Abstract
Background
Metabolite levels within an individual vary over time. This within-individual variability, coupled with technical variability, reduces the power for epidemiological studies to detect associations with disease. Here, the authors assess the variability of a large subset of metabolites and evaluate the implications for epidemiologic studies.
Methods
Using LC-MS and GC-MS platforms, 385 metabolites were measured in 60 women at baseline and year-1 of the Shanghai Physical Activity Study, and observed patterns were confirmed in the Prostate, Lung, Colorectal, and Ovarian Cancer Screening study.
Results
Although the authors found high technical reliability (median intra-class correlation = 0.8), reliability over time within an individual was low. Taken together, variability in the assay and variability within the individual accounted for the majority of variability for 64% of metabolites. Given this, a metabolite would need, on average, a Relative Risk of 3 (comparing upper and lower quartiles of “usual” levels) or 2 (comparing quartiles of observed levels) to be detected in 38%, 74% and 97% of studies including 500, 1000, and 5000 individuals. Age, gender, and fasting status, factors which are often of less interest in epidemiological studies were associated with 30%, 67%, and 34% of metabolites, respectively, but the associations were weak, and explained only a small proportion of the total metabolite variability.
Conclusion
Metabolomics will require large, but feasible, sample sizes to detect the moderate effect sizes typical for epidemiological studies.
Impact
We offer guidelines for determining the sample sizes needed to conduct metabolomic studies in epidemiology.
Keywords: metabolomics, power, variance components, measurement error
Introduction
Metabolomics is the assessment of small molecules [1], often defined to be only those molecules participating in cellular metabolism, within a given biological system [2]. Modern methods, such as Nuclear Magnetic Resonance (NMR) and Mass Spectroscopy (MS) coupled with liquid chromatography (LC) or gas chromatography (GC) [3], can identify and quantify a large number of metabolites simultaneously within a biospecimen, capturing its metabolomic profile. These profiles have been used to predict the risk of diabetes [4, 5], diagnose prostate cancer [6], and identify biomarkers of Crohn’s disease [7]. While these initial studies have demonstrated the potential of metabolomics, several important issues need to be resolved before considering metabolomics as a tool for large epidemiological studies.
A common goal in epidemiology is to relate a “usual” [8] level of an exposure, such as blood pressure, vitamin D levels, or smoking status, with the risk of disease. Usual is an ambiguous term, but it might be loosely translated as the average level over the last month or, perhaps, year. To assess the potential association, epidemiological studies often rely on only a single measurement in time as an estimate or surrogate for an individual’s usual level. For characteristics that have large day-to-day variation or are measured with low technical reliability, the surrogate may poorly reflect the desired quantity. Given that it is the usual level that is likely to be associated with the disease, within-individual and technical variability will reduce the study’s power to detect and quantify the tested association [9–11].
There is a potential concern that a single metabolomic profile may poorly reflect usual levels. Several metabolites are already known to vary within an individual over time. For example, vitamin D levels vary with the seasons [12],estrogen levels vary with the menstrual cycle in pre-menopausal women [13, 14], and aldosterone, cortisol, and rennin levels follow a circadian rhythm [15]. On a shorter time scale, carbohydrate, lipid, and amino acids levels in the blood respond to dietary patterns, spiking sharply in the post-prandial period (1–2 hours after eating) [16]. However, recent studies have suggested that metabolomic profiles may be relatively stable [11, 17–20]. Floegel and colleagues found that the median intra-class correlation (ICC), over a fourth month interval, of 163 serum metabolites measured by MS was 0.57 [20] and Nicholson and colleagues found the stable proportion of biological variation, over a similar period, for 38 annotated plasma metabolites measured by NMR was, on average, 0.68 [11], with similar ICCs for a larger number of spectral peaks. Here, we extend this research by studying a larger set of 385 metabolites, by including non-fasting samples so as to represent samples typically collected in epidemiological studies, and by considering measurements separated by 1 year so as to capture the variability around persistent exposures, which are more likely to affect the risk of many diseases.
Our overarching goal is to provide key information needed to design metabolomics analyses in the context of large-scale epidemiological studies. Our first objective is to estimate the within-individual, technical, and between-individual variability in 385 plasma metabolites measured by LC/MS and GC/MS, when samples are collected as part of an epidemiological study. Higher between individual variability is desirable, because that encompasses the measurable differences that can be associated with disease. We assess these three sources of variability in 184 individuals from the Shanghai Physical Activity (SPA) study and confirm our observations in a smaller subsample from the Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial. Our second objective is to translate these estimates of variability into estimates of the study power [11] that can be expected for epidemiological studies, with a specific focus on large case/control and case/cohort studies. While our conclusions are based on our results observed for LC/GC MS, our methodological framework can evaluate other metabolomic platforms such as NMR.
Materials and Methods
Studies and Sample Collection
The Shanghai Women’s Health Study (SWHS) and Shanghai Men’s Health Study (SMHS) are prospective cohort studies that include 74,943 women (ages 40–70 years at study baseline) and 61,582 men (ages 40–75 years at baseline) from 8 communities in Shanghai, China between 1997 and 2006. The SPA study included a randomly selected subcohort residing in two communities [21, 22]. Participants were each enrolled for 1 year and provided EDTA plasma samples at the beginning (T0) and end of the study year (T1). Samples were stored at −70°C [23]. Our analysis includes all 106 women and 78 (out of 100) men who were enrolled in the first wave of recruitment, donated T1 plasma samples and have a valid Actigraph accelerometer measurement, a requirement for a complementary study of physical activity. The study included the 60 men with the most extreme levels of physical activity (30 high; 30 low) and 18 randomly selected men. The median age at T1 was 55 and 52 for men and women respectively, and 55% of the women were post-menopausal.
Metabolite levels were measured for all 184 individuals at T1 and a randomly selected subset of 60 women at T0. Although fasting was not required, 6, 4, and 21 of these 60 women reported to be fasting during the morning of sample collection at both T0 and T1, T0 only, and T1 only. Two replicate samples, needed to assess technical variability, were measured on 8 of the T0 samples.
The PLCO screening trial is a large randomized trial, starting in 1993, that examines the effects of screening on cancer-related outcomes in the United States [24]. Biological specimen were collected under a uniform protocol and placed in long term storage at −70°C in a common PLCO Biorepository at Frederick, MD [25]. Our analysis focused on 254 individuals, collected as healthy age and gender matched controls for 254 colorectal cancer cases. At baseline (T0), the median age for the 143 men and 111 women were 65 and 63 respectively, and 98% of the women reported being post menopausal. Metabolites were measured in serum samples from all 254 individuals at T0 and a randomly selected group of 30 individuals (14 women, 16 men) at year 1 (T1). To evaluate technical variability in PLCO, we used EDTA plasma samples collected as part of a separate pilot study that measured replicate samples collected during the fourth year (T4) of follow-up from 15 randomly selected healthy men. Previous studies have already demonstrated that plasma and serum metabolite profiles behave similarly [26]. Because of differences in the study populations, we do not perform a combined analysis, and instead, use our observations from PLCO, which has fewer individuals with multiple measurements, to confirm the SPA results.
Metabolite Measurement
Study samples were analyzed at the laboratory of Metabolon Inc. using ultra high performance liquid-phase chromatography and gas chromatography coupled with mass spectrometry and tandem mass spectrometry, as described previously [27, 28]. A non-targeted single extraction was used, followed by protein precipitation, to recover a diversity of metabolites. Relative quantities were obtained from MS peaks, and peaks were linked by informatics methods to metabolite identities. The list of measured metabolites includes, but is not limited to, amino acids, carbohydrates, fatty acids, androgens, and xenobiotics (Supplementary Table 1). Metabolites were individually normalized according to test-day.
Between-individual, Within-individual, and Technical Variability
For each metabolite, we can estimate the variance across all measurements. We consider the variance of the transformed quantity, the log of the peak intensity, as it is the quantity most commonly used in association studies. After log-transformation, metabolites were approximately normally distributed (supplementary figure 4). One goal is to decompose this total variance, , into three different components: the between subject variance, , which can also be considered the variance of the “usual” level in a population; the within subject variance, , which reflects the true year-to-year variability around the “usual” level within an individual; the technical variance or lab reproducibility, , which is the expected variance from two identical samples.
These three variance components can be combined into other quantities of interest.
The biological variance [29, 30] is .
The technical ICC is the proportion of the total variation that is attributable to biological variance, as opposed to random laboratory error. The technical ICC is a common measure of laboratory accuracy or reproducibility.
We denote the proportion of the population’s biological variability that is due to the variation across individuals, by
The usual measurement “error”, or the variation around an individual’s “usual” level, is . Larger values of this “error” in the usual measurement often imply lower power for an epidemiological study to detect associations. Therefore, we desire that the proportion, , of total variability attributable to between subject differences to be large. This proportion, , has also been known as the ICC [11, 20] in previous literature, but use here to avoid confusion with the technical ICC.
We can estimate each of the three variance components, and the other relevant quantities, by using linear mixed models with the normalized log-transformed metabolite level, Y, as the outcome and random effects for subject, S, and year, T, (nested within subject)[9]. Estimates of in PLCO are based only on individuals with samples collected at year T0 and T1, while estimates of technical ICC are based only on the 15 individuals with samples collected at T4.
| Equation (1) |
Evaluating Associations with Age, Fasting Status, and Gender
For each metabolite, we can further partition the sources of variation by expanding upon equation (1). We now consider covariates for subject i: Gi = gender, Fi = fasting status, and Ai = age quartile, with both gender and fasting status being binary, 0 or 1, variables.
We expand equation (1) by including fixed effects for age quartile, gender, and fasting status, which are respectively represented by α, γ, and ϕ in equation (2). The subscripts, Gi, Fi, and Ai, indicate that we are including the fixed effect appropriate for subject i. By adjusting out these factors, we can assess the percentage of variability attributable to between-subject differences within specific demographic subcohorts.
| Equation (2) |
We can now define the proportion of unexplained variability attributed to between subject differences using equation (2) as
We will also identify the variance attributable to age, gender, and fasting, denoted by σ2(age), σ2(gender), and σ2(fasting) to get a better idea of their overall influence on the metabolomic profile. Exact definitions of these variance components are provided in Appendix A, but we can now define the total variance by
and examine the proportion of the variance attributable to each of the three covariates
Furthermore, we can assess whether the covariates are significantly associated with metabolite levels, and obtain p-values, by performing an analysis of variance (anova) on the mixed models described by equation (2).
Global Summaries
Until now, we have focused on statistics that describe the behavior of a single metabolite. For each metabolite, we have described calculating , and ICCest, our estimates for and ICC, by fitting linear mixed models. However, we are also interested in statistics that can describe the global behavior of all metabolites that were reported in at least 90% of the samples. We will therefore report the proportion of these metabolites where the estimated parameters exceed 0.2, 0.5, or 0.8, and treat these proportions as estimates of the proportion of metabolites exceeding the corresponding threshold. As a global summary, we also estimate the proportion of these metabolites that are associated with age, gender, and fasting status by the maximum False Discovery Rate estimated among all metabolites.
Estimates of Power
Our objective is to estimate the expected power for a case/control study focused on a single disease. Specifically, we estimate the average power, or the proportion of true metabolite-disease associations that are expected to be discovered, accounting for the three sources of variability and the testing of multiple metabolites.
We assume that a study will collect n individuals, equally split between cases and controls and use a t-test, with the appropriate Bonferroni-corrected significance threshold, to test for an association between the disease and each metabolite. We then estimate the power to detect each metabolite given its variance components measured in SPA and an assumed effect size. Study-level power averaged these values over all metabolites. We also consider the scenario where there are 1 to 5 samples per individual.
For purposes of interpretation and to enable comparisons with previously reported studies, we define the effect size for a given metabolite to be the relative risk, RR, of disease for an individual in the top quartile of the usual metabolite levels, as compared to the bottom quartile. Note, we still presume the metabolites are normally distributed and assume a t-test is used in the study (Appendix B).
RESULTS
Measurement/Technical Variability
Within the 252 SPA samples, there were 567 observed metabolites. Of those 567 metabolites, 385 metabolites were observed in at least 90% of all samples and 341 were observed in 95% of all samples. We consider only those 385 most common metabolites for the remainder of this paper. Of those 385 metabolites, the identities of 254 had already been determined.
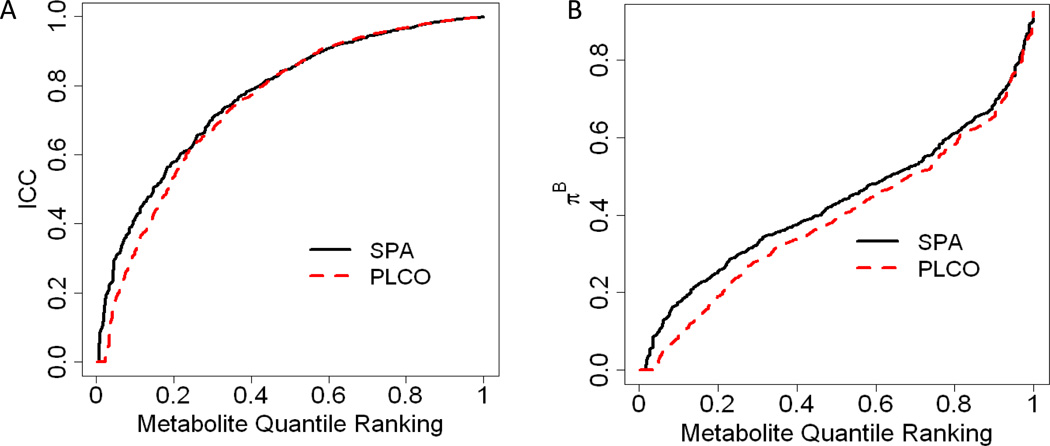
The majority of technical ICCs, a measure of the similarity between replicate samples, were high (figure 1A). With the SPA samples, the estimated ICCs for 57%, 85%, and 97% of the metabolites exceeded 0.8, 0.5, and 0.2 respectively (table 1). The distribution of ICCs were similar for all categories of metabolites (supplementary table 2). The distribution of CVs is illustrated in supplementary figure 3. When the analysis was repeated using the T4 samples from 15 men in PLCO, the distribution of estimated ICCs was nearly identical and is depicted by the red line in figure 1A. Among metabolites common to both studies, the reported ICCs were highly correlated (ρ=0.48) but far from identical (supplementary figure 1), as expected for two distinct populations, one from China and one from the US, and given the limited sample size.
Figure 1.
The plots illustrate the distributions of the technical ICCs, a measure of laboratory variability (figure A) and , a measure of between individual variability (figure B). The x-axis represents the metabolite quantile ranking (e.g. 0.5 represents the median), the y-axis represents the actual ICC (fig A) or (fig B) quantile, and the curves (black for SPA and dashed red for PLCO) show the ICC (fig A) or (fig B) quantile for the specified metabolite quantile ranking (e.g. the median ICC is 0.84 and is illustrated by the height of the black curve in figure A being 0.84 above the quantile ranking of 0.5).
Table 1.
The first row lists the percentage of metabolites in the Shanghai Physical Activity study with an estimated Intraclass Correlation Coefficient (ICC) that exceeds thresholds of 0.2, 0.5, and 0.8. The second row lists the percentages of metabolites where the estimated proportion of biological variability attributable to between-subject differences exceeds these same thresholds. The third row lists the percentages of metabolites where the estimated proportion of total variability attributable to between-subject differences exceeds these same thresholds.
| Parameter Threshold | ||||
|---|---|---|---|---|
| 0.2 | 0.5 | 0.8 | ||
| ICC | 97% | 85% | 57% | |
| 93% | 61% | 23% | ||
|
|
87% | 36% | 3.6% | |
Within and Between Individual Variability
Given only a single measurement, a study’s power to detect long-term epidemiological associations tends to be higher when , the proportion of total variability attributed to between subject differences, is larger. The estimates of were generally lower than the estimated ICC’s, with only 3.6%, 36%, and 87% of metabolites having exceeding 0.8, 0.5, and 0.2 respectively (figure 1B, table 1). The distribution of was not unimodal, with 23 identified and 13 unidentified metabolites with high values of above 0.7. The majority of these metabolites were in the biosynthesis pathway of androsterone or markers of specific dietary habits. The metabolites with lowest values of , among all metabolites with an ICCest > 0.8, were more heterogeneous and included multiple markers for episodically consumed foods (supplementary table 1).
Within specific age and gender demographic groups, study power will be limited by , the proportion of variability attributed to between subject differences after adjusting for the covariates in equation 2. Similar to the distribution of , 3.1%, 31%, and 83% of metabolites had values exceeding 0.8, 0.5, and 0.2. When estimating in a subgroup of only women and among only metabolites measured in women, results, again, were nearly unchanged with 3.7%, 33%, and 88% of metabolites having values of exceeding 0.8, 0.5, and 0.2. Even among the 36 metabolites with the highest estimates of , many of which were strongly associated with age and gender, the proportion of variation attributable to between subject differences after these adjustments only decreased minimally (table 2).
Table 2.
A list of the identified metabolites with the highest values of between-subject variability, (e.g. the lowest within-subject variability), among all metabolites. Rows include metabolite name, , the equivalent value from the age and gender adjusted (A.G.A) model, the equivalent from a female-only model, p-value for the metabolite’s association with age, and p-value for the metabolite’s association with gender.
|
A.G.A. |
Women |
p-value Age |
p-value Gender |
|||
|---|---|---|---|---|---|---|
| 1,5-anhydroglucitol (1,5-AG) | 0.91 | 0.91 | 0.92 | 0.88 | 0.32 | |
| 4-androsten-3β,17 β -diol disulfate 1 | 0.9 | 0.86 | 0.88 | 0.085 | <0.0001 | |
| pregnen-diol disulfate* | 0.9 | 0.87 | 0.89 | 0.018 | <0.0001 | |
| DHEA-S | 0.89 | 0.86 | 0.89 | 0.00039 | <0.0001 | |
| 4-androsten-3 β,17 β -diol disulfate 2 | 0.85 | 0.82 | 0.86 | 0.076 | <0.0001 | |
| pyroglutamine | 0.83 | 0.68 | 0.74 | <0.0001 | <0.0001 | |
| androsterone sulfate | 0.82 | 0.77 | 0.82 | 0.022 | <0.0001 | |
| andro steroid monosulfate 2 | 0.81 | 0.81 | 0.84 | 0.92 | 0.19 | |
| 5alpha-androstan-3 β,17 β -diol disulfate | 0.8 | 0.7 | 0.72 | 0.0064 | <0.0001 | |
| epiandrosterone sulfate | 0.79 | 0.73 | 0.78 | 0.033 | <0.0001 | |
| pseudouridine | 0.78 | 0.68 | 0.8 | <0.0001 | 0.43 | |
| pregn steroid monosulfate | 0.76 | 0.72 | 0.75 | 0.014 | <0.0001 | |
| 3-(4-hydroxyphenyl)lactate | 0.76 | 0.7 | 0.69 | 0.0044 | <0.0001 | |
| 21-hydroxypregnenolone disulfate | 0.76 | 0.74 | 0.77 | 0.13 | 0.00067 | |
| alpha-hydroxyisovalerate | 0.76 | 0.72 | 0.67 | 0.4 | <0.0001 | |
| C-glycosyltryptophan | 0.74 | 0.63 | 0.75 | <0.0001 | 0.15 | |
| urate | 0.74 | 0.72 | 0.73 | 0.14 | <0.0001 | |
| glutaroyl carnitine | 0.72 | 0.69 | 0.65 | 0.0037 | 0.00081 | |
| creatine | 0.72 | 0.62 | 0.55 | 0.00026 | <0.0001 | |
| 3-dehydrocarnitine | 0.72 | 0.71 | 0.71 | 0.87 | 0.32 | |
| 1-arachidonoylglycerophosphocholine | 0.72 | 0.7 | 0.74 | 0.3 | 0.15 | |
| 2-hydroxybutyrate (AHB) | 0.71 | 0.71 | 0.71 | 0.43 | 0.24 | |
| undecanoate (11:0) | 0.7 | 0.65 | 0.64 | 0.53 | <0.0001 |
We also measured in the individuals from PLCO. Although these measurements were from serum samples, the distribution of was nearly identical in this population (figure 1B), and there was high correlation (ρ=0.49) when comparing the estimates of between-subject variability among metabolites common to both groups. Supplementary Figure 2 confirms that those metabolites with high values of in SPA have similarly high values in PLCO.
Our study design permits the estimation of , the proportion of biological variability that can be attributed to between individual differences. Although these results are limited by our ability to distinguish technical and within-individual variability using only 8 replicate samples, the majority of natural variability appeared to be attributable to between-subject differences: 23%, 61%, and 93% of the metabolites having estimated values of exceeding 0.8, 0.5, and 0.2 (table 1). Again, adjusting for age and gender did not alter our estimates much: 22%, 62%, and 92% of metabolites had estimates of exceeding 0.8, 0.5, and 0.2.
Age, Gender, and Fasting Status
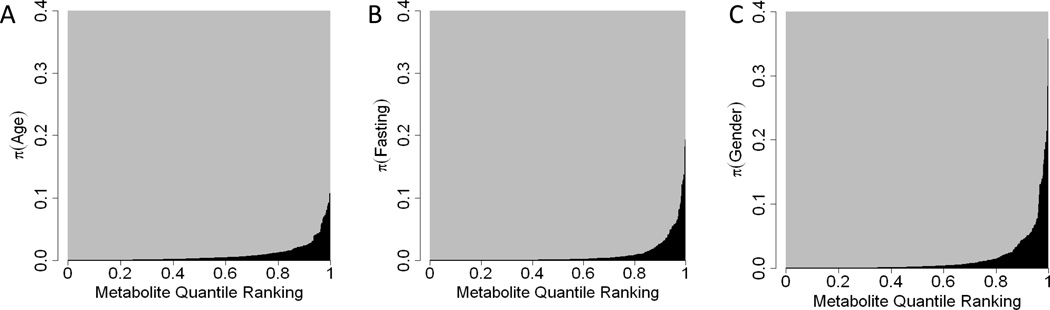
Those covariates suspected to have associations with metabolite levels were able to explain small, but statistically significant, proportions of the variation in many of the metabolites. We found that age, fasting status, and gender were correlated with 30%, 34%, and 67% of metabolites, respectively. Using the Bonferonni adjusted alpha level of 0.05/385, we find that 9.1%, 14.3%, and 7.3% of metabolites have a statistically significant association with age, fasting status, and gender, respectively. However, the proportion of the variability attributable to each metabolite was small, explaining why changed little after adjusting for covariates. Figure 2 shows the proportion of total variability attributed to these covariates.
Figure 2.
Figures A, B, and C illustrate the distribution of π̂(Age), π̂(Fasting), and π̂(Gender). The x-axis represents the metabolite quantile ranking (e.g. 0.5 is the median), the y-axis represents π̂, and the curves show the π̂ for the specified metabolite quantile ranking (e.g. the median π̂ is well below 0.01 for all three covariates).
Power
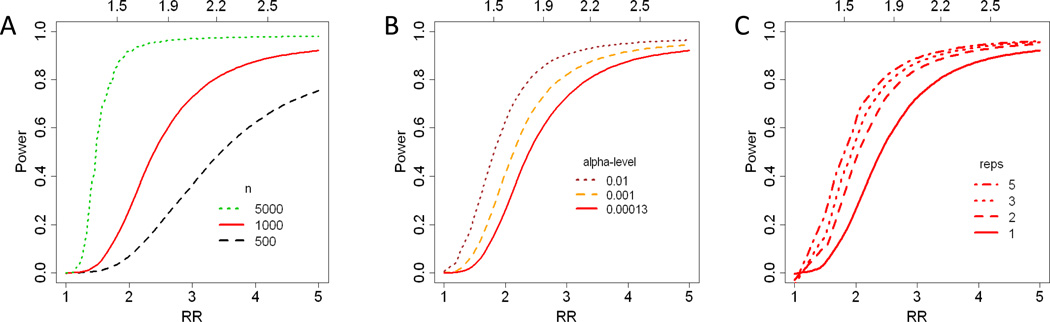
We quantified the effect size as the relative risk of disease when comparing individuals in the top and bottom quartiles of the usual metabolite level. However, when calculating power, we presumed a t-test comparing cases and controls. Given this definition of effect size and the assumption that all measured metabolites are equally likely to be associated with the disease, a case/control study with a total of 500 individuals is expected to detect <1%, 38%, and 75% of the metabolites with a RR of 1.5, 3.0, and 5.0 (figure 3A). Similarly, a study with 1000 individuals should detect 3%, 74%, and 92% and a study with 5000 individuals should detect 55%, 97%, and 98% of metabolites with a RR of 1.5, 3.0, and 5.0. All estimates assume a conservative Bonferroni-adjusted alpha level of 0.0013 = 0.05/385 (figure 3A, table 3). Although these relative risks are larger than typically reported in epidemiological studies, the naïve or observed relative risks would be lower and in-line with typical values. When the true relative risks are 1.5, 3.0, and 5.0, the naïve relative risks are expected to be 1.3, 2.0, and 2.8.
Figure 3.
The curves show the proportion of metabolites likely to be detected in a case/control study as a function of effect size. Effect size is defined by the relative risk (RR, x-axis) of disease when comparing individuals within the highest quartile of the “usual” metabolite level, as compared to the lowest quartile. The top axis indicates the “naïve” relative risk that would be observed in the specified case/control study when not adjusting for measurement error. Each figure varies one parameter: sample size, alpha level, or number of samples/individual. Figure A) Power for studies with 500 (black), 1000 (red), or 5000 (green) individuals (alpha-level = 0.00013 = 0.05/385). Figure B) Power for studies that define significance as a p-value below a threshold or alpha-level of 0.0013 = 0.05/385 (red), 0.001 (orange), and 0.01 (brown) (1000 individuals).Figure C) Power for studies with 1, 2, 3, or 5 distinct blood samples (1000 individuals). All measured metabolites are assumed equally likely to be associated with disease.
Table 3.
The entries list the average power to detect associations between metabolites and disease in a case/control study that has 500, 1000, and 5000 individuals and where the metabolites have true relative risks of 1.5, 3.0, and 5.0. These true values translate to naïve estimates for RR of 1.3, 2.0, and 2.8.
| Relative Risk | |||
|---|---|---|---|
| N | 1.5 | 3.0 | 5.0 |
| 500 | <1% | 38% | 75% |
| 1000 | 2.9% | 74% | 92% |
| 5000 | 55% | 97% | 98% |
We will detect higher proportions of those metabolites that have higher ICCs. Considering only those 36 metabolites with a above 0.7, a case/control study with 1000 individuals should detect 25%, 50%, and 80% of metabolites with a true RR of 1.7, 1.9, and 2.2. Focusing on only the 287, 142, and 36 metabolites with a exceeding 0.3, 0.5, and 0.7 would be equivalent to setting the alpha-threshold at 0.00017, 0.00035, and 0.0014.
If the most promising set of metabolites can be evaluated in a second stage of the study or if a complementary study can limit candidate pathways, requiring a family-wise-error rate of 0.05 would be unnecessarily strict. If we raise the alpha level to 0.001, a case/control study with 1000 individuals should detect 25%, 50%, and 80% of metabolites with a true RR of 1.8, 2.1, and 2.8. The corresponding naïve RR would be respectively 1.5, 1.6, and 2.0. Figure 3B compares the power for studies with an alpha-threshold of 0.01, 0.001, and 0.00013.
Power would be improved by collecting multiple samples from each individual. Additional samples reduce the within-individual and technical variability. Figure 3C illustrates the gains in power from taking 2, 3, or 5 samples throughout the year for a 1000 subject study, while assuming the correlation between any two measures is independent of time. Collecting a second sample increases the study’s power by 1.84×, 1.15×, and 1.05× when the RR are 2, 3, and 4 respectively.
Discussion
Our objective was to assess the potential role of metabolomics in large epidemiological studies, with a specific focus on case/control studies. We first demonstrated that although LC- and GS-MS produced reliable and reproducible results, there was also considerable within-individual variability. Approximately 40% of the biological variability, on average, could be attributed to variation occurring within an individual over time. Using our estimates of technical and within-individual variability, we then estimated the power for detecting metabolite-disease associations in epidemiological studies.
Although we assume associations will be tested by a t-test or linear regression, we quantify a metabolite’s effect size by the RR comparing individuals within the top and bottom quartiles of the metabolite’s distribution. We demonstrate the need for a large number of samples in case/control studies, and expect our figures relating RR to power, as well as the distributions of , σ̂2 and ICCest used to calculate that relationship, to serve as a guide for studies considering metabolomic profiling of samples. If laboratory variability can be reduced, perhaps by using a targeted approach of only a few metabolites, similar levels of power could be achieved with fewer individuals.
Our results corroborate and expand upon previous studies measuring metabolomic variability and estimating power for epidemiological studies [11, 20]. Our median 1 year of 0.43 was similar to an earlier targeted analysis of 163 metabolites that found a median 4 month (or ICC using their definition) to be 0.57 and an NMR analysis of 38 metabolites that also found a median 4 month around 0.57 (0.68/1.19), after accounting for technical variability. Our are likely lower, but perhaps more pertinent for planning epidemiological studies, because of the eight additional months between measurements and the fewer requirements imposed on study participants (e.g. no fasting). Unlike many metabolomic focused studies which control for diet [11, 20] or behavior [31], our samples were collected as part of SPA and PLCO and therefore the observed variability will likely be more similar to that reported in future epidemiological studies. Even with our slightly lower values, we reached the same qualitative conclusion as Nicholson [11], that studies will require large sample sizes, upwards of 1000 subjects, to detect metabolomic associations. We have further expanded upon Nicholson’s results by relating power to RR, considering different significance thresholds, and discussing the power from repeat measurements.
Studies should plan for, but not be discouraged by, the potentially high intra-individual variability. Strong associations between usual exposure levels and disease risk have allowed previous epidemiological studies to overcome imprecision of this magnitude. For example, in post-menopausal women, insulin and estradiol have of 0.68 and 0.59, respectively, over a span of 1–3 years [32, 33], but studies, nonetheless, have successfully detected their associations with breast cancer [34, 35]. Similarly, for heart-disease [36] and diabetes [4], studies have recently identified metabolites related to branch chain amino acids as important predictors of disease risk, with odds ratios ranging from 2 to 4 for top vs. bottom quartile comparisons. For smoking-related cancers, a comparison of high vs. low cotinine levels would be expected to yield odds ratios of up to 20+ [37].
Moreover, we demonstrated that although a reasonably high proportion of metabolites were associated with age, fasting status, and gender, these three covariates only accounted for a small proportion of the total variability. Therefore, metabolomic profiles can still be useful for distinguishing risks within specific demographic cohorts, in that within-cohort variation is still high. Similarly, metabolomic profiles can still be useful even when epidemiological studies did not impose dietary restrictions (e.g. fasting) before blood draws. These factors do little to affect our overall conclusions about detectable RRs.
Our study had five main limitations. First, the SPA dataset only contained measurements on women at two time points, and therefore within-subject variability results are gender specific. However, similar results were seen in the PLCO data set, which includes equal numbers of men and women. The second limitation is that we only calculated power for identifying disease associations with individual metabolites. There is also interest in identifying metabolic profiles, such as those created by partial least squares regression (PLS) [38], that can differentiate cases and controls and be used as a diagnostic tool. Third, LC/MS and GC/MS platforms do not report the actual metabolite levels, but peak intensities, and our estimates of parameters can be slightly sensitive to scale. Fourth, we only had a total of 23 replicates for assessing technical variability. While this may introduce imprecision for estimating the for individual metabolites, their distributions, across all metabolites, should be more accurate. Also, our power calculations combined within-individual and technical variability, and therefore were based on the larger sample set of 60 samples repeated over time. Finally, the may be underestimated if storage at −70°C had a variable effect on samples or if the additional year of storage had a significant impact on biomarker levels [39–41].
Even given the limitations of the study, we were able to assess the magnitude of each of the three variance components, and estimate the power for large case/control studies. Because the likely relative risks will depend on the disease, time between sample collection and disease diagnosis, and specimen type, we do not offer any universal conclusion about the utility of metabolomics in epidemiological studies. We do, however, strongly suggest considering our analysis of power when planning such studies.
Supplementary Material
Acknowledgments
This study is, in part, supported by the Intramural Research Program of the National Institutes of Health and the Breast Cancer Research Stamp Fund, awarded through competitive peer review. SWHS was supported by R37CA070867. We thank Dr. Mitch Gail (NCI) for valuable discussions.
Appendix A
We discuss the “variance” attributable to age, fasting status, and gender. However, we caution against any literal interpretation of their values, which is our reason for the quotes. We define the “variance” as the proportion of the total variance that can be explained by that covariate in a population where all categories are equally represented (e.g. 50% men/50% women and 50% fasting/50% non-fasting) and there is one sample per individual. Note that the variances are highly dependent on how we chose to categorize the variables and their distribution within SPA.
Dropping the quotes, we now define the variances for the three covariates to be
Where αk is the fixed effect for age quartile k, γ1 and γ0 are the fixed effects for gender, ϕ1 and ϕ0 are the fixed effects for fasting, and:
These variances, and their corresponding proportions π(Age), π(Fasting), and π(Gender), provide a measure of the influence of these three covariates on metabolite profiles.
Appendix B
This appendix provides the details for estimating power. If we define the effect of a metabolite in terms of its standard deviation and the mean difference between cases and controls, this calculation would be trivial. The appendix is only needed because we choose to define the effect by the more interpretable RR. Again, the relative risk (RR) is defined as the probability of disease for an individual in the top quartile of the usual metabolite levels, as compared to an individual in the bottom quartile:
| Equation (3) |
where D and X are random variables respectively indicating disease status and metabolite level, t0 is the threshold for the top quartile of X, and t1 is the threshold for the bottom quartile of X, i.e.:
| Equation (4) |
We assume that the usual metabolite level within cases and controls are each normally distributed with respective means at μRR/2 and −μRR/2 and a common variance, . Thus equation (3) can be reformulated as
where Z is a normal variable with mean = 0 and variance = 1. We further assume that the disease has a prevalence of 0.1, and equation (4) can be reformulated as
We can thus solve for μRR, t0, and t1 from the set of three equations above. For a given value of μRR, and false positive rate, α, the power for a case- control study is the probability that a chi-squared variable with non-centrality parameter and 1 degree of freedom (df) exceeds the 1− α quantile of a central chi-squared distribution with 1 df.
We have defined the effect size for a given metabolite in terms of the “usual” metabolite level. The listed relative risks will therefore be substantially higher than those reported in previous epidemiological studies that did not correct for measurement error. To assess whether the listed relative risks are reasonably in line with previous studies, we also report the naïve relative risk, RR’, or the uncorrected estimate. For each metabolite and each specified Relative Risk, we estimate the naïve relative risk by solving the following set of equations for RR’, t0’, and t1’, where μRR is calculated above and σB is replaced by σT:
We then estimate the average naïve relative risk for a given true relative risk across all metabolites.
Footnotes
The authors have no relationships that they believe could be construed as resulting in an actual, potential, or perceived conflict of interest.
References
- 1.Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, et al. HMDB: the Human Metabolome Database. Nucleic Acids Research. 2007;35:D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nicholson JK, Wilson ID. Understanding 'Global' Systems Biology: Metabonomics and the Continuum of Metabolism. Nat Rev Drug Discov. 2003;2:668–676. doi: 10.1038/nrd1157. [DOI] [PubMed] [Google Scholar]
- 3.Dettmer K, Aronov PA, Hammock BD. Mass spectrometry-based metabolomics. Mass Spectrometry Reviews. 2007;26:51–78. doi: 10.1002/mas.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suhre K, Meisinger C, Döring A, Altmaier E, Belcredi P, Gieger C, et al. Metabolic Footprint of Diabetes: A Multiplatform Metabolomics Study in an Epidemiological Setting. PLoS ONE. 2010;5:e13953. doi: 10.1371/journal.pone.0013953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abate-Shen C, Shen MM. Diagnostics: The prostate-cancer metabolome. Nature. 2009;457:799–800. doi: 10.1038/457799a. [DOI] [PubMed] [Google Scholar]
- 7.Jansson J, Willing B, Lucio M, Fekete A, Dicksved J, Halfvarson J, et al. Metabolomics Reveals Metabolic Biomarkers of Crohn's Disease. PLoS ONE. 2009;4:e6386. doi: 10.1371/journal.pone.0006386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dodd KW, Guenther PM, Freedman LS, Subar AF, Kipnis V, Midthune D, et al. Statistical Methods for Estimating Usual Intake of Nutrients and Foods: A Review of the Theory. Journal of the American Dietetic Association. 2006;106:1640–1650. doi: 10.1016/j.jada.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 9.Laird NM, Ware JH. Random-Effects Models for Longitudinal Data. Biometrics. 1982;38:963–974. [PubMed] [Google Scholar]
- 10.Carroll RJ. Encyclopedia of Biostatistics. John Wiley & Sons, Ltd; 2005. Measurement Error in Epidemiologic Studies. [Google Scholar]
- 11.Nicholson G, Rantalainen M, Maher AD, Li JV, Malmodin D, Ahmadi KR, et al. Human metabolic profiles are stably controlled by genetic and environmental variation. Mol Syst Biol. 2011;7 doi: 10.1038/msb.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Norman AW. Sunlight, season, skin pigmentation, vitamin D, and 25-hydroxyvitamin D: integral components of the vitamin D endocrine system. The American Journal of Clinical Nutrition. 1998;67:1108–1110. doi: 10.1093/ajcn/67.6.1108. [DOI] [PubMed] [Google Scholar]
- 13.Lee SJ, Lenton EA, Sexton L, Cooke ID. The effect of age on the cyclical patterns of plasma LH, FSH, oestradiol and progesterone in women with regular menstrual cycles. Human Reproduction. 1988;3:851–855. doi: 10.1093/oxfordjournals.humrep.a136796. [DOI] [PubMed] [Google Scholar]
- 14.Wallace M, Hashim YZHY, Wingfield M, Culliton M, McAuliffe F, Gibney MJ, et al. Effects of menstrual cycle phase on metabolomic profiles in premenopausal women. Human Reproduction. 2010;25:949–956. doi: 10.1093/humrep/deq011. [DOI] [PubMed] [Google Scholar]
- 15.Katz FH, Romfh P, Smith JA, Roper EF, Barnes JS, Boyd JB. Diurnal Variation of Plasma Aldosterone, Cortisol and Renin Activity in Supine Man. Journal of Clinical Endocrinology & Metabolism. 1975;40:125–134. doi: 10.1210/jcem-40-1-125. [DOI] [PubMed] [Google Scholar]
- 16.Secor S. Specific dynamic action: a review of the postprandial metabolic response. Journal of Comparative Physiology B: Biochemical, Systemic, and Environmental Physiology. 2009;179:146–152. doi: 10.1007/s00360-008-0283-7. [DOI] [PubMed] [Google Scholar]
- 17.Kaplan RC, Ho GYF, Xue X, Rajpathak S, Cushman M, Rohan TE, et al. Within-Individual Stability of Obesity-Related Biomarkers among Women. Cancer Epidemiology Biomarkers & Prevention. 2007;16:1291–1293. doi: 10.1158/1055-9965.EPI-06-1089. [DOI] [PubMed] [Google Scholar]
- 18.Kotsopoulos J, Tworoger SS, Campos H, Chung F-L, Clevenger CV, Franke AA, et al. Reproducibility of Plasma, Red Blood Cell, and Urine Biomarkers among Premenopausal and Postmenopausal Women from the Nurses' Health Studies. Cancer Epidemiology Biomarkers & Prevention. 2010;19:938–946. doi: 10.1158/1055-9965.EPI-09-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shah SH, Hauser ER, Bain JR, Muehlbauer MJ, Haynes C, Stevens RD, et al. High heritability of metabolomic profiles in families burdened with premature cardiovascular disease. Mol Syst Biol. 2009;5 doi: 10.1038/msb.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Floegel A, Drogan D, Wang-Sattler R, Prehn C, Illig T, Adamski J, et al. Reliability of Serum Metabolite Concentrations over a 4-Month Period Using a Targeted Metabolomic Approach. PLoS ONE. 2011;6:e21103. doi: 10.1371/journal.pone.0021103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peters TM, Moore SC, Xiang YB, Yang G, Shu XO, Ekelund U, et al. Accelerometer-Measured Physical Activity in Chinese Adults. American Journal of Preventive Medicine. 2010;38:583–591. doi: 10.1016/j.amepre.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peters TM, Shu X-O, Moore SC, Xiang YB, Yang G, Ekelund U, et al. Validity of a Physical Activity Questionnaire in Shanghai. Medicine & Science in Sports & Exercise. 2010;42:2222–2230. doi: 10.1249/MSS.0b013e3181e1fcd5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng W, Chow W-H, Yang G, Jin F, Rothman N, Blair A, et al. The Shanghai Women's Health Study: Rationale, Study Design, and Baseline Characteristics. American Journal of Epidemiology. 2005;162:1123–1131. doi: 10.1093/aje/kwi322. [DOI] [PubMed] [Google Scholar]
- 24.Prorok PC, Andriole GL, Bresalier RS, Buys SS, Chia D, David Crawford E, et al. Design of the prostate, lung, colorectal and ovarian (PLCO) cancer screening trial. Controlled Clinical Trials. 2000;21:273S–309S. doi: 10.1016/s0197-2456(00)00098-2. [DOI] [PubMed] [Google Scholar]
- 25.Hayes RB, Reding D, Kopp W, Subar AF, Bhat N, Rothman N, et al. Etiologic and early marker studies in the prostate, lung, colorectal and ovarian (PLCO) cancer screening trial. Controlled Clinical Trials. 2000;21:349S–355S. doi: 10.1016/s0197-2456(00)00101-x. [DOI] [PubMed] [Google Scholar]
- 26.Yu Z, Kastenmüller G, He Y, Belcredi P, Möller G, Prehn C, et al. Differences between Human Plasma and Serum Metabolite Profiles. PLoS ONE. 2011;6:e21230. doi: 10.1371/journal.pone.0021230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 28.Suhre K, Shin S-Y, Petersen A-K, Mohney RP, Meredith D, Wagele B, et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477:54–60. doi: 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lacher DA, Hughes JP, Carroll MD. Estimate of Biological Variation of Laboratory Analytes Based on the Third National Health and Nutrition Examination Survey. Clinical Chemistry. 2005;51:450–452. doi: 10.1373/clinchem.2004.039354. [DOI] [PubMed] [Google Scholar]
- 30.Fraser CG, Petersen PH. Desirable standards for laboratory tests if they are to fulfill medical needs. Clinical Chemistry. 1993;39:1447–1453. [PubMed] [Google Scholar]
- 31.Nieman DC, Gillitt ND, Henson DA, Sha W, Shanely RA, Knab AM, et al. Bananas as an Energy Source during Exercise: A Metabolomics Approach. PLoS ONE. 2012;7:e37479. doi: 10.1371/journal.pone.0037479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kabat GC, Kim M, Caan BJ, Chlebowski RT, Gunter MJ, Ho GYF, et al. Repeated measures of serum glucose and insulin in relation to postmenopausal breast cancer. International Journal of Cancer. 2009;125:2704–2710. doi: 10.1002/ijc.24609. [DOI] [PubMed] [Google Scholar]
- 33.Hankinson SE, Manson JE, Spiegelman D, Willett WC, Longcope C, Speizer FE. Reproducibility of plasma hormone levels in postmenopausal women over a 2–3-year period. Cancer Epidemiology Biomarkers & Prevention. 1995;4:649–654. [PubMed] [Google Scholar]
- 34.Gunter MJ, Hoover DR, Yu H, Wassertheil-Smoller S, Rohan TE, Manson JE, et al. Insulin, Insulin-Like Growth Factor-I, and Risk of Breast Cancer in Postmenopausal Women. Journal of the National Cancer Institute. 2009;101:48–60. doi: 10.1093/jnci/djn415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.James RE, Lukanova A, Dossus L, Becker S, Rinaldi S, Tjønneland A, et al. Postmenopausal Serum Sex Steroids and Risk of Hormone Receptor–Positive and -Negative Breast Cancer: a Nested Case–Control Study. Cancer Prevention Research. 2011;4:1626–1635. doi: 10.1158/1940-6207.CAPR-11-0090. [DOI] [PubMed] [Google Scholar]
- 36.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, DuGar B, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boffetta P, Clark S, Shen M, Gislefoss R, Peto R, Andersen A. Serum Cotinine Level as Predictor of Lung Cancer Risk. Cancer Epidemiology Biomarkers & Prevention. 2006;15:1184–1188. doi: 10.1158/1055-9965.EPI-06-0032. [DOI] [PubMed] [Google Scholar]
- 38.Trygg J, Wold S. Orthogonal projections to latent structures (O-PLS) Journal of Chemometrics. 2002;16:119–128. [Google Scholar]
- 39.Shih WJ, Bachorik PS, Haga JA, Myers GL, Stein EA. Estimating the Long-Term Effects of Storage at −70 °C on Cholesterol, Triglyceride, and HDL-Cholesterol Measurements in Stored Sera. Clinical Chemistry. 2000;46:351–364. [PubMed] [Google Scholar]
- 40.Rundle AG, Vineis P, Ahsan H. Design Options for Molecular Epidemiology Research within Cohort Studies. Cancer Epidemiology Biomarkers & Prevention. 2005;14:1899–1907. doi: 10.1158/1055-9965.EPI-04-0860. [DOI] [PubMed] [Google Scholar]
- 41.Tworoger SS, Hankinson SE. Collection, Processing, and Storage of Biological Samples in Epidemiologic Studies: Sex Hormones, Carotenoids, Inflammatory Markers, and Proteomics as Examples. Cancer Epidemiology Biomarkers & Prevention. 2006;15:1578–1581. doi: 10.1158/1055-9965.EPI-06-0629. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.