Background: The ssSPTs activate serine palmitoyltransferase and specify its acyl-CoA selectivity.
Results: Both properties are contained within a 33-amino acid core that spans the membrane.
Conclusion: A single amino acid difference between ssSPTa and ssSPTb is responsible for the acyl-CoA preference of heterotrimers containing each isoform.
Significance: The ssSPTs are critical regulatory components of the rate-limiting enzyme in sphingolipid biosynthesis.
Keywords: Enzyme Mutation, Lipids, Membrane Enzymes, Membrane Lipids, Sphingolipid, HSAN1, Long Chain Bases, Serine Palmitoyltransferase, ssSPTs
Abstract
The topological and functional organization of the two isoforms of the small subunits of human serine palmitoyltransferase (hssSPTs) that activate the catalytic hLCB1/hLCB2 heterodimer was investigated. A variety of experimental approaches placed the N termini of the ssSPTs in the cytosol, their C termini in the lumen, and showed that they contain a single transmembrane domain. Deletion analysis revealed that the ability to activate the heterodimer is contained in a conserved 33-amino acid core domain that has the same membrane topology as the full-length protein. In combination with analysis of isoform chimera and site-directed mutagenesis, a single amino acid residue in this core (Met25 in ssSPTa and Val25 in ssSPTb) was identified which confers specificity for palmitoyl- or stearoyl-CoA, respectively, in both yeast and mammalian cells. This same residue also determines which isoform is a better activator of a mutant heterodimer, hLCB1S331F/hLCB2a, which has increased basal SPT activity and decreased amino acid substrate selectivity. This suggests that the role of the ssSPTs is to increase SPT activity without compromising substrate specificity. In addition, the observation that the C-terminal domains of ssSPTa and ssSPTb, which are highly conserved within each subfamily but are the most divergent regions between isoform subfamilies, are not required for activation of the heterodimer or for acyl-CoA selectivity suggests that the ssSPTs have additional roles that remain to be discovered.
Introduction
The α-oxoamine synthases are a family of related proteins almost all of whose members are soluble head-to-tail homodimers with two symmetrical active sites located at the subunit interface (1–3). This is also true for bacterial SPT2 (4–9). However, eukaryotic SPT is an integral ER membrane protein that contains a head-to-tail heterodimer of two related but distinct subunits, LCB1 and LCB2, with a single catalytic site (10–12). Furthermore, it is now clear that there are additional subunits necessary for maximal activity as well as associated negative regulatory components. In yeast, it has been known for some time that maximal activity requires Tsc3p (13). More recently, it has been demonstrated that the Lcb1p/Lcb2p/Tsc3p heterotrimer is associated with Orm1p, Orm2p, and Sac1p to form the SPOTS complex (14) that regulates long chain base (LCB) synthesis in response to sphingolipid availability (14, 15). Whereas in higher eukaryotes there are no homologs of Tsc3p, we have identified two functional orthologs, ssSPTa and ssSPTb, that activate both the human and yeast Lcb1p/Lcb2p heterodimers (16). In addition, the ORMDLs, which are homologous to the Orms, bind to SPT, and silencing their expression results in elevated ceramide levels (14, 17). Thus, it is likely that they also regulate SPT activity.
The presence of two ssSPT isoforms, along with the identification by Hornemann et al. of two isoforms of hLCB2 (18), indicates that higher eukaryotic SPT is far more complex than was previously appreciated and suggests that there might be functionally distinct forms of SPT. This hypothesis was confirmed by the demonstration that SPT isozymes containing different combinations of subunits have distinct acyl-CoA preferences (16). Specifically, the presence of ssSPTa confers a preference for condensation of serine with palmitoyl-CoA whereas ssSPTb confers a preference for condensation of serine with stearoyl-CoA. The highest homology between the ssSPTa and ssSPTb subfamilies resides in a 33-amino acid centrally located domain, suggesting that this region mediates membrane association and binding to and activation of the hLCB1/hLCB2a/b heterodimers. Although less homologous, the N termini of the two proteins are also related. In contrast, whereas the C-terminal domains are highly conserved within each subfamily, there is little homology between the C-terminal domains of the ssSPTa and ssSPTb subfamilies.
To confirm that the central domain is responsible for binding and activation of the heterodimer and to determine which region of the ssSPTs specifies substrate selectivity, we have constructed and analyzed a series of N- and C-terminal deletion mutants and generated ssSPT chimera. In addition, single amino acid substitutions were used to precisely map the residue responsible for the distinct acyl-CoA selectivities conferred by the ssSPT subunits. To more fully characterize this novel family of activator proteins, we also analyzed their membrane topology. The results of these experiments, as well as the fact that coccolithal virus-encoded SPT is a single-chain LCB2/LCB1 heterodimer (19, 20) and our previous success at expressing active yeast and mammalian LCB2-LCB1 fusions, suggested that it might also be possible to express heterotrimeric SPT isoforms as single-chain fusion proteins. Remarkably, not only were single-chain heterotrimers active (21) but they retained the same acyl-CoA preferences as heterotrimers comprised of individual subunits. Taken together, these results suggest that the ssSPTs are essential components of eukaryotic SPT that not only activate the enzyme but contribute to sphingolipid diversity.
EXPERIMENTAL PROCEDURES
Cells and Cell Growth
The yeast strain TDY9103 (Mata lcb1Δ::KAN lcb2Δ::KAN tsc3Δ::NAT ura3 leu2 lys2 his3 trp1Δ::HIS3) lacking all endogenous SPT subunits was constructed using standard methods and used for all heterologous expression studies. The mutant was cultured in medium containing 15 μm phytosphingosine (PHS) and 0.2% tergitol. To assay growth, 0.2 A600 nm of exponentially growing cultures were serially diluted 1:5 in a microtiter plate. The cells were transferred to plates and incubated 4–5 days at the indicated temperatures. CHO-Ly-B cells (RIKEN Bioresource Center, Japan) deficient in LCB1 expression (22, 23) were maintained in 95:5 Ham's F-12:Dulbecco's modified Eagle's medium containing 4.5 g/liter glucose supplemented with 10% fetal bovine serum (JRH Biosciences, Lenexa, KS) and penicillin and streptomycin. Cells were transfected using Lipofectamine 2000 (Invitrogen) as described previously (24).
Plasmids
The ssSPT N-terminal deletion mutants were constructed by inserting SfiI-ended PCR fragments encoding residues 10–68 or 19–68 of ssSPTa or residues 10–76 or 19–76 of ssSPTb into the SfiI site of pPR3-N (Dualsystems Biotech, Zurich, Switzerland). The ssSPT C-terminal deletions were constructed by introducing stop codons into the pPR3-N-NubG-HA-ssSPTa or pPR3-N-NubG-HA-ssSPTb plasmids (16) or into the NΔ10 ssSPT deletion mutant plasmids by QuikChange (QC) mutagenesis (Agilent, Santa Clara, CA). For construction of the mammalian ssSPT-GC expression plasmids, a fragment containing the Nub-HA-ssSPTs (from the pPR3-N plasmids) was ligated into pcDNA, an NheI site was inserted immediately before the stop codon by QC mutagenesis, and a SpeI-ended PCR fragment containing residues 80–133 of yeast invertase (the GC cassette (24)) was ligated into the NheI site. For expression of the C-terminally GC-tagged ssSPTs in yeast, the SfiI fragment from the mammalian ssSPT-GC expression plasmids was moved into pPR3-N. Construction of the N-terminally HA-tagged ssSPT-expressing pGH316-based plasmids used in the protease accessibility studies was described previously (21). Plasmids expressing the C-terminally HA-tagged ssSPTs were constructed in the same way, with the exception that the SpeI-ended HA cassettes were inserted into an XbaI site immediately preceding the stop codon. The plasmids containing the N-terminally HA-tagged CΔ21 and CΔ25 ssSPTa truncation mutants were used for construction of the HA-ssSPTa-Myc and the HA-ssSPTa-GC expressing plasmids by inserting either a SpeI-ended 3×-Myc or GC cassette into NheI sites placed immediately upstream of the stop codons. Yeast and mammalian expression plasmids containing the single-chain fusion SPTs were described previously (21); all mutations were then introduced by QC mutagenesis. The ssSPTaba chimera was created by in vivo recombination using a gapped (at codon 33) pPR3-N-NubG-HA-ssSPTa plasmid and a PCR fragment comprising the ssSPTa open reading frame into which residues 27–54 from ssSPTb were substituted. The ssSPTab chimera was constructed by substituting the BstZ17I fragment from pPR3-N-NubG-HA-ssSPTb for the same fragment in the plasmid containing the aba chimera. The ssSPTba chimera was made by substituting the BstZ17I fragment from the plasmid containing the aba chimera into pPR3-N-NubG-HA-ssSPTb.
Preparation of Microsomes
Yeast microsomes were prepared from exponentially growing cells that were pelleted, washed in TEGM (50 mm Tris-HCl, pH 7.5, 1 mm EGTA, 1 mm β-mercaptoethanol) and resuspended in TEGM containing 1 mm PMSF, 2 mg/ml pepstatin A, 1 mg/ml leupeptin, and 1 mg/ml aprotinin. Glass beads were added to the meniscus, and cells were disrupted by repeated (four times, 1 min each) cycles of vortexing with cooling on ice between. Unbroken cells, beads, and debris were removed by centrifugation (10,000 × g, 10 min), and the low speed supernatant was centrifuged at 100,000 × g for 40 min. The crude microsomal pellet was homogenized in TEGM and spun at 100,000 × g for 40 min to obtain the microsomal pellet. The pellet was homogenized at 5–8 mg/ml in TEGM containing 33% glycerol and stored at −80 °C. Microsomes were prepared from CHO-Ly-B cells as described previously (24).
SPT Assay
SPT was assayed in 300 μl of 50 mm HEPES, pH 8.1, containing 50 μm pyridoxal phosphate, 10 mm [3H]serine (3 Ci/mol), 0.02 mm BSA, and 0.1–0.2 mm palmitoyl- or stearoyl-CoA. The reaction was initiated by adding 0.2–0.3 mg of microsomal protein and terminated after 10 min at 37 °C by the sequential addition of 100 μl of 2 n NH4OH and 2 ml of CHCl3:methanol (1:2). After vortexing, an additional 1 ml of CHCl3 and 2 ml of 0.5 n NH4OH were added, with vortexing after each addition. After brief centrifugation to separate the phases, the upper phase was removed, and the lower phase was washed two times with 60 mm KCl. A fixed volume was dried under N2 and resuspended in scintillation fluid for determination of radioactivity. SPT activity, expressed in pmoles of serine converted/mg of microsomal protein per min, was calculated from triplicate determinations.
LCB Extraction and Analysis
Total LCBs were isolated from 5 A600 nm of exponentially growing yeast cells by HCl methanolysis. Briefly, washed cells were resuspended in 1 ml of 1 n methanolic HCl (Supelco, Bellefonte, PA), and the tubes were placed in boiling water for 30 min. After cooling on ice, 1 ml of 0.9% NaCl and 2 ml of hexane:diethyl ether (1:1) were added, and following centrifugation the upper phase was removed and the LCBs extracted from the lower phase by adding 0.25 ml of 10 n NaOH, vortexing, and then adding 2 ml of hexane and vortexing. Following centrifugation, 1.5 ml of the upper phase was dried under N2 and resuspended in 80 μl of MeOH:190 mm triethylamine (2:0.3, v/v), and 20 μl of AccQ reagent (Waters, Milford, MA) was added and allowed to react for 60 min. Derivatized LCBs from 1.0 A600 nm of yeast cells were analyzed by HPLC as described previously (25).
Glycosylation Cassette Mobility Shift Assays
Mobility shift assays of proteins containing the GC cassettes were performed using EndoH according to the protocol provided by New England Biolabs (Beverly, MA).
Preparation of Rightside-out Vesicles
Sealed vesicles were prepared as described previously (24) with the exception that following zymolyase 100-T treatment the spheroplasts were purified on a 2 m sorbitol cushion.
RESULTS
Membrane Topology of the ssSPTs
The mammalian ssSPTs are predicted to have two transmembrane domains (TMDs) residing within the conserved core as do other vertebrate ssSPTs (Fig. 1A). However, candidate ssSPTs from invertebrates and plants, identified based on their homologies to the human ssSPTs, only have a single predicted TMD (Fig. 1B). The same is true for the yeast ortholog of the ssSPTs, Tsc3p. To determine whether the mammalian ssSPTs have one or two TMDs, their membrane topology and organization were investigated.
FIGURE 1.
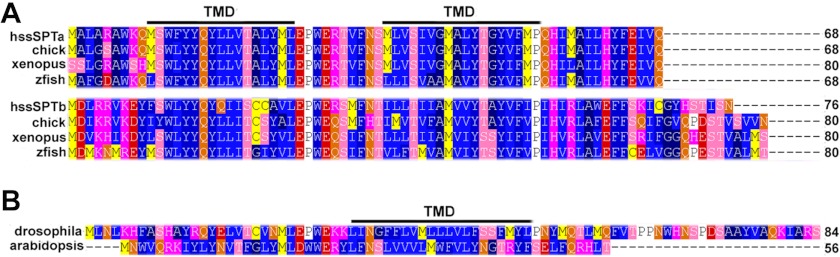
Alignment of vertebrate and candidate invertebrate ssSPTs. A, vertebrate ssSPTs are predicted by hydropathy analysis to have two TMDs. Shown in the alignment are members of the ssSPTa and ssSPTb subfamilies from Homo sapiens (hssSPTa or hssSPTb), Gallus domesticus (chick), Xenopus laevis (xenopus) and Danio rerio (zebrafish). hssSPTa is aligned and numbered without its three N-terminal residues (MAG) that the other vertebrate ssSPTa subunits lack. B, in contrast to the vertebrate ssSPTs, candidate invertebrate ssSPTs including the Drosophila melanogaster (cg32038) and Arabidopsis thaliana (At2g30942) ssSPTs are predicted to have only one TMD.
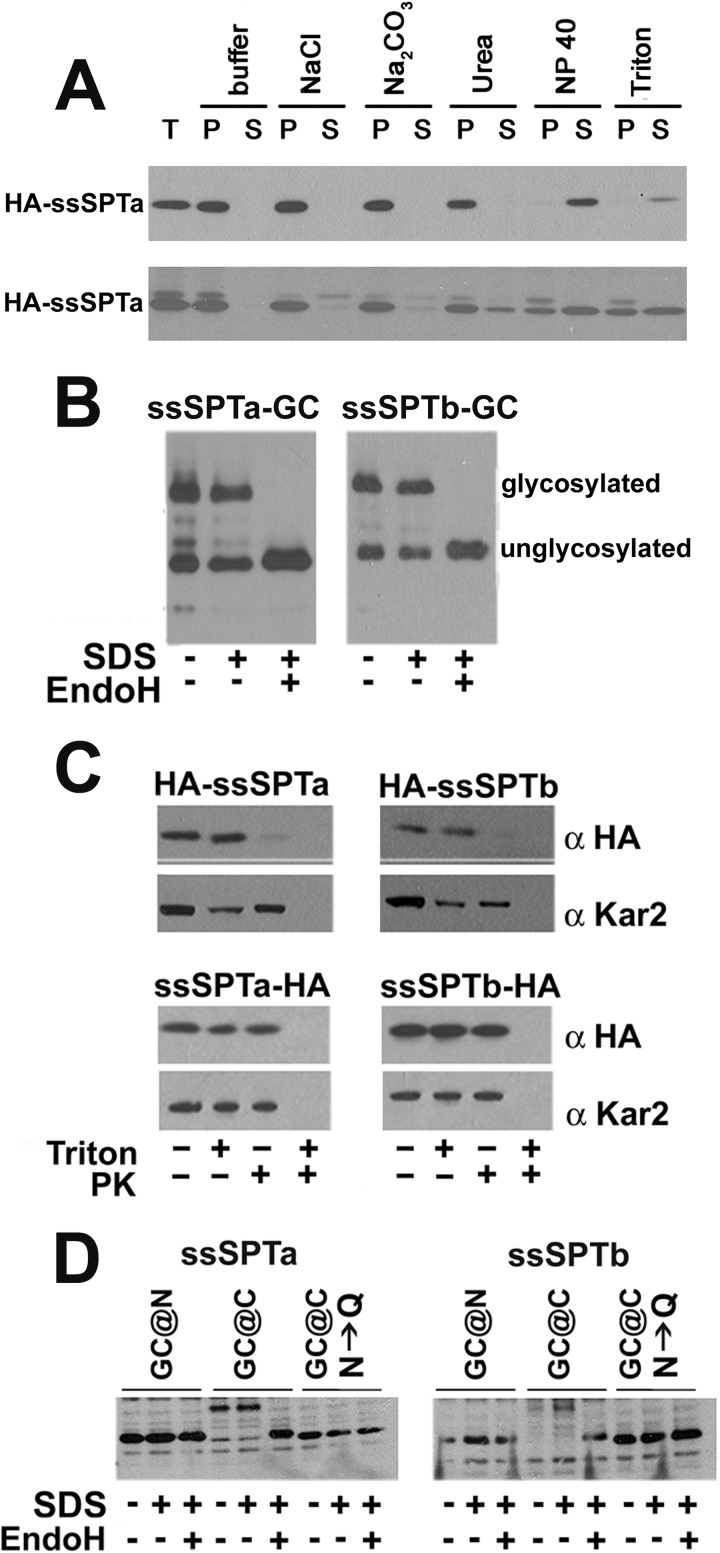
The ssSPTs behave like integral membrane proteins when coexpressed with hLCB1 and hLCB2a in both yeast (Fig. 2A, upper) and mammalian cells (Fig. 2A, lower); in either case, the proteins are solubilized with detergents, but not by salt, bicarbonate, or urea. In addition, a positive split ubiquitin two-hybrid interaction between Cub-ssSPTa or Cub-ssSPTb and hLCB1-Nub, whose C terminus is known to be cytoplasmic (24), demonstrated that their N termini are cytoplasmic (16). To directly address the location of the C terminus, a glycosylation cassette (GC) containing three consensus NX(S/T) glycosylation sites was appended to the C terminus of ssSPTa or ssSPTb and tagged proteins assessed for glycosylation based on their sensitivity to EndoH treatment. The increased electrophoretic mobility of both proteins following EndoH treatment (Fig. 2B) provides strong evidence that their C termini are in the ER lumen. In addition, limited proteolysis of rightside-out vesicles containing N- and C-terminally epitope-tagged ssSPTa or ssSPTb showed that the N termini were accessible to proteinase K in the absence of detergent (Fig. 2C, upper) whereas the C termini were protease-resistant unless the vesicles were detergent-solubilized (Fig. 2C, lower). These results indicate that the ssSPTs, when expressed in yeast, have an odd number of TMDs. To determine the topology of the ssSPTs in mammalian cells, N- and C-terminally GC-tagged ssSPTa or ssSPTb was expressed in CHO-Ly-B cells. EndoH treatment showed that the GC-ssSPTs were not glycosylated whereas the ssSPT-GCs were and that glycosylation was eliminated by mutating the three consensus glycosylation sites in the C-terminal GC from NXS/T to QXS/T (N→Q) (Fig. 2D). These results establish that, as in yeast, the N termini of the ssSPTs are cytoplasmic and the C termini luminal. Given these results and the length of the minimal activating domain (see below), we conclude that, as predicted for their invertebrate counterparts, the human ssSPTs contain a single TMD.
FIGURE 2.
The hssSPTs are integral ER membrane proteins with their N termini in the cytosol and C termini in the lumen. A, microsomes prepared from yeast (upper) or CHO-Ly-B cells (lower) expressing HA-hssSPTa, hLCB1 and hLCB2a were extracted on ice with an equal volume of TEGM or TEGM containing 1 m NaCl, 5 m urea, 0.2 m Na2CO3, 0.4% Nonidet P-40 or 2% Triton X-100 for 60 min. The samples were subjected to centrifugation at 100,000 × g for 30 min, and equal proportions of the supernatants and pellets were resolved by SDS-PAGE. HA-ssSPTa was detected by immunoblotting with anti-HA antibody. B, ssSPTa and ssSPTb containing an N-terminal HA tag and a C-terminal GC cassette were expressed in yeast along with hLCB1 and hLCB2a. Microsomal protein (12.5 μg with or without EndoH treatment) was resolved by SDS-PAGE, and ssSPTa was detected by immunoblotting. C, sealed microsomal vesicles were prepared from yeast expressing N- or C-terminally HA-tagged ssSPTa or ssSPTb, hLCB1, and hLCB2a. The microsomes were mock digested or digested with proteinase K (PK) in the absence or presence of Triton X-100. The luminal ER protein, Kar2p, served as a control for vesicle integrity. D, the N- or C-terminally GC-tagged ssSPTs were expressed along with hLCB1 and hLCB2a in CHO-Ly-B cells and assessed for glycosylation as described above. GC cassettes in which the asparagines in the consensus glycosylation sites were mutated to glutamines (N→Q) were used as controls.
Activity and Acyl-CoA Selectivity of the ssSPTs Reside in the Conserved Core
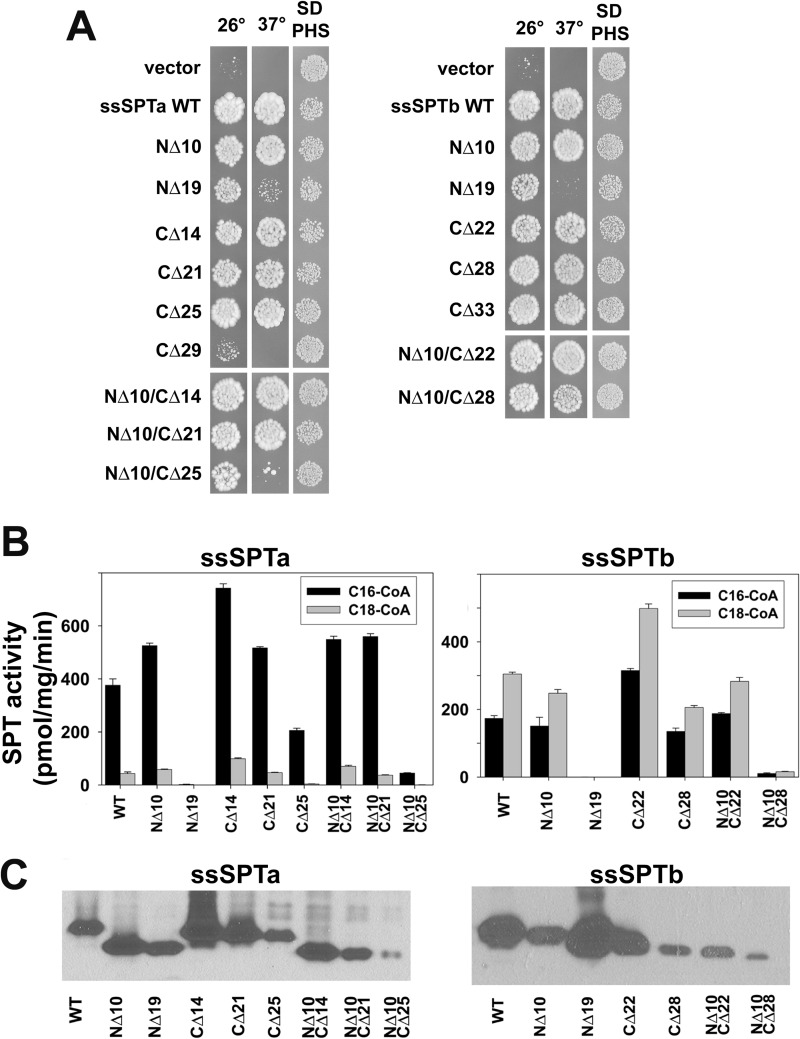
We have previously shown that the human ssSPTs confer distinct acyl-CoA preferences on the SPT heterotrimers (16). Whereas ssSPTa confers a preference for C16-CoA as substrate, heterotrimers containing ssSPTb use both C16- and C18-CoA. Inspection of the alignments in Fig. 1A show that the most highly conserved portion of the candidate members of the ssSPT family is located in a central domain, suggesting that the acyl-CoA preference is determined by the divergent N- or C-terminal domains of the proteins. To test this hypothesis, N- and C-terminal deletion mutants of ssSPTa and ssSPTb were generated and their acyl-CoA preferences determined. The results revealed that deletion of the N-terminal 10 amino acids of ssSPTa or ssSPTb had no effect on the ability of the proteins to activate hLCB1/hLCB2a heterodimers sufficiently to complement growth of yeast lacking endogenous SPT, even at 37 °C where the requirement for SPT activity is relatively high (Fig. 3A). In addition, deletion of the N-terminal 10 amino acids of ssSPTa or ssSPTb had no effect on in vitro SPT activity or acyl-CoA preference of heterotrimers (Fig. 3B). More extensive deletion of the first 19 amino acids of ssSPTa or ssSPTb yielded stable proteins (Fig. 3C) with activities too low to measure accurately, making it impossible to assess their acyl-CoA preference. Analogous results were obtained when C-terminal deletions were examined. As many as 21 residues from ssSPTa and 22 from ssSPTb could be removed without affecting SPT activity or altering their ability to confer acyl-CoA preference (Fig. 3B). Thus, neither the N nor C terminus of the ssSPTs is required for activity or specification of acyl-CoA preference. Interestingly, even a CΔ25 truncation in ssSPTa and a CΔ28 truncation in ssSPTb retained the ability to significantly activate the hLCB1/hLCB2a heterodimer (Fig. 3B) and were capable of supporting growth at both 26 °C and 37 °C (Fig. 3A), indicating that a core region of as few as 33 residues is functional.
FIGURE 3.
Identification of a functional core domain of the ssSPTs. A, Lcb1Δlcb2Δtsc3Δ mutant yeast expressing the indicated HA-ssSPTa (left) and HA-ssSPTb (right) mutant proteins along with hLCB1and hLCB2a were tested for growth at 26 °C or 37 °C or with PHS. B, SPT activities with either C16- or C18-CoA as substrate were measured using microsomes prepared from cells expressing the wild-type and N-, C-, or doubly truncated ssSPTa (left) or ssSPTb (right) proteins as described under “Experimental Procedures.” C, expression of the truncated HA-ssSPTa mutant (left) or HA-ssSPTb mutant (right) subunits was assessed by immunoblotting with anti-HA.
Because the longest functional C-terminal truncations extend into the distal predicted TMD (Fig. 1A), the data in Fig. 3 also support the conclusion that the ssSPTs contain a single TMD. To establish that the C-terminally truncated proteins span the membrane, rightside-out vesicles were prepared from yeast expressing HA-ssSPTaΔ21-Myc or HA-ssSPTaΔ25-Myc, and accessibility of the epitope tags to protease digestion was assessed. The results showed that the N-terminal HA tag was accessible to protease digestion whether or not the vesicles were detergent-solubilized, whereas digestion of the C-terminal Myc tag required detergent solubilization (Fig. 4A). The increased mobility seen in the anti-Myc immunoblots following digestion in the absence of detergent is consistent with the interpretation that the N-terminal portion of the protein extends into the cytoplasm. Additional evidence that the C termini of the truncated ssSPTs are luminal was obtained from analysis of truncated ssSPTs with GCs appended to their C termini; as was seen for the full-length ssSPTs (Fig. 2, B and D), the GCs on the truncated proteins were glycosylated (Fig. 4B). Because truncated ssSPTs lacking both the N and C termini were also stably expressed (Fig. 3C) and active (Fig. 3, A and B), a central core of 33 amino acids in ssSPTa and 38 amino acids in ssSPTb is sufficient to span the membrane, activate the heterodimer, and specify acyl-CoA preference. Thus, it is not the N or C terminus of the ssSPTs that determines acyl-CoA preference.
FIGURE 4.
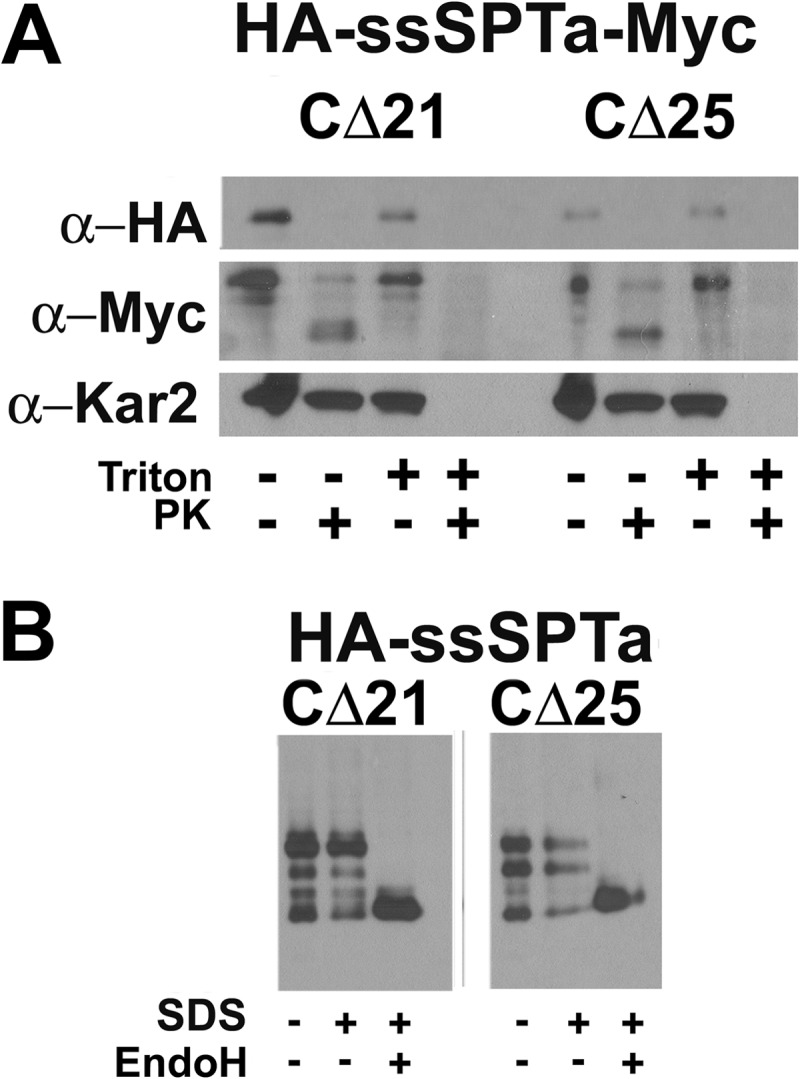
hssSPTs lacking the second predicted TMD retain their native membrane topology. A, sealed microsomal vesicles were prepared from yeast expressing hLCB1, hLCB2a, and N-terminally HA-tagged ssSPTa with either the C-terminal 21 or 25 amino acids replaced by a Myc tag. The microsomes were mock digested or digested with proteinase K (PK) in the absence or presence of Triton X-100. The luminal ER protein, Kar2p, served as a control for vesicle integrity. B, the C-terminal 21 or 25 amino acids of ssSPTa were replaced with a GC cassette. After expression in yeast, glycosylation was assessed as described in Fig. 2.
A Single Amino Acid in the Core Domain Determines Acyl-CoA Selectivity of ssSPTa and ssSPTb
The deletion studies demonstrated that the acyl-CoA preference of the ssSPTs resides in a central core. To map the acyl-CoA-specifying region more precisely, ssSPT chimera were constructed and tested for their ability to confer preference for C-16 and C18-CoAs. Accordingly, a chimera (aba) in which residues Glu27 to Pro54 of ssSPTa were replaced with residues Glu27 to Pro54 of ssSPTb or a chimera (ab) in which residues Glu27 to Gln68 of ssSPTa were replaced by residues Glu27 to Asn76 of ssSPTb was expressed in yeast, along with hLCB1 and hLCB2a. Microsomal SPT assays showed that both chimeric heterotrimers preferred palmitoyl-CoA as a substrate (Fig. 5). In combination with the analysis of the N-terminal deletions, this places the region responsible for the acyl-CoA selectivity of ssSPTa between residues Ser11 and Glu27. The conclusion that the acyl-CoA selectivity does not lie in the C-terminal region of ssSPTb was confirmed by analysis of a chimera (ba) in which residues Pro54 to Asn76 of ssSPTb were replaced by residues Pro54 to Gln68 of ssSPTa; like the full-length ssSPTb, this chimera conferred a preference for stearoyl-CoA (Fig. 5).
FIGURE 5.
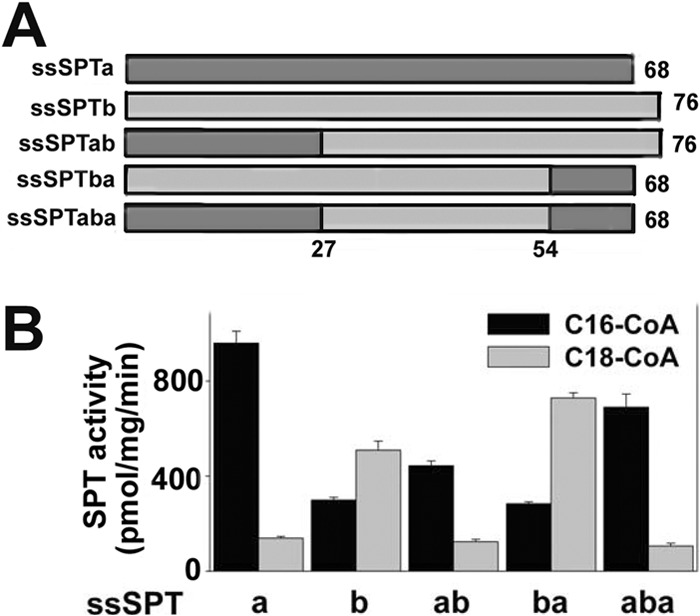
ssSPT chimera reveal that residues 11–26 specify acyl-CoA selectivity. A, schematic represents ssSPT chimera. B, microsomes were prepared from lcb1Δlcb2Δtsc3Δ mutant yeast expressing hLCB1, hLCB2a, and either ssSPTa (a), ssSPTb (b), a chimera that contains the first 27 residues of ssSPTa and the last 49 residues of ssSPTb (ab), a chimera that contains the first 54 residues of ssSPTb and the last 14 residues of ssSPTa (ba), or a chimera in which residues 27–54 of ssSPTa were replaced by residues 27–54 from ssSPTb (aba). SPT activities were measured using C16- and C18-CoA as substrate.
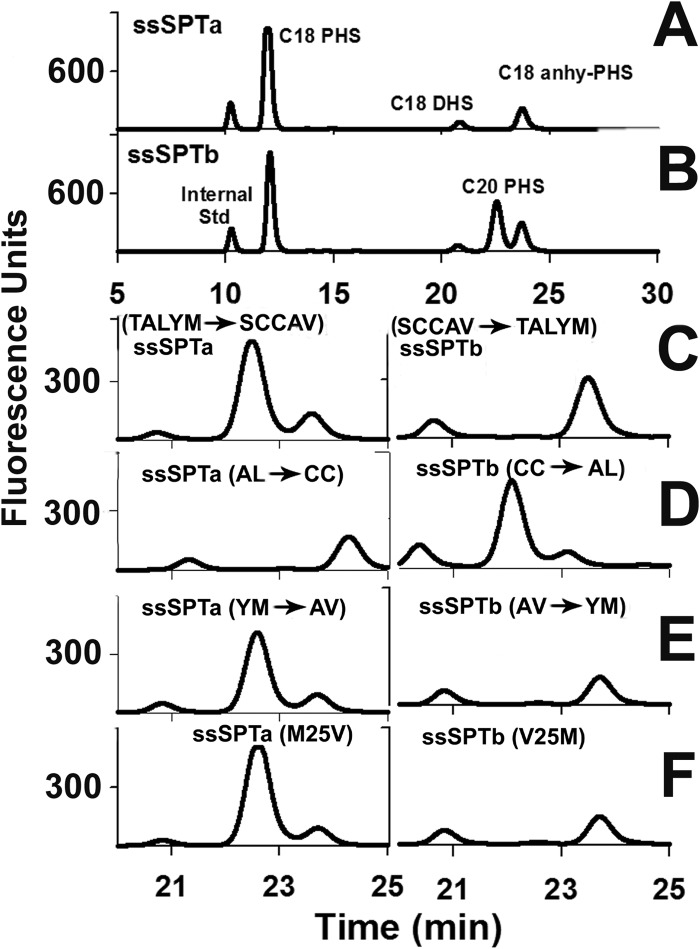
To identify precisely the residues responsible for conferring acyl-CoA selectivity, we focused on residues 18–25, all of which differ between ssSPTa and ssSTPb. Accordingly, a series of mutant ssSPTs in which nonconserved residues were exchanged between ssSPTa and ssSPTb was generated and expressed in yeast along with hLCB1 and hLCB2a. Extracts prepared from these cells were analyzed for the presence of C20-PHS (eluting at 22.5 min), the defining characteristic of heterotrimers containing ssSPTb (compare Fig. 6, A and B). Substitution of TALYM in ssSPTa with the corresponding residues (SCCAV) from ssSPTb resulted in a clear increase in the presence of C20-PHS (Fig. 6C, left). Conversely, the reciprocal substitution in which SCCAV was replaced with TALYM in ssSPTb eliminated the accumulation of C20-PHS (Fig. 6C, right). Thus, acyl-CoA selectivity is specified within this nonconserved pentapeptide. Swapping the second two amino acids (AL to CC or CC to AL) did not alter the amount of C20-PHS synthesized in cells expressing either mutant ssSPT isoform (Fig. 6D). However, swapping the last two amino acids (YM to AV or AV to YM) resulted in an increase in C20-PHS in cells expressing the mutant ssSPTa isoform and a decrease in C20-PHS in cells expressing the mutant ssSPTb isoform (Fig. 6E). Remarkably, acyl-CoA preference could be completely reversed by swapping only Met25 of ssSPTa with Val25 of ssSPTb; heterotrimers containing the ssSPTaM25V mutant synthesized C20-PHS at levels comparable with heterotrimers containing wild-type ssSPTb whereas heterotrimers containing the ssSPTbV25M mutant accumulated no C20-PHS (Fig. 6F). Thus, a single amino acid is responsible for the difference in acyl-CoA substrate preference of the two ssSPT isoforms.
FIGURE 6.
Residue 25 is responsible for the difference in acyl-CoA preference of ssSPTa and ssSPTb. Total LCBs prepared from lcb1Δlcb2Δtsc3Δ mutant yeast expressing hLCB1, hLCB2a and the indicated wild-type or mutant ssSPT subunit were analyzed by HPLC as described under “Experimental Procedures.” A and B, complete elution profiles containing all of the C18 and C20 LCBs. C–F, only the regions of the elution profiles containing C18-dihydrosphinganine (DHS), C20-PHS, and C18-anhydrous PHS since the C20-PHS, eluting at 22.5 min, is diagnostic for the presence of an active ssSPTb subunit.
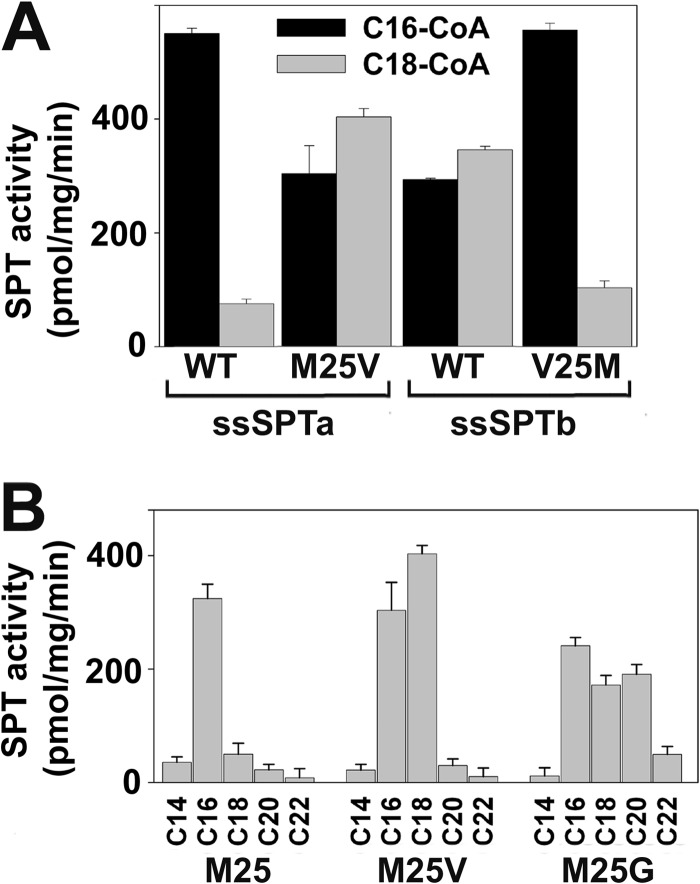
The effects of mutating residue 25 of the ssSPTs on acyl-CoA selectivity were also seen in in vitro SPT assays using microsomes from cells expressing the mutant subunits (Fig. 7A). One explanation for these results is that the size of the side chain helps to determine the selectivity of the heterotrimer for acyl-CoAs of various chain lengths. Indeed, this appears to be the case; a heterotrimer containing ssSPTa in which Met25 has been replaced by glycine is capable of efficiently condensing C20-CoA with serine (Fig. 7B).
FIGURE 7.
In vitro SPT activity and acyl-CoA selectivity of ssSPT residue 25 mutants. A, microsomes prepared from lcb1Δlcb2Δtsc3Δ mutant yeast cells expressing hLCB1, hLCB2a, and wild-type or mutant ssSPTs were assayed for SPT activity using either C16- or C18-CoA as described in Fig. 3. B, microsomes prepared from cells expressing hLCB1, hLCB2a, and the indicated ssSPTa mutants were assayed for SPT activity using C14-, C16-, C18-, C20- or C22-CoA as substrate.
The Single Amino Acid Residue Also Controls Acyl-CoA Selectivity in Mammalian Cells
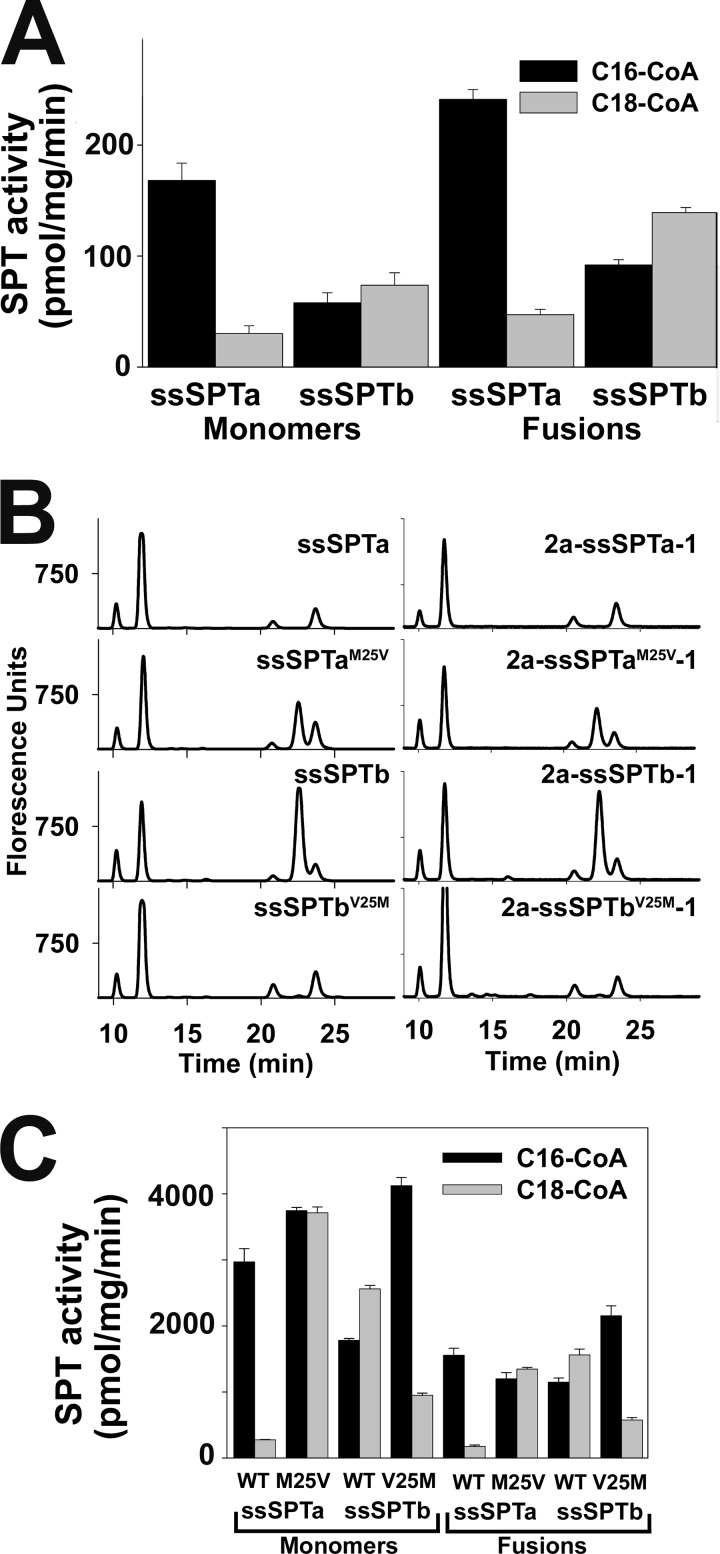
Single-chain heterotrimeric SPTs (hLCB2-ssSPT-hLCB1) have been used to study the effects of HSAN1 mutations (which increase the ability of SPT to condense alanine with acyl-CoAs thereby generating 1-deoxySA (26)) on amino acid substrate preference (21). The single-chain SPT results in stoichiometric expression of the three subunits and ensures that every heterodimer in transfected CHO-Ly-B cells, which lack endogenous hLCB1, contains the same wild-type or mutant isoform of ssSPT regardless of their endogenous levels of expression. To determine whether residue 25 of ssSPT also controls acyl-CoA preference in mammalian cells, single-chain fusions containing either ssSPTa or ssSPTb were constructed, tested in yeast for acyl-CoA selectivity, and then transfected into CHO-Ly-B cells. Microsomes prepared from yeast cells lacking endogenous SPT but expressing hLCB2a-ssSPT-hLCB1 fusion proteins had SPT activities comparable with those expressing the individual subunits (Fig. 8A). More importantly, the fusion SPTs displayed the same acyl-CoA preferences as did heterotrimers of the individual subunits in vitro (Fig. 8A) and in vivo (Fig. 8B). Furthermore, the single amino acid residues responsible for acyl-CoA preference in heterotrimers containing individual subunits also controlled preference in the fusion proteins (Fig. 8B). Not surprisingly, similar results were obtained when the fusion SPTs were expressed in CHO-Ly-B cells (Fig. 8C).
FIGURE 8.
Activity and acyl-CoA selectivity of monomeric and triple-fusion SPTs in yeast and CHO-Ly-B cells. A, the hLCB1 and hLCB2a genes were expressed in lcb1Δlcb2Δtsc3Δ yeast cells along with wild-type ssSPTa or ssSPTb as described under “Experimental Procedures.” Alternatively, single-chain hLCB2a-ssSPTa-hLCB1 or hLCB2a-ssSPTb-hLCB1 fusions were expressed in yeast cells to ensure stoichiometric expression of the three subunits. Microsomal SPT activities were measured as described under “Experimental Procedures” using either C16- or C18-CoA as substrate. B, monomeric subunits (left) or the triple fusions (right) containing the indicated ssSPTs were expressed in yeast. LCBs were prepared and analyzed by HPLC as described under “Experimental Procedures.” C, monomeric subunits or triple fusions containing the indicated ssSPTs were expressed in CHO-Ly-B cells, and microsomal SPT activities were determined as described under “Experimental Procedures.”
Residue 25 of the ssSPTs Also Influences Their Ability to Activate the hLCB1S331F/hLCB2 HSAN1 Mutant Heterodimer
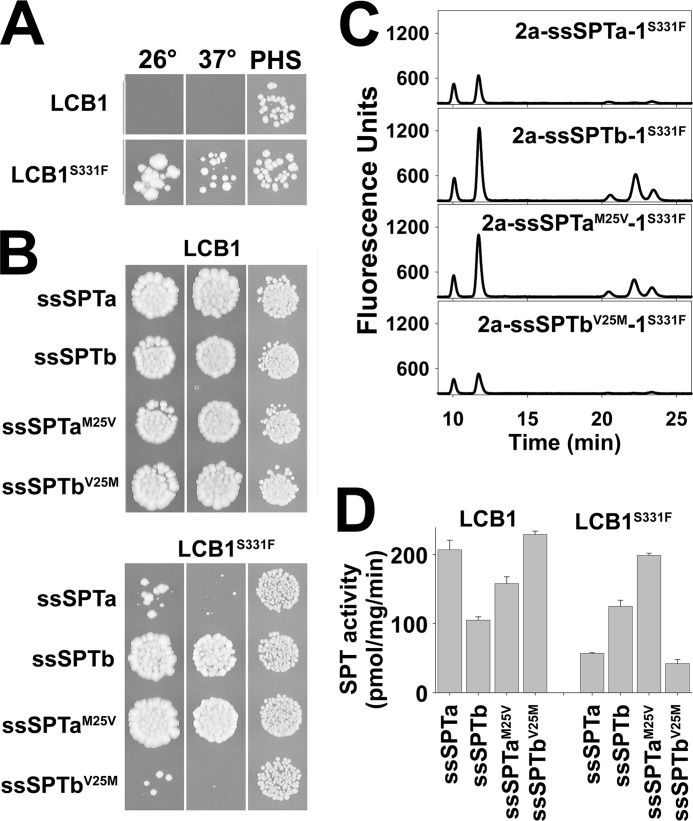
In the course of these studies, we identified a spontaneous mutation in hLCB1 (S331F) that results in elevated basal activity of the hLCB1/hLCB2a heterodimer. Whereas yeast lacking endogenous SPT and expressing the wild-type hLCB1/hLCB2a heterodimer fail to grow at 26 °C (Figs. 3A and 9A), the hLCB1S331F/hLCB2a heterodimer supports robust growth at 26 °C (Fig. 9A). In addition, in vitro enzyme assays revealed that the hLCB1S331F/hLCB2 heterodimer had severalfold higher SPT activity than the wild-type heterodimer (data not shown). It was therefore of interest to determine whether the mutant heterodimers would be responsive to the ssSPTs.
FIGURE 9.
A disease-causing mutation in hLCB1 results in decreased activation by ssSPTa but not by ssSPTb. A, the lcb1Δlcb2Δtsc3Δ mutant yeast cells expressing hLCB2a and wild-type or mutant (S331F) hLCB1 were grown on YPD medium at 26 °C or 37 °C, or on SD medium at 26 °C with PHS. B, yeast cells expressing triple fusion SPTs containing the indicated ssSPT were grown as in A. C, LCBs were prepared from yeast expressing the indicated triple fusion SPTs and analyzed by HPLC as in Fig. 8. D, microsomes were prepared from yeast expressing the indicated wild-type hLCB1- and hLCB1S331F-containing fusions, and SPT activity was measured using palmitoyl-CoA as described above.
To assess the ability of the ssSPT subunits to activate the mutant hLCB1S331F/hLCB2a heterodimer, various combinations of mutant and wild-type subunits were expressed in yeast as single-chain fusions. Not surprisingly, expression of hLCB2a-ssSPTb-hLCB1S331F allowed growth at both 26 °C and 37 °C, presumably reflecting the ability of ssSPTb to activate the mutant heterodimer (Fig. 9B). However, whereas ssSPTa activates the wild-type enzyme to a greater extent than ssSPTb (Fig. 9D and Ref. 16), the opposite is seen for the mutant; yeast expressing hLCB2a-ssSPTa-hLCB1S331F barely grow at 26 °C and fail to grow at 37 °C (Fig. 9B). Strikingly, introducing the Met to Val substitution into the ssSPTa component of hLCB2a-ssSPTa-hLCB1S331F increased the ability of the fusion SPT to support growth, whereas introduction of Val to Met in the ssSPTb component of hLCB2a-ssSPTb-hLCB1S331F resulted in decreased growth (Fig. 9B). These results were confirmed in vivo and in vitro by comparison of the LCB profiles (Fig. 9C) and microsomal SPT activities (Fig. 9D) from cells expressing the four single-chain fusion SPTs. Rotthier et al. have shown that the LCB1S331F mutation results in the human hereditary sensory neuropathy, HSAN1 (27). Thus, residue 25 of the ssSPTs also results in differential activation of heterodimers containing a disease-causing mutation. It seems unlikely that our identification of a spontaneous hLCB1 mutation that increases basal heterodimeric SPT activity and their identification of the same mutation in HSAN1 patients are simply coincidence. Indeed, the counterparts of two recently identified HSAN1 mutations in hLCB2a (G382A and I504F) (28) were originally identified in yeast Lcb2p based on their ability to elevate basal activity of the Lcb1p/Lcb2p heterodimer (29).
DISCUSSION
Recent studies from our laboratory and others have demonstrated that SPT is a far more complex enzyme than was previously appreciated. Contributing to this complexity is the presence of the ssSPTs, small proteins that increase basal activity nearly 100-fold and confer acyl-CoA preference (16). Bioinformatic analysis has revealed the presence of putative activating subunits across the entire eukaryotic evolutionary spectrum.3 These proteins show the greatest degree of conservation within a central domain of ∼40 amino acids (Fig. 1). The least conserved region of the vertebrate isoforms is in the C-terminal domain. However, within the subfamilies of the ssSPTa and ssSPTb isoforms, there is remarkable conservation across a wide range of species. This suggested that the C termini might specify acyl-CoA preference. The purpose of the present study was to identify the regions responsible for activation of the heterodimer and acyl-CoA selectivity and to characterize the topology of the ssSPTs.
Deletion analyses of both isoforms identified minimal activating domains of ∼35 amino acids. In contrast to the dispensable N- and C-terminal domains, the core domains are highly homologous to each other. It was therefore surprising that not only did the core domains contain the information necessary for activation of the heterodimer but they also contained the information necessary for specification of acyl-CoA preference. Even more surprising was the discovery that a single residue, Met25 in ssSPTa and Val25 in ssSPTb, dictates acyl-CoA selectivity. Like the C termini of the ssSPT isoforms, residue 25 is conserved in the respective ssSPT subfamilies. This would predict that the distinct acyl-CoA preference of the ssSPTa and ssSPTb isoforms is also conserved. The ability of heterotrimers containing the ssSPTaM25G mutant to utilize C20-CoA further suggests that the length of the preferred acyl-CoA substrate is inversely proportional to the size of the amino acid in this position of the ssSPTs. Consistent with this hypothesis, although there are two isoforms of ssSPT in Arabidopsis, both contain a methionine, and each confers a strong preference for palmitoyl-CoA. More importantly, mutation of the Met to Val resulted in a significantly increased ability to use stearoyl-CoA as substrate.4
It is remarkable that all information required for the function of the ssSPTs in yeast is contained within a 33-amino acid core. This raises the question of how the core, or even the full-length ssSPTs, is targeted to the membrane. In this regard, it is noteworthy that when the full-length protein is expressed in the absence of its heterotrimeric partners, a significant fraction of the protein is detergent-insoluble,5 suggesting that even the full-length protein requires association with the heterodimer for proper membrane insertion. We have previously shown that coexpression of Nub-ssSPT and hLCB1-Cub yields a positive split-ubiquitin two-hybrid interaction. Thus, we hypothesize that interaction of ssSPTs with LCB1 is sufficient for proper membrane insertion.
The data in Fig. 4 show that the longest C-terminal deletions retain their native topology; although exactly which residues comprise the TMD is unclear, given that the average membrane spanning domain contains ∼20 amino acids, the majority of the minimal activating core must reside within the membrane. Consequently, it is likely that the ssSPTs interact with one or more of the TMDs of LCB1. Topological analysis of yeast Lcb1p identified three TMDs, the most N-terminal of which can be deleted without altering targeting or reducing enzymatic activity (24). Similarly, the N-terminal TMD of mammalian LCB1 can be deleted without apparent functional consequence (24). Thus, it seems that the ssSPTs interact with one or more of the distal TMDs. Interestingly, a mutation in hLCB1, S331F, increases the basal activity of the heterodimer. Because this disease-causing mutation in mammalian SPT results in increased condensation of alanine with palmitoyl-CoA (30), it appears that the penalty for increasing SPT activity of the heterodimer through mutation in one of the catalytic subunits is loss of amino acid substrate specificity. This is consistent with the observation that mutations in hLCB2a corresponding to those we identified in Lcb2p that increased the activity of the yeast heterodimer also result in relaxed amino acid selectivity (28). We therefore hypothesize that a major role of the ssSPTs is to increase enzymatic activity while allowing the enzyme to maintain high amino acid selectivity.
Because of the decreased response of the hLCB1S331F-containing mutant heterodimer to ssSPTa but not ssSPTb or ssSPTa in which Met25 is replaced with valine, it is tempting to speculate that mutation of residue Ser331 of hLCB1 to the bulkier phenyalanine results in a steric clash with Met25 of ssSPTa. In addition, whereas hLCB1S331F-containing mutant heterotrimers synthesize 1-deoxySA when expressed in mammalian cells (30), unlike other HSAN1-causing mutations in hLCB1, such as C133W and V144D, heterotrimers containing hLCB1S331F do not synthesize 1-deoxySA in yeast (data not shown). One possible explanation for this difference is that the structural organization of mammalian SPT is not entirely recapitulated in yeast. Alternatively, there may be post-translational modifications in mammalian cells affecting the catalytic site that do not occur in yeast. This question will need to be resolved in future experiments.
The results presented here show that the N- and C-terminal domains are not essential for function when human heterotrimers are expressed in yeast. However, the high degree of conservation in the C-terminal domains of the ssSPTa and ssSPTb subfamilies is strongly suggestive of a regulatory role in higher eukaryotes. Experiments are in progress to determine whether this is the case.
Acknowledgments
We thank Edgar Cahoon, Dominic Campopiano, and Florian Eichler for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant R01NS072446. This work was also supported by Uniformed Services University Grants R071KD and CO75PI.
S. Majumder, J. M. Harmon, and T. M. Dunn, unpublished data.
A. N. Kimberlin, S. Majumder, M. Chen, G. Han, J. M. Stone, T. M. Dunn, and E. B. Cahoon, unpublished data.
S. D. Gupta, D. Bacikova, J. M. Harmon, and T. M. Dunn, unpublished data.
- SPT
- serine palmitoyltransferase
- 1-deoxySA
- 1-deoxysphinganine
- EndoH
- endoglycosidase H
- ER
- endoplasmic reticulum
- GC
- glycosylation cassette
- HSAN1
- hereditary sensory autonomic neuropathy type I
- LCB
- long chain base
- PHS
- phytosphingosine
- QC
- QuikChange
- ssSPT
- small subunit of SPT
- TMD
- transmembrane domain.
REFERENCES
- 1. Alexeev D., Alexeeva M., Baxter R. L., Campopiano D. J., Webster S. P., Sawyer L. (1998) The crystal structure of 8-amino-7-oxononanoate synthase: a bacterial PLP-dependent, acyl-CoA-condensing enzyme. J. Mol. Biol. 284, 401–419 [DOI] [PubMed] [Google Scholar]
- 2. Astner I., Schulze J. O., van den Heuvel J., Jahn D., Schubert W. D., Heinz D. W. (2005) Crystal structure of 5-aminolevulinate synthase, the first enzyme of heme biosynthesis, and its link to XLSA in humans. EMBO J. 24, 3166–3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schmidt A., Sivaraman J., Li Y., Larocque R., Barbosa J. A., Smith C., Matte A., Schrag J. D., Cygler M. (2001) Three-dimensional structure of 2-amino-3-ketobutyrate CoA ligase from Escherichia coli complexed with a PLP-substrate intermediate: inferred reaction mechanism. Biochemistry 40, 5151–5160 [DOI] [PubMed] [Google Scholar]
- 4. Ikushiro H., Hayashi H., Kagamiyama H. (2001) A water-soluble homodimeric serine palmitoyltransferase from Sphingomonas paucimobilis EY2395T strain: purification, characterization, cloning, and overproduction. J. Biol. Chem. 276, 18249–18256 [DOI] [PubMed] [Google Scholar]
- 5. Ikushiro H., Hayashi H., Kagamiyama H. (2003) Bacterial serine palmitoyltransferase: a water-soluble homodimeric prototype of the eukaryotic enzyme. Biochim. Biophys. Acta 1647, 116–120 [DOI] [PubMed] [Google Scholar]
- 6. Lowther J., Naismith J. H., Dunn T. M., Campopiano D. J. (2012) Structural, mechanistic and regulatory studies of serine palmitoyltransferase. Biochem. Soc. Trans. 40, 547–554 [DOI] [PubMed] [Google Scholar]
- 7. Raman M. C., Johnson K. A., Clarke D. J., Naismith J. H., Campopiano D. J. (2010) The serine palmitoyltransferase from Sphingomonas wittichii RW1: an interesting link to an unusual acyl carrier protein. Biopolymers 93, 811–822 [DOI] [PubMed] [Google Scholar]
- 8. Raman M. C., Johnson K. A., Yard B. A., Lowther J., Carter L. G., Naismith J. H., Campopiano D. J. (2009) The external aldimine form of serine palmitoyltransferase: structural, kinetic, and spectroscopic analysis of the wild-type enzyme and HSAN1 mutant mimics. J. Biol. Chem. 284, 17328–17339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yard B. A., Carter L. G., Johnson K. A., Overton I. M., Dorward M., Liu H., McMahon S. A., Oke M., Puech D., Barton G. J., Naismith J. H., Campopiano D. J. (2007) The structure of serine palmitoyltransferase: gateway to sphingolipid biosynthesis. J. Mol. Biol. 370, 870–886 [DOI] [PubMed] [Google Scholar]
- 10. Gable K., Han G., Monaghan E., Bacikova D., Natarajan M., Williams R., Dunn T. M. (2002) Mutations in the yeast LCB1 and LCB2 genes, including those corresponding to the hereditary sensory neuropathy type I mutations, dominantly inactivate serine palmitoyltransferase. J. Biol. Chem. 277, 10194–10200 [DOI] [PubMed] [Google Scholar]
- 11. Hanada K. (2003) Serine palmitoyltransferase, a key enzyme of sphingolipid metabolism. Biochim. Biophys. Acta 1632, 16–30 [DOI] [PubMed] [Google Scholar]
- 12. Zhao C., Beeler T., Dunn T. (1994) Suppressors of the Ca2+-sensitive yeast mutant (csg2) identify genes involved in sphingolipid biosynthesis: cloning and characterization of SCS1, a gene required for serine palmitoyltransferase activity. J. Biol. Chem. 269, 21480–21488 [PubMed] [Google Scholar]
- 13. Gable K., Slife H., Bacikova D., Monaghan E., Dunn T. M. (2000) Tsc3p is an 80-amino acid protein associated with serine palmitoyltransferase and required for optimal enzyme activity. J. Biol. Chem. 275, 7597–7603 [DOI] [PubMed] [Google Scholar]
- 14. Breslow D. K., Collins S. R., Bodenmiller B., Aebersold R., Simons K., Shevchenko A., Ejsing C. S., Weissman J. S. (2010) Orm family proteins mediate sphingolipid homeostasis. Nature 463, 1048–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Han S., Lone M. A., Schneiter R., Chang A. (2010) Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proc. Natl. Acad. Sci. U.S.A. 107, 5851–5856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Han G., Gupta S. D., Gable K., Niranjanakumari S., Moitra P., Eichler F., Brown R. H., Jr., Harmon J. M., Dunn T. M. (2009) Identification of small subunits of mammalian serine palmitoyltransferase that confer distinct acyl-CoA substrate specificities. Proc. Natl. Acad. Sci. U.S.A. 106, 8186–8191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Siow D. L., Wattenberg B. W. (2012) Mammalian ORMDL proteins mediate the feedback response in ceramide biosynthesis. J. Biol. Chem. 287, 40198–40204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hornemann T., Richard S., Rütti M. F., Wei Y., von Eckardstein A. (2006) Cloning and initial characterization of a new subunit for mammalian serine-palmitoyltransferase. J. Biol. Chem. 281, 37275–37281 [DOI] [PubMed] [Google Scholar]
- 19. Han G., Gable K., Yan L., Allen M. J., Wilson W. H., Moitra P., Harmon J. M., Dunn T. M. (2006) Expression of a novel marine viral single-chain serine palmitoyltransferase and construction of yeast and mammalian single-chain chimera. J. Biol. Chem. 281, 39935–39942 [DOI] [PubMed] [Google Scholar]
- 20. Wilson W. H., Schroeder D. C., Allen M. J., Holden M. T., Parkhill J., Barrell B. G., Churcher C., Hamlin N., Mungall K., Norbertczak H., Quail M. A., Price C., Rabbinowitsch E., Walker D., Craigon M., Roy D., Ghazal P. (2005) Complete genome sequence and lytic phase transcription profile of a Coccolithovirus. Science 309, 1090–1092 [DOI] [PubMed] [Google Scholar]
- 21. Gable K., Gupta S. D., Han G., Niranjanakumari S., Harmon J. M., Dunn T. M. (2010) A disease-causing mutation in the active site of serine palmitoyltransferase causes catalytic promiscuity. J. Biol. Chem. 285, 22846–22852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hanada K., Nishijima M. (2000) Selection of mammalian cell mutants in sphingolipid biosynthesis. Methods Enzymol. 312, 304–317 [DOI] [PubMed] [Google Scholar]
- 23. Momin A. A., Park H., Allegood J. C., Leipelt M., Kelly S. L., Merrill A. H., Jr., Hanada K. (2009) Characterization of mutant serine palmitoyltransferase 1 in LY-B cells. Lipids 44, 725–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Han G., Gable K., Yan L., Natarajan M., Krishnamurthy J., Gupta S. D., Borovitskaya A., Harmon J. M., Dunn T. M. (2004) The topology of the Lcb1p subunit of yeast serine palmitoyltransferase. J. Biol. Chem. 279, 53707–53716 [DOI] [PubMed] [Google Scholar]
- 25. Tsegaye Y., Richardson C. G., Bravo J. E., Mulcahy B. J., Lynch D. V., Markham J. E., Jaworski J. G., Chen M., Cahoon E. B., Dunn T. M. (2007) Arabidopsis mutants lacking long chain base phosphate lyase are fumonisin-sensitive and accumulate trihydroxy-18:1 long chain base phosphate. J. Biol. Chem. 282, 28195–28206 [DOI] [PubMed] [Google Scholar]
- 26. Penno A., Reilly M. M., Houlden H., Laurá M., Rentsch K., Niederkofler V., Stoeckli E. T., Nicholson G., Eichler F., Brown R. H., Jr., von Eckardstein A., Hornemann T. (2010) Hereditary sensory neuropathy type 1 is caused by the accumulation of two neurotoxic sphingolipids. J. Biol. Chem. 285, 11178–11187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rotthier A., Baets J., De Vriendt E., Jacobs A., Auer-Grumbach M., Lévy N., Bonello-Palot N., Kilic S. S., Weis J., Nascimento A., Swinkels M., Kruyt M. C., Jordanova A., De Jonghe P., Timmerman V. (2009) Genes for hereditary sensory and autonomic neuropathies: a genotype-phenotype correlation. Brain 132, 2699–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rotthier A., Auer-Grumbach M., Janssens K., Baets J., Penno A., Almeida-Souza L., Van Hoof K., Jacobs A., De Vriendt E., Schlotter-Weigel B., Loscher W., Vondráček P., Seeman P., De Jonghe P., Van Dijck P., Jordanova A., Hornemann T., Timmerman V. (2010) Mutations in the SPTLC2 subunit of serine palmitoyltransferase cause hereditary sensory and autonomic neuropathy type I. Am. J. Hum. Genet. 87, 513–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Monaghan E., Gable K., Dunn T. (2002) Mutations in the Lcb2p subunit of serine palmitoyltransferase eliminate the requirement for the TSC3 gene in Saccharomyces cerevisiae. Yeast 19, 659–670 [DOI] [PubMed] [Google Scholar]
- 30. Rotthier A., Penno A., Rautenstrauss B., Auer-Grumbach M., Stettner G. M., Asselbergh B., Van Hoof K., Sticht H., Lévy N., Timmerman V., Hornemann T., Janssens K. (2011) Characterization of two mutations in the SPTLC1 subunit of serine palmitoyltransferase associated with hereditary sensory and autonomic neuropathy type I. Hum. Mutat. 32, E2211–2225 [DOI] [PubMed] [Google Scholar]