Abstract
Background
Studies suggest that vitamin D status may be associated with prostate cancer risk, although the direction and strength of this association differs between experimental and observational studies. Genome-wide association studies have identified genetic variants associated with 25-hydroxyvitamin D (25(OH)D) status. We examined prostate cancer risk in relation to SNPs in four genes shown to predict circulating levels of 25(OH)D.
Methods
SNP markers localized to each of four genes (GC, CYP24A1, CYP2R1, and DHCR7) previously associated with 25(OH)D were genotyped in 10,018 cases and 11,052 controls from the NCI Breast and Prostate Cancer Cohort Consortium. Logistic regression was used to estimate the individual and cumulative association between genetic variants and risk of overall and aggressive prostate cancer.
Results
We observed a decreased risk of aggressive prostate cancer among men with the allele in rs6013897 near CYP24A1 associated with lower serum 25(OH)D (per A allele, OR=0.86, 95%CI=0.80–0.93, p-trend=0.0002), but an increased risk for non-aggressive disease (per a allele: OR=1.10, 95%CI=1.04–1.17, p-trend=0.002). Examination of a polygenic score of the four SNPs revealed statistically significantly lower risk of aggressive prostate cancer among men with a greater number of low vitamin D alleles (OR for 6–8 vs. 0–1 alleles = 0.66, 95% CI = 0.44 – 0.98; p-trend=0.003).
Conclusions
In this large, pooled analysis, genetic variants related to lower 25(OH)D were associated with a decreased risk of aggressive prostate cancer.
Impact
Our genetic findings do not support a protective association between loci known to influence vitamin D levels and prostate cancer risk.
Keywords: Vitamin D, prostatic neoplasms, data pooling, genes, SNPs
Introduction
There is evidence that vitamin D compounds promote prostate cell differentiation and inhibit prostate cancer cell growth and invasion (1–3). In contrast to this basic research, a meta-analysis of epidemiologic studies including a total of 3,124 cases and 4,682 controls concluded there was no evidence that higher vitamin D status assessed by circulating 25-hydroxyvitamin D (25(OH)D) levels is associated with a reduced risk of prostate cancer (4). Furthermore, men with higher circulating 25(OH)D were recently reported to have a statistically significantly elevated risk of prostate cancer in one nested case-control analysis of 1,000 cases and 1,000 controls (5).
Two recent genome-wide association studies (GWAS) identified SNPs (including two not previously well-known) in or near four genes related to circulating 25(OH)D (6, 7), the primary circulating form of vitamin D. Considered the best indicator of vitamin D status (8), 25(OH)D is converted to its active form, 1,25-dihydroxyvitamin D (1,25(OH)2D), in the kidney and other organs (8). The four genes identified in the GWAS were: GC, which encodes vitamin D binding protein (DBP), the major carrier of vitamin D compounds in circulation; CYP24A1, which encodes the cytochrome p450 (CYP) 24-hydroxylase that initiates intracellular metabolism of 25(OH)D and 1,25(OH)2D to less bioactive species; CYP2R1, which encodes a key 25-hydroxylase responsible for conversion of vitamin D to 25(OH)D in the liver; and, DHCR7, which encodes the enzyme that catalyzes the conversion of 7-dehydrocholesterol, a vitamin D3 precursor, to cholesterol (6, 7).
In order to further elucidate the vitamin D-prostate cancer association, we examined prostate cancer risk in relation to genetic variants associated with 25(OH)D status identified in genome-wide association (GWAS) studies in a pooled analysis of 10,000 cases and 11,000 controls within the Breast and Prostate Cancer Cohort Consortium (BPC3).
Methods
Study Sample
Details of the BPC3 have been reported previously (9). Briefly, the BPC3 is a consortium effort encompassing nested case-control sets from the following cohort studies: the Alpha-Tocopherol, Beta-Carotene Cancer Prevention (ATBC) Study, the American Cancer Society Cancer Prevention Study II (CPS-II), the European Prospective Investigation into Cancer and Nutrition Cohort (EPIC – includes cohorts from Denmark, Great Britain, Germany, Greece, Italy, the Netherlands, Spain, and Sweden), the Health Professionals Follow-up Study (HPFS), the Multiethnic Cohort (MEC), the Physicians’ Health Study (PHS), and the Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial. Within each cohort, controls were matched to cases based on age, race/ethnicity, and region of recruitment, depending on the study. Because of the small number of non-white participants, the present analysis was restricted to men who reported being of Caucasian ancestry.
Written informed consent was obtained for all participants and each study was approved by its respective institutional review board (IRB). The IRBs for each study were: US National Cancer Institute and Finnish National Institute for Health and Welfare (ATBC Study), the Emory University School of Medicine IRB (CPS-II), Ethikkommission -Medizinische Fakultät Heidelberg and Imperial College Research Ethics Committee (EPIC), the IRB of Harvard School of Public Health (HPFS), the IRB at the University of Southern California and the IRB at the University of Hawaii (MEC), The Human Subjects Committee at Brigham and Women’s Hospital (PHS) and NCI Special Studies IRB (PLCO).
SNP selection and genotyping
We chose SNPs that were identified from GWAS of circulating 25(OH)D levels (6, 7): rs2282679 (GC), rs10741657 (CYP2R1), rs12785878 (DHCR7), and rs6013897 (CYP24A1). A recent paper reported that, collectively, these four SNPs explained a greater degree of the variation in circulating vitamin D levels (i.e., 5.2%) than a polygenic score that included 9,000 SNPs and explained only 0.16% of the variation (10). TaqMan genotyping was conducted at the Core Genotyping Research Laboratory of the U.S. National Cancer Institute, DKFZ (Heidelberg, Germany), and the University of Southern California for 8,881 cases and 9,265 controls from all seven cohorts. An additional 1,137 cases and 1,787 controls with previous BPC3 data for these genes from GWAS analyses were included. Details of the genome-wide scans have been described previously (11). Of the four primary SNPs, only rs2282679 was available from GWAS data, and surrogate SNPs with R2 values of 1.0 were selected for each of the other three SNPs: rs17217119 for rs6013897, rs2060793 or rs1993116 for rs10741657, and rs3794060, rs12800438, or rs7944926 for rs12785878. Because the primary findings were unchanged when the GWAS participants were excluded, the latter were retained in the final analysis.
Outcome assessment
Cases of prostate cancer were identified through cohort linkage with a population-based registry or from self-reports verified through medical records and pathology reports. Genotype data were available for 10,018 prostate cancer cases and 11,052 controls. Information on cancer stage was available for 86% of the cases and information on tumor grade was available for 88%. High stage cancer was defined as stage C or D at diagnosis (n=1,834) and high grade was defined as cases with Gleason sum >7 or cases that were histologically diagnosed as poorly differentiated or undifferentiated (n=1,843). Aggressive prostate cancer (n=3,066) was defined as a case that was either of high stage or high grade at diagnosis.
Collection and harmonization of non-genetic data
Each cohort collected self-reported information on baseline (pre-diagnostic) medical and lifestyle characteristics. These data were assembled by the data coordinating center using a common protocol for variable formatting aimed to retain the most detailed data without resulting in missing data for any study. The data collected and variable formats agreed upon were: age at diagnosis or selection as a control (except for MEC which provided the age at blood draw for controls) (years, continuous), height (cm, continuous), body mass index (BMI) (kg/m2, continuous), history of diabetes (yes, no), smoking status (never, current, former), and family history of prostate cancer (yes, no). Previously measured serum or plasma 25(OH) D concentrations were available for 6,030 participants included in this analysis. Any inconsistencies in the data were resolved through discussion between the data coordinating center and the individual cohorts. All data elements have been used in analyses published by the individual cohorts (as well as in prior BPC3 publications), and details of their collection and quality control can be found in these previous reports.
Statistical methods
Unconditional logistic regression was used to estimate the association between each SNP and risk of prostate cancer. The SNPs were coded based on the number of alleles (0,1,2) associated with lower circulating 25(OH)D levels (i.e., low vitamin D alleles) in the published GWAS studies (6, 7), rather than on the number of minor alleles. The mean circulating 25(OH)D levels by genotype of each individual SNP for the 6,030 individuals with previously measured plasma or serum 25(OH)D are as follows (in nmol/L): rs2282679: GG=56.6, TT=64.6; rs6013987: AA=58.9, TT=62.2; rs10741657: GG=58.6, AA=64.2; rs12785878: GG=56.1, TT=65.3). Circulating 25(OH)D was statistically significantly linearly associated with genotype for each of the SNPs examined with the exception of rs6013897 (p values for correlation: rs2282679 < 1.0 × 10−30, rs6013897 = 0.45, rs10741657 = 9.6 × 10−14, rs12785878 = 9.7 × 10−10). Additionally, the four SNPs were combined to create a polygenic score that ranged from 0–8 low vitamin D alleles. Because few men had 0, 7, or 8 low vitamin D alleles, those with 6, 7, or 8 alleles were merged into one category, as were those with 0 or 1 alleles. The polygenic score, ranging from 0–8, was linearly associated with circulating 25(OH)D, with median concentrations (nmol/L) of 65, 61, 58, 54, 53, and 43 for men with score values of 0–1, 2, 3, 4, 5, and 6–8, respectively, which represents 44% lower average levels for men with the highest versus the lowest score (p for correlation < 1.0 × 10−30). Individual SNPs and the genetic score were analyzed in two ways. First, by entering separate indicator variables for the number of low vitamin D alleles into the regression model using 0 alleles as the referent group for the individual SNP analyses and using 0–1 alleles as the referent group for the score analysis. Second, by including in the model the ordinal variable for the number of low vitamin D alleles ranging from 0–2 each for the individual SNP analyses and from 0–8 for the polygenic score analysis to estimate the per-allele difference in risk of prostate cancer. All models were adjusted for study cohort and age in 5-year categories.
Exploratory subgroup analyses were conducted for strata based on the medians of age, BMI, and height, and by family history of prostate cancer (yes, no), history of diabetes (yes, no), and smoking status (never, ever). Statistical interaction was assessed using the likelihood ratio test. The statistical test for heterogeneity across studies is based on the test for interaction between study and the genetic variable. For our exploratory subgroup analyses, we established a significance threshold of 0.002, given that we conducted 30 tests for interaction without any a priori hypotheses. For all other analyses, p<0.05 was considered statistically significant.
Results
Characteristics of the study sample are shown in Table 1. A notable difference across cohorts is the proportion of aggressive cancers diagnosed, with all studies except EPIC having ascertained more non-aggressive than aggressive cases (particularly in HPFS, MEC, and PLCO) (Table 1).
Table 1.
Characteristics of the study populations
| ATBC | CPS2 | HPFS | MEC | PHS | EPIC | PLCO | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||
| Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | |
| N | 929 | 1,584 | 2,184 | 2,278 | 1,367 | 1,349 | 667 | 734 | 1,447 | 1,361 | 1,173 | 1,443 | 2,251 | 2.303 |
|
| ||||||||||||||
| Age* | ||||||||||||||
| Mean (sd) | 69.6 (5.6) | 70.9 (5.8) | 70.2 (5.8) | 70.2 (5.8) | 69.6 (7.4) | 67.6 (7.6) | 69.0 (7.5) | 70.4 (7.3) | 70.3 (7.6) | 70.0 (7.6) | 65.3 (6.3) | 65.5 (6.3) | 67.5 (5.6) | 68.9 (6.7) |
|
| ||||||||||||||
| Family History | ||||||||||||||
| Yes | 51 (5.5) | 41 (2.6) | 466 (21.3) | 308 (13.5) | 215 (15.7) | 151 (11.2) | 76 (11.4) | 59 (8.0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 256 (11.4) | 154 (6.7) |
| Missing | 121 (13.0) | 175 (11.1) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 37 (5.6) | 35 (4.8) | 1,447 (100) | 1,361 (100) | 1,173 (100) | 1,443 (100) | 51 (2.3) | 51 (2.2) |
|
| ||||||||||||||
| BMI | ||||||||||||||
| Mean (sd) | 26.2 (3.5) | 26.3 (3.6) | 26.0 (3.3) | 26.3 (3.5) | 25.7 (3.3) | 25.8 (3.6) | 26.3 (3.4) | 26.5 (3.9) | 24.7 (2.4) | 24.7 (2.6) | 26.5 (3.3) | 26.7 (3.4) | 27.3 (3.9) | 27.5 (4.1) |
| Missing (%) | 0 (0) | 1 (0.6) | 28 (1.3) | 31 (1.4) | 180 (13.2)) | 167 (12.4) | 0 (0) | 1 (0.1) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 32 (1.4) | 31 (1.4) |
|
| ||||||||||||||
| Height | ||||||||||||||
| Mean (sd) | 173.7 (6.3) | 173.6 (6.4) | 178.4 (6.5) | 178.7 (6.7) | 178.2 (6.7) | 178.2 (6.6) | 177.4 (6.8) | 177.4 (7.3) | 178.8 (6.2) | 178.3 (6.5) | 173.5 (6.8) | 173.5 (6.9) | 178.5 (6.6) | 178.3 (6.7) |
| Missing (%) | 0 (0) | 1 (0.6) | 19 (0.9) | 15 (0.7) | 105 (7.7) | 98 (7.3) | 0 (0) | 1 (0.1) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 22 (1.0) | 14 (0.6) |
|
| ||||||||||||||
| Smoking (%) | ||||||||||||||
| Never | 0 (0) | 0 (0) | 788 (36.1) | 837 (36.7) | 622 (45.5) | 557 (41.3) | 233 (34.9) | 243 (33.1) | 698 (48.2) | 682 (50.1) | 423 (36.1) | 498 (34.5) | 934 (41.5) | 730 (31.7) |
| Current | 929 (100) | 1,584 (100) | 126 (5.8) | 99 (4.4) | 62 (4.5) | 51 (3.8) | 66 (9.9) | 83 (11.3) | 128 (8.9) | 112 (8.2) | 250 (21.3) | 339 (23.5) | 186 (8.3) | 441 (19.2) |
| Former | 0 (0) | 0 (0) | 1,262 (57.8) | 1,336 (58.7) | 616 (45.1) | 659 (48.9) | 363 (54.4) | 404 (55.0) | 619 (42.3) | 567 (41.7) | 477 (40.7) | 585 (40.5) | 1,129 (50.2) | 1,132 (49.2) |
| Missing | 0 (0) | 0 (0) | 8 (0.4) | 6 (0.3) | 67 (4.9) | 82 (6.1) | 5 (0.8) | 4 (0.5) | 2 (0.1) | 0 (0) | 23 (2.0) | 21 (1.5) | 2 (0.1) | 0 (0) |
|
| ||||||||||||||
| Diabetes (%) | ||||||||||||||
| Ever | 53 (5.7) | 93 (5.9) | 338 (15.5) | 440 (19.3) | 105 (7.7) | 94 (7.0) | 55 (8.3) | 62 (8.5) | 0 (0) | 0 (0) | 45 (3.8) | 79 (5.5) | 296 (13.2) | 398 (17.3) |
| Missing | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1,447 (100) | 1,361 (100) | 216 (18.4) | 29 (20.6) | 46 (2.0) | 43 (1.9) |
|
| ||||||||||||||
| High Grade †(%) | ||||||||||||||
| No | 568 (75.1) | NA | 1,521 (79.6) | NA | 1,039 (89.3) | NA | 461 (69.1) | NA | 1,103 (78.4) | NA | 511 (78.1) | NA | 1,750 (78.2) | NA |
| Yes | 188 (24.9) | NA | 391 (20.4) | NA | 124 (10.7) | NA | 206 (30.9) | NA | 303 (21.6) | NA | 143 (21.9) | NA | 488 (21.8) | NA |
| Missing | 173 | NA | 272 | NA | 204 | NA | 0 | NA | 41 | NA | 519 | NA | 13 | NA |
|
| ||||||||||||||
| High Stage‡(%) | ||||||||||||||
| No | 492 (67.7) | NA | 1,673 (78.4) | NA | 1,010 (87.1) | NA | 567 (87.0) | NA | 600 (71.2) | NA | 506 (57.9) | NA | 1,956 (86.9) | NA |
| Yes | 235 (32.3) | NA | 460 (21.6) | NA | 149 (12.9) | NA | 85 (13.0) | NA | 243 (28.8) | NA | 368 (42.1) | NA | 294 (13.1) | NA |
| Missing | 202 | NA | 51 | NA | 208 | NA | 15 | NA | 604 | NA | 299 | NA | 1 | NA |
|
| ||||||||||||||
| Aggressive§ (%) | ||||||||||||||
| No | 385 (53.5) | NA | 1,231 (62.7) | NA | 912 (79.7) | NA | 408 (73.4) | NA | 519 (54.6) | NA | 259 (37.5) | NA | 1,589 (70.8) | NA |
| Yes | 335 (46.5) | NA | 732 (37.3) | NA | 232 (20.3) | NA | 248 (26.6) | NA | 432 (45.4) | NA | 431 (62.5) | NA | 656 (29.2) | NA |
| Missing | 209 | NA | 221 | NA | 223 | NA | 11 | NA | 496 | NA | 483 | NA | 6 | NA |
Age is calculated at age of diagnosis/selection as control except for MEC (at blood draw for controls).
Defined as Gleason sum 8–10 or WHO grade III. Percentages calculated based on participants without missing information.
Defined as stage CD. Percentages calculated based on participants without missing information.
Defined as either high stage or high grade. Percentages calculated based on participants without missing information.
We observed no statistically significant associations between prostate cancer risk and the genetic variants in rs2282679 in GC, rs6013897 near CYP24A1, or rs12785878 near DHCR7 (Table 2). The data suggest that men carrying one or two copies of the allele of rs10741657 near CYP2R1 that has been associated with lower vitamin D status have a borderline decrease in prostate cancer risk (GG vs. AA: OR=0.92, 95% CI=0.84 – 1.00, p=0.05, uncorrected for multiple testing; Table 2). There was no observed heterogeneity across studies for the prostate cancer – SNP associations with the exception of rs6013897 (p=0.001, Table 2), which could be attributed to the EPIC cohort. Excluding data from that study did not, however, materially alter the finding for this SNP (AT vs. TT, OR=1.09, 95% CI=1.02 – 1.17; AA vs. TT, OR=1.01, 95% CI=0.87 – 1.16; additive OR=1.05, 95% CI=1.00 – 1.11; p for heterogeneity=0.19). Assuming no directionality in the vitamin D-prostate cancer association using the likelihood ratio test to compare a logistic regression model that included all four SNPs with one that included none, we did not observe a statistically significant association with overall risk (p=0.15).
Table 2.
Individual SNP associations with risk of overall prostate cancer
| SNP | Gene | Low vitamin D allele frequency | # Low vitamin D alleles | Mean serum 25(OH)D§ | # Cases | # Controls | OR (95% CI)* | p-value† | p for heterogeneity across studies |
|---|---|---|---|---|---|---|---|---|---|
| rs2282679 | GC | ||||||||
| TT | 0.27 | 0 | 64.6 | 5,127 | 5,585 | 1.0 (ref) | 0.24 | 0.49 | |
| GT | 1 | 59.4 | 3,773 | 4,213 | 0.95 (0.90 – 1.01) | ||||
| GG | 2 | 56.6 | 721 | 774 | 0.98 (0.88 – 1.10) | ||||
| Additive (per allele) | 0.97 (0.93 – 1.02) | 0.22 | |||||||
|
| |||||||||
| rs6013897 | CYP24A1 | ||||||||
| TT | 0.50 | 0 | 62.2 | 6,026 | 6,433 | 1.0 (ref) | 0.26 | 0.001 | |
| AT | 1 | 63.0 | 2,754 | 2,774 | 1.05 (0.99 – 1.12) | ||||
| AA | 2 | 58.9 | 436 | 481 | 0.98 (0.86 – 1.12) | ||||
| Additive (per allele) | 1.02 (0.97 – 1.07) | 0.39 | |||||||
|
| |||||||||
| rs10741657 | CYP2R1 | ||||||||
| AA | 0.61 | 0 | 64.2 | 1,505 | 1,530 | 1.0 (ref) | 0.14 | 0.75 | |
| AG | 1 | 62.1 | 4,392 | 4,667 | 0.95 (0.87 – 1.03) | ||||
| GG | 2 | 58.6 | 3,481 | 3,789 | 0.92 (0.84 – 1.00) | ||||
| Additive (per allele) | 0.96 (0.92 – 1.00) | 0.05 | |||||||
|
| |||||||||
| rs12785878 | DHCR7 | ||||||||
| TT | 0.29 | 0 | 65.3 | 4,979 | 5,221 | 1.0 (ref) | 0.57 | 0.73 | |
| GT | 1 | 59.1 | 3,816 | 4,047 | 1.02 (0.96 – 1.09) | ||||
| GG | 2 | 56.1 | 825 | 957 | 0.97 (0.88 – 1.08) | ||||
| Additive (per allele) | 1.00 (0.96 – 1.05) | 0.97 | |||||||
Adjusted for age (5-year groups) and study cohort.
For categorical analyses of genotype this is a 2 degree of freedom test comparing a model containing indicator variables for genotype with a model containing only the covariates. For the per allele analyses, this is a 1 degree of freedom test comparing a model containing the ordinal genotype variable to a model containing only the covariates.
In nmol/L; among the 6,030 participants who had measured serum 25(OH)D available.
The estimated magnitude of the association for rs10741657 was greater for non-aggressive disease (GG vs. AA, OR=0.88, 95% CI=0.80 – 0.98, p=0.03) than for aggressive disease (GG vs. AA, OR=0.97, 95%CI=0.85 – 1.10, p=0.61) (Table 3). Our findings for the other SNPs in relation to aggressive disease were similar to those for overall prostate cancer with the exception of rs6013897 which showed an additive per-allele positive association with non-aggressive disease and an inverse relation with aggressive disease (per A allele: non-aggressive OR=1.10, 95% CI=1.04 – 1.17, p-trend=0.002; aggressive OR=0.86, 95% CI=0.80 – 0.93, p-trend=0.0002; Table 3). Exploratory subgroup analyses showed no statistically significant (i.e., p<0.002) interactions between the vitamin D genetic variants, prostate cancer, and any of the factors examined, including age, family history of prostate cancer, and body mass index (data not shown).
Table 3.
Individual SNP associations with risk of aggressive and non-aggressive prostate cancer
| SNP | Gene | # Controls | Non-Aggressive | Aggressive | ||||
|---|---|---|---|---|---|---|---|---|
| # Cases | OR (95% CI) * | p-value† | # Cases | OR (95% CI)* | p-value† | |||
| rs2282679 | GC | |||||||
| TT | 5,585 | 2,650 | 1.0 (ref) | 0.30 | 1,585 | 1.0 (ref) | 0.52 | |
| GT | 4,213 | 1,949 | 0.94 (0.88 – 1.02) | 1,189 | 0.98 (0.90 – 1.06) | |||
| GG | 774 | 376 | 0.97 (0.85 – 1.11) | 239 | 1.07 (0.92 – 1.25) | |||
| Additive (per allele) | 0.97 (0.92 – 1.02) | 0.23 | 1.01 (0.95 – 1.08) | 0.81 | ||||
|
| ||||||||
| rs6013897 | CYP24A1 | |||||||
| TT | 6,433 | 3,141 | 1.0 (ref) | 2.6 × 10−5 | 1,867 | 1.0 (ref) | 1.7 × 10−8 | |
| AT | 2,774 | 1,620 | 1.19 (1.11 – 1.29) | 596 | 0.74 (0.66 – 0.82) | |||
| AA | 481 | 227 | 1.00 (0.84 – 1.18) | 141 | 1.02 (0.84 – 1.24) | |||
| Additive (per allele) | 1.10 (1.04 – 1.17) | 0.002 | 0.86 (0.80 – 0.93) | 0.0002 | ||||
|
| ||||||||
| rs10741657 | CYP2R1 | |||||||
| AA | 1,530 | 797 | 1.0 (ref) | 0.07 | 447 | 1.0 (ref) | 0.27 | |
| AG | 4,667 | 2,229 | 0.92 (0.83 – 1.02) | 1,438 | 1.04 (0.92 – 1.18) | |||
| GG | 3,789 | 1,782 | 0.88 (0.80 – 0.98) | 1,080 | 0.97 (0.85 – 1.10) | |||
| Additive (per allele) | 0.94 (0.90 – 0.99) | 0.03 | 0.97 (0.92 – 1.03) | 0.34 | ||||
|
| ||||||||
| rs12785878 | DHCR7 | |||||||
| TT | 5,221 | 2,614 | 1.0 (ref) | 0.40 | 1,561 | 1.0 (ref) | 0.90 | |
| GT | 4,047 | 1,962 | 1.04 (0.97 – 1.12) | 1,183 | 1.00 (0.91 – 1.09) | |||
| TT | 957 | 411 | 0.96 (0.85 – 1.10) | 263 | 0.97 (0.83 – 1.12) | |||
| Additive (per allele) | 1.01 (0.95 – 1.06) | 0.86 | 0.98 (0.93 – 1.05) | 0.71 | ||||
djusted for age (5-year groups) and study cohort.
For categorical analyses of genotype this is a 2 degree of freedom test comparing a model containing indicator variables for genotype with a model containing only the covariates. For the per allele analyses, this is a 1 degree of freedom test comparing a model containing the ordinal genotype variable to a model containing only the covariates.
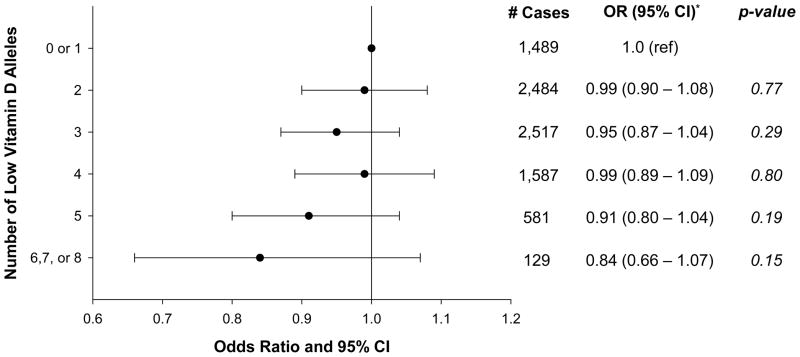
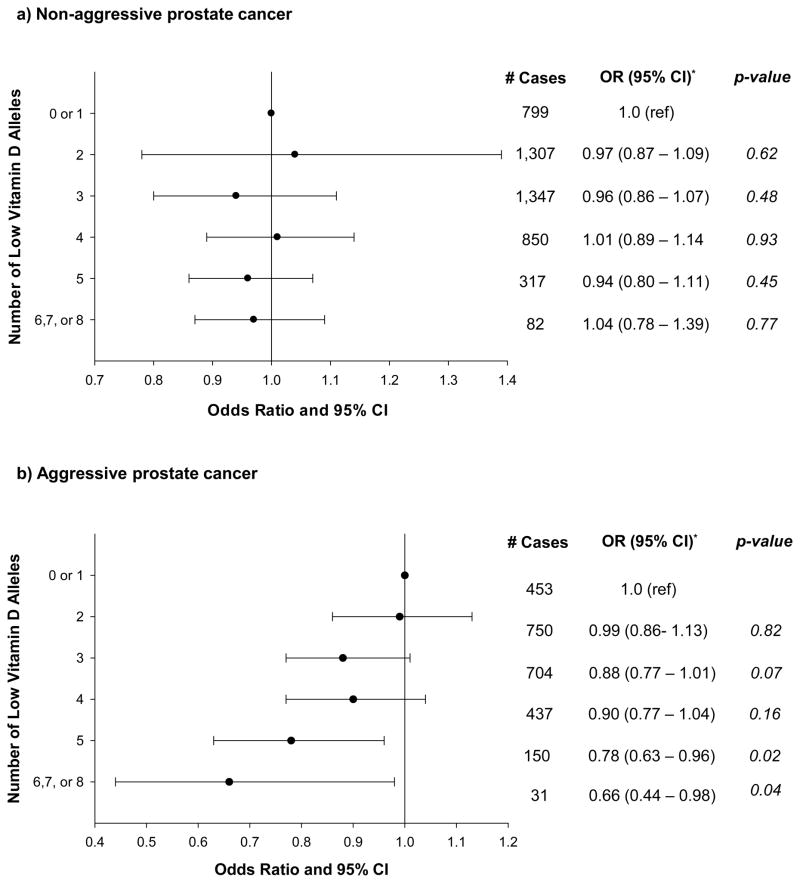
Overall prostate cancer risk was non-statistically significantly lower among men with a greater number of low vitamin D alleles (OR for 6–8 vs. 0–1 alleles = 0.84, 95%CI = 0.66 – 1.07; per-allele OR=0.98, 95% CI = 0.96 – 1.01; p-trend=0.17; Figure 1), an association that was similar across cohorts (p for heterogeneity in trend = 0.11). The magnitude of the association was greater, however, and the association was statistically suggestive for aggressive disease (OR for 6–8 vs. 0–1 alleles = 0.66, 95% CI = 0.44 – 0.98; per-allele OR=0.95, 95%CI = 0.92 – 0.98; p-trend=0.003; Figure 2). In a sensitivity analysis, men with 6 low vitamin D alleles had a statistically significantly decreased risk of prostate cancer compared to men with 0–1 alleles (OR=0.72, 95% CI=0.55 – 0.94), and those with 7–8 alleles had an increased risk (OR=1.91, 95% CI=1.02–3.58); the latter category included only 44 men (29 prostate cancer cases), however. We observed a similar pattern between the vitamin D genetic score and aggressive prostate cancer (6 vs. 0–1, OR=0.51, 95% CI=0.32 – 0.81; 7–8 vs. 0–1, OR=2.17, 95% CI=0.94 – 5.03). Excluding rs6013897 from the score resulted in an attenuated association with aggressive disease (6 vs. 0–1 alleles, OR=0.88, 95% CI=0.63–1.22; p-trend=0.39), and a statistically significant inverse relation with overall prostate cancer (6 vs. 0–1 alleles, OR=0.78, 95% CI=0.62–0.96), although the trend test was marginally not statistically significant (p=0.06). The results in Figures 1 and 2 were not altered by the removal of any of the other three SNPs from the full score or by adjustment for family history of prostate cancer.
Figure 1.
Association* Between 4-SNP Score and Risk of Overall Prostate Cancer
* - Adjusted for age (5-year groups) and study
Figure 2.
Association* Between 4-SNP Score and Risk of Aggressive and Non-Aggressive Prostate Cancer
*-Adjusted for age (5-year groups) and study
Discussion
In this large, pooled analysis, we found evidence that genetic variants previously related to lower vitamin D status are associated with a decreased risk of prostate cancer. A SNP near CYP2R1 was marginally associated with risk of overall prostate cancer, while one near CYP24A1 was associated with aggressive disease. The 4-SNP polygenic score was related to both overall and aggressive prostate cancer. These genetic findings indirectly support a role for vitamin D in the etiology of prostate cancer.
We observed a borderline, nominally statistically significant association for rs10741657 near CYP2R1, the gene encoding a key vitamin D 25-hydroxylation enzyme, such that men with alleles conferring lower vitamin D status were at decreased risk of overall prostate cancer. Similarly, the low vitamin D allele in rs6013897 near CYP24A1, the gene encoding the 24-hydroxylase that initiates intracellular catabolism of 25(OH)D and 1,25(OH)2D, was associated with a reduced risk of aggressive, but not non-aggressive, disease. Study heterogeneity in our findings for rs6013897 could be explained by the differences in the proportion of aggressive disease diagnosed across cohorts, a conclusion supported by the disappearance of heterogeneity following exclusion of the one study with substantially more aggressive prostate cancers having been diagnosed (i.e., EPIC).
Our study also examined the relation between vitamin D and prostate cancer risk using a genetic score proxy for vitamin D that was based on the number of alleles across the four SNPs in or near GC, CYP24A1, CYP2R1, and DHCR7 previously associated with 25(OH)D levels in GWAS. We found an association with overall prostate cancer risk that was stronger and statistically significant for aggressive disease wherein men with a greater number of low vitamin D alleles were at decreased risk compared to men with only 0 or 1 low vitamin D alleles. The stronger association with all prostate cancer, and the weaker association with aggressive disease, for the three SNP score that excluded rs6013897 near CYP24A1 is consistent with the latter being the only examined SNP contributing to lower risk of aggressive disease. A non-linear relation similar to that reported in a previous serologic study of vitamin D and risk of prostate cancer (12) was suggested, with low risk among men with 6 low vitamin D alleles but substantially elevated risk among men with 7 or 8 low vitamin D alleles; however, the latter category was sparsely populated (0.2% of the study sample), and the finding may be due to chance. Examination of this and other vitamin D risk scores in relation to prostate cancer in other studies will be informative.
Most studies of prostate cancer risk and genetic variants in the vitamin D pathway focused on the vitamin D receptor (VDR) gene (13, 14), with few investigations of other relevant loci (15, 16). Five studies examined variants in CYP24A1 and found either no association (15–19) or a statistically significant association in Hispanic Caucasians (20), and three studies of GC variants were null (15, 17, 21). CYP2R1 and DHCR7, genes newly identified in the aforementioned GWAS of circulating 25(OH)D levels (6, 7), have only been examined in relation to prostate cancer risk in two recent studies that found no association with overall (15, 16) or fatal prostate cancer (15). However, the number of cases was relatively small in both studies (overall prostate cancer n=1,260 and 375; fatal prostate cancer n=114). Collectively, these studies have provided little evidence in support of a vitamin D-cancer association, and to our knowledge, no SNPs in vitamin D pathway genes have been associated with prostate cancer at the genome-wide level of significance in GWAS analyses (22, 23).
Our findings for vitamin D genetic variants are consistent with our recent investigation showing an increased risk of prostate cancer for men with higher serum 25(OH)D status (5). A meta-analysis published prior to that study concluded that there was no association between serum 25(OH)D and risk of prostate cancer; however, when we calculated a summary point estimate that included the studies from the meta-analysis by Yin et al. and our data using inverse variance weighting, the summary odds ratio for a 10 ng/mL increase in serum 25(OH)D was borderline statistically significant (OR=1.05, 95%=1.00 – 1.10, p=0.058). The positive associations observed between circulating vitamin D and prostate cancer, as well as the inverse associations for genetic variants that promote lower 25(OH)D levels, are contrary to experimental evidence and do not support the notion that higher vitamin D status should have a preventive role in this malignancy (1–3). Although it remains unclear why lower vitamin D status might be related to decreased risk of prostate cancer, it is known that 1,25(OH)2D stimulates the insulin receptor and increases insulin synthesis (24), and that elevated circulating insulin has been associated with higher prostate cancer risk (25). Alternatively, the fact that the strongest signals we found were for two mixed-function oxidases (i.e., CYP2R1 and CYP24A1) leaves open the possibility that some of the genetic associations observed here may be reflecting effects on the metabolism of other molecular species relevant to prostate cancer risk and progression unrelated to vitamin D (e.g., androgens). In support of this, a recent study found higher 25(OH)D levels to be associated with increased levels of total and free testosterone in men (26). Additional mechanistic studies of these findings in humans are needed.
The present investigation is the largest to examine prostate cancer risk in relation to a score of genetic variants that have been associated with vitamin D status from GWAS studies (16). Our analysis is based on a large, multi-cohort sample, which enabled us to detect more modest risk associations. Studying vitamin D-related genes mitigates some of the limitations of serologic analyses of circulating 25(OH)D, which include inter-laboratory differences, variable season of blood collection, and fasting status, and genetic association studies do not suffer from the effects of reverse causation or residual confounding that are of concern in biomarker studies. Furthermore, the variants in these genes may better represent the potential for higher or lower vitamin D status over the life course than measurement of circulating vitamin D at one point in time. It should be noted, however, that the GWAS reports identifying these genes as predicting 25(OH)D levels estimated that they only explain between 4–5% of the variation in 25(OH)D concentration (6, 7, 10). These genes may, therefore, have pleiotropic effects on prostate cancer that do not operate through vitamin D-related mechanisms. For example, serum transport of vitamin D metabolites has been historically considered the primary function of the vitamin D binding protein (Gc globulin), but there is now evidence that its other biological activities include a role in inflammation and immunity (27, 28). Thus, the observed genetic associations may be acting through biologic mechanisms independent of an association with circulating vitamin D concentration.
Conclusions
In this large, pooled analysis of men of European ancestry, we found that genetic variants near CYP24A1 related to lower vitamin D status could be associated with a decreased risk of aggressive prostate cancer, and a polygenic vitamin D score was similarly related to both overall and aggressive prostate cancer. Our findings do not support a protective association between higher vitamin D status and lower risk of prostate cancer, and point to the possibility of a positive association.
Acknowledgments
Financial Support: This work was supported by the U.S. NIH, National Cancer Institute cooperative agreements U01-CA98233-07 to D.J. Hunter, U01-CA98710-06 to M.J. Thun, U01-CA98216-06 to E. Riboli and R. Kaaks, and U01-CA98758-07 to B.E. Henderson and Intramural Research Program of NIH/National Cancer Institute, Division of Cancer Epidemiology and Genetics). I. M. Shui was supported by a National Research Service Award (T32 CA09001) from the National Cancer Institute, NIH and a US Army Department of Defense Prostate Cancer Post-doctoral Fellowship. The ATBC Study was supported by the Intramural Research Program of the National Cancer Institute at the National Institutes of Health. Additionally, this research was supported by U.S. Public Health Service contracts (N01-CN-45165, N01-RC-45035, N01-RC-37004, HHSN261201000006C, and HHSN261200800001E) from the National Cancer Institute, National Institutes of Health. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Conflict of Interest: The authors have nothing to disclose
References
- 1.Bouillon R, Eelen G, Verlinden L, Mathieu C, Carmeliet G, Verstuyf A. Vitamin D and cancer. J Steroid Biochem Mol Biol. 2006;102:156–62. doi: 10.1016/j.jsbmb.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 2.Holick MF. Vitamin D: its role in cancer prevention and treatment. Prog Biophys Mol Biol. 2006;92:49–59. doi: 10.1016/j.pbiomolbio.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 3.Fleet JC. Molecular actions of vitamin D contributing to cancer prevention. Mol Aspects Med. 2008;29:388–96. doi: 10.1016/j.mam.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yin L, Raum E, Haug U, Arndt V, Brenner H. Meta-analysis of longitudinal studies: Serum vitamin D and prostate cancer risk. Cancer Epidemiol. 2009;33:435–45. doi: 10.1016/j.canep.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 5.Albanes D, Mondul AM, Yu K, Parisi D, Horst RL, Virtamo J, et al. Serum 25-Hydroxy Vitamin D and Prostate Cancer Risk in a Large Nested Case-Control Study. Cancer Epidemiol Biomarkers Prev. 2011;20:1850–60. doi: 10.1158/1055-9965.EPI-11-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahn J, Yu K, Stolzenberg-Solomon R, Simon KC, McCullough ML, Gallicchio L, et al. Genome-wide association study of circulating vitamin D levels. Hum Mol Genet. 19:2739–45. doi: 10.1093/hmg/ddq155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang TJ, Zhang F, Richards JB, Kestenbaum B, van Meurs JB, Berry D, et al. Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet. 376:180–8. doi: 10.1016/S0140-6736(10)60588-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giovannucci E. The epidemiology of vitamin D and cancer incidence and mortality: a review (United States) Cancer Causes Control. 2005;16:83–95. doi: 10.1007/s10552-004-1661-4. [DOI] [PubMed] [Google Scholar]
- 9.Hunter DJ, Riboli E, Haiman CA, Albanes D, Altshuler D, Chanock SJ, et al. A candidate gene approach to searching for low-penetrance breast and prostate cancer genes. Nat Rev Cancer. 2005;5:977–85. doi: 10.1038/nrc1754. [DOI] [PubMed] [Google Scholar]
- 10.Hiraki LT, Major JM, Chen C, Cornelis MC, Hunter DJ, Rimm EB, et al. Exploring the Genetic Architecture of Circulating 25-Hydroxyvitamin D. Genetic Epidemiology. 2012 doi: 10.1002/gepi.21694. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schumacher FR, Berndt SI, Siddiq A, Jacobs KB, Wang Z, Lindstrom S, et al. Genome-wide association study identifies new prostate cancer susceptibility loci. Hum Mol Genet. 2011;20:3867–75. doi: 10.1093/hmg/ddr295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tuohimaa P, Tenkanen L, Ahonen M, Lumme S, Jellum E, Hallmans G, et al. Both high and low levels of blood vitamin D are associated with a higher prostate cancer risk: a longitudinal, nested case-control study in the Nordic countries. Int J Cancer. 2004;108:104–8. doi: 10.1002/ijc.11375. [DOI] [PubMed] [Google Scholar]
- 13.Berndt SI, Dodson JL, Huang WY, Nicodemus KK. A systematic review of vitamin D receptor gene polymorphisms and prostate cancer risk. J Urol. 2006;175:1613–23. doi: 10.1016/S0022-5347(05)00958-4. [DOI] [PubMed] [Google Scholar]
- 14.McCullough ML, Bostick RM, Mayo TL. Vitamin D gene pathway polymorphisms and risk of colorectal, breast, and prostate cancer. Annu Rev Nutr. 2009;29:111–32. doi: 10.1146/annurev-nutr-080508-141248. [DOI] [PubMed] [Google Scholar]
- 15.Shui IM, Mucci LA, Kraft P, Tamimi RM, Lindstrom S, Penney KL, et al. Vitamin D-related genetic variation, plasma vitamin D, and risk of lethal prostate cancer: a prospective nested case-control study. J Natl Cancer Inst. 2012;104:690–9. doi: 10.1093/jnci/djs189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jorde R, Schirmer H, Wilsgaard T, Joakimsen RM, Mathiesen EB, Njolstad I, et al. Polymorphisms related to the serum 25-hydroxyvitamin d level and risk of myocardial infarction, diabetes, cancer and mortality. The tromso study PLoS One. 2012;7:e37295. doi: 10.1371/journal.pone.0037295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahn J, Albanes D, Berndt SI, Peters U, Chatterjee N, Freedman ND, et al. Vitamin D-related genes, serum vitamin D concentrations and prostate cancer risk. Carcinogenesis. 2009;30:769–76. doi: 10.1093/carcin/bgp055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holick CN, Stanford JL, Kwon EM, Ostrander EA, Nejentsev S, Peters U. Comprehensive association analysis of the vitamin D pathway genes, VDR, CYP27B1, and CYP24A1, in prostate cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:1990–9. doi: 10.1158/1055-9965.EPI-07-0487. [DOI] [PubMed] [Google Scholar]
- 19.Holt SK, Kwon EM, Peters U, Ostrander EA, Stanford JL. Vitamin D pathway gene variants and prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2009;18:1929–33. doi: 10.1158/1055-9965.EPI-09-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beuten J, Gelfond JA, Franke JL, Weldon KS, Crandall AC, Johnson-Pais TL, et al. Single and multigenic analysis of the association between variants in 12 steroid hormone metabolism genes and risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:1869–80. doi: 10.1158/1055-9965.EPI-09-0076. [DOI] [PubMed] [Google Scholar]
- 21.Kidd LC, Paltoo DN, Wang S, Chen W, Akereyeni F, Isaacs W, et al. Sequence variation within the 5′ regulatory regions of the vitamin D binding protein and receptor genes and prostate cancer risk. Prostate. 2005;64:272–82. doi: 10.1002/pros.20204. [DOI] [PubMed] [Google Scholar]
- 22.Ioannidis JP, Castaldi P, Evangelou E. A compendium of genome-wide associations for cancer: critical synopsis and reappraisal. J Natl Cancer Inst. 2010;102:846–58. doi: 10.1093/jnci/djq173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kote-Jarai Z, Olama AA, Giles GG, Severi G, Schleutker J, Weischer M, et al. Seven prostate cancer susceptibility loci identified by a multi-stage genome-wide association study. Nat Genet. 2011;43:785–91. doi: 10.1038/ng.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pittas AG, Dawson-Hughes B. Vitamin D and diabetes. J Steroid Biochem Mol Biol. 2010;121:425–9. doi: 10.1016/j.jsbmb.2010.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Albanes D, Weinstein SJ, Wright ME, Mannisto S, Limburg PJ, Snyder K, et al. Serum insulin, glucose, indices of insulin resistance, and risk of prostate cancer. J Natl Cancer Inst. 2009;101:1272–9. doi: 10.1093/jnci/djp260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nimptsch K, Platz EA, Willett WC, Giovannucci E. Association between plasma 25-OH vitamin D and testosterone levels in men. Clinical endocrinology. 2012;77:106–12. doi: 10.1111/j.1365-2265.2012.04332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pike JW, Feldman D, Glorieux FH. Vitamin D. San Diego: Academic Press; 1997. [Google Scholar]
- 28.Speeckaert M, Huang G, Delanghe JR, Taes YE. Biological and clinical aspects of the vitamin D binding protein (Gc-globulin) and its polymorphism. Clin Chim Acta. 2006;372:33–42. doi: 10.1016/j.cca.2006.03.011. [DOI] [PubMed] [Google Scholar]