearby genes can then be characterized to determine the mechanism of oncogenesis. The mouse Retrovirus Tagged Cancer Gene Database has >500 retroviral CISs [1].
In early studies using retroviral vectors as molecular tags, it was realized that these vectors might also be used to identify nearby genes involved in hematopoiesis [2], analogous to the use of viruses to identify oncogenes. This is because vector integrants near genes involved in hematopoietic proliferation can alter the phenotype of these cells. This prediction has been confirmed. In addition to proto-oncogenes, the Retrovirus Tagged Cancer Gene Database also includes genes associated with hematopoietic diseases such as Ras, Notch, Jak/Stat, and nuclear factor–κβ [3]. We and others have shown that vectors are found more frequently in and near proto-oncogenes in primate repopulating cells and that genes involved in growth or survival are over-represented near retroviral integrants [4,5]. Thus, analysis of retroviral integrants in primate repopulating cells can be used to identify dysregulated genes that may alter the process of engraftment. Here we describe a CIS in baboon repopulating cells that suggests that leupaxin and/or ZFP91 may affect the engraftment of hematopoietic repopulating cells. To our knowledge this is the first report that suggests these genes may play a role in hematopoietic stem cells (HSCs).
Materials and methods
Animal care
Healthy juvenile baboons (Papio cynocephalus anubis) were housed at the University of Washington National Primate Research Center under conditions approved by the American Association for Accreditation of Laboratory Animal Care. Study protocols were approved by the Institutional Review Board and the Institutional Animal Care and Use Committee. The transplantation of gammaretrovirally transduced CD34+ cells in these baboons has been described previously [6].
Analysis of the CIS and real-time polymerase chain reaction to evaluate clonal contribution
Methods and criteria used to identify the chromosomal positions of gammaretroviral vector proviruses have been described previously [5]. The University of California Santa Cruz Web site (http://genome.ucsc.edu) was used to localize integrants on human genome assembly hg18 [7]. To analyze the clonal contribution of cells with vector proviruses in the CIS, real-time polymerase chain reaction (PCR) was performed as described previously [8] using the LTR–specific primer 5′-CTCTGTTCCTGACCTTGATCC-3′, and locus-specific primers 5′-TCCCAACAGGGTCTAGTAGAAAA-3′ (monkey J01174), 5′-CGACTTACCTTGGTTTGGTCA-3′ (monkey J00116), or 5-AGGAGAAGCGACAGCTCAAA-3′ (monkey M01277). The internal LTR-specific probe was 5′-FAM-CCTAC AGGTGGGGTCTTTCA-TAMRA-3′.
Results and discussion
A common retroviral vector integration site in baboon repopulating cells
We describe here a retroviral vector CIS in baboon hematopoietic repopulating cells that has not been described previously. In three different animals, gammaretroviral vectors integrated within a window of 664 base pairs in the promoter region between two genes, leupaxin and the zinc finger protein 91 (ZFP91). The gammaretroviral LTR-chromosome junctions and the chromosomal sequence adjacent to the LTR that were used to identify the position of vector proviruses, and the approach used to localize their position on the human genome are provided in Supplementary Figure E1 (online only, available at www.exphem.org). The three vector integration sites were each found in different baboons, so repopulation by a clone with an integrant at this CIS occurred independently in three animals. The source of transplanted (autologous) cells for each animal was different. The second closest three-integrant CIS occurred in a window of 20,360 base pairs and was near two genes called CCNJL (cyclin J-like cyclin) and C1QTNF2 (C1q and tumor necrosis factor–related protein 2). The third three-integrant CIS occurred in a window of 134,208 base pairs and was near two genes: TMEM217 (transmembrane protein 217), and PIM1, a well-characterized anti-apoptotic proto-oncogene. These CISs spanned 30- and 202-fold larger windows, respectively, than the LPXN-ZFP91 CIS.
It is highly unlikely that the three integrations of a total of 380 identified sites [5] occurred within this 664-bp window by chance. We compared the CIS to 10 random datasets of the same size (380 sites). In these 10 datasets, the mean distance between the three closest sites was 476,976 bp (±66,518 bp standard error) and the closest three sites in all 10 random datasets was 129,237 bp. The distance between the three sites in the CIS, 664 bp, is 718-fold smaller than the mean, 198-fold smaller than the smallest CIS in all datasets, and well outside a 99% confidence interval of the mean for these 10 control datasets (476,976 ± 216,183). Instead, we hypothesize that dysregulation of leupaxin and/or ZFP91 may have led to this over-representation. This CIS is between two genes, leupaxin and ZFP91, which are in opposite orientations on human chromosome 11 (Fig. 1). All three integrants are within a CpG island, which suggests that this intergenic region contains a promoter for either or both genes. This CpG region is conserved between all mammals and the entire locus is highly conserved between primates and humans (Fig. 1).
Figure 1.
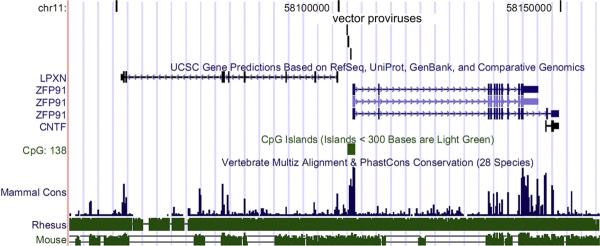
The common integration site (CIS) between leupaxin and zinc finger protein 91 (ZFP91) in repopulating cells from three different baboons. The CIS is shown on human chromosome 11 with the positions of the three different integration sites indicated by vertical lines as generated by the University of California, Santa Cruz genome browser. This region has high homology between mammals and is within a CpG island suggesting this region contains a promoter. LPXN is leupaxin, and CNTF is ciliary neurotrophic factor.
No evidence for clonal expansion after engraftment
These three monkeys had normal hematopoiesis and no signs of expansion of vector-marked cells as determined by trans-gene expression (data not shown). We next investigated whether repopulating cell clones with these specific integrants had expanded over time. For each baboon, we designed primers specific for the chromosome-LTR junction (Fig. 2A) and performed real-time PCR of peripheral blood mononuclear cells containing both myeloid and lymphoid lineages at different time points to determine if there was clonal expansion. Although the marking varied over time, DNA from cells with all three integrations could be detected over background, and there was no evidence of clonal expansion of these specific engrafted gene-modified clones in any monkey (Fig. 2B). These insertions were first detected by linear amplification-modified PCR at days 364, 372, and 398 post-transplantation for baboons J01174, J00116, and M01277, respectively. A retrospective analysis by allele-specific PCR (Fig. 2) showed they were present at days 266, 224, and 224 post-transplantation, respectively, at low levels just above background. Cells with these integrants are thus true long-term repopulating cells, but they have not undergone a premalignant expansion, and represent only a small fraction of the polyclonal repopulating cells in these animals. Based on this lack of evidence for clonal expansion, we hypothesize that retroviral integration may have dysregulated leupaxin and/or ZFP91 and given infused primate HSCs a competitive engraftment advantage. Both leupaxin and ZFP91 are likely candidates to influence the engraftment of repopulating cells. Leupaxin has a high degree of homology with paxillin, an adaptor protein that facilitates assembly of multiprotein complexes involved in signaling, migration, and proliferation in many cell types [9,10]. Leupaxin expression is more restricted than paxillin, is expressed mainly in leukocytes [11], and interacts with α4 integrin [12], which is known to be involved in HSC migration and retention through α4β1 (very late antigen 4)–intracellular adhesion molecule-1 interactions. Leupaxin was identified as a translocation partner of RUNX1 in acute myeloid leukemia [13] and overexpression studies with wild-type leupaxin showed it can confer transformation characteristics to NIH 3T3 cells, including increased growth and the ability to form colonies in soft agar [14].
Figure 2.
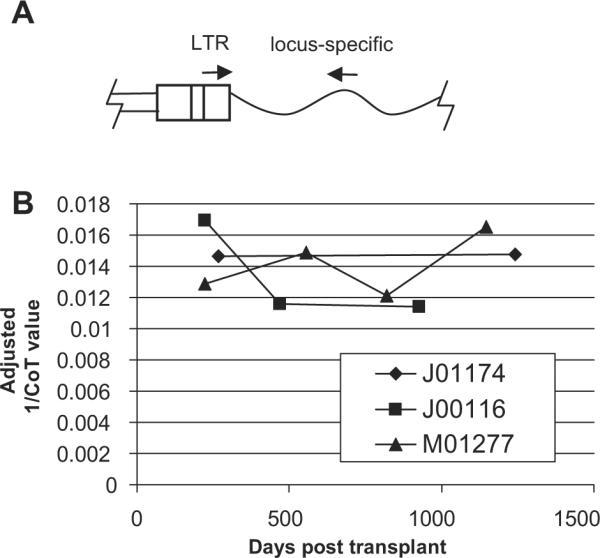
No evidence for clonal expansion of cells with integrants at the common integration site (CIS) after engraftment. (A) To track the clonal contribution of clones with an integrant at the CIS real-time polymerase chain reaction (PCR) was performed using one primer specific to the gammaretroviral vector LTR and a second primer specific to the CIS locus. (B) Real-time PCR was performed on peripheral blood mononuclear cells and the relative marking at different time points was compared based on the CoT value after adjustment for template DNA concentration based on control globin primers.
ZFP91 has been less studied, but it is overexpressed in 90% of acute myeloid leukemia patient cells and inhibits apoptosis in acute myeloid leukemia cell lines [15], suggesting dysregulation may have impacted the survival of repopulating cells. Both migration and inhibition of apoptosis play key roles in HSC engraftment and may explain why we observed this hot spot of vector integration. There are two isoforms of ZFP91, but in acute myeloid leukemia, the overexpressed form of ZFP91 is the 5.2-kb form that does not contain ciliary neurotrophic factor, and expression of the ZFP91 –ciliary neurotrophic factor 2.2-kb variant appears to be restricted to the testis [15].
In summary, we have described a CIS in baboon repopulating cells that suggests leupaxin and/or ZFP91 may play a role in HSCs. Future studies will be needed to confirm this and to examine the mechanism of their action but, to our knowledge, our study is the first to suggest that these genes may play a role in hematopoietic repopulating cells. If leupaxin or ZFP91 are involved in the engraftment and/or survival of repopulating cells, they may be attractive targets to develop small molecule drugs to enhance migration or survival for improved HSC transplantation therapies.
Supplementary Material
Acknowledgments
We would like to thank the staff at the University of Washington National Primate Center (Seattle, WA, USA) and Robert Wu and Brian Swanson for providing technical assistance. This research was supported in part by National Institutes of Health (Bethesda, MD, USA) grants AI63959, AI61839, DK77806, DK56456, DK47754, HL66947, and HL53750. H.P.K. is a Markey Molecular Medicine Investigator and the recipient of the Jose Carreras/E. Donnall Thomas Endowed Chair for Cancer Research.
Footnotes
The authors have no conflicts of interest to declare.
Conflict of Interest Disclosure No financial interest/relationships with financial interest relating to the topic of this article have been declared.
Supplementary data associated with this article can be found in the online version, at doi:10.1016/j.exphem.2010.04.014.
References
- 1.Akagi K, Suzuki T, Stephens RM, Jenkins NA, Copeland NG. RTCGD: retroviral tagged cancer gene database. Nucleic Acids Res. 2004;32:D523–D527. doi: 10.1093/nar/gkh013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dick JE, Magli MC, Huszar D, Phillips RA, Bernstein A. Introduction of a selectable gene into primitive stem cells capable of long-term reconstitution of the hemopoietic system of W/Wv mice. Cell. 1985;42:71–79. doi: 10.1016/s0092-8674(85)80102-1. [DOI] [PubMed] [Google Scholar]
- 3.Suzuki T, Shen H, Akagi K, et al. New genes involved in cancer identified by retroviral tagging. Nat Genet. 2002;32:166–174. doi: 10.1038/ng949. [DOI] [PubMed] [Google Scholar]
- 4.Hematti P, Hong BK, Ferguson C, et al. Distinct genomic integration of MLV and SIV vectors in primate hematopoietic stem and progenitor cells. PLoS Biol. 2004;2:e423. doi: 10.1371/journal.pbio.0020423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beard BC, Dickerson D, Beebe K, et al. Comparison of HIV-derived lentiviral and MLV-based gammaretroviral vector integration sites in primate repopulating cells. Mol Ther. 2007;15:1356–1365. doi: 10.1038/sj.mt.6300159. [DOI] [PubMed] [Google Scholar]
- 6.Beard BC, Mezquita P, Morris JC, Kiem HP. Efficient transduction and engraftment of G-CSF-mobilized peripheral blood CD34+ cells in nonhuman primates using GALV-pseudotyped gammaretroviral vectors. Mol Ther. 2006;14:212–217. doi: 10.1016/j.ymthe.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 7.Kent WJ, Sugnet CW, Furey TS, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trobridge GD, Beard BC, Gooch C, et al. Efficient transduction of pigtailed macaque hematopoietic repopulating cells with HIV-based lentiviral vectors. Blood. 2008;111:5537–5543. doi: 10.1182/blood-2007-09-115022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schaller MD. Paxillin: a focal adhesion-associated adaptor protein. Oncogene. 2001;20:6459–6472. doi: 10.1038/sj.onc.1204786. [DOI] [PubMed] [Google Scholar]
- 10.Turner CE. Paxillin interactions. J Cell Sci. 2000;113(Pt 23):4139–4140. doi: 10.1242/jcs.113.23.4139. [DOI] [PubMed] [Google Scholar]
- 11.Lipsky BP, Beals CR, Staunton DE. Leupaxin is a novel LIM domain protein that forms a complex with PYK2. J Biol Chem. 1998;273:11709–11713. doi: 10.1074/jbc.273.19.11709. [DOI] [PubMed] [Google Scholar]
- 12.Liu S, Thomas SM, Woodside DG, et al. Binding of paxillin to alpha4 integrins modifies integrin-dependent biological responses. Nature. 1999;402:676–681. doi: 10.1038/45264. [DOI] [PubMed] [Google Scholar]
- 13.Dai H, Xue Y, Pan J, et al. Two novel translocations disrupt the RUNX1 gene in acute myeloid leukemia. Cancer Genet Cytogenet. 2007;177:120–124. doi: 10.1016/j.cancergencyto.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 14.Dai HP, Xue YQ, Zhou JW, et al. LPXN, a member of the paxillin superfamily, is fused to RUNX1 in an acute myeloid leukemia patient with a t(11;21)(q12;q22) translocation. Genes Chromosomes Cancer. 2009;48:1027–1036. doi: 10.1002/gcc.20704. [DOI] [PubMed] [Google Scholar]
- 15.Unoki M, Okutsu J, Nakamura Y. Identification of a novel human gene, ZFP91, involved in acute myelogenous leukemia. Int J Oncol. 2003;22:1217–1223. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


