Background: Glycosylation regulates the activities of plant metabolites and is mediated by glycosyltransferases (GT), glycoside hydrolases (GH), and transglycosidases (TG).
Results: The vacuolar TG Os9BGlu31 transfers glucose between phenolic acid esters and other compounds.
Conclusion: Os9BGlu31 equilibrates phenolic acids, phytohormones, and their glucosyl conjugates.
Significance: Os9BGlu31 and similar TG can broaden glycoconjugate diversity in planta.
Keywords: Carbohydrate Glycoconjugate, Enzyme Catalysis, Enzyme Mutation, Glycoconjugate, Glycoside Hydrolases, Natural Products, Plant Biochemistry, Transglycosidase
Abstract
Glycosylation is an important mechanism of controlling the reactivities and bioactivities of plant secondary metabolites and phytohormones. Rice (Oryza sativa) Os9BGlu31 is a glycoside hydrolase family GH1 transglycosidase that acts to transfer glucose between phenolic acids, phytohormones, and flavonoids. The highest activity was observed with the donors feruloyl-glucose, 4-coumaroyl-glucose, and sinapoyl-glucose, which are known to serve as donors in acyl and glucosyl transfer reactions in the vacuole, where Os9BGlu31 is localized. The free acids of these compounds also served as the best acceptors, suggesting that Os9BGlu31 may equilibrate the levels of phenolic acids and carboxylated phytohormones and their glucoconjugates. The Os9BGlu31 gene is most highly expressed in senescing flag leaf and developing seed and is induced in rice seedlings in response to drought stress and treatment with phytohormones, including abscisic acid, ethephon, methyljasmonate, 2,4-dichlorophenoxyacetic acid, and kinetin. Although site-directed mutagenesis of Os9BGlu31 indicated a function for the putative catalytic acid/base (Glu169), catalytic nucleophile residues (Glu387), and His386, the wild type enzyme displays an unusual lack of inhibition by mechanism-based inhibitors of GH1 β-glucosidases that utilize a double displacement retaining mechanism.
Introduction
Glycosylation is a ubiquitous mechanism by which plants regulate or establish the functionality of various compounds. Aside from oligo- and polysaccharides, glycoconjugates consist primarily of O-glycosides, glycosyl esters, S-glycosides, glycosylamines, and C-glycosyl compounds (1). These glycoconjugates may serve as active compounds themselves; as reversibly inactivated forms of phytohormones, defense compounds, monolignols, and volatiles; or as donors for acyl and glycosyl transfer reactions. The glycosyl moiety can serve as a blocking group to prevent undesirable reactions during metabolism, a tag to allow transport to a membranous organelle, such as a vacuole, and a polar group to increase water solubility and prevent diffusion across membranes, thereby allowing sequestration of otherwise lipid-soluble molecules.
The creation and breakage of the glycosyl linkages of these conjugates is mediated by glycosyltransferases (GTs),6 glycoside hydrolases (GHs), and transglycosidases (TGs) (2). GHs and GTs have been grouped in families by sequence similarity, and these families were joined into clans by structural and mechanistic similarities (3–5), as catalogued at the Carbohydrate Active eZYme online resource (CAZy) (6). The TGs that have been described to date fall within the GH families.
The creation of glycoconjugates is primarily catalyzed by GTs, which are enzymes that transfer a sugar from a nucleotide phosphate sugar or phospholipid sugar donor to an acceptor to form a glycosidic or ester linkage (2, 7). For synthesis of most glucosides and 1-O-acyl glucosyl esters in plants, these enzymes use uridine diphosphate α-d-glucoside as the donor. Although the vast majority of glycosylation reactions in plants are catalyzed by GTs (1), TGs also play a role. TGs are enzymes that transfer a sugar from a donor other than a nucleotide phosphate or phospholipid phosphate to an acceptor to form a new glycosidic linkage (2). Although many GHs have TG activities under conditions of high donor and acceptor substrate concentrations, TGs catalyze mainly transglycosylation with little hydrolysis, such as in the transfer of xyloglucan linkages between strands by xyloglucan endotransferases (xyloglucan:xyloglucosyl transferases; EC 2.4.1.207) (8).
Transfer and hydrolysis of glycosyl moieties on glycoconjugates can occur with either retention or inversion of the stereochemistry at the anomeric carbon at the glycosidic bond that is broken in the process (2, 9). The inverting mechanism of both GH and GT is thought to occur through a single displacement SN2 mechanism, in which the glycosidic bond is broken while the acceptor (water in the case of hydrolysis) attacks from the opposite face of the sugar. In GH, the retaining mechanism is thought to occur via a double displacement mechanism, which occurs in two SN2 steps: glycosylation and deglycosylation. In the glycosylation step, the catalytic acid/base protonates the leaving group aglycon, and the catalytic nucleophile (usually a nucleophilic amino acid residue side chain) attacks the anomeric carbon of the sugar to form a glycosyl-enzyme intermediate (10). In the deglycosylation step, water or another nucleophile displaces the enzyme from the sugar with basic assistance from the catalytic acid/base. Transglycosylation can occur by the same mechanism in GH and TG (11). In principle, retaining GT could use this mechanism, but it was recently demonstrated that trehalose phosphate synthase uses a front end attack SNi-type mechanism instead (12). Thus, glycon transfer can occur through multiple mechanisms.
Although most small glucoconjugates in plants, including those of phytohormones, defense compounds, monolignols, and pigments, are thought to be formed by a branch of GT family 1 (1), they are primarily hydrolyzed by GH family 1 (GH1) β-glycosidases (13). Plant GH1 hydrolases have a wide range of activities, including β-d-glucosidases (EC 3.2.1.21), β-d-mannosidases (EC 3.2.1.25), β-disaccharidases, thioglucosidases (myrosinases; EC 3.2.1.147), and even the purine hydrolase hydroxyisourate hydrolase (HIUH; EC 3.5.2.17), which is not a glycosidase at all (13, 14). The GH1 β-d-glycosidases also have a range of specificities for aglycon or saccharide leaving groups, with substrates including oligosaccharides, cyanogenic glucosides, phytohormone glycoconjugates, flavonoid and isoflavonoid glycosides, the monoterpenoid indole alkaloid precursor strictosidine, and benzoxanoids.
In addition to hydrolases, GH1 has recently been found to include TGs (15, 16). The chloroplastic galactolipid:galactolipid galactosyltransferase was found to correspond to the GH1 enzyme SFR2 (sensitive to freezing 2), which is necessary for freezing tolerance in Arabidopsis (Arabidopsis thaliana) (16). Galactolipid:galactolipid galactosyltransferase disproportionates the galactosyl residues to produce diacyl glycerol and β-linked oligogalactosyl diacyl glyceride from monogalactosyl diacyl glycerides. GH1 enzymes were also found to act as 1-O-acyl-β-d-glucose-dependent anthocyanin glucosyltransferases that transfer glucose from aromatic acid esters to the five or seven hydroxyls of anthocyanin-3-glucoside to make the 3,5-diglucoside in carnation (Dianthus caryophyllus) and the 3,7-diglucoside in delphinium (Delphinium grandiflorum) (15). Interestingly, 1-O-acyl β-d-glucose esters can also serve as acyl donors for the vacuolar serine carboxypeptidase-like acyltransferases (17, 18) and for intracellular feruloylation of arabinoxylans (19).
Forty Arabidopsis and thirty-four rice (Oryza sativa) genes encoding apparently functional GH1 proteins have been identified in genome sequences, and these genes were grouped into eight amino acid sequence-based phylogenetic clusters that contain both Arabidopsis and rice genes, At/Os1–8 (20, 21). In the current work, we have characterized Os9BGlu31, a member of the At/Os6 cluster, which also contains HIUH, DcAA5GT, and DgAA7GT. We show that Os9BGlu31 is a vacuolar TG with novel mechanistic properties that acts on a number of natural acceptors and donors, including aromatic acids and phytohormones and their glucoconjugates, suggesting a broader role for GH1 TG than previously realized.
EXPERIMENTAL PROCEDURES
Protein Expression
A plasmid construct containing a full-length cDNA encoding the Os9BGlu31 protein was acquired from the Rice Genome Resource Center (Tsukuba, Japan; GenBankTM accession AK121679). The cDNA fragment encoding mature Os9BGlu31 was amplified with the BGlu31F and BGlu31R primers (supplemental Table S1) and Pfu DNA polymerase (Promega). The PCR product was cloned into the pENTRTM/d-TOPO Gateway® system entry vector (Invitrogen) and subcloned into the pET32a/DEST expression vector (21) by LR Clonase (Invitrogen) reaction to make pET32a/DEST/Os9BGlu31.
Protein Purification
Thioredoxin-Os9BGlu31 fusion protein expression was induced in Escherichia coli strain Origami B (DE3) containing the recombinant pET32a/DEST/Os9BGlu31 plasmid with 0.4 mm isopropyl β-d-thiogalactopyranoside at 20 °C overnight, and the cells were collected and lysed as previously described (22), except that 0.1 mg/ml soybean trypsin inhibitor was added to the lysis buffer. The Os9BGlu31 fusion protein was purified with three steps. First, crude protein was mixed with CoCl2 pre-equilibrated immobilized metal affinity chromatography resin (GE Healthcare) with equilibration buffer (150 mm NaCl, 20 mm Tris-HCl, pH 8.0) at 4 °C for 30 min. The resin with crude protein was loaded into a column and washed sequentially with 10 column volumes (CV) of 5 mm imidazole in equilibration buffer and 5 CV each of 10 mm imidazole and 20 mm imidazole in equilibration buffer. Os9BGlu31 fusion protein was eluted with elution buffer (250 mm imidazole in equilibration buffer). The fractions with activity were pooled, and imidazole was removed by dialysis with 50 mm Tris-HCl, pH 8.0, at 4 °C. Next, the recombinant protein was loaded onto a Q-Sepharose (GE Healthcare), unbound protein was washed from the column with 10 CV of 50 mm Tris-HCl, pH 8.0, and Os9BGlu31 was eluted with a linear gradient of 0–0.5 m NaCl in 50 mm Tris-HCl, pH 8.0. The fractions containing activity were pooled, and the NaCl concentration was adjusted to 2 m. The protein was loaded onto a phenyl-Sepharose (GE Healthcare) column, and unbound protein was washed from the column with 10 CV of 2 m NaCl in 50 mm Tris-HCl, pH 8.0. Os9BGlu31 fusion protein was eluted with a linear gradient of 2 to 0 m NaCl in 50 mm Tris-HCl, pH 8.0, followed by 0–50% ethylene glycol in 50 mm Tris-HCl, pH 8.0. Finally, the buffer of the Os9BGlu31-containing fraction pool was changed to 150 mm NaCl in 20 mm Tris-HCl, pH 8.0, by dialysis.
pH Optimum Determination
The activity of Os9BGlu31 (1.5 μg) was measured with 5 mm 4-nitrophenyl-β-d-glucoside (4NPGlc) as substrate. Two buffer systems of overlapping pH buffer (citrate buffer, pH 3.0–4.0; acetate, pH 4.0–5.5; MES, pH 5.5–7.0; Tris-HCl, pH 7.0–9.0; and sodium phosphate, pH 9.0–12.0) and McIlvaine buffer (0.1 m citric acid and 0.2 m disodium hydrogen phosphate) were used. In additional, the activity of enzyme was determined against 5 mm 4NPGlc in McIlvaine buffer in the presence of 5 mm azide, acetate, formate, fluoride, or ascorbate or 0.2 mm ferulic acid. The reaction was incubated at 30 °C for 1 h and stopped by the addition of 2 m sodium carbonate. The 4-nitrophenol (4NP) released was quantified from the absorbance at 405 nm.
To determine the optimum pH of Os9BGlu31 mutant enzymes, 3 μg of E169Q, 6 μg of E169A, 4.5 μg of H386T, and 42.5 μg of E387A were assayed with 5 mm 4NPGlc and 0.2 mm ferulic acid as acceptor in citrate/phosphate buffer, pH 3.0–10.0. The reactions were incubated at 30 °C for 1 h (E169Q), 2 h (E169A), 1.5 h (H386T), or 24 h (E387A). 4NP release was measured spectrophotometrically, as described above, except release of 4NP by E387A was measured by HPLC (see below).
Enzymatic Characterization of Os9BGlu31
1-O-Gibberellin GA4-β-d-glucose ester (GA4-GE) was synthesized as described by Hiraga et al. (23), and the structure was confirmed by NMR. Production of other 1-O-acyl glucose esters was described previously (15). To investigate the substrate specificity of Os9BGlu31, 5 mm donor substrates, including various phenolic glucose esters and glucosides, were assayed with 5 mm naphthalene acetic acid as acceptor, whereas acceptor substrates for Os9BGlu31 were assayed with 5 mm 4NPGlc as donor and 5 mm acceptor (see supplemental Table S2 for substrate structures). The reactions containing 1–3 μg enzyme in 50 mm citrate, pH 4.5, was incubated at 30 °C for 1 h. The products and substrates were separated on silica gel F254 TLC with chloroform:methanol:ammonia (7:2.8:0.2). The developed plates were observed under UV and then stained with 10% H2SO4 in ethanol and charred at 110 °C.
Preference for glucose donor (see Table 1) was quantified by incubating 0.5 mm glucose donor with 0.2 mm 4-hydroxybenzoic acid (4HB) as glucose acceptor with 1 μg of Os9BGlu31 in 50 mm citrate, pH 4.5, at 30 °C. For 1-O-(4-hydroxybenzoyl)-β-d-glucose (4HBG) donor, 0.2 mm ferulic acid was used as acceptor instead of 4HB. After 10 min, the reactions were stopped by adding phosphoric acid to 1% final concentration. Transglycosylation products were detected by HPLC on an Agilent 1100 system with an XDB-C18 HPLC column (Agilent). The substrate and products were separated at 1 ml/min with a linear gradient from 5 to 12% acetonitrile in 1.5% phosphoric acid for 20 min, followed by 12 to 80% acetonitrile in 1.5% phosphoric acid in 5 min and detected by absorbance at 254 nm on a diode array detector.
TABLE 1.
Relative activities of Os9BGlu31 with various donors
| Donor | Relative activitya |
|---|---|
| % | |
| Phenol glucose esters and glucosides | |
| 1-O-Feruloyl-β-d-glucose | 100.0 |
| 1-O-(4-Coumaroyl)-β-d-glucose | 96.3 ± 1.0 |
| 1-O-Vanillyl-β-d-glucose | 93.4 ± 1.9 |
| 1-O-Vanillyl-α-d-glucose | NDb |
| 1-O-(4-Hydroxybenzoyl)-β-d-glucose | 82.0 ± 2.1 |
| 1-O-Sinapoyl-β-d-glucose | 51.4 ± 0.5 |
| Hormone glucose esters | |
| Gibberellin A4-glucose | 12.5 ± 0.5 |
| Flavonoid/isoflavone/flavone glucosides | |
| Phloridizin | 49.2 ± 1.7 |
| Apigenin 7-glucoside | 32.8 ± 0.9 |
| Quercetin 3-β-d-glucoside | ND |
| Naringin | ND |
| Arbutin | ND |
| Gossypin | ND |
| Daidzin | Lowc |
| Genistin | Lowc |
| Cytokinin glucoside | |
| trans-Zeatin glucoside | ND |
| Nucleoside diphosphate sugar | |
| Uridine 5′-diphosphoglucose | ND |
| Coumarin glucoside | |
| Esculin | ND |
| 4NP-glucosides and synthetic glucoside | |
| 4NP-β-d-glucoside | 3.8 ± 0.05 |
| 4NP-β-d-fucoside | 2.3 ± 0.03 |
| 4NP-β-d-xyloside | 1.8 ± 0.03 |
| 4NP-β-d-galactoside | ND |
| 4NP-β-d-mannoside | ND |
| 4NP-α-d-glucoside | ND |
| 4NP-α-d-galactoside | ND |
| 4NP-β-d-glucosiduronide | ND |
| 4NP-α-l-arabinoside | ND |
| 4NP-n-acetylglucosaminide | ND |
| 2NP-β-d-glucoside | ND |
| 4-Methylumbelliferyl β-d-glucoside | ND |
| n-Octyl glucoside | ND |
| n-Heptyl glucoside | ND |
a The donor preferences of Os9BGlu31 were determined with 4HB as acceptor or ferulic acid when 4HBG was donor. The rate on FG was set at 100% and corresponds to 3.9 nanokatals.
b ND, not detected.
c Daidzin and genistin products could not be detected under the standard assay conditions, but small amounts of product were detected by thin layer chromatography when 5 mm of these glucose donors was incubated in reactions with enzyme and 4HB overnight. Even under these conditions, there was no activity with those compounds listed as “ND” above and the monolignol glucosides 4-coumary alcohol glucoside and coniferin, and the cyanogenic glucosides linamarin, d-amygdalin, and dhurrin as glucose donors.
Activities with various glucose acceptors (see Table 2) were assayed with 0.2 mm glucose acceptor, 5 mm 4NPGlc as glucose donor, and 2 μg of Os9BGlu31 in 50 mm citrate, pH 4.5. The reactions were incubated at 30 °C for 1 h and then stopped by adding phosphoric acid to 1% as described above. Because some of the acceptors have absorbance spectra overlapping that of 4NP, the released 4NP was separated from other reaction components by HPLC with an XDB-C18 column and elution with a linear gradient from 20 to 55% acetonitrile in 1.5% phosphoric acid for 20 min (1 ml/min) and then from 55 to 80% acetonitrile in 5 min. Released 4NP was monitored with a diode array detector and quantified by its absorbance (peak area) at 360 nm. For the assay of 0.2 mm 4NP as an acceptor, 0.5 mm 1-O-feruloyl β-d-glucose (FG) was used as the donor instead of 5 mm 4NPGlc, and the relative activity of 4NP was compared with 0.2 mm 4HB as acceptor with 0.5 mm FG as donor. The 4NPGlc formed in this reaction was detected by HPLC with the same solvent system as the glucose donor preference determination. For those acceptors for which the products were previously synthesized and verified by MS and NMR (15), the product elution positions were confirmed to match those of their respective 1-O-acyl glucosyl esters under the HPLC conditions described for donor identification. The product peak elution position from the reaction with 4HB acceptor was compared with those of 4HBG and 4HB-4-O-β-d-glucoside. To determine activities of Os9BGlu31 mutant enzymes with glucosyl acceptors, 3 μg of E169Q, 20 μg of E169A, 4.5 μg of H386T, and 64 μg of E387A were assayed at the optimum pH. The reactions were incubated at 30 °C for 1 h (E169Q, E169A, and H386T) or 24 h (E387A).
TABLE 2.
Relative activities of Os9BGlu31 on various acceptors
| Acceptora | Relative activity |
|---|---|
| % | |
| Citrate buffer alone | 9.3 ± 0.4 |
| Phenolic compounds | |
| Ferulic acidb | 100.0 |
| Vanillic acidb | 85.5 ± 0.4 |
| 4-Hydroxybenzoic acidb | 78.3 ± 2.6 |
| Syringic acid | 78.3 ± 2.1 |
| trans-Cinnamic acid | 78.3 ± 0.4 |
| Caffeic acid | 78.3 ± 1.9 |
| Sinapic acidb | 64.1 ± 0.04 |
| Benzoic acid | 64.1 ± 0.7 |
| 4-Coumaric acidb | 57.0 ± 0.1 |
| Isovanillic acid | 49.9 ± 1.1 |
| Dihydroxybenzoic acid | 42.7 ± 0.3 |
| Scopoletin | 28.5 ± 0.2 |
| Vanillin | 14.2 ± 0.1 |
| 1-Naphthol | 14.2 ± 0.3 |
| 2-Naphthol | NDd |
| Apocynin | 7.1 ± 0.5 |
| Vanillyl alcohol | 7.1 ± 0.02 |
| Dihydroxybenzaldehyde | 7.1 ± 0.1 |
| 4-Nitrophenolc | 2.0 ± 0.01 |
| Phytohormones | |
| Napthalene acetic acid | 42.7 ± 1.0 |
| Indole acetic acid | 42.7 ± 0.6 |
| 2,4-Dichlorophenoxyacetic acid | 29.9 ± 0.4 |
| Abscisic acid | 14.2 ± 0.1 |
| Gibberellin A3 | 7.1 ± 0.02 |
| Gibberellin A4b | 14.2 ± 0.1 |
| Coumarin | |
| Esculetin | 28.5 ± 3.0 |
| Flavonoids/flavonoid glucoside | |
| (−)-Catechin | 35.6 ± 0.1 |
| Quercitin 3-glucoside | 28.5 ± 1.0 |
| Apigenin | 28.5 ± 1.7 |
| Kaemferol | 21.4 ± 1.2 |
| Nariginin | 14.2 ± 0.3 |
| Anthocyanidin glucoside/vitamin glucoside/monolignol glucoside | |
| Pyridoxine 5-glucoside | 14.2 ± 0.1 |
| Cyanidin 3-glucoside | ND |
| 4-Coumaryl alcohol 4-glucoside | 16.9 ± 0.3 |
| Alkyl acids and chemical nuclophiles | |
| Butyric acid | 28.5 ± 0.3 |
| Propinoic acid | 21.4 ± 0.2 |
| Azide | 9.7 ± 0.08 |
| Acetate | 6.2 ± 0.04 |
| Formate | 4.2 ± 0.04 |
| Thiosulfate | ND |
| Thiocyanate | ND |
| Cyanide | ND |
a Acceptor preferences of Os9BGlu31 with 4NPGlc as donor. Relative activity is shown in comparison with ferulic acid, which is arbitrarily set as 100% and corresponds to 1.9 nanokatals. In addition, alcohol glucosides could be observed by thin layer chromatography in reactions containing 10% (v/v) alcohols (methanol, ethanol, 1-butanol, 1,3-butanediol, 4-methyl-2-pentanol, 2-methyl-1-pentanol, 1-hexanol, 2-hexanol, and 1-octanol) as acceptor when they were incubated at 30 °C overnight.
b The products of these reactions were verified to elute from the HPLC at the positions of the respective 1-O-acyl β-d-glucose esters, which were previously characterized by MS and NMR (Ref. 15 and data not shown).
c 4NP was assayed with 0.5 mm feruloyl glucose rather than 5 mm 4NPGlc as a donor, so the comparison is not equitable. However, it is included to show that the reaction can go in reverse.
d ND, not detected.
The relative activity toward 4NP-glycoside donor substrates was determined by incubating 10 μg of Os9BGlu31 with 0.5 mm 4NP-glycoside and 0.2 mm 4HB in 140 μl of 50 mm citrate, pH 4.5, at 30 °C for 1 h. The reaction was stopped by adding 100 μl of 2 m Na2CO3, and the 4NP released was quantified by the absorbance at 405 nm.
Kinetic Parameter Determination
The apparent Km and Vmax values of 4NPGlc in the presence of various acceptors were determined by varying the concentration of 4NPGlc in the range 2–30 mm with 0.2 mm glucose acceptor and 1.2–2.4 μg of Os9BGlu31 in 50 mm citrate, pH 4.5. The apparent Km and Vmax values of the glucose acceptors were determined by varying their concentrations between 0.02 and 0.5 mm in the presence of 30 mm 4NPGlc with between 1.2 and 2.4 μg of Os9BGlu31 in 50 mm citrate, pH 4.5. The release of 4NP product was quantified as described above. The kinetic parameters were determined by nonlinear regression of the Michaelis-Menten plots with the Grafit 5.0 computer program (Erithacus Software, Horley, UK).
Effect of Metals and Inhibitors
The effects of 1 mm EDTA, CaCl2, CoCl2, CuSO4, FeCl3, HgCl2, MgCl2, MnCl2, NiSO4, and ZnSO4, and 7 μm to 5 mm inhibitors on enzyme activity were determined by preincubating the enzyme with an individual chemical in 50 mm sodium acetate, pH 4.5, at 30 °C for 10 min for EDTA and metal ions or 2 h for organic inhibitors. Activity was then assayed by incubating 2.5 μg of pretreated enzyme with 5 mm 4NPGlc in 140 μl of 50 mm sodium acetate, pH 4.5, at 30 °C for 15 min. The reactions were stopped with 100 μl of 2 m Na2CO3 and released 4NP measured as described above. The release of 2,4-dinitrophenylate from 1 mm 2,4-dinitrophenyl-β-d-2-deoxy-2-fluoro-glucopyranoside (2,4dNP2FG) was assessed in 3-h reactions with 10 μg of Os9BGlu31 in 50 mm citrate buffer, pH 4.5, with and without 0.2 mm ferulic acid. The reactions were stopped and detected as with 4NPGlc.
Mutation of Os9BGlu31
Site-directed mutagenesis was performed following the instructions of the QuikChange® site-directed mutagenesis kit (Stratagene). The pET32a/DEST/Os9BGlu31 vector was used as template with the E169Qf and E169Qr primers for the E169Q mutation, the E169Af and E169Ar primers for the E169A mutation, the E387Af and E387Ar primers for the E387A mutation, and the H386Tf and H386Tr primers for the H386T mutation (supplemental Table S1).
Northern Blot Analysis
Rice (O. sativa L. cv. Yukihikari) seeds were germinated in the dark for 4 days at 27 °C and then grown in a 12-h light/12-h dark cycle from days 4 to 10 at 28 °C and moistened with sterile distilled water. Ten-day-old seedlings were harvested and dissected into shoot, root, and endosperm. Other seedlings were transferred to soil and grown for an additional 4–5 weeks to reach the flowering stage. Rice plants were harvested and separated to flower, stem, root, node, leaf blade, and leaf sheath.
Ten-day-old rice seedlings were exposed to various abiotic stresses and plant hormones for an additional 2 days. The abiotic conditions were: 1) cold temperature, at 5 °C and 12 °C; 2) drought, by supplying no water; 3) salt stress, with 0.3 m NaCl solution; and 4) flooding, by submerging the seedlings in distilled water. Phytohormone treatments were: 1) 0.1 mm abscisic acid (ABA); 2) ethylene (10 mm ethephon); 3) 0.1 mm methyl jasmonic acid; 4) 0.1 mm gibberellin A3 (GA3); 5) 0.1 mm kinetin; and 6) 10 mm 2,4-dichlorophenoxyacetic acid (2,4-D). To further characterize the effect of ethephon, 10-day-old rice seedlings were treated with ethephon for 2 days, and the seedlings were dissected into shoot, root, and endosperm. All of the plant samples were kept at −70 °C for RNA isolation. A gene-specific probe for Os9BGlu31 was amplified from rice genomic DNA with the BGlu31–3′UTRf and BGlu31–3′UTRr primers derived from the 3′-untranslated region of the gene (supplemental Table S1) and Taq DNA polymerase (Roche Diagnostics).
Total RNA was isolated from rice tissues by the method of Bachem et al. (24). Thirty micrograms of total RNA was denatured and electrophoresed on 1.5% formaldehyde-agarose gels and transferred onto Hybond N+ nylon membrane (GE Healthcare) by standard procedures (25). The Os9BGlu31 probe was labeled by Rediprime II random priming with [α-32P]dCTP (GE Healthcare) and was hybridized with RNA blots for 16 h at 42 °C. The blots were then washed once in 0.1% SDS, 2× SSC for 30 min at 65 °C, washed twice in 0.1% SDS, 0.1× SSC for 15 min at 65 °C, and then exposed to a Fuji film imaging plate for 16 h at 20–25 °C. The positions of radioactive bands were visualized with a Fuji Film BAS 1000 BioImaging Analyzer (Fuji Photo Film Co., Ltd, Tokyo, Japan).
Real Time PCR
For quantitative real time PCR, all samples were collected from greenhouse-grown japonica rice cultivar Dongjin, as described previously (26–28). Flag leaves were harvested at four different developmental stages, 15 days before flowering, 15 days after flowering (DAF), 30 DAF, and 40 DAF, respectively. Total RNA was isolated from harvested samples with TRIzol reagent (Invitrogen) and reverse-transcribed with the iScript cDNA synthesis kit (Bio-Rad). The expression level of the rice ubiquitin 5 (OsUBQ5) gene was used to normalize the cDNA quantity (29). Gene-specific primers used for quantitative real time PCR were the BGlu31RT-f and BGlu31RT-r primers for Os9BGlu31 and OsUBQ5RT-f and OsUBQ5RT-r primers for OsUBQ5 (supplemental Table S1). All of the experiments were conducted in triplicate with the SYBR Premix Ex Taq (Takara) and an ABI PRISM 7500 sequence detector (Applied Biosystems) according to the manufacturer's instructions, and changes in gene expression were calculated by the comparative cycle threshold (ΔΔCt) method with Sequence Detector Systems version 1.2 software (Applied Biosystems).
Subcellular Localization Assay
The Os9BGlu31 gene was amplified by PCR with proofreading DNA polymerase (SolGent, Daejeon, Korea) and the BGlu31SL-f and BGlu31SL-r primers (supplemental Table S1). The PCR products were cloned into the pENTR/d-TOPO vector (Invitrogen) and sequenced. Validated insert was subcloned into the transient expression vector p2GWF7 and the binary expression vector pH7FWG2 for C-terminal GFP fusion constructs by LR clonase (Invitrogen) reaction (30). The resulting Os9BGlu31-GFP fusion was placed under the control of the CaMV35S promoter and introduced into maize mesophyll protoplasts by polyethylene glycol-mediated transformation (31). The binary Os9BGlu31-GFP fusion construct was transformed by Agrobacterium-mediated co-cultivation with rice calli, as described previously (32). Expression of the fusion constructs in maize protoplasts and rice calli was monitored using a confocal microscope (LSM 510 META; Carl Zeiss) at various times after transformation.
Sequence Analysis
Plant amino acid sequences of rice GH1 β-glycosidases, TG, and Glycine max HIUH were taken from the GenBankTM database. Secretory signal sequence prepeptides were predicted by SignalP 4.0 (33) and signal sequence and location with Predotar (INRA) (34). The predicted N-terminal signal peptides were removed prior to protein alignment analysis by the MUSCLE program (35). Phylogenetic trees were constructed by the neighbor-joining and minimum evolution methods in MEGA 5.05 (36), which gave similar branching.
Accession Numbers
The Os9BGlu31 gene was taken from the Rice Genome Resource Center clone J033069H08, with the accession number AK121679 in the GenBankTM database. Sequence data for amino acid sequence alignment can be found in the GenBankTM database under accession numbers Q8L7J2 (Os3BGlu6), U28047 (Os3BGlu7), BAH00605 (Os9BGlu31), Q0J0G2 (Os9BGlu32), Q0J0G1 (Os9BGlu33), AAL92115 (HIUH), BAJ33501 (Dc AA5GT), and BAJ33502 (Dg AA7GT).
RESULTS
Characterization of Recombinant Os9BGlu31
The rice genome project locus Os09g0511600 encodes a GH1 precursor protein of 523 amino acid residues designated Os9BGlu31, based on its genomic position and putative activity, and three full-length mRNA clones (GenBankTM accession numbers AK121679, AK102869, and AK121935) and several expressed sequence tags in UniGene accession Os.9105 verify that it is an active gene (21). Two closely related genes are found at the loci Os09g0511700, encoding Os9BGlu32, and Os09g0511900, encoding Os9BGlu33. The three encoded proteins all contain N-terminal secretory targeting sequences and the sequence IHENG around the catalytic nucleophile (Glu387 in Os9BGlu31) instead of the typical ITENG sequence seen in GH1 β-d-glucosidases, which is the same as soybean HIUH and DcAA5GT and DgAA7GT. We introduced the cDNA encoding Os9BGlu31, Os9BGlu32, and Os9BGlu33 into pET32a-derived expression vectors, but only Os9BGlu31 could be produced as an active fusion protein with N-terminal thioredoxin and 6× histidine tags, with best expression in E. coli strain Origami B (DE3). Attempts to express Os9BGlu32 and Os9BGlu33 from a secreted system in Pichia pastoris (37) also gave no active proteins.
The Os9BGlu31 fusion protein was purified by immobilized metal affinity chromatography, Q-Sepharose anion exchange chromatography, and phenyl-Sepharose chromatography to yield protein that was approximately 90% pure on SDS-PAGE (data not shown). When assayed in acetate buffer, the Os9BGlu31 enzyme released 4NP from 4NPGlc, but no glucose release could be detected from any natural substrate by glucose-oxidase/peroxidase assay. The activity versus pH profile in a set of different buffers showed that the enzyme had much higher activity in acetate than citrate or MES buffers at the same pH (data not shown). Because this was reminiscent of the behavior of a GH1 acid/base mutant that can be rescued by transglycosylation, we investigated the reaction product. Upon inspection of products of the reaction of 4NPGlc catalyzed by Os9BGlu31 by TLC, no glucose could be detected, but a product spot that stained for carbohydrate was detected below the 4NPGlc spot (data not shown). This product appeared to be a transglycosylation product of acetate with glucose from the 4NPGlc.
When the pH optimum was determined in citrate/phosphate buffer, in which no transglycosylation product was detected, the activity of Os9BGlu31 in the absence of an additional nucleophile was highest at pH 4.5, the activity decreased to approximately 70% at pH 3.5 and pH 5.0, and there was no activity at pH 6.5 or higher (data not shown). The release of 4NP increased approximately 2.9-, 3.3-, and 4.2-fold in the presence of 5 mm azide, formate, and acetate, respectively, which gave pH optima in the range of 4–4.5, whereas fluoride and ascorbate had no effect on enzyme activity (data not shown).
Substrate Preferences for Donors and Acceptors
Based on the TLC analysis of the above reactions, Os9BGlu31 acts to transfer the glucosyl moiety from a donor substrate to an acceptor. The glycon (sugar) specificity of Os9BGlu31 was determined by transfer from 4NP-glycosides to 4HB. Os9BGlu31 transferred glucose from 4NPGlc better than β-d-fucose and β-d-xylose from their respective 4NP-glycosides (Table 1) and could not use 4NP-β-d-galactoside, 4NP-β-d-mannoside, 4NP-N-acetyl-β-d-glucosaminide, 4NP-β-d-glucuronide, 4NP-α-d-glucoside, 4NP-α-d-galactoside, or 4NP-α-l-arabinoside as substrates.
In addition to 4NP-glycosides, Os9BGlu31 used phenolic 1-O-acyl-β-d-glucose esters as donor substrates with highest activity to FG, 1-O-(4-coumaroyl)-β-d-glucose, 4HBG, 1-O-sinapoyl-β-d-glucose, and 1-O-vanillyl-β-d-glucose (Table 1 and Fig. 1A). Os9BGlu31 also used the flavonoid glucosides, phloridizin and apigenin 7-O-glucoside, and the GA4-GE as glucose donors, although with lower efficiency.
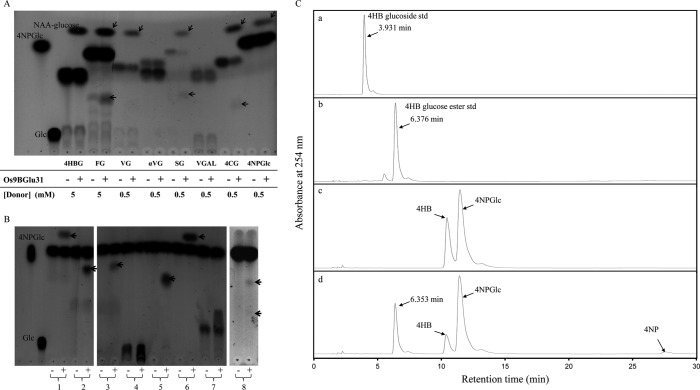
FIGURE 1.
Transglucosidase activity of Os9BGlu31. A, thin layer chromatogram of Os9BGlu31 transfer of glucose from 1-O-phenolic glucose esters to 1-naphthalene acetic acid acceptor. 4HBG, FG, 1-O-vanillyl-β-d-glucose (VG), 1-O-vanillyl-α-d-glucose (αVG), 1-O-sinapoyl-β-d-glucose (SG), 1-O-vanillyl-β-d-galactose (VGAL), 1-O-(4-coumaroyl)-β-d-glucose (4CG), and 4NPGlc were used as donors. The transglycosylation product 1-O-(naphthalene acetic acid) β-d-glucose ester (NAA-glucose, marked by slanted arrows) released phenolic compounds (marked by horizontal arrows) and glucose (Glc) are indicated. B, thin layer chromatography analysis of Os9BGlu31 transfer of glucose from 4NPGlc to acceptors. The acceptors 1-naphthalene acetic acid (lane 1), syringic acid (lane 2), 4-coumaric acid (lane 3), gallic acid (lane 4), 4-hydroxybenzoic acid (lane 5), trans-cinnamic acid (lane 6), caffeic acid (lane 7), and esculetin (lane 8) were incubated in reactions with (+) and without (−) Os9BGlu31 enzyme. The products from A and B were stained with 10% H2SO4 in ethanol, followed by charring. C, detection of Os9BGlu31 transglucosidase activity using 4NPGlc as donor and 4HB as acceptor by HPLC. HPLC elution profiles of 4HB-4-O-β-d-glucoside (panel a) and 1-O-(4-hydroxybenzoyl)-β-d-glucose ester standards (std, panel b), substrate reaction blank (panel c), and transglucosidase reaction containing Os9BGlu31 (panel d).
The TG activity of Os9BGlu31 determined with a range of potential acceptors is summarized in Table 2. When transglycosylation reactions of various acceptors with Os9BGlu31 and 4NPGlc donor were inspected by TLC, spots of 4NP and a single transglycosylation product were seen in each case with little or no glucose detectable (Fig. 1B). The one exception was esculetin, which had two transglycosylation products. One esculetin product migrated to a position similar to esculin (esculetin-6-O-β-d-glucoside), whereas the other product was located between esculetin and esculin and a monoglucoside mass peak at 339.0 m/z, that expected for esculin, and no diglucoside mass could be detected on LCMS (data not shown), suggesting that the other product was esculetin-7-O-β-d-glucoside. The products of the phenolic acids for which 1-O-acyl-β-d-glucose esters were available (Tables 1 and 2) were confirmed to co-elute with these 1-O-esters in reverse phase HPLC. To verify that the glucoside (glucosylation of the hydroxyl group) did not co-elute, the product with 4HB was compared with both the ester and glucoside standards, and only product co-eluting with the 1-O-ester, 4HBG, was detected (Fig. 1C). Similarly, the product with vanillic acid co-eluted with 1-O-vanillyl β-d-glucose and not 1-O-vanillyl α-d-glucose (data not shown).
Os9BGlu31 could glycosylate the auxins indole acetic acid and naphthalene acetic acid with 43% the relative activity of ferulic acid and had significant activity toward ABA and GA4 as well. Glucose was also transferred to the carboxylic group of alkyl organic acids, with increasing relative activity from formic to butyric acid, and to hydroxyl groups of alcohols, especially aromatic alcohols like catechin and esculetin, and certain flavonoid alcohols. Glycoside products of methanol, ethanol, 1-butanol, 1,3-butanediol, 4-methyl-2-pentanol, 2-methyl-1-pentanol, 1-hexanol, 2-hexanol, and 1-octanol could also be detected on TLC, although the addition of these alcohols did not increase the release of 4NP from the donor more than the buffer alone (data not shown).
Quantification of 4NP released from 4NPGlc in the presence of different acceptors by HPLC allowed the apparent kinetic parameters of transglucosylation to be determined, as shown in Table 3. Although a small amount of hydrolysis may occur in these reactions, Fig. 1 (A and B) shows that little glucose is released in the presence of acceptor substrates, and much of this may be attributed to the breakdown of labile substrates and products, such as 4HBG. Thus, the apparent kinetic parameters should primarily reflect the transglycosylation reactions. Os9BGlu31 had the highest relative activity and apparent kcat (1.21 s−1) for ferulic acid but had the highest apparent kcat/Km value for 4-coumaric acid (33.3 mm−1 s−1), because of its low apparent Km, followed by ferulic acid (25.4 mm−1 s−1) and sinapic acid (14.2 mm−1 s−1), respectively.
TABLE 3.
Apparent kinetic parameters of Os9BGlu31with aromatic acid acceptors
| Acceptor | Kinetic parameters for acceptor at 30 mm 4NPGlc |
Kinetic parameters for 4NPGlc at 0.25 mm acceptor |
||||
|---|---|---|---|---|---|---|
| Km | kcat | kcat/Km | Km | kcat | kcat/Km | |
| mm | s−1 | mm−1 s−1 | mm | s−1 | mm−1 s−1 | |
| Phenolic compounds | ||||||
| Ferulic acid | 0.05 ± 0.004 | 1.21 ± 0.06 | 25.42 ± 1.52 | 9.33 ± 0.62 | 1.21 ± 0.2 | 0.13 ± 0.01 |
| Vanillic acid | 0.11 ± 0.012 | 0.43 ± 0.04 | 3.82 ± 0.4 | 5.92 ± 0.28 | 0.36 ± 0.023 | 0.06 ± 0.005 |
| 4-Hydroxybenzoic acid | 0.21 ± 0.02 | 0.49 ± 0.002 | 2.31 ± 0.22 | 2.88 ± 0.27 | 0.33 ± 0.01 | 0.21 ± 0.01 |
| Syringic acid | 0.13 ± 0.012 | 0.35 ± 0.03 | 2.70 ± 0.25 | 1.18 ± 0.13 | 0.24 ± 0.02 | 0.20 ± 0.02 |
| trans-Cinnamic acid | 0.10 ± 0.01 | 0.32 ± 0.02 | 3.26 ± 0.08 | 1.62 ± 0.17 | 0.12 ± 0.004 | 0.07 ± 0.002 |
| Caffeic acid | 0.03 ± 0.003 | 0.25 ± 0.005 | 7.62 ± 0.6 | 7.70 ± 0.6 | 0.25 ± 0.02 | 0.03 ± 0.0004 |
| Sinapic acid | 0.02 ± 0.002 | 0.29 ± 0.01 | 14.15 ± 0.4 | 7.06 ± 0.74 | 0.28 ± 0.02 | 0.05 ± 0.001 |
| Benzoic acid | 0.12 ± 0.014 | 0.35 ± 0.038 | 2.87 ± 0.5 | 2.00 ± 0.17 | 0.28 ± 0.02 | 0.14 ± 0.04 |
| 4-Coumaric acid | 0.01 ± 0.001 | 0.34 ± 0.03 | 33.3 ± 3.9 | 3.31 ± 0.3 | 0.30 ± 0.03 | 0.10 ± 0.008 |
| Isovanillic acid | 0.042 ± 0.003 | 0.22 ± 0.001 | 5.13 ± 0.5 | 1.91 ± 0.2 | 0.17 ± 0.02 | 0.10 ± 0.007 |
| Dihydroxybenzoic acid | 0.05 ± 0.004 | 0.12 ± 0.01 | 2.59 ± 0.2 | 0.64 ± 0.006 | 0.12 ± 0.01 | 0.19 ± 0.02 |
| Phytohormones | ||||||
| Napthalene acetic acid | 0.09 ± 0.007 | 0.12 ± 0.003 | 1.46 ± 0.02 | 0.27 ± 0.03 | 0.11 ± 0.001 | 0.39 ± 0.034 |
| Indole acetic acid | 0.05 ± 0.005 | 0.09 ± 0.001 | 2.05 ± 0.21 | 0.33 ± 0.02 | 0.1 ± 0.002 | 0.31 ± 0.01 |
Effects of EDTA, Metal Salts, and Inhibitors on Activity
Little or no inhibition of Os9BGlu31 activity was seen with 1 mm EDTA, Ni2+, Zn2+, Mg2+, Mn2+, Co2+, and Ca2+, whereas 1 mm Fe3+ and Cu2+ decreased the activity by 25 and 34%, respectively (data not shown). Hg2+ had a greatest effect on Os9BGlu31, decreasing activity by approximately 90% at 1 mm. Surprisingly, Os9BGlu31 was not inhibited when it was preincubated with 1 mm of the covalent inhibitors conduritol B epoxide, cyclophellitol, and 2,4dNP2FG and the β-glucosidase transition state mimic inhibitors glucono δ-lactone at 5 mm, 1-deoxynojirimycin at 7 μm, isofagomine at 1 mm, and phenylethyl glucoimidazole at 30 μm, which completely inhibit most GH1 β-glucosidases (data not shown) (13, 22, 38). No 2,4-dinitrophenolate was detectable in 3-h reactions with 1 mm 2,4dNP2FG, verifying that 2,4dNP2FG was not rapidly turned over as a substrate.
Effects of Mutations of Glu169, His386, and Glu387 on Os9BGlu31 Activity
We analyzed the effects of single substitutions of three amino acid residues: the putative catalytic acid/base, E169Q and E169A; the putative catalytic nucleophile, E387A; and the residue preceding the putative catalytic nucleophile, H386T, on the activity of mutant enzymes. As shown in Table 4, the activities of the E169Q, E169A, H386T, and E387A mutants to transglycosylate ferulic acid using 4NPGlc as donor were decreased 7.4-, 78.5-, 2.2-, and 19,000-fold compared with wild type, respectively. The E169Q, E169A, and H386T mutants shifted the pH optimum for this transglycosylation from pH 4.5 in wild type to pH 6.0, 6.5, and 5.5, respectively (data not shown).
TABLE 4.
Relative activities of Os9BGlu31 wild type and the catalytic acid/base mutants E169Q and E169A, catalytic nucleophile mutant E387A, and H386T mutant
Os9BGlu31 wild type and mutants were purified with immobilized metal affinity chromatography and Q-Sepharose chromatography, respectively.
| Acceptor | Relative activity |
||||
|---|---|---|---|---|---|
| Wild type | E169Q | E169A | E387A | H386T | |
| nmol of 4NP min−1 mg−1 protein | |||||
| Buffer alone | 14.8 ± 0.6 | 3.4 ± 0.3 | 0.3 ± 0.02 | 0.008 ± 0.0001 | 2.7 ± 0.3 |
| Ferulic acid | 133.4 ± 8.3 | 18 ± 1.1 | 1.7 ± 0.05 | 0.007 ± 0.0003 | 61.3 ± 0.4 |
| Caffeic acid | 100.8 ± 0.1 | 13.6 ± 0.9 | 1.6 ± 0.07 | 0.007 ± 0.0004 | 49.4 ± 5.1 |
| Sinapic acid | 93.3 ± 3.2 | 23.6 ± 1.6 | 1.8 ± 0.10 | 0.012 ± 0.0003 | 38.8 ± 1.2 |
| 4-Hydroxybenzoic acid | 86.4 ± 6.1 | 11.6 ± 0.5 | 1.5 ± 0.01 | 0.008 ± 0.0004 | 30.0 ± 0.6 |
| Vanillic acid | 81.5 ± 5.7 | 10.2 ± 1.0 | 1.6 ± 0.06 | 0.011 ± 0.0004 | 19.9 ± 1.5 |
| trans-Cinnamic acid | 78.7 ± 7.6 | 11.1 ± 1.1 | 1.5 ± 0.02 | 0.008 ± 0.0001 | 19.5 ± 2.1 |
| Syringic acid | 83.1 ± 6.4 | 10.4 ± 0.5 | 1.3 ± 0.03 | 0.012 ± 0.001 | 19.0 ± 2.0 |
| Benzoic acid | 77.8 ± 7.8 | 11.0 ± 0.1 | 1.4 ± 0.02 | 0.01 ± 0.001 | 14.4 ± 0.2 |
| 4-Coumaric acid | 81.1 ± 7.3 | 14.1 ± 1.1 | 1.6 ± 0.03 | 0.01 ± 0.0003 | 28.3 ± 2.5 |
| Isovanillic acid | 68.3 ± 6.8 | 10.0 ± 0.7 | 1.1 ± 0.03 | NMa | 22.0 ± 1.0 |
| Catechin | 38.8 ± 3.9 | 4.8 ± 0.2 | 0.5 ± 0.01 | NM | NDb |
| Dihydroxybenzoic acid | 50.6 ± 5.1 | 2.7 ± 0.1 | 0.8 ± 0.01 | NM | 10.7 ± 1.1 |
| 1-Napthalene acetic acid | 53.9 ± 5.4 | 2.5 ± 0.1 | 1.1 ± 0.004 | NM | 6.8 ± 0.2 |
| Scopoletin | 16.8 ± 0.5 | 0.8 ± 0.02 | 0.7 ± 0.003 | NM | 5.0 ± 0.2 |
| Indole acetic acid | 43.9 ± 1.8 | 5.1 ± 0.5 | 0.9 ± 0.03 | NM | 4.9 ± 0.1 |
| Apigenin | 39.6 ± 2.0 | 11.2 ± 1.2 | 0.5 ± 0.03 | NM | 6.9 ± 0.4 |
| 2,4-Dichlorophenoxyacetic acid | 44.8 ± 3.0 | 9.7 ± 0.3 | 1.1 ± 0.01 | NM | 3.8 ± 0.1 |
a NM, not measured.
b ND, not detected.
The putative catalytic nucleophile mutant (E387A) was rescued by 0.5 m formate, which increased the activity of the mutant by 20-fold compared with buffer alone with a pH optimum at 4.5, but 0.5 m azide and acetate did not rescue the activity (data not shown). No difference in 4NP release from 4NPGlc by the E387A mutant was observed when 0.2 mm ferulic acid was added to the reaction with 0.5 mm formate. No transglycosylation product was observed with formate, suggesting either that it promoted hydrolysis by the E387A mutant or the that transglycosylation product rapidly degraded. Enhancement of 4NP release by formate was also observed for Os9BGlu31 wild type, E169Q, E169A, and H386T mutants in the absence of other nucleophiles (data not shown). Because glucose could be detected, but no transglycosylation products could be detected by TLC or HPLC in reactions with buffer alone, we expect that the release of 4NP from 4NPGlc in buffer alone indicates the hydrolysis activity of the enzyme. Os9BGlu31 wild type had GH activity (without acceptor) that was approximately 11% of its TG activity to ferulic acid (Table 4). However, when His386 was replaced with Thr, the residue most commonly seen in this position in GH1 β-glucosidases, no increase in hydrolysis activity was seen (hydrolysis was approximately 4.4% the rate of TG to ferulic acid). Replacement of Glu169 with Gln and Ala resulted in hydrolysis rates of approximately 19 and 18% relative to the rates of transfer to ferulate, but activity was decreased variably for all acceptors, including water. As seen in Table 4, the mutations of the putative catalytic residues and His386 also affected the relative rates of use of different acceptors.
Expression of Os9BGlu31 in Rice Tissues and in Seedlings in Response to Environmental Conditions
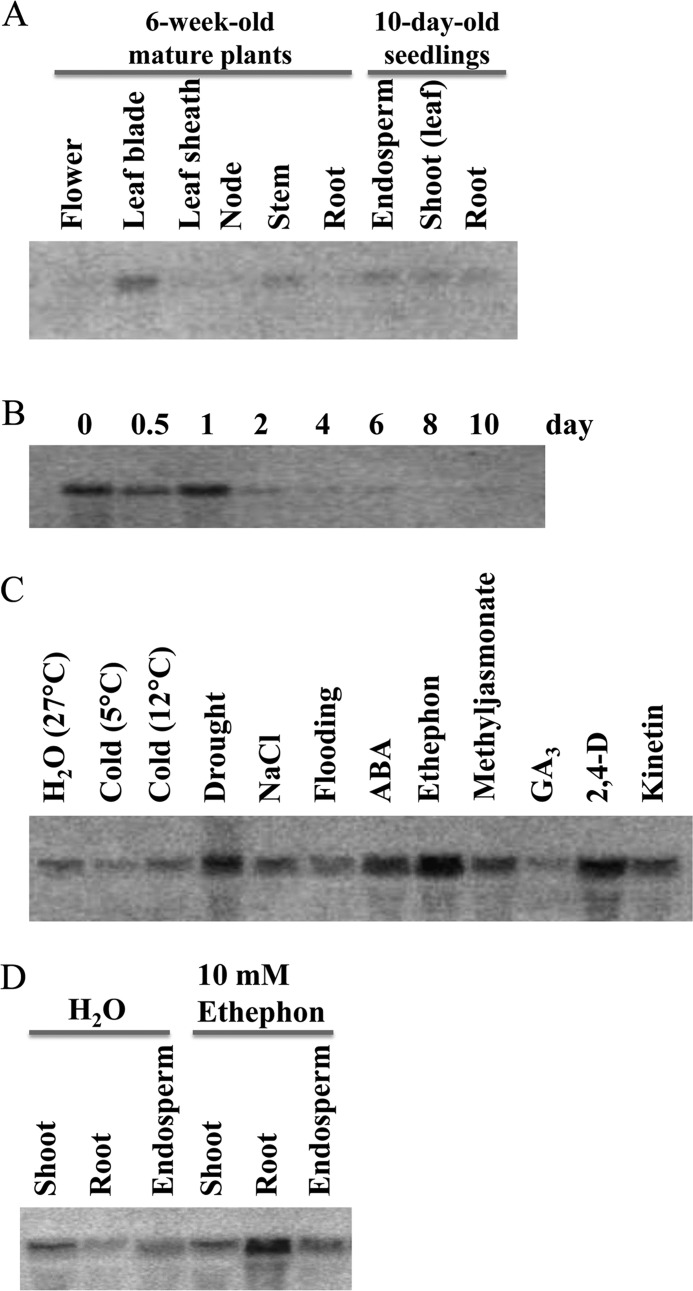
Initially, Os9BGlu31 expression was analyzed by Northern blotting. The Os9BGlu31 mRNA was detected at a relatively high level in the leaf blade and at a low level in the stem of mature plants (6 weeks old), whereas low levels were detected in endosperm, shoot, and root of 10-day-old seedlings (Fig. 2A). When rice seed was germinated, Os9BGlu31 expression was highest during the first day (0, 12, and 24 h) and then decreased at day 2 (Fig. 2B). When 10-day-old seedlings were subjected to 2 days of low temperature (5 or 12 °C), drought, salinity, or flooding, Os9BGlu31 transcript abundance increased in drought and increased slightly in NaCl (0.3 m) compared with the control condition (Fig. 2C). Additionally, treatments with the phytohormones ABA, ethephon (which releases ethylene), and 2,4-D (a synthetic auxin) significantly increased Os9BGlu31 mRNA in rice seedlings, whereas methyljasmonate and kinetin induced small increases. Fig. 2D shows that ethephon appears to induce the gene in roots of seedlings, but not in shoot and endosperm.
FIGURE 2.
Northern blot analysis of Os9BGlu31 gene expression in rice cv. Yukihikari plants and seedlings. A, blot of RNA extracted from various tissues of 6-week-old mature plants and 10-day-old rice seedlings. B, total RNA was extracted from whole seeds or seedlings germinated and grown at 27 °C for the indicated periods of time. C, effects of stresses and phytohormones on Os9BGlu31 gene expression in 10-day-old rice seedlings. RNA gel blots of 10-day-old rice seedlings treated for an additional 2 days with the indicated stresses or phytohormones: H2O (control), cold (5 °C and 12 °C), drought, 0.3 m NaCl, flooding, 0.1 mm ABA, 10 mm ethephon, 0.1 mm methyljasmonate, 0.1 mm gibberellin A3 (GA3), 10 mm 2,4-D, and 0.1 mm kinetin. D, effects of ethephon on different tissues of 10-day-old rice seedlings. As in C, 10-day-old rice seedlings were treated an additional 2 days with water (control) or 10 mm ethephon.
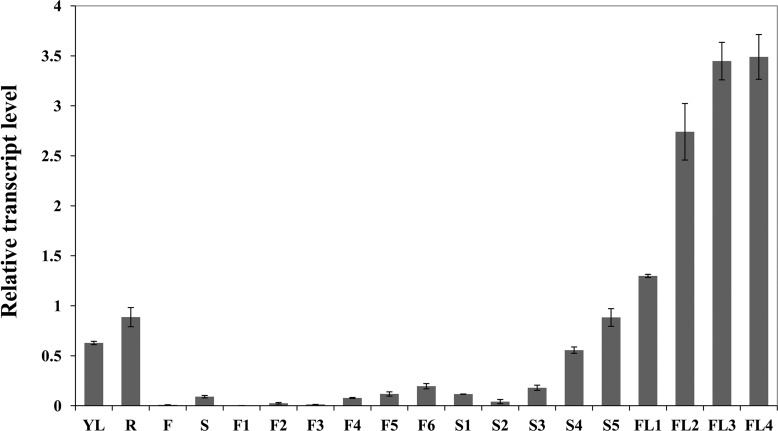
The spatiotemporal expression pattern of Os9BGlu31 was further examined by real time PCR throughout rice development (Fig. 3). The relative expression levels of Os9BGlu31 normalized to the OsUBQ5 gene were high in young leaf, root, developing seed at 7–8 DAF (stage S4) and 9–10 DAF (S5), and flag leaves (FL1 to FL4) and displayed the highest expression in samples harvested from senescing flag leaves, FL3 (30 DAF flag leaf) and FL4 (40 DAF flag leaf), suggesting that Os9BGlu31 may have a function in mature and senescent leaf tissues.
FIGURE 3.
Expression pattern of the Os9BGlu31 gene in rice cv. Dongjin, determined by real time PCR. The relative expression level of Os9BGlu31 was compared with that of OsUBQ5, and error bars indicate standard deviations from three separate experiments. YL, young leaf; FL, flag leaf; R, root; F, flower; S, seed; F1, 0–4 cm inflorescence; F2, 4–8 cm inflorescence; F3, 8–12 cm inflorescence; F4, 12–16 cm inflorescence; F5, 16–20 cm inflorescence; F6, >20 cm inflorescence; S1, 1–2 DAF seeds; S2, 3–4 DAF seeds; S3, 5–6 DAF seeds; S4, 7–8 DAF seeds; S5, 9–10 DAF seeds; FL1, 15 days before flowering flag leaf; FL2, 15 DAF flag leaf; FL3, 30 DAF flag leaf; FL4, 40 DAF flag leaf.
Subcellular Localization of Os9BGlu31
Subcellular localization is an important clue to elucidate function. Os9BGlu31 was previously predicted to be targeted by its N-terminal signal sequence to the secretory pathway (21). The Predotar program (34) confirmed that Os9BGlu31 has an N-terminal signal peptide of 22 residues: 1MTPARVVFICCVVLLAAAAAAA22, which should direct it to enter the endoplasmic reticulum for either retention in the endoplasmic reticulum or for export to the Golgi apparatus, vacuole or secretion. To determine the subcellular localization of Os9BGlu31, we generated the GFP fusion construct of Os9BGlu31-GFP under the control of CaMV35S promoter. The Os9BGlu31-GFP construct gave a poor signal in maize protoplasts, but the stable expression in transgenic rice calli gave clear vacuolar localization (Fig. 4). This suggests that the Os9BGlu31 protein is most likely to be localized to the vacuole in the plant, which is consistent with its low pH optimum of 4.5.
FIGURE 4.
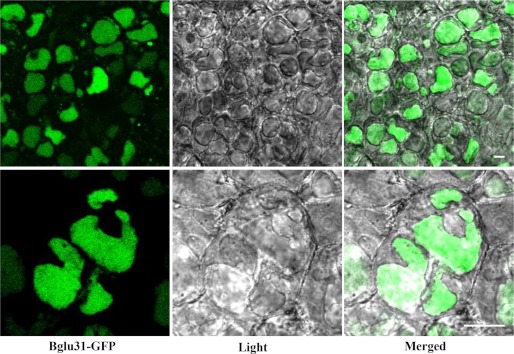
Subcellular localization of Os9BGlu31-GFP in rice calli. The rice calli were produced by Agrobacterium-mediated transformation with the Os9BGlu31-GFP construct and viewed with GFP fluorescence (left column) and light microscopy (center column), and the images are merged in the right column at low (top panel) and high magnification (bottom panel). The scale bars indicate 10 μm.
DISCUSSION
Biochemical Function of Os9BGlu31
Os9BGlu31 shows more efficient TG than GH activity. It transfers glucosyl groups from glucose ester and glucoside donors to nucleophilic acceptors. This activity is similar to GT activity, but it does not use nucleoside diphosphate sugar (uridine 5′-diphosphoglucose) as the glucose donor, which is required in the definition of GT (2, 39, 40). In contrast to previously characterized GH1 TG enzymes (15, 16), Os9BGlu31 displays a broad specificity and could glycosylate free phenolic acids, phytohormones, and flavonoids with phenolic 1-O-β-d-glucose esters acting as better glucose donors than glucosides, whereas the free phenolic acids of these esters are also excellent acceptor substrates. This suggests that Os9BGlu31 may help equilibrate the free phenolic acids and phenolic acid conjugate levels in plants.
The phenolic acids and their 1-O-acyl β-d-glucose esters that were the best substrates for Os9BGlu31 play important roles in plant metabolism. The 1-O-acyl β-d-glucose esters are transported into the vacuole, where they serve as transient intermediates in the formation of other metabolites (41). In the diploid oat (Avena strigosa) vacuolar serine carboxypeptidase-like acyltransferases use benzoyl and anthraniloyl glucoses as acyl donors in avenocide synthesis, and genes encoding similar serine carboxypeptidase-like acyltransferases have been identified in rice and other grasses (17). Because 1-O-hydroxycinnamoyl and 1-O-hydroxybenzoyl β-d-glucoses also appear to act as glucosyl donors for the glycosylation of anthocyanin glucosides in vacuoles in carnation and delphinium (15), as well as in intracellular ferulation of arabinoxylan in wheat cells (19), the ability of vacuolar TG like Os9BGlu31 to equilibrate these glucoconjugates may provide a means to replenish a glucosyl/acyl donor that is being rapidly depleted, provided there is a vacuolar supply of the aglycon and donor glucoconjugates. This activity may help explain the high diversity of glycoconjugates in plants, because expression of a single GT could result in production of 1-O-acyl β-d-glucose esters beyond its range of substrate specificity. In fact, GTs have also been shown to catalyze reversible reactions, so a vacuolar GT might play a similar role (42). Given this possibility, care must be taken in interpreting assays of GT and GH activities in plant extracts.
Recently, Xu et al. (43) demonstrated that a vacuolar β-glucosidase in A. thaliana (BG2) plays a role in osmotic stress response by hydrolyzing ABA-glucose ester to increase ABA. Whether Os9BGlu31 and similar vacuolar TG can act to modulate these responses by transfer of glucose from other 1-O-acyl β-d-glucose esters remains to be investigated, but its transfer of glucose to and from the carboxyl groups on the auxins indole acetic acid, naphthalene acetic acid, and 2,4-D, GA4, and ABA is intriguing. In fact, AtBGlu10, which falls in the same phylogenetic cluster at Os9BGlu31, has recently been suggested to act in 1-O-ABA glucosyl ester metabolism and as an 1-O-acyl glucose-dependent anthocyanin TG (47, 48). The induction of the Os9BGlu31 gene under the stress conditions of drought, ABA, ethephon, methyl jasmonate, 2,4-D, and kinetin is at least consistent with Os9BGlu31 involvement in adaptation to changes in the environment.
Sequence Comparison with Other GH1 Enzymes and Roles of Active Site Amino Acids
When Opassiri et al. (21) analyzed protein sequences of rice and other plant GH1 enzymes, the phylogenetic cluster At/Os6 contained rice Os1BGlu2, Os1BGlu3, Os1BGlu5, Os5BGlu19, Os5BGlu21, Os5BGlu22, Os5BGlu23, Os9BGlu31, Os9BGlu32, Os9BGlu33, and HIUH (approximately 44% sequence identity), as shown in supplemental Fig. S1. However, Os9BGlu31 has no HIUH activity.7 Carnation DcAA5GT and delphinium DgAA7GT also fall in the At/Os6 phylogenetic cluster (15) and share approximately 47% amino acid sequence identity with Os9BGlu31. Although Os9BGlu31, DcAA5GT, and DgAA7GT have the sequence IHENG around the catalytic nucleophile like HIUH, unlike HIUH, they also contain all the conserved glucose binding amino acid residues at the −1 subsite, except Tyr442 was found in place of the conserved Trp at Trp441 in rice Os3BGlu7 (supplemental Fig. S2). The obvious difference of the His in place of Thr or Ser in the position before the catalytic nucleophile (IHENG instead of ITENG) does not cause this functional distinction, because changing His386 to Thr in Os9BGlu31 caused 2–8-fold decreases in transglycosylation, depending on the donor, and a 5.5-fold decrease in hydrolysis.
In contrast to the glycon-binding site, amino acid residues at the aglycon-binding subsite are different in Os9BGlu31 compared with GH1 enzymes with known structures. For instance, the Trp residue corresponding to Trp358 in Os3BGlu7 is conserved in all GH1 enzymes with known structures, in which it serves as a platform for aglycon binding, but it is replaced with Phe359 in Os9BGlu31 and is not conserved in any of the GH1 cluster At/Os6 sequences.
Although the sequence alignment of Os9BGlu31 and the other GH1 TG DcAA5GT and DgAA7GT with GH1 β-glucosidases shows that they have conserved glutamates in the positions of the catalytic acid/base and nucleophile, the roles of these residues in TG activities was not previously shown. Because the best substrates for Os9BGlu31 are acids, the mild decrease in activity when the putative catalytic acid/base was mutated to Ala and Gln was not unusual, and the shift in the pH optimum in these mutants is consistent with the acid/base role (44, 45). This suggests that Os9BGlu33, which has 63% amino acid sequence identity with Os9BGlu31 but has Gln in this acid/base position, may in fact be an active TG.
The 19,000-fold decrease in activity when Glu387 was converted to Ala is consistent with its proposed role as a catalytic nucleophile, as is its rescue by formate. On the other hand, the lack of rescue by azide and the lack of inhibition of the wild type Os9BGlu31 by the putative oxocarbenium ion-like transition state analogs glucono δ-lactone, isofagomine, 1-deoxynojirimycin, and glucoimidazole, and mechanistic covalent inhibitors 2,4dNP2FG, conduritol B epoxide, and cyclophellitol that could inhibit plant GH1 β-glucosidases (13, 22, 38, 46), are unusual for enzymes following this mechanism.
Although the in vivo role of Os9BGlu31 requires further investigation, the unique broad activity points to a potential in vitro application. Broad specificity glycosyltransferases have been used in glycodiversification of drugs and other compounds (42). Although these GTs can use small glucosyl donors, they generally require a nucleotide sugar intermediate before transfer of the glycosyl moiety to another acceptor. The use of TGs like Os9BGlu31 eliminates this requirement, thereby providing a less complicated route to synthesis of glycoconjugates, such as the 1-O-acyl-glucosyl esters and glucosides produced in this work.
In conclusion, we have shown that the vacuolar Os9BGlu31 transglucosidase acts to transfer glucose between a broad range of phenolic acids and their 1-O-acyl β-d-glucose esters. As such, Os9BGlu31 and similar TGs provide a means to equilibrate these plant secondary metabolites. Os9BGlu31 and the two closely related isoenzymes, Os9BGlu32 and Os9BGlu33, may play similar roles in rice, whereas the activity of other GH1 cluster At/Os6 is yet to be determined. These enzymes may also be applied to in vitro glycodiversification without the need for a nucleotide sugar intermediate (42). Os9BGlu31 contains the conserved acid/base and nucleophile residues found in GH1 β-glucosidases, but the acid/base does not have a critical role because of the acidic nature of its substrates, and although Glu387 is essential to catalysis, its exact role remains to be shown.
Acknowledgments
We thank Peter A. Tipton and Annirudha Raychaudhuri (University of Missouri, Columbia) for testing for HIUH activity assay. Tassanee Onkoksoong is thanked for initial work on cloning and protein expression. Stephen G. Withers (University of British Columbia) and Maria Hrmova (University of Adelaide) are thanked for helpful suggestions and for providing cyclophellitol and phenylethyl glucoimidazole, respectively. We are grateful to Spencer Williams and Zalhe Hakki (University of Melbourne) for providing isofagamin and to Waturu Saburi and Hirokazu Matsui (Hokkaido University) for providing 4HB-β-d-glucoside standard.
This work was supported by Suranaree University of Technology and the National Research Council, along with the Higher Education Research Promotion and National Research University Project, Office of the Higher Education Commission of Thailand; by Next-Generation BioGreen 21 Program Grant PJ008114022011 from the Rural Development Administration; and by World Class University Program Grant R33-2008-000-10168-0.

This article contains supplemental Tables S1 and S2 and Figs. S1 and S2.
A. Raychaudhuri and P. A. Tipton, personal communication.
- GT
- glycosyltransferase
- 2,4-D
- 2,4-dichlorophenoxyacetic acid
- 2,4dNP2FG
- 2,4-dinitrophenyl-β-d-2-deoxy-2-fluoro-glucopyranoside
- 4HB
- 4-hydroxybenzoic acid
- 4HBG
- 1-O-(4-hydroxybenzoyl)-β-d-glucose
- 4NP
- 4-nitrophenol
- 4NPGlc
- 4-nitrophenyl-β-d-glucoside
- ABA
- abscisic acid
- CV
- column volume
- DAF
- days after flowering
- FG
- 1-O-feruloyl-β-d-glucose
- GA4-GE
- 1-O-gibberellin GA4-β-d-glucose ester
- GH
- glycoside hydrolase
- HIUH
- purine hydrolase hydroxyisourate hydrolase
- OsUBQ5
- rice ubiquitin 5
- TG
- transglycosidase
- GH1
- GH family 1.
REFERENCES
- 1. Bowles D., Lim E. K., Poppenberger B., Vaistij F. E. (2006) Glycosyltransferases of lipophilic small molecules. Annu. Rev. Plant Biol. 57, 567–597 [DOI] [PubMed] [Google Scholar]
- 2. Lairson L. L., Withers S. G. (2004) Mechanistic analogies amongst carbohydrate modifying enzymes. Chem. Commun. (Camb.) 20, 2243–2248 [DOI] [PubMed] [Google Scholar]
- 3. Henrissat B. (1991) A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280, 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Campbell J. A., Davies G. J., Bulone V., Henrissat B. (1997) A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem. J. 326, 929–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Henrissat B., Davies G. J. (2000) Glycoside hydrolases and glycosyltransferases. Families, modules, and implications for genomics. Plant Physiol. 124, 1515–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) The Carbohydrate-Active EnZymes database (CAZy). An expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lairson L. L., Henrissat B., Davies G. J., Withers S. G. (2008) Glycosyltransferases. Structures, functions, and mechanisms. Annu. Rev. Biochem. 77, 521–555 [DOI] [PubMed] [Google Scholar]
- 8. Eklöf J. M., Brumer H. (2010) The XTH gene family. An update on enzyme structure, function, and phylogeny in xyloglucan remodeling. Plant Physiol. 153, 456–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Koshland D. E. (1953) Stereochemistry and the mechanism of enzymatic reactions. Biol. Rev. Camb. Philos. Soc. 28, 415–436 [Google Scholar]
- 10. Withers S. G., Warren R. A., Street I. P., Rupitz K., Kempton J. B., Aebersold R. (1990) Unequivocal demonstration of the involvement of a glutamate residue as a nucleophile in the mechanism of a “retaining” glycosidase. J. Am. Chem. Soc. 112, 5887–5889 [Google Scholar]
- 11. Piens K., Fauré R., Sundqvist G., Baumann M. J., Saura-Valls M., Teeri T. T., Cottaz S., Planas A., Driguez H., Brumer H. (2008) Mechanism-based labeling defines the free energy change for formation of the covalent glycosyl-enzyme intermediate in a xyloglucan endo-transglycosylase. J. Biol. Chem. 283, 21864–21872 [DOI] [PubMed] [Google Scholar]
- 12. Lee S. S., Hong S. Y., Errey J. C., Izumi A., Davies G. J., Davis B. G. (2011) Mechanistic evidence for a front-side, SNi-type reaction in a retaining glycosyltransferase. Nat. Chem. Biol. 7, 631–638 [DOI] [PubMed] [Google Scholar]
- 13. Ketudat Cairns J. R., Esen A. (2010) β-Glucosidases. Cell Mol. Life Sci. 67, 3389–3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Raychaudhuri A., Tipton P. A. (2002) Cloning and expression of the gene for soybean hydroxyisourate hydrolase. Localization and implications for function and mechanism. Plant Physiol. 130, 2061–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Matsuba Y., Sasaki N., Tera M., Okamura M., Abe Y., Okamoto E., Nakamura H., Funabashi H., Takatsu M., Saito M., Matsuoka H., Nagasawa K., Ozeki Y. (2010) A novel glucosylation reaction on anthocyanins catalyzed by acyl-glucose-dependent glucosyltransferase in the petals of carnation and delphinium. Plant Cell 22, 3374–3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moellering E. R., Muthan B., Benning C. (2010) Freezing tolerance in plants requires lipid remodeling at the outer chloroplast membrane. Science 330, 226–228 [DOI] [PubMed] [Google Scholar]
- 17. Mugford S. T., Qi X., Bakht S., Hill L., Wegel E., Hughes R. K., Papadopoulou K., Melton R., Philo M., Sainsbury F., Lomonossoff G. P., Roy A. D., Goss R. J., Osbourn A. (2009) A serine carboxypeptidase-like acyltransferase is required for synthesis of antimicrobial compounds and disease resistance in oats. Plant Cell 21, 2473–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stehle F., Brandt W., Stubbs M. T., Milkowski C., Strack D. (2009) Sinapoyltransferase in the light of molecular evolution. Phytochemistry 70, 1652–1662 [DOI] [PubMed] [Google Scholar]
- 19. Obel N., Porchia A. C., Scheller H. V. (2003) Intracellular feruloylation of arabinoxylan in wheat evidence for feruloyl-glucose as precursor. Planta 216, 620–629 [DOI] [PubMed] [Google Scholar]
- 20. Xu Z., Escamilla-Treviño L., Zeng L., Lalgondar M., Bevan D., Winkel B., Mohamed A., Cheng C. L., Shih M. C., Poulton J., Esen A. (2004) Functional genomic analysis of Arabidopsis thaliana glycoside hydrolase family 1. Plant Mol. Biol. 55, 343–367 [DOI] [PubMed] [Google Scholar]
- 21. Opassiri R., Pomthong B., Onkoksoong T., Akiyama T., Esen A., Ketudat Cairns J. R. (2006) Analysis of rice glycosyl hydrolase family I and expression of Os4bglu12 β-glucosidase. BMC Plant Biol. 6, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Opassiri R., Ketudat Cairns J. R., Akiyama T., Wara-Aswapati O., Svasti J., Esen A. (2003) Characterization of a rice β-glucosidase highly expressed in flower and germinating shoot. Plant Sci. 165, 627–638 [Google Scholar]
- 23. Hiraga K., Kawabe S., Yokota T., Murofushi N., Takahashi N. (1974) Isolation and characterization of plant growth substances in immature seeds and etiolated seedlings of Phaseolus vulgaris. Agric. Biol. Chem. 38, 2521–2527 [Google Scholar]
- 24. Bachem C. W., van der Hoeven R. S., de Bruijn S. M., Vreugdenhil D., Zabeau M., Visser R. (1996) Visualisation of differential gene expression using a novel method of RNA fingerprinting based on AFLP. Analysis of gene expression during potato tuber development. Plant J. 9, 745–753 [DOI] [PubMed] [Google Scholar]
- 25. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harborm NY [Google Scholar]
- 26. Cho J. I., Lee S. K., Ko S., Kim H. K., Jun S. H., Lee Y. H., Bhoo S. H., Lee K. W., An G., Hahn T. R., Jeon J. S. (2005) Molecular cloning and expression analysis of the cell-wall invertase gene family in rice (Oryza sativa L.). Plant Cell Rep. 24, 225–236 [DOI] [PubMed] [Google Scholar]
- 27. Cho J. I., Ryoo N., Ko S., Lee S. K., Lee J., Jung K. H., Lee Y. H., Bhoo S. H., Winderickx J., An G., Hahn T. R., Jeon J. S. (2006) Structure, expression, and functional analysis of the hexokinase gene family in rice (Oryza sativa L.). Planta 224, 598–611 [DOI] [PubMed] [Google Scholar]
- 28. Kwon Y., Yu S.-I., Park J.-H., Li Y., Han J.-H., Alavilli H., Cho J.-I., Kim T.-H., Jeon J.-S., Lee B.-H. (2012) OsREL2, a Rice TOPLESS homolog functions in axillary meristem development in rice inflorescence. Plant Biotechnol. Rep. 6, 213–224 [Google Scholar]
- 29. Jain M., Nijhawan A., Tyagi A. K., Khurana J. P. (2006) Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 345, 646–651 [DOI] [PubMed] [Google Scholar]
- 30. Karimi M., Inzé D., Depicker A. (2002) Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195 [DOI] [PubMed] [Google Scholar]
- 31. Cho J. I., Ryoo N., Eom J. S., Lee D. W., Kim H. B., Jeong S. W., Lee Y. H., Kwon Y. K., Cho M. H., Bhoo S. H., Hahn T. R., Park Y. I., Hwang I., Sheen J., Jeon J. S. (2009) Role of the rice hexokinases OsHXK5 and OsHXK6 as glucose sensors. Plant Physiol. 149, 745–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jeon J.-S., Lee S., Jung K.-H., Jun S.-H., Jeong D.-H., Lee J.-W., Kim C., Jang S., Lee S.-Y., Yang K., Nam J., An K., Han M.-J., Sung R.-J., Choi H.-S., Yu J.-H., Choi J.-H., Cho S.-Y., Cha S.-S., Kim S.-I., An G. (2000) T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 22, 561–570 [DOI] [PubMed] [Google Scholar]
- 33. Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011) SignalP 4.0. Discriminating signal peptides from transmembrane regions. Nat. Methods. 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 34. Paetzel M., Karla A., Strynadka N. C., Dalbey R. E. (2002) Signal peptidases. Chem. Rev. 102, 4549–4580 [DOI] [PubMed] [Google Scholar]
- 35. Edgar R. C. (2004) MUSCLE. Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011) MEGA5. Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tanthanuch W., Chantarangsee M., Maneesan J., Ketudat-Cairns J. (2008) Genomic and expression analysis of glycosyl hydrolase family 35 genes from rice (Oryza sativa L.). BMC Plant Biol. 8, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hösel W., Tober I., Eklund S. H., Conn E. E. (1987) Characterization of β-glucosidase with high specificity for the cyanogenic glucoside dhurrin in Sorghum bicolor (L.) Moenchh seedlings. Arch. Biochem. Biophys. 252, 152–162 [DOI] [PubMed] [Google Scholar]
- 39. Lim E. K., Bowles D. J. (2004) A class of plant glycosyltransferases involved in cellular homeostasis. EMBO J. 23, 2915–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lim E. K. (2005) Plant glycosyltransferases. Their potential as novel biocatalysts. Chemistry 11, 5486–5494 [DOI] [PubMed] [Google Scholar]
- 41. Mock H.-P., Strack D. (1993) Energetics of the uridine 5′-diphosphoglucose. Hydroxycinnamic acid acyl-glucosyltransferase reaction. Phytochemistry 32, 575–579 [Google Scholar]
- 42. Gantt R. W, Peltier-Pain P., Cournoyer W. J., Thorson J. S. (2011) Using simple donors to drive the equilibria of glycosyltransferase-catalyzed reactions. Nat. Chem. Biol. 7, 685–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xu Z. Y., Lee K. H., Dong T., Jeong J. C., Jin J. B., Kanno Y., Kim D. H., Kim S. Y., Seo M., Bressan R. A., Yun D. J., Hwang I. (2012) A vacuolar β-glucosidase homolog that possesses glucose-conjugated abscisic acid hydrolyzing activity plays an important role in osmotic stress responses in Arabidopsis. Plant Cell 24, 2184–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang Q., Withers S. G. (1995) Substrate-assisted catalysis in glycosidases. J. Am. Chem. Soc. 117, 10137–10138 [Google Scholar]
- 45. Chuenchor W., Pengthaisong S., Robinson R. C., Yuvaniyama J., Svasti J., Ketudat Cairns J. R. (2011) The structural basis of oligosaccharide binding by rice BGlu1 β-glucosidase. J. Struct. Biol. 173, 169–179 [DOI] [PubMed] [Google Scholar]
- 46. Zechel D. L., Withers S. G. (2000) Glycosidase mechanisms. Anatomy of a finely tuned catalyst. Acc. Chem. Res. 33, 11–18 [DOI] [PubMed] [Google Scholar]
- 47. Wang P. T., Liu H., Hua H. J., Wang L., Song C. P. (2011) A vacuole localized β-glucosidase contributes to drought tolerance in Arabidopsis. Chinese Sci. Bull. 56, 3538–3546 [Google Scholar]
- 48. Miyahara T., Sakiyama R., Ozeki Y., Sasaki N. (2013) Acyl-glucose-dependent glucosyltransferase catalyzes the final step of anthocyanin formation in Arabidopsis. J. Plant Physiol. doi.org/ 10.1016/j.jplph.2012.12.001 [DOI] [PubMed] [Google Scholar]