Background: Phosphatidylserine binds specifically and stabilizes the Na,K-ATPase.
Results: Neutral phospholipids with unsaturated fatty acyl chains increase the turnover rate of purified recombinant human Na,K-ATPase (α1β1 or α1β1FXYD1).
Conclusion: Acid and neutral phospholipids may bind in separate pockets and either stabilize or stimulate Na,K-ATPase activity, respectively.
Significance: The findings demonstrate distinct roles of specifically bound acid or neutral phospholipids.
Keywords: Membrane Proteins, Molecular Modeling, NAK-ATPase, Phosphatidylcholine, Phosphatidylethanolamine
Abstract
Membrane proteins interact with phospholipids either via an annular layer surrounding the transmembrane segments or by specific lipid-protein interactions. Although specifically bound phospholipids are observed in many crystal structures of membrane proteins, their roles are not well understood. Na,K-ATPase is highly dependent on acid phospholipids, especially phosphatidylserine, and previous work on purified detergent-soluble recombinant Na,K-ATPase showed that phosphatidylserine stabilizes and specifically interacts with the protein. Most recently the phosphatidylserine binding site has been located between transmembrane segments of αTM8–10 and the FXYD protein. This paper describes stimulation of Na,K-ATPase activity of the purified human α1β1 or α1β1FXYD1 complexes by neutral phospholipids, phosphatidylcholine, or phosphatidylethanolamine. In the presence of phosphatidylserine, soy phosphatidylcholine increases the Na,K-ATPase turnover rate from 5483 ± 144 to 7552 ± 105 (p < 0.0001). Analysis of α1β1FXYD1 complexes prepared with native or synthetic phospholipids shows that the stimulatory effect is structurally selective for neutral phospholipids with polyunsaturated fatty acyl chains, especially dilinoleoyl phosphatidylcholine or phosphatidylethanolamine. By contrast to phosphatidylserine, phosphatidylcholine or phosphatidylethanolamine destabilizes the Na,K-ATPase. Structural selectivity for stimulation of Na,K-ATPase activity and destabilization by neutral phospholipids distinguish these effects from the stabilizing effects of phosphatidylserine and imply that the phospholipids bind at distinct sites. A re-examination of electron densities of shark Na,K-ATPase is consistent with two bound phospholipids located between transmembrane segments αTM8–10 and TMFXYD (site A) and between TM2, -4, -6, -and 9 (site B). Comparison of the phospholipid binding pockets in E2 and E1 conformations suggests a possible mechanism of stimulation of Na,K-ATPase activity by the neutral phospholipid.
Introduction
The Na,K-ATPase is a member of the P-type ATPase family of cation pumps (types I, II, and III) and phospholipid flippase (type IV) or proteins of unknown function (type V) (1). Na,K-ATPase maintains the Na+ and K+ gradients across animal cell membranes by coupling hydrolysis of one molecule of ATP to exchange of three intracellular Na+ for two extracellular K+ ions. Na,K-ATPase is inhibited by vanadate and cardiac glycosides like ouabain. The minimal active unit of the Na,K-ATPase consists of α and β subunits. The α subunit cycles between the E1/E2 conformations and contains the catalytic residues, ion occlusion and transport pathway, and inhibitor binding sites (2, 3). The β subunit is important for protein folding, trafficking to the plasma membrane, and stabilization, and it also affects K+ occlusion. Recently, it has also been implicated in cell adhesion (4). FXYD proteins interact directly with the Na,K-ATPase and regulate it but are not essential for activity (5, 6). There are four isoforms of the α subunit, three of the β subunit, and seven FXYD proteins that are expressed in a cell- and tissue-specific manner to form αβFXYD complexes with distinct Na+/K+ pumping properties.
The crystallographic structures of sarcoplasmic reticulum Ca-ATPase, H-ATPase, and Na,K-ATPase have been determined in the last decade (1, 7, 8). In particular the structures of Ca-ATPase have been resolved in various conformations providing invaluable insight into the molecular mechanism of this ATPase and P-type pumps in general (7, 9–11). Molecular structures of the pig kidney and shark rectal gland Na,K-ATPase in the E2:2K and E2:2K:ouabain and E2P:ouabain conformations have been published recently, revealing unique features such as K+ occlusion sites, β and FXYD subunits, and cardiac glycoside binding site (8, 12–15).
All membrane proteins interact with bilayer lipid components. Membrane lipids form a surrounding annulus that solvates the exposed transmembrane segment of membrane proteins. Annular lipids readily exchange with the bulk bilayer lipids, yet their mobility relative to “free” lipids is restricted, and their variable hydrophobic acyl chains must distort to accommodate the width of the hydrophobic surface of the protein (16, 17). Membrane physical characteristics like curvature (lateral pressure) and thickness (hydrophobic matching) can affect membrane protein function. Examples consistent with the importance of hydrophobic matching or lateral pressure of the lipid bilayer include a strong correlation of activity of Na,K-ATPase reconstituted into proteoliposomes with fatty acyl chain length, which can be modulated by cholesterol (optimal C22:1 without and C18:1 with cholesterol) (18, 19) or inhibition of Na,K-ATPase by polyunsaturated phospholipids (16:0–22:6 or 22:6–22:6), which is modulated by cholesterol (20). Similarly, for Ca-ATPase reconstituted into proteoliposomes C18:1 is an optimal chain length (21). Annular lipids do not usually appear in crystal structures of detergent-solubilized proteins, as they are largely replaced by detergent molecules, although they have been seen in some cases (22).
By contrast to annular lipids, specifically bound phospholipids and cholesterol have now been observed in many crystal structures of membrane proteins (21–23). Specifically bound lipids are usually located in crevices between transmembrane segments or between subunits, and positively charged residues interact with the phosphate or other negatively charged head groups (22, 24). As discussed below, Ca-ATPase structures have revealed a specific binding site for phosphatidylethanolamine (25). In principle, specifically bound phospholipids and cholesterol could affect stability and folding or catalytic activity of membrane proteins or both stability and activity. In practice, there are few examples of membrane proteins in which definitive roles can be attributed to particular specifically bound lipids.
The Na,K-ATPase requires an acid phospholipid for activity, particularly phosphatidylserine (PS).3 The requirement of the native Na,K-ATPase for PS has been known for many years (26–28), although the detailed role of the PS was not well characterized. Extensive studies of purified detergent-soluble recombinant human Na,K-ATPase expressed in Pichia pastoris, including α1–3/β1 isoform complexes, have provided strong evidence for a specific PS-Na,K-ATPase interaction and especially that the optimal phospholipid is SOPS (29–31). SOPS stabilizes the protein against thermal or detergent-mediated inactivation but does not per se alter the Na,K-ATPase activity by comparison with poorly stabilizing phospholipids (such as DMPS (1,2-dimyristoyl-sn-glycero-3-phospho-l-serine) or DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine)) (30). In addition, FXYD proteins stabilize the Na,K-ATPase by enhancing the PS-pump interaction, protecting it from thermal and detergent denaturation, and FXYD1 also protects from phosphatidylserine decarboxylase-mediated inactivation (31, 32). These findings indicate that the bound PS is in close proximity to FXYD1. Recent mutagenesis studies and use of a novel “lipid antagonist” locate the putative binding site of the PS between 8, 9, and 10 TM helices of the α subunit, in proximity to the FXYD TM helix (33). This biochemical data clearly indicate where the SOPS binds to the Na,K-ATPase and the consequence of its binding. However, direct structural data on this crucial interaction is still missing.
Native Na,K-ATPase is strongly dependent on cholesterol (34, 35), and cholesterol also strongly stabilizes the recombinant Na,K-ATPase in the presence of SOPS, although it is not essential for maintaining activity of the recombinant protein (29, 30). The molecular structure of the shark rectal gland Na,K-ATPase in the E2 conformation reveals a key cholesterol molecule bound at the interface of the β TM helix and helices 3 and 7 of α (13). However, thus far cholesterol is the only lipid membrane component resolved in Na,K-ATPase structures.
The compelling biochemical evidence for a specific phosphatidylserine-Na,K-ATPase interaction prompted us to enquire whether there might be additional specific lipid interactions. In this paper we describe an additional phospholipid effect, namely stimulation of the Na,K-ATPase activity of the detergent-soluble recombinant human Na,K-ATPase (α1β1) by neutral phospholipids (PC and phosphatidylethanolamine (PE)). This effect seems to be mediated by a specifically bound phospholipid molecule(s). Both specific lipid effects have prompted a further search in the existing high resolution structure for electron density that might correspond to phospholipid molecules as described here.
EXPERIMENTAL PROCEDURES
Materials
DDM (catalogue no. D310) and C12E8 (25% w/w, catalogue no. 0330) were purchased from Anatrace. Natural PCs (soy, bovine heart, porcine brain, bovine liver) and synthetic lipids (SOPS Na+ salt, DLPS Na+ salt, dilinoleoyl-PE (DLPE), dilinoleoyl-PC (DLPC), stearoyl-linoleoyl-PC (SLPC), and SOPC) were obtained from Avanti Polar Lipids and stored as chloroform solution. Cholesterol was obtained from Sigma. [γ-32P]Adenosine 5′-triphosphate was purchased from PerkinElmer Life Sciences. BD Talon metal affinity resin (catalogue no. 635503) was obtained from Clontech.
Recombinant Human Na,K-ATPase Expression and Purification
Transformation of P. pastoris yeast cells, fermentation, and membrane preparation are described in detail elsewhere (36). Recombinant human α1β1/FXYD1 complex was purified as described in Refs. 29, 30, and 33) with modifications. Briefly, P. pastoris membranes harboring the α1β1 complex are solubilized with DDM (DDM/protein ratio 2:1) and incubated with cobalt beads. Beads are separated and washed with buffer containing 100 mm NaCl, 30 mm imidazole, 10% glycerol, 20 mm MOPS/Tris, pH 7.4, 0.3 mg/ml C12E8, 0.05 mg/ml cholesterol, 0.17 or 0.07 mg/ml SOPS plus 0.1 mg/ml phosphatidylcholine. Complexes were eluted in the same buffer containing 200 mm imidazole. For experiments with increasing concentrations of DLPC, DLPE, and DLPS the total phospholipid concentration was maintained at 0.17 mg/ml by varying the concentration of SOPS.
For column purification, beads loaded with Na,K-ATPase complexes were washed once in wash buffer and loaded on a column. After sedimentation by gravity the beads were washed again, one bead volume of elution buffer was added, and fractions were collected. The fractions containing the highest Na,K-ATPase activity were mixed and used for further analysis.
Recombinant Human FXYD1 Expression, Purification and Reconstitution
Expression of human FXYD1 is also described in Ref. 33. CD41 Escherichia coli cells expressing FXYD1 (phospholemman) (pET28-TevH vector) were disrupted by passage through a high pressure cell. The lysate was harvested by centrifugation at 200,000 × g for 30 min at 4 °C. The pellet was resuspended at 1 mg/ml in 50 mm Tricine/Tris, pH 7.4, 10% glycerol, 500 mm NaCl, 2 mg/ml DDM, 8 m urea, 30 mm imidazole and homogenized. After incubating the homogenate for 30 min, it was diluted 1:2 with 50 mm Tricine/Tris, pH 7.4, 250 mm NaCl, 10% glycerol, and unbroken material was removed by ultracentrifugation (200,000 × g for 30 min). The supernatant was incubated with BD Talon beads for 30 min and subsequently washed 3 times with 50 mm Tricine/Tris, pH 7.4, 10% glycerol, 250 mm NaCl, 1 mg/ml DDM, 4 m urea, 30 mm imidazole. FXYD1 was eluted with 1 bead volume of 50 mm Tricine/Tris, pH 7.4, 10% glycerol, 500 mm NaCl, 0.1 mg/ml C12E8, 4 m urea, 250 mm imidazole and stored at −80 °C.
For reconstitution of α1β1FXYD1 complexes, FXYD1 was dialyzed in a buffer containing 500 mm NaCl, 0.1 mm DTT, 10% glycerol, 50 mm MOPS/Tris, pH 7.4. The N-terminal His tag was cleaved using tobacco etch virus protease. Solubilized yeast membranes expressing α1β1 were incubated with the BD Talon beads for 4 h, the beads were then centrifuged at 700 × g for 10 min, and the supernatant was discarded. The cleaved FXYD1 was then added to the beads at a molar ratio of 10:1 (FXYD1:Na,K-ATPase) and incubated overnight at 4 °C. The beads were then washed twice, and the α1β1FXYD1 complex was eluted as described above.
Activity and Stability Assays
ATPase activity was measured in medium containing 130 mm NaCl, 20 mm KCl, 3 mm MgCl2, 1 mm EGTA, 25 mm histidine, and 1 mm ATP at 37 °C. Using a malachite green dye, released phosphate was detected at three time points, and the Na,K-ATPase activity was calculated from the linear fit-slope of the time course (37).
For thermal inactivation assays, samples were diluted to equal concentrations in elution buffer and incubated at 45 °C. Aliquots were taken at the indicated time points and diluted in cold reaction medium without ATP. Activity was measured and calculated as percent of control from the activity of an untreated sample.
Determination of Phosphoenzyme
Phosphoenzyme was determined by incubating on ice for 15 s, 5–10-μg aliquots of the purified recombinant Na,K-ATPase in a reaction medium containing 17 μm ATP (plus [γ-32P]ATP), 5 mm MgCl2, 35 mm MOPS/Tris buffer at pH 7.4, and 100 mm NaCl. The reaction was stopped with ice-cold 10% trichloroacetic acid (TCA) and 1 mm cold ATP. After 1 h on ice, precipitated protein was adsorbed on GF/B filters, and filters were washed 3 times with 6 ml of ice-cold TCA. 32P was measured, and the amount of phosphoenzyme was calculated as nmol/mg of protein.
Identification and Modeling of Lipids Bound to Na,K-ATPase
The crystal structure and structure factor of shark rectal gland Na,K-ATPase in E2·2K+·MgF42− (13) were used in lipid modeling (PDB code 2ZXE). Phosphate head groups of Na,K-ATPase-bound lipids were searched on the |Fo| − |Fc| electron density map contoured at 3σ. First, phospholipid molecules were modeled as PE and refined with CNS (38). Then, depending on the electron density, the atomic model for the head group was replaced with that of PS or phosphatidylcholine (PC). One PS molecule was placed in the cleft surrounded by M8–10 and FXYD10, and one PC molecule in the cleft surrounded by M4, M6, and M9. The atomic model was refined to Rwork of 0.244 and Rfree of 0.276 at 2.4 Å resolution.
The E1 model of the shark enzyme was prepared with the online protein structure and function prediction server I-Tasser (39) using SERCA structure in the E1·2Ca2+·ATP conformation (PDB code 1VFP) as template. The model produced was then superimposed on 2ZXE using Swiss-PDBViewer. Structure figures were prepared with PyMOL (The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC)
RESULTS
Phosphatidylcholine Increases the Turnover Rate of the Recombinant Human Na,K-ATPase
Table 1 presents evidence for an ∼50% increase in Na,K-ATPase activity of human α1β1 or α1β1FXYD1 detergent-soluble complexes when soy phosphatidylcholine replaced part of the SOPS that is normally added with cholesterol to the wash and elution buffers to preserve activity. The total phospholipid (SOPS plus soy PC, 0.17 mg/ml), cholesterol (0.05 mg/ml), and detergent (C12E8, 0.3 mg/ml) concentrations were maintained constant so as to avoid a change in free C12E8 concentration that might itself alter activity. In these conditions, the lowest concentration of SOPS, 0.07 mg/ml, should more than satisfy the SOPS requirement for stabilizing the protein (32). Stimulation of activity by PC was observed with the human Na,K-ATPase α1β1 whether reconstituted with FXYD1 or not (α1β1FXYD1 or α1β1). The stimulation of Na,K-ATPase activity was observed with enzymes made either by column or batch procedures that produce higher or lower levels of purity and Na,K-ATPase activities, respectively. Although stimulation by the PC is independent of FXYD1, subsequent experiments were done with the α1β1FXYD1 complexes, as FXYD1 strongly stabilizes the preparations against thermal- or detergent-mediated inactivation (32). Fig. 1 shows SDS-PAGE gels of column-purified preparations prepared with SOPS alone or with SOPS/PC and reconstituted with FXYD1 or not reconstituted. The preparations are 80–90% pure and are essentially identical except for the presence or absence of FXYD1.
TABLE 1.
Stimulation of Na,K-ATPase activity by Soy PC
All procedures are described under “Experimental Procedures.”
| Conditions | PS | PS/PC |
|---|---|---|
| μmol/min/mg | mol/min/mg | |
| α1β1FXYD1 | ||
| Column | 20.8 ± 1.3 (n = 6) | 30.7 ± 1.1 (n = 7) p < 0.001 |
| Batch | 14.6 ± 1.0 (n = 7) | 21.1 ± 1.4 (n = 6) p < 0.01 |
| α1β1 | ||
| Column | 21.1 ± 1.2 (n = 3) | 30.6 ± 1.8 (n = 4) p < 0.01 |
| Batch | 13.3 (n = 1) | 19.9 (n = 1) |
FIGURE 1.
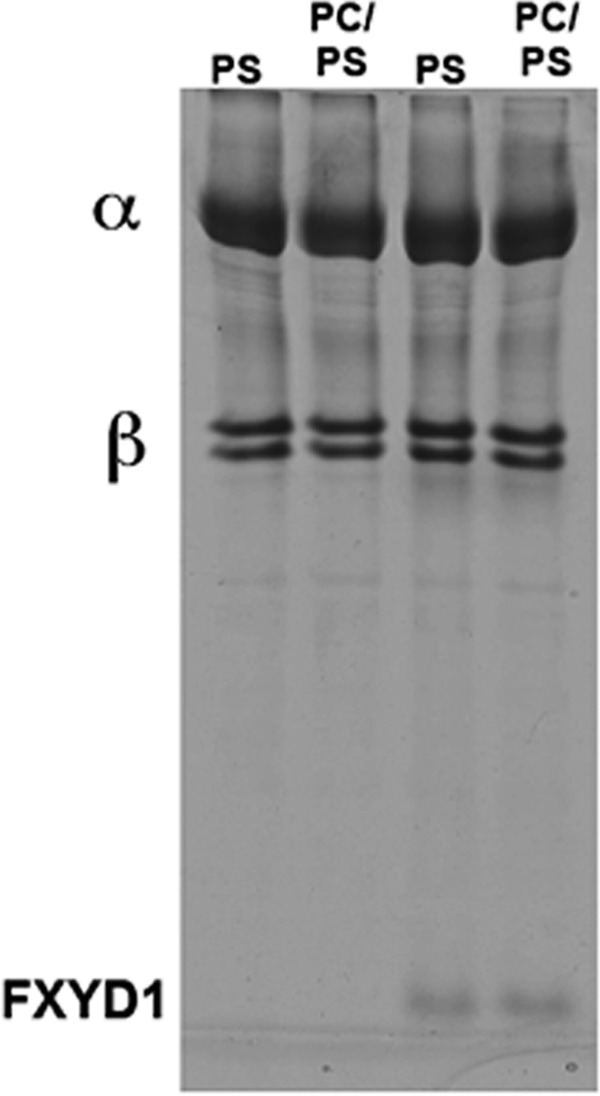
SDS-PAGE of purified α1β1 and α1β1FXYD1 complexes prepared with SOPS or SOPS/soy PC
Table 2 shows that soy PC increases the Na,K-ATPase turnover rate calculated from ATPase-specific activities and steady-state phosphoenzyme levels to quantify the site concentrations. The phosphoenzyme levels were between 4–5 and 2.5–3 nmol/mg protein for column- or batch-purified preparations, respectively, and were not significantly different in preparations made with SOPS/PC or with SOPS alone. The calculated average turnover rates at 37 °C for SOPS/PC versus SOPS were 7552 ± 105 and 5453 ± 106 s−1, respectively. The difference (1.39-fold) is highly significant (p < 0.0001). The high Na,K-ATPase-specific activities, phosphoenzyme levels, and turnover rates for the column-purified preparations of recombinant human α1β1 complexes made with SOPS/PC are quite similar to those for purified native kidney Na,K-ATPase (α1β1) preparations (40).
TABLE 2.
Soy PC increases the turnover rate of Na,K-ATPase
All procedures are described under “Experimental Procedures.”
| Conditions | Specific activity | Phosphoenzyme | Turnover rate at 37 °C |
|---|---|---|---|
| μmol/min/mg | nmol/mg | min− | |
| SOPS/ Soy PC | |||
| Column | 32.2 | 4.3 | 7508 |
| 32.9 | 4.4 | 7483 | |
| 35.8 | 4.9 | 7363 | |
| Batch | 21.6 | 2.75 | 7854 |
| Average, 7552 ± 105 | |||
| SOPS | |||
| Column | 22.4 | 4.0 | 5567 |
| 21.1 | 4.1 | 5202 | |
| 23.6 | 4.41 | 5364 | |
| Batch | 14.6 | 2.57 | 5680 |
| Average, 5453 ± 106 (p < 0.0001) | |||
| Soy PC | |||
| Column | 29.9 | 4.57 | 6534 |
| 33.8 | 5.28 | 6402 | |
| Average, 6468 | |||
Stimulation of Na,K-ATPase activity by soy PC in the presence of SOPS raises an interesting question of whether the PC replaces the SOPS in its specific binding site or whether, perhaps, it is bound at a separate site. An initial pointer to a separate PC site has come from experiments in which α1β1FXYD1 complexes were prepared as in Table 1 with 0.3 mg/ml C12E8, 0.05 mg/ml cholesterol, and 0.17 mg/ml soy PC without SOPS, and then Na,K-ATPase activity phosphoenzyme levels and thermal stability at 37 °C were compared with data for complexes made with SOPS or SOPS/PC (Table 2, Fig. 2). It is striking that the soy PC alone is sufficient to sustain a high Na,K-ATPase activity similar to that with SOPS/PC and significantly higher than for SOPS alone even though the phosphoenzyme levels are similar in all conditions. Accordingly, the turnover rate with PC alone is also substantially higher than with SOPS alone. The slightly lower turnover rate for PC alone compared with SOPS/PC (6468 v 7552 min−1) could imply that the PC alone sample is slightly inactivated during the preparation (10–15%).
FIGURE 2.
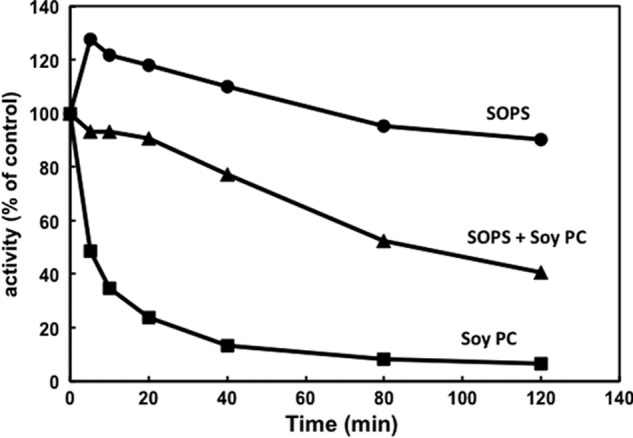
Thermal stability of recombinant Na,K-ATPase preparations made with SOPS, Soy PC, or SOPS plus Soy PC. Na,K-ATPases (α1β1FXYD1) was prepared in 0.05 mg/ml cholesterol, 0.3 mg/ml C12E8 and 0.17 mg/ml SOPS, 0.17 mg/ml Soy PC, or 0.07 mg/ml SOPS plus 0.1 mg/ml Soy PC. Complexes were incubated at 45 °C for the indicated times before activity measurement. vt/v0 represents activity at time t relative to activity v0 before inactivation.
On the other hand there is a remarkable difference of thermal stability (Fig. 2) in that the preparation with PC alone is highly unstable compared with that with SOPS.4 In addition, the preparation made with both SOPS and soy PC is less stable than that made with SOPS alone but much more stable than that made with soy PC alone. Thus, effects of phospholipids on thermal stability (SOPS ≫ PC) and Na,K-ATPase activity (PC > SOPS) are dissociated and seem to reflect separate phenomena, possibly indicative of distinct sites for the two types of phospholipids.
Structural Selectivity of Phospholipid Effects
If the stimulatory effect of soy PC reflects binding to a specific site, then as in the case of the PS, one might be able to detect structural selectivity of PC variants. Because soy PC is of plant origin, preparations were also made with PCs from animal origins, namely from brain, heart, and liver. The specific Na,K-ATPase activities were compared with preparations made with soy PC (Fig. 3). Noticeably, brain PC has little or no stimulatory effect, whereas the stimulatory effect of heart PC is quite similar to that of soy PC, and liver PC lies between brain and heart. The relative composition of saturated, monounsaturated, and polyunsaturated fatty acyl chains in PCs of different origins is recorded below the figure. It is striking that soy and heart PCs contain a high percentage of polyunsaturated fatty acyl chains, whereas brain PC has a low percentage, and the percentage in liver PC lies between that in brain and heart.
FIGURE 3.
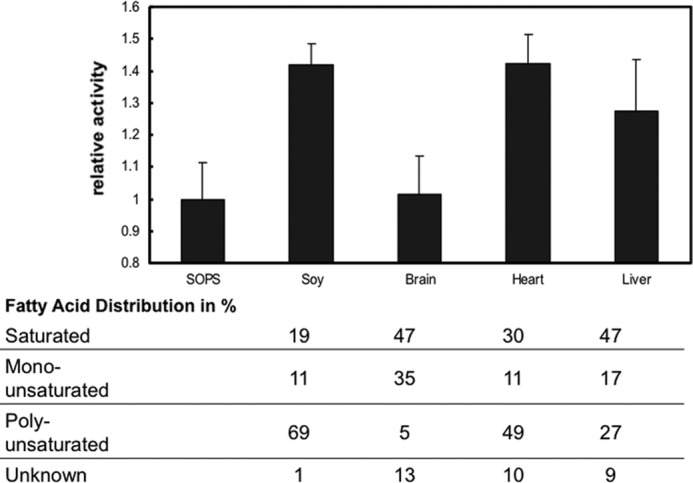
Na,K-ATPase activities of preparations made with soy, brain, heart, and liver PC. α1β1FXYD1 complexes were prepared by batch purification in 0.07 mg/ml SOPS, 0.05 mg/ml cholesterol, 0.1 mg/ml concentrations of the indicated PC, and 0.3 mg/ml C12E8. The SOPS sample contained 0.17 mg/ml SOPS to maintain the concentration of free detergent constant in all samples. The table indicates the fatty acid distribution of the PC tissue extracts. Data represent averages from three experiments ± S.E.
The result in Fig. 3 could imply that stimulation of Na,K-ATPase activity is mediated by PCs with polyunsaturated fatty acyl chains. To test this possibility and also the role of the phospholipid head group, we have made preparations of α1β1FXYD1 with different synthetic PCs, PEs, and also PSs in the wash and elution buffers (in addition to the standard SOPS/cholesterol). Fig. 4 shows that DLPC was as effective or even better than soy PC, whereas SLPC was about half as effective, and SOPC had no effect on the specific Na,K-ATPase activity. Furthermore, DLPE was quite similar to DLPC, whereas DLPS was much less effective, producing about one-third of the stimulatory effect. Further comparison of DLPC, DLPE, and DLPS were made in experiments comparing preparations of α1β1FXYD1 made with increasing concentrations of the different phospholipids (Fig. 5). The experiments confirm that DLPC and DLPE have similar maximal effects (∼15 → 25 μmol/min/mg), whereas that of DLPS is much smaller (∼15 → 18 μmol/min/mg). In addition, the concentration for a half-maximal effect for DLPC and DLPE is essentially the same (c.0.015 mg/ml) and is about 2-fold lower than that of DLPS (∼0.03 mg/ml). This structural selectivity for the stimulatory effect of the phospholipid (DLPC > SLPC ≫ SOPC and DLPC = DLPE > DLPS) is suggestive of a specific neutral phospholipid (PC or PE)-Na,K-ATPase interaction within the mixed detergent-phospholipid-protein micelles. If a mobile equilibrium indeed exists between a specifically bound PC or PE molecule and the Na,K-ATPase protein complex, it should be possible to bind soy PC or DLPC by adding it after elution of the protein from the beads and show stimulation of Na,K-ATPase activity. The experiment in Fig. 6 illustrates such an effect. The experiment was done in preparations made at higher detergent and SOPS concentration than usual (1.2 and 0.4 mg/ml, respectively) to avoid inactivating effects of the detergent added with the PC. The enzyme was incubated with soy PC at 0 °C for 12–48 h, and then ATPase activity was measured. As predicted from a binding equilibrium, at increasing PC concentrations, Na,K-ATPase activity increased by >50% at the saturating PC concentration (0.2–0.4 mg/ml). This experiment was done with soy PC, but an essentially similar result was obtained with DLPC.
FIGURE 4.

Na,K-ATPase activities of preparations made with soy PC, DLPC, SLPC, SOPC, DLPE, and DLPS. Na,K-ATPase was prepared as in Fig. 3, and the specific activity was compared with the sample containing only SOPS/cholesterol (dotted lines).
FIGURE 5.
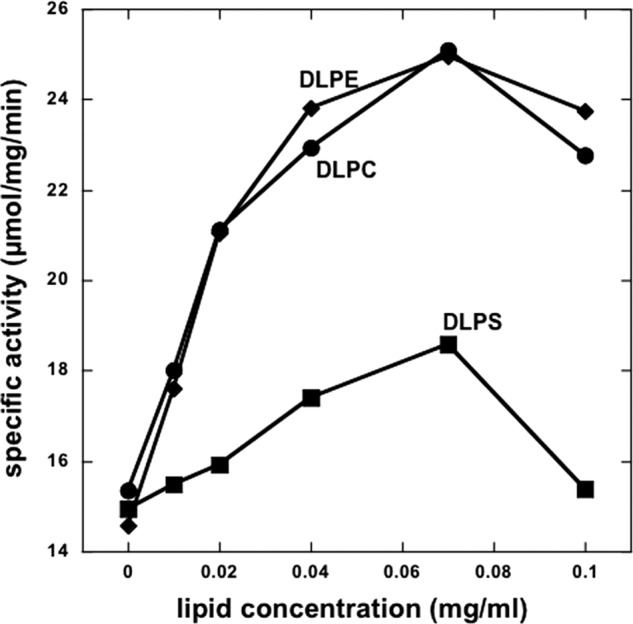
Effect of increasing concentrations of DLPC, DLPE, and DLPS. Na,K-ATPase α1β1FXYD1 complexes were purified with increasing concentrations of the indicated lipids (0–0.1 mg/ml). The total phospholipid concentration (SOPS plus dileneoyl lipid) was maintained constant at 0.17 mg/ml. A representative of two experiments is shown.
FIGURE 6.
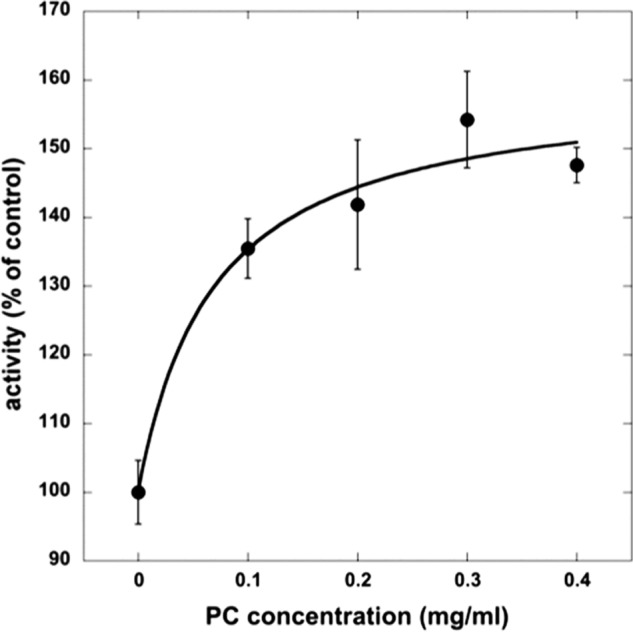
Addition of PC to Na,K-ATPase preparations made with SOPS stimulates activity. α1β1FXYD1 was prepared with 1.2 mg/ml C12E8 and 0.5 mg/ml SOPS and supplemented with increasing concentrations of soy PC. The curve fit was done in Kaleidagraph. vmax = 159.7 ± 10.0, k1/2 = 0.7 ± 0.4, R = 0.98. Data represent averages from four experiments ±S.E.
The experiment in Fig. 7 compared the thermal stability of preparations made without or with different PCs or PEs (and the standard SOPS/cholesterol) and reveals large differences between the preparations. First, all preparations made with PC (or PE) are much less thermally stable than those made only with SOPS. In particular, this includes the sample made with SOPC, which does not stimulate Na,K-ATPase activity. Second, within the group of preparations made with PC (or PE), there is also a discernable order of decreasing thermal stabilities namely DPLC = DLPE > soy PC > SLPC > SOPC, which is the same as for stimulation of Na,K-ATPase activity (see Fig. 3). Third, the DLPS sample shows features characteristic of both SOPS and DLPC, including an initial rise, like SOPS, and then rapid decrease, like DLPC. As discussed below, these results support the hypothesis that there are two specific binding sites, for the acid (PS) or neutral phospholipids (PC/PE), respectively.
FIGURE 7.
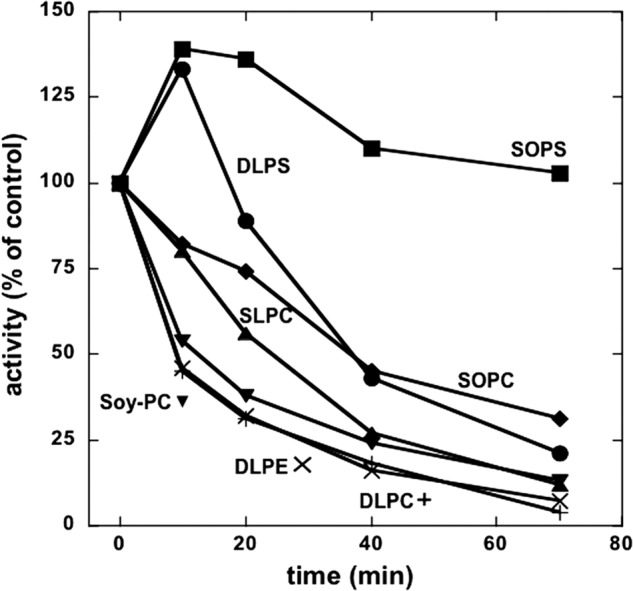
Thermal stability of Na,K-ATPase preparations made with different phospholipids. Na,K-ATPase was prepared as in Figs. 2 and 3. Samples were incubated at 45 °C for the indicated times, and the activity was calculated as percent of the untreated sample (t = 0 min). An average of two experiments is shown.
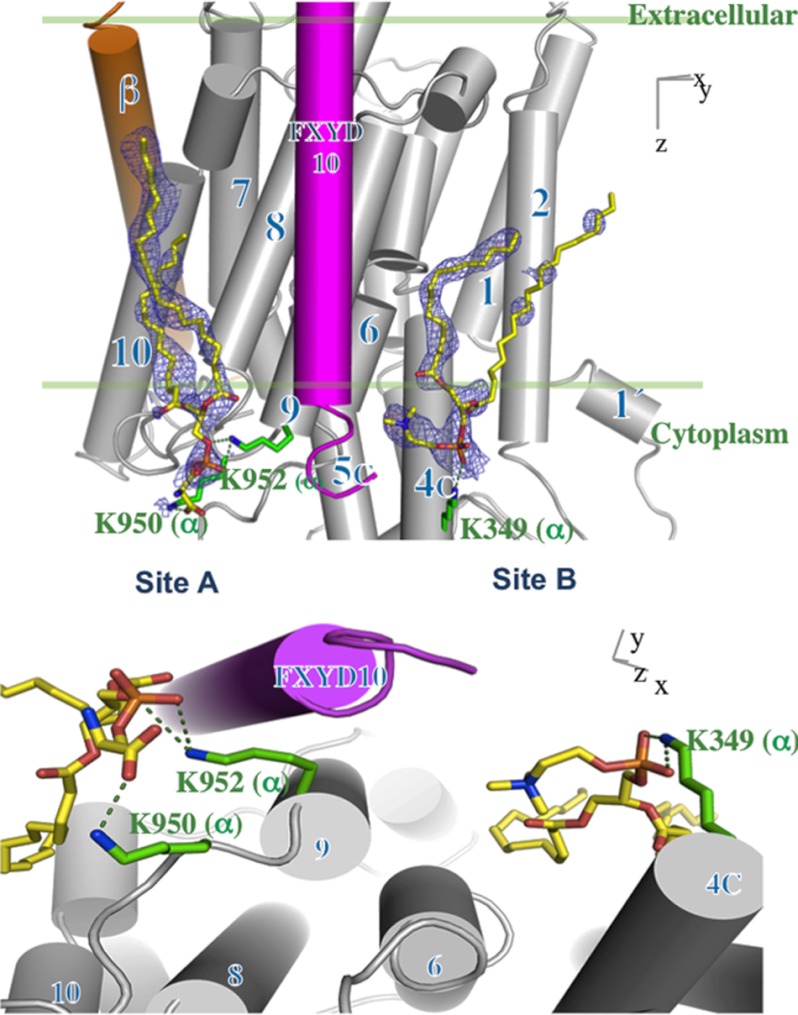
Two Bound Phospholipids Detected in the Structure of the Shark Na,K-ATPase
In view of the biochemical evidence for relatively specific PS and PC/PE interactions with the Na,K-ATPase, we have reexamined the high resolution electron density map of shark Na,K-ATPase in an E2:2K conformation (PDB code 2ZXE) (13) for indications of specifically bound lipids. We have sought unexplained electron density pertaining to phospholipid phosphate head groups (which usually have a distinct form) and other putative lipid features around the trans-membrane helices. We identified two putative phosphate head groups; one positioned in a cleft between TM helices 8, 9, and 10 of the α subunit and the TM helix of FXYD10 (site A), and the second in proximity to the C-terminal end of the TM helix of FXYD10 and TMs 4, 6, and 9 of the α subunit (site B) (Fig. 8). Two phospholipid molecules, assumed to be PS or PC, are modeled into the electron densities in site A or site B, respectively. The electron density map of the phospholipid in site A shows part of the head group and distinct elongated stretches corresponding to acyl chains. The side chains of Lys-950 and Lys-952 on L9/10 form hydrogen bonds to the carboxylate and phosphate groups of the PS. The density in site B shows the head group, the beginning of one acyl chain, and a significant part of another acyl chain. Lys-349 forms electrostatic interaction with the phosphate group of the PC (see “Discussion” and Fig. 8).
FIGURE 8.
Two bound phospholipids in the Na,K-ATPase. Shown are |2Fo| − |Fc| electron density maps (blue mesh, contoured at 1σ) of the two phospholipids (yellow sticks). TM helices are rendered as cylinders for α (gray), β (orange), and FXYD10 (magenta) and numbered according to the shark sequence. PS is modeled in the site designated Site A, and PC is modeled in site designated Site B, as indicated by biochemical data. The crystals are prepared in a medium containing a high concentration of soy PC, but the lipid in site A is assumed to be PS carried over from the original Na,K-ATPase.
DISCUSSION
Two sites for phospholipids; Biochemical and Structural Data
We propose here that there are two specific phospholipid binding sites on the α subunit of Na,K-ATPase (at least), one primarily for acid phospholipids, SOPS, (site A) that stabilizes the protein, and the other for neutral phospholipids PC or PE that mediates increased Na,K-ATPase activity (site B). The findings provide an example of distinct roles of different specifically bound phospholipids that previously have not been well defined for this or other membrane proteins.
The biochemical evidence for the SOPS site within the crevice located between TM8–10 of the α subunit and the transmembrane segment of the FXYD protein has been presented and extensively discussed in previous publications (29–33). The salient feature of this interaction is that the SOPS stabilizes the protein but does not itself affect the Na,K-ATPase activity. By contrast, the present work shows that in the presence or absence of SOPS, neutral phospholipids such as DLPC and DLPE stimulate Na,K-ATPase activity by increasing the turnover rate. Compared with SOPS alone, the neutral phospholipids do not stabilize but somewhat destabilize the protein to thermal inactivation. These features, and the structural selectivity for this effect, DLPC = DLPE > SLPC > SOPC, suggest strongly that there is a separate site for the neutral phospholipids. Despite these distinctions between sites A and B, they are not absolutely selective for acid and neutral phospholipids, respectively. For example, when the protein is prepared with soy PC alone, presumably PC replaces SOPS in site A and also binds to site B, producing an unstable but highly active Na,K-ATPase. SOPC does not stimulate Na,K-ATPase activity in the presence of SOPS but strongly destabilizes, suggesting that SOPC is able to compete with SOPS and destabilize from site A but does not stimulate activity from site B. The moderate additional degree of destabilization seen in the order SOPC < SLPC < DLPC = DLPE may be associated with site B, as this is the same order as for stimulating Na,K-ATPase activity. DLPS may also be able to replace the neutral phospholipids in site B with low efficiency, as seen from the low degree of stimulation of Na,K-ATPase activity and lower affinity compared with either DLPC or DLPE.
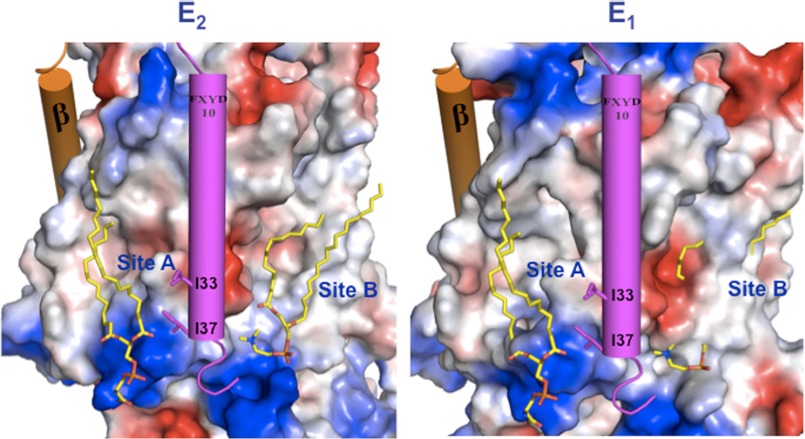
Fig. 8 (upper) shows the two phospholipid binding sites on the surface of the α subunit of shark Na,K-ATPase, with the transmembrane segments of β (orange) and FXYD10 (magenta) as cylinders, and Fig. 8 (lower) shows a detail of the two binding sites. It is noticeable that both phospholipid sites A and B lie in quite deep crevices bounded by the transmembrane segments: site A by αTM8, -9, -and 10 and TMFXYD and site B by αTM2, -4, -6, and -9. Significantly, one acyl chain of the PS in site A is close enough to the transmembrane helix of FXYD10 to interact with hydrophobic side chains of residues, Ile-33 and Ile-37. This fits well with observations that FXYD1 interacts with and strengthens the binding of SOPS and so stabilizes the protein (32). By contrast, both the acyl chains and head group of PC in site B are too far away to interact with the FXYD protein. This is consistent with the finding (Table 1) that the stimulatory effect of PC/PE is independent of FXYD1. Another point is that the negatively charged phosphate and carboxyl of the SOPS head group in site A are close to two lysine residues (Lys-950, Lys-952 shark numbering) near the entrance of TM9, and the phosphate of the PC in site B is close to Lys-439. The increase in the Na,K-ATPase turnover number induced by the PC (Table 2) begs the question as to the mechanism of this effect. Fig. 9 (left) shows a surface charge representation of the lipids bound in the E2(K)MgF4 conformation, and Fig. 9 (right) shows a homology model of the α subunit of the Na,K-ATPase based on Ca-ATPase in an E1P conformation (PDB code 1VFP), with the lipids bound hypothetically in the same positions as in the E2(K)MgF4 conformation. There are two obvious points of significance. First, the PC/PE within site B is largely covered by αTM2, which has moved into the crevice and should, therefore, sterically hinder the PC/PE from binding. One implication of the greater accessibility of the site in E2 is that the PC/PE could bind more tightly and stabilize the E2 conformation and that the lipid does not bind or binds differently in the E1 conformation. This type of effect might increase the turnover rate by accelerating a rate-limiting step of the cycle (e.g. E1P → E2P). The second point is that in site A, binding of SOPS is essentially the same in E1 and E2 due to little or no change in the disposition of TM8–10 and TMFXYD between the conformations. The implication would be that SOPS does not favor either conformation, and consequently, SOPS cannot affect Na,K-ATPase turnover rate, exactly consistent with our experimental observations that SOPS stabilizes the protein but does not affect the Na,K-ATPase activity (30). We are conducting detailed experiments to examine the hypothesis of a PC/PE-induced shift in the conformational equilibrium toward E2. Preliminary kinetic data, on apparent affinities of K+ ions and vanadate, are consistent with this notion.5
FIGURE 9.
Phospholipid binding sites in E2 and E1 conformations. Surface charge representation (positive in blue, negative in red, calculated using PyMOL) of the α subunit for the E2 (a) and homology-modeled E1 (b) conformations of the Shark Na,K-ATPase. Lipids are modeled as yellow sticks, and FXYD10 and β TMs are rendered as magenta and orange cylinders, respectively.
The movement of TM2 shown in Fig. 9 (right) is one of several changes in disposition of TM1–6 in the E1-E2 conformational transition with little or no relative movement of TM7–10, as described extensively for sarcoplasmic reticulum Ca-ATPase (7, 11). In fact, crystal structures of the Ca-ATPase provide an interesting precedent for binding of a neutral phospholipid (PC/PE) within the crevice bounded by TM2, -4, -6, and -9. In E2 conformations of Ca-ATPase, the head group of a PE molecule has been observed between TM2 and TM4 (25, 41, 42), whereas in E1 conformations the TM2 occupies the position of the PE in the E2 conformation and presumably hinders its binding. In E1 structures a PC molecule has been observed between TM2 and TM9 in a more superficial position than in the E2 conformation (43). Accordingly, it has been proposed that the PE molecule stabilizes the E2 conformation and is displaced in the E2-E1 transition (25), i.e. a hypothesis that is similar to that proposed here for Na,K-ATPase.
Physiological Relevance?
Whether stimulation of Na,K-ATPase turnover rate by neutral phospholipids (PC or PE) with polyunsaturated fatty acyl chains has physiological significance can only be speculated on at present. The Na,K-ATPase turnover number in the presence of PS/cholesterol/PC is close to the turnover number observed in native mammalian membranes (e.g. pig kidney) and can be taken as a positive hint in that direction. To prove this hypothesis, it would be necessary to conduct a more detailed study of the structural selectivity for the phospholipid in our system and demonstrate that particular phospholipid requirements are correlated with the presence of those phospholipids in native membranes and also that changes in fatty acyl composition of PC or PE (e.g.18:2, 20:4, and 22:6) are associated with changes in Na,K-ATPase activity. In an interesting series of studies the turnover rates of native membrane-bound Na,K-ATPase of various animal species with a wide range of basal metabolic rates have been shown to vary and to be positively correlated with the polyunsaturated fatty acyl chain content of phospholipids (particularly 22:6) (44, 45). This effect was attributed to different packing density (i.e. lateral pressure) of the polyunsaturated fatty acyl phospholipids within the lipid bilayer (46). In other studies dietary modulation of the ω-6:ω-3 fatty acyl ratio in heart or brain phospholipids has also been reported to alter kinetic properties of the different Na,K-ATPase isoforms (47–49). Although it is quite possible that kinetic effects are mediated by changes in the physical environment of the Na,K-ATPase, as suggested in these papers, in an intact membrane or reconstituted proteoliposomes the exact mechanism of effects of phospholipids on activity is not easily defined. It is equally likely that changes in fatty acid saturation of PC/PE affect Na,K-ATPase activity via a specific phospholipid-protein interaction such as that inferred from this work. Toward an understanding of possible physiological relevance of the effects described here, we are currently undertaking an extensive screen of the phospholipid species associated with Na,K-ATPase complexes isolated from native tissues and effects of ω-6 and ω-3 fatty acyl containing PC and PE species (polyunsaturated fatty acyl phospholipids) on activity and stability of the purified Na,K-ATPase complexes (α1β1FXYD1, α2β1FXYD1, α3β1FXYD1).
A similar result was obtained previously for preparations in which 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) replaced DOPS (30).
M. Habeck, H. Haviv, and J. D. Karlish, unpublished information.
- PS
- phosphatidylserine
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- SOPS
- 1-stearoyl-2-oleoyl-sn-glycero-3-[phosphor-l-serine]
- DLPS
- 1,2-dilinoleoyl-sn-glycero-3-phospho-l-serine
- DLPE
- 1,2-dilinoleoyl-sn-glycero-3-phosphoethanolamine
- DPLC
- 1,2-dilinoleoyl-sn-glycero-3-phosphocholine
- SLPC
- 1-stearoyl-2-linoleoyl-sn-glycero-3-phosphocholine
- SOPC
- 1-stearoyl-2-oleoyl-sn-glycero-3-phosphocholine
- DDM
- n-dodecyl-β-d-maltopyranoside
- C12E8
- octaethylene glycerol mondodecyl
- TM
- transmembrane
- DOPS
- 1,2-dioleoyl-sn-glycero-3-phospho-l-serine.
REFERENCES
- 1. Palmgren M. G., Nissen P. (2011) P-type ATPases. Annu. Rev. Biophys. 40, 243–266 [DOI] [PubMed] [Google Scholar]
- 2. Jorgensen P. L., Hakansson K. O., Karlish S. J. (2003) Structure and mechanism of Na,K-ATPase. Functional sites and their interactions. Annu. Rev. Physiol. 65, 817–849 [DOI] [PubMed] [Google Scholar]
- 3. Kaplan J. H. (2002) Biochemistry of Na,K-ATPase. Annu. Rev. Biochem. 71, 511–535 [DOI] [PubMed] [Google Scholar]
- 4. Geering K. (2008) Functional roles of Na,K-ATPase subunits. Curr. Opin. Nephrol. Hypertens. 17, 526–532 [DOI] [PubMed] [Google Scholar]
- 5. Garty H., Karlish S. J. (2006) Role of FXYD proteins in ion transport. Annu. Rev. Physiol. 68, 431–459 [DOI] [PubMed] [Google Scholar]
- 6. Geering K. (2006) FXYD proteins. New regulators of Na-K-ATPase. Am. J. Physiol. Renal Physiol. 290, F241–F250 [DOI] [PubMed] [Google Scholar]
- 7. Toyoshima C. (2009) How Ca2+-ATPase pumps ions across the sarcoplasmic reticulum membrane. Biochim. Biophys. Acta 1793, 941–946 [DOI] [PubMed] [Google Scholar]
- 8. Toyoshima C., Kanai R., Cornelius F. (2011) First crystal structures of Na+,K+-ATPase. New light on the oldest ion pump. Structure 19, 1732–1738 [DOI] [PubMed] [Google Scholar]
- 9. Toyoshima C. (2008) Structural aspects of ion pumping by Ca2+-ATPase of sarcoplasmic reticulum. Arch. Biochem. Biophys. 476, 3–11 [DOI] [PubMed] [Google Scholar]
- 10. Møller J. V., Nissen P., Sørensen T. L., le Maire M. (2005) Transport mechanism of the sarcoplasmic reticulum Ca2+-ATPase pump. Curr. Opin. Struct. Biol. 15, 387–393 [DOI] [PubMed] [Google Scholar]
- 11. Olesen C., Picard M., Winther A. M., Gyrup C., Morth J. P., Oxvig C., Møller J. V., Nissen P. (2007) The structural basis of calcium transport by the calcium pump. Nature 450, 1036–1042 [DOI] [PubMed] [Google Scholar]
- 12. Morth J. P., Pedersen B. P., Toustrup-Jensen M. S., Sørensen T. L., Petersen J., Andersen J. P., Vilsen B., Nissen P. (2007) Crystal structure of the sodium-potassium pump. Nature 450, 1043–1049 [DOI] [PubMed] [Google Scholar]
- 13. Shinoda T., Ogawa H., Cornelius F., Toyoshima C. (2009) Crystal structure of the sodium-potassium pump at 2.4 Å resolution. Nature 459, 446–450 [DOI] [PubMed] [Google Scholar]
- 14. Ogawa H., Shinoda T., Cornelius F., Toyoshima C. (2009) Crystal structure of the sodium-potassium pump (Na+,K+-ATPase) with bound potassium and ouabain. Proc. Natl. Acad. Sci. U.S.A. 106, 13742–13747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yatime L., Laursen M., Morth J. P., Esmann M., Nissen P., Fedosova N. U. (2011) Structural insights into the high affinity binding of cardiotonic steroids to the Na+,K+-ATPase. J. Struct. Biol. 174, 296–306 [DOI] [PubMed] [Google Scholar]
- 16. Lee A. G. (2003) Lipid-protein interactions in biological membranes. A structural perspective. Biochim. Biophys. Acta 1612, 1–40 [DOI] [PubMed] [Google Scholar]
- 17. Marsh D. (2008) Protein modulation of lipids and, vice versa, in membranes. Biochim. Biophys. Acta 1778, 1545–1575 [DOI] [PubMed] [Google Scholar]
- 18. Cornelius F. (2001) Modulation of Na,K-ATPase and Na-ATPase activity by phospholipids and cholesterol. I. Steady-state kinetics. Biochemistry 40, 8842–8851 [DOI] [PubMed] [Google Scholar]
- 19. Cornelius F., Turner N., Christensen H. R. (2003) Modulation of Na,K-ATPase by phospholipids and cholesterol. II. Steady-state and presteady-state kinetics. Biochemistry 42, 8541–8549 [DOI] [PubMed] [Google Scholar]
- 20. Cornelius F. (2008) Cholesterol-dependent interaction of polyunsaturated phospholipids with Na,K-ATPase. Biochemistry 47, 1652–1658 [DOI] [PubMed] [Google Scholar]
- 21. Lee A. G. (2004) How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta 1666, 62–87 [DOI] [PubMed] [Google Scholar]
- 22. Lee A. G. (2011) Biological membranes. The importance of molecular detail. Trends Biochem. Sci. 36, 493–500 [DOI] [PubMed] [Google Scholar]
- 23. Hunte C., Richers S. (2008) Lipids and membrane protein structures. Curr. Opin. Struct. Biol. 18, 406–411 [DOI] [PubMed] [Google Scholar]
- 24. Adamian L., Naveed H., Liang J. (2011) Lipid binding surfaces of membrane proteins. Evidence from evolutionary and structural analysis. Biochim. Biophys. Acta 1808, 1092–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Obara K., Miyashita N., Xu C., Toyoshima I., Sugita Y., Inesi G., Toyoshima C. (2005) Structural role of countertransport revealed in Ca2+ pump crystal structure in the absence of Ca2+. Proc. Natl. Acad. Sci. U.S.A. 102, 14489–14496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wheeler K. P., Whittam R. (1970) The involvement of phosphatidylserine in adenosine triphosphatase activity of the sodium pump. J. Physiol. 207, 303–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. de Pont J. J., van Prooijen-van Eeden A., Bonting S. L. (1978) Role of negatively charged phospholipids in highly purified (Na+ + K+)-ATPase from rabbit kidney outer medulla studies on (Na+ + K+)-activated ATPase, XXXIX. Biochim. Biophys. Acta 508, 464–477 [DOI] [PubMed] [Google Scholar]
- 28. Hayashi Y., Mimura K., Matsui H., Takagi T. (1988) High-performance gel chromatography of active solubilized Na+,K+-ATPase maintained by exogenous phosphatidylserine. Prog. Clin. Biol. Res. 268A, 205–210 [PubMed] [Google Scholar]
- 29. Cohen E., Goldshleger R., Shainskaya A., Tal D. M., Ebel C., le Maire M., Karlish S. J. (2005) Purification of Na+,K+-ATPase expressed in Pichia pastoris reveals an essential role of phospholipid-protein interactions. J. Biol. Chem. 280, 16610–16618 [DOI] [PubMed] [Google Scholar]
- 30. Haviv H., Cohen E., Lifshitz Y., Tal D. M., Goldshleger R., Karlish S. J. (2007) Stabilization of Na+,K+-ATPase purified from Pichia pastoris membranes by specific interactions with lipids. Biochemistry 46, 12855–12867 [DOI] [PubMed] [Google Scholar]
- 31. Lifshitz Y., Petrovich E., Haviv H., Goldshleger R., Tal D. M., Garty H., Karlish S. J. (2007) Purification of the human a2 isoform of Na,K-ATPase expressed in Pichia pastoris. Stabilization by lipids and FXYD1. Biochemistry 46, 14937–14950 [DOI] [PubMed] [Google Scholar]
- 32. Mishra N. K., Peleg Y., Cirri E., Belogus T., Lifshitz Y., Voelker D. R., Apell H. J., Garty H., Karlish S. J. (2011) FXYD proteins stabilize Na,K-ATPase. Amplification of specific phosphatidylserine-protein interactions. J. Biol. Chem. 286, 9699–9712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kapri-Pardes E., Katz A., Haviv H., Mahmmoud Y., Ilan M., Khalfin-Penigel I., Carmeli S., Yarden O., Karlish S. J. (2011) Stabilization of the α2 isoform of Na,K-ATPase by mutations in a phospholipid binding pocket. J. Biol. Chem. 286, 42888–42899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yeagle P. L., Young J., Rice D. (1988) Effects of cholesterol on (Na+,K+)-ATPase ATP hydrolyzing activity in bovine kidney. Biochemistry 27, 6449–6452 [DOI] [PubMed] [Google Scholar]
- 35. Cornelius F. (1995) Cholesterol modulation of molecular activity of reconstituted shark Na+,K+-ATPase. Biochim. Biophys. Acta 1235, 205–212 [DOI] [PubMed] [Google Scholar]
- 36. Strugatsky D., Gottschalk K. E., Goldshleger R., Bibi E., Karlish S. J. (2003) Expression of Na+,K+-ATPase in Pichia pastoris. Analysis of wild type and D369N mutant proteins by Fe2+-catalyzed oxidative cleavage and molecular modeling. J. Biol. Chem. 278, 46064–46073 [DOI] [PubMed] [Google Scholar]
- 37. Katz A., Lifshitz Y., Bab-Dinitz E., Kapri-Pardes E., Goldshleger R., Tal D. M., Karlish S. J. (2010) Selectivity of digitalis glycosides for isoforms of human Na,K-ATPase. J. Biol. Chem. 285, 19582–19592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography & NMR system. A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 39. Roy A., Kucukural A., Zhang Y. (2010) I-TASSER. A unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jørgensen P. L. (1988) Purification of Na+,K+-ATPase. Enzyme sources, preparative problems, and preparation from mammalian kidney. Methods Enzymol. 156, 29–43 [DOI] [PubMed] [Google Scholar]
- 41. Toyoshima C., Nomura H., Tsuda T. (2004) Lumenal gating mechanism revealed in calcium pump crystal structures with phosphate analogues. Nature 432, 361–368 [DOI] [PubMed] [Google Scholar]
- 42. Winther A. M., Liu H., Sonntag Y., Olesen C., le Maire M., Soehoel H., Olsen C. E., Christensen S. B., Nissen P., Møller J. V. (2010) Critical roles of hydrophobicity and orientation of side chains for inactivation of sarcoplasmic reticulum Ca2+-ATPase with thapsigargin and thapsigargin analogs. J. Biol. Chem. 285, 28883–28892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Toyoshima C., Yonekura S., Tsueda J., Iwasawa S. (2011) Trinitrophenyl derivatives bind differently from parent adenine nucleotides to Ca2+-ATPase in the absence of Ca2+. Proc. Natl. Acad. Sci. U.S.A. 108, 1833–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Turner N., Else P. L., Hulbert A. J. (2003) Docosahexaenoic acid (DHA) content of membranes determines molecular activity of the sodium pump. Implications for disease states and metabolism. Naturwissenschaften 90, 521–523 [DOI] [PubMed] [Google Scholar]
- 45. Turner N., Haga K. L., Else P. L., Hulbert A. J. (2006) Scaling of Na+,K+-ATPase molecular activity and membrane fatty acid composition in mammalian and avian hearts. Physiol. Biochem. Zool. 79, 522–533 [DOI] [PubMed] [Google Scholar]
- 46. Else P. L., Wu B. J., Storlien L. H., Hulbert A. J. (2003) Molecular activity of Na+,K+-ATPase relates to the packing of membrane lipids. Ann. N.Y. Acad. Sci. 986, 525–526 [DOI] [PubMed] [Google Scholar]
- 47. Gerbi A., Maixent J. M., Barbey O., Jamme I., Pierlovisi M., Coste T., Pieroni G., Nouvelot A., Vague P., Raccah D. (1998) Alterations of Na,K-ATPase isoenzymes in the rat diabetic neuropathy. Protective effect of dietary supplementation with n-3 fatty acids. J. Neurochem. 71, 732–740 [DOI] [PubMed] [Google Scholar]
- 48. Gerbi A., Barbey O., Raccah D., Coste T., Jamme I., Nouvelot A., Ouafik L., Lévy S., Vague P., Maixent J. M. (1997) Alteration of Na,K-ATPase isoenzymes in diabetic cardiomyopathy. Effect of dietary supplementation with fish oil (n-3 fatty acids) in rats. Diabetologia 40, 496–505 [DOI] [PubMed] [Google Scholar]
- 49. Gerbi A., Zerouga M., Debray M., Durand G., Chanez C., Bourre J. M. (1993) Effect of dietary α-linolenic acid on functional characteristic of Na+/K+-ATPase isoenzymes in whole brain membranes of weaned rats. Biochim. Biophys. Acta 1165, 291–298 [DOI] [PubMed] [Google Scholar]