Abstract
The malignant transformation in several types of cancer, including lung cancer, results in a loss of growth inhibition by transforming growth factor-β (TGF-β). Here, we show that SMAD6 expression is associated with a reduced survival in lung cancer patients. Short hairpin RNA (shRNA)–mediated knockdown of SMAD6 in lung cancer cell lines resulted in reduced cell viability and increased apoptosis as well as inhibition of cell cycle progression. However, these results were not seen in Beas2B, a normal bronchial epithelial cell line. To better understand the mechanism underlying the association of SMAD6 with poor patient survival, we used a lentivirus construct carrying shRNA for SMAD6 to knock down expression of the targeted gene. Through gene expression analysis, we observed that knockdown of SMAD6 led to the activation of TGF-β signaling through up-regulation of plasminogen activator inhibitor-1 and phosphorylation of SMAD2/3. Furthermore, SMAD6 knockdown activated the c-Jun NH2-terminal kinase pathway and reduced phosphorylation of Rb-1, resulting in increased G0-G1 cell arrest and apoptosis in the lung cancer cell line H1299. These results jointly suggest that SMAD6 plays a critical role in supporting lung cancer cell growth and survival. Targeted inactivation of SMAD6 may provide a novel therapeutic strategy for lung cancers expressing this gene.
Introduction
Transforming growth factor-β (TGF-β) belongs to a superfamily of structurally related polypeptides that are involved in various biological processes, including cell growth, differentiation, angiogenesis, apoptosis, and extracellular matrix remodeling (1). Alterations in TGF-β signaling are linked to a variety of human diseases, including cancer, inflammation, and tissue fibrosis (2, 3). The disruption of TGF-β signaling occurs in several human cancers and the pathway generally possesses a tumor suppressor function (4). However, as carcinogenesis proceeds, tumor cells acquire resistance to TGF-β–induced growth arrest.
TGF-β and its superfamily member, bone morphogenesis protein (BMP), activate their respective intracellular signaling cascades by binding to the type II receptor followed by the recruitment of the type I receptor. The activated type I receptor phosphorylates the receptor SMADs (R-SMAD), such as SMAD2 and SMAD3, which then form a heteromeric complex with the Co-SMAD, SMAD4. The R-SMAD/SMAD4 complex translocates into the nucleus, where it regulates the transcription of target genes (1, 5, 6). Among the TGF-β/BMP target genes are two inhibitory SMAD proteins, SMAD6 and SMAD7. SMAD6 is generally thought to mediate BMP signals, whereas SMAD7 mediates TGF-β signaling. Both proteins regulate the TGF-β signaling pathway through a negative feedback mechanism (7–9). Recently, SMAD6 and SMAD7 have been shown to play a role in tumorigenesis. SMAD7 overexpression causes malignant conversion in a multistage cancer model (10) and enhanced tumorigenicity in pancreatic cancer (11). Otherwise, stable overexpression of SMAD7 in human melanoma cells impairs bone metastasis by blocking the TGF-β signal pathway (12). Similarly, adenoviral delivery of SMAD7 to JygMC(A) breast cancer cells significantly impairs their capacity to metastasize to lung and liver, possibly by altering their adhesive and migratory properties; however, overexpression of SMAD6 had no effect on metastasis (13). The expression of SMAD6 and SMAD7 was inversely correlated with the depth of invasion in the early stages of carcinogenesis, but there was a significant correlation between the expression of SMAD6 and SMAD7 to poor survival esophageal squamous cell carcinoma (14).
In this study, we observed that SMAD6 expression was associated with poor survival in non–small cell lung cancer (NSCLC) patients. Knockdown of SMAD6 restored TGF-β signaling pathway by increasing SMAD2/3 phosphorylation and plasminogen activator inhibitor-1 (PAI-1) activation in lung cancer cell lines but not minimally transformed normal bronchial epithelial cells, Beas2B. We propose that SMAD6 contributes to lung cancer progression by limiting TGF-β signaling-mediated growth inhibition and that SMAD6 down-regulation restores the TGF-β sensitivity, which led to reduced viability, proliferation, and increased apoptosis in lung cancer.
Materials and Methods
Cell lines and culture
All lung cancer cell lines and normal bronchial epithelial cell line, Beas2B, were obtained directly from the American Type Culture Collection. All lung cancer cell lines were cultured in RPMI 1640 supplemented with 10% fetal bovine serum (FBS; Life Technologies). Beas2B was cultured in BEGM and growth supplements (Cambrex Bio Sciences, Inc.) in a humidified atmosphere with 5% CO2.
Tissue arrays and immunohistochemistry
Tissue arrays were prepared as previously described (15). It included 300 NSCLCs (150 adenocarcinomas and 150 squamous cell carcinomas) from the archives of the Armed Forced Institute of Pathology (Washington, DC). All patient information was obtained and used in accordance with approved protocols from the institutional review boards of the participating institutions. The clinical characteristics of the cohort are as previously described (15). We used rabbit anti-Smad6 antibody from Zymed and anti-rabbit secondary antibody from EnVision+ System and Liquid DAB (DAKO) to visualize the immunohistochemistry staining signal. Sections were counterstained lightly with Mayer’s hematoxylin and scored exactly as previously described (15). For survival analyses, the samples were considered as “negative” when the total score was 0 and “positive” when total score was 1 or higher.
Production of lentivirus-containing SMAD6 short hairpin RNA
Two SMAD6 short hairpin RNAs (shRNA #1 and #2) were purchased from Open Biosystems. SMAD6 shRNA #3 was produced in the lab as follows: shRNA #3 oligonucleotides were synthesized (IDT) and annealed by hybridization, and then double-stranded fragments were cloned into the AgeI and EcoRI sites of pLKO.1 (Open Biosystems) and transformed into Escherichia coli. The shRNA sequences were described in Supplementary Table S1. Lentivirus was produced using the lentiviral packaging system (ViraPower Lentiviral Expression System) from Invitrogen. Each lentivirus was harvested at 4 d after transfection. Cells were plated in monolayer at different densities and infected with lentivirus constructs using 8 ng/mL polybrene. After Smad6 shRNA lentiviral infection, expression of SMAD6 was analyzed by Western blotting.
Cell viability, soft-agar, and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assays
The shRNA for Smad6 lentivirus-transduced cells and mock-transduced cells was plated at 2 × 1041 × 1042 × 103and 1 × 103 per well of a 12-well plate. After 10 d in culture, medium was discarded, and the remaining viable adherent cells were washed with 1× PBS before staining with crystal violet (Sigma). We used the Cell Transformation Detection kit (Chemicon) to evaluate for colony formation ability on soft agar. Briefly, 0.5 mL underlayers consisting of 0.8% agar medium were prepared in 24-well plates. SMAD6 shRNA–transduced and mock lentivirus–transduced cells were trypsinized, centrifuged, resuspended in 0.4% agar medium (equal volumes of 0.8% Noble agar and culture medium), and plated onto the top agar at 1,500 per well. The cells were kept wet by adding a small amount of RPMI 1640 (Life Technologies) with 10% FBS and incubated for 3 wk at 37°C. Colonies were visualized using cell staining solution (Chemicon) and counted under the microscope.
To measure the effects of knockdown on cell proliferation, Smad6 shRNA–transduced cells were plated at concentrations of 1 × 1021 × 103and 1 × 104 per well in 96-well plates. After 4 d, cell proliferation was measured by the cell growth determination kit 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)–based assay according to the manufacturer’s protocol (Sigma). In brief, 10 µL of the MTT solution (5 mg/mL) were added to each well, and the cells were cultured for another 3 h at 37°C. At the end of incubation, 100 µL of 0.1 N HCl in isopropanol were added and mixed vigorously to solubilize colored crystals produced within the cells. The absorbance at 570 nm (690 nm as background) was measured using a SpectraMax Plus (Molecular Devices). Experiments were performed thrice each in triplicates.
TGF-β inhibitor treatment
Mock-transduced, shRNA #3–transduced, and shRNA #2–transduced H1299 cells were cultured in RPMI 1640 without FBS for 2 d after infection followed by treatment with SB431542 (10 µmol/L in DMSO) to cells for 48 h. For control experiments, DMSO was added at the same concentration. The status of PAI-1, the known TGF-β effector, was determined by Western blot.
Flow cytometry and terminal deoxynucleotidyl transferase–mediated dUTP nick end labeling assays
Lentivirus-transduced cells were harvested and stained with propidium iodide (20 µg/mL; Sigma), and the DNA content of the cells was analyzed with a flow cytometer (FACSCalibur, Becton Dickinson).
Apoptosis was detected using terminal deoxynucleotidyl transferase–mediated dUTP nick end labeling (TUNEL) apoptosis detection kit (Upstate). H1299 and Beas2B cells were infected with Smad6 shRNA and mock lentivirus. Four days after infection, the TUNEL assay was performed according to the manufacturer’s protocol. Cells were counterstained with propidium iodide and examined under fluorescent microscopy.
Western analysis
Cells were plated in 100-mm2 tissue culture dishes at 60% confluency and incubated overnight. Cell lysates were obtained from transduced cells using cold radioimmunoprecipitation assay buffer [20 mmol/L Tris-HCl (pH 8.0), 100 mmol/L NaCl, 10% glycerol, 1% NP40, 0.5% sodium deoxycholate]. Twenty micrograms of protein were separated on precasted Bis-Tris NuPAGE gels (Invitrogen) and electroblotted to polyvinylidene difluoride membranes (GE Healthcare Life Sciences) and then blocked for 1 h at room temperature in TBS-T [50 mmol/L Tris-HCl (pH 7.5), 150 mmol/L NaCl, 0.1% Tween 20] buffer containing 5% nonfat milk. Membranes were then incubated overnight at 4°C or 1 h at room temperature with the respective primary antibodies: phospho-SMAD2/3 (1:500), SMAD2 (1:1,000), phospho-c-Jun (1:1,000), and c-Jun NH2-terminal kinase (JNK; 1:1,000; all from Santa Cruz Biotechnology); SMAD6 (Imgenex and Santa Cruz Biotechnology); caspase-3, caspase-3, poly(ADP-ribose) polymerase (PARP), and phospho-JNK (all from Cell Signaling Technology); phospho-RB and underphospho-RB (both from BD Pharmingen); and β-actin (product AC-15; Sigma-Aldrich). Anti-mouse or anti-rabbit secondary antibody conjugated to horseradish peroxidase (Santa Cruz Biotechnology) was used to visualize the stained bands with an enhanced chemiluminescence visualization kit (GE Healthcare Life Sciences).
Gene expression analysis using cDNA microarray
Total RNA (10 µg) was extracted from the transduced cells using Trizol and RNeasy MiniPrep (Qiagen) according to the manufacturers’ protocols. The quality of the total RNA was checked with denaturing formamide gel electrophoresis. Biotinylated cRNA was amplified with a double in vitro transcription in accordance with the Affymetrix small sample labeling protocol VII (Affymetrix). The total RNA was then hybridized onto Affymetrix GeneChip HG-U133A according to standard protocols (Affymetrix user guide). Fluorescence intensities were quantified and analyzed using the GeneChip operating software (Affymetrix). For signal pathway analysis, we identified transcripts whose log-transformed expression ratios differed by at least 2-fold in both shRNA #1 and shRNA #3 compared with the mock control–infected cells. Of the identified transcripts, 274 genes were used for network classification by Ingenuity Pathways Analysis (IPA) and were mapped based on the functions and/or canonical pathways from the literature.
Statistical analyses
We used Kaplan-Meier survival curve method to show the prognostic difference in SMAD6 protein expression. Statistical significance was assessed by log-rank (Mantel-Cox) test. Analysis was adjusted by factors of gender and pathologic stage, which represent independent prognostic significance. For colony formation assay, statistical comparisons were made using one-way ANOVA. Statistical significance was indicated by P < 0.05. Data are presented as mean ± SD.
Results
SMAD6 status and its correlation with patient survival in NSCLC
Immunohistochemistry staining was carried out with an anti–SMAD6-specific antibody using a NSCLC tissue microarray. As shown in Supplementary Table S2 and Fig. 1A, 122 cases were positive and 120 cases were negative among the cases present on the tissue microarray. The frequency of positive staining was similar between adenocarcinoma and squamous cell carcinomas of the lung. There was no statistically significant difference in SMAD6 staining status scores between tumor types and gender. However, when staining intensities of the tumors were correlated with the survival status of the corresponding patients, the tumor-related survival was significantly better for tumors negative of SMAD6 protein staining compared with those whose tumors were positive (P = 0.007; Fig. 1B). Five years after surgery, 62% of patients whose tumors expressed SMAD6 were recurrence-free compared with 86% for patients whose tumors were negative for SMAD6 (P = 0.005; Fig. 1B).
Figure 1.
SMAD6 status and tumor-related survival in NSCLC patients. A, representative images of immunohistochemistry staining for SMAD6 in lung cancer patients. AD, adenocarcinoma; Sq, squamous cell carcinoma. Staining status is as indicated. B, Kaplan-Meier curve for tumor-related survival status based on SMAD6 status. Patients with SMAD6-negative tumors (total score = 0) were compared with patients with moderate/high SMAD6-expressing tumors (total score = 1 or higher).
SMAD6 expression in normal and lung cancer cell lines
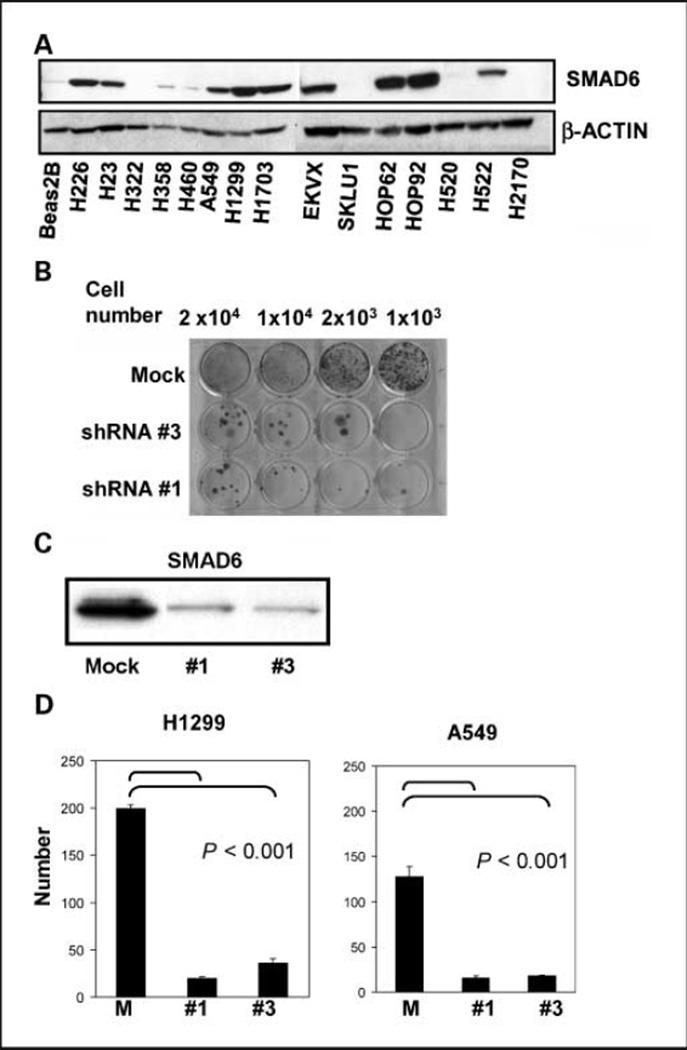
We performed Western blot analysis to examine the level of SMAD6 expression in 15 lung cancer cell lines and Beas2B, a normal bronchial cell line. As shown in Fig. 2A, SMAD6 expression was high in most lung cancer cell lines (H226, H23, A549, H1299, H1703, EKVX, HOP62, HOP92, and H522), minimally expressed in H358 and H460 cell lines, and undetectable in H322, SKLU1, H520, H2170, and Beas2B cells.
Figure 2.
SMAD6 expression and knockdown in lung cancer and normal cell lines. A, expression of SMAD6 in normal lung cells (Beas2B) and indicated lung cancer cell lines. B, cell viability assay in H1299 cells after SMAD6 knockdown. Control (mock) lentivirus or SMAD6 shRNA #1–transduced and shRNA #3–transduced H1299 cells were seeded at indicated densities and stained with crystal violet 10 d after infection. C, Western blot analysis showing SMAD6 levels in transduced cells as indicated. D, colony formation on soft agar in H1299 and A549 cells that were transduced with SMAD6 shRNA #1, shRNA #3, and mock control. Histograms indicate stained soft agar colonies counted from three independent experiments. Bars, SD. P values are as indicated when compared with mock control. Lentiviruses are the same as for B.
Reduced cancer cell growth by SMAD6 knockdown
We generated three different lentivirus constructs carrying Smad6 shRNA and used these constructs to transduce lung cancer cell line, H1299. Cells transduced with shRNA #1 and #3 had significantly reduced cell viability compared with mock-transduced cells (Fig. 2B). Consistent with this, SMAD6 protein levels were drastically reduced in H1299 cells transduced with shRNA #1 and #3, but not the mock-transduced cells (Fig. 2C).
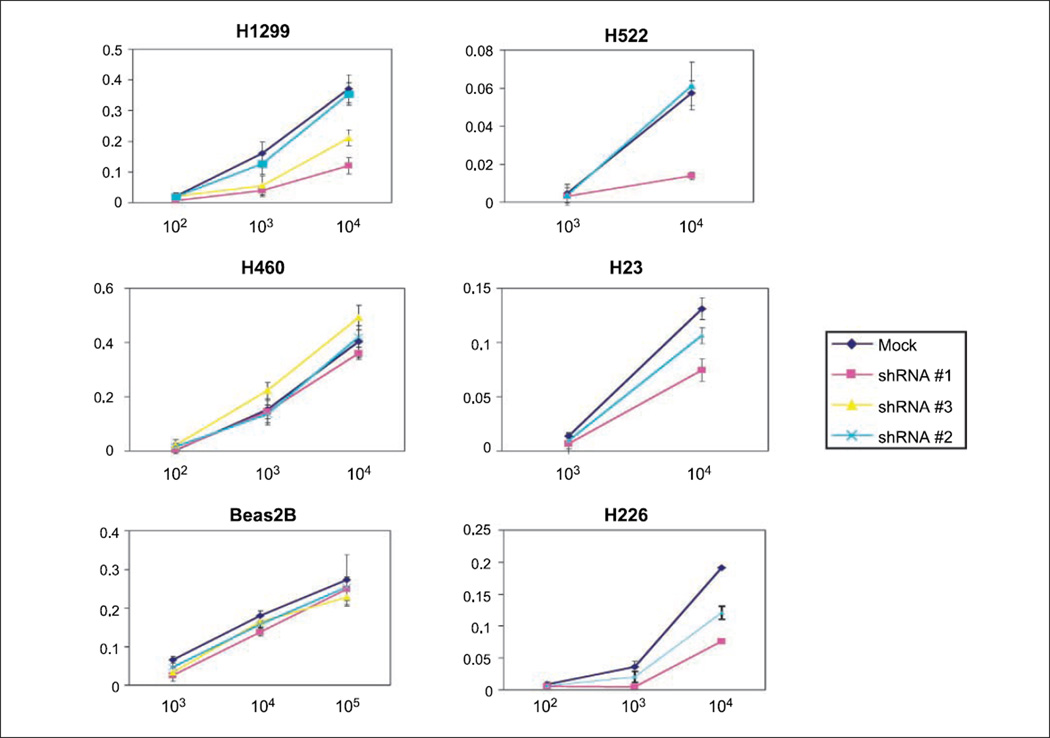
We also tested SMAD6 shRNA–transduced and mock control–transduced cells for their growth on soft agar (Fig. 2D). Mock control–transduced H1299 and A549 cells formed 199 and 127 colonies, whereas SMAD6 shRNA #1–transduced and shRNA #3–transduced H1299 and A549 cells had 5- to 10-fold less colonies (P < 0.001; Fig. 2D). To investigate the possible antiproliferative effects of SMAD6 knockdown, we performed MTT assay 6 days after infection. SMAD6 shRNA #1–transduced and shRNA #3–transduced H1299 cells showed significantly reduced viability relative to mock-transduced H1299 cells. In contrast, no growth inhibition was observed in mock-transduced cells (Fig. 3). The viability of normal bronchial epithelial cell line Beas2B was unchanged regardless of the shRNA constructs transduced. We performed the same test using three additional lung cancer cell lines that expressed SMAD6 (H226, H23, and H522) and one cell line that did not (H460). Reduced cell viability was observed in shRNA #1–transduced compared with mock-transduced cells for all three cell lines expressing SMAD6 but not in H460. Transducing cells with shRNA #2 into H1299 and H522 cells resulted in no growth inhibition, but intermediate inhibition was observed in H23 and H226 cells (Fig. 3).
Figure 3.
SMAD6 knockdown reduces lung cancer cell growth. Cell viability was determined by MTTassay in six cell lines and transduced with mock or SMAD6 shRNA as indicated. X axis, numbers of cells plated. Cell proliferation was determined absorbance at A570 using the Cell Growth Determination kit (Sigma) 4 d after transduction. Left, mock, shRNA #1, shRNA #2, and shRNA #3 were used; right, mock, shRNA #1, and shRNA #2 were used.
Knockdown of SMAD6 promotes cell cycle arrest and apoptosis
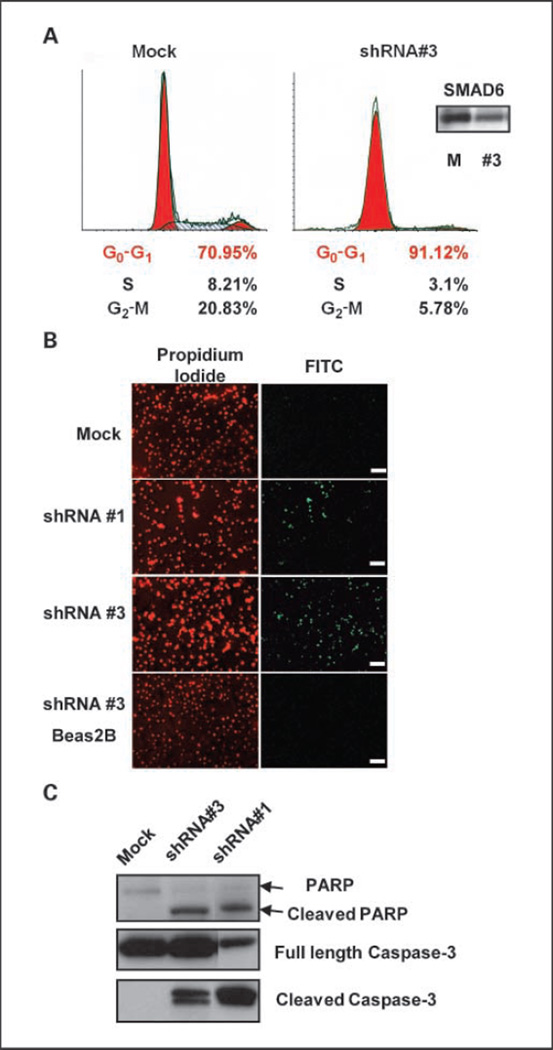
Using fluorescence-activated cell sorting (FACS) analysis, we observed that more than 90% of Smad6 shRNA #3–transduced H1299 cells arrested in G1 phase, whereas mock-transduced cells did not (Fig. 4A). We also investigated the potential apoptotic effect of SMAD6 knockdown using the TUNEL assay. As shown in Fig. 4B, shRNA #1–transduced and shRNA #3–transduced H1299 cells underwent apoptosis, whereas mock-transduced H1299 and Beas2B cells did not. Consistent with this observation, H1299 cells transduced with shRNA #1 and #3 had cleaved the inactive form of procaspase-3 and increased cleavage of the 113-kDa PARP to the 89-kDa fragment compared with the mock-transduced cells (Fig. 4C).
Figure 4.
Knockdown of SMAD6 induces cell arrest and increases apoptosis in H1299 cells. A, FACS analysis was done 4 d after transduction. A total of 10,000 cells were sorted. Proportional changes in S and G2-M phases are as indicated for mock-transduced and shRNA #3–transduced cells. Inset, levels of SMAD6 in the transduced cells. B, TUNEL analysis for H1299 cells transduced with mock, shRNA #1, shRNA #3, and Beas2B cells transduced with shRNA #3. Left, cells were counterstained with propidium iodide; right, apoptotic cells are labeled and visible under FITC channel. Slides were examined under a fluorescence microscope at ×40 magnification. Scale bar, 40 µm. C, Western analysis for PARP, full-length, and cleaved caspase-3. PARP antibody detects the full-length PARP and the large fragment as indicated.
SMAD6 knockdown restores TGF-β signaling in lung cancer cell lines
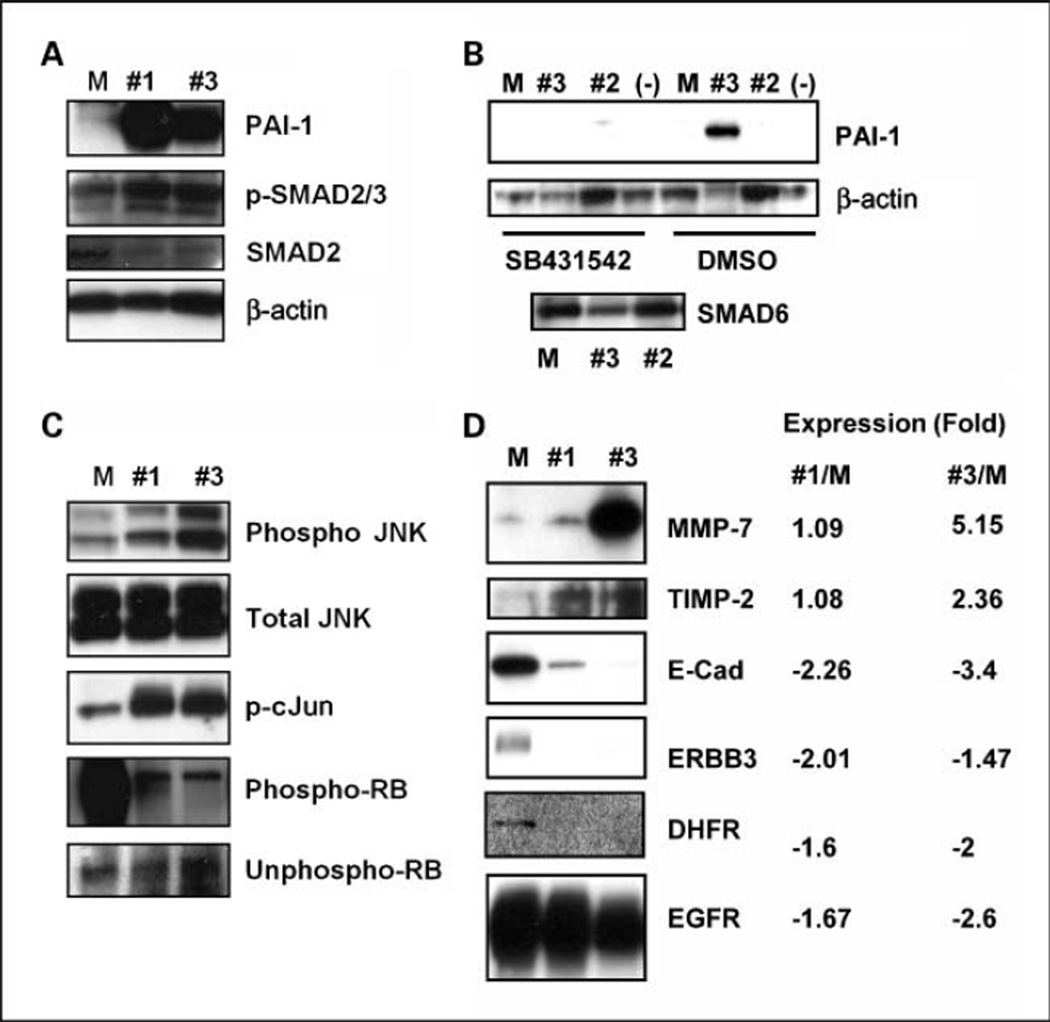
To determine if TGF-β signaling is restored after SMAD6 knockdown, we tested the induction of PAI-1 and phosphorylation of SMAD2/3 as these events are regulated by the TGF-β pathway (16). In shRNA #1–transduced and shRNA #3–transduced H1299 cells, PAI-1 was highly expressed and SMAD2/3 was hyperphosphorylated compared with the mock-transduced cells (Fig. 5A). To confirm these effects, we treated the transduced cells with a TGF-β receptor inhibitor, SB431542. PAI-1 expression was significantly reduced after SB431542 treatment in shRNA #2–transduced, shRNA #3–transduced, and mock-transduced H1299 cells; however, PAI-1 expression in shRNA #3–transduced H1299 cells was not affected by DMSO treatment (Fig. 5B).
Figure 5.
Restoration of TGF-β signaling in H1299 cells on SMAD6 knockdown. A, Western blot analysis was performed using PAI-1, phospho-SMAD2/3, SMAD2, and β-actin–specific antibodies in SMAD6 shRNA #1–transduced, shRNA #3–transduced, and mock-transduced cells. B, expression of PAI-1 after SB431542 treatment in SMAD6 shRNA #1–transduced, shRNA #3–transduced, and mock-transduced H1299 cells. Western blot analysis for transduced cells that were treated with SB431542 (left four lanes) or DMSO (right four lanes). M, mock control lentivirus–infected cells; #3, Smad6 shRNA #3–transduced cells; #2, Smad6 shRNA #2–transduced cells; (−), untreated parental cells. C, phosphorylation changes in JNK, c-Jun, and RB after the transduction of shRNA to Smad6. Antibodies to the corresponding proteins are as indicated. D, protein Western and gene expression analysis for genes affected by SMAD6 knockdown. Each value represents relative expression in log2 ratio when mRNA from SMAD6 shRNA #1–transduced and shRNA #3–transduced H1299 cells was compared with those from mock-transduced H1299 cells (M).
We also tested the phosphorylation status of Jun NH2-terminal kinase (JNK) and the retinoblastoma susceptibility genes (17). Hyperphosphorylation of JNK and c-Jun was observed in shRNA #1–transduced and shRNA #3–transduced cells but not in mock-transduced cells (Fig. 5C). A significant reduction in RB phosphorylation was seen in the shRNA #1–transduced and shRNA #3–transduced cells, whereas unphosphorylated RB was unchanged (Fig. 5C).
Transcriptional effects of SMAD6 knockdown in lung cancer cells
To determine the effect of SMAD6 knockdown on the overall transcriptome in lung cancer, we examined the expression profiles of the shRNA #1–transduced, shRNA #3–transduced, and mock-transduced H1299 cells in three independent experiments. Supplementary Tables S3 and S4 list genes that were differentially expressed by Samd6 knockdown using both shRNA #1 and shRNA #3. We were able to confirm the microarray observation for selected genes at the protein levels using commercially available antibodies (Fig. 5D). Among these genes, ERBB3, E-cadherin, and DHFR genes were down-regulated, whereas MMP7, TIMP-2, THROMBOSPONDIN1, and interleukin-8 (IL-8) were up-regulated by the knockdown of SMAD6. Affymetrix microarray gene expression data and the relative ratios between shRNA-transduced and mock-transduced cells are shown in Supplementary Tables S5 and S6.
We further delineate the molecular pathways that are affected by SMAD6 knockdown in H1299 cells. In our pathway analysis with IPA, there were 58 significant functional categories (P < 0.01). The 20 highly significant categories are shown in Supplementary Fig. S1. The cancer, cell cycle, cell death, cellular growth, and proliferation categories showed highly significant enrichment. Enrichments were also observed for expression in the tissue development, cell morphology, cellular development, and cellular movement categories (Supplementary Fig. S1A). There were 11 significant canonical pathway categories (P < 0.05) that include cell cycle, IL-6 signaling, death receptor signaling, TGF-β signaling, and neuregulin signaling (Supplementary Fig. S1B). The mRNA level of Smad7 was unchanged in the tested cells (Supplementary Table S5) and no genes associated with the induction of BMP signaling were observed (18).
Discussion
In patients with lung cancer, blood levels of TGF-β1 are elevated when compared with normal patients (19) and increased production of TGF-β by cancer cells during tumor progression can promote tumor growth, angiogenesis, and metastasis (20). The malignant transformation in lung cancer results in a loss of tumor suppressor effects of TGF-β, although most lung cancer cells secrete TGF-β (21). Loss of the TGF-β response has been associated with tumor development and/or tumor progression in several cancer cell lines (22–24). Resistance to TGF-β in cancer has been attributed to reduced expression of TGF-β receptor I (TGFRI) and/or TGF-β receptor II (TGFRII; refs. 25, 26), as well as the inactivation of mutations in SMAD2 and SMAD4 (27, 28). TGF-β receptor defects in cancer cells contribute to malignant progression through an interruption in TGF-β–mediated autocrine growth inhibition by the methylation of the TGFRI promoter or mutation of the TGFRII promoter (25, 26). Inhibitory SMADs (SMAD6/7) are thought to play a role in the regulation of TGF-β–mediated growth inhibition. However, the contribution of inhibitory SMADs (Smad6/7) to the loss of TGF-β responsiveness in cancer is not well understood.
To elucidate the role that SMAD6 may have in lung cancer progression, we used small interfering RNA technique to knock down SMAD6 in normal and lung cancer cell lines. We observed that down-regulation of SMAD6 by shRNA inhibited cell growth and induced apoptosis in lung cancer cells but not in the normal cell line Beas2B. In addition, SMAD6 shRNA #3–transduced cells accumulated in G1 phase compared with mock-transduced cells and SMAD6 knockdown affects cell cycle and induces apoptosis in cells overexpressing SMAD6 but have no growth-inhibitory effect on cells with no protein (Figs. 2–4). These observations suggest that lung cancer cells can become dependent on SMAD6 for survival.
TGF-β is a strong and fast-acting regulator of PAI-1 through SMAD2/3 phosphorylation (29, 30). shRNA-mediated knockdown of SMAD6 increased PAI-1 expression and this expression was abolished when Smad6 shRNA-transduced cells were treated with a TGF-β inhibitor, SB431542 (Fig. 5A and B). In our study, hyperphosphorylation of SMAD2/3 as well as hyperphosphorylation of JNK and c-Jun and dephosphorylation of RB were observed in cells with shRNA-mediated SMAD6 knockdown (Fig. 5C). These changes most likely contributed to the overall negative growth-inhibitory effects we observed because JNK pathway is implicated in multiple biological processes, including cellular proliferation, survival, and apoptosis (31). The growth arrest in G0-G1 phase that we observed in lung cancer cells by SMAD6 knockdown is, at least in part, due to RB hypophosphorylation because RB normally represses E2F, which blocks transcription of cyclins necessary for cell cycle progression (32). The results we have obtained in this study combined with previously published data suggest that the removal of SMAD6 negatively affects cell proliferation and increases apoptosis through the JNK/RB pathway.
Several studies have shown that SMAD6 effectively inhibits BMP signaling but only weakly inhibits that of TGF-β and activin (33, 34), whereas Smad7 ubiquitously inhibits TGF-β family signaling. Our Western experiments showed that SMAD7 is expressed uniformly and at much higher levels than SMAD6 in both normal and cancer cells derived from the lung but is induced to a much less extent on TGF-β and BMP stimulation (data not shown). Similarly, down-regulation of SMAD6 by shRNA did not affect the expression of Smad7 or affected genes commonly involved in BMP induction (18).
In our canonical pathway analysis using IPA, many of TGF-β signaling genes were affected by Smad6 knockdown. However, other pathways, including cell cycle, IL-6 signaling, and death receptor signaling pathways, were also affected (Supplementary Fig. S1). Recently, SMAD6 is shown to be a critical mediator of the TGF-β/BMP pathway that mediates anti-inflammatory activity (35). A role of SMAD6 in the nucleus has also been reported, showing that SMAD6 represses target genes through binding with a corepressor or the inhibition of DNA binding (36, 37). These data jointly show that SMAD6 can function to not only suppress the TGF-β signal pathway but also affect other growth regulatory pathways in lung cancer cells. This differential cellular effect could be exploited to benefit the patient with lung cancers that overexpress SMAD6. Consistent with our data, differential regulation by TGF-β signaling in normal and tumor lung has also been shown in a recent study using gene expression profiling (38).
In summary, we show that the reduced expression of SMAD6 is associated with increased tumor-related survival in NSCLC patients. Knockdown of SMAD6 results in transcriptional changes and signal transduction on TGF-β–related genes, such as the overexpression of PAI-1 and phosphorylation of SMAD2/3, JNK, and c-Jun (Fig. 5). SMAD6 reduction inhibits cancer cell growth and induces apoptosis in lung cancer cells. Our results reveal for the first time that SMAD6 plays a key role in the tolerance of lung cancer to inhibitory effects of TGF-β signaling and it could potentially be used as a therapeutic target for lung cancers with SMAD6 overexpression. We propose a model that SMAD6 expression contributes to TGF-β responsiveness in lung cancer cell, and knockdown of SMAD6 restores TGF-β–induced growth inhibition and reduces tumor burden in the lung cancer patient (Fig. 6).
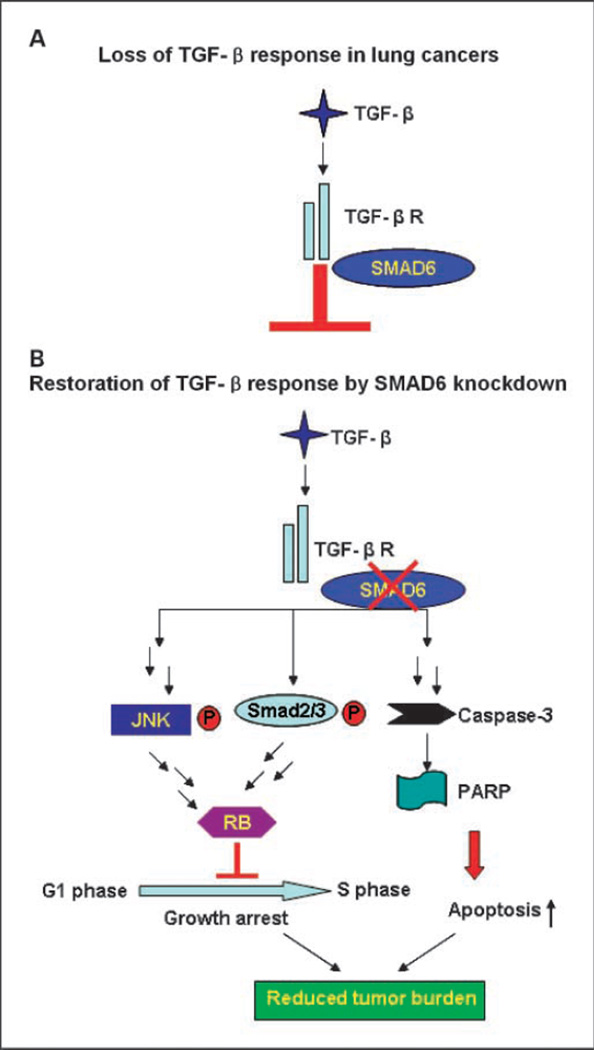
Figure 6.
Model of TGF-β restoration by SMAD6 knockdown. A, down-regulation of TGF-β signal pathway by SMAD6 in lung cancer. B, TGF-β signal pathway reactivation on SMAD6 knockdown. JNK and SMAD2/3 were phosphorylated by TGF-β signal pathway, and apoptosis pathway was also activated by caspase-3 and PARP activation leading to the reduction of tumor burden observed in lung cancer patients.
Supplementary Material
Acknowledgments
Grant support: Center for Cancer Research, National Cancer Institute.
We thank Drs. Lucy Anderson, Katy Flander, and members of Laboratory of Human Carcinogenesis for helpful comments; Dr. Hayoung Hwang for technical advice; and the NIH Fellows Editorial Board and Dorothea Dudek-Creaven for editorial assistance.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Massague J. TGF-β signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 2.Blobe GC, Schiemann WP, Lodish HF. Role of transforming growth factor β in human disease. N Engl J Med. 2000;342:1350–1358. doi: 10.1056/NEJM200005043421807. [DOI] [PubMed] [Google Scholar]
- 3.de Caestecker MP, Piek E, Roberts AB. Role of transforming growth factor-β signaling in cancer. J Natl Cancer Inst. 2000;92:1388–1402. doi: 10.1093/jnci/92.17.1388. [DOI] [PubMed] [Google Scholar]
- 4.Kato Y, Habas R, Katsuyama Y, Naar AM, He X. A component of the ARC/mediator complex required for TGFh/nodal signalling. Nature. 2002;418:641–646. doi: 10.1038/nature00969. [DOI] [PubMed] [Google Scholar]
- 5.Heldin CH, Miyazono K, ten Dijke P. TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 6.Miyazono K, Kusanagi K, Inoue H. Divergence and convergence of TGF-β/BMP signaling. J Cell Physiol. 2001;187:265–276. doi: 10.1002/jcp.1080. [DOI] [PubMed] [Google Scholar]
- 7.Imamura T, Takase M, Nishihara A, et al. Smad6 inhibits signalling by the TGF-β superfamily. Nature. 1997;389:622–626. doi: 10.1038/39355. [DOI] [PubMed] [Google Scholar]
- 8.Nakao A, Afrakhte M, Moren A, et al. Identification of Smad7, a TGFh-inducible antagonist of TGF-β signalling. Nature. 1997;389:631–635. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- 9.Hayashi H, Abdollah S, Qiu Y, et al. The MAD-related protein Smad7 associates with the TGFβ receptor and functions as an antagonist of TGFβ signaling. Cell. 1997;89:1165–1173. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- 10.Liu X, Lee J, Cooley M, Bhogte E, Hartley S, Glick A. Smad7 but not Smad6 cooperates with oncogenic ras to cause malignant conversion in a mouse model for squamous cell carcinoma. Cancer Res. 2003;63:7760–7768. [PubMed] [Google Scholar]
- 11.Kleeff J, Ishiwata T, Maruyama H, et al. The TGF-β signaling inhibitor Smad7 enhances tumorigenicity in pancreatic cancer. Oncogene. 1999;18:5363–5372. doi: 10.1038/sj.onc.1202909. [DOI] [PubMed] [Google Scholar]
- 12.Javelaud D, Mohammad KS, McKenna CR, et al. Stable overexpression of Smad7 in human melanoma cells impairs bone metastasis. Cancer Res. 2007;67:2317–2324. doi: 10.1158/0008-5472.CAN-06-3950. [DOI] [PubMed] [Google Scholar]
- 13.Azuma H, Ehata S, Miyazaki H, et al. Effect of Smad7 expression on metastasis of mouse mammary carcinoma JygMC(A) cells. J Natl Cancer Inst. 2005;97:1734–1746. doi: 10.1093/jnci/dji399. [DOI] [PubMed] [Google Scholar]
- 14.Osawa H, Nakajima M, Kato H, Fukuchi M, Kuwano H. Prognostic value of the expression of Smad6 and Smad7, as inhibitory Smads of the TGF-β superfamily, in esophageal squamous cell carcinoma. Anticancer Res. 2004;24:3703–3709. [PubMed] [Google Scholar]
- 15.Fukuoka J, Fujii T, Shih JH, et al. Chromatin remodeling factors and BRM/BRG1 expression as prognostic indicators in non-small cell lung cancer. Clin Cancer Res. 2004;10:4314–4324. doi: 10.1158/1078-0432.CCR-03-0489. [DOI] [PubMed] [Google Scholar]
- 16.Liu X, Sun Y, Constantinescu SN, Karam E, Weinberg RA, Lodish HF. Transforming growth factor β-induced phosphorylation of Smad3 is required for growth inhibition and transcriptional induction in epithelial cells. Proc Natl Acad Sci U S A. 1997;94:10669–10674. doi: 10.1073/pnas.94.20.10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Atfi A, Djelloul S, Chastre E, Davis R, Gespach C. Evidence for a role of Rho-like GTPases and stressactivated protein kinase/c-Jun N-terminal kinase (SAPK/JNK) in transforming growth factor β-mediated signaling. J Biol Chem. 1997;272:1429–1432. doi: 10.1074/jbc.272.3.1429. [DOI] [PubMed] [Google Scholar]
- 18.Nishanian TG, Kim J-S, Foxworth A, Waldman T. Suppression of tumorigenesis and activation of Wnt signaling by bone morphogenetic protein 4 in human cancer cells. Cancer Biol Ther. 2004;3:667–675. doi: 10.4161/cbt.3.7.965. [DOI] [PubMed] [Google Scholar]
- 19.Kong FM, Washington MK, Jirtle RL, Anscher MS. Plasma transforming growth factor-β1 reflects disease status in patients with lung cancer after radiotherapy: a possible tumor marker. Lung Cancer. 1996;16:47–59. doi: 10.1016/s0169-5002(96)00611-3. [DOI] [PubMed] [Google Scholar]
- 20.Akhurst RJ, Derynck R. TGF-β signaling in cancer—a double-edged sword. Trends Cell Biol. 2001;11:S44–S51. doi: 10.1016/s0962-8924(01)02130-4. [DOI] [PubMed] [Google Scholar]
- 21.Yanagisawa K, Osada H, Masuda A, et al. Induction of apoptosis by Smad3 and down-regulation of Smad3 expression in response to TGF-β in human normal lung epithelial cells. Oncogene. 1998;17:1743–1747. doi: 10.1038/sj.onc.1202052. [DOI] [PubMed] [Google Scholar]
- 22.Masui T, Wakefield LM, Lechner JF, LaVeck MA, Sporn MB, Harris CC. Type β transforming growth factor is the primary differentiation-inducing serum factor for normal human bronchial epithelial cells. Proc Natl Acad Sci U S A. 1986;83:2438–2442. doi: 10.1073/pnas.83.8.2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arteaga CL, Tandon AK, Von Hoff DD, Osborne CK. Transforming growth factor β: potential autocrine growth inhibitor of estrogen receptor-negative human breast cancer cells. Cancer Res. 1988;48:3898–3904. [PubMed] [Google Scholar]
- 24.Kimchi A, Wang XF, Weinberg RA, Cheifetz S, Massague J. Absence of TGF-β receptors and growth inhibitory responses in retinoblastoma cells. Science. 1988;240:196–199. doi: 10.1126/science.2895499. [DOI] [PubMed] [Google Scholar]
- 25.Kang SH, Bang YJ, Im YH, et al. Transcriptional repression of the transforming growth factor-β type I receptor gene by DNA methylation results in the development of TGF-β resistance in human gastric cancer. Oncogene. 1999;18:7280–7286. doi: 10.1038/sj.onc.1203146. [DOI] [PubMed] [Google Scholar]
- 26.Munoz-Antonia T, Li X, Reiss M, Jackson R, Antonia S. A mutation in the transforming growth factor β type II receptor gene promoter associated with loss of gene expression. Cancer Res. 1996;56:4831–4835. [PubMed] [Google Scholar]
- 27.Miyaki M, Iijima T, Konishi M, et al. Higher frequency of Smad4 gene mutation in human colorectal cancer with distant metastasis. Oncogene. 1999;18:3098–3103. doi: 10.1038/sj.onc.1202642. [DOI] [PubMed] [Google Scholar]
- 28.Xu J, Attisano L. Mutations in the tumor suppressors Smad2 and Smad4 inactivate transforming growth factor β signaling by targeting Smads to the ubiquitinproteasome pathway. Proc Natl Acad Sci U S A. 2000;97:4820–4825. doi: 10.1073/pnas.97.9.4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lund LR, Riccio A, Andreasen PA, et al. Transforming growth factor-β is a strong and fast acting positive regulator of the level of type-1 plasminogen activator inhibitor mRNA in WI-38 human lung fibroblasts. EMBO J. 1987;6:1281–1286. doi: 10.1002/j.1460-2075.1987.tb02365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM. Direct binding of Smad3 and Smad4 to critical TGFh-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103:239–252. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 32.Harbour JW, Dean DC. Rb function in cell-cycle regulation and apoptosis. Nat Cell Biol. 2000;2:E65–E67. doi: 10.1038/35008695. [DOI] [PubMed] [Google Scholar]
- 33.Hanyu A, Ishidou Y, Ebisawa T, Shimanuki T, Imamura T, Miyazono K. The N domain of Smad7 is essential for specific inhibition of transforming growth factor-β signaling. J Cell Biol. 2001;155:1017–1027. doi: 10.1083/jcb.200106023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hata A, Lagna G, Massague J, Hemmati-Brivanlou A. Smad6 inhibits BMP/Smad1 signaling by specifically competing with the Smad4 tumor suppressor. Genes Dev. 1998;12:186–197. doi: 10.1101/gad.12.2.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi K-C, Lee YS, Lim S, et al. Smad6 negatively regulates interleukin 1-receptor-Toll-like receptor signaling through direct interaction with the adaptor Pellino-1. Nat Immunol. 2006;7:1057–1065. doi: 10.1038/ni1383. [DOI] [PubMed] [Google Scholar]
- 36.Lin X, Liang Y-Y, Sun B, et al. Smad6 recruits transcription corepressor CtBP to repress bone morphogenetic protein-induced transcription. Mol Cell Biol. 2003;23:9081–9093. doi: 10.1128/MCB.23.24.9081-9093.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berghorn KA, Clark-Campbell PA, Han L, McGrattan M, Weiss RS, Roberson MS. Smad6 represses Dlx3 transcriptional activity through inhibition of DNA binding. J Biol Chem. 2006;281:20357–20367. doi: 10.1074/jbc.M603049200. [DOI] [PubMed] [Google Scholar]
- 38.Ranganathan P, Agrawal A, Bhushan R, et al. Expression profiling of genes regulated by TGF-β: differential regulation in normal and tumour cells. BMC Genomics. 2007;8:98. doi: 10.1186/1471-2164-8-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.