Background: Little is known about the enzymes involved in O-glycosylation of arabinogalactan proteins (AGPs) in plants.
Results: Heterologously expressed AtGALT2 (At4g21060) catalyzed the addition of galactose to hydroxyproline in AGP peptide substrates.
Conclusion: AtGALT2 is a galactosyltransferase responsible for initial galactosylation of AGPs.
Significance: This work broadens our understanding of plant cell wall biosynthesis and provides an access point to identify other AGP glycosyltransferases.
Keywords: Arabidopsis, Galactose, Glycosylation, Hydroxyproline, Plant Cell Wall, AGP Biosynthesis, Arabinogalactan Proteins, Galactosyltransferase
Abstract
Although plants contain substantial amounts of arabinogalactan proteins (AGPs), the enzymes responsible for AGP glycosylation are largely unknown. Bioinformatics indicated that AGP galactosyltransferases (GALTs) are members of the carbohydrate-active enzyme glycosyltransferase (GT) 31 family (CAZy GT31) involved in N- and O-glycosylation. Six Arabidopsis GT31 members were expressed in Pichia pastoris and tested for enzyme activity. The At4g21060 gene (named AtGALT2) was found to encode activity for adding galactose (Gal) to hydroxyproline (Hyp) in AGP protein backbones. AtGALT2 specifically catalyzed incorporation of [14C]Gal from UDP-[14C]Gal to Hyp of model substrate acceptors having AGP peptide sequences, consisting of non-contiguous Hyp residues, such as (Ala-Hyp) repetitive units exemplified by chemically synthesized (AO)7 and anhydrous hydrogen fluoride-deglycosylated d(AO)51. Microsomal preparations from Pichia cells expressing AtGALT2 incorporated [14C]Gal to (AO)7, and the resulting product co-eluted with (AO)7 by reverse-phase HPLC. Acid hydrolysis of the [14C]Gal-(AO)7 product released 14C-radiolabel as Gal only. Base hydrolysis of the [14C]Gal-(AO)7 product released a 14C-radiolabeled fragment that co-eluted with a Hyp-Gal standard after high performance anion-exchange chromatography fractionation. AtGALT2 is specific for AGPs because substrates lacking AGP peptide sequences did not act as acceptors. Moreover, AtGALT2 uses only UDP-Gal as the substrate donor and requires Mg2+ or Mn2+ for high activity. Additional support that AtGALT2 encodes an AGP GALT was provided by two allelic AtGALT2 knock-out mutants, which demonstrated lower GALT activities and reductions in β-Yariv-precipitated AGPs compared with wild type plants. Confocal microscopic analysis of fluorescently tagged AtGALT2 in tobacco epidermal cells indicated that AtGALT2 is probably localized in the endomembrane system consistent with its function.
Introduction
Plant cell walls are complex, dynamic structures composed of polysaccharides and glycosylated proteins (1, 2). Proteins are important components in plant cell walls because of their contribution to cell wall architecture and function. Hydroxyproline-rich glycoproteins are one such structural cell wall protein. They are represented by a spectrum of molecules ranging from highly glycosylated arabinogalactan proteins (AGPs)2 to the moderately glycosylated extensins (EXTs) and finally to the lightly glycosylated proline-rich proteins (PRPs) (3). Bioinformatic analysis has revealed the presence of 166 hydroxyproline-rich proteins from Arabidopsis, including 85 AGPs, 59 EXTs, 18 PRPs, and 4 AGP/EXT hybrid proteins (3).
AGPs are the most structurally complex hydroxyproline-rich protein and are implicated to function in plant growth, development, signaling, and plant-pathogen interactions (4, 5). They are found in plasma membranes, cell walls, and plant exudates (6). AGPs are defined by three criteria: the presence of type II arabino-3,6-galactan chains, a hydroxyproline-rich protein backbone, and the ability of most AGPs to bind to a class of synthetic phenylazo dyes, the β-glycosyl Yariv reagents (7). AGP protein backbones are typically rich in hydroxyproline (Hyp) alternating with Ala, Thr, and Ser, whereas their carbohydrate moieties are mostly composed of galactose and arabinose with varying amounts of rhamnose, fucose, and glucuronic acid (8). The polysaccharide chains of AGPs are composed of β-(1,3)-galactan chains interrupted with β-(1,6)-galactopyranose (Gal) side chains and terminated mostly with arabinofuranose residues (9–11). Recently, Tryfona et al. (12) reported that Arabidopsis AGPs are similarly decorated with a linear β-(1,3)-galactan backbone with β-(1,6)-d-galactan side chains.
Although substantial progress has been made in elucidating glycosyltransferases (GTs) responsible for biosynthesis of many cell wall polysaccharides, little is known about the mechanisms and enzymes involved in the biosynthesis of AGPs. Unraveling the biosynthesis of these glycoproteins remains a daunting scientific challenge given that isolating and characterizing the enzymes involved in glycosylation is difficult. One critical aspect of this challenge is to isolate these enzymes, which are most likely integral membrane proteins, in their active form. In other cases, the lack of a robust and reproducible enzyme assay to validate their function presents another challenge. Liang et al. (13) proposed that as many as 15 different GTs may be involved in the biosynthesis of AGPs, of which only two GTs, specifically two fucosyltransferases, have been successfully characterized and shown to add terminal fucose (Fuc) residues on AGPs (14). Another candidate gene, encoding a putative transferase, has been recently demonstrated to transfer Gal to an O-methylated Gal-β-(1,3)-Gal disaccharide acceptor, an analog of the β-(1,3)-galactan chains found in AGPs (8).
Of all of the GTs involved in O-glycosylation of AGPs, the hydroxyproline-O-galactosyltransferase (Hyp-O-GALT) that adds the first Gal onto the protein backbone is crucial because it produces the acceptor for further glycosylation events. In mammals, GALTs are extensively studied, and their activity and biological functions are well characterized (15–17). AGPs are analogous to animal proteoglycans and mucins (18). Hassan et al. (19) suggested that lectin domain-containing GTs are a large family of N-acetyl galactosaminyltransferases (GalNAc-Ts) that add N-acetylgalactosamine (GalNAc) to mammalian mucins and other protein backbones initiating O-glycosylation on either the nascent polypeptide or on a glycopeptide acceptor. Thus, it is hypothesized that the GTs responsible for adding the first sugar to the protein core of mammalian proteoglycans should be similar to the GTs responsible for adding the first sugar to the AGP protein backbone. Recently, Liang et al. (13) and Oka et al. (20) reported on novel in vitro assays using synthetic AGP peptides for detecting Hyp-O-GALT activities in Arabidopsis microsomal membranes. Using the protocol published by Liang et al. (13), we report here on the identification and functional characterization of an Arabidopsis At4g21060 gene (named AtGALT2) that encodes a β-GALT involved in the biosynthesis of the glycan chain of AGPs.
EXPERIMENTAL PROCEDURES
Identification of Putative GALTs Involved in AGP Biosynthesis
A DELTA-BLAST search was performed using human β-(1,3)-GALT1 (Hs-B3GALT2; O43825), human β-(1,3)-N-acetylgalactosaminyltransferase (O75752), human UDP-Gal:glycoprotein-N-acetylgalactosamine β-(1,3)-GALT5 (Hs-B3GALT5; Q9Y2C3), mouse β-(1,3)-GALT1 (Mm-B3GALT1; O54904), and Caenorhabditis elegans GALT (Ce-T09F5.1; O62375) for identification of 20 Arabidopsis, 20 rice, one Medicago truncatula, one Sorghum bicolor, eight Vitis vinifera, seven Poplus, nine Brachypodium, and 11 Zea mays proteins (21). All of these proteins contain the structural motif pfam 01762, which represents the GALT domain, and all of these proteins except those from Poplus, Brachypodium, and Z. mays, which are yet to be included in the CAZy database, belong to the GT31 family as defined by Henrissat and Davies (22). The Poplus, Brachypodium, and Z. mays proteins were instead retrieved from ARAMEMNON (23). Phylogenetic analysis was performed with 68 sequences from the GT31 family using the online Web service Phylogeny.fr (24). Multiple sequence alignments were performed by MUSCLE and PhylML for tree building, whereas TreeDyn was used for tree rendering. Accession numbers presented in this study are available through the CAZy database GT31, the National Center for Biotechnology Information, or the ARAMEMNON Web site. For prediction of transmembrane domains, sequences were submitted to the TMHMM 2.0 server (25). GALECTIN and GALT domains were predicted from Pfam. In order to characterize the catalytic motif (DXD) of AtGALT2, hydrophobic cluster analysis was performed using the drawhca server. Homology modeling of AtGALT2 was done by the Protein Homology/Analogy Recognition Engine (PHYRE) version 2.0 (26) and also by the I-TASSER server (27). First, the full-length sequence of the AtGALT2 protein was analyzed, and then to test for a sugar nucleotide binding site, only the sequence corresponding to the GALT domain (amino acid residues 450–639) was analyzed. These protein modeling tools used the structure of the catalytic domain of mouse manic fringe in2 complexed with UDP and manganese (Protein Data Bank code c2j0bA) as the template.
Cloning and Expression of AtGALTs in Pichia pastoris
The cDNAs of the coding region of four candidate AtGALTs (AtGALT1, -3, -4, and -5) were obtained from the RIKEN Bioresource center. The cDNA for AtGALT6 was obtained from CNRGV, the French Plant Genomic Resource Center. A cDNA of the coding region for At4g21060 (AtGALT2) was graciously provided by Dr. Richard Strasser. The open reading frame of AtGALT2 was amplified with primers with a 5′ restriction site for SacII followed by a His6 tag and a 3′ restriction site for ApaI (forward, GCCGCGGATGCATCATCATCATCATCACATGAAAAGAGTAAAAAGCGAATCTTTTA; reverse, TCATCTGAAATTGCAACATTGTGGGGCCC. The boldface letters denote the restriction sites, the italic type denotes the His6 tag, and the underlined region denotes the translational start site. Amplified products were sequenced, cloned in the shuttle vector pPICZ A by a traditional “cut and paste” strategy, and transformed into E. coli (DH5α) for zeocin resistance. Transformed plasmids were electroporated into competent P. pastoris X-33 cells following manufacturer's instructions (Invitrogen). Twenty individual Pichia clones were selected, and the presence of the gene was confirmed by PCR using genomic DNA isolated from transformants and gene-specific primers. Genomic DNA was isolated from Pichia cells as described previously (28). A similar strategy was adopted for cloning and expressing other AtGALTs in Pichia. Primers for the respective AtGALTs are listed in supplemental Table S1. Ten of the 20 independent transformants were screened for expression of the recombinant AtGALT2 protein as follows. Twenty-five ml of buffered minimal glycerol medium supplemented with 100 mg/liter of zeocin in a 250-ml flask was inoculated with a single colony and grown at 28 °C in a shaking incubator at 260 rpm for ∼24 h to obtain an A600 reading of ∼2. Cells were harvested by centrifugation at 2,500 × g for 5 min and resuspended in ∼75 ml of buffered minimal methanol medium to obtain an A600 of ∼1. Protein expression was induced by adding 0.5% (v/v) methanol (final concentration) every 24 h, and 2 ml of cell cultures were harvested every 24 h for 5 days. Cells were pelleted by centrifugation at 2,500 × g for 5 min at 4 °C and stored at −80 °C until analysis.
Preparation of Pichia Microsomes and Immunoblot Analysis
Transformed Pichia cells from a 75-ml culture grown in an Erlenmeyer flask in the presence of methanol for 5 days were centrifuged at 2,500 × g for 5 min at 4 °C and resuspended in 10 ml of homogenization buffer (0.1 m HEPES-KOH, pH 7, 0.4 m sucrose, 1 mm dithiothreitol, 5 mm MgCl2, 5 mm MnCl2, 1 mm phenylmethylsulfonyl fluoride, and one tablet of Roche Applied Science EDTA-free complete protease inhibitor mixture and 100 μl of RPI protease inhibitor IV). Cells were disrupted by vortexing eight times for 1 min each, with 2 min on ice between each vortexing, in the presence of acid-washed 425–600-μm glass beads (Sigma-Aldrich). The supernatant was centrifuged at 2,500 × g for 5 min at 4 °C to remove the beads and then at 150,000 × g for 60 min at 4 °C to obtain the membrane fraction (29). This microsomal pellet was resuspended in 50 μl of homogenization buffer. For immunoblot analysis, 5 μg of microsomal protein from Pichia transformants was denatured, subjected to 10% SDS-PAGE, and electroblotted onto PVDF Immobilon membranes (Millipore) using the Mini Protean3 system according to manufacturer's recommendations. Blots were probed with an anti-His primary antibody (Clontech) at a 1:10,000 dilution and a secondary goat anti-mouse IgG antibody conjugated to horseradish peroxidase (HRP) (Clontech) at a 1:20,000 dilution. West Femto Maximum Sensitivity Substrate (Thermo Scientific) was used for HRP detection. Pichia cell lines transformed with the empty expression vector were used as the negative control (NC). Protein quantification was done using the Bradford reagent (Sigma). Blots were stained with Coomassie Brilliant Blue R-250 following HRP detection to ensure equal loading.
Galactosyltranferase Assay with Microsomal Preparations from Pichia Expressing AtGALT2
The standard GALT reaction (100 μl) consisted of detergent-permeabilized microsomal membranes (250 μg of total protein), acceptor substrate peptide (20 μg), and ∼3 μm UDP-[14C]Gal (90,000 cpm, 465 cpm/pmol; MP Biomedical Sciences). Permeabilization was achieved in two steps. Fifty μl of microsomal protein was first treated with 0.3% Triton X-100 (15 min, 4 °C), followed by ultracentrifugation at 100,000 × g for 45 min. The pellet obtained was resuspended in 50 μl of extraction buffer and subjected to a second permeabilization step with 1% Triton X-100 for 15 min at 4 °C, followed by ultracentrifugation at 100,000 × g for 45 min. (AO)7 and d(AO)51 were the two substrate acceptors used in the standard GALT assay. The reaction mixture was incubated for 2 h at room temperature and was terminated by mixing with 400 μl of anion-exchange resin (DOWEX 1X8-100 resin; Sigma-Aldrich; 1:1 (v/v) in double-distilled water). The resin mixture was loaded on a Zeba spin column (Pierce) and centrifuged at 15,000 × g for 1 min to remove unreacted UDP-[14C]Gal retained by the ion-exchange resin. The flow-through contained the incorporated 14C-radiolabeled product and was analyzed with an LS6500 multipurpose scintillation counter (Beckman). Two reactions were included as controls, one with no substrate acceptor and one with permeabilized microsomal membranes from the Pichia line (X33) transformed with the empty expression vector (pPICZ A) to serve as NC.
Purification of Hyp-GALT2 Reaction Products by Reverse-phase HPLC
The GALT reaction product was purified by RP-HPLC as described by Liang et al. (13).
Analysis of the Hyp-[14C]galactoside Profile by Gel Permeation Chromatography and High Performance Anion-exchange Chromatography (HPAEC)
25 standard GALT reactions were fractionated by RP-HPLC and combined to generate enough 14C-radiolabeled product for base hydrolysis and separation on a Biogel P2 column (13). The radioactive peak eluting at degree of polymerization 4 (DP4) on a Biogel P2 column was analyzed along with a chemically synthesized Hyp-Gal standard by HPAEC on a CarboPac PA-20 column using 20 mm NaOH as the elution buffer to provide additional confirmation of this DP4 peak as Hyp-Gal. trans-4-(β-d-Galactopyranosyloxy)-l-proline (i.e. the Hyp-Gal standard) was chemically synthesized from commercially available galactopyranosyl bromide and hydroxyproline methyl ester as described with minor modifications (30).
Monosaccharide Composition Analysis of GALT Reaction Products by High Performance Anion-exchange Chromatography
Fifteen standard GALT assays were pooled to generate sufficient 14C-products for acid hydrolysis and monosaccharide composition analysis as described by Liang et al. (13).
Determination of Substrate Specificity of the AtGALT2 Enzyme Activity
A standard GALT assay was performed using 20 μg of various peptide substrate acceptors, (AO)7, (AO)14, and d(AO)51 (containing 7, 14, and 51 AO repeating dipeptide units, respectively), an extensin peptide (ExtP) containing repetitive SO4 units, and a (AP)7 peptide containing seven AP units as described by Liang et al. (13). Rhamnogalactan I from potato and rhamnogalactan from soybean (100 μg each) were used as potential pectin substrates. Permeabilized microsomal membranes (250 μg) from the NC Pichia line and the C2 Pichia line expressing His6-AtGALT2 served as the enzyme source in the GALT reactions. For all of the peptide substrate acceptors, the standard GALT assay was performed, and the reaction products were fractionated by RP-HPLC before monitoring incorporation of radiolabeled 14C in a liquid scintillation counter (Beckman Coulter LS 6500). For the pectin substrate acceptors, reactions were incubated at room temperature for 2 h, terminated by adding 1 ml of cold 70% ethanol, and precipitated overnight at −20°C. Reaction products were collected by centrifugation at 10,000 × g for 10 min, and pellets were washed five times with 1 ml of cold 70% ethanol to remove excess UDP-[14C]Gal. The 14C-radiolabel incorporation was estimated by resuspending the pellets in 300 μl of water before counting in a liquid scintillation counter.
Biochemical Characterization of AtGALT2 Enzyme Activity
The standard GALT assay was modified for AtGALT2 characterization using (AO)7 peptide as the acceptor substrate. Assay products from each reaction were fractionated by RP-HPLC to measure incorporated 14C-radiolabel into acceptor substrates.
The optimum pH for AtGALT2 activity was determined using permeabilized microsomal membranes (250 μg) from the C2 Pichia line expressing His6-GALT2 dissolved in test buffers at a final concentration of 100 mm. Test buffers included MES-KOH buffer at pH 4, 5, 6, and 7; HEPES-KOH buffer at pH 6, 6.5, 7, 7.5, and 8; Tris-HCl buffer at pH 8, 9, and 10; and CAPS-KOH buffer at pH 10.
To examine the effect of divalent cations on AtGALT2 activity, microsomal membranes were extracted with homogenizing buffer lacking divalent ions. MnCl2, MgCl2, CaCl2, CuCl2, NiCl2, or ZnSO4 was added to the GALT assay (at a final concentration of 5 mm) when tested. Two controls were added, one with no ions in the buffer used for resuspending the detergent permeabilized membrane fraction and the other with EDTA (5 mm) to chelate any residual divalent cations trapped in the membranes. An equal volume of deionized distilled water was added instead of divalent ions in the control reaction.
To analyze the enzyme specificity for nucleotide sugar donors, the standard activity assay was performed with (AO)7 as the acceptor substrate and various 14C-radiolabeled nucleotide sugar donors (90,000 cpm). The nucleotide sugars tested included UDP-[14C]Glc (MP Biomedicals), UDP-[14C]Xyl (PerkinElmer Life Sciences), and GDP-[14C]Fuc (PerkinElmer Life Sciences). Four separate GALT reactions with no substrate acceptors were performed as controls.
AtGALT2 Mutant Analysis
Two T-DNA insertional lines for At4g21060-AtGALT2 (galt2-1 (SALK_117233) and galt2-2 (SALK_141126)) were selected using the SIGnaL database and were obtained from the Arabidopsis Biological Research Centre. The wild type plants were Columbia (Col-0), and galt2 mutants were in the Columbia (Col-0) genetic background. Homozygous mutants were identified by PCR analysis using primer sequences obtained with the T-DNA Primer Design Tool provided by the Salk Institute Genomics Analysis Laboratory (supplemental Table S2). To confirm homozygous plants at the transcript level, RNA was extracted and analyzed by RT-PCR. RNA was isolated using a Qiagen RNeasy plant minikit followed by DNase I digestion using Qiagen RNase-free DNase to remove traces of DNA. The Qiagen One-Step RT-PCR kit was used for first-strand synthesis and subsequent PCR steps (primers are listed in supplemental Table S2).
Plants were germinated after 4 days of stratification in darkness at 4 °C and grown on soil at 22 °C and 60% relative humidity. Plants were grown under long-day conditions (16-h photoperiod and 8-h dark, 120 μmol m−2 s−1 fluorescent light).
Plant microsomal membranes were prepared and assayed according to Liang et al. (13). Specifically, 8 g of leaf tissue from 14-day-old wild type and galt2 mutant plants were used to perform GALT reactions with (AO)7 as the peptide substrate acceptor and UDP-[14C]Gal as the sugar donor.
AGPs were extracted from WT, galt2-1, and galt2-2 plants as described by Schultz et al. (33). Specifically, 5 g of aerial tissue from 14-day-old plants were used for each line to obtain β-Yariv-precipitable AGPs, which were quantified spectrophotometrically as described by Gao et al. (34).
Transient Expression and Subcellular Localization of AtGALT2 in Nicotiana tabacum Leaves
The AtGALT2 coding region was subcloned into the pVKH18En6-vYFP plasmid to generate the AtGALT2:vYFP construct by a traditional cut and paste strategy using XbaI and SalI (forward, CAGGACTCTAGAATGAAAAGAGTAAAAAGCGAATCTTTTAGAGGAG and reverse, CATGACGTCGACTCTGAAATTGCAACATTGTGATCGACCTTTC), respectively. The italic type denotes restriction sites, and the underlined region denotes the translational start site. Agrobacterium-mediated transient expression was performed in the leaves of 3–4-week-old tobacco plants (N. tabacum cv. Petit Havana) grown at 22–24 °C using a bacterial optical density (A600) of 0.05 for single infiltrations and 0.025 each for co-infiltrations (31). The AtGALT2-vYFP construct was co-expressed with either the ER marker mGFP5-HDEL (31) or the Golgi marker sialic acid transferase (ST)-mGFP5 (32) to ascribe subcellular localization. Transformed plants were incubated under normal growth conditions and sampled daily for 2–7 days postinfiltration.
Leaf epidermal sections were imaged using an upright Zeiss LSM 510 META laser-scanning microscope (Jena, Germany), using a ×40 oil immersion lens and an argon laser. For imaging the expression of vYFP constructs, the excitation line was 514 nm, and emission data were collected at 535–590 nm, whereas for mGFP5 constructs, the excitation line was 458 nm, and the emission data were collected at 505–530 nm. Singly infiltrated controls were analyzed to optimize gain and pinhole settings for each channel and to exclude any bleed-through fluorescence between channels. Postacquisition image processing was done using the LSM Image Browser 4 (Zeiss).
RESULTS
Identification of Putative AGP GALTs in Arabidopsis thaliana by in Silico Analysis
A bioinformatics approach was adopted for identifying putative Hyp-GALT genes (Hyp GALTs) involved in adding the first Gal to Hyp residues in AGPs. First, a phylogenetic tree was generated by submitting homologous animal and plant sequences encoding a GALT catalytic domain (i.e. pfam 06712) (supplemental Fig. S1). Only 20 of the 33 members of Arabidopsis GALTs in the GT31 family were used in this phylogenetic analysis because the remaining 13 accessions do not contain a GALT domain but instead have a domain of unknown function (DUF604). Two of these 20 family members have been characterized; At1g26810 (GALT1) was identified as a β-(1,3)-GALT involved in biosynthesis of a Lewis a epitope on N-linked glycans (35), and At1g77810 was reported to be a β-(1,3)-GALT that catalyzes transfer of Gal to an O-methylated Gal-β-(1,3)-Gal disaccharide, which mimics a partial structure of AGP side chains (8). Interestingly, only six Arabidopsis proteins (At1g26810-GALT1, At4g21060-GALT2, At3g06440-GALT3, At1g27120-GALT4, At1g74800-GALT5, and At5g62620-GALT6) contain a GAL-LECTIN (GALECTIN) binding domain (pfam 00337) in addition to the GALT domain (pfam 06712). This finding was consistent with that reported by Qu et al. (8). Moreover, this GALECTIN domain is absent in all mammalian β-1,3-GALTs in the GT31 family and in all other plant glycosyltransferases in the CAZy database. Interestingly, previous studies found that a lectin domain is present in polypeptide GalNAc-Ts. These enzymes belong to GT27 and are involved in catalyzing the first step of O-glycosylation of mucins (15–17). Consequently, it was hypothesized that plant GALTs contain analogous lectin domains and function in initiating O-glycosylation of AGPs (8, 36, 37). Thus, bioinformatics analysis indicated that AtGALT1 to -6 represent six promising candidates for having Hyp-enzymatic activity.
Heterologous Expression of Putative Hyp-GALT Genes in Pichia Cells
Six recombinant proteins (AtGALT1, AtGALT2, AtGALT3, AtGALT4, AtGALT5, and AtGALT6) fused with a His6 tag were expressed in Pichia. Microsomal proteins from these recombinant lines were examined by immunoblotting with antibodies against the His6 tag and demonstrated the presence of recombinant fusion proteins of the predicted sizes. For example, AtGALT2 recombinant lines had the expected 78 kDa protein band reacting with the His6 antibody (data not shown). For the AtGALT2 transformants, as well as the other recombinant lines, an additional smaller protein band (∼50 kDa) was often detected that may be attributed to protein degradation by endogenous proteases in Pichia. Pichia cells transformed with the empty expression vector served as NC and lacked the recombinant protein band.
Heterologously Expressed AtGALT2 Demonstrates Hyp-GALT Activity
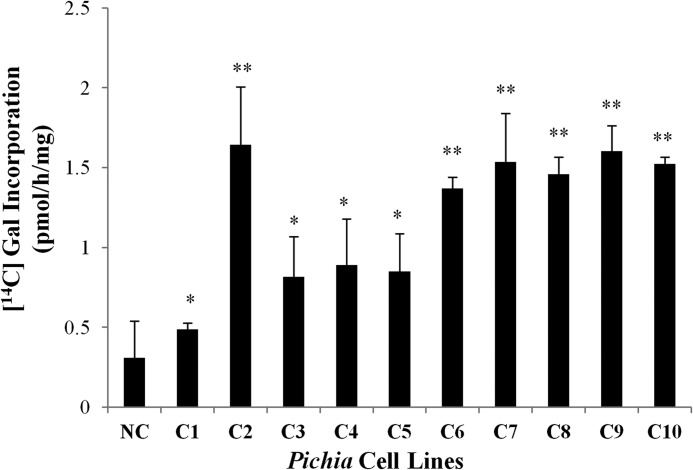
An in vitro GALT assay developed by Liang et al. (13) was used to test for activity of the recombinant AtGALTs expressed in Pichia cells. The components of the GALT assay were detergent-permeabilized microsomal membranes from the transformed Pichia cell lines expressing one of the six AtGALT proteins as the enzyme source, UDP-[14C]Gal as the sugar donor, and two AGP peptide analogs (d(AO)51 and (AO)7) as the substrate acceptors. Only AtGALT2 showed activity to date; consequently, further product characterization and biochemical analysis has focused on AtGALT2. The amount of GALT activity varied in the 10 independent cell lines (C1–C10) of Pichia cells expressing AtGALT2 based on the rate of [14C]Gal incorporation using the (AO)7 substrate acceptor (Fig. 1). The C2 clone demonstrated the highest enzyme activity (Fig. 1).
FIGURE 1.
Screening for AtGALT2 expressed in Pichia cell lines. (AO)7-dependent GALT activity tests of the 10 transgenic Pichia cell lines using Triton X-100-permeabilized microsomal membranes. For each line, 250 μg of total microsomal membrane protein was used for the assay. [14C]Gal radiolabel incorporation is expressed as pmol/h/mg protein and reflects the difference between total incorporation obtained in reaction products in the presence versus the absence of (AO)7 acceptor substrate. Reactions were done in triplicate, and mean values are presented. All cell lines tested had AtGALT2 activity but varied in the rate of incorporation. Student's t test was performed using GraphPad Quickcalcs, and significant differences in GALT activity were detected with respect to NC. * and **, p < 0.05 and p < 0.01, respectively.
(AO)7 and d(AO)51 Are Substrate Acceptors for AtGALT2
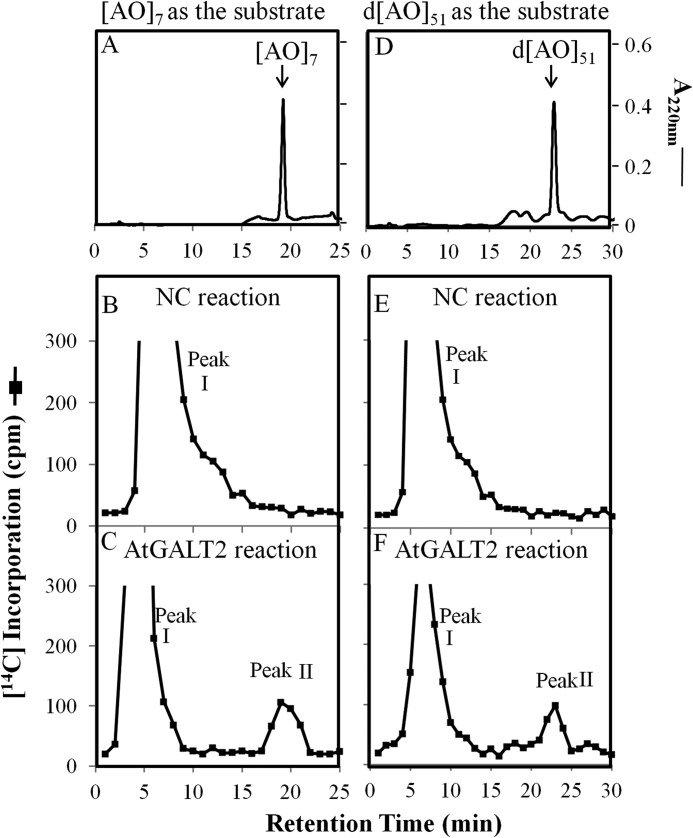
Total microsomal membranes from Pichia transformants expressing AtGALT2 were analyzed for Hyp-GALT activity using two substrate acceptors: (AO)7 (a synthetic peptide) and d(AO)51 (a transgenically expressed and chemically deglycosylated protein). Incorporation of [14C]Gal from UDP-[14C]Gal onto the two substrate acceptors was observed by HPLC fractionation (Fig. 2, C and F) and by comparison with the non-radioactive (AO)7 and d(AO)51 substrate acceptor peaks (Fig. 2, A and D). Two 14C-radioactive peaks were detected, of which peak II has the same retention times as their respective substrate acceptors ((AO)7 and d(AO)51). The identity of peak I is not known; it may represent free [14C]Gal released by an endogenous galactosidase (38) or be composed of oligosaccharides with [14C]Gal incorporated into endogenous sugar acceptors, as suggested previously (13). Microsomal preparations from a Pichia cell line transformed with the empty expression vector were used as NCs (Fig. 2, B and E). Thus, HPLC fractionation provided evidence for incorporation of the 14C-radiolabel from UDP-[14C]Gal onto the (AO)7 and d(AO)51 acceptors with relatively higher AtGALT2 enzyme activity demonstrated with the (AO)7 substrate acceptor compared with d(AO)51. Consequently, the (AO)7:AtGALT2 reaction product was subjected to further characterization.
FIGURE 2.
RP-HPLC fractionation of the (AO)7:AtGALT2 and d(AO)51:AtGALT2 reaction products on a PRP-1 reverse-phase column. Acceptor substrate alone (A and D), GALT reaction with microsomal membranes from the NC Pichia line transformed with the empty expression vector (B and E), and the GALT reaction with microsomal membranes from the transgenic Pichia C2 line (C and F) were fractionated by RP-HPLC using identical elution conditions. Radioactive peak II coeluted with the (AO)7 and d(AO)51 acceptor substrates in the GALT2 reaction and was used for subsequent product analysis.
Product Characterization by Acid and Base Hydrolysis Shows That AtGALT2 Transfers Gal to Hyp Residues
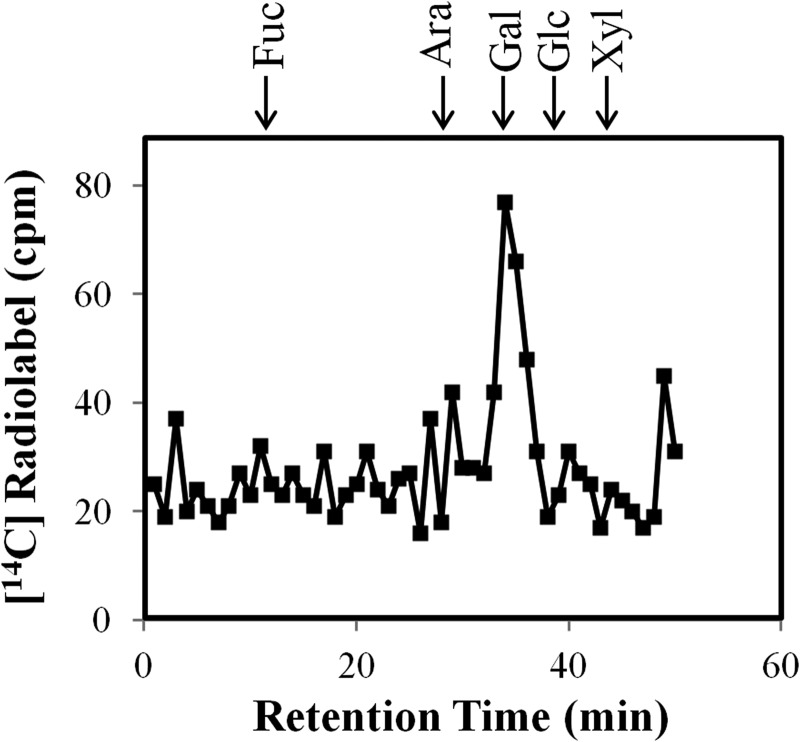
To confirm that the 14C-radiolabel remained associated with Gal, RP-HPLC fractions containing the 14C-radiolabeled (AO)7:AtGALT2 reaction products were pooled and subjected to total acid hydrolysis. The resulting acid-hydrolyzed 14C-radiolabeled monosaccharide was fractionated by HPAEC and showed that 14C-label co-eluted with Gal, thereby confirming incorporation of [14C]Gal onto the (AO)7 peptide (Fig. 3).
FIGURE 3.
Monosaccharide analysis of the RP-HPLC purified (AO)7:GALT reaction product following acid hydrolysis. Permeabilized microsomal membranes from the transgenic Pichia C2 line expressing His6-AtGALT2 served as the enzyme source in the (AO)7:GALT reaction. The 14C-labeled monosaccharides were analyzed by HPAEC on a CarboPac PA-10 column. Elution times of monosaccharide standards are as indicated with arrows at the top.
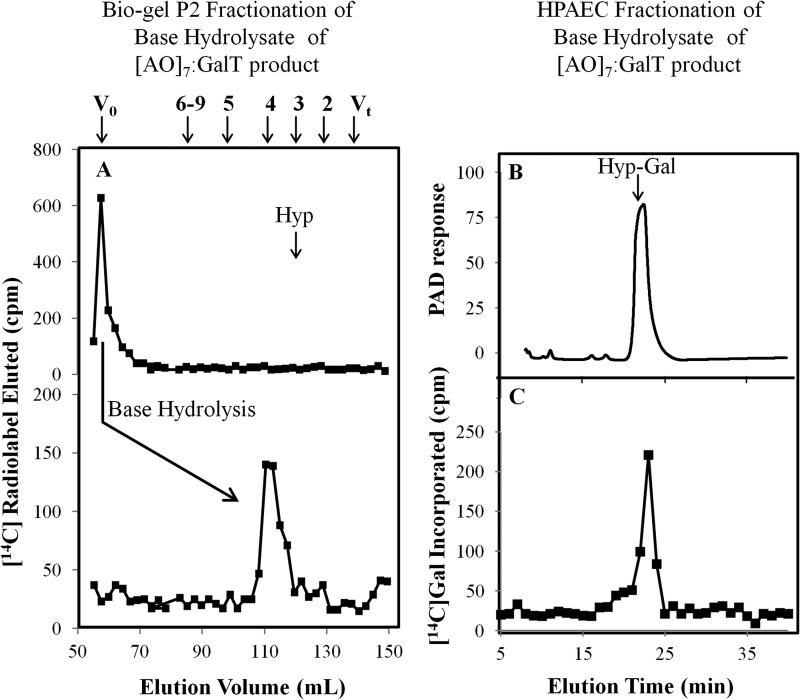
In another set of experiments, base hydrolysis was used to confirm that the [14C]Gal residues are added to Hyp residues and to examine the extent of galactosylation of the (AO)7 peptide acceptor. Base hydrolysis degrades peptide bonds but keeps Hyp-glycosidic bonds intact (39). The intact 14C-radiolabeled (AO)7 peptide product eluted in the void volume (V0) on the P2 column, whereas the base hydrolysate of this product eluted at DP4 (Fig. 4A). Given that Hyp residues alone elute as a DP3 sugar on a P2 column, it was concluded that AtGALT2 catalyzes the addition of one Gal onto the (AO)7 peptide, consistent with our previous work (13). Further confirmation of this conclusion was provided by fractionation of the base hydrolysate on a CarboPac PA-20 column (Dionex), demonstrating that 14C-radiolabel co-eluted with a Hyp-Gal standard (Fig. 4, B and C).
FIGURE 4.
Bio-gel P2 fractionation of the RP-HPLC-purified (AO)7:AtGALT2 reaction product and HPAEC of the resulting base hydrolysis product. A, Bio-gel P2 fractionation of the RP-HPLC-purified (AO)7:AtGALT2 reaction product before and after base hydrolysis. Permeabilized microsomal membranes from the Pichia C2 line expressing His6-GALT2 served as the enzyme source in the (AO)7:AtGALT2 reaction. Elution profiles of the reaction product before and after base hydrolysis are shown. The column was calibrated with high molecular mass dextran (V0), galactose (Vt), xylo-oligosaccharides with DP2 to -5, and xyloglucan-oligosaccharides (DP6 to -9); their elution positions are indicated with arrows at the top. The elution position of free Hyp amino acid (corresponding to DP3) is shown with an arrow in the panel. Base hydrolysis produces a radioactive peak eluting at DP4, which corresponds to Hyp-Gal. B, HPAEC profile of a chemically synthesized Hyp-Gal standard detected as a PAD response. C, the radioactive peak eluting at DP4 coelutes with the chemically synthesized Hyp-Gal standard following HPAEC. Both the Hyp-Gal standard and the radioactive peak eluting at DP4 were fractionated in 20 mm NaOH elution buffer on a CarboPac PA-20 column.
AtGALT2 Is Specific for AGPs
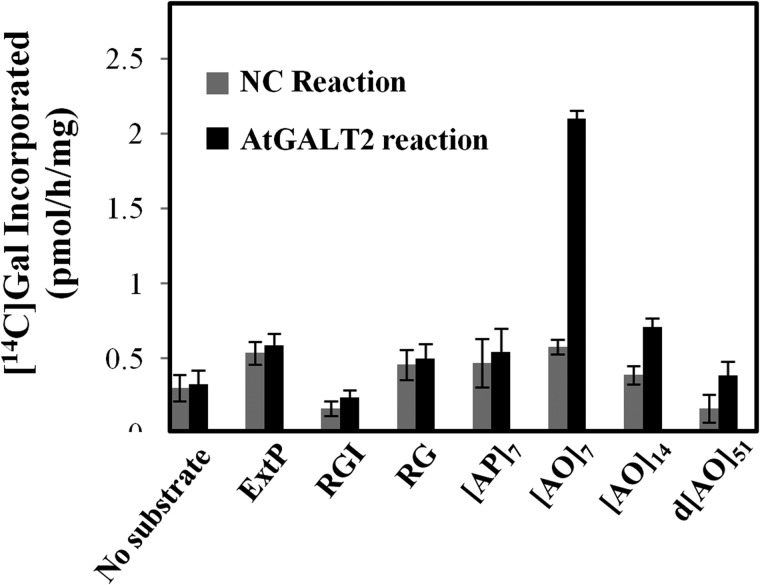
Various substrates that might act as potential substrate acceptors for a β-(1,3)-GALTs were tested to investigate AtGALT2 enzyme specificity. Namely, (AO)7, (AO)14, and d(AO)51, consisting of non-contiguous peptidyl Hyp residues, were used to examine AGP peptide sequences of various lengths. (AP)7, consisting of alternating Ala and Pro residues, was used to test the requirement of peptidyl Hyp for galactosylation. ExtP, a chemically synthesized extensin peptide consisting of contiguous peptidyl Hyp residues, was used to test whether contiguous peptidyl Hyp residues act as potential acceptors. Two pectic polysaccharides, rhamnogalactan I from potato and rhamnogalactan from soybean, were also used as potential substrates acceptors. All of the non-AGP substrate acceptors, including (AP)7, failed to incorporate [14C]Gal, indicating the AtGALT2 activity was specific for AGP sequences containing non-contiguous peptidyl Hyp. It was also observed that the incorporation of 14C-radiolabel decreased with increasing lengths of these AO acceptor substrates (Fig. 5).
FIGURE 5.
Effect of various peptide and polysaccharide acceptor substrates on incorporation of 14C-radiolabeled galactose. A, permeabilized microsomal membranes from the NC Pichia line transformed with the empty expression vector and the C2 Pichia line expressing His6-AtGALT2 served as the enzyme source in the GALT reactions. (AO)7, (AO)14, and d(AO)51 contain 7, 14, and 51 AO units, respectively. A chemically synthesized extensin peptide (ExtP) contains repetitive SO4 units. (AP)7 contains seven AP units. Rhamnogalactan I (RGI) from potato and rhamnogalactan (RG) from soybean represent pectin polymer substrates. Enzyme reactions were done in triplicate, and mean values are presented. Error bars, S.E.
Biochemical Characteristics of the AtGALT2 Enzyme
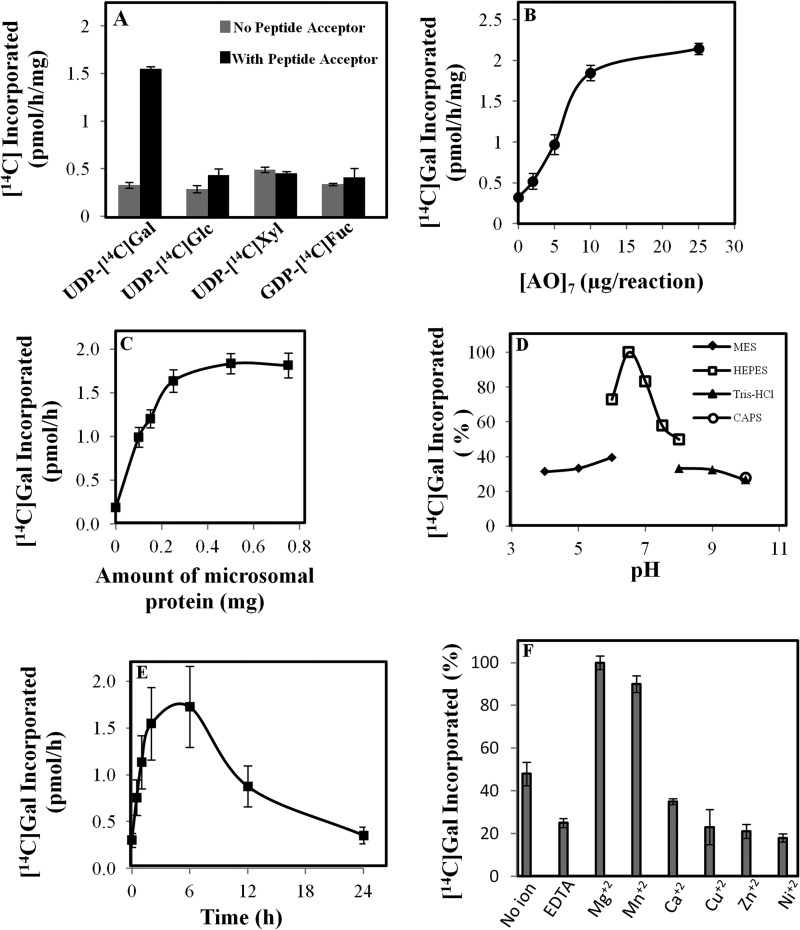
To determine the preference of nucleotide sugar donors, the standard GALT assay was performed with other potential sugar nucleotides, including UDP-[14C]Glc, UDP-[14C]Xyl, and GDP-[14C]Fuc, in the presence and absence of the (AO)7 peptide acceptor. Hyp-GALT activity was only detected with UDP-[14C]Gal as the sugar donor (Fig. 6A).
FIGURE 6.
Biochemical characterization of the (AO)7:AtGALT2 activity. Data presented are an average of duplicate assays. A, specificity of AtGALT2 enzyme for nucleotide sugar donors was analyzed by monitoring the incorporation of 14C-radiolabeled galactose onto (AO)7 substrate acceptor in the presence of UDP-[14C]Glc, UDP-[14C]Gal, UDP-[14C]Xyl, and GDP-[14C]Fuc. B, relationship between (AO)7 concentration and incorporation of [14C]Gal into (AO)7. C, relationship between microsomal protein concentration and incorporation of [14C]Gal into (AO)7. D, effect of pH on enzyme activity. E, effect of increasing the reaction time on enzyme activity. F, effect of different divalent ions (5 mm) on enzyme activity. Error bars, S.E.
The effects of pH and divalent cations as well as the concentrations of enzyme and substrate acceptor on the enzyme reaction were determined. With a total of 250 μg of microsomal proteins in the assay system, (AO)7:AtGALT2 activity approached saturation when 20 μg of (AO)7 was included in the reaction mixture (Fig. 6B). With 20 μg of (AO)7 in the GALT assay, incorporation of [14C]Gal increased proportionally with respect to the amount of microsomal protein up to 250 μg using an incubation time of 2 h (Fig. 6C). The (AO)7:AtGALT2 activity had a pH optimum of 6.5 with a HEPES-KOH buffer (Fig. 6D). The recombinant AtGALT2 was relatively stable because [14C]Gal incorporation into product increased over the first 6 h before decreasing significantly (Fig. 6E). Finally, a divalent cation requirement for optimal enzyme activity was also observed. Mg2+ followed by Mn2+ significantly enhanced AtGALT2 activity, whereas the presence of Ca2+, Cu2+, Zn2+, and Ni2+ had inhibitory effects to different extents (Fig. 6F).
AtGALT2 Mutants Have Lower GALT Activity and Reduced β-Yariv-precipitable AGPs
To provide additional in vivo evidence that AtGALT2 encodes an AGP GALT, two allelic AtGALT2 homozygous knock-out mutants, galt2-1 (SALK_117233) and galt2-2 (SALK_141126), were obtained (supplemental Fig. S2A). RT-PCR confirmed that the AtGALT2 transcripts were absent in these mutants (supplemental Fig. S2B). The mutants appeared to be phenotypically indistinguishable from wild type under normal growth conditions. Biochemical analysis of the mutants, however, revealed that galt2-1 and galt2-2, respectively, contained 21 and 13% less GALT activity compared with wild type plants (Table 1). In addition, both mutants contained ∼33% less β-Yariv-precipitable AGPs compared with wild type plants (Table 1).
TABLE 1.
GALT activity and the amount of β-Yariv-precipitated AGPs in wild type and galt2 mutant plants
Detergent-solubilized microsomal fractions were used for performing a standard GALT assay, and AGPs were extracted, precipitated by β-Yariv reagent, and quantified from 14-day-old plants. The values are the averages of at least two experiments from two biological replicates. The S.E. values are indicated.
| Genetic line | GALT activity | β-Yariv-precipitated AGPs |
|---|---|---|
| pmol/h/mg | μg/g | |
| Wild type | 6.7 ± 0.8 | 14.1 ± 3.5 |
| galt2-1 | 5.3 ± 1.2 | 9.6 ± 3.5 |
| galt2-2 | 5.8 ± 1.0 | 9.2 ± 2.8 |
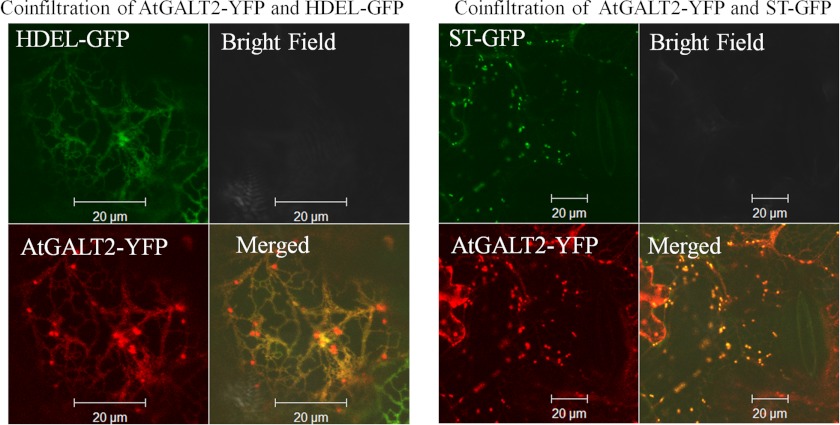
AtGALT2 Is Probably Localized to the Endomembrane System
To establish the subcellular localization of AtGALT2, live cell confocal imaging of fluorescently tagged AtGALT2 protein was performed. An AtGALT2-vYFP fusion was constructed and transiently co-expressed with either a Golgi marker protein, ST-mGFP5, or an ER marker, HDEL-mGFP5, in tobacco leaves. Upon co-infiltration with the Golgi marker, AtGALT2-vYFP was not only observed as discrete punctate structures typical of a Golgi-localized staining pattern but also observed in a reticulate ER localization pattern (Fig. 7). Co-infiltration of AtGALT2-vYFP with the ER marker revealed characteristic reticulate structures typical of an ER localization but also showed punctate Golgi localization (Fig. 7). There is a concern regarding transient expression experiments about overburdening the secretory system as well as specifically identifying ER versus Golgi subcellular localization because these two membrane systems are highly connected in plants (40). To address this inherent problem, a time course of co-localization was performed, where co-infiltrated leaf sections were observed consecutively over 4 days starting from the second day of infiltration. The hypothesis is that if the localization is an outcome of overburdening the endomembrane system, then over time, the amount of the transient AtGALT2 may decrease considerably from ER and accumulate in the Golgi. However, no significant difference in co-localization between ER and Golgi over time was observed here, indicating that AtGALT2 is probably present in both ER and Golgi compartments (supplemental Fig. S3, A–E). Additionally, control images of tobacco cells expressing only ST-mGFP5, only HDEL-mGFP5, and only AtGALT2-vYFP at day 2 postinfiltrations were observed to exclude spectral overlaps between YFP and GFP channels (supplemental Fig. S3, G–I). AtGALT2 was also examined using multiple subcellular localization prediction programs, TargetP and Golgi Predictor, and the TMHMM server (25) for the prediction of transmembrane domains. Based on these analyses and consistent with the live cell imaging data, AtGALT2 is targeted to the secretory pathway and has a single N-terminal transmembrane domain (supplemental Table S3).
FIGURE 7.
Subcellular localization of AtGALT2 in tobacco leaf epidermal cells observed after 5 days of infiltration. Transiently expressed AtGALT2-vYFP co-localized with ST-mGFP5 fusion protein (a Golgi marker) as well as with HDEL-mGFP5 fusion protein (an ER marker). These constructs were examined by laser-scanning confocal microscopy under fluorescent and white light, and the fluorescent images were merged to observe co-localization.
Computational Modeling of AtGALT2 Predicts UDP-Sugar Binding
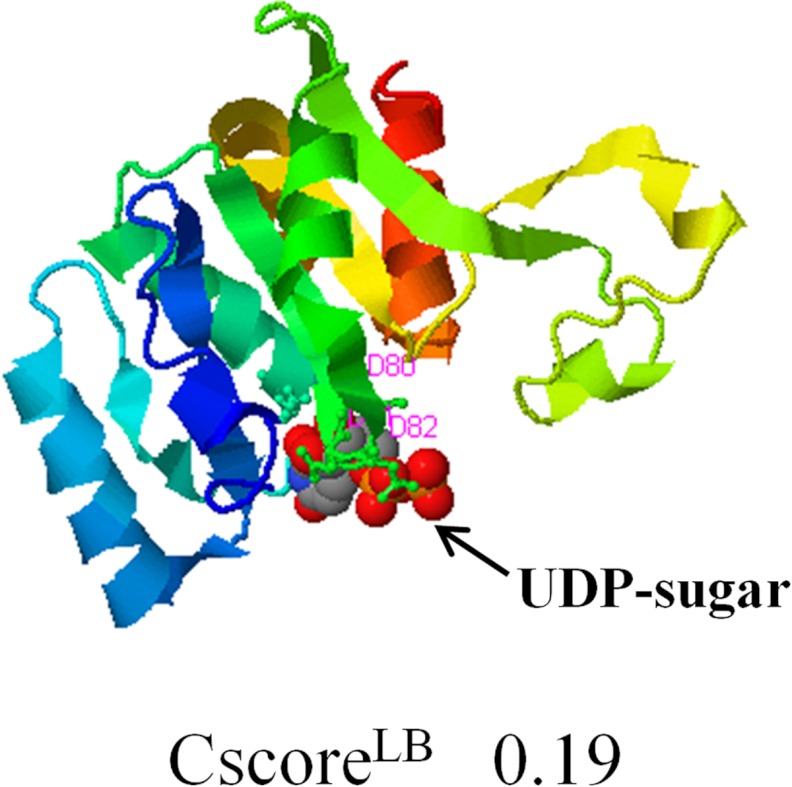
A three-dimensional structural model of AtGALT2 was created using I-TASSER and corroborated by Phyre2 (supplemental Fig. S4) (41, 26). I-TASSER and Phyre2 identified mouse manic fringe protein (2j0aA) as the closest structural homolog; this protein is a β-1,3-N-acetylglucosaminyltransferase. COFACTOR was then used to identify putative molecular functions of AtGALT2 based on the predicted three-dimensional structure by I-TASSER (41). COFACTOR analysis revealed that three aspartic acid residues at positions 80, 81, and 82 of AtGALT2 are involved in the binding and catalysis of a UDP-sugar donor substrate (Fig. 8).
FIGURE 8.
A three-dimensional structural model of AtGALT2 as predicted by the COFACTOR server available in the I-TASSER program. This three-dimensional model shows the predicted ligand and its binding site with a confidence score (CscoreLB) of 0.19. The highlighted residues correspond to the DXD motif, with the numbers denoting the positions of the three aspartic acid residues at positions 80, 81, and 82.
DISCUSSION
In contrast to the considerable knowledge about the biosynthesis of cell wall polysaccharides and lignin, relatively little is known about the mechanisms involved in biosynthesis of AGPs (2, 36, 42). A bioinformatics approach was used to identify six putative AGP-GALTs (named GALT1–GALT6) that act directly on the AGP protein backbone based on the finding that these are the only Arabidopsis proteins that contain both a GALECTIN domain and a GALT domain, similar to certain mammalian GTs that O-glycosylate mucins and are composed of analogous lectin and GT domains. These six candidate genes were heterologously expressed in Pichia cells and tested for Hyp-GALT activity using an in vitro Hyp-GALT assay previous developed in our laboratory (13). Only AtGALT2 has shown activity in this assay to date and thus became the focus of this investigation.
Microsomal preparations obtained from Pichia cells expressing recombinant AtGALT2 exhibited Hyp-GALT activity catalyzing the transfer of [14C]Gal from UDP-[14C]Gal onto a chemically synthesized peptide (AO)7 substrate acceptor (Figs. 2 and 3). Further product characterization revealed that a single Gal residue is transferred to Hyp residues; there was no evidence for additional Gal units being added to the substrate acceptor (Figs. 2 and 4). This observation is consistent with the hypothesis that O-glycosylation in plants occurs by the stepwise addition of sugar residues, as opposed to en block transfer. The recent identification and characterization of two AGP fucosyltransferases that have the ability to fucosylate AGPs lacking terminal Fuc residues is also consistent with sequential sugar addition in plants (14). These observations are consistent with O-glycosylation in animals, which is viewed to occur by the sequential addition of single sugar residues to the polypeptide, as exemplified by two well defined processes, O-mannosylation (43) and mucin type O-glycosylation (44). In addition, glycoengineered mammalian mucin-type O-glycosylation in transgenic plants demonstrates stepwise sugar addition (45, 46).
Furthermore, the observation that a single Gal residue is transferred to Hyp is also consistent with the hypothesis that AtGALT2 is specific for peptidyl Hyp and lacks the ability to transfer additional Gal units onto peptidyl Hyp-Gal units; however, given the relatively small amount of product produced, the possibility that insufficient amounts of peptidyl Hyp-Gal substrate are available for further enzyme action cannot be excluded. It should be noted that the Hyp-GALT activity observed here for heterologously expressed AtGALT2 in Pichia was considerably lower than that observed using plant microsomes (13). One possible explanation for this could be that multiple Hyp-GALT enzymes, multienzyme complexes, and/or plant-specific cofactors are involved in the biosynthesis of AGP glycans, which are absent in Pichia cells.
AtGALT2 is specific for AGP sequences and not for other related protein sequences, including extensin with its characteristic Ser-(Hyp)4 repeat units and a non-hydroxylated AGP-like sequence containing Pro in place of Hyp (Fig. 5). Pectic polysaccharides contain Gal residues (47, 48), but pectic substrate acceptors also failed to serve as substrate acceptors for AtGALT2. In addition, Gal-(1,3)-β-Gal-O-Me, which mimics the β-(1,3)Gal sugar backbone of AGPs, also failed to serve as a substrate acceptor for AtGALT2 (data not shown). These findings are consistent with the Hyp contiguity hypothesis, which states that non-contiguous Hyp residues are sites of arabinogalactan polysaccharide addition, whereas contiguous Hyp residues are sites for the addition of arabinofuranose oligosaccharides (37, 49). Interestingly, shorter AGP peptides served as more effective substrate acceptors. Although this observation lacks an explanation, it is consistent with previous findings with plant microsomes (13). It should also be noted that Strasser et al. (35) tested GALT2 as well as GALT1, -3, -4, -5, and -6 for N-glycosylation activity, and only GALT1 was found to have such activity.
Heterologously expressed AtGALT2 in Pichia microsomes has similar biochemical properties to the GALT(s) present in Arabidopsis microsomal membranes (Fig. 6) (13, 20). AtGALT2 is specific for UDP-Gal as the sugar donor, has a pH optimum of 6.5 (in contrast to 7 for plant microsomes) and has a requirement for Mg2+ and Mn2+ (in contrast to Mn2+ for plant microsomes) for high activity. These differences are probably a reflection of studying the properties of a single GALT enzyme in yeast microsomes in contrast to the more complex GALT enzyme mixture in Arabidopsis microsomes that includes plant-specific factors. The observed divalent cation requirement agrees with the structural conformation of all CAZy GT31 members, which share a catalytic domain containing a DXD motif in the GT-A superfamily. In addition, the three-dimensional protein structure of AtGALT2 predicted by I-TASSER and Phyre had as its closest match the catalytic domain of the mouse manic fringe in2 complexed with UDP and manganese (supplemental Fig. S4C).
Biochemical analysis of the AtGALT2 mutants provided additional in vivo evidence that AtGALT2 is indeed an AGP GALT. The absence of a mutant phenotype under normal growth conditions and the reduced GALT activity and lower β-Yariv-precipitable AGPs are consistent with gene redundancy. Other Hyp-GALTs probably compensate for the loss of AtGALT2. Apparently, the reduced GALT activity and the reduced β-Yariv-precipitable AGPs in these mutants are not sufficient to bring about a phenotypic change under normal growth conditions. Examination of these mutants under non-physiological conditions or the production of multigene mutants within this gene family may reveal novel phenotypes in the future.
The Golgi apparatus is not only a central sorting point within the secretory pathway but also plays a central biosynthetic role in processing of complex carbohydrate structures. Subcellular localization of AtGALT2 to the ER and Golgi in tobacco leaf epidermal cells is consistent with the localization of Hyp-GALT enzyme activity to the endomembrane system in tobacco and Arabidopsis cell cultures (Fig. 7) (13, 20). In addition, bioinformatics analysis predicts that AtGALT2 is a type II membrane protein localized to the Golgi (supplemental Table S3). Based on these data, AtGALT2 may initiate Hyp galactosylation of AGP protein backbone in the ER following the action of prolyl hydroxylase and continue to act in the Golgi to ensure that Hyp galactosylation is complete so as to allow for subsequent glycosylation and elongation of the arabinogalactan polysaccharide.
An unrooted phylogenetic analysis of animal and plant GT31 members revealed three distinct clusters: clade I composed of plant-specific GALTs devoid of a GALECTIN domain; clade II consisting of animal GALTs involved in core β-(1,3) O-glycosylation and β-(1,3) N-acetylglucosamine or α-N-acetylgalactosaminyltransferase (GlcNAc-T; GalNAc-T); and clade III consisting of plant-specific GALTs with a Gal-binding lectin domain (supplemental Fig. S1). The association of a GALECTIN lectin domain with a GALT domain was conserved across several plant GT31 members, including both dicots (Arabidopsis, M. truncatula, Poplus, and V. vinifera) and monocots (Brachypodium, Z. mays, rice, and S. bicolor). The GALECTIN domain is defined by the presence of a conserved carbohydrate recognition domain that specifically binds to β-galactosides, although they can display a wide range of substrate specificities due to structural heterogeneity in the carbohydrate recognition domain (50). Although lectin domains are common in mammalian GT27 members, they are absent in mammalian GT31 members. By analogy to the mammalian GTs containing a lectin domain, the GALECTIN domain in plants may modulate the GALT activity (17). Future experiments can be designed to test this hypothesis.
Two independent homology modeling methods were used to generate a predicted structure for AtGALT2 (Fig. 8 and supplemental Fig. S4). First, AtGALT2 was submitted to the protein fold recognition PHYRE server (26), which was used to generate a predicted structural model (supplemental Fig. S4F). In the second approach, the automated homology-modeling server I-TASSER was utilized to generate five predicted structures for AtGALT2 (supplemental Fig. S4, A–E). Both PHYRE and I-TASSER generated similar homology model predictions for AtGALT2. The outputs of these predictions were then used as a template to guide further structure-function analyses. The resulting three-dimensional structure revealed the interaction of AtGALT2 with a UDP-nucleotide sugar in a hydrophobic pocket containing a DXD motif (Fig. 8).
In summary, this study indicates that AtGALT2 (At4g21060) catalyzes galactosylation of Hyp residues in AGP protein backbones and thus represents the initial step in the biosynthesis of the polysaccharide side chains that decorate AGPs. Moreover, transient expression of fluorescently tagged AtGALT2 and bioinformatics analysis indicates that this enzyme is a membrane-bound protein localized in the endomembrane system, consistent with its established biochemical function. Future studies will now focus on examining galt2 knock-out mutants in Arabidopsis and testing for the existence of AtGALT2-containing enzyme complexes involved in AGP biosynthesis and expression of AtGALT2 and other putative Hyp-GALTs in other host systems to test for Hyp-GALT and additional GALT enzymatic activities.
Acknowledgments
We thank Vijayanand Nadella of the genomic facility at Ohio University for sequencing our AtGALT constructs. We thank Dr. Richard Strasser (University of Natural Resources and Life Sciences, Vienna) for providing the cDNA clone of AtGALT2.
This work was supported by Ohio Plant Biotechnology Consortium Grant 020340004090 GR0017687.01 and National Science Foundation Grant 0918661.

This article contains supplemental Figs. S1–S4 and supplemental Tables S1–S3.
- AGP
- arabinogalactan protein
- Hyp or O
- hydroxyproline
- EXT
- extensin
- PRP
- proline-rich protein
- GT
- glycosyltransferase
- GALT
- galactosyltransferase
- Fuc
- fucose
- GalNAc-T
- N-acetylgalactosaminyltransferase
- RP-HPLC
- reverse-phase high performance liquid chromatography
- HPAEC
- high pH anion exchange chromatography
- NC
- negative control
- ExtP
- extensin peptide
- CAPS
- 3-(cyclohexylamino)propanesulfonic acid
- ER
- endoplasmic reticulum
- ST
- sialic acid transferase
- DPn
- degree of polymerization n.
REFERENCES
- 1. Cosgrove D. J. (2005) Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6, 850–861 [DOI] [PubMed] [Google Scholar]
- 2. Somerville C., Bauer S., Brininstool G., Facette M., Hamann T., Milne J., Osborne E., Paredez A., Persson S., Raab T., Vorwerk S., Youngs H. (2004) Toward a systems approach to understanding plant cell walls. Science 306, 2206–2211 [DOI] [PubMed] [Google Scholar]
- 3. Showalter A. M., Keppler B., Lichtenberg J., Gu D., Welch L. R. (2010) A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiol. 153, 485–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ellis M., Egelund J., Schultz C. J., Bacic A. (2010) Arabinogalactan proteins. Key regulators at the cell surface? Plant Physiol. 153, 403–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu C., Mehdy M. (2007) A nonclassical arabinogalactan protein gene highly expressed in vascular tissues, AGP31, is transcriptionally repressed by methyl jasmonic acid in Arabidopsis. Plant Physiol. 145, 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Seifert G. J., Roberts K. (2007) The biology of arabinogalactan proteins. Annu. Rev. Plant Biol. 58, 137–161 [DOI] [PubMed] [Google Scholar]
- 7. Clarke A., Gleeson P., Harrison S., Knox R. B. (1979) Pollen-stigma interactions. Identification and characterization of surface components with recognition potential. Proc. Natl. Acad. Sci. U.S.A. 76, 3358–3362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Qu Y., Egelund J., Gilson P. R., Houghton F., Gleeson P. A., Schultz C. J., Bacic A. (2008) Identification of a novel group of putative Arabidopsis thaliana β-(1,3)-galactosyl transferases. Plant Mol. Biol. 68, 43–59 [DOI] [PubMed] [Google Scholar]
- 9. Bacic A., Churms S. C., Stephen A. M., Cohen P. B., Fincher G. B. (1987) Fine-structure of the arabinogalactan protein form Lolium multiflorum. Carbohydr. Res. 162, 85–93 [Google Scholar]
- 10. Tan L., Qiu F., Lamport D. T., Kieliszewski M. J. (2004) Structure of a hydroxyproline (Hyp)-arabinogalactan polysaccharide from repetitive Ala-Hyp expressed in transgenic Nicotiana tabacum. J. Biol. Chem. 279, 13156–13165 [DOI] [PubMed] [Google Scholar]
- 11. Tan L., Varnai P., Lamport D. T., Yuan C., Xu J., Qiu F., Kieliszewski M. J. (2010) Plant O-hydroxyproline arabinogalactans are composed of repeating trigalactosyl subunits with short bifurcated side chains. J. Biol. Chem. 285, 24575–24583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tryfona T., Liang H. C., Kotake T., Tsumuraya Y., Stephens E., Dupree P. (2012) Structural characterisation of Arabidopsis leaf arabinogalactan polysaccharides. Plant Physiol. 160, 653–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liang Y., Faik A., Kieliszewski M., Tan L., Xu W. L., Showalter A. M. (2010) Identification and characterization of in vitro galactosyltransferase activities involved in arabinogalactan protein glycosylation in tobacco and Arabidopsis. Plant Physiol. 154, 632–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wu Y., Williams M., Bernard S., Driouich A., Showalter A. M., Faik A. (2010) Functional identification of two nonredundant Arabidopsis (1,2) fucosyltransferases specific to arabinogalactan proteins. J. Biol. Chem. 285, 13638–13645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wandall H. H., Irazoqui F., Tarp M. A., Bennett E. P., Mandel U., Takeuchi H., Kato K., Irimura T., Suryanarayanan G., Hollingsworth M. A. (2007) The lectin domains of polypeptide GalNAc-transferases exhibit carbohydrate binding specificity for GalNAc. Lectin binding to GalNAc-glycopeptide substrates is required for high density GalNAc-O-glycosylation. Glycobiology 17, 374–387 [DOI] [PubMed] [Google Scholar]
- 16. Pedersen J. W., Bennett E. P., Schjoldager K. T., Meldal M., Holmér A. P., Blixt O., Cló E., Levery S. B., Clausen H., Wandall H. H. (2011) Lectin domains of polypeptide GalNAc-Ts exhibit glycopeptide binding specificity. J. Biol. Chem. 286, 32684–32696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bennett E. P., Mandel U., Clausen H., Gerken T. A., Fritz T. A., Tabak L. A. (2012) Control of mucin-type O-glycosylation. A classification of the polypeptide GalNAc-transferase gene family. Glycobiology 22, 736–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nothnagel E. A. (1997) Proteoglycans and related components in plant cells. Int. Rev. Cytol. 174, 195–291 [DOI] [PubMed] [Google Scholar]
- 19. Hassan H., Reis C. A., Bennett E. P. (2000) The lectin domain of UDP-N-acetyl-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase-T4 directs its glycopeptide specificities. J. Biol. Chem. 275, 38197–38205 [DOI] [PubMed] [Google Scholar]
- 20. Oka T., Saito F., Shimma Y., Yoko-o T., Nomura Y., Matsuoka K., Jigami Y. (2010) Characterization of endoplasmic reticulum-localized UDP-d-galactose. Hydroxyproline O-galactosyltransferase using synthetic peptide substrates in Arabidopsis. Plant Physiol. 152, 332–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boratyn G. M., Schäffer A. A., Agarwala R., Altschul S. F., Lipman D. J., Madden T. L. (2012) Domain enhanced lookup time accelerated BLAST. Biol. Direct. 7, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Henrissat B., Davies G. J. (2000) Glycoside hydrolases and glycosyltransferases. Families, modules, and implications for genomics. Plant Physiol. 124, 1515–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schwacke R., Schneider A., van der Graaff E., Fischer K., Catoni E., Desimone M., Frommer W. B., Flügge U. I., Kunze R. (2003) ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 131, 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J. F., Guindon S., Lefort V., Lescot M., Claverie J. M., Gascuel O. (2008) Phylogeny.fr. Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001) Predicting transmembrane protein topology with a hidden Markov model. Application to complete genomes. J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 26. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web. A case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 27. Zhang Y. (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hoffman C. S., Winston F. (1987) A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57, 267–272 [DOI] [PubMed] [Google Scholar]
- 29. Mølhøj M., Verma R., Reiter W. D. (2004) The biosynthesis of d-galacturonate in plants. Functional cloning and characterization of a membrane-anchored UDP-d-glucuronate 4-epimerase from Arabidopsis. Plant Physiol. 135, 1221–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Strahm A., Amado R., Neukom H. (1981) Hydroxyproline-galactoside as a protein polysaccharide linkage in a water soluble arabinogalactan-peptide from wheat endosperm. Phytochemistry 20, 1061–1063 [Google Scholar]
- 31. Saint-Jore C. M., Evins J., Batoko H., Brandizzi F., Moore I., Hawes C. (2002) Redistribution of membrane proteins between the Golgi apparatus and endoplasmic reticulum in plants is reversible and not dependent on cytoskeletal networks. Plant J. 29, 661–678 [DOI] [PubMed] [Google Scholar]
- 32. Batoko H., Zheng H. Q., Hawes C., Moore I. (2000) A Rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell 12, 2201–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schultz C. J., Johnson K. L., Currie G., Bacic A. (2000) The classical arabinogalactan protein gene family of Arabidopsis. Plant Cell 12, 1751–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gao M., Kieliszewski M. J., Lamport D. T., Showalter A. M. (1999) Isolation, characterization, and immunolocalization of a novel, modular tomato arabinogalactan protein corresponding to the LeAGP-1 gene. Plant J. 18, 43–55 [DOI] [PubMed] [Google Scholar]
- 35. Strasser R., Bondili J. S., Vavra U., Schoberer J., Svoboda B., Glössl J., Léonard R., Stadlmann J., Altmann F., Steinkellner H., Mach L. (2007) A unique 1,3-galactosyltransferase is indispensable for the biosynthesis of N-glycans containing Lewis a structures in Arabidopsis thaliana. Plant Cell 19, 2278–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tan L., Showalter A. M., Egelund J., Hernandez-Sanchez A., Doblin M. S., Bacic A. (2012) Arabinogalactan-proteins and the research challenges for these enigmatic plant cell surface proteoglycans. Front. Plant Sci. 3, 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Egelund J., Ellis M. A., Doblin M. S., Qu Y., Bacic A. (2011) Genes and enzymes of the GT31 family. Towards unraveling the function(s) of the plant glycosyltransferase family members. in Plant Polysaccharides: Biosynthesis and Bioengineering, pp. 213–234, Oxford University Press, Hoboken, NJ [Google Scholar]
- 38. Kato H., Takeuchi Y., Tsumuraya Y., Hashimoto Y., Nakano H., Kovác P. (2003) In vitro biosynthesis of galactans by membrane-bound galactosyltransferase from radish (Raphanus sativus L.) seedlings. Planta 217, 271–282 [DOI] [PubMed] [Google Scholar]
- 39. Shpak E., Barbar E., Leykam J. F., Kieliszewski M. J. (2001) Contiguous hydroxyproline residues direct hydroxyproline arabinosylation in Nicotiana tabacum. J. Biol. Chem. 276, 11272–11278 [DOI] [PubMed] [Google Scholar]
- 40. Boevink P., Oparka K., Santa Cruz S., Martin B., Betteridge A., Hawes C. (1998) Stacks on tracks. The plant Golgi apparatus traffics on an actin/ER network. Plant J. 15, 441–447 [DOI] [PubMed] [Google Scholar]
- 41. Roy A., Kucukural A., Zhang Y. (2010) I-TASSER A unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Boerjan W., Ralph J., Baucher M. (2003) Lignin biosynthesis. Annu. Rev. Plant Biol. 54, 519–546 [DOI] [PubMed] [Google Scholar]
- 43. Goto M. (2007) Protein O-glycosylation in fungi. Diverse structures and multiple functions. Biosci. Biotechnol. Biochem. 71, 1415–1427 [DOI] [PubMed] [Google Scholar]
- 44. Hanisch F. G. (2001) O-Glycosylation of the mucin type. Biol. Chem. 382, 143–149 [DOI] [PubMed] [Google Scholar]
- 45. Yang Z., Drew D. P., Jørgensen B., Mandel U., Bach S. S., Ulvskov P., Levery S. B., Bennett E. P., Clausen H., Petersen B. L. (2012) Engineering mammalian mucin-type O-glycosylation in plants. J. Biol. Chem. 287, 11911–11923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Castilho A., Neumann L., Daskalova S., Mason H. S., Steinkellner H., Altmann F., Strasser R. (2012) Engineering of sialylated mucin-type O-glycosylation in plants. J. Biol. Chem. 287, 36518–36526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Geshi N., Pauly M., Ulvskov P. (2002) Solubilization of galactosyltransferase that synthesizes 1,4-β-galactan side chains in pectic rhamnogalacturonan I. Physiol. Plant 114, 540–548 [DOI] [PubMed] [Google Scholar]
- 48. Peugnet I., Goubet F., Bruyant-Vannier M. P., Thoiron B., Morvan C., Schols H. A., Voragen A. G. (2001) Solubilization of rhamnogalacturonan I galactosyltransferases from membranes of a flax cell suspension. Planta 213, 435–445 [DOI] [PubMed] [Google Scholar]
- 49. Kieliszewski M. J., Shpak E. (2001) Synthetic genes for the elucidation of glycosylation codes for arabinogalactan-proteins and other hydroxyproline-rich glycoproteins. Cell Mol. Life Sci. 58, 1386–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dodd R. B., Drickamer K. (2001) Lectin-like proteins in model organisms. Implications for evolution of carbohydrate-binding activity. Glycobiology 11, 71R-79R [DOI] [PubMed] [Google Scholar]