Abstract
The 5’ and 3’ untranslated regions (UTRs) of messenger RNAs (mRNAs) function as platforms that can determine the fate of each mRNA individually and in aggregate. Multiple mRNAs that encode proteins that are functionally related often interact with RNA-binding proteins (RBPs) and noncoding RNAs (ncRNAs) that coordinate their expression in time and space as RNA regulons within the ribonucleoprotein (RNP) infrastructure we term the ribonome. Recent ribonomic methods have emerged that can determine which mRNAs are bound and regulated by RBPs and ncRNAs, some of which act in combination to determine global outcomes. ELAV/Hu proteins bind to AU-rich elements (ARE) in mRNAs and regulate their stability from splicing to translation, and the ubiquitous HuR protein has been implicated in cancerous cell growth. Recent work is focused on mechanistic models of how ELAV/Hu proteins increase mRNA stability and translation by repressing microRNAs (miRs) and the RNA induced silencing complex (RISC) via ARE-based ribonucleosomes that may affect global functions of mRNA regulons.
Post-transcriptional gene regulation (PTR) became a dominant process during the evolution of eukaryotes, presumably because of the origin of the nucleus that sequesters the chromosomes and the transcriptional apparatus. While PTR begins in the nucleus with mRNA splicing, polyadenylation and capping, and export, once a mature mRNA reaches the cytoplasm its fate largely determines how much protein will be generated. Many studies indicate that nascent mRNAs are bound to RBPs in the nucleus and conveyed to cellular sites of mRNA processing, eventually arriving at locations in the cytoplasm where they are capable of being translated into proteins [1,2]. Indeed, functionally related groups of mRNAs are tagged in their coding and noncoding regions within the ribonome early in their lives such that their subsequent fates are organized and coordinated at the steps of splicing, export, stabilization, localization and translation [1,3,4]. Many techniques and procedures have been devised to examine the coordinated changes in mRNAs. These methods include Selex based on natural sequences [5], RIP-chip/seq [6], CLIP [7], PAR-CLIP [8] and other methods of RNP enrichment and RNA turnover [reviewed in 4,9,10]. However, the detailed mechanisms that determine how RBPs bind to coding and noncoding regions of multiple mRNAs allowing them to orchestrate global outcomes of protein production are poorly understood. For example, one could ask how RBPs, ncRNAs and their associated trans-acting factors cooperate or compete to coordinate PTR and protein production in time and space. This question is beginning to be addressed in eukaryotic species with a few of the hundreds of known RBPs. This article will discuss mechanisms by which the ELAV/Hu family proteins bind to mRNAs and regulate PTR on a global level.
ELAV/Hu proteins bind A/G-UUU rich RNA sequences while stabilizing and/or activating translation of targeted mRNAs
The highly conserved ELAV/Hu family of RBPs consists of four family members, including three that are predominantly cytoplasmic and neuron-specific (HuB /Hel-N1, HuC and HuD) and one that is expressed primarily in the nucleus of all human cells (HuA / HuR) [reviewed in 11-14]. Each Hu protein consists of three RNA recognition motifs (RRMs) and a flexible hinge/linker region between RRM2 and RRM3 [11]. Using several assays including UV crosslinking procedures, our lab discovered that HuB binds directly to ARE sequences in 3’ UTRs of c-myc, c-fos and GM-CSF [5,15], and that HuB stabilizes as well as activates translation of the mRNA encoding glucose transporter 1 (GLUT1) [16,17]. In addition, we devised an in vitro selection procedure involving total brain mRNA 3’ UTRs and found about 100 novel mRNA binding targets of HuB, representing the first demonstration of multi-targeting by an RBP other than polyA-binding protein [5]. Most of these early in vitro mRNA targets were subsequently confirmed in ours and other laboratories to bind multiple ELAV family members [6,10,12-14,18]. Many of these findings were unexpected because we had assumed that ELAV/Hu proteins would destabilize ARE-containing mRNAs since AREs were known destabilizing sequences. However, as it turned out ELAV/Hu proteins are one of the few RBPs found to stabilize U-rich mRNAs under most conditions. Subsequently, HuD and HuR were shown to also bind AREs [19,20] and to stabilize a bound mRNA [21-24]. Thus, the functional similarities between the four ELAV/Hu proteins appear to be greater than their differences.
While the level of translation of any mRNA can increase in turn by stabilization of the mRNA, studies of HuB binding to GLUT1 mRNA demonstrated a direct effect on both stability and translation [16,17]. Jain et. al., [17] determined mRNA stability by measuring decay following inhibition of transcription, and they determined effects on translation by measuring a shift of the GLUT1 mRNA from unassembled to assembled polysomes following induction of adipocyte differentiation. These experiments indicated that recruitment of Glut1 to active polysomes occurs independent of mRNA stabilization, resulting in a dramatic increase in GLUT1 protein production. Moreover, subsequent studies demonstrated that HuB can increase translation of neurofilament M (NFM) mRNA [24] and HuR can increase translation of p53 mRNA by binding the 3’ UTRs [25], both without any detectable effect on mRNA stability. To date, ELAV/Hu proteins are among the few RBPs demonstrated to activate the expression of mRNAs that contain AU-rich sequences in their 3’UTRs, as most other ARE binding proteins function in their degradation. However, depending on the mRNA target of interest, many examples of mRNA stabilization followed by translational activation are documented, as well as a few examples of increased translation without detectable changes in the levels of the mRNA target [12,13]. Thus, more studies are needed to distinguish the mechanisms responsible for these differences.
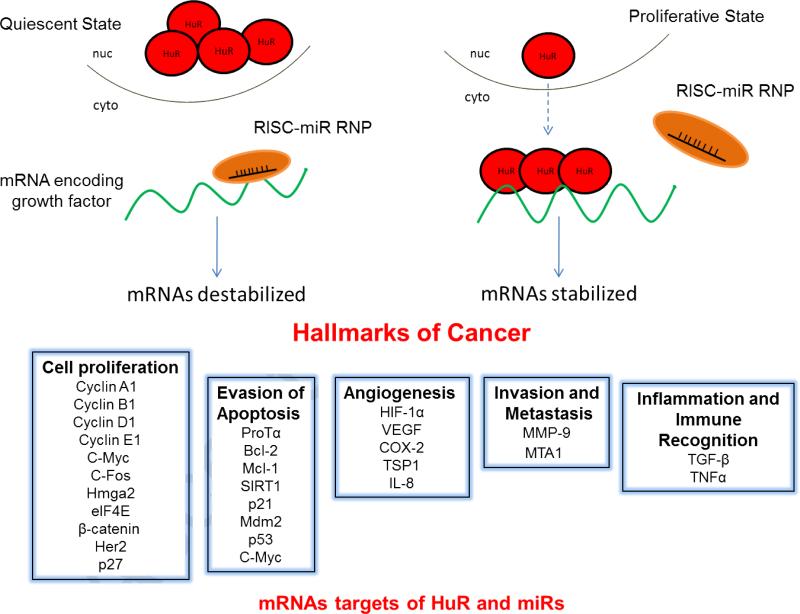
The neuronal Hu proteins were originally identified as specific tumor antigens in lung cancer patients that had developed paraneoplastic neuropathies [12], suggesting a role for this protein family in cell differentiation. Studies have been primarily focused on the role of HuR, as opposed to the neuronal-specific ELAV proteins, in cellular proliferation. The increased cytoplasmic accumulation of HuR observed in both patient tumors and cancer cells correlates with an increased stabilization of mRNAs encoding cancer-related proteins, including regulators of the cell cycle, cell proliferation, cell survival, angiogenesis as well as factors involved in reducing immune recognition and enhancing invasion and metastasis [reviewed in 26]. Many of these cancer-related HuR targets are also subject to miR-mediated regulation based upon the presence of miRNP binding sites [27] (Figure 1). Indeed, our data have shown that HuR mRNA targets are among the most concentrated mRNA targets of microRNAs [28].
Figure 1. Depiction of the cytoplasmic mobilization of HuR known to occur during various activation conditions as well as the proliferative states of transformed cells [21, 26].
In quiescent and non-transformed cells, HuR protein (red circles) is primarily nuclear, and ARE-containing mRNAs are rapidly destabilized by the RISC-miR complex or other destabilizing RBPs [21]. In highly proliferating cells (including cancer cells), as well as in response to certain environmental stresses, HuR protein is shuttled to the cytoplasm where it can stabilize normally labile mRNA targets, potentially through antagonizing destabilizing factors [21,69,71-73,77,78]. The boxes at the bottom show established mRNA targets of HuR and many miRs [26]. HuR and miRs regulate overlapping subsets of mRNAs encoding proteins involved in the acquisition of cancer-related phenotypes [27].
While it is clear that HuR functions as a modulator of the proliferative gene expression program through regulating the stability and translation of mRNAs encoding growth-related proteins, the molecular mechanisms underlying these functional effects remain unclear. However, the neuron-specific ELAV/Hu proteins that have been shown to promote differentiation also bind to many of these same mRNAs. Therefore, while the mechanisms are likely dependent upon the cellular context and environmental conditions, clues can be obtained from studies of the neuronal Hu proteins. As discussed below, data are consistent with the possibility that Hu proteins may antagonize the effects of microRNAs and RISC that are known to affect both the stability and translation of targeted mRNAs [27, 29, 30], with some exceptions. Indeed, recent global analysis of mRNA stability using RIP-chip and UV crosslinking with PAR-CLIP demonstrated that miRs appear to be suppressed by ELAV/HuR when binding sites are close together or overlapping [27]. These and other data suggest a model by which Hu proteins stabilize globally targeted mRNAs by competing with the miR-RISC (miRNP).
Given that HuR and miRs significantly overlap in cancer-related mRNA targets and typically have opposing functional outcomes, it is reasonable to suggest that functional antagonism between HuR the RISC-microRNP is an important determinant of proliferation and possibly tumorigenesis. HuR may rescue the mRNA from miR-mediated suppression via a model whereby HuR multimerizes on targets and either physically occludes or displaces the microRNP and allows the transcripts to be translated. Experimental evidence for this model is discussed in subsequent sections.
ELAV/Hu proteins cooperatively form multimeric “ribonucleosomes”
An early observation regarding the functions of ELAV/Hu RBPs was that they bind to A/G-U rich sequences and form multimeric (or oligomeric) RNPs when binding to RNA targets [15,31]. For example, a uniform array of progressively shifted HuB-RNA complexes was observed using gel mobility shifts suggesting that multiple protein molecules bind to a targeted mRNA in a concentration-dependent manner [15]. Similar findings were subsequently reported for HuD using an in vivo chemical crosslinking method [32]. However, when ELAV/HuB was bound in vitro to a 154 nucleotide fragment from the 3’ UTR of c-myc mRNA, multimers formed as expected but were disrupted by competition with recombinant HuB RRM3 [31]. While RRM3 is highly conserved among the ELAV/Hu family, its ability to block formation of Hu RBP ribonucleosomes is a clue to the underlying mechanism of their functions.
A more recent study suggested that RRM3 does not participate in high affinity binding by HuR but does play a role in multimer formation [33]. Also, in studies by White and coworkers of the Drosophila ELAV prototype, RNP multimers were formed with an ARE-containing RNA fragment of the ewg gene splicing substrate [34,35]. Further examination of protein-RNA and protein-protein interactions using the Drosophila ELAV RRM3 led to a dodecamer model of multimerization that extends over 135 nucleotides of AU-rich sequence [35]. This model suggested a switching mechanism by which RRM3 flipped from binding the RNA substrate to binding RRMs1 and 2 of the protein [34,35]. While this model has not been confirmed using mammalian ELAV-like proteins, it presents an intriguing feature of multimer formation. However, it does not adequately explain how far multimers may extend along the RNA or whether there are structural boundaries to these putative ribonucleosomes [36]. Indeed, it is important to know mechanistically whether cooperative generation of ELAV/Hu multimers encounters boundary limitations or simply grows until the supply of the RBP is exhausted (Figure 2).
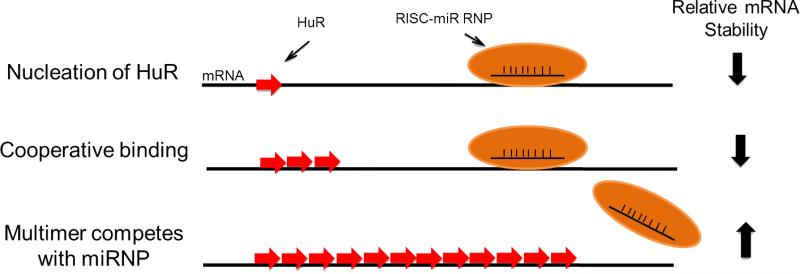
Figure 2. Ribonucleosome model depicting antagonism of the miR/RISC mRNA destabilizing complex by proximal binding of HuR and possibly other members of the ELAV/Hu RBP family [27,82].
In the example, a single molecule of HuR binds at a U-rich RNA-recognition element followed by cooperative binding of multiple HuR molecules [15,19-21,31-37]. Depending on the exact spacing between the nucleation site and the miR/RISC, multimeric binding of HuR could result in steric hindrance or physical displacement of the RISC. This event would logically antagonize the destabilization functions of miR/RISC [29] resulting in increased stability of the mRNA target by HuR, and plausibly, all ELAV/Hu RBPs as demonstrated [16-17,21-23]. The model suggests that a HuR ribonucleosome may displace the miRNP to stabilize mRNA targets. However, the precise binding motifs and coding rules that determine nucleation and multimerization of HuR are poorly understood and may differ with each message [15,31,34,35].
In addition, ElrB, the HuB homolog in Xenopus, was shown to form multimers, but curiously, the HuR homolog, ElrA, could not form multimers in this species [37]. In addition to RRM3, several papers have suggested that the linker region between RRM2 and RRM3 plays a role in the formation of RNP multimers. Known splicing factors are believed to form multimeric RNP complexes in the vicinity of exon-intron junctions and this mechanism is consistent with our recent finding that HuR associates with mRNA targets very early in their life and appears to affect their stability and splicing [27]. This may be a general mechanism by which all ELAV/Hu proteins, including the Drosophila ELAV, function at splice junctions and remain bound throughout the life of the mRNAs, including at sites of alternative polyadenylation [14,38]. It remains to be determined whether HuR functions in this mode by forming a dodecamer that would cover approximately 135 nucleotides [35]. In any case, data to date are consistent with the proposal that multimerization is a general mechanism by which the entire family of ELAV/Hu proteins function from splicing to translation, including their ability to suppress the destabilization property of the miR-RISC [27,30]. Whether this involves the proposed switch of RRM3 from binding RNA to binding protein is to be determined [34].
The troika of coordinated gene expression
Gene expression is coordinated by three known mechanisms: i) DNA operons/regulons, ii) promoter-based transcription initiation, and iii) RNA operons/regulons [4,9,39-41]. In bacteria, DNA operons represent genes that function together and are physically grouped on the chromosome, but also by transcription factors that function at each promoter site. Eukaryotes also use promoter-based gene coordination, but they do not generally have traditional DNA operons. Genes that function together in DNA operons in bacteria are instead dispersed across the mammalian chromosomes rather than being clustered at a single location. Historically, transcription factor initiation at specific promoters was assumed to be the predominant means of mammalian gene coordination [42]. While transcriptional initiation is still believed to be the predominant coordinating process of gene expression, the advent of post-transcriptional RNA regulons led to the idea that PTR events can be coordinated as well [1,4,9,41,43]. Moreover, mRNA regulons can change dynamically under different conditions of growth and development, thus, demonstrating significant biological agility that affects regulation of functionally related mRNA subpopulations [28].
While many studies have supported the concept of RNA regulons, among the strongest evidence is found in trypanosomes where groups of functionally related mRNAs can be coordinated even when transcriptional control is dramatically disrupted [44,45]. For example, several recent studies demonstrated coordinated kinetics of functionally related mRNAs during trypanosome differentiation in blood of the host. Interestingly, the parasite has significantly reduced transcription during these phases of its life cycle. The explanation for the coordination of these functionally related mRNAs is that they are regulated as RNA regulons given the kinetics of decay, and many RBPs have been shown to regulate these events [46,47]. For example, trypanosome RNA regulons were demonstrated to coordinate many pathways and biological responses, including parasite transmission and variation of surface antigens, an established mode to escape the host immune system. These findings represent very strong evidence that RNA regulons function in the development and pathogenesis of this important kinetoplastid parasite.
Transcriptomic and ribonomic analysis of gene expression
One of the inherent problems with simple transcriptomic methods such as RNA-seq is that they only detect the net accumulation of each type of mRNA. In fact, such “steady state” or accumulated levels depend not only on rates of RNA synthesis but also on rates of decay. Recent methods, such as 4-thiouridine pulsing [48], allow these two competing processes to be discriminated because newly synthesized mRNA can be separated from “old” partially decayed mRNA. These and other ribonomic methods have demonstrated that RBPs coordinate the expression of distinct sets of functionally related mRNAs in homeostatic as well as stressed cells. Moreover, these studies have demonstrated that functionally coherent outcomes of global gene expression can be discerned with PTR data that are not apparent from the transcriptomic data including mRNA decay kinetics, RBP and miR binding, and translational activation [1,4,49-54]. Therefore, various experimental approaches have been used to discover post-transcriptional RNA operons and regulons.
In addition to the coordinated changes in mRNA subpopulations, as identified in trypanosomes and by the RIP-chip/seq procedures, CLIP and PAR-CLIP, polysome profiling methods and genetic approaches have also revealed functionally related mRNA targets that encode pathways and macromolecular complexes [4,9,10,28]. Most importantly, the RIP-chip procedure and the later devised CLIP and PAR-CLIP methods have advanced the discovery of functionally related mRNA subsets and pinpointed precise cross-linked nucleoside adducts [7,8,27,28,55]. In the course of these experiments many developmental, environmental and pathogen related RNA regulons have been reported [4,9,10,39,56]. Examples include: a) analysis of mRNA decay rates combined with nuclear run-on and mRNA decay assays using microarrays [1,57-59]; b) identification of mRNA components of ribonucleoproteins by RIP-chip/seq [6,58,60-62]. Indeed, both methods have provided functional insights into these underlying processes that determine the transcriptome [4,9,10].
Comparisons of high-throughput sequence analyses of RNA-binding sites with RIP data have shown that RIP procedures detect stable binding sites while ultraviolet light crosslinking procedures, such as PAR-CLIP, detect both transient and stable interactions [27,55,63,64]. For example, several hundred mRNA binding targets are usually detected for HuR by the RIP procedure, but several thousands of mRNAs are detected by the PAR-CLIP procedures. One interpretation of these surprising results is that UV crosslinking covalently locks in transient binding events, while the simple and direct RIP conditions wash away most transient mRNA targets, leaving the stably bound subsets of mRNA [27].
Studies in the Keene and Tuschl Labs used RIP-chip and PAR-CLIP comparisons before and after global HuR knockdown by RNA interference [27]. Results showed that the most stable RIP-derived HuR mRNA targets were more functionally responsive to effects on RNA stability than were the transient PAR-CLIP-derived mRNA targets. Of interest, the three neuronal ELAV/Hu family members all appear to bind the same mRNA targets when expressed in HEK293 cells [27,38,(T. Farazi, T. Tuschl, unpublished)]. These data suggested an interactive model of RBP-RNA searching or scanning by which the HuR RBP searches for potential AU-rich RNA binding sites for HuR and can be locked in by UV radiation. Without crosslinking, the majority of transient interactions are not detected due to weak binding affinity. Thus, RIP-chip/seq detects high affinity RNP interactions leading to functionally assembled complexes that lead to mRNA target stabilization and increased translation into protein. The stepwise combinatorial RNP assembly model further explains how RNA regulons can be remodeled, and at the same time vary in target specificity depending on the combination of binding components available in the cellular environment [28]. For example, one can imagine an ordered process of assembly advancing in a hierarchy from low complexity ribonucleosomes toward progressively more complex sequence motifs until reaching states of higher stability. In this sense, the “RIP code” may involve increasing complexity of sequence elements recognized by combinations of RBPs or noncoding RNAs in trans, or RNA folding structures in cis, as sequence interactions become increasingly complex (i.e. progressively less homopolymeric and more colorful sequence-binding elements). Naturally, such states would involve dynamic protein-protein, protein-RNA and RNA-RNA remodeling, making the elucidation of definitive mechanisms challenging.
Several studies reporting global remodeling of HuR RNPs during tumorigenesis have been reported. Array analysis of colon cancer cells either over- or under-expressing HuR demonstrated that HuR differentially regulates genes encoding proteins involved in proliferation, angiogenesis and tissue invasion, suggesting that it plays an important role in cancer by altering the stability/translation of mRNAs encoding proteins involved in malignant progression [65]. Ribonomic analyses have demonstrated extensive remodeling of HuR RNPs during tumorigenesis and in response to DNA damage [66-68]. Interestingly, mRNAs encoding proteins involved in anti-cancer functions, such as thrombospondin1, lost association with HuR and was thus destabilized, and mRNAs encoding proteins involved in tumorigenesis, such as members of the Ras pathway and PI3K/AKT pathway, gained association with HuR and thus are presumably stabilized during malignant progression. Thus, HuR RNPs may play a role in tumorigenesis, and likely other disease states as well, by remodeling and thus altering the stability of normally labile mRNAs encoding cancer-related proteins (Figure 1).
Mechanistic models of ELAV/Hu effects on miRNPs and implications in oncogenesis
As noted above, early indications that over expression of ELAV proteins HuB and HuR stabilize ARE-containing mRNAs helped explain how synthesis of the encoded target proteins increases [16,17,21-25,65]. There have been many attempts to address how HuR may affect mRNA functions through interactions with microRNAs. One of the first proposed mechanistic explanations for increased translation of a HuR mRNA target was discovered using the cationic amino acid transporter 1 (CAT1) protein [69,70]. The data indicated that miR-122-mediated repression of CAT1 could be reversed in a HuR-dependent manner during induced stress. The mechanistic interpretation was that miR-122 confines the CAT1 mRNA to processing bodies (PBs); but following induced stress, HuR translocates CAT1 mRNA from PBs to active polysomes where it can escape the miR repression and be translated. A similar mechanism was recently reported by the Gorospe lab demonstrating that HuR attenuates the miR-548c-3p-mediated repression of TOP2A through the recruitment of TOP2A mRNA from PBs to actively translating polysomes [71]. Alterations in TOP2A levels have been observed as a mechanism of TOP2A-targeted drug resistance. Their lab also showed that HuR can outcompete and displace miR-494 from nucleolin mRNA, which encodes an RBP that positively regulates mRNAs encoding anti-apoptotic and proliferation factors [72].
One could propose that the increased expression of CAT1 and TOP2A be explained by a more direct mechanism of HuR competition with the miRNP as demonstrated in our global analysis of multiple miRs [27]. Still, there does not appear to be a single general mechanism by which HuR antagonizes miRNPs. For example, the Gorospe Lab also reported a very different mechanism by which HuR can actually decrease mRNA stability and translation by recruiting the let-7 miR and AGO2 (RISC) to the 3’UTR of c-myc mRNA [73]. HuR can act synergistically with let-7 to reduce the expression of c-Myc, a potent inducer of apoptosis. This suggests that HuR may act to prevent apoptosis and promote tumorigenesis by negatively regulating c-Myc expression. The precise actions of HuR on c-Myc, and likely other targets, can be variable and context-dependent. For example, HuR appears to positively regulate c-Myc both directly and indirectly [reviewed in 74]. The contrary effects of HuR-miRNP interactions and the different outcome observed in the studies by Gorospe and others are likely contextual depending on cell types and stresses as well as the combination of binding factors present and the RNA substrates used.
While the direct mechanism of HuR competition with the miRNP that we reported may not be generalizable [27], other studies of HuR-miRNP interactions have also been reported and may have implications for cell proliferation and tumorigenesis as well. For example, the let-7 family was shown to target several HuR-regulated oncogenes, including Ras, Myc and HmgA2 [reviewed in 75], suggesting that disruption of let-7 activity, either through reduced biogenesis or direct disruption of function by an antagonizing factor, may be an important event in tumorigenesis. Likewise, miR-15a and miR-16 can function as tumor suppressors by targeting the anti-apoptotic gene BCL-2, which is positively regulated by HuR [reviewed in 76]. Another recent study demonstrated that cytoplasmic HuR binds directly to miR-16 to prevent degradation of Cox-2 mRNA, a prostaglandin synthase commonly elevated in several types of cancer [77]. HuR also appears to rescue the mRNA encoding the ERBB-2 receptor tyrosine kinase (Her2/neu) from miR-331-3p-mediated suppression in prostate cancer cells through hindering mRNA association with the miRNP complex but not the miR itself [78].
It has also been suggested that HuR itself may be a direct functional target of tumor suppressive miRs. For example, miR-125a and miR-519 negatively regulate HuR, are inversely correlated with HuR levels, and enforced expression of these miRs decreases both HuR protein levels and tumorigenicity in athymic mice [79-81]. Therefore, not only does HuR alter miR-mediated suppression, but miRs also can mediate HuR-regulated gene expression, and thus, cell proliferation and potentially tumorigenesis, indicating a dynamic post-transcriptional regulatory network [26,30]. Therefore, these experimental outcomes appear to be complex and multifactorial, and suggest that there are many different mechanisms for combinatorial interactions by the HuR RBP and miRs.
Collectively, these studies demonstrate that miRNP and HuR regulation are dynamic and widespread. Both HuR and miRs are responsible for the post-transcriptional regulation of mRNAs encoding cancer-related proteins, and interplay between the two types of factors is potentially a determinant of a proliferative gene expression program. HuR may be a downstream effector of critical cancer mutations, allowing a cell to increase its malignant potential through the increased expression of normally suppressed growth-related proteins. It will be interesting to elucidate the precise conditions that allow HuR to act synergistically or antagonistically to alter miR-mediated repression to promote tumorigenesis, as well as the interplay between HuR and other RBPs. Elucidating mechanisms of competition and cooperation between RBPs and miRs will be important in order to fully understand the combinatorial codes of RNA regulons and other complex post-transcriptional networks in various disease states.
In conclusion, our demonstration that HuR can globally repress the degradative functions of microRNAs associated with RISC suggests a mechanism of direct competition when the binding sites are proximal [27]. We found that HuR binding proximal to many microRNAs-RISC binding sites effectively competes against the RISC complex and counters its mRNA degradative function. But these data were cumulative across multiple mRNA targets and a defined group of miRs, and were not designed to detect mRNA derepression occurring over longer distances. Whether the derepression of RISC-miRNP on the CAT1 mRNA as observed in the Filipowicz Lab [69] requires proximal overlap or can work at various distances in vivo via the multimerization of HuR remains to be determined. However, their more recent in vitro study [82] suggests that HuR can stabilize mRNA at a distance of up to 50 nucleotides from the RISC. Thus, regardless of the precise distances over which HuR appears to functionally repress RISC-miRNPs, the logical model is that multimerization of HuR, and likely other members of the ELAV family, can either block or displace the miRNP complex, allowing mRNA targets to be stabilized (Figure 2), albeit within a context that may not be universal. Our data [27] are consistent with the hypothesis that RNA regulons use ribonucleosome competition as a mechanism to coordinate functionally related mRNA subpopulations associated with ELAV/Hu RBPs and to dynamically remodel RNPs in a contextual manner in response to developmental and environmental signals. Further investigation will be necessary to address this ribonucleosome model.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Keene JD. Ribonucleoprotein infrastructure regulating the flow of genetic information between the genome and the proteome. Proc Natl Acad Sci USA. 2001;98:7018–7024. doi: 10.1073/pnas.111145598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nat. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 3.Moore MJ. From Birth to Death: The Complex Lives of Eukaryotic mRNAs. Sci. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 4.Keene JD. RNA Regulons: coordination of posttranscriptional events. Nat Rev Gen. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 5.Gao F, Carson C, Levine TD, Keene JD. Selection of a subset of mRNAs from 3'UTR combinatorial libraries using neuronal RNA-binding protein, Hel-N1. Proc Natl Acad Sci USA. 1994;91:11207–11211. doi: 10.1073/pnas.91.23.11207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tenenbaum SA, Carson CC, Lager PJ, Keene JD. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc Natl Acad Sci USA. 2000;97:14085–14090. doi: 10.1073/pnas.97.26.14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Sci. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 8**.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [Using a nondamaging RNA-protein UV crosslinking procedure of high efficiency, this paper describes the first global method to determine where specific nucleotides bind to various RNA-binding proteins, including the AGO/RISC RBPs. It advances the RIP-seq methods that determine functionally related mRNA targets with stable RNA-protein interactions but do not definitively identify the UV adduct site.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9**.Imig J, Kanitz A, Gerber AP. RNA regulons and the RNA-protein interaction network. BioMol Concepts. 2012 doi: 10.1515/bmc-2012-0016. (doi: 10.1515/bmc-2012-0016). [This up to date article reviews the field of ribonomics and how RNA operons/regulons are dynamically remodeled and how knowledge of RNA/RNP codes is emerging. The authors also describe advantages and limitations of the RIP-chip/seq, and the global RNA crosslinking procedures.] [DOI] [PubMed] [Google Scholar]
- 10.Morris AD, Mukherjee N, Keene JD. Systematic analysis of post-transcriptional gene expression. Wiley Interdiscip Rev WIRES:Systems Biology and Medicine. 2009 doi: 10.1002/wsbm.54. [doi:10.1002/wsbm.54] [DOI] [PubMed] [Google Scholar]
- 11.Antic D, Keene JD. Embryonic lethal abnormal visual RNA-binding proteins involved in growth, differentiation, and posttranscriptional gene expression. Am J Hum Genet. 1997;61:273–278. doi: 10.1086/514866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keene JD. Why is Hu Where? Shuttling of Early-Response Gene Messenger RNA Subsets. Proc Natl Acad Sci USA. 1999;96:5–7. doi: 10.1073/pnas.96.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell Mol Life Sci. 2008;65:3168–3181. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levine TD, Gao F, King PH, Andrews LG, Keene JD. Hel-N1: an autoimmune RNA-binding protein with specificity for 3' uridylate-rich untranslated regions of growth factor mRNAs. Mol Cell Biol. 1993;13:3494–3504. doi: 10.1128/mcb.13.6.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jain RG, Andrews LG, McGowan KM, Gao F, Keene JD, Pekala PP. Hel-N1, an RNA-binding protein, is a ligand for an A + U rich region of the GLUT1 3' UTR. Nuc Acids Symp Ser. 1995;33:209–211. [PubMed] [Google Scholar]
- 17.Jain RG, Andrews LG, McGowan KM, Pekala P, Keene JD. Ectopic expression of Hel-N1, an RNA-binding protein, increases glucose transporter (GLUT1) expression in 3T3- L1 adipocytes. Mol Cell Biol. 1997;17:954–962. doi: 10.1128/mcb.17.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc Natl Acad Sci USA. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chung S, Jiang L, Cheng S, Furneaux H. Purification and properties of HuD, a neuronal RNA-binding protein. J Biol Chem. 1996;271:11518–11524. doi: 10.1074/jbc.271.19.11518. [DOI] [PubMed] [Google Scholar]
- 20.Abe R, Sakashita E, Yamamoto K, Sakamoto H. Two different RNA binding activities for the AU-rich element and the poly(A) sequence of the mouse neuronal protein mHuC. Nuc Acids Res. 1996;24:4895–4901. doi: 10.1093/nar/24.24.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan XC, Steitz JA. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng SS, Chen CY, Xu N, Shyu AB. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levy NS, Chung S, Furneaux H, Levy AP. Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J Biol Chem. 1998;273:6417–6423. doi: 10.1074/jbc.273.11.6417. [DOI] [PubMed] [Google Scholar]
- 24.Antic D, Lu N, Keene JD. ELAV tumor antigen, Hel-N1, by increases translation of neurofilament M mRNA and induces formation of neurites in human teratocarcinoma cells. Gen Dev. 1999;13:449–461. doi: 10.1101/gad.13.4.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mazan-Mamczarz K, Galban S, Lopez de Silanes I, Martindale JL, Atasoy U, Keene JD, Gorospe M. RNA-binding protein HuR enhances p53 translation after ultraviolet light irradiation. Proc Natl Acad Sci USA. 2003;100:8354–8359. doi: 10.1073/pnas.1432104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abdelmohsen K, Gorospe M. Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip Rev WIRES: RNA. 2010;1:214–229. doi: 10.1002/wrna.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27**.Mukherjee N, Corcoran DL, Nusbaum J, Reid D, Georgiev S, Hafner M, Ascano M, Jr, Tuschl T, Ohler U, Keene JD. Integrative regulatory mapping indicates that RNA-binding protein HuR (ELAVL1) couples pre-mRNA splicing and mRNA stability. Mol Cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [This paper combines RIP-chip and PAR-CLIP to reveal novel functions for the HuR RBP including global stabilization and splicing of pre-mRNAs as well as HuR's ability to repress the mRNA destabilizing functions of microRNAs with the RISC when their binding sites are proximal to one another.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mukherjee N, Lager PJ, Friedersdorf M, Thompson M, Keene JD. Coordinated post-transcriptional mRNA population dynamics during T-cell activation. Mol Sys Biol. 2009;5:288. doi: 10.1038/msb.2009.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartel DP. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 30.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Gen. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 31.Gao FB, Keene JD. Hel-N1/Hel-N2 proteins are bound to polyA+ mRNA in granular RNP structures and are implicated in neuronal differentiation. J Cell Sci. 1996;109:579–589. doi: 10.1242/jcs.109.3.579. [DOI] [PubMed] [Google Scholar]
- 32.Kasashima K, Sakashita E, Saito K, Sakamoto H. Complex formation of the neuron-specific ELAV-like Hu RNA-binding proteins. Nuc Acids Res. 2002;30:4519–4526. doi: 10.1093/nar/gkf567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fialcowitz-White EJ, Brewer BY, Ballin JD, Willis CD, Toth EA, Wilson GM. Specific protein domains mediate cooperative assembly of HuR oligomers on AU-rich mRNA-destabilizing sequences. J Biol Chem. 2007;282:20948–20959. doi: 10.1074/jbc.M701751200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toba G, White K. The third RNA recognition motif of Drosophila ELAV protein has a role in multimerization. Nucleic Acids Res. 2008;36:1390–1399. doi: 10.1093/nar/gkm1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soller M, White K. ELAV multimerizes on conserved AU4-6 motifs important for ewg splicing regulation. Mol Cell Biol. 2005;25:7580–7591. doi: 10.1128/MCB.25.17.7580-7591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chung SY, Wooley J. Set of novel, conserved proteins fold pre-messenger RNA into ribonucleosomes. Protein: Structure, Function, and Genetics. 1986;1:195–210. doi: 10.1002/prot.340010302. [DOI] [PubMed] [Google Scholar]
- 37.Devaux A, Colegrove-Otero LJ, Standart N. Xenopus ElrB, but not ElrA, binds RNA as an oligomer: possible role of the linker. FEBS Lett. 2006;580:4947–4952. doi: 10.1016/j.febslet.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 38.Mansfield KD, Keene JD. Neuron-specific ELAV/Hu proteins suppress HuR mRNA during neuronal differentiation by alternative polyadenylation. Nuc Acids Res. 2012;40:2734–2746. doi: 10.1093/nar/gkr1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Halbeisen RE, Galgano A, Scherrer T, Gerber AP. Post-transcriptional gene regulation: From genome-wide studies to principles. Cell Mol Life Sci. 2008;65:798–813. doi: 10.1007/s00018-007-7447-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hieronymus H, Silver PA. A systems view of mRNP biology. GenDev. 2004;18:2845–2860. doi: 10.1101/gad.1256904. [DOI] [PubMed] [Google Scholar]
- 41.Keene JD, Tenenbaum SA. Eukaryotic mRNPs May Represent Posttranscriptional Operons. Mol Cell. 2002;9:1161–1167. doi: 10.1016/s1097-2765(02)00559-2. [DOI] [PubMed] [Google Scholar]
- 42.Orphanides G, Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- 43*.Anderson P. Post-transcriptional regulons coordinate the initiation and resolution of inflammation. Nat Rev Immun. 2010;10:24–35. doi: 10.1038/nri2685. [This timely review of several of the RNA operons/regulons known to coordinate RNAs involved in immune responses focuses on inflammatory responses of T cells and macrophage.] [DOI] [PubMed] [Google Scholar]
- 44.Clayton CE. Life without transcriptional control? From fly to man and back again. EMBO J. 2002;21:1881–1888. doi: 10.1093/emboj/21.8.1881. Erratum in: EMBO J2002, 21: 3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fernández-Moya SM, Estévez AM. Posttranscriptional control and the role of RNA-binding proteins in gene regulation in trypanosomatid protozoan parasites. Wiley Interdiscip Rev:RNA. 2010;1:34–46. doi: 10.1002/wrna.6. [DOI] [PubMed] [Google Scholar]
- 46.Queiroz R, Benz C, Fellenberg K, Hoheisel JD, Clayton C. Transcriptome analysis of differentiating trypanosomes reveals the existence of multiple post-transcriptional regulons. BMC Genom. 2009;10:495–505. doi: 10.1186/1471-2164-10-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ouellette M, Papadopoulou B. Coordinated gene expression by post-transcriptional regulons in African trypanosomes. J Biol. 2009;8:100–104. doi: 10.1186/jbiol203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dolken L, Ruzsics Z, Radle B, Friedel CC, Zimmer R, Mages J, Hoffmann R, Dickinson P, Forster T, Ghazal P, Koszinowski UH. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA. 2008;14:1959–1972. doi: 10.1261/rna.1136108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y, Liu CL, Storey JD, Tibshirani RJ, Herschlag D, Brown PO. Precision and functional specificity in mRNA decay. Proc Natl Acad Sci USA. 2002;99:5860–5865. doi: 10.1073/pnas.092538799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grigull J, Mnaimneh S, Pootoolal J, Robinson MD, Hughes TR. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol Cell Biol. 2004;24:5534–5547. doi: 10.1128/MCB.24.12.5534-5547.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hao S, Baltimore D. The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat Immun. 2009;10:281–288. doi: 10.1038/ni.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anderson P. Intrinsic mRNA stability helps compose the inflammatory symphony. Nat Immun. 2009;10:233–234. doi: 10.1038/ni0309-233. [DOI] [PubMed] [Google Scholar]
- 53.Raghavan A, Dhalla M, Bakheet T, Ogilvie RL, Vlasova IA, Khabar KSA, Williams BRG, Bohjanen PR. Patterns of coordinate down-regulation of ARE-containing transcripts following immune cell activation. Genom. 2004;84:1002–1013. doi: 10.1016/j.ygeno.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 54.Vlasova IA, McNabb J, Raghavan A, Reilly C, Williams DA, Bohjanen KA, Bohjanen PR. Coordinate stabilization of growth-regulatory transcripts in T cell malignancies. Genom. 2005;86:159–171. doi: 10.1016/j.ygeno.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 55**.Lebedeva S, Jens M, Theil K, Schwanhausser B, Selback M, Landthaler M, Rajewsky N. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein, HuR. Mol Cell. 2011;43:340–352. doi: 10.1016/j.molcel.2011.06.008. [This study used PAR-CLIP and RIP-seq to demonstrate that HuR functions in pre-mRNA splicing and in microRNA repression by upregulation of Let-7 when HuR was knocked down using RNA interference.] [DOI] [PubMed] [Google Scholar]
- 56.Mansfield KD, Keene JD. The Ribonome: a dominant force in gene expression. Biol Cell. 2009;101:169–181. doi: 10.1042/BC20080055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fan J, Yang X, Wang W, Wood WH, Becker KG, et al. Global analysis of stress-regulated mRNA turnover by using cDNA arrays. Proc Natl Acad Sci USA. 2002;99:10611–10616. doi: 10.1073/pnas.162212399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tenenbaum SA, Carson CC, Atasoy U, Keene JD. Genome-wide regulatory analysis using en masse nuclear run-ons and ribonomic profiling with autoimmune sera. Gene. 2003;317:79–87. doi: 10.1016/s0378-1119(03)00661-9. [DOI] [PubMed] [Google Scholar]
- 59.Perez-Ortin JE, de Miguel-Jimenez L, Chavez S. Genome-wide studies of mRNA synthesis and degradation in eukaryotes. Biochem Biophys Acta. 20121819:604–615. doi: 10.1016/j.bbagrm.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 60.Tenenbaum SA, Lager PJ, Carson CC, Keene JD. Ribonomics: Identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods. 2002;26:191–198. doi: 10.1016/S1046-2023(02)00022-1. [DOI] [PubMed] [Google Scholar]
- 61.Gerber AP, Herschlag D, Brown PO. Extensive Association of Functionally and Cytotopically Related mRNAs with Puf Family RNA-Binding Proteins in Yeast. PLoS Biol. 2004;e79:342–354. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jovanovic M, Reiter L, Clark A, Weiss M, Picotti P, Rehrauer H, Frei A, Neukomm LJ, Kaufman E, Wollscheid B, Simard MJ, Miska EA, Aebersold R, Gerber AP, Hengartner MO. RIP-chip-SRM--a new combinatorial large-scale approach identifies a set of translationally regulated bantam/miR-58 targets in C. elegans. Genom Res. 2012;22:1360–1371. doi: 10.1101/gr.133330.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63*.Ascano M, Mukherjee N, Bandaru P, Miller JB, Nusbaum J, Corcoran D, Langlois C, Munschauer M, Hafner M, Williams Z, Ohler U, Tuschl T. FMR1 targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012 doi: 10.1038/nature11737. in press. [This exceedingly important PAR-CLIP study, assisted by RIP-chip, of the Fragile X Mental Retardation protein establishes the precise motifs of binding for the RGG and KH domains. Several previous studies using the CLIP procedure reported different motifs but were not able to be confirmed in other studies. In addition, both techniques identified nearly 100 mRNAs encoding proteins deemed important in autism.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Erhard F, Dolken L, Zimmer R. RIP-chip enrichment analysis. Bioinform. 2012 doi: 10.1093/bioinformatics/bts631. Epub ahead of print [PMID: 23104891] [DOI] [PubMed] [Google Scholar]
- 65.Lopez de Silanes I, Fan J, Galban CJ, Spencer RG, Becker KG, Gorospeb M. Global analysis of HuR-regulated gene expression in colon cancer systems of reducing complexity. Gene expr. 2004;12:49–59. doi: 10.3727/000000004783992215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mazan-Mamczarz K, Hagner PR, Corl S, Srikantan S, Wood WH, Becker KG, Gorospe M, Keene JD, Levenson AS, Gartenhaus RB. Post-transcriptional gene regulation by HuR promotes a more tumorigenic phenotype. Oncogene. 2008;27:6151–6163. doi: 10.1038/onc.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mazan-Mamczarz K, Hagner PR, Dai B, Wood WH, Zhang Y, Becker KG, Liu Z, Gartenhaus RB. Identification of transformation-related pathways in a breast epithelial cell model using a ribonomics approach. Cancer res. 2008;68:7730–7735. doi: 10.1158/0008-5472.CAN-08-2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mazan-Mamczarz K, Hagner PR, Zhang Y, Dai B, Lehrmann E, Becker KG, Keene JD, Gorospe M, Liu Z, Gartenhaus RB. ATM regulates a DNA damage response posttranscriptional RNA operon in lymphocytes. Blood. 2011;117:2441–2450. doi: 10.1182/blood-2010-09-310987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 70.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Gen. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 71.Srikantan S, Abdelmohsen K, Lee EK, Tominaga K, Subaran SS, Kuwano Y, Kulshrestha R, Panchakshari R, Kim HH, Yang X, Martindale JL, Marasa BS, Kim MM, Wersto RP, Indig FE, Chowdhury D, Gorospe M. Translational control of TOP2A influences doxorubicin efficacy. Mol Cell Biol. 2011;31:3790–3801. doi: 10.1128/MCB.05639-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tominaga K, Srikantan S, Lee EK, Subaran SS, Martindale JL, Abdelmohsen K, Gorospe M. Competitive regulation of nucleolin expression by HuR and miR-494. Mol Cell Biol. 2011;31:4219–4231. doi: 10.1128/MCB.05955-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. GenDev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer. 2011;11:644–56. doi: 10.1038/nrc3107. [DOI] [PubMed] [Google Scholar]
- 75.Hammond SM. MicroRNAs as tumor suppressors. Nat Genet. 2007;39:582–583. doi: 10.1038/ng0507-582. [DOI] [PubMed] [Google Scholar]
- 76.Hammond SM. MicroRNAs as oncogenes. Curr Opin Genet Dev. 2006;16:4–9. doi: 10.1016/j.gde.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 77.Young LE, Moore AE, Sokol L, Meisner-Kober N, Dixon DA. The mRNA stability factor HuR inhibits microRNA-16 targeting of COX-2. Mol Cancer Res. 2012;10:167–180. doi: 10.1158/1541-7786.MCR-11-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Epis MR, Barker A, Giles KM, Beveridge DJ, Leedman PJ. The RNA-binding protein HuR opposes the repression of ERBB-2 gene expression by microRNA miR-331-3p in prostate cancer cells. J Biol Chem. 2011;286:41442–52. doi: 10.1074/jbc.M111.301481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Abdelmohsen K, Srikantan S, Kuwano Y, Gorospe M. miR-519 reduces cell proliferation by lowering RNA-binding protein HuR levels. Proc Natl Acad Sci USA. 2008;105:20297–302. doi: 10.1073/pnas.0809376106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guo X, Wu Y, Hartley RS. MicroRNA-125a represses cell growth by targeting HuR in breast cancer. RNA Biol. 2009;6:575–593. doi: 10.4161/rna.6.5.10079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Abdelmohsen K, Kim MM, Srikantan S, Mercken EM, Brennan SE, Wilson GM, Cabo R, Gorospe M. miR-519 suppresses tumor growth by reducing HuR levels. Cell Cycle. 2010;9:1354–1359. doi: 10.4161/cc.9.7.11164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82**.Kundu P, Fabian MR, Sonenberg N, Bhattacharyya SN, Filipowicz W. HuR protein attenuates miRNA-mediated repression by promoting miR/RISC dissociation from the target RNA. Nuc Acids Res. 2012;40:5088–5100. doi: 10.1093/nar/gks148. [This in vitro study of a model mRNA suggested that HuR appears to repress miR-122/RISC at distance as far as 50 nucleotides.] [DOI] [PMC free article] [PubMed] [Google Scholar]