Background: The TXNRD1_v3 (“v3”) protein is a rare variant of human thioredoxin reductase 1.
Results: Membrane targeting of v3 occurs by N-terminal myristoylation and palmitoylation, and its overexpression triggers induction of filopodia independently of its redox active site integrity.
Conclusion: The v3 protein is targeted to membrane rafts.
Significance: These results imply that v3, shown to be targeted to membrane rafts, may be involved in signaling events.
Keywords: Alternative Splicing, Lipid Raft, Membrane Proteins, Protein Acylation, Protein Myristoylation, Protein Palmitoylation, Thioredoxin Reductase, Filopodia, Membrane Raft
Abstract
The human selenoprotein thioredoxin reductase 1 (TrxR1), encoded by the TXNRD1 gene, is a key player in redox regulation. Alternative splicing generates several TrxR1 variants, one of which is v3 that carries an atypical N-terminal glutaredoxin domain. When overexpressed, v3 associates with membranes and triggers formation of filopodia. Here we found that membrane targeting of v3 is mediated by myristoylation and palmitoylation of its N-terminal MGC motif, through which v3 specifically targets membrane rafts. This was suggested by its localization in cholera toxin subunit B-stained membrane areas and also shown using lipid fractionation experiments. Utilizing site-directed mutant variants, we also found that v3-mediated generation of filopodia is independent of the Cys residues in its redox active site, but dependent upon its membrane raft targeting. These results identify v3 as an intricately regulated protein that expands TXNRD1-derived protein functions to the membrane raft compartment.
Introduction
Thioredoxin reductase (TrxR)2 and thioredoxin (Trx) together with NADPH comprise the Trx system, which is involved in a wide range of cellular processes, including cell proliferation and differentiation, antioxidant defense, maintenance of deoxyribonucleotide synthesis, signaling of apoptosis, redox control of protein function, transcription factor activity, and cancer development (1–7). The Trx system orchestrates its many functions mainly through redox reactions, whereby Trx reduces disulfides in target proteins for the support or modulation of their activities, whereas Trx in turn is kept reduced and active by TrxR using NADPH.
Human cells carry three genes encoding three distinct TrxR isoenzymes. The TXNRD1 gene encodes the classical and most abundant, predominantly cytosolic, TrxR1, which is expressed in most human cells and uses Trx1 as its prime substrate (3, 4, 8, 9). The predominantly mitochondrial TrxR2 enzyme is encoded by TXNRD2 and mainly reduces mitochondrial Trx2 (10–12). The TXNRD3 gene encodes a thioredoxin glutathione reductase isoenzyme that contains a monothiol glutaredoxin (Grx) domain as an N-terminal addition to the TrxR module, which otherwise is similar in domain structure to TrxR1 and TrxR2. The thioredoxin glutathione reductase isoenzyme was found to be involved in the maturation of sperm cells and is mainly expressed in early spermatids in testis (13–15). Both the cytoplasmic and the mitochondrial Trx systems are essential for mammals, as demonstrated by the embryonically lethal phenotype of knock-out mice for any one of the enzymes TrxR1, Trx1, TrxR2, or Trx2 (16–19).
The human TXNRD1 gene on chromosome 12 (12q23-q24.1) displays a complex genomic organization. It gives rise to numerous transcripts that can undergo extensive splicing, in particular at the 5′-end, producing several different protein isoforms (8, 9, 20–22). One of these isoforms, TXNRD1_v3 (“v3”), is peculiar by utilizing three additional exons encoding an atypical dithiol active site Grx domain, which is expressed in N-terminal fusion to the classical TrxR1 module (8, 20, 23, 24). These three exons, termed β-VIII, β-VI, and β-V, are unique to v3 and are encoded by a genomic region upstream of the more commonly transcribed TXNRD1 exons. Therefore, transcription of v3 must initiate upstream of the previously characterized core promoter of TrxR1 (8, 21, 22, 25, 26) and must thus be regulated by an alternative promoter, which hitherto has remained uncharacterized. Intriguingly, humans, chimpanzees, and dogs express v3, but mice or rats do not (20). Endogenous expression of v3 has been demonstrated in human testis by Northern blot analyses as well as using immunohistochemistry, with the latter displaying particularly strong staining in Leydig cells (23). Immunoblotting and mass spectrometry also indicated v3 protein expression in a human mesothelioma cell line (24), and v3 could furthermore be detected in extracts of bovine and dog testis (20). In addition, several human cancer cell lines show expression of v3-encoding transcripts, as detected by first-strand reverse transcription-polymerase chain reaction (PCR), with v3 expression also found to be induced by estradiol or testosterone treatment (23). However, transcripts for v3 are rarely found in the form of expressed sequence tag clones with only few such clones currently found in the National Center for Biotechnology Information (NCBI) databases (including five from testis, accession numbers BG772375, AY057105, BG717223, DC401599, and DC400412; four from trachea, accession numbers AK304241, DC417264, DB230289, and DB233566; two from glioblastoma, accession numbers BF342747 and AW027910; one from squamous cell carcinoma, accession number BP355955; and one from astrocytes, accession number DA033928). This should be compared with more than 1,700 expressed sequence tag clones found to encode the other forms of TrxR1. It should be noted, however, that some of those other sequences could also be derived from v3-encoding transcripts, although they will not be discovered as such if they have incomplete 5′-ends, which is the case with many expressed sequence tag clones.
The Grx domain of v3 has an atypical CTRC redox active site motif (8, 20) and lacks activity in any of the classical Grx assays (20). However, when mutated to CPYC, the motif commonly found in Grx proteins (27), the altered v3 protein also gained classical Grx activity (20). The v3 isoform, when overexpressed in human cells either as the isolated Grx domain or in fusion with the TrxR1 module as its C-terminal partner, triggers rapid changes in cell shape and a dynamic formation of cell membrane protrusions (23). GFP fusion variants of v3 were found to locate along the length and growing tips of these protrusions, in close proximity to actin. Furthermore, v3 seemed to lead actin into these protrusions followed by β-tubulin (23). These cell membrane protrusions were later characterized as having all features typical of filopodia (28). In the present study, we wished to further characterize the features of v3 that trigger these changes of the cellular phenotype and to understand how the protein is targeted to the membrane compartment. Because it was previously found that expression of either the complete TXNRD1_v3 protein or only the isolated v3(Grx) domain was sufficient for membrane targeting and induction of filopodia (23), we focused here on this property as held by the v3(Grx) domain.
By mutating the two v3(Grx) active site Cys residues to Ser and thus converting its CTRC motif to STRS, thereby incapacitating any potential redox activity of this motif, we show herein that association of v3(Grx) with actin polymerization as well as its membrane targeting is independent upon the integrity of its active site. Instead we found that N-acylation of the N terminus of v3(Grx) is both required and sufficient to target the protein to the plasma membrane. We furthermore found that it is specifically targeted to membrane rafts. These membrane structures have commonly also been called “lipid rafts,” but are in the present study named and defined according to the 2006 consensus of the Keystone Symposium on Lipid Rafts and Cell Function (29).
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
All regular chemicals or reagents were of high purity and obtained from Sigma-Aldrich, unless otherwise specified.
Cell Lines
Human A549 lung carcinoma (A549 cells) (CCL-185; ATCC) were cultured in Dulbecco's modified Eagle's medium (DMEM; Life Technologies) containing 4.5 g/liter glucose at 37 °C in a humidified atmosphere with 5% CO2. Human embryonic kidney cells (HEK293) (CRL-1573; ATCC) were cultured in Eagle's minimum essential medium (ATCC), and human ovarian SKOV3 cells (HTB-77) were cultured in McCoy's 5A medium, modified with l-glutamine and sodium bicarbonate (Sigma-Aldrich) at 37 °C in a humidified atmosphere with 5% CO2. Cell culture media were supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mm l-glutamine, 100 μg/ml streptomycin, and 100 units/ml penicillin (all from PAA Laboratories).
Vectors for Expression of the Glutaredoxin Domain of v3 in Fusion with GFP and Mutants Thereof
The construct expressing the wild type Grx domain of v3 with GFP as a C-terminal fusion partner (here called “v3(Grx)”) was kindly provided by Dr. Anastasios E. Damdimopoulos (Karolinska Institutet, Stockholm, Sweden) and was previously described in detail (23). Using standard cloning techniques with that plasmid as template, we created vectors expressing the active site double mutant C76S/C79S and the G2A and C3S mutants, as well as a variant encompassing only the first 14 amino acids of v3 in fusion with GFP (1–14), as further described under “Results.” We also used a pure GFP control. Primers were purchased from Thermo Scientific, and all constructs were sequenced by GATC Biotech to confirm the desired mutations.
Transfection and Immunocytochemistry
Cells were grown on glass chamber slides (Lab-Tek II chamber slide system, Nalge Nunc International) and transiently transfected using Lipofectamine 2000 (Invitrogen) or TurboFect transfection reagent (Thermo Scientific) according to the manufacturer's instructions. About 18 h after transfection, the slides were washed with PBS or treated with 2-hydroxymyristic or 2-bromopalmitic acid (see below) before fixation in 4% paraformaldehyde solution for 15 min. For experiments that involved subsequent actin staining, the slides were washed two times with PBS and cells were permeabilized using PBS containing 0.5% Triton X-100 and 2% BSA for 20 min at ∼20 °C whereupon they were washed two times in PBS and incubated with rhodamine-conjugated phalloidin (1:500; Molecular Probes/Invitrogen) in PBS for 1 h at ∼20 °C. For experiments involving subsequent staining of membrane rafts/caveolae, slides were then washed two times with PBS and subsequently incubated with Alexa Fluor 555-conjugated cholera toxin subunit-B (CT-B; Molecular Probes/Invitrogen) (1 mg/ml) for 20 min in chilled complete growth medium on ice. Finally, all slides were washed two times with PBS and mounted with glass coverslips (Menzel-Gläser/Thermo Scientific) using ProLong Gold antifade reagent with DAPI (4′,6-diamidino-2-phenylindole, Invitrogen). Confocal imaging was carried out on a LSM700 (Zeiss). In all cases, multicolor imaging was performed sequentially to minimize cross-talk between the channels.
Treatment with 2-Hydroxymyristic Acid (2-HMA) and 2-Bromopalmitic Acid (2-BPA)
2-Hydroxymyristic acid and 2-bromopalmitic acid (Santa Cruz Biotechnology) were stored as 100 mm stock solutions in ethanol and delivered to cells as complexes with BSA. To prepare each complex, the fatty acid compound was incubated at a concentration of 2 mm in serum-free medium containing 2 mm fatty acid-free BSA for 2 h at 37 °C. Subsequently, transfected cells were treated with the fatty acid-BSA solution for 2 h before adding serum containing full medium to final concentrations of 1 mm fatty acid, 1% (v/v) ethanol, and 5% (w/v) serum. The cells were thereupon incubated for 24 h with 2-bromopalmitic acid or for 48 h with 2-hydroxymyristic acid and subsequently prepared for immunocytochemistry as described above.
Isolation of Membrane Rafts
HEK293 cells transfected with the wild type v3(Grx), G2A, or C3S expression plasmids were harvested for membrane extractions 48–64 h after transfection. Membrane rafts were extracted by “flotation” ultracentrifugation according to methods described by Alexander et al. (30), with minor modifications. Briefly, ∼6 × 106 transfected cells were harvested and resuspended in 400 μl of flotation buffer (25 mm Tris-HCl, pH 7.4, 150 mm NaCl, 5 mm EDTA, 10 mm β-glycerol phosphate disodium salt pentahydrate, 30 mm sodium pyrophosphate, and 1% Triton X-100). The suspension was centrifuged at 2,000 × g for 5 min, the pellet was washed with 100 μl of fresh flotation buffer, and sucrose was added to the combined supernatants to a concentration of 45% (final volume of 2 ml in flotation buffer). The 2 ml of sucrose-containing lysate was transferred to the bottom of a 12.5-ml polycarbonate centrifuge tube (Beckman), over which 30% (5.5-ml) and then 5% (4.5-ml) sucrose solutions in flotation buffer were gently and sequentially layered. The tubes containing the sucrose cushions were then centrifuged at 200,000 × g for 18 h at 4 °C. 1-ml fractions (12 in total) were gently collected from the top of the gradient, snap-frozen in liquid nitrogen, and stored at −80 °C for further analysis.
Biochemical Analysis of Grx-GFP Association with Detergent-resistant Membrane Fractions
The level of expression and distribution of v3(Grx)-GFP variants in transfected HEK293 cells was assessed by Western and slot blotting approaches, respectively. Firstly, to confirm that all three variants were uniformly expressed, total cell lysates for each transfected culture were generated by pooling equal quantities of each fraction (1–12) from the membrane raft preparations. The pooled samples were diluted in PBS, reconcentrated, and analyzed by SDS-PAGE and Western blot using an HRP-conjugated monoclonal anti-GFP antibody (Rockland Immunochemicals) and standard procedures. In the experiments designed to gain insights into the oligomeric state of the raft-associated Grx-GFP proteins, only fractions 4–6 and 10–12 from each sample were pooled, and the samples were then either subjected to conventional SDS/DTT/heat treatment or treated with a DTT-free loading buffer and not heated, before loading onto gels. To control for the specificity of GFP detection, mock-transfected cells were processed and analyzed identically. To evaluate the extent of localization of the Grx-GFP variants in detergent-resistant membrane fractions, all of the fractions 1–12 from each preparation were analyzed individually by a slot blot procedure. Briefly, a 50-μl sample of each fraction was diluted 5-fold in Tris-buffered saline (TBS, pH 7.4) containing 10% methanol, with 100 μl subsequently adsorbed onto a nitrocellulose membrane using a Minifold II slot blot apparatus (Schleicher & Schuell). Detection of GFP fusion protein was carried out using the anti-GFP antibody as described above. To confirm consistency of raft enrichment for each sample, the same fractions were immobilized and probed in parallel with an HRP-conjugated CT-B (Life Technologies), which binds the exclusively membrane raft-residing GM1 ganglioside with high affinity.
RESULTS
Membrane Targeting of v3 by Myristoylation and Palmitoylation at Its N-terminal MGC Motif
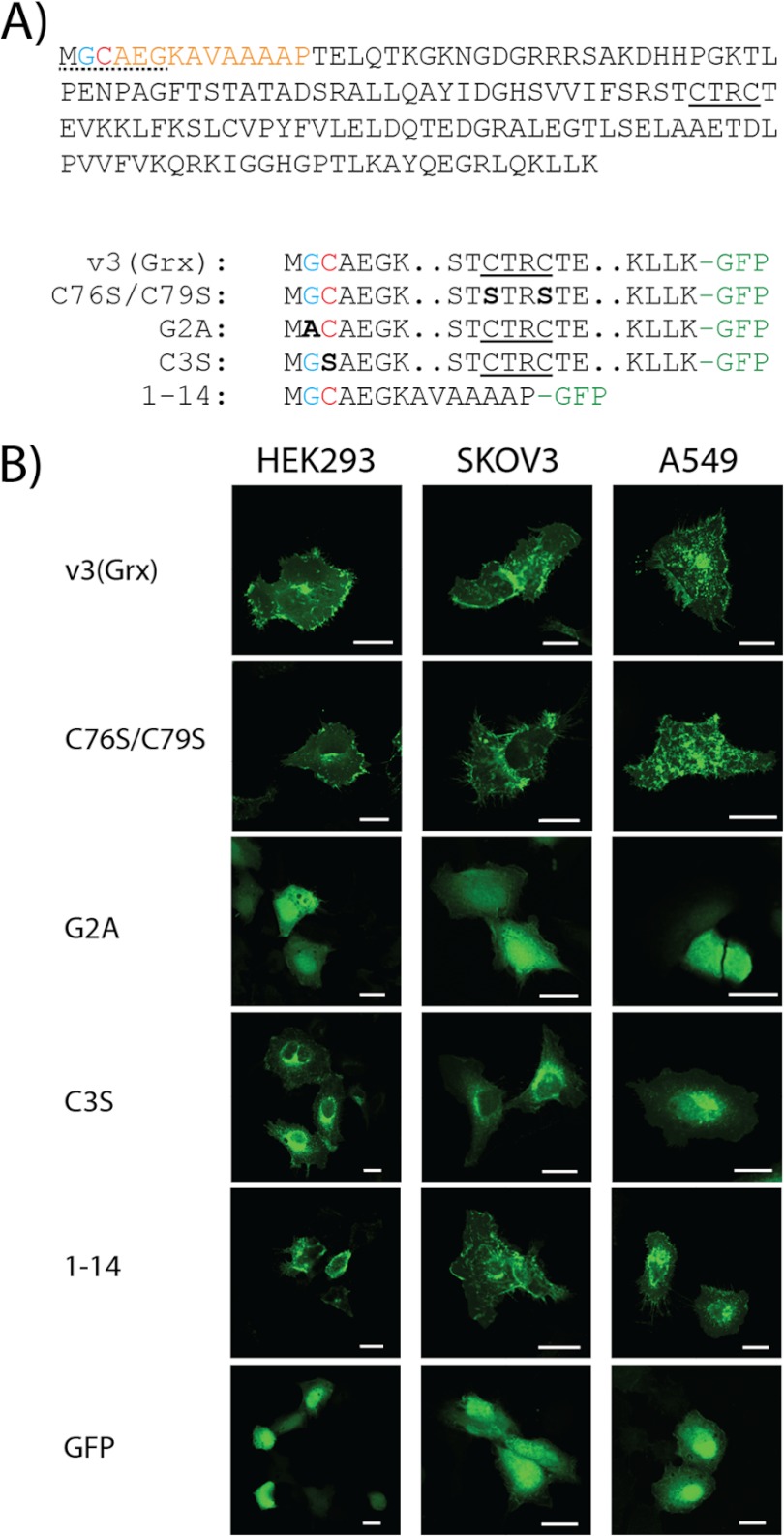
When overexpressed in cancer cells, the glutaredoxin domain of v3 (v3(Grx)) in fusion with GFP at its C-terminal end displays a distinct localization pattern that is characterized by strong staining of the perinuclear region and cytosolic speckles as well as accumulation of the protein at the plasma membrane (23, 28). An N-terminal myristoylation motif of v3 was suggested using ExPASy Prosite and NMT – The MYR Predictor, as reported earlier (8, 28, 31, 32), but this has not yet been experimentally studied. The Grx domain of v3 also carries an atypical dithiol active site motif, as discussed above. To characterize the importance of these motifs for v3 targeting to cell membrane regions, we expressed a number of v3(Grx)-derived mutant variants in fusion with GFP (see Fig. 1A for a scheme of the constructs) using three different human cell lines (A549, HEK293, and SKOV3) (Fig. 1B). This revealed that a variant with the two Cys moieties of the redox active site changed to redox inactive Ser residues (C76S/C79S) yielded an identical phenotype of membrane association as seen with wild type v3(Grx), which was highly reminiscent of that reported earlier for the wild type protein (23, 28). Both variants strongly accumulated in the perinuclear area and showed distinct cytosolic structures in a dotted pattern, as well as a pronounced plasma membrane association in all three cell types (Fig. 1B). Interestingly, however, a single substitution of the Gly residue at position 2 with Ala (G2A) completely abolished the membrane association of the protein. This G2A mutant, destroying the myristoylation consensus motif (33–35), showed a diffuse cytosolic and nuclear distribution similar to that of pure GFP (Fig. 1B). Thus, the prominent features of plasma membrane association, cytosolic speckles and strong perinuclear accumulation, were all impeded by this single amino acid substitution. In contrast, substituting solely the Cys residue at position 3 with Ser, yielding the (C3S) construct that is expected to eliminate the possibility of palmitoylation (35, 36) while maintaining the myristoylation site at the Gly-2 residue (35, 37, 38), lowered the extent of plasma membrane association and the amount of cytosolic speckles, but maintained a strong compartmentalization of the protein, with mainly perinuclear localization (Fig. 1B). To study whether the membrane targeting could indeed be guided solely by acylation of the N-terminal motif of v3, we also analyzed a construct encompassing only the first 14 amino acids of v3(Grx) in fusion with GFP. Cells expressing this protein displayed the same phenotype as seen with wild type v3(Grx) (Fig. 1B). Together, these results revealed that the N-terminal MGC motif of v3(Grx) is both required and sufficient for the targeting of this protein to specific membrane structures of the transfected cells.
FIGURE 1.
The redox active site dithiol motif of v3 is dispensable, whereas its N-terminal acylation motif is required for membrane association of the v3(Grx) domain. A, the Grx domain of v3 is here shown with the proposed consensus sequence for myristoylation underlined (dotted; presumed myristoylated Gly residue shown in blue) at the beginning of a 14-amino acid stretch at the N terminus of the protein (orange). A Cys-3 residue may potentially be palmitoylated (red). The dithiol active site motif of v3(Grx) is underlined (solid). Final amino acid sequences of the v3(Grx)-derived constructs studied herein are schematically indicated below. Mutated residues are marked bold and in black with the names of the variants to the left. Two dots indicate omitted residues (for full sequence, see top panel), and -GFP in green indicates a C-terminal GFP fusion partner. B, A549, HEK293, and SKOV3 cells were transfected using either of the v3(Grx)-derived GFP fusion constructs: wild type v3(Grx), the active site mutant C76S/C79S, the N-terminal G2A or C3S mutants, or a truncated variant solely containing the first 14 amino acids of v3 (1–14). Control transfections were performed using a construct expressing GFP alone (GFP). Fluorescence was recorded 18 h after transfection using confocal microscopy. Scale bar = 20 μm.
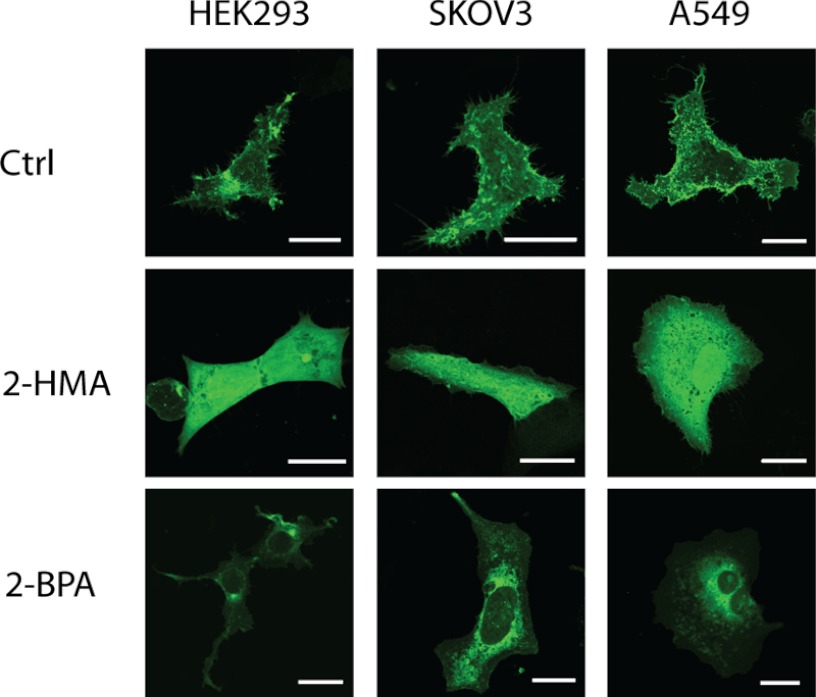
To further study the dependence of the v3 membrane targeting upon myristic and palmitic acid, we incubated A549, HEK293, or SKOV3 cells expressing v3(Grx) with 2-hydroxymyristic acid (2-HMA) and 2-bromopalmitic acid (2-BPA), two competitive inhibitors of myristoylation and palmitoylation, respectively (39–41). After treatment with 2-BPA, the subcellular localization of the protein changed to a mainly perinuclear localization with significantly reduced plasma membrane staining and fewer cytosolic speckles (Fig. 2). This pattern coincides with the expression profile of the palmitoylation-impeded C3S variant (compare Fig. 1B and Fig. 2). The 2-HMA treatment, on the other hand, nearly completely abolished compartmentalization of the protein and gave a diffuse cytosolic distribution closely reminiscent of pure GFP or the G2A variant of v3 (compare Fig. 1B and Fig. 2).
FIGURE 2.
Inhibition of myristoylation and palmitoylation by 2-HMA and 2-BPA. A549, HEK293, and SKOV3 cells were transfected using the wild type v3(Grx)-GFP fusion construct. 18 h after transfection, cells were treated with either 1% EtOH or 2-hydroxymyristate for 48 h or with 2-bromopalmitate for 24 h. GFP fluorescence was recorded using confocal microscopy. Scale bar = 20 μm. Ctrl, control.
Co-localization of v3(Grx) with the Membrane Raft Marker CT-B
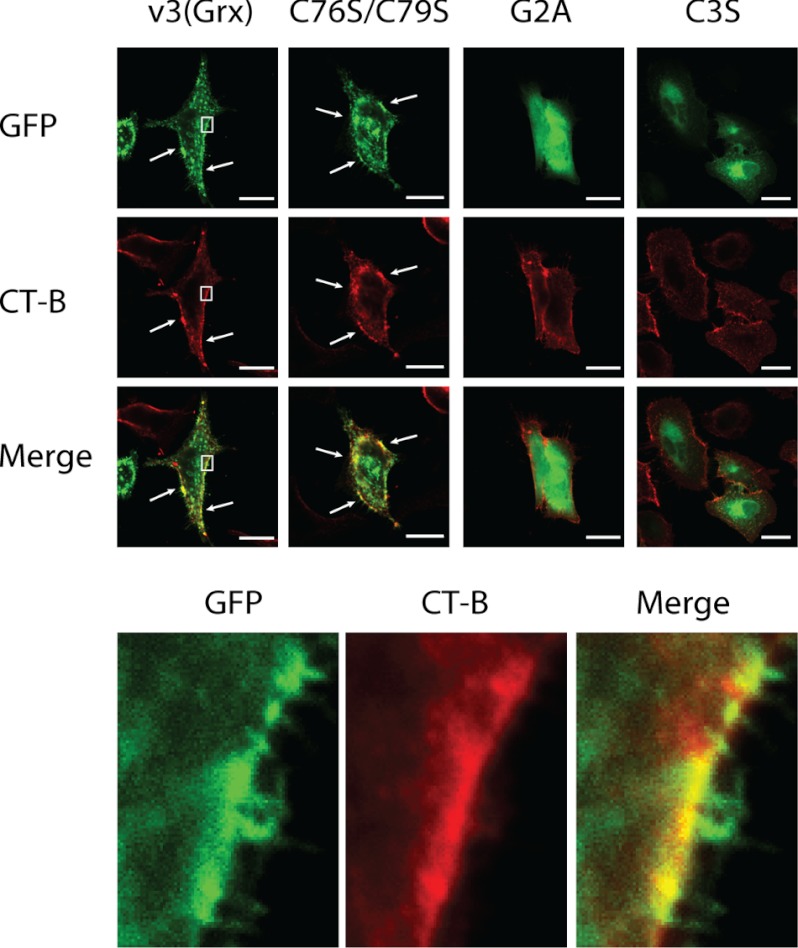
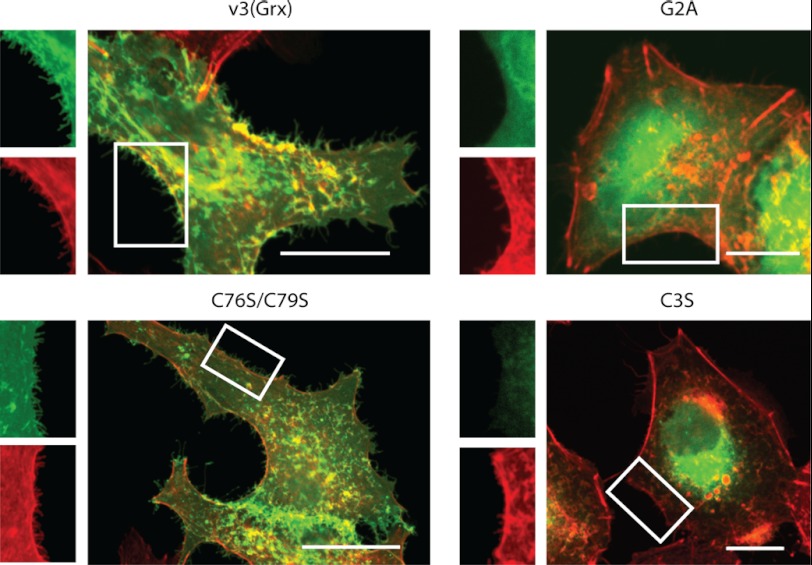
Next we investigated the subcellular localization of v3(Grx) in relation to membrane structures binding CT-B, a marker often used for membrane rafts (42–44). For this, A549 cells were transfected with the GFP fusion constructs expressing wild type v3(Grx), the active site mutant C76S/C79S, or the N-terminal mutants G2A and C3S. To subsequently assess localization in relation to the CT-B marker for membrane rafts, the cells were fixed 24 h after transfection and incubated with Alexa Fluor 555-conjugated CT-B. Incubation with CT-B did not affect the overall cellular appearance nor the subcellular GFP fluorescence patterns obtained with the various v3(Grx) variants. CT-B showed a similar pattern in all of the cells, with plasma membrane staining in selected localized areas, as well as dotted cytosolic and perinuclear distribution. The latter compartments were previously identified as being early endosomes and Golgi apparatus or endoplasmic reticulum, respectively (45, 46). The GFP signal of the v3(Grx) and C76S/C79S variants closely overlapped with that of CT-B staining, although the overlap was not exclusive, and fractions of the cells also showed staining for only one of the fluorophores (Fig. 3, arrows and magnified lower panel). In contrast, the G2A variant of v3(Grx), which displayed a diffuse cytosolic distribution, lacked subcellular proximity with CT-B in all cellular compartments (Fig. 3). Devoid of the palmitoylation site (Cys-3) but maintaining an intact myristoylation site at Gly-2, the C3S variant displayed mainly its perinuclear distribution, where it showed some overlap with CT-B, but appeared only minimally at the plasma membrane (Fig. 3) as also shown above.
FIGURE 3.
Co-localization of v3(Grx)-GFP fusion variants with the membrane raft marker CT-B. A549 cells were transfected with constructs expressing either the wild type v3(Grx) domain or mutants thereof (C76S/C79S, G2A, or C3S) fused to GFP, as indicated. 18 h after transfection, the cells were fixated and stained using Alexa Fluor 555-conjugated CT-B for 20 min on ice to visualize membrane rafts. Fluorescence was acquired using confocal microscopy. The lower panel shows a higher magnification of the selected area in the cells transfected with the wild type v3(Grx) (white rectangle). Regions with a high degree of co-localization are indicated by arrows. Scale bar = 20 μm.
These results strongly suggested that overexpressed v3 in transfected cells becomes targeted to cell membranes through myristoylation and palmitoylation, where it furthermore closely associates with the membrane raft marker CT-B. We next wished to confirm the localization of v3(Grx) in membrane rafts by the alternative method of membrane fractionation.
Appearance of v3(Grx) in CT-B-positive Purified Membrane Raft Fractions
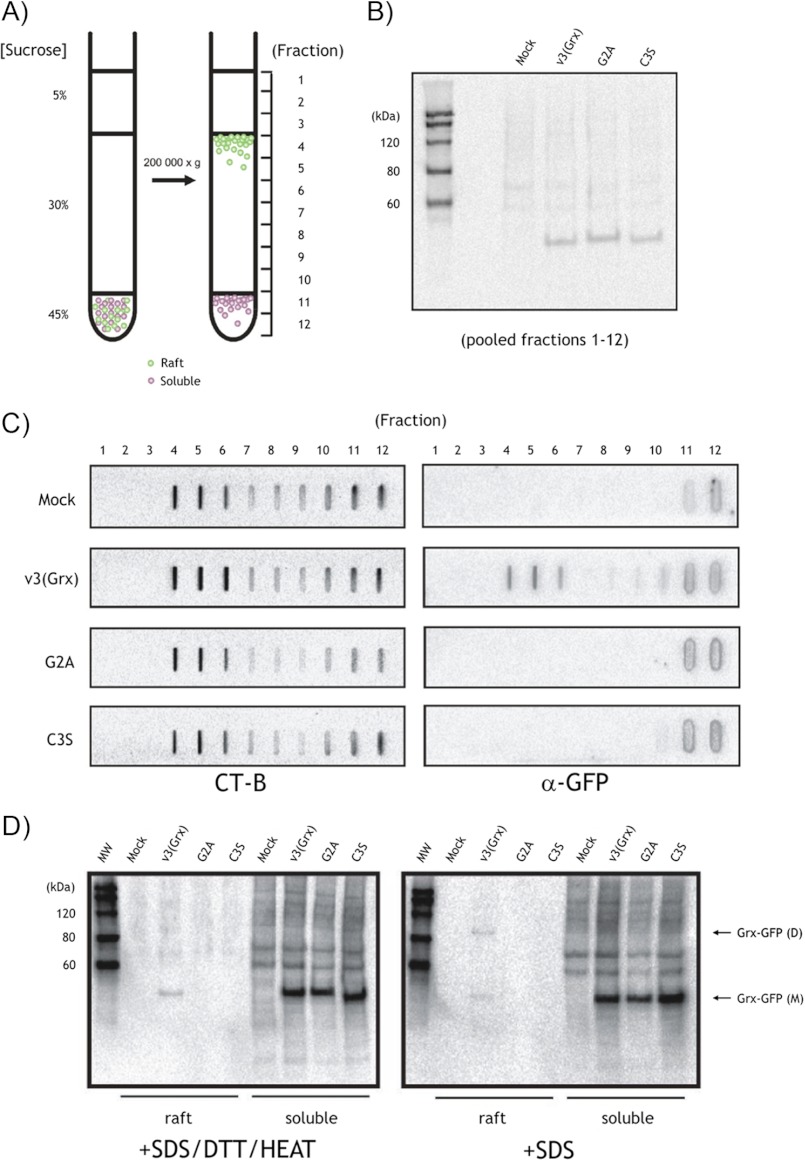
The proposed sizes of membrane raft microdomains are below 100 nm and are thus not resolvable by conventional confocal microscopy, thereby limiting the interpretation of co-localization studies by microscopy. To further validate co-localization of v3(Grx) with CT-B, we therefore purified membrane rafts from HEK293 cells expressing either the wild type v3(Grx) or the G2A or C3S variants and analyzed these by slot and Western blot approaches. The membrane rafts were purified by conventional flotation methods, which involve separation of intact, detergent-resistant rafts from Triton X-100 solubilized membranes and cytosolic proteins, based on their unique “buoyancy” in sucrose-containing media subjected to high speed centrifugation (Fig. 4A, left). The v3(Grx), G2A, and C3S variants were all expressed as similar levels and recovered to the same extent in this centrifugation, as visualized using immunoblotting with antibodies directed against GFP, which were used to probe pooled fractions (Fig. 4B). In agreement with the co-localization data suggested by fluorescent microscopy, only the wild type v3(Grx) variant showed association with membrane rafts (fractions 4–6), whereas the G2A and C3S variants were not detected in these membrane microdomain fractions (Fig. 4C, right panel). This effect was specific and directly related to the amino acid sequences of the N-terminal raft-localization domain of the proteins because the amounts of purified membrane rafts, measured by enrichment of CT-B-binding lipids in fractions 4–6, were comparable between the samples (Fig. 4C, left panel).
FIGURE 4.
Localization of wild type v3(Grx) in purified, CT-B-positive membrane raft fractions. A, principle of membrane raft purification by centrifugation and flotation. HEK293 cells expressing the v3(Grx)-GFP fusion variants were harvested and lysed 48 h upon transfection. The Triton X-100-solubilized cell lysate was overlaid sequentially with the indicated sucrose cushions, and the samples were centrifuged at 200,000 × g for 18 h. Consecutive 1-ml fractions (fractions 1–12) were collected from the top of the centrifuge tubes. B, Western blot analysis of pooled fractions (fractions 1–12) showing uniform expression of the ∼48-kDa v3(Grx)-GFP variants in all transfected cultures. C, slot blot analysis of individual fractions 1–12, probed with an HRP-conjugated CT-B (left panel) or GFP antibody (α-GFP, right panel). MOCK, mock-transfected. D, fractions 4–6 (raft) or fractions 11–12 (soluble) of each sample were pooled and analyzed by Western blot for the presence of v3(Grx)-GFP fusion variants using α-GFP. The samples were either treated with SDS, DTT, and heat or treated only with SDS. The monomeric v3(Grx)-GFP variants (Grx-GFP (M)) as well as the dimeric form of the wild type variant v3(Grx) (Grx-GFP (D)) are indicated by arrows. MW, molecular size markers.
To additionally confirm the findings and gain further insights into the nature of the raft-associated v3(Grx)-GFP, we next analyzed fractions 4–6 (rafts) and fractions 11–12 (soluble) by conventional and “mildly denaturing, nonreducing” SDS-PAGE/Western blot protocols. For this, we pooled fractions 4–6 and 11–12 and then treated them with SDS without or with DTT and heat, before separation on SDS-PAGE and subsequent Western blot analyses. Although all v3(Grx) variants displayed a clear and equally strong signal at the expected size of ∼48 kDa in the soluble fractions, again confirming comparable total expression levels, only the wild type v3(Grx) variant was seen in the membrane raft fractions. This membrane raft-associated protein appeared partly in the form of a dimer in the absence of DTT and heat, whereas reducing and denaturing conditions resolved the dimeric band into a solely monomeric protein (Fig. 4D).
Induction of Filopodia, Changes of Cell Morphology, and Effects on Actin Polymerization
Concomitant with its compartmentalized membrane targeting, v3(Grx) overexpression was previously found to be correlated with actin polymerization and induction of cell membrane protrusions, identified as filopodia (23, 28). Here we analyzed how these features compared between wild type v3(Grx) and the C76S/C79S, G2A, or C3S mutants, as visualized using detection of the GFP fusion partner and co-staining of actin with rhodamine-conjugated phalloidin. Both v3(Grx) and actin appeared localized in close proximity with each other at the cell membrane, as found earlier (28). This was also seen with the redox active site mutant C76S/C79S (Fig. 5). Particularly strong accumulation could be seen at cell-to-cell contact sites (Fig. 5, see C76S/C79S variant). As also found above, cell membrane accumulation required the Gly-2 and Cys-3 residues (Fig. 5), i.e. uncompromised N-terminal myristoylation and palmitoylation motifs, respectively (Fig. 1A). Expression of v3(Grx) as well as C76S/C79S triggered high increases in the number of filopodia-like membrane protrusions, and at higher magnification, both proteins could be seen directly associated with these protrusions (Fig. 5). In contrast, the mainly cytosolic G2A and preferentially perinuclear C3S variants did not induce this filopodia-enriched cellular phenotype, although a few membrane protrusions were also seen in cells expressing these proteins (Fig. 5).
FIGURE 5.
The relation of v3(Grx) to cell morphology and actin polymerization is independent of the active site but requires a functional N-acylation motif. A549 cells were transfected using the indicated v3(Grx)-GFP fusion constructs. Cells were additionally stained for actin using rhodamine-conjugated phalloidin, and fluorescence was recorded using confocal microscopy. The pictures show merged fluorescent signals of GFP (green) and rhodamine (red) after excitation at 488 and 555 nm, respectively. Higher magnifications of selected areas, showing only GFP or rhodamine signal, are displayed for all variants as indicated. The intensity and contrast of the magnified areas were optimized for visualization of filopodia. Scale bar = 20 μm.
DISCUSSION
Here we found that the association of v3 with cell membranes was independent of its dithiol redox active site motif and fully governed by targeting to membrane rafts through its N-terminal MGC myristoylation and palmitoylation motif. This targeting of the protein to membrane rafts was also sufficient and required to support v3-stimulated increases in the number of cell membrane filopodia.
The N-terminal MGCAEG sequence of v3 meets the general consensus motif for myristoylation, even if the second Gly residue at position 6 deviates from more commonly seen Ser or Thr residues in myristoylated proteins (8, 31, 32). Upon further inspection of this sequence, we noted that the Cys-3 residue might potentially be palmitoylated, which led us to the construction and characterization of the C3S variant. The results presented herein indeed strongly suggest that the Cys-3 residue of v3 is palmitoylated, as judged from the typical subcellular targeting patterns of the different fusion variant proteins and their changes in localization upon treatment with the palmitoylation inhibitor 2-BPA. N-Acylation of proteins is a rather well characterized process. It occurs predominantly co-translationally with myristic acid (C14:0) linked via an amide bond to the N-terminal Gly residue in N-acylation motifs of targeted proteins. This myristoylation will, however, not provide stable membrane attachment but serves to increase the hydrophobicity of the N-terminal end of the modified protein to facilitate transient membrane association (47). Myristoylated membrane-associated proteins thus gain spatial access to membrane-bound DHHC (Asp-His-His-Cys) domain proteins that may subsequently catalyze the addition of palmitic acid (C16:0) to Cys residues located adjacent to the myristoylation site, which further increases the hydrophobicity of a target protein. In this manner, palmitoylation yields stable binding or association to membranes of N-acylated proteins (48). Our substitution of Gly-2 in v3(Grx) with Ala as well as the treatment with 2-HMA resulted in diffuse cellular distribution, similar to sole expression of GFP. We suggest that myristoylation of v3 at the Gly-2 residue is the only explanation for the observed protein localization phenotypes. Importantly, all of the unique subcellular targeting features of v3(Grx)-GFP, including plasma membrane association, formation of cytosolic speckles, and strong perinuclear accumulation, were completely impeded by this single Gly-for-Ala amino acid substitution or by use of the inhibitor of myristoylation, hence strongly suggesting that v3 is indeed N-acylated at its MGC motif. The C3S mutant also showed a clear reduction of plasma membrane association and cytosolic speckles, whereas maintaining strong compartmentalization with mainly perinuclear localization. This phenotype was also highly similar to the change in subcellular targeting of the wild type v3(Grx) if the cells were treated with the palmitoylation inhibitor 2-BPA and also exactly mimicked the phenotype of other N-acylated proteins, having eliminated N-palmitoylation but maintained N-myristoylation (35, 37, 38). We thereby suggest that v3 is myristoylated at its Gly-2 residue and palmitoylated at its Cys-3 residue.
The permanently strong perinuclear staining of wild type v3(Grx) and the C76S/C79S variant should be the result of highly regulated cellular palmitoylation processes (49). The dynamics of palmitoylation and depalmitoylation have been studied for several N-acylated proteins, including Ras, endothelial nitric-oxide synthase (eNos), GAP43, and Giα1, or a number of model peptides, which all display subcellular localization patterns highly similar to those found here for v3 (49–51). As an example, H- and N-Ras need to acquire palmitoylation to achieve stable membrane association and trafficking between the Golgi and plasma membrane, thereby yielding patterns of localization highly reminiscent of those seen here for v3 (51). Also, expressing a synthetic protein with a consensus myristoylation motif including a palmitoylable Cys residue (MGCTLS-), Navarro-Lérida et al. (35) found very similar distribution of that model protein as that seen here with v3; a G2A mutant completely impeded membrane association, whereas a C3S mutant showed perinuclear distribution representing accumulation in the Golgi apparatus. Thus, here we found that v3 displays typical compartmentalization properties as previously shown for other proteins that are myristoylated and palmitoylated at their N-acylation motifs.
The close overlap of v3(Grx) with CT-B-stained specific substructures of the plasma membrane and in perinuclear areas, but not in the intracellular vesicles, was a striking finding. With CT-B being a well recognized probe for membrane rafts, the overlap in signal with wild type v3(Grx) strongly suggested to us that the protein was targeted to membrane rafts and the Golgi, where raft-specific gangliosides to which CT-B bind are known to accumulate (45, 46). The intracellular vesicles that solely showed CT-B-coupled fluorescence were likely endosomes (45, 46), and it should therefore not be surprising that they lacked the v3(Grx)-GFP signal. Because the v3-derived fusion proteins were intracellularly expressed, they would not co-localize with CT-B in endosomes, carrying extracellular proteins as taken up from the medium. The resolution of conventional confocal microscopy, however, cannot resolve membrane rafts as these are thought to be dynamic membrane domains with sizes of less than 100 nm in length (29, 52). We therefore purified CT-B-binding membrane raft fractions from HEK293 cells expressing v3(Grx) and probed for co-localization using immunoblotting. It was thereby notable that we could not only validate membrane raft association of wild type v3(Grx), but additionally detected a loosely associated dimeric form of the protein, which was specifically seen in the membrane raft fractions. We propose that this dimeric variant could have been formed by the GFP domain, which has a tendency to dimerize at high concentrations (53). Overexpression of v3(Grx)-GFP in combination with the partitioning into membrane rafts should likely create a high local concentration of the protein, thereby triggering dimerization. Such effects have previously been reported by Zacharias et al. (37) who studied partitioning of GFP variants into membrane microdomains by FRET. Expressing myristoylated and palmitoylated 13-amino acid NH2-terminal fragments of the Lyn protein in fusion with either CFP or YFP, they could detect strong clustering at the plasma membrane (37). They also showed that hydrophobic residues of the GFP dimer interface contributed to dimer formation specifically at the membrane microdomains (37).
Partitioning of v3 into membrane rafts may also give further insights to the close association of v3 with actin polymerization and the stimulated generation of filopodia. With regards to signaling via glycosylphosphatidylinositol-anchored proteins, it is well known that actin plays a central role in organization of sphingolipid- and cholesterol-rich membrane domains and is tightly linked to these structures (43, 54–56). Actin has indeed been described as a stabilizer of membrane rafts (57), with proteins targeted to these structures being able to affect the organization of the cytoskeleton (58, 59). Additionally, several studies link induction of filopodia to stimulation or patching of membrane raft microdomains (43, 60–63). An example that is reminiscent of expression of v3(Grx) with induction of filopodia was given by Gauthier-Campbell and co-workers (61, 62), studying phenotypes of Cos-7 cells as induced by expression of diverse palmitoylated peptides. In particular, a 14-amino acid doubly palmitoylated peptide derived from GAP43 (GAP-1–14) induced formation of filopodia when expressed in fusion with GFP (61, 62). Another example reminiscent of our findings herein is the neuronal glycoprotein M6a, which also associates with membrane rafts and induces formation of filopodia (63). Thus, our findings open the possibility that v3 is first targeted to membrane rafts through N-acylation, whereupon the protein can interact with actin in a compartmentalized manner, directly or indirectly, which may trigger the generation of filopodia as observed.
The role(s) of endogenous v3 in membrane rafts are still unknown, but its targeting to these structures by N-acylation clearly expands the possible spectrum of TXNRD1-derived protein functions. The Trx and Grx systems are generally not well studied in terms of signal regulation within membrane rafts, although these structures are known as key players in redox signaling events (64). However, a few studies have reported upon Trx1 association with membrane rafts, in leukocyte-endothelial cell interaction during inflammation (65) or when internalized through endocytosis (66). Interestingly, Volonte and Galbiati (67) showed that caveolin 1, a key protein of caveolae, is an inhibitor of TrxR1 through direct binding via a proposed caveolin-binding motif of TrxR1 (amino acids 454–463). They also showed that a constitutively active variant of TrxR1, lacking the caveolin-binding motif, could inhibit oxidative-stress-mediated activation of p53/p21waf1/Cip1 and induction of premature senescence (67). It is not yet clear whether or how those findings relate to targeting of v3 to membrane rafts. A large number of proteins important for cellular signaling events are myristoylated and located to membrane rafts, including several Src family tyrosine kinases and other protein kinases, phosphatases, Ca2+-binding proteins, cytoskeleton-binding proteins, viral proteins, and specific redox-related proteins such as NO synthases (31). The v3 protein should hereby also be considered in the context of cellular signaling through membrane rafts, and its possible relation to cellular signaling events clearly deserves further study.
In conclusion, we have herein identified the mechanisms for targeting of v3 to membrane rafts to be dependent upon its N-terminal motif and likely to involve myristoylation at Gly-2 and palmitoylation at its Cys-3 residue. We also showed that the induction of filopodia triggered by overexpression of v3(Grx) was independent of its redox active site motif, but required its ability to associate with membrane microdomains.
Acknowledgment
Dr. Anastasios E. Damdimopoulos is acknowledged for kindly providing plasmid constructs.
This work was supported by the Swedish Research Council (Medicine), the Swedish Cancer Society, The Wallenberg Foundations, and the Karolinska Institutet. This work was also supported in part by the Medical Research Council (MRC) and Polio Research Foundation (PRF) of South Africa (to A. C. and N. M.).
- TrxR
- thioredoxin reductase
- Trx
- thioredoxin
- TXNRD1_v3
- splice variant of human TrxR1 carrying an additional N-terminal glutaredoxin domain
- TXNRD1
- human gene encoding TrxR1
- CT-B
- cholera toxin subunit-B
- Grx
- glutaredoxin
- v3
- short notation for the TXNRD1_v3 splice variant
- v3(Grx)
- glutaredoxin domain of v3
- 2-HMA
- 2-hydroxymyristic acid
- 2-BPA
- 2-bromopalmitic acid
- GAP
- growth-associated protein.
REFERENCES
- 1. Nakamura H., Nakamura K., Yodoi J. (1997) Redox regulation of cellular activation. Annu. Rev. Immunol. 15, 351–369 [DOI] [PubMed] [Google Scholar]
- 2. Nordberg J., Arnér E. S. (2001) Reactive oxygen species, antioxidants, and the mammalian thioredoxin system. Free Rad. Biol. Med. 31, 1287–1312 [DOI] [PubMed] [Google Scholar]
- 3. Arnér E. S., Holmgren A. (2000) Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 267, 6102–6109 [DOI] [PubMed] [Google Scholar]
- 4. Rundlöf A. K., Arnér E. S. (2004) Regulation of the mammalian selenoprotein thioredoxin reductase 1 in relation to cellular phenotype, growth, and signaling events. Antioxid. Redox Signal. 6, 41–52 [DOI] [PubMed] [Google Scholar]
- 5. Gromer S., Urig S., Becker K. (2004) The thioredoxin system–from science to clinic. Med. Res. Rev. 24, 40–89 [DOI] [PubMed] [Google Scholar]
- 6. Arnér E. S., Holmgren A. (2006) The thioredoxin system in cancer. Semin. Cancer Biol. 16, 420–426 [DOI] [PubMed] [Google Scholar]
- 7. Arnér E. S. (2009) Focus on mammalian thioredoxin reductases–important selenoproteins with versatile functions. Biochim. Biophys. Acta 1790, 495–526 [DOI] [PubMed] [Google Scholar]
- 8. Rundlöf A. K., Janard M., Miranda-Vizuete A., Arnér E. S. (2004) Evidence for intriguingly complex transcription of human thioredoxin reductase 1. Free Rad. Biol. Med. 36, 641–656 [DOI] [PubMed] [Google Scholar]
- 9. Sun Q. A., Zappacosta F., Factor V. M., Wirth P. J., Hatfield D. L., Gladyshev V. N. (2001) Heterogeneity within animal thioredoxin reductases: evidence for alternative first exon splicing. J. Biol. Chem. 276, 3106–3114 [DOI] [PubMed] [Google Scholar]
- 10. Rigobello M. P., Callegaro M. T., Barzon E., Benetti M., Bindoli A. (1998) Purification of mitochondrial thioredoxin reductase and its involvement in the redox regulation of membrane permeability. Free Rad. Biol. Med. 24, 370–376 [DOI] [PubMed] [Google Scholar]
- 11. Miranda-Vizuete A., Damdimopoulos A. E., Pedrajas J. R., Gustafsson J. A., Spyrou G. (1999) Human mitochondrial thioredoxin reductase cDNA cloning, expression, and genomic organization. Eur. J. Biochem. 261, 405–412 [DOI] [PubMed] [Google Scholar]
- 12. Lee S. R., Kim J. R., Kwon K. S., Yoon H. W., Levine R. L., Ginsburg A., Rhee S. G. (1999) Molecular cloning and characterization of a mitochondrial selenocysteine-containing thioredoxin reductase from rat liver. J. Biol. Chem. 274, 4722–4734 [DOI] [PubMed] [Google Scholar]
- 13. Su D., Novoselov S. V., Sun Q. A., Moustafa M. E., Zhou Y., Oko R., Hatfield D. L., Gladyshev V. N. (2005) Mammalian selenoprotein thioredoxin-glutathione reductase: roles in disulfide bond formation and sperm maturation. J. Biol. Chem. 280, 26491–26498 [DOI] [PubMed] [Google Scholar]
- 14. Sun Q. A., Kirnarsky L., Sherman S., Gladyshev V. N. (2001) Selenoprotein oxidoreductase with specificity for thioredoxin and glutathione systems. Proc. Natl. Acad. Sci. U.S.A. 98, 3673–3678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sun Q. A., Su D., Novoselov S. V., Carlson B. A., Hatfield D. L., Gladyshev V. N. (2005) Reaction mechanism and regulation of mammalian thioredoxin/glutathione reductase. Biochemistry 44, 14528–14537 [DOI] [PubMed] [Google Scholar]
- 16. Jakupoglu C., Przemeck G. K., Schneider M., Moreno S. G., Mayr N., Hatzopoulos A. K., de Angelis M. H., Wurst W., Bornkamm G. W., Brielmeier M., Conrad M. (2005) Cytoplasmic thioredoxin reductase is essential for embryogenesis but dispensable for cardiac development. Mol. Cell. Biol. 25, 1980–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Conrad M., Jakupoglu C., Moreno S. G., Lippl S., Banjac A., Schneider M., Beck H., Hatzopoulos A. K., Just U., Sinowatz F., Schmahl W., Chien K. R., Wurst W., Bornkamm G. W., Brielmeier M. (2004) Essential role for mitochondrial thioredoxin reductase in hematopoiesis, heart development, and heart function. Mol. Cell. Biol. 24, 9414–9423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Matsui M., Oshima M., Oshima H., Takaku K., Maruyama T., Yodoi J., Taketo M. M. (1996) Early embryonic lethality caused by targeted disruption of the mouse thioredoxin gene. Dev. Biol. 178, 179–185 [DOI] [PubMed] [Google Scholar]
- 19. Nonn L., Williams R. R., Erickson R. P., Powis G. (2003) The absence of mitochondrial thioredoxin 2 causes massive apoptosis, exencephaly, and early embryonic lethality in homozygous mice. Mol. Cell. Biol. 23, 916–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Su D., Gladyshev V. N. (2004) Alternative splicing involving the thioredoxin reductase module in mammals: a glutaredoxin-containing thioredoxin reductase 1. Biochemistry 43, 12177–12188 [DOI] [PubMed] [Google Scholar]
- 21. Rundlöf A. K., Carlsten M., Giacobini M. M., Arnér E. S. (2000) Prominent expression of the selenoprotein thioredoxin reductase in the medullary rays of the rat kidney and thioredoxin reductase mRNA variants differing at the 5′ untranslated region. Biochem. J. 347, 661–668 [PMC free article] [PubMed] [Google Scholar]
- 22. Osborne S. A., Tonissen K. F. (2001) Genomic organisation and alternative splicing of mouse and human thioredoxin reductase 1 genes. BMC Genomics 2, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dammeyer P., Damdimopoulos A. E., Nordman T., Jiménez A., Miranda-Vizuete A., Arnér E. S. (2008) Induction of cell membrane protrusions by the N-terminal glutaredoxin domain of a rare splice variant of human thioredoxin reductase 1. J. Biol. Chem. 283, 2814–2821 [DOI] [PubMed] [Google Scholar]
- 24. Rundlöf A. K., Fernandes A. P., Selenius M., Babic M., Shariatgorji M., Nilsonne G., Ilag L. L., Dobra K., Björnstedt M. (2007) Quantification of alternative mRNA species and identification of thioredoxin reductase 1 isoforms in human tumor cells. Differentiation 75, 123–132 [DOI] [PubMed] [Google Scholar]
- 25. Rundlöf A. K., Carlsten M., Arnér E. S. (2001) The core promoter of human thioredoxin reductase 1: cloning, transcriptional activity, and Oct-1, Sp1, and Sp3 binding reveal a housekeeping-type promoter for the AU-rich element-regulated gene. J. Biol. Chem. 276, 30542–30551 [DOI] [PubMed] [Google Scholar]
- 26. Hintze K. J., Wald K. A., Zeng H., Jeffery E. H., Finley J. W. (2003) Thioredoxin reductase in human hepatoma cells is transcriptionally regulated by sulforaphane and other electrophiles via an antioxidant response element. J. Nutr. 133, 2721–2727 [DOI] [PubMed] [Google Scholar]
- 27. Lillig C. H., Holmgren A. (2007) Thioredoxin and related molecules–from biology to health and disease. Antioxid. Redox Signal. 9, 25–47 [DOI] [PubMed] [Google Scholar]
- 28. Damdimopoulou P. E., Miranda-Vizuete A., Arnér E. S., Gustafsson J. A., Damdimopoulos A. E. (2009) The human thioredoxin reductase-1 splice variant TXNRD1_v3 is an atypical inducer of cytoplasmic filaments and cell membrane filopodia. Biochim. Biophys. Acta 1793, 1588–1596 [DOI] [PubMed] [Google Scholar]
- 29. Pike L. J. (2006) Rafts defined: a report on the Keystone Symposium on Lipid Rafts and Cell Function. J. Lipid Res. 47, 1597–1598 [DOI] [PubMed] [Google Scholar]
- 30. Alexander M., Bor Y. C., Ravichandran K. S., Hammarskjöld M. L., Rekosh D. (2004) Human immunodeficiency virus type 1 Nef associates with lipid rafts to downmodulate cell surface CD4 and class I major histocompatibility complex expression and to increase viral infectivity. J. Virol. 78, 1685–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Resh M. D. (1999) Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta 1451, 1–16 [DOI] [PubMed] [Google Scholar]
- 32. van't Hof W., Resh M. D. (2000) Targeting proteins to plasma membrane and membrane microdomains by N-terminal myristoylation and palmitoylation. Methods Enzymol. 327, 317–330 [DOI] [PubMed] [Google Scholar]
- 33. Towler D. A., Adams S. P., Eubanks S. R., Towery D. S., Jackson-Machelski E., Glaser L., Gordon J. I. (1987) Purification and characterization of yeast myristoyl CoA:protein N-myristoyltransferase. Proc. Natl. Acad. Sci. U.S.A. 84, 2708–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bhatnagar R. S., Fütterer K., Waksman G., Gordon J. I. (1999) The structure of myristoyl-CoA:protein N-myristoyltransferase. Biochim. Biophys. Acta 1441, 162–172 [DOI] [PubMed] [Google Scholar]
- 35. Navarro-Lérida I., Alvarez-Barrientos A., Gavilanes F., Rodriguez-Crespo I. (2002) Distance-dependent cellular palmitoylation of de-novo-designed sequences and their translocation to plasma membrane subdomains. J. Cell Sci. 115, 3119–3130 [DOI] [PubMed] [Google Scholar]
- 36. Grassie M. A., McCallum J. F., Guzzi F., Magee A. I., Milligan G., Parenti M. (1994) The palmitoylation status of the G-protein G(o)1α regulates its activity of interaction with the plasma membrane. Biochem. J. 302, 913–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zacharias D. A., Violin J. D., Newton A. C., Tsien R. Y. (2002) Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296, 913–916 [DOI] [PubMed] [Google Scholar]
- 38. Wolven A., Okamura H., Rosenblatt Y., Resh M. D. (1997) Palmitoylation of p59fyn is reversible and sufficient for plasma membrane association. Mol. Biol. Cell 8, 1159–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nadler M. J., Harrison M. L., Ashendel C. L., Cassady J. M., Geahlen R. L. (1993) Treatment of T cells with 2-hydroxymyristic acid inhibits the myristoylation and alters the stability of p56lck. Biochemistry 32, 9250–9255 [DOI] [PubMed] [Google Scholar]
- 40. Saeki K., Miura Y., Aki D., Kurosaki T., Yoshimura A. (2003) The B cell-specific major raft protein, Raftlin, is necessary for the integrity of lipid raft and BCR signal transduction. EMBO J. 22, 3015–3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yap M. C., Kostiuk M. A., Martin D. D., Perinpanayagam M. A., Hak P. G., Siddam A., Majjigapu J. R., Rajaiah G., Keller B. O., Prescher J. A., Wu P., Bertozzi C. R., Falck J. R., Berthiaume L. G. (2010) Rapid and selective detection of fatty acylated proteins using ω-alkynyl-fatty acids and click chemistry. J. Lipid Res. 51, 1566–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gri G., Molon B., Manes S., Pozzan T., Viola A. (2004) The inner side of T cell lipid rafts. Immunol. Lett. 94, 247–252 [DOI] [PubMed] [Google Scholar]
- 43. Harder T., Simons K. (1999) Clusters of glycolipid and glycosylphosphatidylinositol-anchored proteins in lymphoid cells: accumulation of actin regulated by local tyrosine phosphorylation. Eur. J. Immunol. 29, 556–562 [DOI] [PubMed] [Google Scholar]
- 44. Blank N., Schiller M., Krienke S., Wabnitz G., Ho A. D., Lorenz H. M. (2007) Cholera toxin binds to lipid rafts but has a limited specificity for ganglioside GM1. Immunol. Cell Biol. 85, 378–382 [DOI] [PubMed] [Google Scholar]
- 45. Lencer W. I., Hirst T. R., Holmes R. K. (1999) Membrane traffic and the cellular uptake of cholera toxin. Biochim. Biophys. Acta 1450, 177–190 [DOI] [PubMed] [Google Scholar]
- 46. Sugimoto Y., Ninomiya H., Ohsaki Y., Higaki K., Davies J. P., Ioannou Y. A., Ohno K. (2001) Accumulation of cholera toxin and GM1 ganglioside in the early endosome of Niemann-Pick C1-deficient cells. Proc. Natl. Acad. Sci. U.S.A. 98, 12391–12396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Maurer-Stroh S., Eisenhaber B., Eisenhaber F. (2002) N-terminal N-myristoylation of proteins: refinement of the sequence motif and its taxon-specific differences. J. Mol. Biol. 317, 523–540 [DOI] [PubMed] [Google Scholar]
- 48. Shahinian S., Silvius J. R. (1995) Doubly-lipid-modified protein sequence motifs exhibit long-lived anchorage to lipid bilayer membranes. Biochemistry 34, 3813–3822 [DOI] [PubMed] [Google Scholar]
- 49. Salaun C., Greaves J., Chamberlain L. H. (2010) The intracellular dynamic of protein palmitoylation. J. Cell Biol. 191, 1229–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Goodwin J. S., Drake K. R., Rogers C., Wright L., Lippincott-Schwartz J., Philips M. R., Kenworthy A. K. (2005) Depalmitoylated Ras traffics to and from the Golgi complex via a nonvesicular pathway. J. Cell Biol. 170, 261–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rocks O., Peyker A., Kahms M., Verveer P. J., Koerner C., Lumbierres M., Kuhlmann J., Waldmann H., Wittinghofer A., Bastiaens P. I. (2005) An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science 307, 1746–1752 [DOI] [PubMed] [Google Scholar]
- 52. Pike L. J. (2009) The challenge of lipid rafts. J. Lipid Res. 50, (suppl.) S323–S328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chalfie M. (1995) Green fluorescent protein. Photochem. Photobiol. 62, 651–656 [DOI] [PubMed] [Google Scholar]
- 54. Suzuki K. G., Fujiwara T. K., Sanematsu F., Iino R., Edidin M., Kusumi A. (2007) GPI-anchored receptor clusters transiently recruit Lyn and Gα for temporary cluster immobilization and Lyn activation: single-molecule tracking study 1. J. Cell Biol. 177, 717–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Goswami D., Gowrishankar K., Bilgrami S., Ghosh S., Raghupathy R., Chadda R., Vishwakarma R., Rao M., Mayor S. (2008) Nanoclusters of GPI-anchored proteins are formed by cortical actin-driven activity. Cell 135, 1085–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lingwood D., Simons K. (2010) Lipid rafts as a membrane-organizing principle. Science 327, 46–50 [DOI] [PubMed] [Google Scholar]
- 57. Mishra S., Joshi P. G. (2007) Lipid raft heterogeneity: an enigma. J. Neurochem. 103, Suppl. 1, 135–142 [DOI] [PubMed] [Google Scholar]
- 58. Saito Y. D., Jensen A. R., Salgia R., Posadas E. M. (2010) Fyn: a novel molecular target in cancer. Cancer 116, 1629–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Staubach S., Hanisch F. G. (2011) Lipid rafts: signaling and sorting platforms of cells and their roles in cancer. Expert Rev. Proteomics 8, 263–277 [DOI] [PubMed] [Google Scholar]
- 60. Larive R. M., Baisamy L., Urbach S., Coopman P., Bettache N. (2010) Cell membrane extensions, generated by mechanical constraint, are associated with a sustained lipid raft patching and an increased cell signaling. Biochim. Biophys. Acta 1798, 389–400 [DOI] [PubMed] [Google Scholar]
- 61. Arstikaitis P., Gauthier-Campbell C., Huang K., El-Husseini A., Murphy T. H. (2011) Proteins that promote filopodia stability, but not number, lead to more axonal-dendritic contacts. PLoS One 6, e16998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gauthier-Campbell C., Bredt D. S., Murphy T. H., El-Husseini Ael-D. (2004) Regulation of dendritic branching and filopodia formation in hippocampal neurons by specific acylated protein motifs. Mol. Biol. Cell 15, 2205–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Scorticati C., Formoso K., Frasch A. C. (2011) Neuronal glycoprotein M6a induces filopodia formation via association with cholesterol-rich lipid rafts. J. Neurochem. 119, 521–531 [DOI] [PubMed] [Google Scholar]
- 64. Patel H. H., Insel P. A. (2009) Lipid rafts and caveolae and their role in compartmentation of redox signaling. Antioxid. Redox Signal. 11, 1357–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hara T., Kondo N., Nakamura H., Okuyama H., Mitsui A., Hoshino Y., Yodoi J. (2007) Cell-surface thioredoxin-1: possible involvement in thiol-mediated leukocyte-endothelial cell interaction through lipid rafts. Antioxid. Redox Signal. 9, 1427–1437 [DOI] [PubMed] [Google Scholar]
- 66. Kondo N., Ishii Y., Kwon Y. W., Tanito M., Sakakura-Nishiyama J., Mochizuki M., Maeda M., Suzuki S., Kojima M., Kim Y. C., Son A., Nakamura H., Yodoi J. (2007) Lipid raft-mediated uptake of cysteine-modified thioredoxin-1: apoptosis enhancement by inhibiting the endogenous thioredoxin-1. Antioxid. Redox Signal. 9, 1439–1448 [DOI] [PubMed] [Google Scholar]
- 67. Volonte D., Galbiati F. (2009) Inhibition of thioredoxin reductase 1 by caveolin 1 promotes stress-induced premature senescence. EMBO Rep. 10, 1334–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]