Abstract
BCL-2 family proteins are key regulators of the apoptotic pathway. Antiapoptotic members sequester the BCL-2 homology 3 (BH3) death domains of proapoptotic members such as BAX to maintain cell survival. The antiapoptotic BH3-binding groove has been successfully targeted to reactivate apoptosis in cancer. We recently identified a geographically distinct BH3-binding groove that mediates direct BAX activation, suggesting a new strategy for inducing apoptosis by flipping BAX’s ‘on switch’. Here we applied computational screening to identify a BAX activator molecule that directly and selectively activates BAX. We demonstrate by NMR and biochemical analyses that the molecule engages the BAX trigger site and promotes the functional oligomerization of BAX. The molecule does not interact with the BH3-binding pocket of antiapoptotic proteins or proapoptotic BAK and induces cell death in a BAX-dependent fashion. To our knowledge, we report the first gain-of-function molecular modulator of a BCL-2 family protein and demonstrate a new paradigm for pharmacologic induction of apoptosis.
The BCL-2 family includes both pro- and antiapoptotic members that form a complex protein-interaction network of checks and balances that dictate cell fate1. The α-helical BH3 domains of proapoptotic members function as death ligands that can be intercepted by the structurally defined surface grooves of antiapoptotic members2. The relative levels of death-activating BH3 domains and antiapoptotic BH3-binding pockets determine the cellular response to stress. Cancer cells hijack the survival circuitry of the BCL-2 family pathway, exploiting pathologic overexpression of antiapoptotic proteins to stymie physiologic and pharmacologic proapoptotic stimuli. The published structures of BCL-2 family antiapoptotic proteins and their complexes with BH3 peptides provided a blueprint for the development of small molecules3–5 and stapled peptides6,7 that indirectly activate the mitochondrial apoptotic pathway by targeting the antiapoptotic groove. Such compounds are being advanced in clinical trials as promising next-generation cancer therapeutics8–10.
BAX is an executioner protein of the BCL-2 family that, when activated, undergoes a structural transformation, which converts it from an inactive cytosolic monomer into a lethal mitochondrial pore11. Oligomerization of BAX and its close homolog BAK within the mitochondrial outer membrane enables the release of apoptogenic factors, such as cytochrome c and Smac/DIABLO, that turn on caspases, the enzymatic effectors of apoptosis12–15. The explicit mechanism by which BAX is triggered and how select proapoptotic BCL-2 proteins directly engage and activate BAX have been key issues in the apoptosis field16. Our recent structural analysis of a BIM BH3 death domain in complex with proapoptotic BAX uncovered a new BH3 interaction site that, when engaged, propels this seminal executioner protein into action17. Whereas the main focus of developmental BCL-2 family therapeutics has been the loss-of-function strategy of inhibiting antiapoptotic proteins, the discovery of the BAX trigger site presented a new opportunity to develop what is to our knowledge the first gain-of-function molecular activator of a proapoptotic member.
A BIM BH3 α-helix, structurally reinforced by hydrocarbon stapling, engages BAX at the opposite side of the protein from the canonical BH3-binding groove of antiapoptotic proteins, which in BAX is occupied by its C-terminal helix 9 (ref. 17) (Fig. 1a). This BH3 trigger site on BAX is formed by the confluence of α-helices 1 and 6 and is structurally defined by a hydrophobic groove comprising amino acids Met20, Ala24, Leu27, Ile31, Ile133, Met137 and Leu141 and a perimeter of charged and hydrophilic residues, including Lys21, Gln28, Gln32, Glu131 and Arg134 (Supplementary Results, Supplementary Fig. 1). The flexible loop between α-helices 1 and 2 partially overlies the binding site, and its displacement by BIM BH3 has been implicated as the first ligand-induced conformational change of the BAX activation mechanism11. As our stapled BIM BH3 helix that directly binds and activates BAX also targets the canonical BH3-binding groove of antiapoptotic members18, we sought to identify a direct and selective small-molecule BAX activator. We pursued an in silico screening strategy guided by the structural topography of the trigger site rather than a biochemical approach because of the challenge of generating sufficient quantities of stable, recombinant, monomeric BAX for high-throughput screening. We report the successful application of computational screening to identify a small molecule that directly activates proapoptotic BAX through selective engagement of the BAX trigger site.
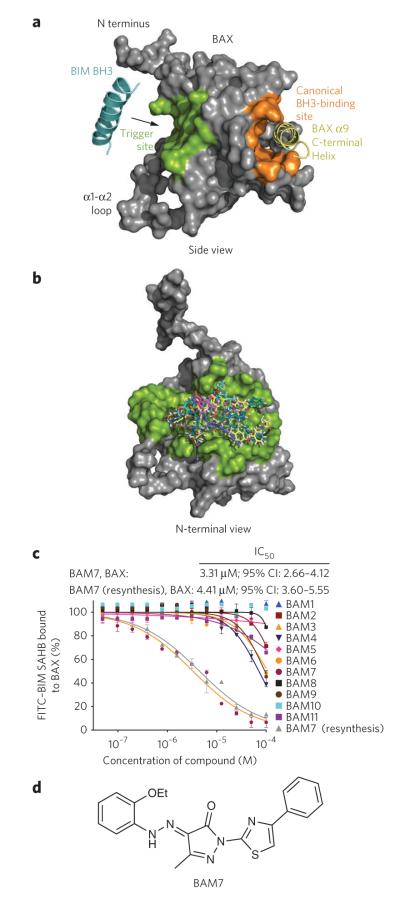
Figure 1. In silico screen for small-molecule binders of the BAX trigger site identifies BAM7.
(a) The BIM BH3 trigger site localizes to the N-terminal face of BAX, as highlighted in green in this side view of the protein. In contrast, the canonical BH3 binding pocket of antiapoptotic proteins (orange) maps to the opposite side of BAX and remains occupied by the C-terminal helix 9 (yellow) when the protein is in the inactive, monomeric state. (b) A computational screening algorithm using an in silico library of 750,000 small molecules docked on average minimized BAX structures yielded a panel of 100 candidate BAMs. A compilation of the docked structures demonstrates how candidate BAMs occupy the topographic landscape of the BAX trigger site (green). (c) BAM7 emerged as the most effective small-molecule binder (IC50, 3.3 μM) in a competitive FPA involving FITC-BIM SAHB and BAX. The structural identity of BAM7 was confirmed by NMR and MS, and the molecule was resynthesized and found to have a similar IC50 (4.4 μM) upon repeat testing by competitive FPA. 95% CI, 95% confidence interval. Data are mean and s.d. for experiments performed in at least triplicate. (d) BAM7 is a 405.5-Da small molecule whose chemical features include a pyrazolone core substituted with ethoxyphenylhydrazono, methyl and phenylthiazol R groups.
RESULTS
Discovery of molecules that bind the BAX trigger site
We generated a diverse in silico compilation of 750,000 small molecules from commercially available libraries and docked the database of three-dimensional molecules on average minimized BAX structures using Glide 4.0 (refs. 19,20) in standard precision mode (Supplementary Fig. 2). The top-ranked 20,000 hits based on the GlideScore function for each BAX structural model were selected and redocked to the BAX structures using extra precision docking mode. The top 1,000 hits from each docking calculation were visualized with the Glide pose viewer and analyzed for their interactions with key BAX binding-site residues. A subset of 100 molecules was selected for experimental analysis on the basis of the presence of favorable hydrogen bonds, hydrophobic contacts and molecular properties. Docking this compilation of putative BAX activator molecules (BAMs) demonstrates how the compounds blanket the surface of the BAX trigger site (Fig. 1b).
To evaluate the capacity of candidate BAMs to bind BAX, we developed a competitive fluorescence polarization assay (FPA) based on the interaction between recombinant BAX and the fluoresceinated stabilized α-helix of BCL-2 domain (SAHB) modeled after BIM BH3 (half-maximum effective concentration (EC50), 283 nM; Supplementary Fig. 3a,b). Small molecules were then benchmarked against the displacement of fluorescein isothiocyanate (FITC)-BIM SAHB by N-terminally acetylated BIM SAHB (half-maximum inhibitory concentration (IC50), 314 nM; Supplementary Fig. 3c,d). Of the 78 molecules that lacked autofluorescence, 11 molecules achieved >55% displacement of FITC–BIM SAHB at the 100-μM screening dose (Supplementary Fig. 3e). The ability of candidate BAMs to dose-responsively compete with FITC–BIM SAHB for BAX binding was then examined by FPA. BAM7 (1) emerged as the most effective competitor, achieving an IC50 of 3.3 μM, which compared favorably with the IC50 of unlabeled BIM SAHB, considering that the molecule is only one-sixth the size of the BIM BH3 α-helical peptide (Fig. 1c). We verified the identity of BAM7 by NMR (Supplementary Fig. 4), resynthesized the molecule and documented a similar IC50 value for competition with FITC–BIM SAHB (Fig. 1c). The molecular structure of BAM7 (molecular weight, 405.5 Da) is characterized by a pyrazolone core substituted with ethoxyphenylhydrazono, methyl and phenylthiazol R groups (Fig. 1d).
BAM7 is selective for the BH3-binding site on BAX
The BH3-binding pocket of antiapoptotic targets shares topographic similarities with the BH3 trigger site on BAX, including a hydrophobic groove that engages the hydrophobic face of BH3 helices and a perimeter of similarly oriented charged and polar residues that are complementary to discrete residues of the hydrophilic BH3 interface. The two BH3-binding sites differ in their geographic location, pocket depth and functionality. Whereas BIM BH3 is compatible with both the BAX trigger site and canonical pockets, we examined whether BAM7 was selective for BCL-2 family targets by competitive FPA. As demonstrated for BAX, direct FPA analyses documented high-affinity interactions between FITC–BIM SAHB and the antiapoptotic proteins BCL-XLΔC (EC50, 13.6 nM), MCL-1ΔNΔC (EC50, 19.8 nM; the homologous N-terminal region relevant to BAX activation is retained in this construct with partial N- and C-terminal deletions (MCL-1ΔNΔC, residues 172–327)) and BFL-1/A1ΔC (EC50, 17.9 nM), which represent the structural diversity of the prosurvival arm of the BCL-2 family (Supplementary Fig. 5a–c). In addition, FITC–BIM SAHB also directly bound BAKΔC (EC50, 173 nM), the proapoptotic, mitochondrial membrane–localized homolog of BAX (Supplementary Fig. 5d). Correspondingly, the N-terminally acetylated analog of BIM SAHB effectively competed with FITC–BIM SAHB for binding to BCL-XLΔC (IC50, 572 nM), MCL-1ΔNΔC (IC50, 136 nM), BFL-1/A1ΔC (IC50, 603 nM) and BAKΔC (IC50, 827 nM) (Supplementary Fig. 6a–d). These binding data highlight that BIM SAHB can readily engage the diversity of BCL-2 family targets. In contrast, BAM7 showed little to no antiapoptotic or BAKΔC competitive binding interactions even at 50-μM dosing, revealing a remarkable selectivity of BAM7 for BAX (Supplementary Fig. 6a–d).
Structural analysis of the BAM7-BAX interaction
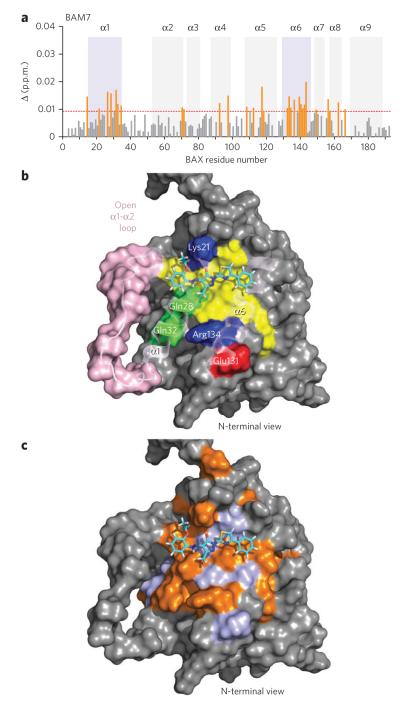
To determine whether BAM7 selectively competed with FITC–BIM SAHB for BAX binding through a direct trigger-site interaction or an indirect allosteric effect, we performed NMR analysis of [15N]BAX upon BAM7 titration. As observed for BIM SAHB17, the addition of BAM7 up to a 1:1 ratio induced significant (P > 0.009 p.p.m.) backbone amide chemical-shift changes in those BAX residues concentrated in the region of the α1-α6 trigger site (Fig. 2a and Supplementary Fig. 7). These data are consistent with a direct interaction between BAM7 and BAX at the very surface used by the BIM BH3 helix to trigger BAX activation.
Figure 2. BAM7 directly engages the BAX trigger site.
(a) Measured chemical shift changes of [15N]BAX upon BAM7 titration up to a ratio of 1:1 BAX:BAM7 are plotted as a function of BAX residue number. Residues with significant backbone amide chemical shift change (calculated significance threshold > 0.009 p.p.m.) are concentrated in the region of the trigger site (violet shading, α1 and α6). (b) the docked structure of BAM7 at the trigger site predicts that the pyrazolone core of BAM7 sits at the base of the 6A7 activation epitope (amino acids 12–24), with the molecule’s carbonyl group engaged in hydrogen-bonding interactions with lys21. The ethoxyphenyl group is positioned at the confluence of residues comprising the α1-α2 loop’s C terminus and the N termini of α1 and α2, a presumed hinge region for loop opening upon initiation of direct BAX activation. The methyl and phenylthiazol R groups are predicted to make hydrophobic contact with the aliphatic residues of α1 and α6, which also form a portion of the BIM BH3–binding groove. Residues: yellow, hydrophobic; green, hydrophilic; blue, positively charged; red, negatively charged. α1-α2 loop residues are colored pink. (c) BAX residues that undergo significant (P > 0.009 p.p.m.) chemical shift change (orange) upon BAM7 titration colocalize with the calculated docking site and its immediate vicinity within the BAX trigger-site region (violet).
We next performed molecular docking analysis to examine the predicted interactions between BAM7 and the BAX trigger site. BAM7 is placed along a crevice formed by residues located at (i) the junction between the α1-α2 loop’s C terminus and the N terminus of α2, (ii) the N terminus of α1 and (iii) the C terminus of α6 (Fig. 2b). Of note, the observed HSQC changes are discretely localized to the docking site and its immediate vicinity within the topographic region of the trigger site (Fig. 2c). This is a remarkable model of the complex from a functional standpoint, as engagement of this region by BIM SAHB is believed to displace the α1-α2 loop and expose an epitope comprising amino acids 12–24, which are recognized by the 6A7 antibody only upon BAX activation21. Indeed, the pyrazolone core of BAM7 is predicted to sit at the base of the 6A7 activation epitope, with the carbonyl group engaged in hydrogen-bonding interactions with Lys21, a key residue that participates in complementary charge-charge interactions with Glu158 of the BIM BH3 helix11,17. Whereas the ethoxyphenyl group abuts the confluence of residues at the α1-α2 loop’s C terminus and the N termini of α1 and α2, a presumed hinge site for loop opening upon initiation of BAX activation, the methyl and phenylthiazol R groups make hydrophobic contact with that portion of the BIM BH3–binding groove formed by aliphatic residues of α1 and α6. Thus, docking analysis positions BAM7 at a critical region of the BAX trigger site implicated in ligand-induced α1-α2 loop displacement and resultant exposure of the 6A7 activation epitope. This binding region is geographically and functionally distinct from the canonical BH3-binding site located at the C-terminal face of multidomain BCL-2 family proteins and may account for the remarkable selectivity of BAM7 for BAX.
To validate the docked structure and develop negative controls for our biochemical and cellular experiments designed to examine the functional consequences of the BAM7-BAX interaction, we performed both BAX mutagenesis and a pilot small-molecule structure-activity relationship analysis. We previously determined that a K21E point mutation in BAX impaired BIM SAHB binding and BH3-induced BAX activation in vitro and in cells11,17. The docked structure suggests that the pyrazolone core of BAM7 also engages Lys21 (Fig. 2b). We conducted NMR analysis of [15N]BAXK21E upon BAM7 titration and observed no chemical shift changes, consistent with abrogation of the small-molecule interaction as a consequence of this single point mutation in BAX (Supplementary Fig. 8). In examining a series of analogs of BAM7 (ANA-BAMs), we observed that derivatization of the phenylhydrazono and phenylthiazole side groups differentially affected competitive BAX binding, with replacement of the ortho ethoxy by meta chloro on the phenylhy-drazono moiety of analog 16 (ANA-BAM16; 2) being the most detrimental (Supplementary Fig. 9a,b). The docking analysis suggests that the bulky chloro group at the meta position would produce steric hindrance at the junction of the α1-α2 loop’s C terminus with the N termini of α1 and α2 (Fig. 2b). Indeed, NMR analysis of [15N]BAX upon ANA-BAM16 titration showed no chemical shift changes (Supplementary Fig. 10), consistent with the inability of this analog to effectively compete with FITC–BIM SAHB for BAX binding by FPA (Supplementary Fig. 9). Taken together, these data provide validation for the docked structure of BAM7 at the BAX trigger site and identify BAXK21E and ANA-BAM16 as protein- and small-molecule–based negative-control reagents, respectively, for evaluating the specificity of action of BAM7 in triggering the functional activation of proapoptotic BAX.
BAM7 activates BAX and BAX-dependent cell death
To transform from an inactive cytosolic monomer into a toxic mitochondrial oligomer, BAX undergoes a major conformational change upon BH3 triggering. We recently demonstrated, using correlative structural and biochemical methods, that these essential changes include ‘opening’ of the α1-α2 loop, mobilization of the C-terminal α9 for mitochondrial translocation and BAX BH3 exposure for propagating BAX activation11. To determine whether the selective binding interaction we documented for BAM7 results in functional BAX activation, we performed a series of structural, biochemical and cellular studies. First, we conducted an NMR analysis of [15N]BAX upon titration with higher concentrations of BAM7 to examine secondary structural changes that ensue upon ligand binding. We observed that increasing the ratio of BAM7 to BAX from 1:1 to 2:1 caused additional chemical shift changes in the α1-α2 loop, the BH3 domain (α2) and the C-terminal α9 helix, three discrete regions that also undergo allosteric changes in response to increased BIM SAHB exposure11 (Supplementary Fig. 11). No such effects were observed upon incubating [15N]BAXK21E with BAM7 or [15N]BAX with ANA-BAM16 at the corresponding dose ratios (Supplementary Figs. 8 and 10).
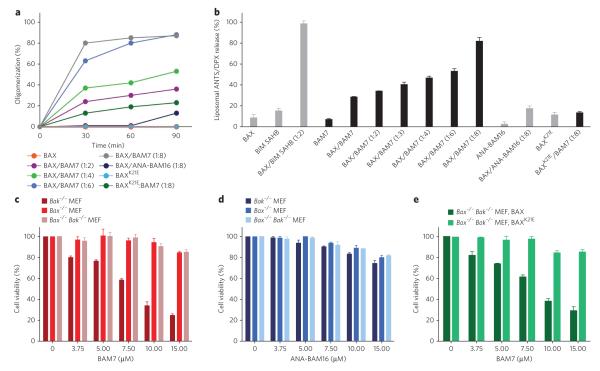
To link the BAM7-induced structural changes to the biochemical conversion of BAX from monomer to oligomer, we performed solution-phase BAX oligomerization assays in which BAX is exposed to increasing quantities of triggering ligand followed by monitoring of BAX species by size-exclusion chromatography (SEC) over time. Like BIM SAHB11, BAM7 triggered the conversion of BAX from monomer to oligomer in a dose- and time-responsive manner, the kinetics of which approach saturation at a 1:8 dose ratio of BAX:BAM7 (Fig. 3a and Supplementary Fig. 6a). Comparatively little to no effect was observed upon incubating BAM7 with BAXK21E or ANA-BAM16 with wild-type BAX, even at the 1:8 dose ratio (Fig. 3a). We subjected the SEC-purified BAM7-induced BAX oligomer to MS analysis and did not detect co-eluted BAM7, suggesting that the interaction between BAM7 and BAX is transient, in accordance with the proposed hit-and-run mechanism for ligand-induced BAX activation22,23; however, small-molecule dissociation as a result of the dilutional conditions of SEC cannot be ruled out. To confirm that the SEC-based detection of BAM7-induced BAX oligomerization reflects functional activation of BAX for its release activity, we performed liposomal assays that explicitly evaluate the capacity of BAM7 to directly trigger BAX pore formation in a membrane environment without the contribution of other factors. Whereas treatment with BAX or BAM7 alone had no effect on the liposomes, the combination of BAM7 and BAX yielded dose-responsive liposomal release of entrapped fluorophore (Fig. 3b). Exposing BAXK21E to BAM7 or wild-type BAX to ANA-BAM16 had no effect on the liposomes even at the highest dose ratio (Fig. 3b). Of note, the comparative potencies of BIM SAHB and BAM7 in the biochemical assays11,17 (Fig. 3a,b and Supplementary Fig. 12) corresponded to the relative BAX-binding activities observed by competitive FPA (Fig. 1c and Supplementary Fig. 3d). Thus, the direct interaction between BAM7 and BAX at the trigger site induces the characteristic structural changes that yield functional BAX oligomerization.
Figure 3. BAM7 triggers in vitro BAX oligomerization, BAX-mediated pore formation and BAX-dependent cell death.
(a) Co-incubation of BAM7 (10 μM, 20 μM, 30 μM and 40 μM) and monomeric BAX (5 μM) induced dose- and time-responsive BAX oligomerization in solution, as monitored by SEC. The combinations of BAX with ANA-BAM16 and BAXK21E with BAM7 had comparatively little to no effect even at the 1:8 dose ratio. The experiment was conducted twice for each condition using two independent preparations of recombinant BAX with similar results. (b) In the presence of ANTS (8-aminonaphthalene-1,3,6-trisulfonic acid, disodium salt)/DPX (p-xylene-bis-pyridinium bromide) loaded liposomes, BAM7 treatment triggers dose-responsive BAX-mediated liposomal release. The exposure of liposomes to BAM7 or BAX alone or the combinations of BAX/ANA-BAM16 or BAXK21E/BAM7 had no effect. BAX/BIM SAHB at a 1:2 dose ratio is shown for comparison. (c) BAM7 selectively impaired the viability of Bak−/− MEFs but had no effect on MEFs that lack BAX (Bax−/−) or both BAX and BAK (Bax−/− Bak−/−). (d) ANA-BAM16, which does not bind or activate BAX in vitro, had no specific effect on the viability of Bak−/−, Bax−/− or Bax−/− Bak−/− MEFs. (e) BAM7 dose-responsively impaired the viability of BAX-reconstituted, but not BAXK21E-reconstituted, Bax−/− Bak−/− MEFs. Data are mean and s.d. for experiments performed in at least triplicate.
We next investigated whether this prototype BAX activator molecule that directly, selectively and functionally activates BAX in vitro could induce BAX-dependent cell death. For these studies, we used genetically defined mouse embryo fibroblasts (MEFs) that express either only BAX (Bak−/−, hereafter referred to as Bak−/−), only BAK (Bax−/−) or neither death effector (Bax−/− Bak−/−). Thus, to undergo apoptosis, Bak−/− MEFs rely on BAX, and Bax−/− MEFs rely on BAK, whereas Bax−/− Bak−/− MEFs are profoundly resistant to apoptosis15. Notably, BAM7 dose- and time-responsively impaired the viability of Bak−/− MEFs that exclusively express BAX but had no effect on Bax−/− MEFs that contain BAK but lack BAX (Fig. 3c and Supplementary Fig. 13). In contrast, standard proapoptotic stimuli such as serum withdrawal, staurosporine and etoposide induced an equivalent apoptotic response in Bax−/− and Bak−/− MEFs (Supplementary Fig. 14). As further evidence of BAM7 specificity of action, (i) BAM7 did not affect the viability of Bax−/− Bak−/− MEFs (Fig. 3c); (ii) ANA-BAM16, which does not bind or activate BAX, had no effect on Bak−/− MEFs (Fig. 3d); and (iii) BAM7 selectively induced cell death of Bax−/− Bak−/− MEFs reconstituted with wild-type BAX but not BAXK21E (Fig. 3e), which bears the mutation that abrogates BAM7 binding (Supplementary Fig. 8).
To further characterize the effect of BAM7 treatment on Bak−/− MEFs, we conducted a series of assays that probe changes in BAX conformational state, intracellular localization, mitochondrial cytochrome c release and cellular morphology. Using the conformation-specific 6A7 antibody that detects exposure of the N-terminal activation epitope of BAX, we observed dose-responsive BAX activation as assessed by 6A7 immunoprecipitation from lysates of BAM7-treated Bak−/− MEFs (Fig. 4a and Supplementary Fig. 15). Because this activation process leads to translocation of BAX from the cytosol to the mitochondria, we evaluated the intracellular distribution of BAX in response to BAM7 treatment. We transfected Bax−/− Bak−/− MEFs with enhanced green fluorescent protein (EGFP)-BAX, labeled mitochondria with MitoTracker and then monitored BAX translocation by confocal fluorescence microscopy. We observed a dose-responsive increase in BAX translocation as evidenced by conversion of the diffuse, cytosolic EGFP-BAX pattern to a mitochondrion-localized distribution (Fig. 4b and Supplementary Fig. 16). To correlate BAX activation and translocation to mitochondrial cytochrome c release, we isolated Bak−/− MEF supernatant and pellet fractions from cells exposed to BAM7 and observed dose-responsive mitochondrial cytochrome c release as assessed by western blot analysis (Fig. 4c and Supplementary Fig. 17). BAM7-treated Bak−/− MEFs likewise showed characteristic microscopic features of apoptosis, including cellular shrinkage and membrane blebbing (Fig. 4d and Supplementary Fig. 18). Taken together, these data highlight that BAM7 selectively induces BAX-mediated apoptosis by triggering the hallmark features of intracellular BAX activation.
Figure 4. BAM7 induces the biochemical and morphologic features of BAX-mediated apoptosis in Bak−/− MEFs.
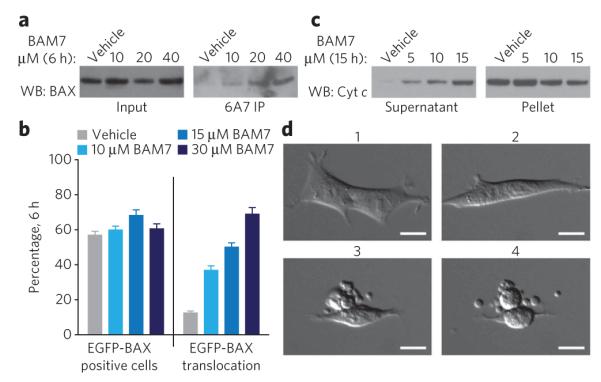
(a) BAM7 treatment of Bak−/− MEFs resulted in dose-responsive exposure of the N-terminal BAX activation epitope, as captured by 6A7 immunoprecipitation (IP) from the corresponding cellular lysates. WB, western blot. (b) Bax−/− Bak−/− MEFs reconstituted with EGFP-BAX (~60% eGFp-positive cells) showed dose-responsive BAX translocation upon exposure to BAM7, as evidenced by the conversion of EGFP-BAX localization from a diffuse pattern to a mitochondrion-localized distribution (Supplementary Fig. 16). Data are mean and s.d. for experiments performed in quadruplicate. (c) BAM7 treatment of Bak−/− MEFs elicited dose-responsive mitochondrial cytochrome c (cyt c) release, as monitored by western blot analysis of the supernatant and pellet fractions. (d) Bak−/− MEFs demonstrate the morphologic features of apoptosis in response to BAM7 treatment (15 μM). The time lapse images reveal progressive cellular shrinkage, membrane blebbing and the formation of apoptotic bodies (Supplementary Fig. 18). 1, 20 min; 2, 6 h; 3, 12 h; 4, 12.5 h. Scale bars, 15 μm.
We find that BAM7 directly binds the BAX trigger site and initiates the characteristic structural changes that lead to functional BAX activation in vitro. When applied to genetically defined MEFs, BAM7 only kills the cell line that contains BAX, eliciting the biochemical and morphologic features of BAX-mediated apoptosis. Thus, these structural, biochemical and cellular studies demonstrate the feasibility of targeting BAX with a selective small molecule to trigger its proapoptotic activity.
DISCUSSION
BAX is a proapoptotic BCL-2 family member that shares structural and functional homology with diphtheria and colichin toxins24,25. Whereas such exogenous toxins self-associate to form pores that destroy cells by piercing the plasma membrane, BAX and its close homolog BAK engage in similar membrane-penetrating and oligomerization activity except that the target organelle is the mitochondrion. By lancing the mitochondrial outer membrane, BAX and BAK effectively destroy the cell’s power plant and release key factors that activate the irreversible enzymatic digestion of the cell. BCL-2 family survival proteins intercept the activated forms of BAX and BAK using a surface groove that traps proapoptotic BH3 helices. Cancer cells overexpress these antiapoptotic proteins to maintain a survival advantage in the face of proapoptotic stimuli. Potent and selective molecules that inhibit antiapoptotic proteins, such as ABT-263 (ref. 26), can resensitize cells to apoptosis through indirect activation or derepression of BAX and BAK.
Direct activation of BAX by select proapoptotic BCL-2 members that only contain a conserved BH3 domain (‘BH3-only’ proteins) has emerged as a physiologically relevant mechanism for inducing mitochondrial apoptosis during development and homeostasis27. A series of biochemical and cellular data document the capacity of select BH3-only proteins, such as BID and BIM, to directly bind and activate BAX and BAK17,18,28–35. Just as solving the structure of the antiapoptotic groove in complex with a BH3 helix led to the development of targeted inhibitors of antiapoptotic proteins3–5,7, our discovery of a distinct BH3 binding site that regulates BAX activation17 provides a new opportunity to modulate apoptosis for therapeutic benefit using a unique gain-of-function strategy. Whereas pharmacologic activation of BAX, whether by indirect activators (loss of antiapoptotic function) or direct activators (gain of proapoptotic function), may seem risky, the preclinical3,4,7 and early clinical data8,10,36 suggest that differences in apoptotic reserve among normal and diseased cells may provide a practical therapeutic window. Whereas unstressed cells and tissues may have a sufficient pool of unoccupied antiapoptotic proteins to tolerate antiapoptotic blockade (for example, ABT-263) and/or trap activated BAX conformers, diseased cells—bombarded by external and internal death signals—may acutely respond to BAX activation because of an overwhelmed antiapoptotic reserve37–39.
Here we identified by in silico screen of the BAX trigger site a 405.5-Da small molecule, BAM7, which directly binds the previously uncharacterized BH3-binding groove at the N-terminal face of BAX. In doing so, the compound triggers functional BAX oligomerization and induces BAX-dependent cell death in a genetically defined context. Notably, BAM7 is selective for the BH3-binding groove at the N-terminal face of BAX and thus may serve as a powerful chemical tool for dissecting the physiologic consequences of direct BAX activation in a variety of homeostatic and pathologic conditions. Thus, we find that small-molecule targeting of the BAX trigger site is achievable and could lead to a new generation of apoptotic modulators that directly activate BCL-2 family executioner proteins in cancer and other diseases driven by pathologic apoptotic blockades.
METHODS
In silico screening
A diverse in silico library was generated from the following commercially available libraries downloaded from the ZINC database40: ACB Blocks, Asinex, Chembridge, Maybridge, Microsource, NCI, Peakdale and FDA-approved. The in silico library was filtered for drug-like features, ADME properties and appropriate functional groups with QikProp. Molecules were converted to three-dimensional all-atom structures, generating a maximum of four stereoisomers, ionization states for pH 7.0 and pH 2.0 and different tautomers with Ligprep. The database of in silico three-dimensional molecules totaled approximately 750,000 compounds. BAX structures for docking were prepared using an averaged BAX closed-loop structure and an averaged BAX open-loop structure with GROMACS software. The two structures were generated in suitable format for docking with MAESTRO. Docking was performed using Glide, with the small-molecule database for each BAX structure in standard precision (SP) mode19. The top 20,000 hits based on GlideScore function were selected and redocked to the BAX structures using extra-precision (XP) docking mode20. The top 1,000 hits from each docking calculation were visualized on the structure and then analyzed for interactions with key BAX residues, leading to selection of 100 compounds for experimental screening.
BCL-2 family protein production
Recombinant and tagless full-length BAX, BCL-XLΔC, MCL-1ΔNΔC, BFL-1/A1ΔC and BAKΔC were expressed and purified as previously reported17,41 and as detailed in the Supplementary Methods.
Fluorescence polarization binding assays
FPAs were performed as previously described41,42 and as detailed in the Supplementary Methods.
BAM7 characterization by MS and 1H-NMR spectroscopy
4-(2-(2-ethoxyphenyl) hydrazono)-3-methyl-1-(4-phenylthiazol-2-yl)-1H-pyrazol-5(4H)-one. LC-MS: ES+ 406 (M+1). 1H NMR (300 MHz, DMSO-d6) d 7.96 (d, 2H, J=8.1 Hz), 7.86 (s, 1H), 7.75 (d, 1H, J=7.8 Hz), 7.46 (t, 2H), 7.37–7.33 (m, 1H), 7.25–7.20 (m, 2H), 7.13–7.07 (m, 1H), 4.29–4.22 (q, 2H), 2.36 (s, 3H), 1.48 (t, 3H).
NMR samples and spectroscopy
Uniformly 15N-labeled full-length human BAX and BAXK21E were generated for titration experiments as previously described17,43. NMR spectra were acquired and analyzed as detailed in the Supplementary Methods.
Structure modeling
Docked structures of BAX and BAM7 were generated using Glide 4.0 (refs. 19,20; Schrodinger, 2006) and analyzed using PYMOL44.
BAX oligomerization assay
Ligand-induced conversion of BAX from monomeric to oligomeric forms was analyzed as previously described11,17 and as detailed in the Supplementary Methods.
Liposomal release assay
Liposomes were prepared and release assays were performed as previously described34,45 and as detailed in the Supplementary Methods.
Cell viability assay
MEF cells were maintained in DMEM high glucose (Invitrogen) supplemented with 10% (v/v) FBS, 100 U ml−1 penicillin, 100 μg ml−1 streptomycin, 2 mM l-glutamine, 50 mM HEPES, 0.1 mM MEM nonessential amino acids and 50 μM β-mercaptoethanol. MEFs (2.5 × 103 cells per well) were seeded in 96-well opaque plates for 18–24 h and then incubated with serial dilutions of BAM7, ANA-BAM16 or vehicle (0.15% (v/v) DMSO) in DMEM at 37 °C in a final volume of 100 μl. Cell viability was assayed at 24 h by addition of CellTiter-Glo reagent according to the manufacturer’s protocol (Promega), and luminescence was measured using a SpectraMax M5 microplate reader (Molecular Devices). Viability assays were performed in at least triplicate, and the data were normalized to vehicle-treated control wells.
BAX conformational change assay
Bak−/− MEFs (5 × 105 cells per well) were seeded in six-well clear-bottom plates for 18–24 h and then incubated with serial dilutions of BAM7 or vehicle (0.4% (v/v) DMSO) in DMEM at 37 °C in a final volume of 500 μl. After 6 h, cells were lysed in 100 μl buffer (20 mM HEPES, pH 7.4, 150 mM sodium chloride, 1.0% (w/v) 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate) with freshly added protease inhibitors (Complete Protease Inhibitors Cocktail, Roche), incubated on ice for 1 h and centrifuged at 15,000g for 10 min. Ten microliters of each supernatant was mixed with SDS sample buffer for input samples, and 90 μl of each supernatant was mixed with 25 μl preclarified Protein A/G–Agarose beads (Santa Cruz Biotechnology) and preincubated at 4 °C with rotation for 1 h. Subsequently, 3 μl of 6A7 antibody (1:1,000, sc-23959, Santa Cruz Biotechnology) was added to each sample and incubated at 4 °C with rotation for an additional 1 h. Beads were collected by brief spin, washed three times with 1 ml of 3% (w/v) BSA/PBS buffer and then solubilized with 25 μl SDS sample buffer. Samples were boiled at 95 °C for 2 min, separated on 10% NuPAGE (Invitrogen) gels and blotted on a PVDF membrane. Western blot analysis was performed using the rabbit polyclonal N20 antibody against BAX (1:1,000, sc493, Santa Cruz Biotechnology) and chemiluminescence-based detection (PerkinElmer).
Mitochondrial cytochrome c release
Bak−/− MEFs (5 × 105 cells per well) were seeded in six-well clear-bottom plates for 18–24 h and then incubated with serial dilutions of BAM7 or vehicle (0.4% (v/v) DMSO) in DMEM at 37 °C in a final volume of 500 μl. After 15 h, cells were lysed in 100 μl buffer (20 mM HEPES, pH 7.2, 10 mM KCl, 5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 250 mM sucrose, 0.025% (w/v) Digitonin) with freshly added protease inhibitors (Complete Protease Inhibitors Cocktail, Roche) and incubated on ice for 10 min. The supernatants were isolated by centrifugation at 15,000g for 10 min, and the mitochondrial pellets were solubilized in 1% (v/v) Triton X-100 in PBS for 1 h at 4 °C. Pellets were solubilized and subjected to a 15,000g spin for 10 min, and 50 ng of protein was mixed with 25 μl SDS sample buffer. The equivalent fractional volume of the corresponding supernatant samples was mixed with 25 μl SDS sample buffer. The mitochondrial supernatant and pellet fractions were then separated by 10% (w/v) polyacrylamide NuPAGE (Invitrogen) gels, blotted on a PVDF membrane and subjected to western analysis with antibody against cytochrome c (1:300, 7H8.2C12, BD PharMingen).
BAX translocation assay
Confocal microscopic analysis of GFP-BAX translocation in treated MEFs was performed as described in the Supplementary Methods.
Light microscopy
Live cell imaging of treated MEFs was performed as described in the Supplementary Methods.
Supplementary Material
ACKNOWLEDGMENTS
We thank E. Smith for editorial and graphics assistance, M. Davis for help with establishing the competitive BAX binding assay, G. Bird for BIM SAHB production and characterization and CreaGen Biosciences for BAM7 resynthesis and characterization. This research program was supported by a grant from the William Lawrence and Blanche Hughes Foundation to L.D.W. Additional funding was provided by US National Institutes of Health (NIH) grant 4R00HL095929 to E.G. and NIH grant 5R01CA050239 and a Stand Up to Cancer Innovative Research Grant to L.D.W.
Footnotes
Author contributions E.G. performed the in silico screen; E.G., D.E.R., J.A.B. and E.S.L. conducted the binding assays; E.G. carried out the structural analyses; E.G. and D.E.R. performed the biochemical studies; and E.G., D.E.R. and J.A.B. conducted the cellular experiments, with guidance from L.D.W. L.D.W. wrote the manuscript, which was reviewed by all authors.
Competing financial interests The authors declare no competing financial interests.
Additional information Supplementary information and chemical compound information is available in the online version of the paper. Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 2.Sattler M, et al. Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science. 1997;275:983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- 3.Nguyen M, et al. Small molecule obatoclax (GX15–070) antagonizes MCL-1 and overcomes MCL-1–mediated resistance to apoptosis. Proc. Natl. Acad. Sci. USA. 2007;104:19512–19517. doi: 10.1073/pnas.0709443104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oltersdorf T, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 5.Wang G, et al. Structure-based design of potent small-molecule inhibitors of anti-apoptotic Bcl-2 proteins. J. Med. Chem. 2006;49:6139–6142. doi: 10.1021/jm060460o. [DOI] [PubMed] [Google Scholar]
- 6.Stewart ML, Fire E, Keating AE, Walensky LD. The MCL-1 BH3 helix is an exclusive MCL-1 inhibitor and apoptosis sensitizer. Nat. Chem. Biol. 2010;6:595–601. doi: 10.1038/nchembio.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walensky LD, et al. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science. 2004;305:1466–1470. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson WH, et al. Navitoclax, a targeted high-affinity inhibitor of BCL-2, in lymphoid malignancies: a phase 1 dose-escalation study of safety, pharmacokinetics, pharmacodynamics, and antitumour activity. Lancet Oncol. 2010;11:1149–1159. doi: 10.1016/S1470-2045(10)70261-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Brien SM, et al. Phase I study of obatoclax mesylate (GX15–070), a small molecule pan–Bcl-2 family antagonist, in patients with advanced chronic lymphocytic leukemia. Blood. 2009;113:299–305. doi: 10.1182/blood-2008-02-137943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gandhi L, et al. Phase I study of Navitoclax (ABT-263), a novel Bcl-2 family inhibitor, in patients with small-cell lung cancer and other solid tumors. J. Clin. Oncol. 2011;29:909–916. doi: 10.1200/JCO.2010.31.6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gavathiotis E, Reyna DE, Davis ML, Bird GH, Walensky LD. BH3-triggered structural reorganization drives the activation of proapoptotic BAX. Mol. Cell. 2010;40:481–492. doi: 10.1016/j.molcel.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X, Kim CN, Yang J, Jemmerson R, Wang X. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- 13.Li P, et al. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 14.Du C, Fang M, Li Y, Li L, Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- 15.Wei MC, et al. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 17.Gavathiotis E, et al. BAX activation is initiated at a novel interaction site. Nature. 2008;455:1076–1081. doi: 10.1038/nature07396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walensky LD, et al. A stapled BID BH3 helix directly binds and activates BAX. Mol. Cell. 2006;24:199–210. doi: 10.1016/j.molcel.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 19.Friesner RA, et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 20.Friesner RA, et al. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 21.Hsu YT, Youle RJ. Nonionic detergents induce dimerization among members of the Bcl-2 family. J. Biol. Chem. 1997;272:13829–13834. doi: 10.1074/jbc.272.21.13829. [DOI] [PubMed] [Google Scholar]
- 22.Wei MC, et al. tBID, a membrane-targeted death ligand, oligomerizes BAK to release cytochrome c. Genes Dev. 2000;14:2060–2071. [PMC free article] [PubMed] [Google Scholar]
- 23.Wang K, Yin XM, Chao DT, Milliman CL, Korsmeyer SJ. BID: a novel BH3 domain-only death agonist. Genes Dev. 1996;10:2859–2869. doi: 10.1101/gad.10.22.2859. [DOI] [PubMed] [Google Scholar]
- 24.García-Sáez AJ, Mingarro I, Perez-Paya E, Salgado J. Membrane-insertion fragments of Bcl-xL, Bax, and Bid. Biochemistry. 2004;43:10930–10943. doi: 10.1021/bi036044c. [DOI] [PubMed] [Google Scholar]
- 25.Muchmore SW, et al. X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death. Nature. 1996;381:335–341. doi: 10.1038/381335a0. [DOI] [PubMed] [Google Scholar]
- 26.Tse C, et al. ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res. 2008;68:3421–3428. doi: 10.1158/0008-5472.CAN-07-5836. [DOI] [PubMed] [Google Scholar]
- 27.Ren D, et al. BID, BIM, and PUMA are essential for activation of the BAX- and BAK-dependent cell death program. Science. 2010;330:1390–1393. doi: 10.1126/science.1190217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cartron PF, et al. The first α helix of Bax plays a necessary role in its ligand-induced activation by the BH3-only proteins Bid and PUMA. Mol. Cell. 2004;16:807–818. doi: 10.1016/j.molcel.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 29.Kim H, et al. Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat. Cell Biol. 2006;8:1348–1358. doi: 10.1038/ncb1499. [DOI] [PubMed] [Google Scholar]
- 30.Kim H, et al. Stepwise activation of BAX and BAK by tBID, BIM, and PUMA initiates mitochondrial apoptosis. Mol. Cell. 2009;36:487–499. doi: 10.1016/j.molcel.2009.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuwana T, et al. BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol. Cell. 2005;17:525–535. doi: 10.1016/j.molcel.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 32.Kuwana T, et al. Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell. 2002;111:331–342. doi: 10.1016/s0092-8674(02)01036-x. [DOI] [PubMed] [Google Scholar]
- 33.Letai A, et al. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell. 2002;2:183–192. doi: 10.1016/s1535-6108(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 34.Lovell JF, et al. Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell. 2008;135:1074–1084. doi: 10.1016/j.cell.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 35.Takeuchi O, et al. Essential role of BAX, BAK in B cell homeostasis and prevention of autoimmune disease. Proc. Natl. Acad. Sci. USA. 2005;102:11272–11277. doi: 10.1073/pnas.0504783102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts AW, et al. Substantial susceptibility of chronic lymphocytic leukemia to BCL2 inhibition: results of a phase I study of navitoclax in patients with relapsed or refractory disease. J. Clin. Oncol. 2012;30:488–496. doi: 10.1200/JCO.2011.34.7898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Certo M, et al. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell. 2006;9:351–365. doi: 10.1016/j.ccr.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 38.Walensky LD. From mitochondrial biology to magic bullet: navitoclax disarms BCL-2 in chronic lymphocytic leukemia. J. Clin. Oncol. 2012;30:554–557. doi: 10.1200/JCO.2011.37.9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walensky LD, Gavathiotis E. BAX unleashed: the biochemical transformation of an inactive cytosolic monomer into a toxic mitochondrial pore. Trends Biochem. Sci. 2011;36:642–652. doi: 10.1016/j.tibs.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Irwin JJ, Shoichet BK. ZINC—a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pitter K, Bernal F, Labelle J, Walensky LD. Dissection of the BCL-2 family signaling network with stabilized α-helices of BCL-2 domains. Methods Enzymol. 2008;446:387–408. doi: 10.1016/S0076-6879(08)01623-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bernal F, et al. A stapled p53 helix overcomes HDMX-mediated suppression of p53. Cancer Cell. 2010;18:411–422. doi: 10.1016/j.ccr.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzuki M, Youle RJ, Tjandra N. Structure of Bax: coregulation of dimer formation and intracellular localization. Cell. 2000;103:645–654. doi: 10.1016/s0092-8674(00)00167-7. [DOI] [PubMed] [Google Scholar]
- 44.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos: 2002. [Google Scholar]
- 45.Yethon JA, Epand RF, Leber B, Epand RM, Andrews DW. Interaction with a membrane surface triggers a reversible conformational change in Bax normally associated with induction of apoptosis. J. Biol. Chem. 2003;278:48935–48941. doi: 10.1074/jbc.M306289200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.