Abstract
The small IQ motif proteins PEP-19 (62 amino acids) and RC3 (78 amino acids) greatly accelerate the rates of Ca2+ binding to sites III and IV in the C-domain of calmodulin (CaM). We show here that PEP-19 decreases the degree of cooperativity of Ca2+ binding to sites III and IV, and we present a model showing that this could increase Ca2+ binding rate constants. Comparative sequence analysis showed that residues 28 to 58 from PEP-19 are conserved in other proteins. This region includes the IQ motif (amino acids 39–62), and an adjacent acidic cluster of amino acids (amino acids 28–40). A synthetic peptide spanning residues 28–62 faithfully mimics intact PEP-19 with respect to increasing the rates of Ca2+ association and dissociation, as well as binding preferentially to the C-domain of CaM. In contrast, a peptide encoding only the core IQ motif does not modulate Ca2+ binding, and binds to multiple sites on CaM. A peptide that includes only the acidic region does not bind to CaM. These results show that PEP-19 has a novel acidic/IQ CaM regulatory motif in which the IQ sequence provides a targeting function that allows binding of PEP-19 to CaM, whereas the acidic residues modify the nature of this interaction, and are essential for modulating Ca2+ binding to the C-domain of CaM.
Calmodulin (CaM)2 is a 17-kDa Ca2+ receptor found in all eukaryotic cells, where it regulates activities ranging from neural transmission to growth and differentiation (for a condensed review see Ref. 1). This daunting task requires that CaM interact with a large number of proteins and that it properly sense Ca2+ signals that vary greatly in frequency and amplitude (for review see Refs. 2 and 3). To effectively fulfill its diverse roles, mechanisms have evolved to regulate, or fine-tune CaM activity over short time frames. For example, phosphorylation of a small protein called ARPP-21 promotes high affinity binding to Ca2+-CaM, (neurogranin or Ng) thereby competitively inhibiting activation of other CaM-dependent enzymes (4).
PEP-19 (Purkinje cell protein 4 or pcp4) and RC3 (neurogranin or Ng) are small proteins expressed primarily in neuronal tissues, but with no known activity other than binding to CaM in the presence or absence of Ca2+. We found that both PEP-19 and RC3 have profound effects on the rate-limiting kinetics of Ca2+ binding to the C-domain of CaM. Specifically, PEP-19 accelerates the rates of both association and dissociation of Ca2+ without greatly affecting the overall KCa of the C-domain (5). RC3 accelerates the rate of Ca2+ dissociation from CaM, but has a lesser effect on the association rate, thereby decreasing the affinity of binding Ca2+ to the C-domain of CaM (6). Importantly, both PEP-19 and RC3 exert these effects even when CaM is bound to CaM-dependent protein kinase II (CKIIα) (5, 6).
These results suggest that PEP-19 and RC3 could have broad extrinsic effects on CaM-related signaling pathways by modulating the Ca2+ binding properties of free or enzyme-bound CaM. This is consistent with the phenotype of RC3 knock-out mice, which show defects in synaptic plasticity (7), attenuated phosphorylation of hippocampal protein kinase A and C substrates (8), and altered Ca2+ dynamics in cortical neurons (9).
Both PEP-19 and RC3 contain an IQ motif. This rather loose consensus sequence (IQXXXRGXXXR) was first identified as the light chain binding site in conventional myosins, but was subsequently recognized as a CaM binding sequence in numerous other proteins (10). IQ motif proteins exhibit diverse modes of interaction with CaM that include Ca2+-dependent or independent binding (10), binding to both or only one domain of CaM (5, 11–13), binding multiple CaMs to multiple IQ motifs (14), and exchange of CaM between the IQ motif and other sites in the same protein (15, 16).
These intriguing structure-function relationships of IQ motifs led us to identify amino acids in PEP-19 that are required to modulate Ca2+ binding to CaM. We show here that the consensus IQ CaM binding motif is necessary, but not sufficient to mimic the effect of intact PEP-19 on CaM. An adjacent highly acidic amino acid sequence acts in synergy with the IQ motif to modulate Ca2+ binding to the C-domain of CaM. We propose that this acidic/IQ sequence constitutes a new CaM regulatory motif.
EXPERIMENTAL PROCEDURES
Recombinant Proteins and Peptides
Recombinant CaM, CaM(K75C), CaM(T110C), CaM(T34C), CaM(T34C, T110C), PEP19, and RC3 were cloned, expressed, and purified as described previously (5, 6, 16–18). The expression plasmid for the C-domain of CaM (residues 78–148) was a generous gift from Dr. Madeline Shea (University of Iowa). Synthetic peptides purchased from Sigma Genosys had greater than 90% purity, and were further purified as necessary by C4 reverse phase HPLC using 0–60% acetonitrile gradient in water, 0.1% trifluoroacetic acid.
NMR Methodology
NMR spectra of isotope-labeled CaM and PEP-19 were generated using Varian Inova 800 MHz and Bruker DRX800 MHz spectrometers with room temperature triple resonance probes, as well as a Bruker DRX 600 MHz spectrometer equipped with 5-mm TXI CryoProbe. Backbone assignments for Ca2+-CaM in the absence and presence of PEP-19 were reported previously (5). Titration of [15N]Ca2+-CaM with PEP-19 or peptides was performed in a buffer containing 10 mM imidazole, pH 6.3, 100 mM KCl, 5 mM CaCl2, 5% D2O at 310 or 320 K. Amide chemical shifts in the 15N,1H-heteronuclear single quantrum coherence spectra for [15N]Ca2+-CaM exhibited fast exchange characteristics during titration with PEP-19, PEP(28–62), or PEP(39–62), which allowed assignments to be made by following chemical shift changes from the free to bound forms of CaM. All titrations were conducted at a CaM concentration of 50 μM. To eliminate dilution and pH effects, titrations were made from a stock solution of concentrated unlabeled PEP-19 or peptides containing 50 μM Ca2+-CaM in the appropriate buffer. At each titration point a measured amount of sample was removed from the NMR tube and replaced with the identical volume from the stock solution. Titrations were carried out until a 10-fold excess of ligand was added. The data were processed using the Auto-screen module in FELIX 2002 software (19). The average amide chemical shift change was calculated using the following formula:
| (Eq. 1) |
where ΔδH = change in 1H chemical shift and Δδn = change in 15N chemical shift.
Chemical shift changes for CaM backbone amide 1H and 15N nuclei were analyzed separately to derive Kd values for binding PEP-19 to CaM because the relative contribution of change in each dimension can vary significantly. In general, backbone 15N chemical shifts experienced the greatest change relative to the spectral window.
Generation of Fluorescently Labeled Proteins
Labeling of CaM(K75C) or CaM(T110C) with either acylodan or IAEDANS was previously reported (18, 20). To obtain the fluorescent CaM used in the FRET study, a double Cys CaM mutant, CaM(T34C, T110C), was labeled with IAEDANS (donor) and DDPM (accepter) to generate CaMD/A. CaM(T34C, T110C) was first reacted with 0.4 mol of IAEDANS/mol of protein in 20 mM Tris-HCl at pH 7.5 and 100 mM KCl for 2 h at 20 °C in the dark. Free IAEDANS was removed using a Bio-Gel P-6DG (Bio-Rad) desalting column. The IAEDANS-labeled CaM averaged 0.3 mol of IAEDANS/mol of protein. A portion of this partially labeled protein was saved as the donor-alone protein (CaMD), whereas the rest was labeled with excess DDPM to give CaMD/A. Free DDPM was removed by desalting. PEP-19 and PEP (39–62) with C-terminal Gly-Cys extensions were labeled with DDPM as described for CaM, but free DDPM was removed using a semi-prep C4 reverse phase HPLC column with a 0–60% acetonitrile gradient. Protein and peptide concentrations were determined using the Pierce BCA protein assay with a bovine serum albumin standard and color developed at 60 °C.
Equilibrium Binding of PEP-19 to CaM
Solutions of fluorescently labeled CaM derivatives were prepared in a buffer of 20 mM MOPS, pH 7.5, 100 mM KCl, and 1 mM dithiothreitol. Concentrated stock solutions of PEP-19, PEP(39–62), or their DDPM-labeled derivatives, were prepared by dissolving lyophilized protein or peptide in the labeled CaM solution to eliminate dilution of CaM during titration. The increase in volume was less than 10%. We assessed potential nonspecific FRET effects using DDPM coupled to free Cys, and found a linear, 5% decrease in fluorescence from donor-labeled CaM per increment of 25 μM Cys-DDPM. The FRET effect between donor-labeled CaM and acceptor-labeled PEP-19 or PEP(39–62) was corrected for this nonspecific effect, and the upper concentration of DDPM-labeled ligands was limited to 50 μM.
Dissociation constants (Kd) were derived from fluorescence or NMR data by fitting titration curves to the following equation, which does not require measurement of free ligand concentrations,
| (Eq. 2) |
Where S = fluorescence or NMR signal at a given titration point; Si = initial signal in the absence of ligand; Sf = final signal in the presence of excess ligand; L = total ligand added at a given titration point; Ct = total CaM concentration; and Kd = dissociation constant. The equation was used to fit plots of S versus L with Si, Sf, and Kd as fitted variables.
Equilibrium Ca2+ Binding
Macroscopic equilibrium Ca2+ binding constants were determined using the competitive binding assay described by Linse et al. (21). Calcium was removed from buffers by passage over a Calcium-Sponge column (Molecular Probes). Residual Ca2+ detected using Indo-1 was typically <10−7 M. CaM was decalcified by adding 1–5 mM BAPTA followed by desalting into Ca2+-free buffers. This effectively removed greater than 95% of Ca2+ from CaM as determined by Tyr fluorescence. All pipette tips, cuvettes, and other labware were rinsed with 0.1 M HCl and MilliQ water to remove Ca2+.
Samples used for equilibrium binding studies contained 30 μM CaM, 30 μM 5,5′-dibromo-BAPTA (Br2BAPTA; Molecular Probes/Invitrogen; Kd = 1.59 μM), with or without 60 μM PEP-19 polypeptides, in a buffer of 20 mM MOPS, pH 7.5, 100 mM KCl. Calcium was added from a stock solution prepared in a buffer that contained both CaM and Br2BAPTA such that only the Ca2+ concentration varied during the titration. Titrations were performed by addition of 2-, 3-, 5-, or 10-μl aliquots of the Ca2+ stock, to an initial sample volume of 0.7 ml. The decrease in absorbance of Br2BAPTA at 263 nm was monitored using a Cary/Varian 100 spectrophotometer. The number and volume of aliquots was adjusted to achieve an even distribution of data points on the binding isotherm. The total Ca2+ concentration was then calculated based on the initial volume and total added volume at each titration point. Macroscopic calcium binding constants were calculated essentially as described by Linse et al. (21) using the following equations,
| (Eq. 3) |
| (Eq. 4) |
Where [Ca]T = total Ca2+; [Ca]F = concentration of free Ca2+; [Pro] = concentration of CaM; [BAP] = concentration of Br2BAPTA; Kd(BAP) = Ca2+ dissociation constant for Br2BAPTA; K = macromolecular binding constants for N number of sites (K1 through K4); and AbsMAX, AbsMIN, and Abs are the absorbance of Br2BAPTA in the absence, at saturating, and at intermediate concentrations of Ca2+, respectively. Because the stoichiometry of Ca2+ binding is very sensitive to small errors in protein concentration, we incorporated factor F to compensate for slight errors in CaM protein concentrations as done by Linse et al. (21). Plots of [Ca]T versus Abs were fit directly to Equation 4 by least squares using Kaleidagraph software. The fitted variables were the macroscopic binding constants, F, AbsMAX, and AbsMIN. Values for F were typically greater than 0.8. The Ca2+ binding constant for Br2BAPTA of 1.59 × 10−6 was experimentally determined at the same pH and ionic strength used in the binding assay.
Stopped-flow Measurements
Stopped-flow fluorescence experiments were performed at 23 °C using an Applied Photo-physics Ltd. (Leatherhead, UK) Model SX.18 MV sequential stopped-flow spectrofluorimeter with a 150 watt Xe/Hg lamp, and a dead time of 1.7 ms. All solutions contained a base buffer of 20 mM MOPS, pH 7.5, 100 mM KCl. The concentration of other reagents in stopped-flow mixing solutions A and B are defined in the figure legends. The final concentration of these reagents in the optical chamber was one-half of these values, because the mixing ratio was 1:1.
Calcium koff rates were determined using 2 μM CaM, 20 μM Ca2+, and 300 μM Quin-2. Fluorescence from Quin-2 was detected using an excitation wavelength of 334.5 nm and Oriel emission cut-off filter 51282. A stopped-flow experiment to measure Ca2+ kon rates was devised using the Ca2+-sensitive chromophore Br2BAPTA as a buffer to maintain free Ca2+levels in the range of 0.5 to 5μM, and as a chromophore to monitor Ca2+binding. The kinetics of binding Ca2+to Br2BAPTA and the N-domain of CaM are very fast and thus, the increase in absorbance observed upon mixing apo-CaM with Ca2+/Br2BAPTA solutions is due to the release of Ca2+from Ca2+/Br2BAPTA as Ca2+binds to the C-domain of CaM. The observed change in absorbance was fitted to a single exponential equation. All buffers contained 20 mM MOPS, pH 7.5, and 100 mM KCl. The assay was performed at 20 °C. Typically, Buffer A contained 5 μM BAPTA with or without 2 to 4 μM decalcified CaM or EGTA, whereas Buffer B contained 250 μM Br2BAPTA and sufficient Ca2+ to achieve a desired free Ca2+ upon mixing equal volumes of Buffers A and B. This allows less than a 3% change in the concentrations of Br2BAPTA and Ca2+-Br2BAPTA as Ca2+ binds to CaM, thereby maintaining a reasonably constant level of free Ca2+.
Free Ca2+ levels of Ca2+ /Br2BAPTA solutions were calculated from the absorbance at 263 nm (Absobs) versus controls in the absence (Absmin) or presence (Absmax) of excess Ca2+ using the following equation.
| (Eq. 5) |
Free Ca2+ levels in the optical chamber of the stopped-flow instrument at 20 °C and pH 7.5 were determined from the observed rates of binding Ca2+ to EGTA present in solution A (Kd = 0.038 μM, koff = 0.53 s−1, kon = 13.8 μM−1 s−1). Free Ca2+ levels were in close agreement when calculated based on Br2BAPTA absorbance or observed rates of binding Ca2+ to EGTA.
Microscopic Kinetic Model for Ca2+ Binding to the C-domain of CaM
Linked differential equations for the forward and reverse microscopic binding events illustrated in Fig. 6A were incorporated into a computational model using Berkeley Madonna software. Initial parameter values were taken from the current study and literature reports. The overall average apparent dissociation constant (Kd or KCa50) of 2.3 ± 0.3 μM was derived from Table 2 and other reports using a variety of techniques at pH 7.4 to 7.5 with 90 to 100 mM KCl (5, 21–30). The macroscopic dissociation constants, Kd1 and Kd2, correspond to sequential binding of the first and second Ca2+ ions, regardless of which site is filled first. Starting values for Kd1 and Kd2 given in Table 2 are very close to those reported with others (21, 22). The microscopic dissociation constants KdIII and KdIV correspond to binding the first Ca2+ to site III or IV, respectively, whereas KdIII/IV and KdIV/III correspond to binding the second Ca2+ when the other site is already occupied. The model includes algebraic relationships that relate macroscopic and microscopic association constants (Ka; Ka = 1/Kd) as follows, where c is the coupling factor (31).
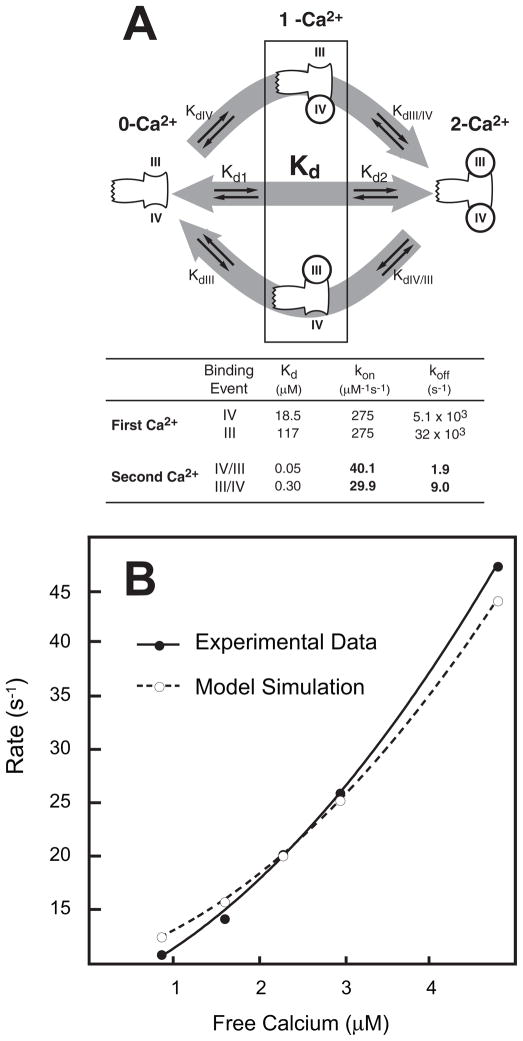
FIGURE 6. Cooperative Ca2+ binding to the C-domain of CaM.
Panel A illustrates the microscopic equilibrium Ca2+ binding reactions associated with transition of the C-domain of CaM from the 0-Ca2+ to 2-Ca2+ states. The table in panel A lists optimized equilibrium and kinetic microscopic Ca2+ binding constants derived as described under “Experimental Procedures.” Panel B shows the relationship between observed Ca2+ association rates at free Ca2+ in the range of 1 to 5 μM. The closed circles are experimental data derived as described in the legend to Fig. 5E and under “Experimental Procedures.” The open circles are derived from a simulation carried out using the model described under “Experimental Procedures” and the binding parameters shown in panel A.
TABLE 2.
Effect of PEP-19, PEP(28–62), and PEP(39–62) on calcium binding affinity and cooperativity
The macroscopic dissociation constants were derived as described in Material and Methods. All values are the average mean ± S.E. of (n) independent titrations. The square root of the product of macroscopic dissociation constants for each domain is equivalent to the KCa50 for binding to each domain. The precision of this value is greater than the individual dissociation constants. The overall decrease in free energy from binding two Ca2+ ions to the N- or C-domains (ΔGtot) was calculated as ΔGtot = −RT ln(K1×K2). The upper limit for the change in free energy due to cooperative Ca2+ binding (ΔΔGc) was calculated as ΔΔGc = −RT ln(4 × K2/K1). Free energy values are in kcal/mole.
| N | C-Domain
|
N-Domain
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Macroscopic Kd (μM)
|
ΔGtot | ΔΔGc | Macroscopic Kd (μM)
|
ΔGtot | ΔΔGc | ||||||
| Kd1 | Kd2 |
|
Kd3 | Kd4 |
|
||||||
| CaM | (15) | 17 ± 3 | 0.4 ± 0.1 | 2.2 ± 0.2 | −15.3 ± 0.3 | −3.4 ± 0.3 | 34 ± 4 | 6.2 ± 1 | 13.1 ± 1.0 | −13.3 ± 0.1 | −1.9 ± 0.2 |
| CaM + PEP-19 | (6) | 4.7 ± 1 | 2.0 ± 0.2 | 2.8 ± 0.2 | −15.0 ± 0.1 | −1.3 ± 0.2 | 35 ± 6 | 3.9 ± 0.6 | 14.0 ± 1.0 | −13.2 ± 0.1 | −2.3 ± 0.2 |
| CaM + PEP(28–62) | (3) | 5.0 ± 1.3 | 1.7 ± 0.3 | 2.8 ± 0.1 | −15.0 ± 0.1 | −1.4 ± 0.3 | 57 ± 25 | 4.1 ± 1.4 | 13.1 ± 0.5 | −13.2 ± 0.1 | −2.3 ± 0.5 |
| CaM + PEP(39–62) | (3) | 1.4 ± 0.3 | 0.4 ± 0.1 | 0.7 ± 0.1 | −16.6 ± 0.1 | −1.6 ± 0.3 | 36 ± 1 | 2.6 ± 1.0 | 8.6 ± 0.7 | −13.7 ± 0.1 | −2.4 ± 0.4 |
These relationships allow for the calculation of microscopic equilibrium binding constants if K1, K2, and the magnitude of difference between microscopic binding constants is known. K1 and K2 are reported in Table 2, and Evenas et al. (32) who used Ca2+ binding mutants to show that the relative Ca2+ binding affinity of site IV is ~6.3-fold greater than site III in both the 0-Ca2+ and 1-Ca2+ states of the C-domain.
Microscopic rate constants were constrained by the results of Malmendal et al. (32) who used NMR relaxation methods to conclude that the first Ca2+ ion binds preferentially to site IV, and that the koff of site IV (koffIV) was 5100 s−1. If the 6.3-fold difference in affinity of binding Ca2+ to sites IV and III were due exclusively to koff rates, then koffIII would be = 32,000 s−1, which is consistent with an exchange rate of 27,000 s−1 determined for transition between the open and closed conformation of the C-domain in which site IV was mutated (33). These koff values would correspond to konIV and konIII rates of around 300 μM−1 s−1, which is consistent with a diffusion-limited event, and similar to the kon for Ca2+ binding to the N-domain of CaM. Microscopic rate constants for binding the second Ca2+ ion were constrained by the fact that the observed koff (koff,Obs) for dissociation of both Ca2+ ions from the C-domain measured using stopped-flow experiments described as above and best fits a single exponential rate between 8.5 s−1 and 12.6 s−1 (5, 25, 34, 35). Because NMR relaxation data show that the rate of dissociation of Ca2+ from the 1-Ca2+ state is very fast, then koff,Obs reflects the rate-limiting dissociation of the first Ca2+ from either site III or IV of the 2-Ca2+ state, followed by very rapid release of the second Ca2+. This means that koff,Obs = koff,IV/III + koff,III/IV, and it allows constraint of microscopic rate constants using the following relationships.
Thus, kdIII/IV and kdIV/III are calculated if koff,Obs is defined within an experimentally observed range, and koff,IV/III is varied between 0 and the defined koff,Obs.
The model includes a Ca2+ buffer based on Br2BAPTA with Kd = 1.59 μM, a diffusion limited kon = 500 μM−1 s−1 and koff = 795 s−1. This allowed simulation of stopped-flow experiments described above to measure the rate of association of Ca2+ with the C-domain of CaM, and to use the built-in curve fit function of Berkeley Madonna to optimize parameter sets against experimental data.
Global parameter optimization and error analysis were also performed in the MATLAB computing environment (The MathWorks). Although the computational model (Fig. 1) has 8 kinetic parameter values, the experimental data and algebraic constraints described above and under “Results ” reduced the unknown parameters to kon,III and koff,IV/III. However, to take into account the different reported values of koff,Obs, we also treated koff,III/IV as an additional unknown parameter for the global optimization. We set the maximum values of kon,III, koff,IV/III, and koff,III/IV (500 μM −1 s−1, 50 s−1, and 50 s−1, respectively) and divided the entire parameter space into 200 × 500 × 500 = 5 × 107 grid points. The error (root mean square difference) was calculated for each of these 5 × 107 grid points by comparing the simulated Ca2+ association and dissociation rates with the experimental data. This systematic parameter optimization revealed a single distinct region of the parameter space in which the computational model best fit the data. The estimate of kon,III resulted in a unique set of parameter values of the model, which was again reconfirmed by the lsqnonlin function of the MATLAB Optimization Toolbox.
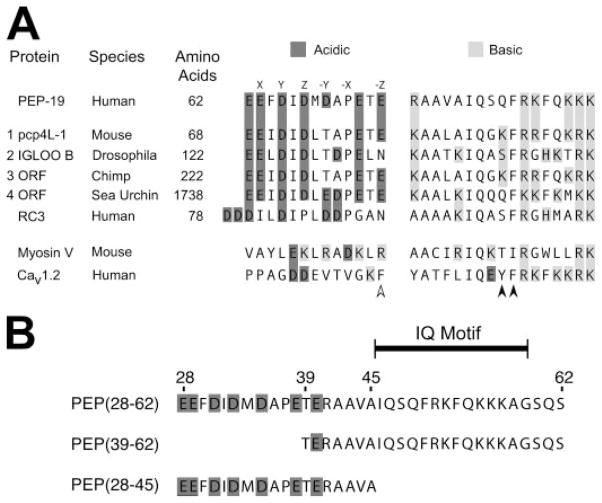
FIGURE 1. Sequence alignment of PEP-19 with residues in other proteins.
A protein BLAST similarity search was performed using the NCBI web server to compare PEP-19 residues 25–62 with all non-redundant sequences in GenBank CDS translations, RefSeq proteins, PDB, SwissProt, PIR, and PRF databases. Panel A shows the sequence similarity between PEP-19 and 4 of 149 BLAST hits. Also shown are the corresponding regions from RC3, myosin V, and voltage-gated Ca2+ channel (Cav1.2). The letters above residues in the PEP-19 sequence are the relative positions of amino acids that coordinate Ca2+ in a consensus EF-hand (see text for details). The accession numbers for BLAST hits 1 to 4 are: AY304481.1, NP_477061, XM_001152960.1, and XM_787004.2. Panel B shows sequences for PEP-19 synthetic peptides used in the current study. The indicated IQ motif is based on a comparison of numerous IQ motif proteins (10), and the minimal region in PEP-19 that has CaM antagonist activity (36).
RESULTS
Identifying Sequences in PEP-19 That Bind to Ca2+-CaM
A BLAST protein similarity search identified PEP-19 orthologs with high degrees of identity throughout their primary sequences. Other proteins of diverse size and from diverse species had sequence similarity to the C-terminal portion of PEP-19 that includes the IQ motif and cluster of adjacent acidic residues (see Fig. 1A). RC3 also has an acidic cluster but with a sequence that differs from PEP-19 (see “Discussion ” for more details). The corresponding sequences from myosin V and the voltage-gated Ca2+ channel Cav1.2 are shown in Fig. 1A to emphasize the absence of acidic clusters in these IQ motif proteins.
The sequence comparisons in Fig. 1A led us to hypothesize that both the IQ motif and the adjacent acidic region in PEP-19 are necessary to modulate the Ca2+ binding of CaM. The peptides shown in Fig. 1B were synthesized to directly test this hypothesis. PEP(28–62) spans the acidic and IQ regions. PEP(39–62) includes the core IQ motif (10), and is the minimal region in PEP-19 shown to have CaM antagonist activity (36). PEP(28–45) encodes the acidic region without the IQ motif.
Intact PEP-19 and Its Consensus IQ Motif Peptide Have Divergent Effects on CaM Amide Chemical Shifts
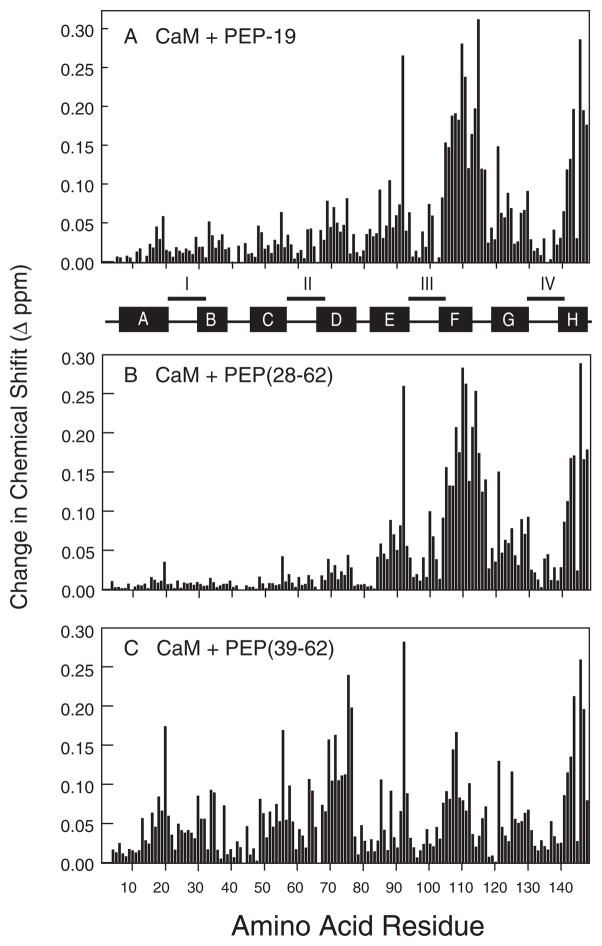
The amide resonances of Ca2+-CaM exhibit fast exchange characteristics on the NMR time scale upon titration with PEP-19, and are maximal at a CaM:PEP-19 ratio of 1:1. Fig. 2A shows that the greatest effects of PEP-19 are localized to the C-domain of Ca2+-CaM, primarily in helix F, the linker between helices F and G, and in helix H. These data indicate a single major binding site for PEP-19 in the C-domain of Ca2+-CaM.
FIGURE 2. Comparative effects of PEP-19 (panel A), PEP(28–62) (panel B), and PEP(39–62) (panel C) on the amide chemical shifts of Ca2+-CaM.
Chemical shift changes were calculated as described under “Experimental Procedures.”
Fig. 2, A and B, summarize the effects of PEP(28–62) and PEP(39–62) on the amide chemical shifts of Ca2+-CaM. Both peptides caused chemical shift changes that were characteristic of fast exchange on the NMR time scale. It is clear that PEP(28–62) induces a pattern of chemical shift changes that is strikingly similar to that of intact PEP-19, with dominant effects on residues in the C-domain of CaM. In contrast, PEP(39–62) has pervasive effects on amide chemical shifts for residues in both the N- and C-domains of CaM, with clusters of perturbation localized to helical segments, especially helices C, D, F, and H.
PEP(28–45) showed only minor effects on the amide chemical shifts of Ca2+-CaM, even when present at a molar excess of 100-fold (5 mM peptide versus 0.05 μM CaM). Thus, both the IQ motif and an adjacent group of acidic residues are necessary to mimic the effect of intact PEP-19 on CaM amide chemical shifts.
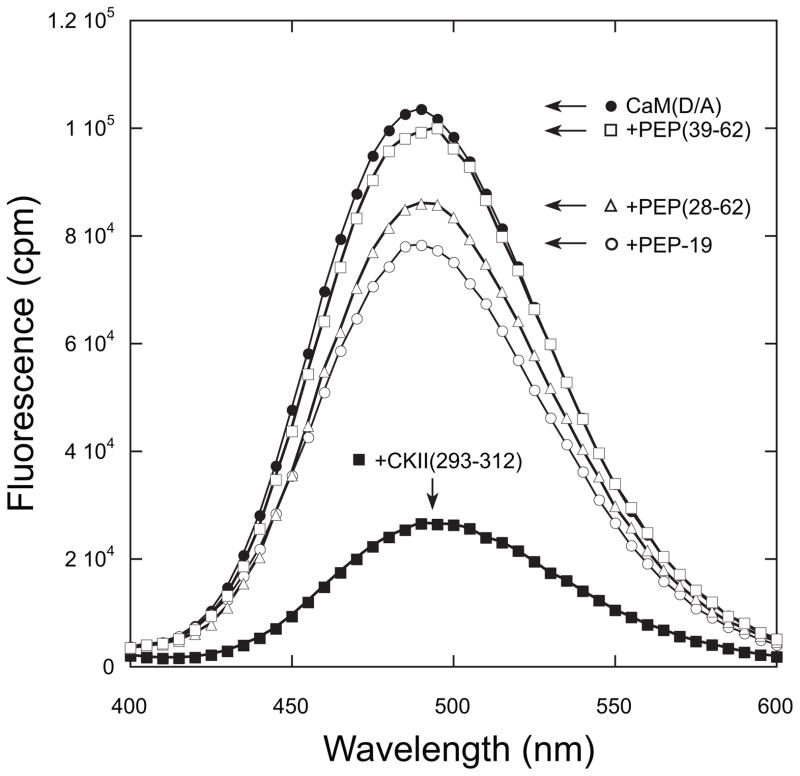
PEP-19 and Its Consensus IQ Motif Have Divergent Effects on the Global Conformation of CaM
CaM(T34C, T110C) with FRET donor and acceptor probes bound to N- and C-domains (CaMD/A) (16, 37) was used to determine the effect of PEP-19 derivatives on the global conformation of CaM. A peptide from CaM kinase II, CKII-(293–312) (20), was used as a positive control because it causes CaM to adopt a compact shape with the N- and C-domains in close proximity (38). Fig. 3 shows that binding CKII-(293–312) to Ca2+-CaMD/A causes a large decrease in fluorescence due to a FRET effect. PEP-19 and PEP(28–62) cause much smaller decreases in fluorescence, indicating they do not induce CaM to adopt a highly compact shape. CaM remains extended when bound to PEP(39–62) because fluorescence from CaMD/A is essentially unaffected by the peptide.
FIGURE 3. Effect of PEP-19 polypeptides on the global conformation of CaM.
The fluorescence emission spectrum of CaMD/A was compared in the absence or presence of PEP-19, PEP(28–62), PEP(39–62), or CKII-(293–312). Spectra were collected in solutions of 10 mM MOPS, pH 7.5, 100 mM KCl, 0.1 mM CaCl2, and 1 μM CaMD/A. All PEP-19 derivatives were present at 60 μM, whereas CKII-(293–312) was present at 4 μM. The excitation wavelength was 335 nm, and spectra were corrected for the effect of the various polypeptides on fluorescence from CaM labeled with the FRET donor alone as described under “Experimental Procedures.”
Relative Affinities of Binding PEP-19 Polypeptides to CaM
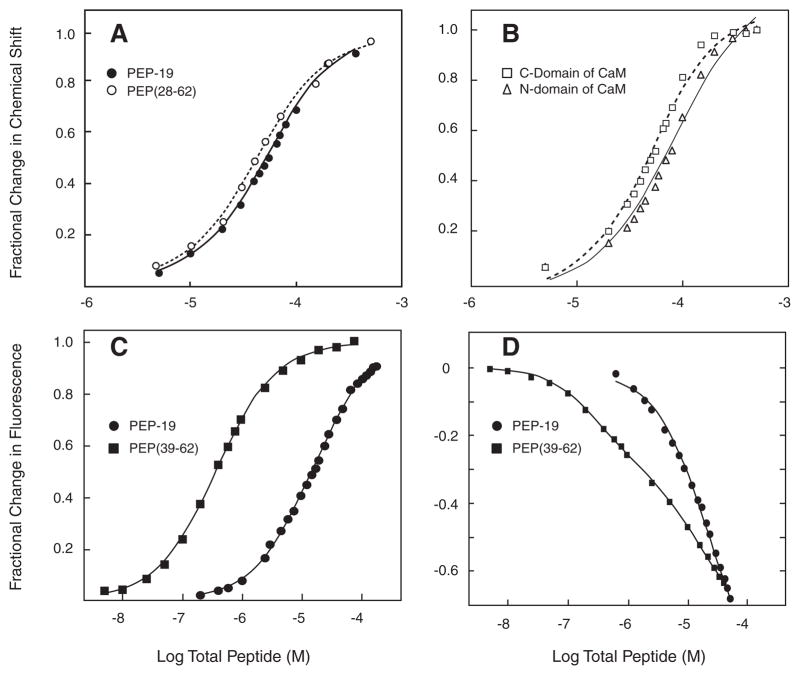
Fig. 4A plots Ca2+-CaM amide chemical shifts as a function of increasing concentrations of PEP-19 or PEP(28–62). Both data sets fit well to a single-site binding model with Kd values of 29 and 24 μM for PEP-19 and PEP(28–62), respectively (see Table 1). Because PEP(39–62) affects amides in the N- and C-domains of Ca2+-CaM, the average responses for selected residues in these domains were analyzed separately as shown in Fig. 4B. Chemical shift changes reached a maximum, but the chemical shift response curve for residues in the N-domain was right shifted relative to residues in the C-domain, suggesting that N-and C-domains sense different binding events. The lines in Fig. 4B show a fit to a single-site binding model, but the fits were poor relative to those shown in Fig. 4A. This was not surprising given the potential complexity of chemical shift changes in response to multiple ligands, and the high concentration of CaM required for NMR (50 μM) is not ideal for analysis of potentially high affinity binding events.
FIGURE 4. Equilibrium binding of PEP-19 polypeptides to CaM.
Panel A shows the fractional change in amide chemical shifts in the C-domain of Ca2+-CaM as a function of increasing concentrations of either PEP-19 (closed circles) or PEP(28–62) (open circles). The data represent the average fractional change for 1H and or 15N nuclei for residues 99, 105, 109, 116, 121, 146, and 147. The line represents the fit to a single-site binding model described under “Experimental Procedures.” Panel B shows the average fractional change in chemical shifts for residues 5, 17, 19, 21 29, 33, 44, 53, 55, 57, 64, 70, and 73 in the N-domain (open squares) and residues 94, 105, 106, 110, 116, 117, 130, 137, 146, 147, and 148 in the C-domain (open triangles) of Ca2+-CaM as a function of increasing concentrations of PEP(39–62). Chemical shift changes in response to PEP(39–62) did not fit well to a single-site binding model. Panel C shows the change in fluorescence from CaMACR upon titration with PEP-19 or PEP(39–62). Panel D shows titration of IAEDANS-labeled CaM(T110C) with DDPM-labeled PEP-19 or PEP(39–62). Labeled CaM for experiments in both panels C and D was present at 0.05 or 0.5 μM for titration with PEP(39–62) or PEP-19, respectively.
TABLE 1.
Dissociation constants (μM) for Binding CaM to PEP-19, PEP(28–62), and PEP(39–62)
| Assay | PEP-19 | PEP(28–62) | PEP(39–62)
|
|
|---|---|---|---|---|
| Kd1 | Kd2 | |||
| NMRa | 29 ± 0.7 | 24 ± 0.8 | —b | — |
| Fluorescencec | 18 ± 3 | NDd | 0.24 ± 0.04 | — |
| FRET | 20 ± 4 | ND | 0.16 ± 0.05 | 27 ± 3 |
Values are the average mean ± S.E. of Kd values derived separately from chemical shift changes for 1H and/or 15N nuclei of amides for residues 99, 105, 109, 116, 117, 146, and 147.
NMR data fit poorly to either single or two-site binding models. Fluorescence data fit best to a single-site binding model.
The values derived from fluorescence and FRET data are the mean ± S.E. of three to four experiments.
ND, not determined.
Fluorescence assays were also used to determine the relative affinities of binding PEP-19 and PEP(39–62) to CaM. Fig. 4C shows the effect of PEP-19 and PEP(39–62) on fluorescence intensity from acrylodan-labeled CaM(K75C), CaMACR. The data for intact PEP-19 fit a single-site model with a Kd of 18 μM (see Table 1). Interestingly, even though Figs. 2C and 3 indicate multiple binding sites for PEP(39–62) on Ca2+-CaM, the response of Ca2+-CaMACR to this peptide fit a single-site model with an apparent Kd of 0.24 μM. Thus, the affinity of binding PEP(39–62) to one site in Ca2+-CaM is about 70-fold greater relative to binding intact PEP-19.
Fig. 4D shows results of a FRET assay using derivatives of PEP-19 and PEP(39–62) with C-terminal Cys residues labeled with the FRET acceptor DDPM, and CaM(T110C) labeled with the FRET donor IAEDANS. A large decrease in fluorescence of up to 70% due to FRET quenching was observed upon binding acceptor-labeled peptides to donor-labeled Ca2+-CaM. Changes in fluorescence upon binding acceptor-labeled PEP-19 to donor-labeled Ca2+-CaM fit a single-site model with a Kd of 20 μM, which is comparable with the values derived from both NMR and CaMACR (see Table 1). Interestingly, changes in fluorescence upon binding PEP(39–62) indicate at least two classes of binding sites. A Kd of 0.16 μM for the higher affinity site is comparable with that derived using CaMACR, whereas the Kd of 27 μM for the low affinity site is consistent with the Kd for binding intact PEP-19 to Ca2+-CaM (see Table 1).
Together, the data in Figs. 2–4 show that PEP-19 and PEP(28–62) bind predominately to a single site in the C-domain of CaM. In contrast, there are at least two binding sites for PEP(39–62) on CaM, located in the N- and C-domains.
Comparative Effects of PEP-19 Peptides on Equilibrium Ca2+ Binding
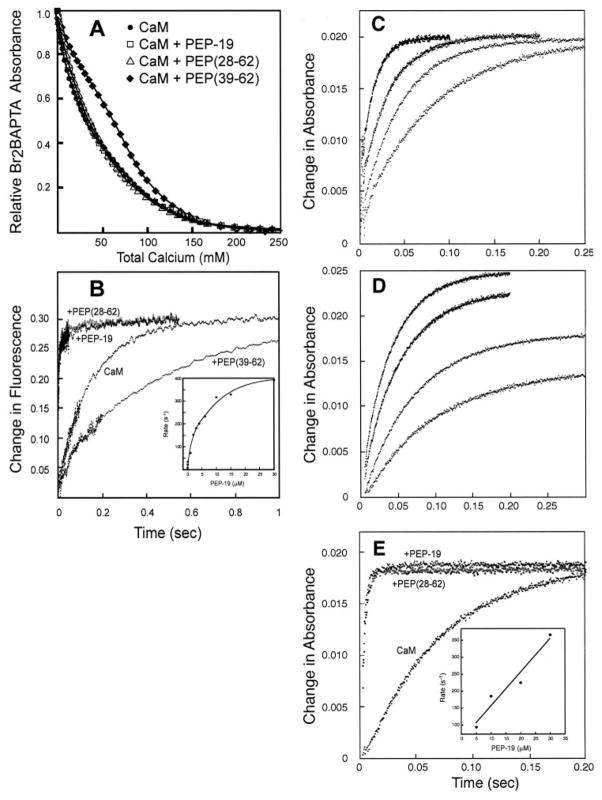
Fig. 5A and Table 2 compare the macroscopic Ca2+ dissociation constants, Kd1 through Kd4, for CaM in the absence or presence of PEP-19 derivatives. Linse et al. (21) assigned Kd1/Kd2 and Kd3/Kd4 to Ca2+ binding sites in the C- and N-domains of free CaM, respectively. We have adopted these assignments because the binding constants in Table 2 for free CaM are in agreement with those reported by other groups using the same technique under similar conditions (21, 22). Similar to previous reports using a variety of techniques (21, 27–29), Table 2 demonstrates strong positive cooperativity of Ca2+ binding to the C-domain of CaM because Kd2 < Kd1/4, with a lower limit for the free energy of cooperativity (ΔΔGc) of −3.4 kcal/mol.
FIGURE 5. Effects of PEP-19 polypeptides on Ca2+ binding to CaM.
Panel A shows equilibrium Ca2+ binding to CaM in the presence of PEP-19, PEP(28–62), or PEP(39–62). The lines show a least-squares fit of the data to the algorithm described under “Experimental Procedures.” Panel B shows the rate of dissociation of Ca2+ from free CaM (2 μM), and in the presence of 30 μM PEP-19, PEP(28–62) (gray circles), or PEP(39–62). The inset to panel B shows the effect of increasing concentrations of PEP-19 on the rate of dissociation of Ca2+ from the C-domain of CaM (2 μM). A single rate was observed at concentrations of PEP-19 > 2 μM, whereas Ca2+ dissociation at concentrations of PEP-19 of <2 μM best fit an exponential decay with two rate constants. For these data points, a weighted average rate was determined from (R1 × A1 + R2 × A2)/(A1 + A2). Panel C shows the rate of change in absorbance when a solution of EGTA (2 μM) was rapidly mixed with Br2BAPTA solutions of increasing free Ca2+ levels as described under “Experimental Procedures.” All data sets best fit a single exponential equation. The observed rate was used to calculate the free Ca2+ levels in the optical chamber (see “Experimental Procedures”) for the various Ca2+ /BrBATPA buffer solutions as 0.9, 1.6, 2.3, and 4.8 μM. Panel D shows the rate of change in absorbance when a solution of apo-CaM (2 μM) was rapidly mixed with Br2BAPTA solutions with free Ca2+ levels of 0.9, 1.6, 2.3, and 2.9 μM Ca2+ levels used in the experiments in panel C. All data sets best fit a single exponential equation. Panel E compares the rate of change in absorbance when a Ca2+ /Br2BAPTA solution with a free Ca2+ level of 3μM was rapidly mixed with apo-CaM with or without 30 μM PEP-19 or PEP(28–62) (gray circles).
Table 2 shows that intact PEP-19 and PEP(28–62) have little effect on Kd3 or Kd4, but have significant effects on Kd1 and Kd2. Because both PEP-19 and PEP(28–62) have relatively small effects on amide chemical shifts in the N-domain (see Fig. 2), we have assigned Kd3 and Kd4 as macroscopic binding constants for the N-domain, and Kd1 to Kd2 to the C-domain of CaM bound to either PEP-19 or PEP(28–62). Both PEP-19 and PEP(28–62) cause a significant decrease in the cooperativity of Ca2+ binding to the C-domain of CaM from a ΔΔGc of −3.4 to −1.3 kcal/mol. PEP(39–62) has little effect on Kd2, Kd3, or Kd4, but increases the affinity of binding the first Ca2+ (Kd1) from 17 μM with free CaM to 1.4 μM in the presence of the peptide. This results in a lower degree of cooperativity of Ca2+ binding to the putative C-domain of CaM, but a 3-fold increase in overall affinity.
Comparative Effects of PEP-19 Peptides on Ca2+ Binding Kinetics
The rate of dissociation of Ca2+ from the N-domain of CaM is very fast and occurs within the dead-time of the stopped-flow fluorimeter at room temperature (1.7 ms). Thus, only the slower release of two Ca2+ ions from the C-domain of CaM can be readily detected. Fig. 5B and Table 3 show that both intact PEP-19 and PEP(28–62) greatly increase the rate of dissociation of Ca2+ from CaM. PEP-19 has a similar effect on a recombinant CaM C-terminal fragment, CaM-(76–148), which further supports its domain-specific effect, and assignment of Kd1 and Kd2 in Table 2 to the C-domain of CaM. In contrast, the shorter PEP(39–62) has the opposite effect of decreasing the observed rate of Ca2+ dissociation by about 3-fold. The magnitude of this slow phase is consistent with the release of 2 Ca2+ ions from the C-domain, and the 3-fold decrease in rate would account for the higher affinity of Ca2+ binding to the C-domain in the presence of PEP(36–62) shown in Table 2.
TABLE 3.
Effect of PEP-19, PEP(28–62), and PEP(39–62) on Ca2+ dissociation from CaM
| Condition | koff rate |
|---|---|
| (s−1) | |
| CaM | 9.9 ± 0.4 |
| CaM-(76–148) | 12.1 ± 0.2 |
| CaM + PEP-19 | 430 ± 50 |
| CaM-(76–148) + PEP-19 | 490 ± 20 |
| CaM + PEP(28–62) | 390 ± 40 |
| CaM + PEP(39–62) | 3.2 ± 0.3 |
Dissociation rates (koff) were derived by fitting data such as that in Fig. 5B to a single exponential equation. The values are the mean ± S.D. of three to six experiments. CaM was present at 2 to 5 μM, and PEP-19 or PEP-19 peptides were present at 30 μM.
“Experimental Procedures” describes an assay to measure Ca2+ kon rates at free Ca2+ levels maintained between 0.5 and 5 μM using Br2BAPTA as both a Ca2+ buffer and a chromophore to monitor Ca2+ binding to CaM. EGTA was used in control experiments, because its high affinity for Ca2+ at pH 7.5 (Kd = 0.038 μM) ensures saturation at all levels of free Ca2+ used in the assay, but the Ca2+ association rate for EGTA is easily measured using stopped-flow techniques. Fig. 5C shows the expected increase in rate of association of Ca2+ with EGTA at increasing free Ca2+ levels.
Fig. 5D shows the rate of Ca2+ binding to CaM at free Ca2+ levels ranging from about 0.5 to 5 μM. The data best fit a single exponential rate at all Ca2+ levels. The increased magnitude of change at higher free Ca2+ levels is consistent with an increased percent saturation of the C-domain with Ca2+, which has an overall Kd of 2.3 μM. We observed no effect of either PEP(39–62) or PEP(29–45) on the rate of Ca2+ binding to CaM, however, Fig. 5E shows that both intact PEP-19 and PEP(28–62) greatly increased the observed Ca2+ kon.
DISCUSSION
The primary goal of the current study was to define residues in PEP-19 that modulate Ca2+ binding to CaM. During the course of these experiments we also showed that PEP-19 attenuates the degree of positive cooperativity of Ca2+ binding to sites III and IV. Positive cooperativity simply means that binding the first Ca2+ ion increases the affinity of binding the second Ca2+. With respect to the macroscopic binding constants Kd1 and Kd2 shown in Fig. 6A, positive cooperativity is implied if Kd2 < Kd1/4. Because Kd = koff/kon, this criteria can be satisfied by a wide range of rate constants. It is therefore not immediately apparent from macroscopic Ca2+ binding constants how changes in cooperativity could account for the large effects of PEP-19 on the rates of Ca2+ binding.
Little is known about the microscopic equilibrium Ca2+ binding and rate constants for CaM, but it is these parameters that would provide the greatest insight into the mechanism of action of PEP-19. Thus, we developed a kinetic model for cooperative Ca2+ binding to CaM that is based on experimental data and algebraic expressions that relate microscopic and macroscopic binding and rate constants (see “Experimental Procedures” for details). The model was used to derive and optimize the microscopic rate constants shown in Fig. 6A for transition of the C-domain of CaM from the 0-Ca2+ to 2-Ca2+ states. The models were tested by comparing experimental data with a simulation using rate constants derived from the model to predict the pseudo first-order rate of association of Ca2+ with the C-domain of CaM. Fig. 6B shows that the simulation closely approximates the experimental data, with both data sets showing a non-linear relationship between free Ca2+ and the rate of Ca2+ binding.
A key feature of the kinetic model described in Fig. 6 is that binding the first Ca2+ ion to either site III or IV is characterized by fast rate constants, whereas binding the second Ca2+ occurs with much slower rates. In essence, binding the first Ca2+ to either site III or IV increases the affinity of binding the second Ca2+, which defines positive cooperativity, but it also drastically slows the rates of this second binding event. This immediately implies that the observed attenuation of cooperativity by PEP-19 could accelerate Ca2+ rate constants by allowing greater expression of rapid rates associated with independent binding of Ca2+ to sites III and IV.
Our results demonstrate that the core IQ sequence (amino acids 39–62) is necessary to promote binding of PEP-19 to CaM, but that it does not mimic other properties of PEP-19. The core IQ motif binds to at least two sites on CaM. One site has a Kd similar to that of binding intact PEP-19, whereas another site binds the IQ motif with higher affinity. This must be considered when evaluating data that utilize IQ peptides taken out of context of the intact protein. For example, our results are consistent with a previous report showing that a synthetic peptide called camstatin, which spans the IQ motif in PEP-19 (residues 36–60), was a more effective inhibitor of nNOS than intact PEP-19 (36). It is likely that the properties of camstatin are similar to those of PEP(39–62). Although intact PEP-19 binds to Ca2+-CaM with relatively low affinity (Kd of 20 to 30 μM), it is present at high concentrations in brain (39), and the inset to Fig. 5 shows that PEP-19 at a concentration of 5 μM significantly increases the rate of dissociation of Ca2+ from CaM.
Our data show that coupling the acidic-rich amino acids 28–40 to the core IQ motif is necessary to mimic intact PEP-19 with respect to preferential binding to the C-domain of CaM, and modulating Ca2+ binding to sites III and IV. This defines the functionally relevant region of PEP-19 to a short acidic/IQ motif of 35 amino acids. The presence of this motif in other proteins (see Fig. 1) implies a functional significance that extends beyond PEP-19. Of particular interest are large proteins that may have intrinsic activity, such as the sea urchin protein in Fig. 1A that also encodes fibronectin type II and multiple PLAT/LH2 domains. The acidic/IQ motif may mediate direct CaM-dependent regulation of larger proteins, with modulation of Ca2+ binding to CaM as an integral feature of this regulation.
A critical role for acidic residues in modulating Ca2+ binding to CaM implies mechanisms involving interactions between Ca2+ and PEP-19. The acidic region may function as a negatively charged antenna that electrostatically “steers” Ca2+ ions to and from sites III and IV. A more specific interaction with Ca2+ is suggested by the primary sequence in Fig. 1A. With the exception of Pro-37, residues Glu-29 to Glu-40 in PEP-19 conform well to the consensus sequence of an EF-hand Ca2+ binding loop, with oxygen-containing side chains at coordination positions X, Y, Z, −Y, and −Z. Interestingly, the acidic region of RC3 does not have a similar distribution of acidic residues. The role of this putative Ca2+ binding loop is currently being studied.
Synergy between the core IQ sequence and adjacent residues to achieve unique functionality is a paradigm that may apply to other IQ motif proteins. For example, residues N-terminal to the IQ motif of CaV1.2 are not highly acidic. Instead, this sequence includes a Phe residue (see open arrow in Fig. 1A) that anchors CaV1.2 to the N-domain of CaM (see open arrow in Fig. 1A) (11, 12). This region of CaV1.2 may play an important regulatory role because channel facilitation and inactivation is thought to be mediated by dynamic differential binding of the N- and C-domains of CaM to the IQ region (15). A corresponding Phe residue is not present in PEP-19. Thus, different modules extending N-terminal to the IQ motifs of PEP-19 versus CaV1.2 appear to confer unique functionalities to these CaM binding proteins.
In summary, this study reports a comprehensive new kinetic model that can account for cooperative Ca2+ binding to the C-domain of CaM and provides a mechanistic model for the effects of PEP-19 at the level of attenuating cooperativity. We also show that the effects of PEP-19 on CaM rely on the synergy between the core IQ motif that targets PEP-19 to CaM, and an adjacent acidic cluster that modulates Ca2+ binding. We propose that this acidic/IQ motif is a regulator of CaM signaling found in diverse proteins and species.
Footnotes
This work was supported in part by National Institutes of Health Grants GM069611 and NS038310 and Robert A. Welch Foundation Grant AU1144.
The abbreviations used are: CaM, calmodulin; Ca2+-CaM, Ca2+-bound calmodulin; CaMACR, acrylodan labeled CaM(K75C); CaMDANS, IAEDANS labeled CaM(K75C); CKII, CaM-dependent protein kinase II; RC3, neurogranin; FRET, fluorescence resonance energy transfer; MOPS, 4-morpholinepropane-sulfonic acid; acrylodan, 6-acryloyl-2-dimethylaminonaphthalene; IAEDANS, 5-((((2-iodoacetyl)amino)ethyl)amino)naphthalene-1-sulfonic acid; DDPM, N-(4-dimethylamino-3,5-dinitrophenyl)maleimide; HPLC, high performance liquid chromatography; Br2, 5,5′-dibromo; BAPTA, 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid.
References
- 1.Chin D, Means AR. Cell Biol. 2000;10:322–328. doi: 10.1016/s0962-8924(00)01800-6. [DOI] [PubMed] [Google Scholar]
- 2.Berridge MJ, Bootman MD, Roderick HL. Nat Rev. 2003;4:517–525. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- 3.Bootman M, Lipp P, Berridge MJ. J Cell Sci. 2002;114:2213–2222. doi: 10.1242/jcs.114.12.2213. [DOI] [PubMed] [Google Scholar]
- 4.Rakhilin SV, Olson PA, Nishi A, Starkove NN, Fienberg AA, Nairn AC, Surmeier DJ, Greengard P. Science. 2004;306:698–701. doi: 10.1126/science.1099961. [DOI] [PubMed] [Google Scholar]
- 5.Putkey JA, Kleerekoper Q, Gaertner TR, Waxham MN. J Biol Chem. 2003;278:49667–49670. doi: 10.1074/jbc.C300372200. [DOI] [PubMed] [Google Scholar]
- 6.Gaertner TR, Putkey JA, Waxham MN. J Biol Chem. 2004;279:9374–9382. doi: 10.1074/jbc.M405352200. [DOI] [PubMed] [Google Scholar]
- 7.Pak JH, Huang FL, Li J, Balschun D, Reymann KG, Chiang C, Westphal H, Huang KP. Proc Natl Acad Sci U S A. 2000;97:11232–11237. doi: 10.1073/pnas.210184697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu J, Li J, Huang KP, Huang FL. J Biol Chem. 2002;277:19498–19505. doi: 10.1074/jbc.M109082200. [DOI] [PubMed] [Google Scholar]
- 9.van Dalen JJ, Gerendasy DD, de Graan PN, Schrama LH, Gruol DL. Eur J Neurosci. 2003;18:13–22. doi: 10.1046/j.1460-9568.2003.02720.x. [DOI] [PubMed] [Google Scholar]
- 10.Rhoads A, Bahler M. FEBS Lett. 2002;513:107–113. doi: 10.1016/s0014-5793(01)03239-2. [DOI] [PubMed] [Google Scholar]
- 11.Fallon JL, Halling DB, Hamilton SL, Quiocho FA. Structure. 2005;13:1881–1886. doi: 10.1016/j.str.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 12.Van Petegem F, Chatelain FC, Minor DL., Jr Nat Struct Mol Biol. 2005;12:1108–1115. doi: 10.1038/nsmb1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui Y, Wen J, Hung Sze K, Man D, Lin D, Liu M, Zhu G. Anal Biochem. 2003;315:175–182. doi: 10.1016/s0003-2697(03)00007-1. [DOI] [PubMed] [Google Scholar]
- 14.Trybus KM, Gushchin MI, Lui H, Hazelwood L, Krementsova EB, Volkmann N, Hanein D. J Biol Chem. 2007;282:23316–23325. doi: 10.1074/jbc.M701636200. [DOI] [PubMed] [Google Scholar]
- 15.Liang H, DeMaria CD, Erickson MG, Mori MX, Alseikhan BA, Yue DT. Neuron. 2003;39:951–960. doi: 10.1016/s0896-6273(03)00560-9. [DOI] [PubMed] [Google Scholar]
- 16.Xiong L, Kleerekoper QK, He R, Putkey JA, Hamilton SL. J Biol Chem. 2005;280:7070–7079. doi: 10.1074/jbc.M410558200. [DOI] [PubMed] [Google Scholar]
- 17.Putkey JA, Donnelly PV, Means AR. Methods Enzymol. 1987;139:303–317. doi: 10.1016/0076-6879(87)39094-9. [DOI] [PubMed] [Google Scholar]
- 18.Putkey JA, Waxham MN. J Biol Chem. 1996;271:29619–29623. doi: 10.1074/jbc.271.47.29619. [DOI] [PubMed] [Google Scholar]
- 19.Peng JW, Wagner G. Biochemistry. 2004;34:16733–16752. doi: 10.1021/bi00051a023. [DOI] [PubMed] [Google Scholar]
- 20.Waxham MN, Tsai AL, Putkey JA. J Biol Chem. 1998;273:17579–17584. doi: 10.1074/jbc.273.28.17579. [DOI] [PubMed] [Google Scholar]
- 21.Linse S, Helmersson A, Forsén S. J Biol Chem. 1991;266:8050–8054. [PubMed] [Google Scholar]
- 22.Bayley PM, Findlay WA, Martin SR. Protein Sci. 1996;5:1215–1228. doi: 10.1002/pro.5560050701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maune JF, Klee CB, Beckingham K. J Biol Chem. 1992;267:5286–5295. [PubMed] [Google Scholar]
- 24.Johnson JD, Snyder C, Walsh C, Flynn M. J Biol Chem. 1996;271:761–767. doi: 10.1074/jbc.271.2.761. [DOI] [PubMed] [Google Scholar]
- 25.Peersen OB, Madsen TS, Falke JJ. Protein Sci. 1997;6:794–807. doi: 10.1002/pro.5560060406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pedigo S, Shea MA. Biochemistry. 1995;34:10676–10689. doi: 10.1021/bi00033a044. [DOI] [PubMed] [Google Scholar]
- 27.VanScyoc WS, Sorensen BR, Rusinova E, Laws WR, Ross JA, Shea MA. Biophys J. 2002;83:2767–2780. doi: 10.1016/S0006-3495(02)75286-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sorensen BR, Shea MA. Biochemistry. 1998;37:4244–4253. doi: 10.1021/bi9718200. [DOI] [PubMed] [Google Scholar]
- 29.Biekofsky RR, Matin SR, Browne JP, Bayley PM, Feeney J. Biochemistry. 1998;37:7617–7629. doi: 10.1021/bi9800449. [DOI] [PubMed] [Google Scholar]
- 30.Ulmer TS, Soelaiman S, Li S, Klee CB, Tang WJ, Bax A. J Biol Chem. 2003;278:29261–29266. doi: 10.1074/jbc.M302837200. [DOI] [PubMed] [Google Scholar]
- 31.Haiech J, Kilhoffer M-C. Calcium-binding Protein Protocols. 173. II. Humana Press; Totowa, NJ: 2002. pp. 25–42. [Google Scholar]
- 32.Evanas J, Thulin E, Malmendal A, Forsen S, Carlstrom G. Biochemistry. 1997;36:3448–3457. doi: 10.1021/bi9628275. [DOI] [PubMed] [Google Scholar]
- 33.Evenas J, Malmendal A, Akke M. Structure. 2001;9:185–195. doi: 10.1016/s0969-2126(01)00575-5. [DOI] [PubMed] [Google Scholar]
- 34.Martin SR, Maune JF, Beckingham K, Bayley PM. Eur J Biochem. 1992;205:1107–1114. doi: 10.1111/j.1432-1033.1992.tb16879.x. [DOI] [PubMed] [Google Scholar]
- 35.Persechini A, White HD, Gansz KJ. J Biol Chem. 1996;271:62–67. doi: 10.1074/jbc.271.1.62. [DOI] [PubMed] [Google Scholar]
- 36.Slemmon JR, Morgan JI, Fullerton SM, Danho W, Hilbush BS, Wengenack TM. J Biol Chem. 1996;271:15911–15917. doi: 10.1074/jbc.271.27.15911. [DOI] [PubMed] [Google Scholar]
- 37.Torok K, Tzortzopoulos A, Grabarek Z, Best SL, Thorogate R. Biochemistry. 2001;40:14878–14890. doi: 10.1021/bi010920+. [DOI] [PubMed] [Google Scholar]
- 38.Meador WE, Means AR, Quiocho FA. Science. 1993;262:1718–1721. doi: 10.1126/science.8259515. [DOI] [PubMed] [Google Scholar]
- 39.Slemmon JR, Feng B, Erhardt JA. Mol Neurobiol. 2001;22:99–113. doi: 10.1385/MN:22:1-3:099. [DOI] [PubMed] [Google Scholar]