Abstract
Exposure to endocrine disruptors is associated with developmental defects. One compound of concern, to which humans are widely exposed, is bisphenol A (BPA). In model organisms, BPA exposure is linked to metabolic disorders, infertility, cancer, and behavior anomalies. Recently, BPA exposure has been linked to DNA methylation changes, indicating that epigenetic mechanisms may be relevant. We investigated effects of exposure on genomic imprinting in the mouse as imprinted genes are regulated by differential DNA methylation and aberrant imprinting disrupts fetal, placental, and postnatal development. Through allele-specific and quantitative real-time PCR analysis, we demonstrated that maternal BPA exposure during late stages of oocyte development and early stages of embryonic development significantly disrupted imprinted gene expression in embryonic day (E) 9.5 and 12.5 embryos and placentas. The affected genes included Snrpn, Ube3a, Igf2, Kcnq1ot1, Cdkn1c, and Ascl2; mutations and aberrant regulation of these genes are associated with imprinting disorders in humans. Furthermore, the majority of affected genes were expressed abnormally in the placenta. DNA methylation studies showed that BPA exposure significantly altered the methylation levels of differentially methylated regions (DMRs) including the Snrpn imprinting control region (ICR) and Igf2 DMR1. Moreover, exposure significantly reduced genome-wide methylation levels in the placenta, but not the embryo. Histological and immunohistochemical examinations revealed that these epigenetic defects were associated with abnormal placental development. In contrast to this early exposure paradigm, exposure outside of the epigenetic reprogramming window did not cause significant imprinting perturbations. Our data suggest that early exposure to common environmental compounds has the potential to disrupt fetal and postnatal health through epigenetic changes in the embryo and abnormal development of the placenta.
Author Summary
BPA is a widely used compound to which humans are exposed, and recent studies have demonstrated the association between exposure and adverse developmental outcomes in both animal models and humans. Unfortunately, exact mechanisms of BPA–induced health abnormalities are unclear, and elucidation of these relevant biological pathways is critical for understanding the public health implication of exposure. Recently, increasing data have demonstrated the ability of BPA to induce changes in DNA methylation, suggesting that epigenetic mechanisms are relevant. In this work, we study effects of BPA exposure on expression and regulation of imprinted genes in the mouse. Imprinted genes are regulated by differential DNA methylation, and they play critical roles during fetal, placental, and postnatal development. We have found that fetal exposure to BPA at physiologically relevant doses alters expression and methylation status of imprinted genes in the mouse embryo and placenta, with the latter tissue exhibiting the more significant changes. Additionally, abnormal imprinting is associated with defective placental development. Our data demonstrate that BPA exposure may perturb fetal and postnatal health through epigenetic changes in the embryo as well as through alterations in placental development.
Introduction
Perturbed gestation affects fetal growth and development, resulting in a predisposition to diseases [1]. The “developmental origin of adult disease” hypothesis was originally formulated based on clinical data linking low birth weight to increased risks for adult onset cardiovascular and metabolic disorders. The hypothesis has been supported by a growing number of human diseases associated with unfortunate events during pregnancy including drug exposure [2], chemical exposure [3], prenatal stress [4], and maternal caloric restriction [5]. The observed phenotypes in the fetus were frequently accompanied by altered gene expression [2]–[5]. Although the available data strongly suggest that the environment significantly impacts fetal development, exact molecular mechanism(s) linking diseases and early life events remain unclear despite significant research.
Accumulating evidence implicates the role of epigenetics in mediating gene-environment interactions. Studies have demonstrated the ability of environmental factors including food constituents [6], prenatal famine [7], and endocrine disruptors [8]–[10] to alter global or gene-specific DNA methylation patterns. DNA methylation, the addition of a methyl group to the cystosine residue of DNA, is an epigenetic mechanism that regulates genes important for a wide range of biological processes. DNA methylation at regulatory regions typically leads to gene repression and altered patterns are associated with developmental defects, tumors and cancer [11], [12]. Furthermore, early mammalian development represents a vulnerable window as the genome undergoes dynamic changes of DNA methylation [13] and environmental factors that alter these epigenetic reprogramming events may adversely impact growth and development.
Several environmental factors that can perturb epigenetic reprogramming events in the embryo have been reported [14]–[17]. These factors include conditions or techniques related to embryo culture used in assisted reproductive technologies (ART). Although the number of babies that are conceived through ART is growing, these individuals represent a relatively small percentage (1–4%) of babies in industrialized countries [18]. In the current study, we investigated a widely used environmental compound that has the ability to alter DNA methylation [9]. The compound, bisphenol A (BPA), is essentially ubiquitous in the environment and can be found in various consumer products including polycarbonate plastics, resin and paper receipts [19]. Most humans are exposed to BPA [19] with estimated blood levels ranging from 0.5 to 10 ng/mL [20]. Importantly, BPA is a significant public health issue as early developmental exposure is linked to abnormal brain function, metabolism, reproduction, behavior, and immune system in model organisms [19]. Aside from its estrogenic properties and ability to bind to estrogen receptors [21], molecular mechanisms related to BPA action are not well elucidated and most likely, are complex based on the broad range of phenotypes related to exposure. Recently, it has been reported that BPA exposure can alter the mouse and rat epigenome [9], [22]–[24]. Jirtle and coworkers [9] reported that BPA exposed mice exhibited decreased DNA methylation at two mouse metastable loci while other studies [22]–[24] found DNA methylation changes at endogenous loci relevant to prostate, brain and uterine development. These studies [22], [24] investigated BPA effects when exposure occurred during mid-gestation or neonatal period. As we hypothesized that the earlier period of post-fertilization development is the most sensitive window due to its inherent epigenetic reprogramming, it would be important to determine potential impacts of BPA-induced epigenetic perturbations on developmentally relevant genes in the conceptus when the exposure occurs during this window.
In this study we ask whether BPA exposure in mice can alter imprinted gene expression and DNA methylation. Genomic imprinting in mammals results in the epigenetic modification of a small number of developmental genes such that a single parental allele is preferentially or exclusively expressed [25]. The use of genomic imprinting to study the developmental effects of BPA on the epigenome is highly suitable as differential DNA methylation is a well-characterized imprinting mechanism [25]. Furthermore, imprinted genes are developmentally critical as they are essential for embryonic, placental and postnatal growth and aberrant imprinting causes cancer and human disorders including Beckwith-Wiedemann Syndrome (BWS) and Angelman Syndrome (AS) [26]. Here we perform extensive characterization of the allelic and total expression of imprinted genes on mouse chromosome 7 and on DNA methylation of the associated imprinting control regions (ICRs) as well as genome-wide methylation in mice exposed to two physiological doses of BPA during early embryonic development. Our data show that BPA disrupts proper expression of imprinted genes in a gene- and tissue-specific manner when exposure occurs early in development. Specifically, we observe significant perturbations at the Snrpn, Ube3a, Igf2, Kcnq1ot1 and Cdkn1c loci but not at the H19 and Peg3 genes. Moreover these effects are DNA methylation-dependent as demonstrated by analysis at the Snrpn ICR and Igf2 differentially methylated region (DMR) 1, although our data also suggest that other epigenetic mechanisms cannot be excluded. Further studies reveal that aberrant imprinting in BPA exposed mice is associated with abnormal placental development. In contrast, BPA treatment of pregnant females outside of the epigenetic reprogramming window, i.e., from embryonic day (E) 5.5 to 12.5, had no observable effect on genomic imprinting. These findings indicate that exposure to the common environmental compound at low, physiologically relevant doses during the earliest stage of embryonic development may disrupt epigenetic reprogramming events and perturb developmental gene expression.
Results
BPA exposure leads to aberrant expression of imprinted genes on mouse chromosome 7
Dolinoy et al. reported that BPA exposure in pregnant viable yellow agouti (Avy) mice resulted in offspring with reduced DNA methylation at 9 CpGs within the intracisternal A particle (IAP) retrotransposon locus and altered Agouti expression [9]. More recent studies reported DNA methylation changes at various loci in BPA exposed rats and mice [22]–[24]. Because expression of imprinted genes is typically regulated by allele-specific methylation at ICRs, we hypothesized that BPA exposure would perturb expression of imprinted genes.
To test the hypothesis, we analyzed F1 hybrid mice generated from reciprocal matings of the C57BL/6 (B6) and the B6 (CAST7) or C7 mice. We treated female mice starting from 2 weeks prior to mating until E9.5 with three different doses of dietary BPA: 0 (control), 10 mg/kg bw/day (upper dose) and 10 µg/kg bw/day (lower dose). The upper dose treatment paradigm was similar to the method employed by Jirtle and coworkers, except that we analyzed the offspring before birth [9]. The lower dose represented safe human exposure level, i.e., at or below 50 µg/kg bw/day [20]. BPA serum measurement revealed that the treatment paradigm resulted in significantly increased level of unconjugated BPA in pregnant mice from the upper dose group compared to controls (2.0±0.4 ng/mL vs. 0.7±0.2 ng/mL, respectively; n = 3; P<0.01; Figure 1). Serum BPA detected in pregnant mice from the lower dose exposure group showed a trend towards increased levels but the difference was not statistically significant compared to controls (data not shown). Embryonic and placental tissues from the F1 hybrid offspring were subsequently analyzed at E9.5 for allele-specific transcription of imprinted genes located in multiple domains, including the Peg3, Snrpn, H19/Igf2 and Kcnq1 domains, on mouse chromosome 7 (Figure 2). We initially analyzed E9.5 because many imprinted genes display parent-of-origin specific expression in the whole embryo at this time. Additionally, the embryo and placenta are sufficiently differentiated so that risk for cross contamination is low, and more importantly, maternal tissue contamination in decidua-removed placenta is minimal compared to later stages.
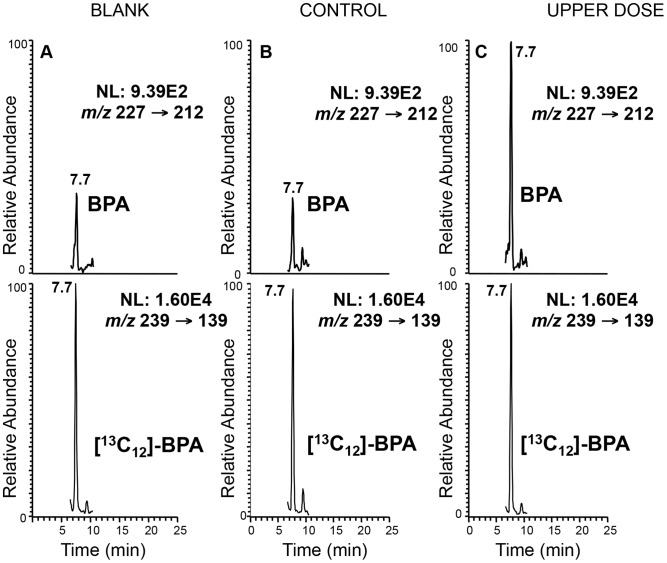
Figure 1. Representative LC-SRM/MS chromatograms demonstrating significantly higher BPA level in serum from treated mice.
Top panels represent the chromatograms for unlabeled BPA and lower panels labeled BPA ([13C12]-BPA) spiked prior to sample extraction to quantify BPA levels in serum from representative control (B) and upper dose treated mice (C). Blank sample used as control for storage and extraction is indicated in (A). Y-axis represents relative abundance of signal intensity and X-axis retention time in minute. Differences in BPA concentrations were determined based on peak heights (see Materials and Methods). NL = normalized levels of intensity; m/z = mass to charge ratio.
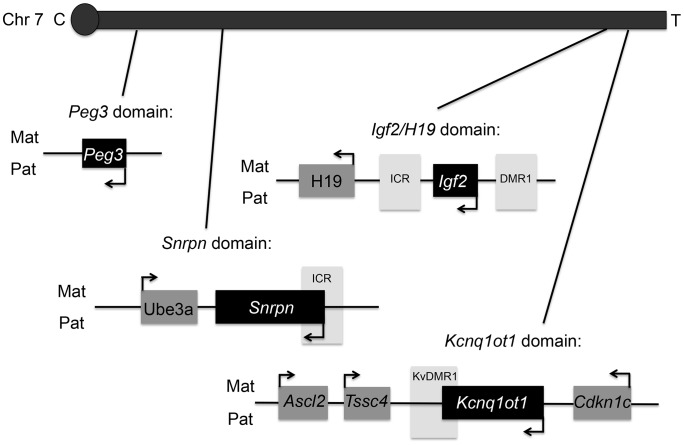
Figure 2. Map of mouse chromosome 7 showing imprinted domains analyzed in the current work is illustrated.
Black = paternally expressed genes; dark grey = maternally expressed genes; and light grey = differentially methylated regions (DMRs). Arrows indicated direction of gene transcription. C = centromere and T = telomere. Not drawn to scale.
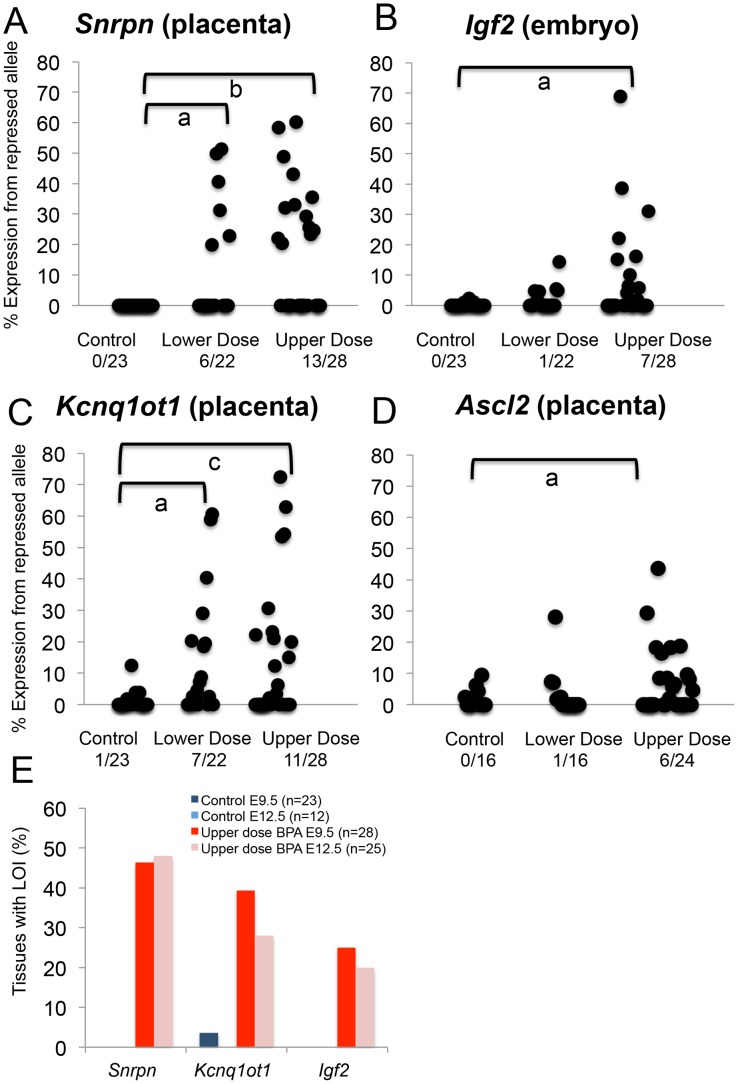
Upper dose BPA exposure disrupted the parental specific, monoallelic expression of the Snrpn, Igf2 and Kcnq1ot1 genes in a tissue-specific manner (Figure 3A–3C). Upper dose BPA exposure resulted in significantly more placentas exhibiting biallelic expression of the paternally expressed Snrpn gene (Figure 3A); no significant effect was observed in the embryo (Table S1). Loss of imprinting (LOI) occurred in 13/28 placentas from upper dose BPA exposed mice compared to 0/23 in controls (P<0.001; Figure 3A). Analysis of the placentas exhibiting LOI in the upper dose BPA group showed that the normally repressed maternal Snrpn allele contributed 20.3–60.2% of total expression (Figure 3A). At the Igf2 locus, upper dose BPA exposure significantly resulted in LOI in 7/28 embryos as compared to 0/23 in controls (Figure 3B; P<0.05), whereas no effects were observed in the placenta (Table S1). Among the upper dose BPA exposed embryos showing LOI, Igf2 expression from the normally repressed maternal allele ranged from 10.0–68.9% (Figure 3B). Analysis of the placenta-specific P0 promoter (Igf2 P0) revealed normal, monoallelic expression in placentas from both control and upper dose exposure groups (data not shown). Interestingly, although Igf2 and H19 were located in the same domain and are coordinately regulated through a shared ICR [27], the maternally expressed H19 gene remained monoallelically expressed (Table S1). We also tested allelic expression of the paternally expressed Kcnq1ot1 (located in the Kcnq1 domain) and found 11/28 placentas showing LOI in the upper dose BPA group vs. 1/23 in the control (Figure 3C; P<0.05). No significant perturbation was detected in the embryo (Table S1). Expression of the normally repressed maternal allele in the placentas showing Kcnq1ot1 LOI ranged from 12.3–72.3%; in the control group, one placenta with LOI had 12.4% of total expression from the normally repressed maternal allele (Figure 3C). At the Peg3 locus, no significant effect was observed in conceptuses exposed to upper dose BPA (Table S1).
Figure 3. Allele-specific expression studies showed increased proportion of tissues with loss of imprinting following BPA exposure.
(A–D) Each black circle represents an individual E9.5 embryo (Igf2) or placenta (Snrpn, Kcnq1ot1 and Ascl2). Y-axis indicates percentage of total mRNA expression derived from the repressed allele. Loss of imprinting (LOI) or biallelic expression was called when the repressed allele exhibited ≥10% of total expression. Data from control (n = 23), lower dose (n = 22) and upper dose (n = 28) exposure groups are shown for (A) Snrpn, (B) Igf2, (C) Kcnq1ot1 and (D) Ascl2; a = P<0.05; b = P<0.01 and c = P<0.001. Number of tissues (out of total examined) showing LOI is indicated below each exposure group. In (E), percentages of placentas (Snrpn and Kcnq1ot1) and embryos (Igf2) showing LOI at E9.5 and E12.5 are compared.
Lower dose BPA exposure resulted in mostly normal, monoallelic expression, except at the Snrpn and Kcnq1ot1 loci in the placenta. At the Snrpn locus, 6/22 placentas exhibited LOI (compared to 0/23 in controls; P<0.05) with expression of the normally repressed maternal allele ranging from 19.8–51.3% (Figure 3A). At the Kcnq1ot1 locus, 7/22 placentas in the lower dose group showed LOI (compared to 1/23 in the control group; P<0.05; Figure 3C) with expression of the repressed maternal allele ranging from 18.5–60.7%. Igf2, H19 and Peg3 (Figure 3B and Table S1) and Igf2 P0 (data not shown) were normally expressed in the embryonic tissues and/or placentas from mice exposed to lower dose BPA.
To determine whether BPA exposure affected genomic imprinting in embryonic organs, we analyzed allele-specific expression of the Snrpn, Igf2 and Kcnq1ot1 genes in E12.5 conceptuses. We first analyzed LOI frequencies at these loci in upper dose BPA and control exposed E12.5 whole embryos and placentas. Similar frequencies of imprinting abnormalities were observed in E12.5 as compared to E9.5 in the placenta: 12/25 and 7/25 exhibited LOI at the Snrpn and Kcnq1ot1 loci, respectively (P>0.05 as compared to 13/28 and 11/28 in E9.5 placentas, respectively; Figure 3E and Table S2) – no significant effect was detected at the Igf2, H19 and Peg3 loci in the placenta (data not shown). In E12.5 whole embryos, 5/25 showed LOI at the Igf2 locus (P>0.05 as compared to 7/28 at E9.5; Figure 3E and Table S2) but no LOI observed at the Snrpn and Kcnq1ot1 genes (data not shown). Our analysis had therefore demonstrated similar LOI rates in E12.5 vs. E9.5 conceptuses.
Next, we analyzed LOI rates in E12.5 embryonic organs including the heart, liver, brain and kidney. No significant imprinting abnormalities were observed in these organs at the Snrpn, Kcnq1ot1 and Igf2 loci (n = 16–17 mice analyzed; data not shown), except in the brain at the Snrpn locus (4/16 in upper dose BPA vs. 0/17 in control; P = 0.05).
Imprinting analyses were conducted in maternal decidua-removed placentas. Nevertheless, a recent study indicated that these tissues contain significant levels of some RNAs from maternal cells that can bias analysis of maternally expressed imprinted genes [28]. As we observed abnormal imprinting of maternally expressed genes (e.g., Snrpn and Kcnq1ot1) in the placenta, we subsequently determined whether BPA exposed placentas had more maternal cell contamination as compared to controls by measuring total expression of the Gatm, Tfpi2 and Ampd3 genes; these genes were shown to be highly expressed in the maternal decidua as compared to placenta [28]. Total expression of the Gatm, Tfpi2 and Ampd3 genes relative to the reference genes Arppo and Gapdh in upper dose BPA exposed (n = 5) and control E12.5 placentas (n = 5) was not significantly different (P>0.05; data not shown) indicating that the increased LOI rates observed in BPA exposed placentas were not associated with increased maternal cell contamination.
In total, we analyzed 4–6 litters per exposure group from both C7XB6 and B6XC7 matings at E9.5 and 2–4 litters at E12.5 (see Table S1, S2 for complete list). We did not observe significant LOI differences between C7XB6 and B6XC7 matings for any of the tested genes (Table S1, S2). Although similar LOI rates were observed in E9.5 (Table S1) and E12.5 (Table S2) whole embryos and placentas, significant litter-specific effects were observed in the latter stage litters (P = 0.02 for Snrpn) but not in the earlier (P>0.05 for all genes tested). In E12.5 brains, however, we did not observe significant litter-specific effects for Snrpn LOI.
BPA exposure outside of the epigenetic reprogramming window does not lead to imprinting defects
Our exposure paradigm coincided with the latter stage of oocyte development during DNA methylation acquisition [29] and the earliest stage of embryonic development during the extensive epigenetic reprogramming in the genome [13]. To determine whether exposure outside of this window leads to aberrant imprinting, we treated pregnant mice with control and upper dose BPA from E5.5 to E12.5 and determined the allele-specific expression of the Snrpn, Igf2 and Kcnq1ot1 genes in the whole embryo and placenta. Our data demonstrated no significantly different rates of LOI at these loci in both the embryonic and placental tissues of upper dose BPA exposed E12.5 mice as compared to the control group (n = 12 and 20 mice analyzed in 2 control litters and 4 upper dose BPA litters, respectively; data not shown).
Aberrant allelic expression of imprinted genes is associated with significant changes in total expression
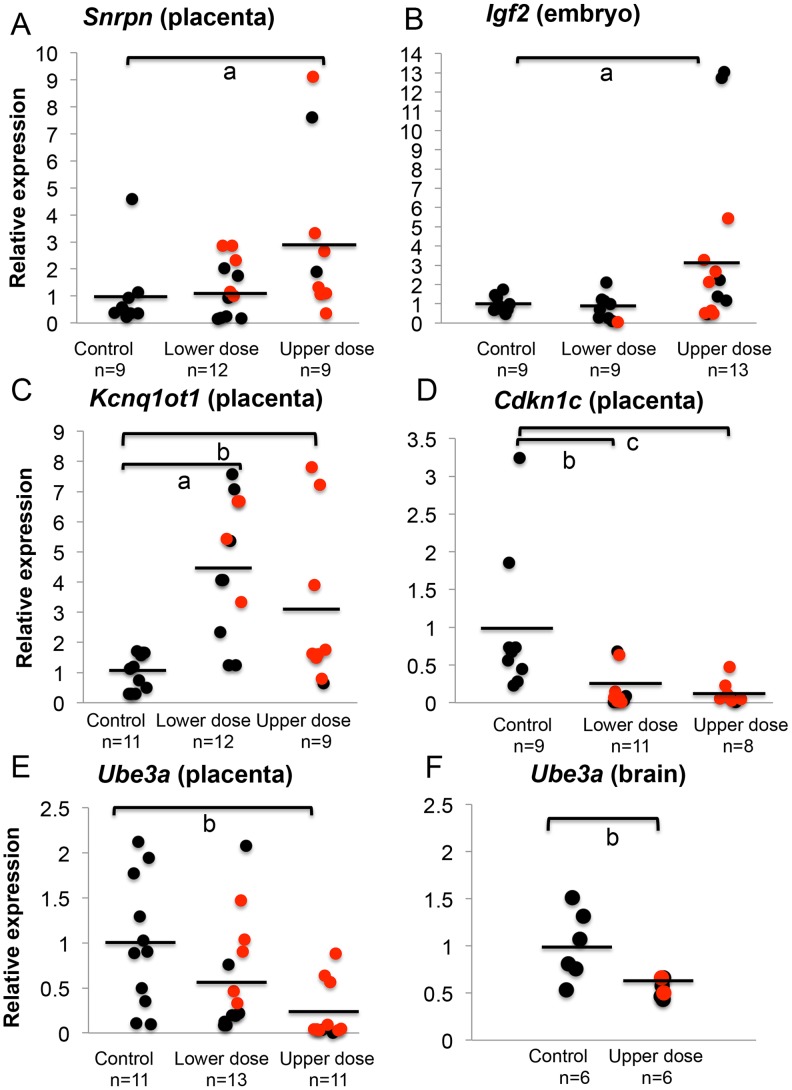
To address whether total expression of imprinted genes was impacted by BPA exposure, we re-analyzed the embryonic and placental tissues from the offspring of pregnant mice treated with control and BPA diets starting from pre-mating until E9.5. We quantified expression of the Snrpn, Igf2, and Kcnq1ot1 genes relative to the reference genes Arppo and Gapdh. Quantitative real time PCR analysis of Snrpn revealed that placentas from mice exposed to upper dose BPA had a significant increase of mean total RNA compared to control placentas (3.2±3.1 vs. 1.0±1.3, respectively; n = 9 in each group; P<0.05; Figure 4A). In the lower dose group, mean total expression was not significantly different when placentas with and without LOI were analyzed together (1.3±1.0; n = 12; P>0.05; Figure 4A); however, lower dose BPA exposed placentas exhibiting LOI had significantly higher Snrpn expression (2.0±0.8; n = 5) when compared to samples with normal, monoallelic expression (0.7±0.6; n = 7; P<0.01). At the Igf2 locus, average total expression was also significantly increased in upper dose BPA exposed embryos (3.6±4.3; n = 13) compared to controls (1.0±0.4; n = 9; P<0.05; Figure 4B); no significant change was observed in embryos from the lower dose group (0.8±0.7; n = 9; P = 0.19; Figure 4B), consistent with their normal allelic expression (Figure 3B). Total H19 expression in both lower and upper dose groups was not different from controls (data not shown). Furthermore, biallelic expression of Kcnq1ot1 in the placentas from the upper dose group was associated with significant increased total expression as compared to controls (3.0±2.7 vs. 1.0±0.5, respectively; n = 9 and 11; P<0.01; Figure 4C); similar observation was found in the lower dose group (4.5±2.2 vs. 1.0±0.5 in controls; n = 12 and 11, respectively; P<0.05; Figure 4C).
Figure 4. BPA exposure altered total mRNA expression of imprinted genes relative to reference genes.
Samples from control, lower dose and upper dose exposure groups were analyzed for total expression of the imprinted (A) Snrpn, (B) Igf2, (C) Kcnq1ot1, (D) Cdkn1c and (E and F) Ube3a genes with sample sizes (n) indicated. Assayed samples included embryos (Igf2), placentas (Snrpn, Kcnq1ot1, Cdkn1c, and Ube3a) or brains (Ube3a) showing normal, monoallelic (black circle) or biallelic expression (red circle). Average total expression in each exposure group is indicated with a black horizontal line; a = P<0.05; b = P<0.01 and c = P<0.001.
One common feature of imprinted genes is that they are typically located in clusters throughout the genome [30]. The clusters contain maternally and paternally expressed genes, a non-coding RNA and an ICR that is subject to differential methylation, hence often called the DMR [30]. Because imprinted genes located in the same domain were often co-regulated [30], we further examined whether BPA also impacted expression of other genes located in the affected domains by analyzing their allelic and/or total expression. At the Kcnq1 domain, we measured total expression of Cdkn1c, a cell cycle inhibitor gene. Placentas from mice exposed to upper dose BPA showed significant reduction in total Cdkn1c expression compared to control placentas (0.1±0.2 vs. 1.0±0.9, respectively; n = 8 and 9; P<0.05; Figure 4D). The lower dose group (n = 11) was similarly impacted (0.2±0.2 vs. 1.0±0.9 in controls; P<0.01; Figure 4D). These observations suggested that effects of BPA were not limited to the Kcnq1ot1 gene in this domain. Analysis of the allelic expression of Cdkn1c in exposed tissues revealed normal monoallelic expression (data not shown).
At the Snrpn domain, we tested expression of the maternally expressed Ube3a gene (∼1110 kb downstream; Figure 2). Interestingly, total expression of Ube3a in upper dose BPA exposed placentas (0.2±0.3; n = 11) was significantly reduced as compared to controls (1.0±0.7; n = 11; P<0.01; Figure 4E). The difference observed in the lower dose group, however, was not statistically significant compared to controls (0.6±0.6; n = 13; P = 0.08). It is unclear why Ube3a was downregulated in the placenta as it is a brain-specific imprinted gene and was transcribed biallelically in both control and BPA exposed placentas (data not shown). We subsequently tested total expression of Ube3a gene in E12.5 brain from control and upper dose BPA exposed mice and found that brain tissues from upper dose BPA exposure group expressed significantly lower levels of Ube3a (0.6±0.1; n = 6) than controls (1.0±0.4; n = 6; P<0.01; Figure 4F).
To account for the potentially non-normal distribution of our total expression measurements (Figure 4A–4F), all data have been alternatively analyzed using a non-parametric statistical test (Materials and Methods). We found that BPA exposure-induced changes in total expression of the imprinted genes were statistically significant using this method (Figure S1A–S1F).
BPA exposure alters average DNA methylation levels at the differentially methylated regions of imprinted loci
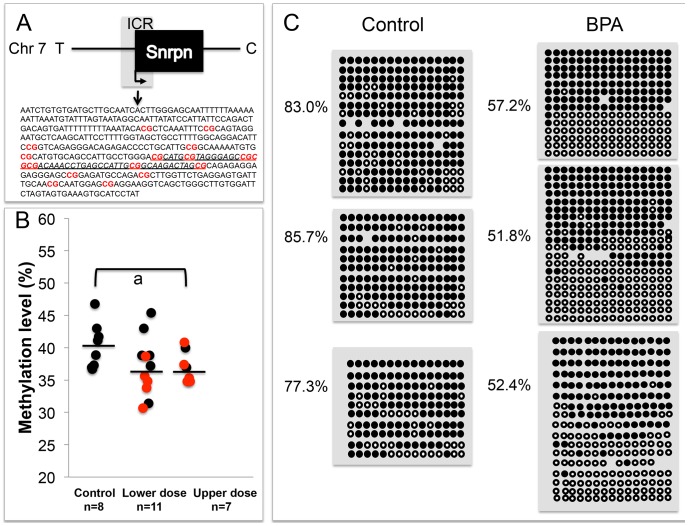
To determine whether LOI observed in tissues from BPA exposed mice was linked to abnormal patterns of DNA methylation, we performed DNA methylation analysis in E9.5 embryos and placentas. We first conducted a pyrosequencing assay testing DNA methylation profiles at 7 CpG sites of the promoter region of Snrpn. The Snrpn promoter is located within an ICR that is hypermethylated on the maternal allele but hypomethylated on the paternal allele [31]. Pyrosequencing analysis of a subset of placentas (that exhibited either normal monoallelic or LOI of the Snrpn gene) revealed a small reduction in mean methylation levels in upper dose BPA exposed placentas (37.2±2.5%; n = 7) as compared to controls (40.2±3.3%; n = 8; P<0.05 through ANOVA; Figure 5A, 5B); however upon non-parametric analysis, this difference was not statistically significant (P>0.05 through Kruskal-Wallis). We subsequently performed bisulfite mutagenesis analysis to assay allele-specific methylation levels at 16 CpG sites at the Snrpn ICR (Figure 5A, 5C). In addition to examining methylation level at more CpG sites as compared to the pyrosequencing assay (16 vs. 7 sites, respectively; Materials and Methods), bisulfite mutagenesis sequencing is a more sensitive assay to detect small methylation level differences as allele-specific methylation, rather than total methylation is measured. We assayed a subset of F1 hybrid placentas from E9.5 upper dose BPA exposed and control mice (n = 3 in each group; all placentas from the upper dose group showed LOI). Consistent with previously published data [15], [31], control placentas showed the expected patterns of differential methylation, i.e., hypomethylated paternal allele (data not shown) and hypermethylated maternal allele with methylation levels at all CpG sites ranging from 77.3 to 85.7% (average of 82.0±4.3%; Figure 5C). Upper dose BPA exposed placentas also exhibited the expected hypomethylated paternal allele (data not shown) and hypermethylated maternal allele, however, methylation levels of all CpG sites of the maternal allele were lower than controls, ranging from 51.8 to 57.2% (average of 53.8±2.4%; Figure 5C; P<0.01).
Figure 5. Analysis in control and upper dose BPA exposed placentas indicated that exposure significantly reduced methylation.
The Snrpn ICR is depicted in (A), including the sequences analyzed in the (B) pyrosequencing and (C) bisulfite sequencing assays. A. 16 CpG sites (highlighted in red and bold) located in a 451 bp region of the Snrpn ICR were assayed by bisulfite sequencing. The underlined genomic sequence containing 7 CpGs represents the region analyzed by pyrosequencing. B. Pyrosequencing data in control, lower dose and upper dose BPA exposure are shown with samples exhibiting monoallelic (black circles) or biallelic expression of Snrpn (red circles). Y-axis represents percentage of total methylation. Black horizontal line in each exposure group indicates average methylation. Sample sizes analyzed in each exposure group are shown; a = P<0.05 when analyzed through ANOVA but not significant through Kruskal-Wallis. C. Three placentas from both control and upper dose BPA exposure groups were analyzed by bisulfite sequencing and shown here is the methylation status of maternal ICR. Each circle represents a CpG site with black = methylated and white = unmethylated. Percentages of methylation at all CpGs are indicated. Average CpG methylation levels are 82.0±4.3% in controls and 53.8±2.4% in upper dose (P<0.01).
In the E9.5 embryos, our pyrosequencing analysis revealed a small reduction of mean methylation levels at the 7 CpG sites in the Snrpn ICR (40.8±2.1% in control vs. 36.8±4.0% in upper dose BPA; n = 7 and 9, respectively; P<0.05 through ANOVA) but this difference was not significant when tested in a non-parametric method (P>0.05 through Kruskal-Wallis). Analysis on the E12.5 brain tissues revealed no difference in mean methylation between control (42.6±2.4%; n = 6) and upper dose BPA exposure (43.9±2.4; n = 9) groups despite the increased LOI rate we observed. No mean methylation difference was detected in embryos from the lower dose (37.1±9.1%; n = 14) and control groups (P = 0.16).
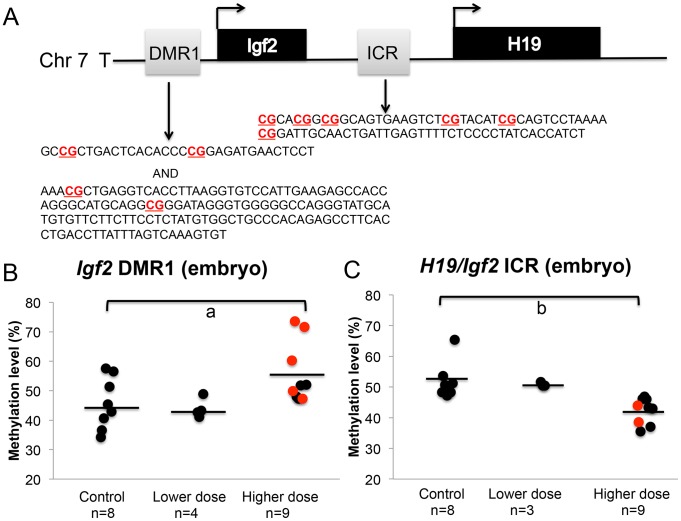
We next determined whether the observed increased proportion of upper dose BPA exposed embryos exhibiting LOI of the Igf2 gene was associated with changes in DNA methylation levels by performing pyrosequencing assays analyzing the H19/Igf2 ICR located between the two genes and a DMR located at the promoter region of Igf2, the Igf2 DMR1 (Figure 6A). Our pyrosequencing analysis for the H19/Igf2 ICR revealed a slight but significantly reduced average methylation of the 6 CpG sites assayed in upper dose BPA exposed embryos (51.6%±5.9% in control vs. 42.3%±4.2% in upper dose BPA; n = 8 and 9, respectively; P<0.001; Figure 6C and Figure S2A). This pattern was unexpected because it was inconsistent with the Igf2 expression profiles; biallelic Igf2 is typically associated with gain of DNA methylation at the ICR [27]. No effect on methylation was observed in embryos from lower dose group (50.9±0.7%; n = 3). Analysis of the Igf2 DMR1, however, was consistent with its expression profiles [32] because it showed significantly increased mean methylation at 4 CpG sites in the embryos from upper dose BPA exposed mice (45.6±8.7% in control vs. 55.7±10.3% in upper dose BPA; n = 8 and 9, respectively; P<0.05; Figure 6B and Figure S2B); no effect was observed in the lower dose group (43.9±3.3%; n = 4; Figure 6B). Neither assay revealed significant methylation changes between BPA exposed (both upper and lower doses) and control placentas (data not shown).
Figure 6. BPA exposure altered DNA methylation at the Igf2 DMR1 and H19/Igf2 ICR in the embryos.
A. Pyrosequencing assays tested methylation of 4 and 6 CpGs (highlighted in bold) in 457 bp and 221 bp genomic regions at the Igf2 DMR1 and H19/Igf2 ICR, respectively. Methylation at the (B) Igf2 DMR1 and (C) H19/Igf2 ICR was analyzed in embryos from controls, lower dose and upper dose exposure groups. Analysis in the upper dose exposure group included embryos that showed monoallelic (black circles) or biallelic (red circles) expression of the Igf2 gene. Sample sizes analyzed in each exposure group are indicated with a = P<0.05 and b = P<0.01. Black horizontal line indicates average methylation level in each exposure group.
Effects of BPA exposure on imprinted gene expression are more severe in the placenta compared to the embryo
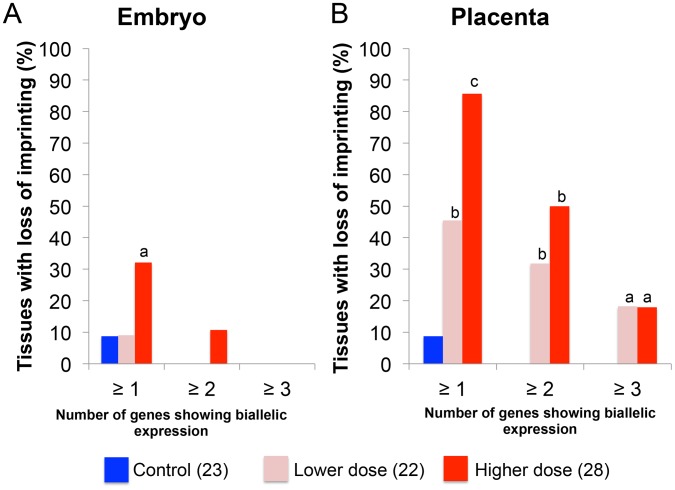
We subsequently determined the severity of impact of BPA exposure in the E9.5 embryo and placenta by comparing the number of imprinted genes exhibiting LOI in each tissue. In the embryo, 32.1% (n = 28) of upper dose BPA samples exhibited LOI of at least 1 imprinted gene compared to 8.7% in control (n = 23; P<0.05; Figure 7A). Impact of lower dose exposure did not differ significantly from controls (8.7%; n = 22; Figure 7A). The percentage of embryos showing LOI of 2 genes or more in control (0.0%) and upper dose BPA (10.7%) exposure groups was not statistically different (Figure 7A).
Figure 7. Impact of BPA exposure is shown as number of genes exhibiting imprinting perturbations.
Effects of BPA exposure were more significant in the (B) placentas compared to (A) embryos. For each exposure group (control [blue], lower dose [pink] or upper dose [red]), percentage of tissues showing LOI (Y-axis) of at least 1 gene (≥1), 2 genes (≥2) or 3 genes (≥3) is indicated; a = P<0.05; b = P<0.01; c = P<0.001.
BPA exposure affected the E9.5 placenta more severely with 85.7% and 45.5% of samples in the upper dose (n = 28) and lower dose (n = 22) exposure groups, respectively, showing LOI of at least 1 imprinted gene compared to 8.7% in controls (n = 23; P<0.0001 and P<0.01, respectively; Figure 7B). The difference between severity of LOI in BPA exposed and control groups was still statistically significant when 2 imprinted genes or more were considered (50.0% and 31.8% of placentas from upper dose and lower dose exposure groups, respectively, compared to 0.0% in controls; P<0.01; Figure 7B) or even for 3 imprinted genes or more (17.9% and 18.2% of placentas from upper dose and lower dose exposure groups, respectively, compared to 0.0% in controls; P<0.05; Figure 7B).
BPA exposed placentas exhibit reduced global DNA methylation
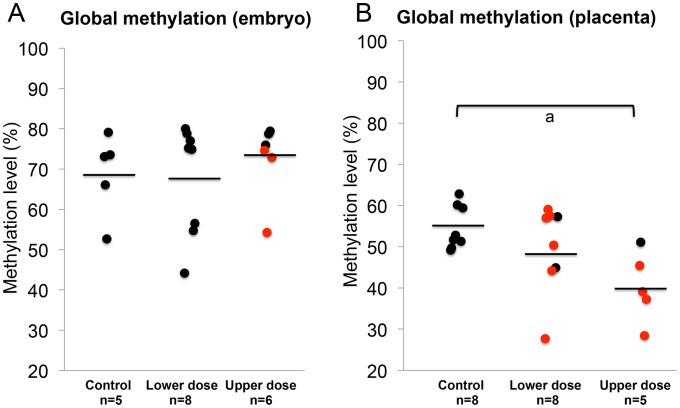
The observed effects of BPA exposure on imprinted gene expression and DNA methylation as well as differences in the severity of BPA effects in the embryo vs. placenta prompted us to investigate effects of exposure on genome wide DNA methylation and to determine whether the embryo and placenta displayed significant differences. We performed experiments using the Luminometric Methylation Assay (LUMA) in control and BPA exposed embryos and placentas. The method estimates genome wide DNA methylation based on combined DNA cleavage by restriction endonucleases followed by a polymerase extension assay using the pyrosequencer [33]. Our data showed that upper dose BPA exposure significantly reduced the average global methylation of the placenta at E9.5 (54.6±5.3% vs. 40.3±5.9% in the control vs. upper dose BPA exposed, P<0.05; with n = 8 and 5, respectively; Figure 8B and Figure S2C). Average DNA methylation difference between lower dose and control groups was not statistically different (49.8±10.6% vs. 54.6±5.3%, respectively; n = 8 and 7; Figure 8B). No significant differences in methylation were seen in the E9.5 embryo (68.9±10.2% in control, 67.7±13.7% in lower dose and 72.7±9.3% in upper dose BPA exposure groups; n = 5, 8 and 6, respectively; Figure 8A). Similar observations were observed in the embryos and placentas at E12.5 (data not shown). We note that our global methylation data in control tissues were consistent with previous publications: Gallou-Kabani et al., (2010) reported similar average DNA methylation in E15.5 placentas using the LUMA although levels in the embryo were not reported [17]. Additionally, Popp et al., (2010) reported similar levels of average DNA methylation in E13.5 embryos and placentas as well as highly variable individual global methylation in their genome-wide bisulfite sequencing analysis [34].
Figure 8. BPA exposure reduced genome-wide DNA methylation in the placenta but not the embryo.
Results of LUMA studies showed genome-wide DNA methylation levels in the (A) embryos and (B) placentas from control, lower dose and upper dose BPA exposure groups. Black circles indicate tissues with normal, monoallelic expression of all imprinted genes tested; red circles are tissues that showed LOI of at least 1 imprinted gene. Black horizontal lines represent average total methylation in each exposure group; a = P<0.05.
Upper dose BPA exposure results in abnormal placentation
Our data suggested that the placenta was a significant tissue target for BPA induced epigenetic perturbations as indicated by aberrant expression and methylation of imprinted genes located in the Snrpn and Kcnq1 domains as well as significantly reduced genome-wide methylation. We further tested whether upper dose BPA exposure affected placenta-specific imprinted genes by studying the allelic expression patterns of the placenta-specific imprinted genes Ascl2 and Tssc4 located in the Kcnq1 domain (Figure 2). Analysis of Ascl2 revealed that a significant proportion of upper dose BPA exposed E9.5 placentas showed biallelic expression (6/24) as compared to controls (0/16; P<0.05) with expression of the normally repressed maternal allele ranging from 16.4–43.7% (Figure 3D). No significant effects were observed at the Ascl2 gene in the lower dose group (1/16; Figure 3D). At the Tssc4 locus, a trend towards increased rate of LOI was observed in the upper dose but was not statistically significant (Table S1).
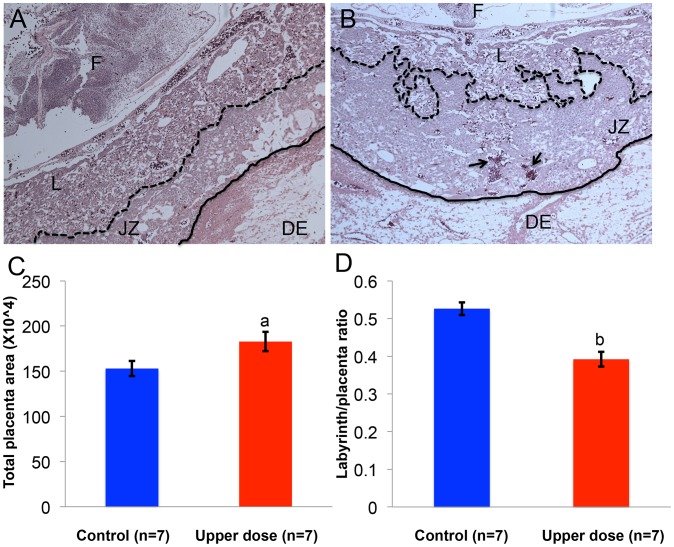
As imprinted genes play a critical role in placental development [35], we determined whether aberrant imprinting induced by BPA exposure was associated with abnormal placental phenotypes. We performed histological and immunohistochemical examinations on placentas from the upper dose and control exposure groups. Because the major placental zones (i.e., the labyrinth and junctional zones) have just begun to develop at E9.5 [36], histology and immunohistochemistry were conducted at E12.5. In both control and upper dose exposure groups, the junctional zone and labyrinth were easily distinguishable (Figure 9A, 9B). We observed, however, that the placentas from upper dose exposure group (n = 7) had significantly larger total area (182.7±10.6 µm2 vs. 152.9±8.3 µm2; P<0.05; Figure 9C) but smaller labyrinth/placenta ratio (0.39±0.02 vs. 0.52±0.02; P<0.001; Figure 9D) compared to controls (n = 7). Additionally, BPA exposed placentas had distorted boundary between the junctional zone and labyrinth and increased accumulation of red blood cells (Figure 9B). There was no significant difference in the maternal decidua/placenta ratio between control and upper dose BPA exposed placentas (data not shown).
Figure 9. BPA exposure was associated with defective placental development.
Hematoxylin and eosin-stained histological sections of representative E12.5 conceptuses from (A) control and (B) upper dose BPA exposure groups are shown. F = fetus; DE = maternal decidua; L = labyrinth zone and JZ = junctional zone. Dotted line indicates the boundary between JZ and L, continuous line the boundary between JZ and DE, and arrows the accumulation of red blood cells. Total placenta area (C) and the ratio of area occupied by the labyrinth zone and total placenta area (D) were measured in both control (blue) and upper dose BPA exposed (red) placentas; a = P<0.05; b = P<0.001.
Discussion
In this work, we performed an extensive characterization of the effects of environmental exposures on epigenetic regulation of developmental genes. We analyzed impact of fetal BPA exposure on imprinted gene expression and methylation in mouse embryos and placentas. As BPA exposure alters DNA methylation status [9], [22]–[24], we hypothesized that exposure perturbed genomic imprinting and disrupted embryonic and/or placental development. We analyzed E9.5 and E12.5 embryos and placentas from BPA exposed conceptuses for evidence of imprinting perturbations. Recently, Szabo and coworkers [37] investigated effects of exposure to 200 µg/kg/day of BPA on genomic imprinting but reported mild perturbations in E13.5 mouse embryonic and extraembryonic tissues. Their exposure, however, occurred between E8.5 and E12.5, during the period when germ cells are migrating and beginning to reprogram [38]. Our maternal exposure initiated 2 weeks prior to mating until pregnancy day 9.5 and 12.5, coincided with the epigenetic reprogramming of the genome during late oocyte maturation and post-fertilization development [13]. This exposure window includes the period when methylation is being established in the oocyte and post fertilization when the majority of the genome is reprogrammed [29]. Importantly, genomic imprints of ICRs are maintained during this time. Because Szabo and coworkers reported no significant effects on the Snrpn, Kcnq1ot1 and Ascl2 genes, the findings of LOI and/or DNA methylation changes at these loci in this current study suggest that imprinting regulation is more vulnerable to environmental exposure during the epigenetic reprogramming period that coincides with the establishment and maintenance of DNA methylation [6]–[10] as compared to during mid-gestation. Consistent with this observation, BPA exposure outside of the epigenetic reprogramming window, i.e., from E5.5–E12.5, in the current study did not lead to significant imprinting perturbations. Additionally, differences in doses, mouse strains and route of administrations between the two studies may explain differences in results. Neither study found significant effects at the H19 and Peg3 loci.
Our work has demonstrated that BPA exposure disrupted imprinted gene regulation, with the Snrpn and Kcnq1 domains significantly affected in the placenta while the Igf2 gene imprinting was disrupted exclusively in the embryo. Our findings support previous studies demonstrating the ability of environmental factors to impact genomic imprinting regulation in early mouse development [14]–[17], although unlike these studies [14]–[16], we did not observe significant effects at the H19 gene. Furthermore, ART-associated environmental perturbations are applicable to patients undergoing ART procedures [14]–[16] and therefore, the potential health consequences are limited to the babies conceived through this technology; in contrast, the environmental factor investigated in the current study is present ubiquitously and the potential risks of exposure are applicable to the general population.
Importantly, our observations at the Snrpn domain recapitulate the molecular signatures of Angelman Syndrome (AS) in humans, of which a significant proportion of cases are caused by absence of the E3A ubiquitin ligase gene UBE3A product due to mutation or epigenetic aberrations at the SNRPN imprinting center (IC) in the brain [39], [40]. In the brain of AS mouse models carrying insertion/duplication mutation of a region analogous to human AS IC, the maternal Snrpn promoter had reduced DNA methylation, biallelic Snrpn gene expression and reduced total expression of the Ube3a gene [40]. Given the potentially reduced DNA methylation of the ICR at this locus in the E9.5 whole embryo, significant increased LOI rate in the E12.5 brain as well as lower expression of Ube3a following BPA exposure in our study, these results raise the possibility that environmental perturbations could contribute to the rare cases of AS caused by loss of methylation at the IC [40]. Although the environment has been postulated to influence incidence of imprinting diseases [41], our study is the first to suggest that exposure to a man-made compound may be implicated.
Similar to previous studies [15], [16], we found the placenta to exhibit more significant imprinting perturbations than the whole embryo (Figure 7A, 7B). One possible explanation is that the placenta is in direct contact with maternal tissue causing it to be more highly exposed to the environment. Blood measurement in humans has found more accumulation of BPA in the placenta compared to the fetus [42]. However, in the current study, the placentas and/or embryos from BPA exposed mice exhibited significant changes in DNA methylation at DMRs, including the Snrpn ICR and Igf2 DMR1 (Figure 5A, 5B), suggesting that the embryo was not protected from environmental insults. Normal, monoallelic expression of Snrpn in BPA exposed embryos observed in our study, despite significantly reduced methylation, could reflect differences in imprinting regulation between the embryo and placenta. It is postulated that imprinting in the embryo depends largely on DNA methylation while both posttranslational histone modifications and DNA methylation play a regulatory role in the placenta [43], [44] - as DNA methylation is the primary regulator of imprinting in the embryo, it is possible that additional layers of repression are present. Alternatively, frequencies of LOI may be variable in different embryonic organs and effects of imprinting perturbations may become less significant when the whole embryo is investigated. Our allele-specific expression analysis on E12.5 brain from upper dose BPA exposure group indeed revealed increased LOI rate at the Snrpn locus as compared to controls, although, we found no changes in DNA methylation levels. Furthermore, it would be interesting to determine whether increased duration of exposure or combined exposure with other endocrine disruptors would produce more significant effects on imprinted gene expression at the Snrpn and Kcnq1 domains in the embryo and/or organs. These paradigms would be relevant to humans as BPA is ubiquitous and exists in the presence of other environmental pollutants, and effects of these different compounds may be additive or synergistic [45].
Furthermore, we observed significantly increased LOI of the Igf2 gene among E9.5 (Figure 3B) and E12.5 embryos (Figure 3E) exposed to 10 mg/kg bw/day of BPA and these effects were associated with increased methylation at the Igf2 DMR1 in E9.5 embryos (Figure 5D and Figure S2B). In humans, altered methylation patterns at the IGF2 gene are linked to environmental perturbation or intervention during early development including smoking [46], maternal starvation [47] and folic acid supplementation [48]. In humans and mice, altered IGF2 or Igf2 expression significantly alters growth [49]–[51]. In the current study, we did not observe significant fetal body weight differences between controls and BPA exposed mice at E9.5 and E12.5 (data not shown), although it was possible that these stages were too early to assess growth effects. Alternatively, the abnormal placental development in our BPA exposed mice may have counteracted the effect of increased Igf2 expression.
We found that although BPA exposure significantly altered average methylation levels at various DMRs, both allelic and total expression data were not perfectly correlated with each other or with methylation changes (e.g., total expression changes may occur without LOI or hypomethylation at the Snrpn ICR was not always associated with LOI or vice versa). The observation, however, has been previously noted in studies of Dnmt1 mouse mutants in which allelic expression changes in the embryos and placentas were not always associated with comparable changes in total expression [52]. Additionally, Weaver et al (2010) also observed that loss of methylation in Dnmt1 mouse mutants did not always cause LOI [52]. Lack of perfect correlation between methylation and imprinted expression in our BPA exposure model suggests that other epigenetic mechanisms in addition to DNA methylation could be involved in mediating BPA-induced imprinting aberrations. Additionally, most imprinted genes are expressed only in a subset of cells in the embryo and placenta, making a 1 to 1 correlation in a complex tissue very difficult.
One important finding of our study is that lower dose BPA exposure was sufficient to induce significant epigenetic changes. We initially employed an exposure level of 10 mg/kg bw/day BPA as a starting point because Dolinoy et al. reported significant effects on DNA methylation in the agouti mice when exposed to this dose [9]. To our surprise, a 1000-fold lower dose of 10 µg/kg bw/day BPA was sufficient to alter imprinting at the Snrpn and Kcnq1 domains in E9.5 placentas (Figure 3A, 3C). This lower dose corresponds to what is considered a safe human exposure level (i.e., determined as ≤50 µg/kg bw/day [20]). Although the lower dose does not result in a significantly different serum BPA level compared to controls (presumably due to the difficulty in assaying this level of BPA), the average serum BPA levels measured in pregnant mice treated with 10 mg/kg bw/day of BPA in this work was consistent with previous studies employing similar paradigm [53]. Moreover, the detected serum levels were within the range of BPA concentrations measured in human blood [20], suggesting that our exposure paradigm was biologically relevant to humans. Thus, our studies are consistent with lower dose BPA scientific literature that reports a wide variety of developmental defects related to metabolism, immune system, reproduction and brain function [21] following exposure at or below the determined “safe” human exposure level.
Our placenta studies revealed that upper dose BPA exposure was associated with abnormal development characterized by significantly larger placenta area and smaller labyrinth to placenta ratio (Figure 9A–9D) that may have arisen due to overgrowth of the junctional zone and/or reduced size of the labyrinth. Tachibana et al (2007) have previously reported effects of BPA exposure in placental development with similar findings of reduced proportion of labyrinth layer in the ICR mouse strain [54]. To our knowledge, however, the current study is the first to suggest that the placental phenotype induced by environmental exposures may be associated with epigenetic perturbations. Our findings are consistent with other mouse studies reporting similar placental phenotypes including overgrowth that are linked to defective epigenetic reprogramming including in mice produced by embryo transfer [55] or mice with aberrations in imprinting due to paternal uniparental disomy of chromosome 12 [56]. Furthermore, loss of function of the cyclin dependent kinase Cdkn1c gene in the mouse causes larger placentas due to overgrowth of the trophoblast cells [57]; the larger placenta area and reduced ratio of labyrinth/placenta observed in the current study may be partly explained by the significantly lower total expression of the Cdkn1c gene in our BPA exposed placentas. As placentomegaly is often observed in the human overgrowth disease, BWS, in which 10% of cases are attributed to loss of function or mutation in the CDKN1C gene [27], results of our work demonstrate that disruption of this pathway due to environmental exposures may be associated with adverse placental development. The placental phenotype observed in the current study may also be the result of perturbed non-imprinted genes that play a role in placental development as our genome wide methylation analysis indicated significantly reduced methylation levels in BPA-exposed placentas. Future studies aiming at the identification of exact mechanistic links between exposure and placental defects as well as detailed examinations at the placenta at various developmental stages would be critical.
In the mouse, maternal-fetal material exchanges including O2 occur in the labyrinth [36]. Because hypoxia is commonly linked to formation of new blood vessels in the placenta [58], the observation of increased accumulation of red blood cells in our BPA exposed placentas (Figure 9B) suggests that the reduced proportion of labyrinth may have affected oxygenation. This observation needs to be investigated carefully in the future as its implication is directly relevant to placental function and fetal health. Lack of O2 at later stages of placental development restricts vascularization of the labyrinth and is potentially associated with intrauterine growth restriction (IUGR) [59]. Although human placenta is structurally distinct from mouse, mouse labyrinth layer is functionally analogous to the chorionic villi of human placenta and certain severe cases of early onset IUGR and spontaneous abortion can be attributed to its dysfunction [36]. The placentation defect in the current study may explain the absence of weight increase in BPA exposed embryos despite increased total expression of Igf2 as discussed above.
The mechanisms involved in BPA induced epigenetic changes are currently under investigation. The best-characterized property of BPA is its ability to act as an estrogen and increasing data have shown that some relevant developmental effects are mediated through estrogen receptors (ERs) [21]. Role of estrogen in DNA methylation-dependent epigenetic regulation has been suggested [60] and BPA exposure has recently been shown to decrease or increase expression levels of the Dnmt3a/b and/or methyl CpG binding proteins (Mbd)2/4 genes [61], [62]. BPA may therefore induce hypomethylation or hypermethylation in exposed rodent models [9], [61], [62] through alterations in the genes responsible for DNA methylation maintenance using estrogen-dependent signaling pathways.
As imprinting regulation between mice and humans shares similar features, results of the current work have important implications for human health. DNA methylation defects are often observed in human imprinting disorders, for example a significant proportion of BWS cases arises from abnormal methylation at the H19/IGF2 ICR or epigenetic defects at the ICR of KCNQ1 [27]. The current study found that environmental exposure induced methylation changes at the DMRs of imprinted domains, suggesting that it is highly likely that the environment may play a role in the etiology of imprinting diseases.
Furthermore, our work suggests that BPA exposure can perturb epigenetic regulation of developmental genes that play significant roles in neurobehavioral development or growth. Increasing evidence has linked imprinted genes to brain function [63], [64] and disrupted imprinting causes neurodevelopmental disorders including AS and PWS. As maternal BPA levels in humans are linked to neurobehavioral development in children including behavioral and emotional functions [65]–[67], it would be critical to determine whether endocrine disruptors including BPA may influence brain and behavior functions through perturbed imprinting regulation or aberrant function of other developmental genes that are subject to epigenetic regulation. Additionally, BPA exposure is linked to alteration in body weight of the offspring in animal studies [68], [69]. These findings are consistent with epidemiological observations in humans suggesting the association between BPA exposure and preterm birth [70], or small for gestation age [71]. Our work demonstrated that BPA exposure can potentially influence growth and development by altering expression of the growth-promoting gene Igf2 or through disruption of placental development.
In summary, our work has demonstrated the ability of exposure to the common chemical BPA to disrupt epigenetic regulation of developmentally relevant genes. We have shown that exposure to physiological relevant doses of BPA perturbs expression and methylation of imprinted genes in the mouse with the most significant effects observed in the placenta and that the effects were associated with abnormal placental development. The current study found that early embryonic development is susceptible to environmental perturbations, and we have shown for the first time that man-made compounds are potentially capable to alter epigenetic reprograming events. In humans, this sensitive developmental window coincides with the earliest stage of pregnancy at the time when it is not yet clinically recognized, thus, to reduce potential BPA exposure-induced health abnormalities, exposure management should begin even before a woman gets pregnant. Unfortunately, due to the ubiquitous nature of BPA, complete avoidance is almost impossible as most individuals in the US are exposed [20]. In the future, studies should be performed to address whether BPA-induced defects can alter placental function and whether the dysfunction leads to changes in body weight and/or other metabolic disorders. Furthermore, studies elucidating mechanisms related to BPA-induced epigenetic alterations including the utilization of the estrogen receptor knock out mice should be undertaken as results from such studies would provide critical insights into BPA related developmental abnormalities and preventive actions.
Materials and Methods
Mouse information
For allele-specific expression and methylation studies, we used two different mouse strains: the C57BL/6 (B6; Jackson Laboratory, Bar Harbor, ME) and the B6 (CAST7) or C7 strain [72] that contains chromosome 7 of the Mus castaneus (Cast) strain in a B6 background. F1 hybrid progeny, which were assayed for allelic expression and methylation, were derived from reciprocal matings between B6 and C7 mice. Polymorphisms between the two mouse strains were used to distinguish the parental origin of expression or methylation of genes of interest. All animal work was conducted with the approval of the Institutional Animal Care and Use Committee.
Virgin females, 6–10 weeks old, were assigned to one of the following diets: a) modified AIN-93G diet (diet TD.95092 with 7% corn oil substituted for 7% soybean oil; Harlan Teklad) as “control”; b) modified AIN-93G diet supplemented with 50 mg/kg diet of BPA (diet TD.06156; Harlan Teklad) as “upper dose”; or c) modified AIN-93G diet supplemented with 50 µg/kg diet of BPA (diet TD.110337; Harlan Teklad) as “lower dose”. Teklad Diets (Harlan Laboratories Inc, Madison, WI) provided all ingredients except BPA (Sigma Aldrich, St. Louis, MO). Drinking water was provided in polypropylene bottles. The estimated daily treatment of BPA in our study was 10 mg/kg body weight (bw) in the “upper dose” and 10 µg/kg bw in the “lower dose” groups. Diet was provided 2 weeks prior to mating and during pregnancy until embryonic day (E) 9.5 and E12.5. At E9.5 and E12.5, pregnant mice were euthanized and tissues from the whole embryo and placenta isolated. Prior to placental tissue isolation, the maternal decidua was carefully removed. For tissue-specific analysis, different embryonic organs including the heart, liver, kidney and brain were isolated from E12.5 mice. Upon isolation, tissues were halved (each half for expression and methylation studies) and frozen at −80°C.
Expression studies
Total RNA was extracted using Trizol according to the manufacturer's instructions and quantified using the Nano Drop spectrophotometer. cDNA was prepared by using Superscript III reverse transcriptase and random hexamers. To test that RNA was free of genomic DNA contamination, a minus reverse transcription control was included.
Allele-specific expression
Allele-specific Snrpn and H19 expression studies were conducted on E9.5 and E12.5 embryonic and placental cDNA using the Light Cycler real time PCR system as described previously [14], [52]. For analysis of Igf2, Kcnq1ot1, Cdkn1c, Ascl2 and Peg3, we conducted RT-PCR followed by restriction enzymes using the conditions described previously [14],[52]. Samples that showed ≥10% of total expression derived from the repressed allele were considered to be biallelic or to exhibit loss of imprinting (LOI).
Total expression
Quantification of mRNA levels was determined in E9.5 embryos and placentas using the Light Cycler Real Time PCR System (Roche); for analysis of the Ube3a gene, E12.5 embryonic brains were also included. The PCR conditions and primers information were described previously [14], [52]. Total expression of imprinted genes was calculated relative to the geometric mean of the reference genes Arpp0 (acidic phosphoprotein P0 subunit) and Gapdh (glyceraldehyde-3-phosphate dehydrogenase). Samples were analyzed at least in duplicates using the Light Cycler 4.0 software as described [52].
Methylation studies
DNA was extracted using the standard phenol chloroform method and amount measured using the Nano Drop spectrophotometer. One microgram of DNA was bisulfite treated using the EpiTect Bisulfite Kit (Qiagen) following manufacturer's protocol.
Pyrosequencing assays
Pyrosequencing was performed to analyze the methylation profiles at the Snrpn ICR (chr7: 67,149,897–67,150,141; NCBI37/MM9), H19/Igf2 ICR (chr7: 149,767,599–149,767,819; NCBI37/MM9) and Igf2 DMR1 (chr7: 149,851,180–149,851,655; NCBI37/MM9). 50 ng of bisulfite treated DNA was used for PCR. For Snrpn ICR, the primer sequences were as follows: forward primer 5′- GGTAGTTGTTTTTTGGTAGGATAT-3′, biotinylated reverse primer 5′- ACTAAAATCCACAAACCCAACTAACCT-3′. For H19/Igf2 ICR, the primer sequences were as follows: forward primer 5′- GGGTAGGATATATGTATTTTTTAGGTTG- 3′ and biotinylated reverse primer 5′- CTCATAAAACCCATAACTATAAAATCAT- 3′. For Igf2 DMR1, the primer sequences were as follows: forward primer 5′- TGAGGTTAGATTAGGTTGTAAGTT-3′ and biotinylated reverse primer 5′- CTTCCCTACCCCTTAAACC -3′. The PyroMark PCR kit (Qiagen) was used in a 25 µL reaction according to the manufacturer's protocol. PCR conditions were: 95°C for 15 minutes followed by 45 cycles of 95°C for 15 seconds, 55°C (or 58°C for Igf2 DMR1) for 30 seconds and 72°C for 15 seconds. 10 µL of the biotinylated PCR product was used for each sequencing assay with the Snrpn sequencing primer 5′- GTGTAGTTATTGTTTGGGA- 3′, or H19/Igf2 ICR sequencing primer 5′- TGTAAAGATTAGGGTTGT- 3′, or Igf2 DMR1 sequencing primers 5′-GGATTTTGTTAGGTAGGA-3′ and 5′-TTTTAGAGGTTTTTGGAGAA-3′. Pyrosequencing was done using the PyroMark Q96MD (Qiagen) system following the manufacturer's protocol and the PyroMark Gold 96 reagents kit (Qiagen). Methylation was analyzed using Qiagen's Pyro Q- CpG software. 7 CpGs were analyzed for Snrpn ICR, 6 CpGs for Igf2/H19 ICR, and 4 CpGs for Igf2 DMR1. The Luminometric Methylation Assay (LUMA) was performed and analyzed as described previously [33] using 500 ng of genomic DNA.
Bisulfite sequencing
Bisulfite treated DNA was subject to nested PCR amplification, subcloning and sequencing of the Snrpn promoter-exon 1 region (2073–2601 bp, AF081460) as previously described in [72], [73]. At least 2 independent PCRs were performed on each sample. Snrpn parental alleles were distinguished by single nucleotide polymorphisms as previously reported [31], [72], [74].
BPA serum measurement
Pregnant females were sacrificed on E9.5 and blood was collected through cardiac puncture, incubated at room temperature for 15 min and centrifuged at 3000 rpm. Serum was collected and stored at −80°C until analysis.
Materials
13C12-BPA (99% isotopic purity) used as internal standard for LC-MS analysis was purchased from Cambridge Isotope Labs (Andover, MA). 2,3,4,5,6-pentafluorobenzyl bromide (PFB-Br), sodium hydroxide and solvents were purchased from Sigma-Aldrich (St. Louis, MO). HPLC-grade hexanes, isopropanol and ethanol were obtained from Fisher Scientific Co. (Fair Lawn, NJ). ACS-grade ethanol was obtained from Pharmco (Brookfield, CT). Gases were supplied by BOC Gases (Lebanon, NJ). Chiralpak AD-H column was obtained from Chiral Technologies (West Chester, PA).
LC/MS conditions for BPA–PFB quantification
BPA was quantified based on a previously described procedure [75]. Normal-phase chiral chromatography for LC/MS experiments was performed using a Waters Alliance 2690 HPLC system (Waters Corp., Milford, MA). Gradient elution was performed in the linear mode. A Chiralpak AD-H column (250×4.6 mm i.d., 5 µm; Daicel Chemical Industries, Ltd., Tokyo, Japan) was employed with a flow rate of 1 mL/min. Solvent A was hexanes and solvent B was 2-propanol/methanol (1/1, v/v). The linear gradient was as follows: 2% B at 0 min, 2% B at 2 min, 50% B at 14 min, 50% B at 16 min, 2% B at 17 min, and 2% B at 25 min. The separation was performed at 30°C.
The following SRM transitions were monitored: BPA-PFB, m/z 227 → 113 (collision energy 28 eV), m/z 227 → 212 (collision energy 25 eV), [13C12]-BPA-PFB, m/z 239 → 139 (collision energy 28 eV) and 13[C12]-BPA-PFB, m/z 239 → 224 (collision energy 25 eV).
BPA extraction from serum samples
Serum collected from pregnant females was measured with a glass syringe and transferred to a 10 mL glass test tube containing 1 mL of Optima water spiked with 2 ng of 13[C12]-BPA (20 µL×100 pg/µL sol in acetonitrile). For blank controls, the same procedures were performed with 40 µL of water. After equilibrating for 10 min at room temperature, the samples were extracted with ethyl ether (3 mL). The organic layer was then evaporated to dryness under nitrogen, and the BPA in the residue was converted to PFB derivatives as described in [75]. The alcohol moiety was derivatized as PFB ether under strong basic conditions (sodium hydroxide). The mass spectrum obtained by CID and MS/MS analysis of [M-PFB]− was identical with the underivatized BPA (spectrum not shown). However, the retention time was different on the LC system, with the PFB derivative eluting earlier (7.7 min vs. 12.7 min) than the underivatized BPA. The detection limit for the BPA-PFB was 3 pg on column.
Data analysis
The final concentration of BPA in serum was determined by measuring the area ratio of BPA over 13C12-BPA peaks, calculated based on the amount of 13C12-BPA added and adjusted based on the volume. All data analysis was performed using Xcalibur software, version 2.0 SR2 (Thermo Electron Corporation) from raw mass spectral data.
Placenta analysis
At E12.5, pregnant mice were sacrificed by CO2 asphyxiation, and the gravid uterine horns removed from the abdominal cavity via laparotomy incision. The two most proximal gestations from each horn were excised en bloc between adjacent implantation sites with the surrounding myometrium intact and immersed in 10% phosphate buffered formalin for histological examination. The remaining conceptuses were carefully dissected from the surrounding myometrium. The amniotic membranes ruptured and separated from the placenta. Fetuses and placentas were snap frozen in liquid nitrogen and stored at −80°C for future studies.
Placental histology and morphometric analysis
Tissues were fixed overnight at 4°C, dehydrated, and then bivalved in sagittal section through the mid placental plane. Care was taken to place all tissues in similar orientation prior to embedding. Tissues were embedded in a paraffin block using standard protocols. Serial tissue sections, 4 µm thick, were cut through the mid placenta in sagittal section and stained with hematoxylin and eosin. Images were collected on a Leica DMRBE upright widefield microscope with a 2.5x, 5x, and 10x objective, a 5.0 MegaPixel color CCD camera, and iVision acquisition software (BioVision Technologies). Micrographs were analyzed with ImageJ v1.4.5 (NIH). All measurements were made on multiple sections through the mid sagittal plane of the placenta. The total cross-sectional area of the placenta was outlined from its base at the chorioallantois to the giant cell layer, identified morphologically. The total cross-sectional area of the labyrinth was outlined based on its characteristic morphological features. Ratios of the area of the labyrinth to the placenta as well as maternal decidua to the placenta were then calculated.
Statistical analysis
Goodness-of-fit analyses were used to assess differences in the proportion of tissues exhibiting loss of imprinting or biallelic expression of imprinted genes. Fisher's exact test of independence was used to evaluate litter-specific effects. To evaluate the statistical differences in serum BPA levels and areas of the major placental zones, we performed standard t-test analyses. To test for differences in total imprinted gene expression and levels of methylation, we used the PRISM software to perform one-way analysis of variance (ANOVA) and Tukey's posttest. When indicated, expression and methylation data had been additionally analyzed using the non-parametric Kruskal-Wallis test and the results were presented as supplementary materials (Figures S1, S2).
Supporting Information
Total expression of imprinted genes analyzed by a non-parametric test. Kruskal-Wallis statistical test was conducted to measure differences among exposure groups on expression of the (A) Snrpn, (C) Kcnq1ot1, (D) Cdkn1c and (E) Ube3a genes in the E9.5 placenta, (B) Igf2 gene in the E9.5 embryo and (F) Ube3a gene in the E12.5 brain. Y axis = relative expression as compared to reference genes; X axis = measurement values of relative expression ranked from lowest (left) to highest (right). P values shown represent all exposure groups analyzed. P values between control and upper dose are as followed: (A) 0.006, (B) 0.05, (C) 0.04, (D) 0.002, and (E) 0.003. P values between control and lower dose are 0.003 in (C) and 0.001 in (D).
(TIF)
Imprinted genes-specific and genome-wide methylation levels analyzed by a non-parametric test. Results from the Kruskal-Wallis test are presented for methylation levels of the (A) H19/Igf2 ICR and (B) Igf2 DMR1 in the E9.5 embryos and (C) genome-wide methylation levels as measured by LUMA in the E9.5 placentas. Y = percentage of methylation level; X axis = measurement values of methylation ranked from lowest (left) to highest (right). P values shown represent all exposure groups analyzed. P values between control and upper dose are as followed: (A) 0.0006, (B) 0.04 and (C) 0.02.
(TIF)
Percentage of total expression of imprinted genes derived from the repressed allele in each exposure group in the E9.5 embryo and placenta.
(XLS)
Percentage of total expression of imprinted genes derived from the repressed allele in each exposure group in the E12.5 embryo and placenta.
(XLS)
Acknowledgments
We thank Rebecca Simmons for discussions and helpful comments on the manuscript and for providing the reagents used in the placental studies described in this work. We also thank Christopher Krapp for designing the pyrosequencing assays and conducting part of the methylation analysis, and Maria Navarro and Martha Stefaniak for technical assistance.
Funding Statement
This publication was made possible, in part, by grant number 1 P30 ES013508-06 from the National Institute of Environmental Health Sciences (NIEHS), NIH, and March of Dimes. MS was supported by NIEHS F32ES019416 and K99ES022244. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Barker DJ (1997) Maternal nutrition, fetal nutrition, and disease in later life. Nutrition 13: 807–813. [DOI] [PubMed] [Google Scholar]
- 2. Harris RM, Waring RH (2012) Diethylstilboestrol-A long-term legacy. Maturitas 72: 108–112. [DOI] [PubMed] [Google Scholar]
- 3. Eubig PA, Aguiar A, Schantz SL (2010) Lead and PCBs as risk factors for attention deficit/hyperactivity disorder. Environ Health Perspect 118: 1654–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Markham JA, Koenig JI (2011) Prenatal stress: role in psychotic and depressive diseases. Psychopharmacology (Berl) 214: 89–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roseboom TJ, Painter RC, van Abeelen AF, Veenendaal MV, de RooijSR (2011) Hungry in the womb: what are the consequences? Lessons from the Dutch famine. Maturitas 70: 141–145. [DOI] [PubMed] [Google Scholar]
- 6. Lillycrop KA, Slater-Jefferies JL, Hanson MA, Godfrey KM, Jackson AA, et al. (2007) Induction of altered epigenetic regulation of the hepatic glucocorticoid receptor in the offspring of rats fed a protein-restricted diet during pregnancy suggests that reduced DNA methyltransferase-1 expression is involved in impaired DNA methylation and changes in histone modifications. Br J Nutr 97: 1064–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tobi EW, Lumey LH, Talens RP, Kremer D, Putter H, et al. (2009) DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum Mol Genet 18: 4046–4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Anway MD, Leathers C, Skinner MK (2006) Endocrine disruptor vinclozolin induced epigenetic transgenerational adult-onset disease. Endocrinology 147: 5515–5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dolinoy DC, Huang D, Jirtle RL (2007) Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci U S A 104: 13056–13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Waterland RA, Jirtle RL (2004) Early nutrition, epigenetic changes at transposons and imprinted genes, and enhanced susceptibility to adult chronic diseases. Nutrition 20: 63–68. [DOI] [PubMed] [Google Scholar]
- 11. Feinberg AP, Tycko B (2004) The history of cancer epigenetics. Nat Rev Cancer 4: 143–153. [DOI] [PubMed] [Google Scholar]
- 12. Arnaud P, Feil R (2005) Epigenetic deregulation of genomic imprinting in human disorders and following assisted reproduction. Birth Defects Res C Embryo Today 75: 81–97. [DOI] [PubMed] [Google Scholar]
- 13. Weaver JR, Susiarjo M, Bartolomei MS (2009) Imprinting and epigenetic changes in the early embryo. Mamm Genome 20: 532–543. [DOI] [PubMed] [Google Scholar]
- 14. Rivera RM, Stein P, Weaver JR, Mager J, Schultz RM, et al. (2008) Manipulations of mouse embryos prior to implantation result in aberrant expression of imprinted genes on day 9.5 of development. Hum Mol Genet 17: 1–14. [DOI] [PubMed] [Google Scholar]
- 15. Mann MR, Lee SS, Doherty AS, Verona RI, Nolen LD, et al. (2004) Selective loss of imprinting in the placenta following preimplantation development in culture. Development 131: 3727–3735. [DOI] [PubMed] [Google Scholar]
- 16. Fortier AL, Lopes FL, Darricarrere N, Martel J, Trasler JM (2008) Superovulation alters the expression of imprinted genes in the midgestation mouse placenta. Hum Mol Genet 17: 1653–1665. [DOI] [PubMed] [Google Scholar]
- 17. Gallou-Kabani C, Gabory A, Tost J, Karimi M, Mayeur S, et al. (2010) Sex- and diet-specific changes of imprinted gene expression and DNA methylation in mouse placenta under a high-fat diet. PLoS ONE 5: e14398 doi:10.1371/journal.pone.0014398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. de Mouzon J, Goossens V, Bhattacharya S, Castilla JA, Ferraretti AP, et al. (2010) Assisted reproductive technology in Europe, 2006: results generated from European registers by ESHRE. Hum Reprod 25: 1851–1862. [DOI] [PubMed] [Google Scholar]
- 19. vom Saal FS, Hughes C (2005) An extensive new literature concerning low-dose effects of bisphenol A shows the need for a new risk assessment. Environ Health Perspect 113: 926–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vandenberg LN, Chahoud I, Heindel JJ, Padmanabhan V, Paumgartten FJ, et al. (2012) Urinary, circulating, and tissue biomonitoring studies indicate widespread exposure to bisphenol a. Cien Saude Colet 17: 407–434. [DOI] [PubMed] [Google Scholar]
- 21. Welshons WV, Nagel SC, vom Saal FS (2006) Large effects from small exposures. III. Endocrine mechanisms mediating effects of bisphenol A at levels of human exposure. Endocrinology 147: S56–69. [DOI] [PubMed] [Google Scholar]
- 22. Ho SM, Tang WY, Belmonte de Frausto J, Prins GS (2006) Developmental exposure to estradiol and bisphenol A increases susceptibility to prostate carcinogenesis and epigenetically regulates phosphodiesterase type 4 variant 4. Cancer Res 66: 5624–5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yaoi T, Itoh K, Nakamura K, Ogi H, Fujiwara Y, et al. (2008) Genome-wide analysis of epigenomic alterations in fetal mouse forebrain after exposure to low doses of bisphenol A. Biochem Biophys Res Commun 376: 563–567. [DOI] [PubMed] [Google Scholar]
- 24. Bromer JG, Zhou Y, Taylor MB, Doherty L, Taylor HS (2010) Bisphenol-A exposure in utero leads to epigenetic alterations in the developmental programming of uterine estrogen response. Faseb J 24: 2273–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bartolomei MS (2009) Genomic imprinting: employing and avoiding epigenetic processes. Genes Dev 23: 2124–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kelsey G (2007) Genomic imprinting–roles and regulation in development. Endocr Dev 12: 99–112. [DOI] [PubMed] [Google Scholar]
- 27. Ideraabdullah FY, Vigneau S, Bartolomei MS (2008) Genomic imprinting mechanisms in mammals. Mutat Res 647: 77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Okae H, Hiura H, Nishida Y, Funayama R, Tanaka S, et al. (2012) Re-investigation and RNA sequencing-based identification of genes with placenta-specific imprinted expression. Hum Mol Genet 21: 548–558. [DOI] [PubMed] [Google Scholar]
- 29. Smallwood SA, Tomizawa S, Krueger F, Ruf N, Carli N, et al. (2011) Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat Genet 43: 811–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Verona RI, Mann MR, Bartolomei MS (2003) Genomic imprinting: intricacies of epigenetic regulation in clusters. Annu Rev Cell Dev Biol 19: 237–259. [DOI] [PubMed] [Google Scholar]
- 31. Lucifero D, Mertineit C, Clarke HJ, Bestor TH, Trasler JM (2002) Methylation dynamics of imprinted genes in mouse germ cells. Genomics 79: 530–538. [DOI] [PubMed] [Google Scholar]
- 32. Constancia M, Dean W, Lopes S, Moore T, Kelsey G, et al. (2000) Deletion of a silencer element in Igf2 results in loss of imprinting independent of H19 . Nat Genet 26: 203–206. [DOI] [PubMed] [Google Scholar]
- 33. Karimi M, Luttropp K, Ekstrom TJ (2011) Global DNA methylation analysis using the luminometric methylation assay. Methods Mol Biol 791: 135–144. [DOI] [PubMed] [Google Scholar]
- 34. Popp C, Dean W, Feng S, Cokus SJ, Andrews S, et al. (2010) Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature 463: 1101–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bartolomei MS, Ferguson-Smith AC (2011) Mammalian genomic imprinting. Cold Spring Harb Perspect Biol 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rossant J, Cross JC (2001) Placental development: lessons from mouse mutants. Nat Rev Genet 2: 538–548. [DOI] [PubMed] [Google Scholar]
- 37. Kang ER, Iqbal K, Tran DA, Rivas GE, Singh P, et al. (2011) Effects of endocrine disruptors on imprinted gene expression in the mouse embryo. Epigenetics 6: 937–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, et al. (2008) Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature 452: 877–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Amor DJ, Halliday J (2008) A review of known imprinting syndromes and their association with assisted reproduction technologies. Hum Reprod 23: 2826–2834. [DOI] [PubMed] [Google Scholar]
- 40. Wu MY, Chen KS, Bressler J, Hou A, Tsai TF, et al. (2006) Mouse imprinting defect mutations that model Angelman syndrome. Genesis 44: 12–22. [DOI] [PubMed] [Google Scholar]
- 41. Jirtle RL, Skinner MK (2007) Environmental epigenomics and disease susceptibility. Nat Rev Genet 8: 253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schonfelder G, Wittfoht W, Hopp H, Talsness CE, Paul M, et al. (2002) Parent bisphenol A accumulation in the human maternal-fetal-placental unit. Environ Health Perspect 110: A703–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lewis A, Mitsuya K, Umlauf D, Smith P, Dean W, et al. (2004) Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat Genet 36: 1291–1295. [DOI] [PubMed] [Google Scholar]
- 44. Umlauf D, Goto Y, Cao R, Cerqueira F, Wagschal A, et al. (2004) Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat Genet 36: 1296–1300. [DOI] [PubMed] [Google Scholar]
- 45. Wingfield JC, Mukai M (2009) Endocrine disruption in the context of life cycles: perception and transduction of environmental cues. Gen Comp Endocrinol 163: 92–96. [DOI] [PubMed] [Google Scholar]
- 46. Murphy SK, Adigun A, Huang Z, Overcash F, Wang F, et al. (2012) Gender-specific methylation differences in relation to prenatal exposure to cigarette smoke. Gene 494: 36–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, et al. (2008) Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A 105: 17046–17049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Steegers-Theunissen RP, Obermann-Borst SA, Kremer D, Lindemans J, Siebel C, et al. (2009) Periconceptional maternal folic acid use of 400 microg per day is related to increased methylation of the IGF2 gene in the very young child. PLoS ONE 4: e7845 doi:10.1371/journal.pone.0007845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Weksberg R, Smith AC, Squire J, Sadowski P (2003) Beckwith-Wiedemann syndrome demonstrates a role for epigenetic control of normal development. Hum Mol Genet 12 Spec No 1: R61–68. [DOI] [PubMed] [Google Scholar]
- 50. DeChiara TM, Efstratiadis A, Robertson EJ (1990) A growth-deficiency phenotype in heterozygous mice carrying an insulin-like growth factor II gene disrupted by targeting. Nature 345: 78–80. [DOI] [PubMed] [Google Scholar]
- 51. Leighton PA, Saam JR, Ingram RS, Stewart CL, Tilghman SM (1995) An enhancer deletion affects both H19 and Igf2 expression. Genes Dev 9: 2079–2089. [DOI] [PubMed] [Google Scholar]
- 52. Weaver JR, Sarkisian G, Krapp C, Mager J, Mann MR, et al. (2010) Domain-specific response of imprinted genes to reduced DNMT1. Mol Cell Biol 30: 3916–3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sieli PT, Jasarevic E, Warzak DA, Mao J, Ellersieck MR, et al. (2011) Comparison of serum bisphenol A concentrations in mice exposed to bisphenol A through the diet versus oral bolus exposure. Environ Health Perspect 119: 1260–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tachibana T, Wakimoto Y, Nakamuta N, Phichitraslip T, Wakitani S, et al. (2007) Effects of bisphenol A (BPA) on placentation and survival of the neonates in mice. J Reprod Dev 53: 509–514. [DOI] [PubMed] [Google Scholar]
- 55. Tanaka S, Oda M, Toyoshima Y, Wakayama T, Tanaka M, et al. (2001) Placentomegaly in cloned mouse concepti caused by expansion of the spongiotrophoblast layer. Biol Reprod 65: 1813–1821. [DOI] [PubMed] [Google Scholar]
- 56. Georgiades P, Watkins M, Surani MA, Ferguson-Smith AC (2000) Parental origin-specific developmental defects in mice with uniparental disomy for chromosome 12. Development 127: 4719–4728. [DOI] [PubMed] [Google Scholar]
- 57. Tunster SJ, Van de Pette M, John RM (2011) Fetal overgrowth in the Cdkn1c mouse model of Beckwith-Wiedemann syndrome. Dis Model Mech 4: 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sherer DM, Abulafia O (2001) Angiogenesis during implantation, and placental and early embryonic development. Placenta 22: 1–13. [DOI] [PubMed] [Google Scholar]
- 59. Tuuli MG, Longtine MS, Nelson DM (2011) Review: Oxygen and trophoblast biology—a source of controversy. Placenta 32 Suppl 2: S109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rodriguez BA, Weng YI, Liu TM, Zuo T, Hsu PY, et al. (2011) Estrogen-mediated epigenetic repression of the imprinted gene cyclin-dependent kinase inhibitor 1C in breast cancer cells. Carcinogenesis 32: 812–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tang WY, Morey LM, Cheung YY, Birch L, Prins GS, et al. (2012) Neonatal Exposure to Estradiol/Bisphenol A Alters Promoter Methylation and Expression of Nsbp1 and Hpcal1 Genes and Transcriptional Programs of Dnmt3a/b and Mbd2/4 in the RatProstate Gland Throughout Life. Endocrinology 153: 42–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Doshi T, Mehta SS, Dighe V, Balasinor N, Vanage G (2011) Hypermethylation of estrogen receptor promoter region in adult testis of rats exposed neonatally to bisphenol A. Toxicology 289: 74–82. [DOI] [PubMed] [Google Scholar]
- 63. Ivanova E, Kelsey G (2011) Imprinted genes and hypothalamic function. J Mol Endocrinol 47: R67–74. [DOI] [PubMed] [Google Scholar]
- 64. Wilkins JF, Ubeda F (2011) Diseases associated with genomic imprinting. Prog Mol Biol Transl Sci 101: 401–445. [DOI] [PubMed] [Google Scholar]
- 65. Braun JM, Kalkbrenner AE, Calafat AM, Yolton K, Ye X, et al. (2011) Impact of early-life bisphenol A exposure on behavior and executive function in children. Pediatrics 128: 873–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sathyanarayana S, Braun JM, Yolton K, Liddy S, Lanphear BP (2011) Case report: high prenatal bisphenol a exposure and infant neonatal neurobehavior. Environ Health Perspect 119: 1170–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Braun JM, Yolton K, Dietrich KN, Hornung R, Ye X, et al. (2009) Prenatal bisphenol A exposure and early childhood behavior. Environ Health Perspect 117: 1945–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Vom Saal FS, Nagel SC, Coe BL, Angle BM, Taylor JA (2012) The estrogenic endocrine disrupting chemical bisphenol A (BPA) and obesity. Mol Cell Endocrinol 354: 74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rubin BS, Soto AM (2009) Bisphenol A: Perinatal exposure and body weight. Mol Cell Endocrinol 304: 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cantonwine D, Meeker JD, Hu H, Sanchez BN, Lamadrid-Figueroa H, et al. (2010) Bisphenol a exposure in Mexico City and risk of prematurity: a pilot nested case control study. Environ Health 9: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chou WC, Chen JL, Lin CF, Chen YC, Shih FC, et al. (2011) Biomonitoring of bisphenol A concentrations in maternal and umbilical cord blood in regard to birth outcomes and adipokine expression: a birth cohort study in Taiwan. Environ Health 10: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mann MRW, Chung YG, Nolen LD, Verona RI, Latham KE, et al. (2003) Disruption of imprinted gene methylation and expression in cloned preimplantation stage mouse embryos. Biol Reprod 69: 902–914. [DOI] [PubMed] [Google Scholar]
- 73. Davis TL, Trasler JM, Moss SB, Yang GJ, Bartolomei MS (1999) Acquisition of the H19 methylation imprint occurs differentially on the parental alleles during spermatogenesis. Genomics 58: 18–28. [DOI] [PubMed] [Google Scholar]
- 74. Tremblay KD, Duran KL, Bartolomei MS (1997) A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol Cell Biol 17: 4322–4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Singh G, Gutierrez A, Xu K, Blair IA (2000) Liquid chromatography/electron capture atmospheric pressure chemical ionization/mass spectrometry: analysis of pentafluorobenzyl derivatives of biomolecules and drugs in the attomole range. Anal Chem 72: 3007–3013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Total expression of imprinted genes analyzed by a non-parametric test. Kruskal-Wallis statistical test was conducted to measure differences among exposure groups on expression of the (A) Snrpn, (C) Kcnq1ot1, (D) Cdkn1c and (E) Ube3a genes in the E9.5 placenta, (B) Igf2 gene in the E9.5 embryo and (F) Ube3a gene in the E12.5 brain. Y axis = relative expression as compared to reference genes; X axis = measurement values of relative expression ranked from lowest (left) to highest (right). P values shown represent all exposure groups analyzed. P values between control and upper dose are as followed: (A) 0.006, (B) 0.05, (C) 0.04, (D) 0.002, and (E) 0.003. P values between control and lower dose are 0.003 in (C) and 0.001 in (D).
(TIF)
Imprinted genes-specific and genome-wide methylation levels analyzed by a non-parametric test. Results from the Kruskal-Wallis test are presented for methylation levels of the (A) H19/Igf2 ICR and (B) Igf2 DMR1 in the E9.5 embryos and (C) genome-wide methylation levels as measured by LUMA in the E9.5 placentas. Y = percentage of methylation level; X axis = measurement values of methylation ranked from lowest (left) to highest (right). P values shown represent all exposure groups analyzed. P values between control and upper dose are as followed: (A) 0.0006, (B) 0.04 and (C) 0.02.
(TIF)
Percentage of total expression of imprinted genes derived from the repressed allele in each exposure group in the E9.5 embryo and placenta.
(XLS)
Percentage of total expression of imprinted genes derived from the repressed allele in each exposure group in the E12.5 embryo and placenta.
(XLS)