Background: Bre1p is required for H2B ubiquitylation that promotes RNA polymerase II association with active genes, and hence transcription.
Results: The RING domain of Bre1p facilitates, but a non-RING domain represses, transcription and RNA polymerase II association with active genes.
Conclusion: Bre1p has a dominant-negative role in addition to its well known stimulatory function in transcription.
Significance: This study unravels a hidden role of Bre1p in transcriptional regulation.
Keywords: Chromatin Histone Modification, Chromatin Structure, Gene Regulation, RNA Polymerase II, Transcription, Bre1p, GAL1, Rad6p
Abstract
H2B ubiquitylation is carried out by Bre1p, an E3 ligase, along with an E2 conjugase, Rad6p. H2B ubiquitylation has been previously implicated in promoting the association of RNA polymerase II with the coding sequence of the active GAL1 gene, and hence transcriptional elongation. Intriguingly, we find here that the association of RNA polymerase II with the active GAL1 coding sequence is not decreased in Δbre1, although it is required for H2B ubiquitylation. In contrast, the loss of Rad6p significantly impairs the association of RNA polymerase II with GAL1. Likewise, the point mutation of lysine 123 (ubiquitylation site) to arginine of H2B (H2B-K123R) also lowers the association of RNA polymerase II with GAL1, consistent with the role of H2B ubiquitylation in promoting RNA polymerase II association. Surprisingly, unlike the Δrad6 and H2B-K123R strains, complete deletion of BRE1 does not impair the association of RNA polymerase II with GAL1. However, deletion of the RING domain of Bre1p (that is essential for H2B ubiquitylation) impairs RNA polymerase II association with GAL1. These results imply that a non-RING domain of Bre1p counteracts the stimulatory role of the RING domain in regulating the association of RNA polymerase II with GAL1, and hence RNA polymerase II occupancy is not impaired in Δbre1. Consistently, GAL1 transcription is impaired in the absence of the RING domain of Bre1p, but not in Δbre1. Similar results are also obtained at other genes. Collectively, our results implicate both the stimulatory and repressive roles of Bre1p in regulation of RNA polymerase II association with active genes (and hence transcription) in vivo.
Introduction
Covalent modifications of histones are strongly associated with transcription (1–7). One such modification is the monoubiquitylation of histone H2B at lysine 123 (Lys-123) in yeast (Lys-120 in mammals). Histone H2B ubiquitylation has been implicated in transcriptional elongation (8–13), and thus, the association of RNA polymerase II with the coding sequence of the active gene is significantly decreased in the absence of histone H2B ubiquitylation (8–12). A complex of three proteins, Rad6p (E2 ubiquitin conjugase), Bre1p (E3 ubiquitin ligase) and Lge1p, is essential for histone H2B Lys-123 ubiquitylation in Saccharomyces cerevisiae (2, 4, 5, 13–19). This complex shows a very specific subset of synthetically fitness or lethal defect interactions with the SWR1 chromatin remodeling complex (9) and Sin3p/Rpd3p histone deacetylase (18). Furthermore, synthetically fitness or lethal defect interaction profiles and DNA damage sensitivity of the Δbre1 and Δlge1 mutants are almost identical (18).
Hypersensitivity of yeast deletion mutants to a drug, brefeldin A (which inhibits growth and secretion in eukaryotes by blocking transport between endoplasmic reticulum and golgi complex), has led to the identification of Bre1p (20). Later on, a null mutation in BRE1 was also identified in a screen for deletion mutants, which are synthetically lethal with Δhtz1 (HTZ1 encodes H2A.Z in S. cerevisiae) (15). Wood et al. (16) have also identified Bre1p in a screen for histone H3 K4 methylation. These two later studies (15, 16) identified BRE1 as an E3 ligase for histone H2B ubiquitylation. Furthermore, Bre1p has been characterized to direct Rad6p for histone H2B ubiquitylation at Lys-123 to promote transcriptional elongation (15, 16, 19). Without Bre1p, Rad6p ubiquitylates all histones nonspecifically (19, 21–23). Although histone H2B ubiquitylation by Rad6p-Bre1p has been implicated to promote transcriptional elongation (8–13), it is also involved in activating the Rad53 kinase and cell cycle arrest (DNA damage checkpoint response), gene silencing, replication stress, and apoptosis (24–32).
Yeast Bre1p forms a homomeric complex through multiple intermolecular interactions (19). It contains a C3HC4 zinc finger RING (really interesting new gene) domain near its C terminus. The RING domain is an E3 signature motif and is essential for histone H2B ubiquitylation (19). In addition to the roles of Rad6p and Bre1p in histone H2B ubiquitylation, polymerase II-associated factor 1 (Paf1)2 complex (Paf1C) is also required for efficient histone H2B ubiquitylation (9, 33, 34). Paf1C that associates with elongating RNA polymerase II promotes the recruitment of Rad6p for efficient histone H2B ubiquitylation (9, 19). Furthermore, a recent study has demonstrated a direct interaction of Paf1C with Bre1p (19). Thus, RNA polymerase II-associated Paf1C plays an important role in promoting histone H2B ubiquitylation. Indeed, Shilatifard and colleagues (34) have demonstrated that Paf1C enhances histone H2B ubiquitylation activity of Rad6p-Bre1p.
As mentioned above, previous studies (8–13) have implicated histone H2B ubiquitylation in transcriptional elongation. Consistently, we have demonstrated that the absence of histone H2B ubiquitylation in the histone H2B-K123R point mutant strain significantly impaired the association of RNA polymerase II with the coding sequence of the GAL1 gene following transcriptional induction (8). Therefore, it is expected that deletion of BRE1 or RAD6 would also lower the association of RNA polymerase II with GAL1, similar to the results in the histone H2B-K123R point mutant strain. Indeed, we find that the loss of Rad6p significantly impairs the association of RNA polymerase II with the GAL1 coding sequence following transcriptional induction. Surprisingly, the deletion of BRE1 does not decrease the association of RNA polymerase II with GAL1, even though Bre1p is essential for histone H2B ubiquitylation. However, deletion of the RING domain of Bre1p significantly impairs the association of RNA polymerase II with GAL1, consistent with the role of histone H2B ubiquitylation in association of RNA polymerase II and hence transcription. Thus, a non-RING domain of Bre1p appears to suppress the association of RNA polymerase II and counteracts the stimulatory role of the RING domain in RNA polymerase II association. Hence, the association of RNA polymerase II with GAL1 is not impaired in the null mutation of BRE1. We also find similar results at several other genes such as GAL10, ADH1, and RPS5. Collectively, our results support both the stimulatory role (as expected based on previous studies; Refs. 8–13) as well as novel repressive function of Bre1p in regulation of RNA polymerase II association (and hence transcription) via its RING and non-RING domains. Thus, this study unravels a hidden function of Bre1p in gene regulation.
EXPERIMENTAL PROCEDURES
Plasmids
Plasmids pFA6a-13Myc-KanMX6 and pFA6a-3HA-His3MX6 (35) were used for genomic tagging of the proteins of interest by Myc and HA epitopes, respectively. The plasmid pRS406 (36) was used in the PCR-based gene disruption.
Strains
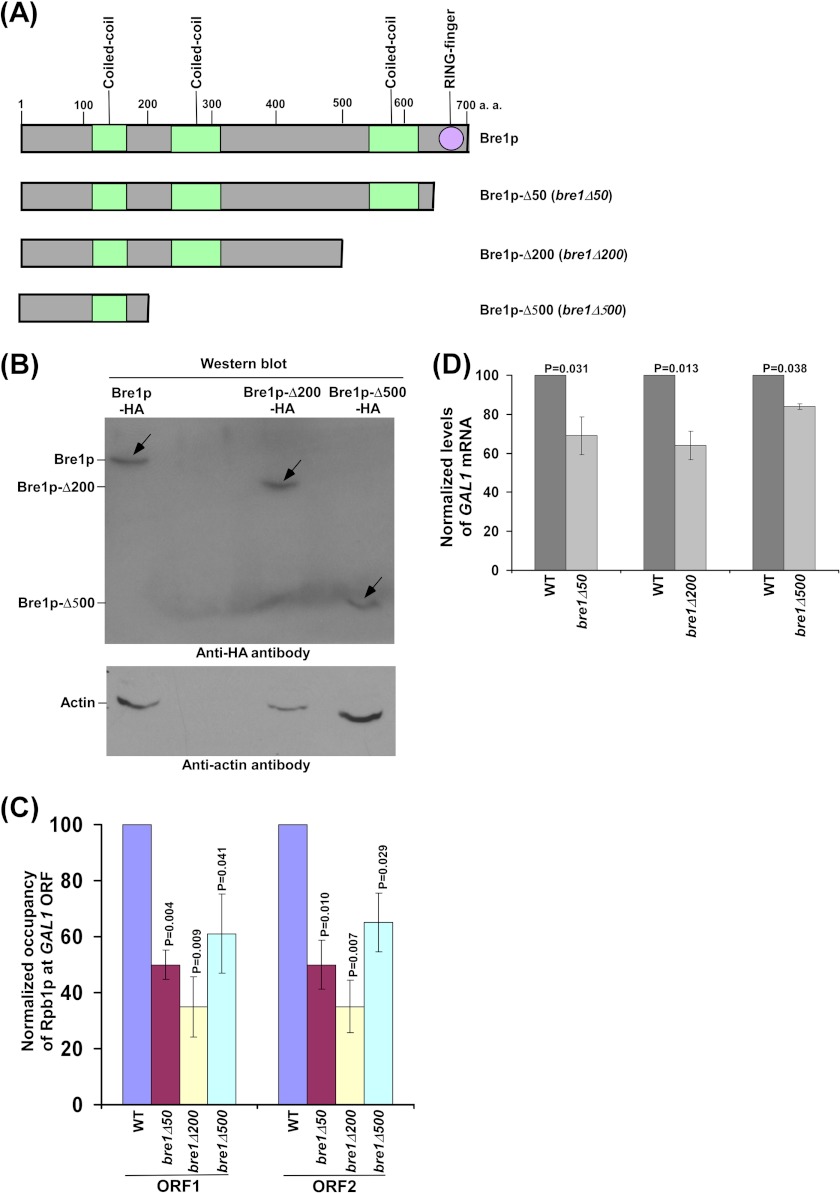
Yeast strain (S. cerevisiae) harboring point mutation in histone H2B at Lys-123 (YKH046) and its isogenic wild-type equivalent (YKH045) were obtained from the Osley laboratory (Mary Ann Osley, University of New Mexico Health Science Center) (31). The yeast strain bearing FLAG-tagged histone H2B (YTT31) was obtained from the Osley laboratory (37). The strain YTT31 was derived from JKM179 (FlagHTB1::LEU2 in JKM179) (38). The yeast strain harboring the null mutation of RAD6 (STY2; Δrad6 in FM392) and the wild-type equivalent (STY1; FM392) were obtained from the Shilatifard laboratory (Ali Shilatifard, Stowers Institute for Medical Research; purchased from Research Genetics). The PAF1 deletion mutant strain (DY7014) in the W303a background was obtained from the Stillman laboratory (David Stillman, University of Utah Health Sciences Center). Wild-type (BY4741) and Δbre1 strains were obtained from the Davie laboratory (Judith K. Davie, SIU School of Medicine; purchased from Open Biosystems). The strain, SLY1a, was generated by deleting the BRE1 gene in the YTT31 strain using the pRS406 plasmid. Multiple Myc epitope tags were added to the original chromosomal loci of DST1 and PAF1 in SLY1a to generate RSY20 and RSY17 strains, respectively, using the pFA6a-13Myc-KanMX6 plasmid. Similarly, multiple Myc epitope tags were added to the original chromosomal loci of DST1 and PAF1 in YTT31 to generate RSY19 and RSY16 strains, respectively, using the pFA6a-13Myc-KanMX6 plasmid. The HA epitope tags were added genomically to different locations toward the C-terminal of Bre1p using the pFA6a-3HA-KanMX6 plasmid in YTT31 to generate RSY12 (Bre1p), RSY13 (Bre1p-Δ50), RSY14 (Bre1p-Δ200), and RSY15 (Bre1p-Δ500) strains as schematically mentioned in Fig. 5A. Multiple Myc epitope tags were added to the original chromosomal locus of PAF1 in the RSY12 and RSY13 strains to generate RSY21 and RSY22 strains, respectively, using the pFA6a-13Myc-Trp1 plasmid. Likewise, multiple Myc epitope tags were added to the original chromosomal locus of DST1 in RSY13 to generate RSY24 strains.
FIGURE 5.
A non-RING domain of Bre1p represses the association of RNA polymerase II with GAL1. A, schematic diagram of different Bre1p mutants. B, Western blot analysis. C, analysis of RNA polymerase II association with GAL1 in different Bre1p mutants. Yeast strains (RSY12, RSY13, RSY14, and RSY15) were grown, cross-linked, and immunoprecipitated as described in the legend to Fig. 1B. The p values for the differences between wild-type and Bre1p mutants are presented on top of the histograms of the mutants. D, analysis of GAL1 transcription by RT-PCR. Yeast cells were grown as described in the legend to Fig. 1B.
Growth Media
The yeast strains were grown in YPR (yeast extract-peptone plus 2% raffinose) up to an A600 (optical density at 600 nm) of 0.9 at 30 °C, and then transferred to YPG (yeast extract, peptone plus 2% galactose) for 60 min to induce GAL1 prior to formaldehyde-based in vivo cross-linking or harvesting for mRNA analysis. The GAL10 gene was similarly induced. For studies at RPS5 and ADH1, yeast cells were grown in YPD (yeast extract, peptone plus 2% dextrose) up to an A600 of 1.0 at 30 °C prior to cross-linking or harvesting for mRNA analysis.
Chromatin Immunoprecipitation (ChIP) Assay
The ChIP assay was performed as described previously (39–43). Briefly, yeast cells were treated with 1% formaldehyde, collected, and resuspended in lysis buffer. Following sonication, cell lysate (400 μl of lysate from 50 ml of yeast culture) was precleared by centrifugation, and then 100 μl of lysate was used for each immunoprecipitation. Immunoprecipitated protein-DNA complexes were treated with proteinase K, the cross-links were reversed, and DNA was purified. Immunoprecipitated DNA was dissolved in 20 μl of TE 8.0 (10 mm Tris-HCl, pH 8.0, and 1 mm EDTA), and 1 μl of immunoprecipitated DNA was analyzed by PCR. PCR contained [α-32P]dATP (2.5 μCi for 25 μl of reaction), and the PCR products were detected by autoradiography after separation on a 6% polyacrylamide gel. As a control, “input” DNA was isolated from 5 μl of lysate without going through the immunoprecipitation step, and dissolved in 100 μl of TE 8.0. To compare the PCR signal arising from the immunoprecipitated DNA with the input DNA, 1 μl of input DNA was used in the PCR analysis.
For analysis of the recruitment of Dst1p and Paf1p, the above ChIP protocol was modified as described previously (43, 44). Briefly, a total of 800 μl of lysate was prepared from 100 ml of yeast culture. Following sonication, 400 μl of lysate was used for each immunoprecipitation (using 10 μl of anti-Myc antibody and 100 μl of protein A/G plus-agarose beads from Santa Cruz Biotechnology, Inc.), and the immunoprecipitated DNA sample was dissolved in 10 μl of TE 8.0 of which 1 μl was used for the PCR analysis. In parallel, the PCR analysis for input DNA was performed using 1 μl of DNA that was prepared by dissolving purified DNA from 5 μl of lysate in 100 μl of TE 8.0. The primer pairs used for PCR analysis were as follows: GAL1 (Core), 5′-ATAGGATGATAATGCGATTAGTTTTTTAGCCTT-3′ and 5′-GAAAATGTTGAAAGTATTAGTTAAAGTGGTTATGCA-3′; GAL1 (ORF1), 5′-CAGTGGATTGTCTTCTTCGGCCGC-3′ and 5′-GGCAGCCTGATCCATACCGCCATT-3′; GAL1 (ORF2), 5′-CAGAGGGCTAAGCATGTGTATTCT-3′ and 5′-GTCAATCTCTGGACAAGAACATTC-3′; GAL10 (ORF1), 5′-CTACGAGATTCCCAAATATGATTCC-3′ and 5′-TAACGCAAGATAGCAAACTTCCAAC-3′; GAL10 (ORF2), 5′-TTAATGCGAATCATAGTAGTATCGG-3′ and 5′-TTACCAATAGATCACCTGGAAATTC-3′; ADH1 (ORF), 5′-CTGGTTACACCCACGACGGTTCTT-3′ and 5′-GCAGACTTCAAAGCCTTGTAGACG-3′; RPS5 (ORF), 5′-AGGCTCAATGTCCAATCATTGAAAG-3′ and 5′-CAACAACTTGGATTGGGTTTTGGTC-3′. Autoradiograms were scanned and quantitated by NIH Image 1.62. Immunoprecipitated DNAs were quantitated as the ratio of immunoprecipitate to input in the autoradiogram (ORF, open reading frame; and core, core promoter). ORF1 and ORF2 represent two different locations in the ORF.
The ChIP experiments were carried out three times. These experiments are biologically independent. The average ChIP signal of these biologically independent experiments is reported with standard deviation (S.D.; Microsoft Excel 2003). The Student's t test of Microsoft Excel 2003 (with tail = 2 and types = 3) was used to determine the p values for statistical significance of the change in the ChIP signals. The changes were considered to be statistically significant at p < 0.05.
Total RNA Preparation
The total RNA was prepared from yeast cell culture as described by Peterson et al. (45). Briefly, 10 ml of yeast culture was harvested, and then suspended in 100 μl of RNA preparation buffer (500 mm NaCl, 200 mm Tris-HCl, 100 mm Na2EDTA, and 1% SDS) along with 100 μl of phenol/chloroform/isoamyl alcohol and a 100-μl volume-equivalent of glass beads (acid washed; Sigma). Subsequently, yeast cell suspension was vortexed with a maximum speed (10 in VWR mini-vortexer; catalog number 58816-121) 5 times (30 s each). Cell suspension was put in ice for 30 s between pulses. After vortexing, 150 μl of RNA preparation buffer and 150 μl of phenol/chloroform/isoamyl alcohol were added to yeast cell suspension followed by vortexing for 15 s with a maximum speed on VWR mini-vortexer. The aqueous phase was collected following a 5-min centrifugation at a maximum speed in a microcentrifuge. The total RNA was isolated from aqueous phase by ethanol precipitation.
Reverse Transcriptase-PCR (RT-PCR) Analysis
RT-PCR analysis was performed according to the standard protocols (46). Briefly, total RNA was prepared from 10 ml of yeast culture. Ten micrograms of total RNA was used in the reverse transcription assay for both wild-type and mutant strains. RNA was treated with RNase-free DNase (M610A, Promega) and then reverse-transcribed into cDNA using oligo(dT) as described in the protocol supplied by Promega (A3800, Promega). PCR was performed using synthesized first strand as template and the primer pairs targeted to GAL1, GAL10, ADH1, and RPS5 ORFs. RT-PCR products were separated by 2.2% agarose gel electrophoresis and visualized by ethidium bromide staining. The primer pairs used in the PCR analysis were as follows: GAL1, 5′-CAGAGGGCTAAGCATGTGTATTCT-3′, 5′-GTCAATCTCTGGACAAGAACATTC-3′; GAL10, 5′-TTAATGCGAATCATAGTAGTATCGG-3′, 5′-TTACCAATAGATCACCTGGAAATTC-3′; ADH1, 5′-CGGTAACAGAGCTGACACCAGAGA-3′, 5′-ACGTATCTACCAACGATTTGACCC-3′; RPS5, 5′-AGGCTCAATGTCCAATCATTGAAAG-3′, 5′-CAACAACTTGGATTGGGTTTTGGTC-3′. The RT-PCR experiments were carried out three times. These experiments are biologically independent. The average signal of these biologically independent experiments is reported with S.D. (Microsoft Excel 2003). The Student's t test (with tail = 2 and types = 3) was used to determine p values for statistical significance of the change in the RT-PCR signals. The changes were considered to be statistically significant at p < 0.05.
Whole Cell Extract Preparation and Western Blot Analysis
For analysis of the global levels of Bre1p mutants, yeast strains expressing the HA epitope-tagged proteins were grown in 25 ml of YPD up to an A600 of 1.0, and then harvested cells were lysed in 100 μl of lysis buffer to prepare the whole cell extract following the protocol as described for the ChIP assay (39–44). The whole cell extract (12.5 μl) was run on a SDS-polyacrylamide gel, and then analyzed by Western blot assay. The anti-HA (Santa Cruz Biotechnology, Inc.) antibody was used as a primary antibody in Western blot analysis. The level of Actin was monitored as a loading control using an anti-actin antibody (A2066; Sigma). For analysis of global levels of Paf1p in the wild-type and Bre1p null mutant strains, yeast strains expressing Myc epitope-tagged Paf1p were grown in 10 ml of YPR up to an A600 of 0.9, and then switched to YPG for 60 min at 30 °C prior to harvesting. The harvested cells were lysed in 100 μl of lysis buffer to prepare the whole cell extract. 10 μl of whole cell extract was used for Western blot analysis using an anti-Myc (9E10; Santa Cruz Biotechnology, Inc.) antibody. For analysis of histone H2B ubiquitylation, yeast cells were grown in YPD at 30 °C up to an A600 of 1.0. 5 ml of yeast culture was harvested, washed, and boiled with 70 μl of 1× SDS-polyacrylamide gel loading buffer for electrophoresis, as described previously (15). Western blot analysis was performed using an anti-FLAG antibody against FLAG-tagged histone H2B.
Growth Analysis following Genotoxic Attacks
The growth of the Δrad6, Δbre1, and wild-type cells was analyzed on plates containing solid YPD plus 0.026% MMS (methyl methanosulfonate; 129925–5G, Sigma) or 100 mm HU (hydroxy urea; H8627-5G; Sigma). Yeast cells were inoculated in YPD, and grown up to an A600 of 1.0 without dilution. Yeast cells were then spotted on solid growth media following serial dilutions. Yeast cells were grown at 30 °C, and photographed after 2 or 3 days.
RESULTS
Association of RNA Polymerase II with GAL1 Is Not Impaired in the Δbre1 Strain, whereas Histone H2B K123 Ubiquitylation or Rad6p Facilitates GAL1 Association of RNA Polymerase II
Previous studies have implicated histone H2B ubiquitylation in promoting RNA polymerase II to passage through nucleosomes at the active coding sequence, and hence transcriptional elongation (8, 10, 11). Thus, the association (or occupancy) of RNA polymerase II with the coding sequence of the active gene would be impaired in the absence of the enzymes (Rad6p and Bre1p) involved in this covalent modification. Indeed, we find that deletion of RAD6 significantly decreased the association of RNA polymerase II with the coding sequence of the GAL1 gene following transcriptional induction in galactose-containing growth medium (Fig. 1, A and B), consistent with the role of histone H2B ubiquitylation in transcriptional elongation (8–12). Similar to the Δrad6 strain, mutation of Lys-123 of histone H2B to arginine (H2B-K123R) also decreased the association of RNA polymerase II with GAL1 (Fig. 1C), consistent with previous studies (8, 10, 12). These results demonstrate that histone H2B ubiquitylation promotes the association of RNA polymerase II with GAL1. Consistently, histone H2B ubiquitylation has been shown to promote GAL1 transcription (8–10). Furthermore, recent genome-wide studies have also implicated histone H2B ubiquitylation in promoting transcription (11). Because Bre1p is essential for histone H2B ubiquitylation (15, 16, 19), the association of RNA polymerase II with GAL1 would also be impaired in the Δbre1 strain, similar to the results obtained in the Δrad6 and H2B-K123R mutant strains (Fig. 1, B and C). Surprisingly, we did not observe the decrease in RNA polymerase II association with the GAL1 coding sequence following transcriptional induction in the Δbre1 strain (Fig. 1D), even though it is essential for histone H2B ubiquitylation (15, 16, 19). Thus, our results (Figs. 1, B–D) demonstrate that the complete deletion of BRE1 does not decrease the association of RNA polymerase II with GAL1, whereas the absence of histone H2B ubiquitylation or E2 conjugase Rad6p significantly impairs RNA polymerase II association.
FIGURE 1.
Analysis of the association of RNA polymerase II with GAL1. A, a schematic diagram showing the locations of the primer pairs (ORF1 and ORF2) at the GAL1 coding sequence for the ChIP analysis. The numbers are presented with respect to the position of the first nucleotide of the initiation codon (+1). B, Rad6p promotes the association of RNA polymerase II with GAL1. Yeast strains (STY1 and STY2) were grown in YPR at 30 °C up to an A600 of 0.9, and then switched to YPG for 60 min prior to formaldehyde-based in vivo cross-linking. The ChIP assay was performed as described under ”Experimental Procedures.“ Immunoprecipitation was performed using 8WG16 antibody (Covance, Inc.) against the carboxyl-terminal domain of the largest subunit (Rpb1p) of RNA polymerase II. Immunoprecipitated DNA was analyzed by PCR using the primer pairs located at two different locations of the GAL1 coding sequence (ORF1 and ORF2). The ratio of the immunoprecipitate over the input in the autoradiogram (termed as a ChIP signal) was measured. The ChIP signal of the wild-type strain was set to 100, and the ChIP signal of the mutant strain was normalized with respect to 100 (represented as normalized or relative occupancy). Error bars denote S.D. from three sets of biologically independent experiments. p values were calculated using the Student's t test. C, association of RNA polymerase II with GAL1 is significantly decreased in the histone H2B-K123R point mutant strain. Yeast strains (YKH045 and YKH046) were grown, cross-linked, and immunoprecipitated as in panel B. D, association of RNA polymerase II with GAL1 is not impaired in the absence of Bre1p. Yeast strains (YTT31 and SLY1a) were grown, cross-linked, and immunoprecipitated as in panel B. E, analysis of the GAL1 transcription by RT-PCR. The wild-type and mutant strains were grown as in panel B. The transcript level of the wild-type strain was set to 100, and the mRNA level in the mutant strain was normalized with respect to 100. F and G, analysis of RNA polymerase II association with GAL1 and transcription in the commercial Δbre1 and isogenic wild-type (BY4741) strains (from Open Biosystems). Both wild-type and mutant strains were grown as in panel B. H, Western blot analysis for histone H2B ubiquitylation in the presence and absence of Bre1p or its RING domain, using an anti-FLAG antibody against FLAG-tagged histone H2B. The bre1Δ50 strain (RSY13) represents Bre1p without its RING domain. UbH2B, ubiquitylated histone H2B; and Flag-H2B, FLAG-tagged histone H2B. I, growth analysis of the Δrad6, Δbre1, and isogenic wild-type strains in solid YPD containing 100 mm HU or 0.026% MMS.
GAL1 Transcription Is Impaired in Δrad6 and H2B K123R Mutant Strains, but Not in the Δbre1 Strain
Because RNA polymerase II association is correlated with transcription, GAL1 transcription would be impaired in Δrad6 and H2B K123R mutant strains. To test this, we analyzed GAL1 mRNA levels in Δrad6 and H2B K123R mutant strains and their isogenic wild-type equivalents. Our RT-PCR analysis revealed that GAL1 transcription was significantly impaired in Δrad6 and H2B K123R mutant strains in comparison to wild-type equivalents (Fig. 1E). However, transcription of GAL1 was not impaired in the Δbre1 strain (Fig. 1E). These transcription results are nicely correlated with RNA polymerase II association with GAL1 in Δbre1, Δrad6, and H2B K123R mutant strains.
The Δbre1 strain used in our study might have contained some hidden mutation, which has reversed the defect in the RNA polymerase II association phenotype with GAL1. To address this issue, we also used a commercial Δbre1 strain and its isogenic wild-type equivalent (from Open Biosystems) for analysis of RNA polymerase II association with GAL1 and transcription. Using this commercial Δbre1 strain, we again found that association of RNA polymerase II with GAL1 was not impaired in the absence of Bre1p (Fig. 1F). Consistently, transcription of GAL1 was not altered in the commercial Δbre1 strain (Fig. 1G). Thus, the Δbre1 strain used in our study does not appear to contain a hidden mutation. Similar to our results, a recent study in S. cerevisiae also demonstrated that association of RNA polymerase II with GAL10 and PMA1 was not decreased in the Δbre1 strain (14).
As presented above, unlike Δrad6 and H2B K123R mutants, we do not observe an impairment of RNA polymerase II association with GAL1 (and hence transcription) in the Δbre1 strain in comparison to the wild-type equivalent. These observations raised the possibility that there may be another redundant histone H2B ubiquitin ligase. However, we rule out this possibility as previous studies (15, 16, 19) have demonstrated a dramatic impairment of histone H2B ubiquitylation in the Δbre1 strain. Consistently, we also do not observe histone H2B ubiquitylation in the absence of Bre1p or its RING domain (Fig. 1H). Furthermore, similar to the Δrad6 strain, the Δbre1 strain shows sensitivity to genotoxic agents such as MMS and HU (Fig. 1I), consistent with previous studies (16, 25, 47).
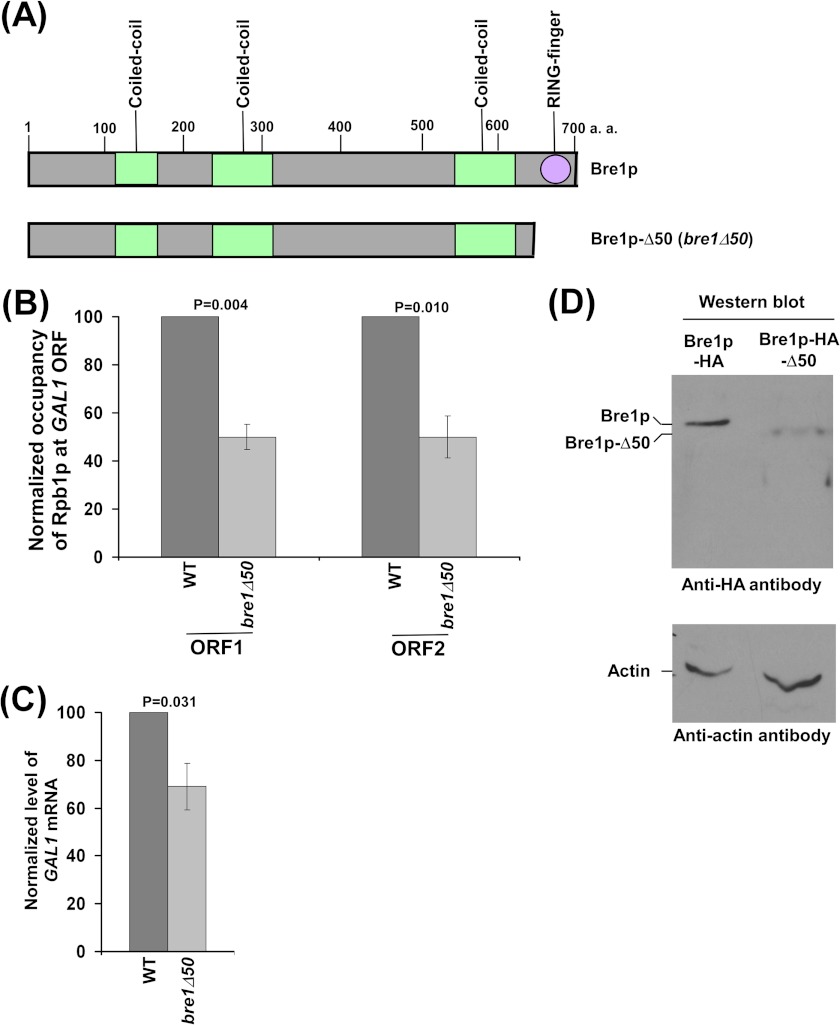
The RING Domain of Bre1p Promotes RNA Polymerase II Association with GAL1 and hence Transcription
Why does the null mutation of BRE1 not show an impaired association of RNA polymerase II with GAL1 (and hence transcription), similar to the results in the Δrad6 and H2B-K123R mutant strains? It is likely that the RING domain of Bre1p (that is essential for histone H2B ubiquitylation; Ref. 19) promotes RNA polymerase II association via histone H2B ubiquitylation, like E2 conjugase Rad6p, and a non-RING domain is involved in repression of the association of RNA polymerase II. Hence, the decrease in association of RNA polymerase II in the Δbre1 strain was not observed due to counteracting of the stimulatory role of the RING domain by the repressive role of a non-RING domain of Bre1p. To test this possibility, we deleted the RING domain of Bre1p (Fig. 2A) following HA-epitope tagging at the genomic locus of BRE1, and then analyzed the effect of such a deletion on RNA polymerase II association with GAL1. Interestingly, we find that the association of RNA polymerase II with the GAL1 coding sequence was significantly decreased in the absence of the RING domain of Bre1p (Fig. 2B). The RING domain is essential for histone H2B ubiquitylation (19) (Fig. 1H). Thus, histone H2B ubiquitylation activity of Bre1p is essential to promote the association of RNA polymerase II with GAL1, similar to the RNA polymerase II association phenotype in Δrad6 and histone H2B-K123R mutant strains (Fig. 1, B and C). Consistently, transcription of GAL1 was significantly impaired in the absence of the RING domain of Bre1p (Fig. 2C). Furthermore, deletion of the RING domain does not alter the interaction of Bre1p with Rad6p (19). However, Bre1p without the RING domain is relatively less stable (Fig. 2D), but does not generate the phenotypes of the Δbre1 strain. Thus, even at the lower level, the Bre1p mutant without the RING domain exhibits a repressive function. Consistently, it has been demonstrated previously that the global level of a protein may not be directly correlated with its targeted function. For example, TATA-box binding protein is not recruited to the GAL1 core promoter in the absence of Gal4p, whereas the global level of TATA-box binding protein is not altered in Δgal4 (39, 42). Similarly, Tra1p is not recruited to the GAL1 upstream activating sequence in the absence of Spt20p, whereas its global level is not altered in Δspt20 (42). Deletion of RING and the adjacent coiled-coil domain does not impair the stability of the mutant (see below; Fig. 5B) and lowers the association of RNA polymerase II and transcription (see below; Fig. 5, C and D), similar to the Δrad6 and H2B K123R mutant strains. Together, these results support that like Rad6p, the RING finger with/without the adjacent coiled-coil domain of Bre1p (or its histone H2B ubiquitylation activity; Ref. 19) (Fig. 1H) is essential for promoting the association of RNA polymerase II with the active GAL1 coding sequence. Interestingly, deletion of the whole BRE1 reverses the defect in RNA polymerase II association caused by the removal of the RING finger of Bre1p (Figs. 1D and 2B) and the adjacent coiled-coil domain (see below; Fig. 5C). These results support that a domain within the non-RING domain at the N-terminal of Bre1p is involved in repression of RNA polymerase II association. As a result of the two opposing effects of the RING and non-RING domains of Bre1p, we do not observe the decrease in RNA polymerase II association with GAL1 (and hence transcription) in the null mutant of BRE1. However, the Δbre1 strain shows MMS and HU sensitivity, similar to the Δrad6 strain (Fig. 1I). MMS causes DNA damage, whereas HU generates replication stress. Thus, the repressive role of Bre1p appears to be restricted to transcription, but not DNA repair or replication. Likewise, a recent study in S. cerevisiae also showed that the association of RNA polymerase II with GAL10 and PMA1 was not decreased in the null mutant of BRE1 in comparison to the wild-type equivalent (14). On the other hand, the association of RNA polymerase II with these genes was significantly impaired in the H2B-K123R point mutant strain (14). Therefore, similar to our data, the results of a recent study (14) also implicated the repressive role of Bre1p in association of RNA polymerase II with the active gene.
FIGURE 2.
The RING domain of Bre1p promotes the association of RNA polymerase II with GAL1. A, schematic diagram of the Bre1p mutant without the RING domain. B, analysis of RNA polymerase II association with GAL1 in the presence and absence of the RING domain of Bre1p. Yeast strains (RSY12 and RSY13) were grown, cross-linked, and immunoprecipitated as described in the legend to Fig. 1B. C, analysis of GAL1 transcription by RT-PCR. Yeast cells were grown as described in the legend to Fig. 1B. D, Western blot analysis.
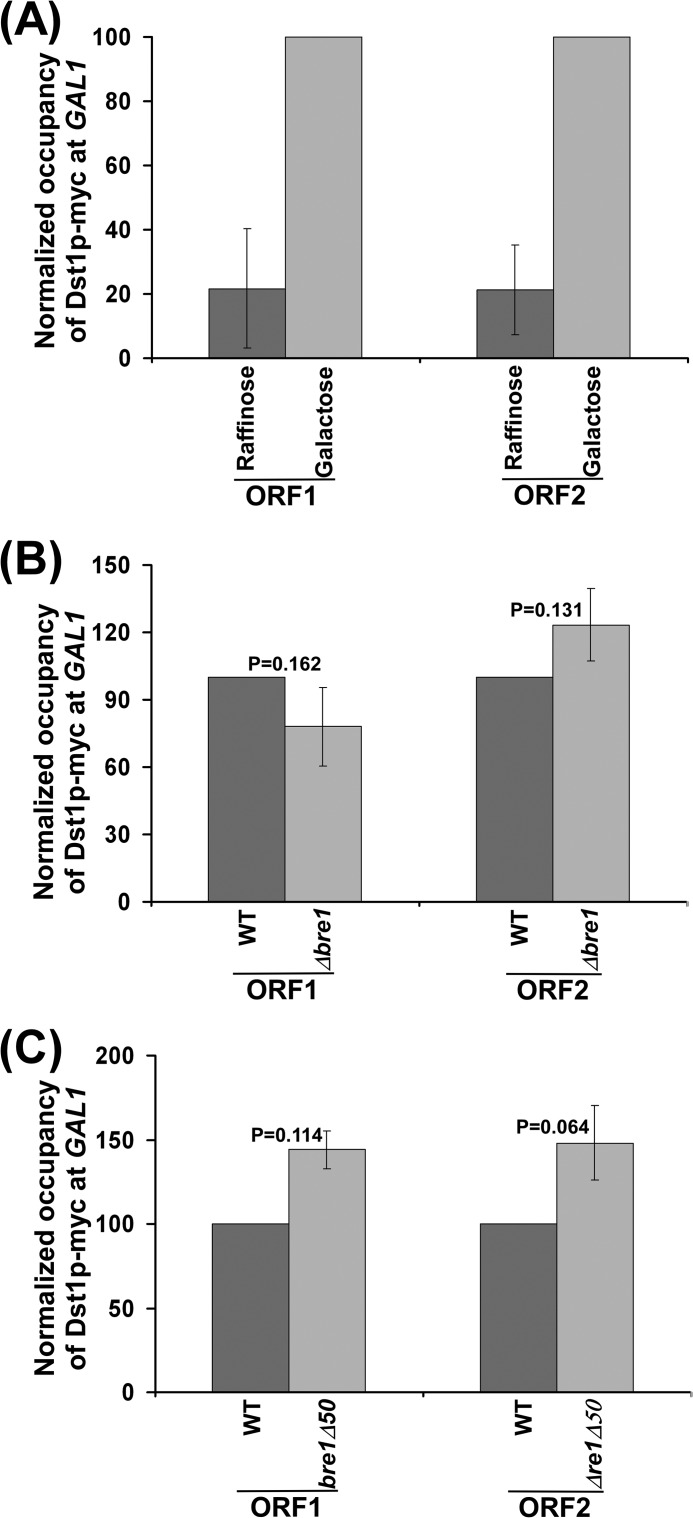
Bre1p or Its RING Domain Does Not Regulate the Recruitment of TFIIS to GAL1
How does Bre1p exert its repressive role in association of RNA polymerase II? A recent study (48) in mammalian cells demonstrated a set of genes whose expression was repressed by RNF20 (human paralog of yeast Bre1p). This study further demonstrates that such suppression is mediated via an impaired recruitment of TFIIS that promotes transcription (48). To test whether Bre1p exerts its repressive role in association of RNA polymerase II by inhibiting the recruitment of TFIIS in yeast, we analyzed the association of TFIIS to GAL1 in the presence and absence of Bre1p. In this direction, we tagged TFIIS (DST1) by the Myc epitope in wild-type and Δbre1 mutant strains. Using these strains, we analyzed the recruitment of TFIIS to GAL1 following transcriptional induction. We find that recruitment of TFIIS to GAL1 was not significantly altered in the absence of Bre1p (Fig. 3, A and B). Similarly, deletion of the RING domain of Bre1p did not impair the recruitment of TFIIS (Fig. 3C). Thus, Bre1p does not appear to function through TFIIS in yeast.
FIGURE 3.
Association of TFIIS (Dst1p) with GAL1 is not altered in the absence of Bre1p or its RING domain. A, analysis of the association of TFIIS with GAL1 under inducible (galactose) and non-inducible (raffinose) conditions. Yeast strain expressing Myc-tagged Dst1p (RSY19) was grown at 30 °C in YPR up to an A600 of 0.9 prior to formaldehyde-based in vivo cross-linking. For GAL1 induction, a yeast strain (RSY19) was grown at 30 °C in YPR up to A600 of 0.9, and then transferred to YPG for 60 min prior to formaldehyde-based in vivo cross-linking. The ChIP assay was performed following the modified protocol as described under ”Experimental Procedures.“ Immunoprecipitation was carried out using an anti-Myc antibody (9E10; Santa Cruz Biotechnology, Inc.) against Myc-tagged Dst1p. The ChIP signals at ORF1 and ORF2 in galactose (or YPG) were set to 100. The ChIP signals at ORF1 and ORF2 in raffinose (or YPR) were normalized with respect to 100 (represented as normalized or relative occupancy). Error bars in YPR denote S.D. from three sets of biologically independent experiments. B, analysis of the association of TFIIS with GAL1 in the presence and absence of Bre1p. Both the wild-type and Δbre1 strains expressing Myc-tagged Dst1p (RSY19 and RSY20) were grown and cross-linked as described in the legend to Fig. 1B. Immunoprecipitation was performed as in panel A. C, analysis of the association of TFIIS with GAL1 in the presence and absence of the RING domain of Bre1p. Yeast cells were grown, cross-linked, and immunoprecipitated as in panel B.
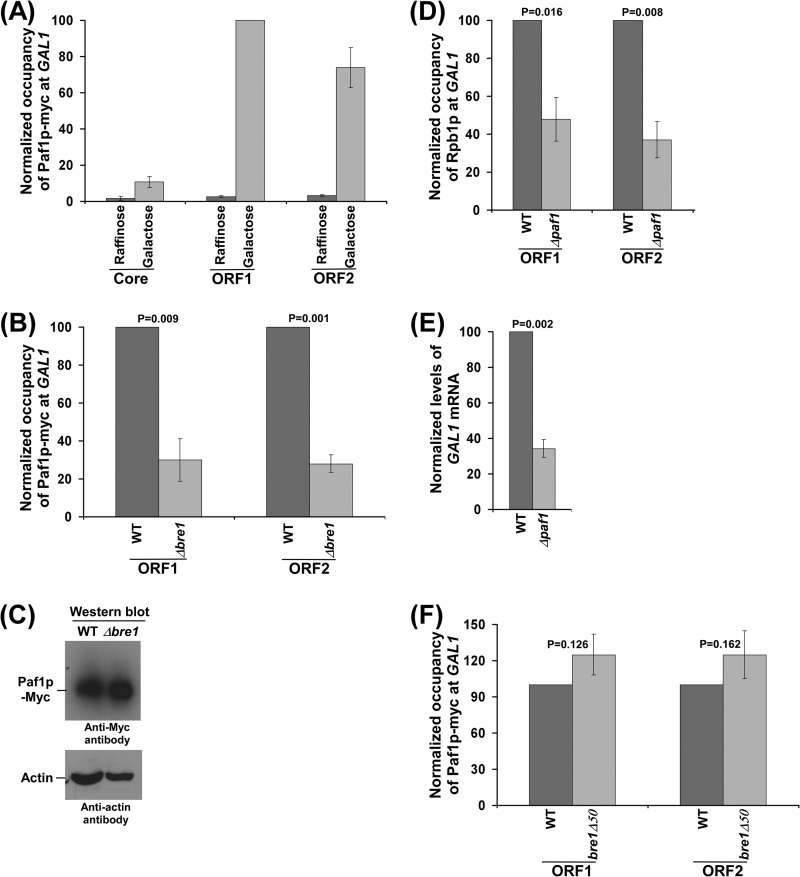
Bre1p Promotes the Recruitment of Paf1C to GAL1
Because the role of Bre1p in regulation of RNA polymerase II association is not mediated via TFIIS, it may be likely that the non-RING domain of Bre1p interacts with other factors that regulate the association of RNA polymerase II. The non-RING domain of Bre1p has been recently shown to interact with Rad6p (19), and such an interaction is essential for histone H2B ubiquitylation in conjunction with the RING domain (19, 34). Thus, deletion of RAD6 or the RING domain of Bre1p significantly impaired the association of RNA polymerase II (Figs. 1B and 2B; see below, Fig. 5C), via the loss of histone H2B ubiquitylation (19). Recently, Kim and Roeder (19) demonstrated that Bre1p interacts with Paf1C. Paf1C associates with elongating RNA polymerase II, and promotes transcriptional elongation (33, 49–53). The Paf1C has also been previously implicated in histone H2B ubiquitylation (9, 33, 34). Therefore, there is a link between the general transcription machinery and histone H2B ubiquitylation. Interestingly, we find here that the recruitment of Paf1C (Paf1p) to GAL1 was significantly decreased in the Δbre1 strain (Fig. 4, A and B). Such a decrease in the recruitment of Paf1C in the absence of Bre1p was not due to an altered stability of Paf1p in the absence of Bre1p as evident from Western blot analysis (Fig. 4C). Thus, our results support that Bre1p promotes the recruitment of Paf1C. It is important to note here that association of RNA polymerase II with GAL1 in the Δbre1 strain was not decreased (Fig. 1D), but recruitment of RNA polymerase II-associated Paf1C was significantly impaired in the absence of Bre1p (Fig. 4B). Thus, the association of Paf1C with the active gene depends on Bre1p in addition to RNA polymerase II. This is further substantiated by the fact that Bre1p interacts with Paf1C (19). Therefore, our results demonstrate the function of Bre1p in targeting the recruitment of Paf1C in addition to the role of RNA polymerase II in vivo.
FIGURE 4.
Analysis of Paf1p recruitment to GAL1 in the presence and absence of Bre1p. A, analysis of Paf1p association with GAL1 under inducible and non-inducible conditions. Wild-type yeast strain expressing Myc-tagged Paf1p (RSY16) was grown, cross-linked, and immunoprecipitated as described in the legend to Fig. 3A. The maximum ChIP signal was set to 100, and other ChIP signals were normalized with respect to 100 (represented as normalized or relative occupancy). Error bars denote S.D. from three sets of biologically independent experiments. B, association of Paf1p with GAL1 is significantly decreased in the Δbre1 strain. Both the wild-type and Δbre1 strains expressing Myc-tagged Paf1p (RSY16 and RSY17) were grown, cross-linked, and immunoprecipitated as in Fig. 3B. C, Western blot analysis. Yeast cells were grown as described in the legend to Fig. 1B. D, Paf1p promotes the association of RNA polymerase II with GAL1. Both the wild-type and Δpaf1 strains (W303a and DY7014) were grown, cross-linked, and immunoprecipitated as described in the legend to Fig. 1B. E, analysis of GAL1 transcription in the Δpaf1 strain by RT-PCR. Yeast cells were grown as described in the legend to Fig. 1B. F, the RING domain of Bre1p does not regulate the recruitment of Paf1C to GAL1. Both the wild-type and mutant strains expressing Myc-tagged Paf1p (RSY21 and RSY22) were grown and cross-linked as described in the legend to Fig. 1B. Immunoprecipitation was performed as in panel A.
Paf1C Promotes RNA Polymerase II Association with GAL1, and hence Transcription
Our results demonstrate that Bre1p promotes the recruitment of Paf1C. Previous studies have demonstrated that Paf1C is required for recruitment of the E2 conjugase Rad6p (9, 19). Therefore, Paf1C has been implicated in histone H2B ubiquitylation (9, 33, 34). Hence, deletion of PAF1 would decrease the association of RNA polymerase II via an impaired histone H2B ubiquitylation. To test this, we analyzed the association of RNA polymerase II with the GAL1 coding sequence in the Δpaf1 and wild-type strains. As expected, we find that association of RNA polymerase II with GAL1 was significantly decreased in the Δpaf1 strain (Fig. 4D). Consistently, GAL1 transcription was impaired in the Δpaf1 strain (Fig. 4E).
The RING Domain of Bre1p Is Dispensable for Recruitment of Paf1C to GAL1
We observe that deletion of Bre1p significantly impairs the recruitment of Paf1C to GAL1. We next analyzed whether the RING domain of Bre1p plays a role in targeting Paf1C to GAL1. We find that recruitment of Paf1C was not altered in the absence of the RING domain of Bre1p (Fig. 4F). Thus, the non-RING domain of Bre1p facilitates the recruitment of Paf1C. Hence, the non-RING domain of Bre1p appears to interact with Paf1C and promotes RNA polymerase II association. Therefore, the non-RING domain of Bre1p has a stimulatory role in addition to its repressive role in controlling the association of RNA polymerase II.
Functional Analysis of Different Domains of Bre1p in Association of RNA Polymerase II with GAL1
To determine the region within the non-RING domain of Bre1p involved in repression of RNA polymerase II association, we generated two Bre1p constructs containing first 200 or 500 amino acids at the N-terminal (Fig. 5A) following HA epitope tagging at the genomic locus of BRE1. These Bre1p mutants with 200 or 500 amino acids at the N-terminal were found to be stable in the Western blot analysis (Fig. 5B). Using these mutants, we analyzed the association of RNA polymerase II with the GAL1 coding sequence in conjunction with the RING-deficient Bre1p mutant and wild-type strains. We find that like the RING-deficient Bre1p mutant, Bre1p with 200 or 500 amino acids at the N-terminal also significantly reduced the association of RNA polymerase II with GAL1 (Fig. 5C). Consistently, GAL1 transcription was significantly impaired in these mutants (Fig. 5D). However, deletion of the whole BRE1 reversed the defect of RNA polymerase II association with GAL1 (and hence transcription), caused by the loss of the RING domain or deletion of 200/500 amino acids at the C-terminal of Bre1p (Figs. 1, D and E, and 5, C and D). These results support that the first ∼200 amino acids at the N-terminal of Bre1p exhibits the repressive role in association of RNA polymerase II with the active gene, and thus transcription.
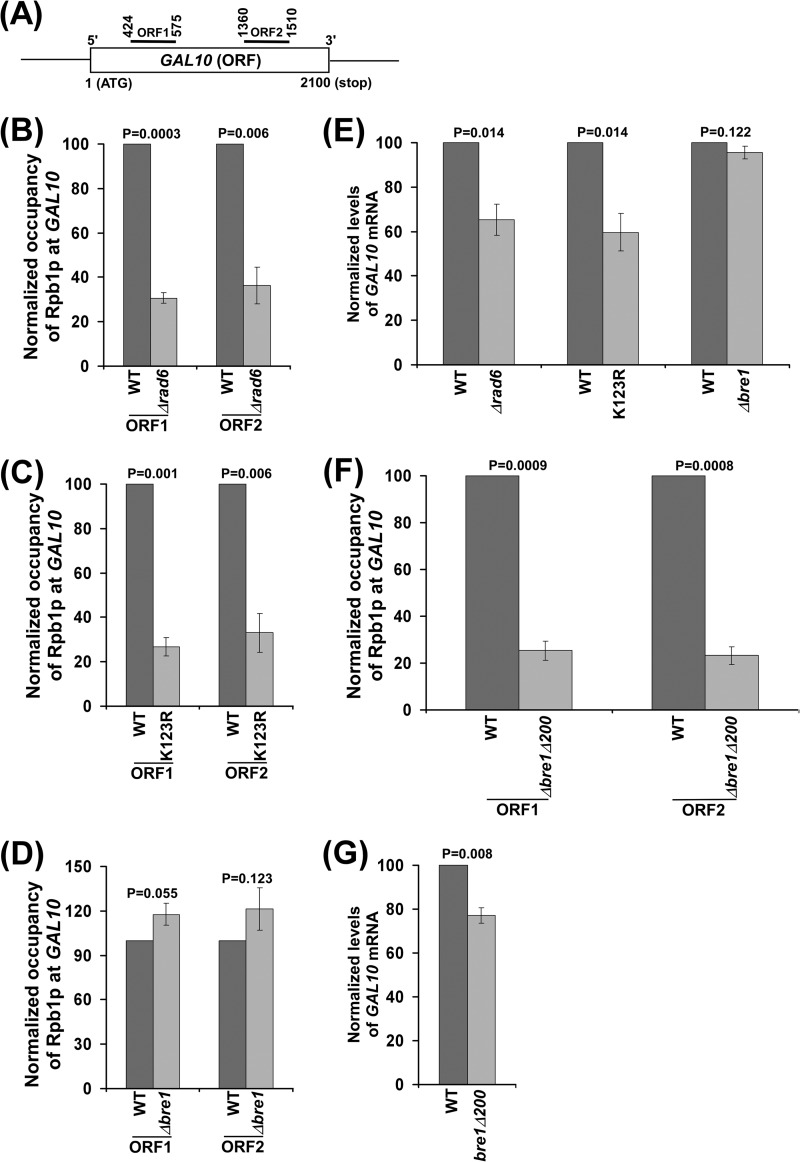
Functional Analysis of the RING and Non-RING Domains of Bre1p in Association of RNA Polymerase II with GAL10, ADH1, and RPS5, and hence Transcription
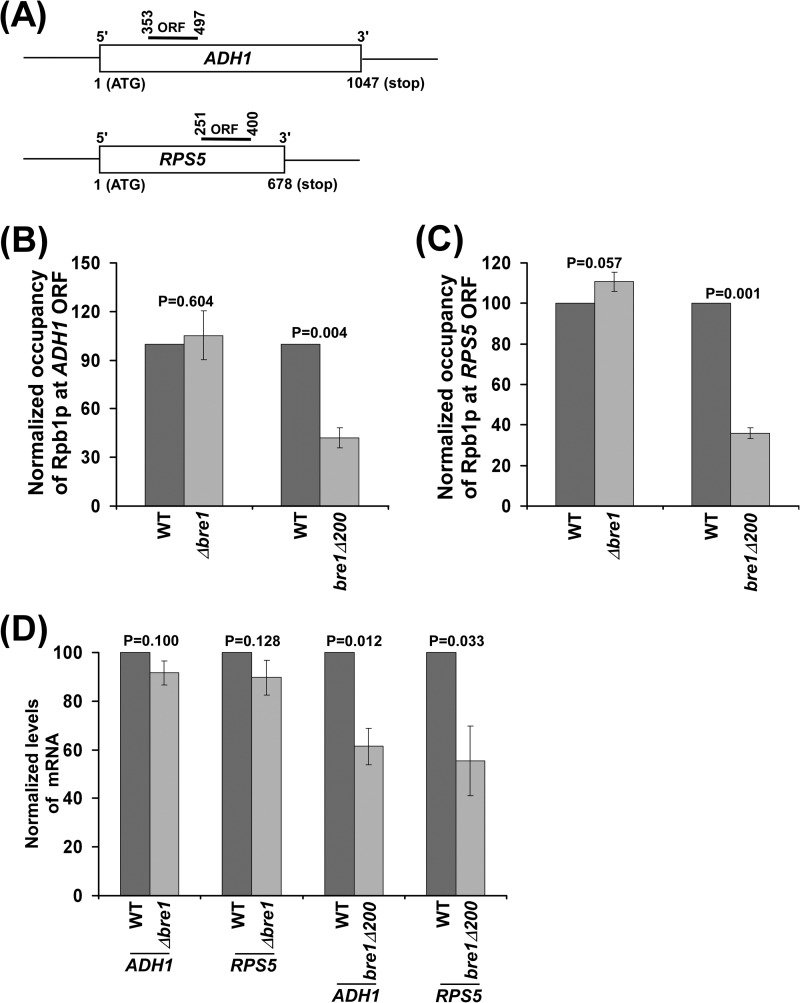
So far, we have demonstrated that the absence of the RING finger with/without a part of the non-RING domain of Bre1p impairs the association of RNA polymerase II with GAL1 (and hence transcription), similar to the Δrad6 and H2B K123R mutants. However, deletion of the whole BRE1 reverses the phenotypes caused by loss of the RING finger with/without a portion of non-RING domain at GAL1. These results support the repressive role of a non-RING domain of Bre1p in association of RNA polymerase II with GAL1 (and hence transcription), as discussed above. To determine whether similar results are found at other genes, we extended our studies to another galactose-inducible gene, GAL10. Similar to the results at GAL1, we found that association of RNA polymerase II with GAL10 (and hence transcription) was significantly impaired in the Δrad6 and H2B K123R mutant strains (Fig. 6, A–C and E). On the other hand, the association of RNA polymerase II with GAL10 (and hence transcription) was not altered in the Δbre1 strain in comparison to the wild-type equivalent (Fig. 6, D and E), as observed at GAL1. However, deletion of the RING finger of Bre1p and adjacent coiled-coil domain significantly impaired the association of RNA polymerase II with GAL10 (Fig. 6F), and consistently, GAL10 transcription was also impaired (Fig. 6G). Thus, like the results at GAL1, we found that the RING finger and adjacent coiled-coil domain of Bre1p promoted association of RNA polymerase II with GAL10, and hence transcription. However, deletion of the whole BRE1 reversed the phenotype of the RING finger and adjacent coiled-coil domain at GAL10 (Fig. 6, D–G). We also found similar results at non-GAL genes, such as ADH1 and RPS5 (Fig. 7, A–D). Thus, a non-RING domain of Bre1p has a repressive role in association of RNA polymerase II with active genes and hence transcription.
FIGURE 6.
Analysis of the association of RNA polymerase II with GAL10. A, a schematic diagram showing the locations of the primer pairs (ORF1 and ORF2) at the GAL10 coding sequence for ChIP analysis. The numbers are presented with respect to the position of the first nucleotide of the initiation codon (+1). B, Rad6p promotes the association of RNA polymerase II with GAL10. Yeast strains (STY1 and STY2) were grown, cross-linked, and immunoprecipitated as described in the legend to Fig. 1B. C, association of RNA polymerase II with GAL10 is significantly decreased in the histone H2B-K123R point mutant strain. Yeast strains (YKH045 and YKH046) were grown, cross-linked, and immunoprecipitated as described in the legend to Fig. 1B. D, association of RNA polymerase II with GAL10 is not impaired in the absence of Bre1p. Yeast strains (YTT31 and SLY1a) were grown, cross-linked, and immunoprecipitated as described in the legend to Fig. 1B. E, analysis of GAL10 transcription by RT-PCR. Yeast strains were grown as described in the legend to Fig. 1B prior to harvesting for mRNA analysis. F and G, analysis of RNA polymerase II association with GAL10 and transcription in the bre1Δ200 and wild-type strains. Both wild-type and mutant strains were grown as described in the legend to Fig. 1B.
FIGURE 7.
Analysis of the association of RNA polymerase II with ADH1 and RPS5. A, the schematic diagrams showing locations of the primer pairs at the ADH1 and RPS5 coding sequences for the ChIP analysis. The numbers are presented with respect to the position of the first nucleotide of the initiation codon (+1). B and C, association of RNA polymerase II with ADH1 and RPS5 is altered in the bre1Δ200 strain, but not in the Δbre1 strain. Wild-type and mutant strains were grown in YPD up to an A600 of 1.0 prior to cross-linking. Immunoprecipitations were performed as described in the legend to Fig. 1B. D, RT-PCR analysis of ADH1 and RPS5 mRNAs. Yeast strains were grown as in panel B.
DISCUSSION
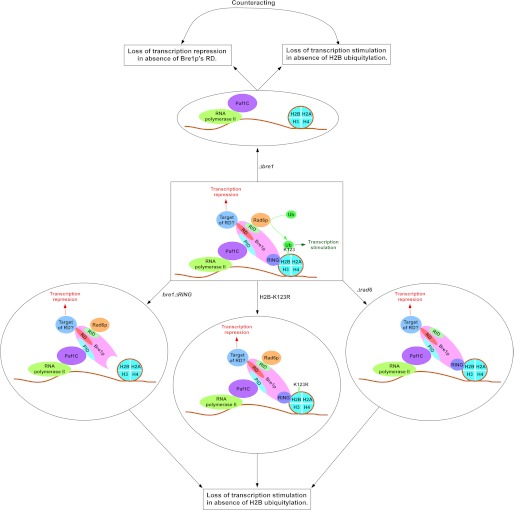
Bre1p is an E3 ubiquitin ligase for histone H2B ubiquitylation that promotes the association of RNA polymerase II with the coding sequence of active gene and hence transcriptional elongation (8–12). Therefore, the deletion of BRE1 is expected to impair the association of RNA polymerase II with the active coding sequence as well as transcription, similar to the Δrad6 and H2B-K123R mutant strains. Surprisingly, we find here that deletion of BRE1 does not impair the association of RNA polymerase II with the coding sequence of GAL1 following transcriptional induction (Fig. 1, D and F). Consistently, GAL1 transcription is not altered in the Δbre1 strain (Fig. 1, E and G). On the other hand, the association of RNA polymerase II with the active GAL1 coding sequence is significantly impaired in Δrad6 and H2B-K123R mutant strains (Fig. 1, B, C, and E). Similar results are also obtained at several other genes such as GAL10, ADH1, and RPS5 (Figs. 6 and 7). These results raise the possibility that there may be a redundant gene that has compensated the function of Bre1p in the Δbre1 strain. However, we rule out this possibility, based on previous studies (15, 16, 19) and our studies (Fig. 1H) that demonstrated a dramatic loss of histone H2B ubiquitylation in the Δbre1 strain. Furthermore, we find that the Δbre1 strain is sensitive to genotoxic agents, such as MMS and HU, similar to the Δrad6 strain (Fig. 1I). Moreover, Rad6p does not complement the function of Bre1p in the Δbre1 strain as there is a drastic impairment of histone H2B ubiquitylation in the Δbre1 strain (Fig. 1H) (15, 16, 19). Therefore, there does not appear to be a gene that is redundant to Bre1p for histone H2B ubiquitylation in yeast. Thus, the invariant association of RNA polymerase II as well as transcription in the Δbre1 strain support the repressive role of Bre1p in addition to its stimulatory function (via H2B ubiquitylation) in association of RNA polymerase II with active gene and hence transcription, as schematically shown in Fig. 8.
FIGURE 8.
The schematic diagram showing the stimulatory and repressive roles of Bre1p in transcription. Bre1p interacts with chromatin via its RING domain at the C-terminal (19). Rad6p interacts with a domain within the first 210 amino acids at the N-terminal (19), and leads to targeted histone H2B ubiquitylation (19). Paf1C interacts with Bre1p (19) via its non-RING domain to promote histone H2B ubiquitylation activity of Rad6p-Bre1p (19, 34). Thus, bre1ΔRING, Δpaf1, Δrad6, and H2B-K123R mutant strains impair histone H2B ubiquitylation (9, 15–17, 19), RNA polymerase II association, and transcription (8–12; this study). The non-RING part of Bre1p has a repression domain (RD) that lowers the association of RNA polymerase II with the active gene and hence transcription (this study), possibly via interaction with an unknown factor. The transcription stimulation is lost in the absence of histone H2B ubiquitylation in the bre1ΔRING, Δrad6, and H2B-K123R mutant strains. When the whole BRE1 is deleted, transcription repression is lost in the absence of the repression domain of Bre1p, in addition to impairment of transcriptional stimulation. These two opposing activities counteract, and hence the defect in transcription (or RNA polymerase II association) is not apparently observed in the Δbre1 strain. The bre1Δ50 mutant is represented by bre1ΔRING (i.e. Bre1p without RING domain). PID, Paf1C interaction domain; RID, Rad6p interaction domain; Ub, ubiquitin.
Although complete deletion of BRE1 does not impair the association of RNA polymerase II with active genes (as well as transcription), loss of the RING finger of Bre1p and a portion of the non-RING domain significantly reduces RNA polymerase II association and hence transcription (Figs. 2, B and C, and 5, C and D), similar to the Δrad6 and H2B K123R mutant strains (Fig. 1, B, C, and E). Thus, the repressive role of Bre1p appears to be confined within a region toward the N-terminal in its non-RING domain. Deletion of the RING domain does not alter the interaction of Bre1p with Rad6p (19), but impairs the targeted histone H2B ubiquitylation (19) (Fig. 1H). Furthermore, the RING domain has been implicated to interact with histone H2B (19). Thus, the RING domain of Bre1p is essential for targeted histone H2B ubiquitylation via its interaction with histone H2B (Fig. 8). The first 210 amino acids at the N-terminal of Bre1p interact with Rad6p for histone H2B ubiquitylation (19). Because the N-terminal region of the non-RING domain of Bre1p interacts with Rad6p for histone H2B ubiquitylation, the non-RING region at the N-terminal of Bre1p also has a transcriptional stimulatory role via histone H2B ubiquitylation in addition to its repressive role (Fig. 8).
Intriguingly, Paf1C that promotes histone H2B ubiquitylation activity of Rad6p-Bre1p (34) has recently been shown to interact biochemically with Bre1p (19). Consistent with recent biochemical data (19), we find here that the recruitment of Paf1C is significantly impaired in the Δbre1 strain (Fig. 4B). Our data further reveal that the RING domain of Bre1p is dispensable for recruitment of Paf1C (Fig. 4F). Thus, Paf1C appears to interact with the non-RING domain of Bre1p to promote histone H2B ubiquitylation. Therefore, the association of RNA polymerase II with GAL1 (as well as transcription) is significantly decreased in the Δpaf1 strain (Fig. 4, D and E), similar to Δrad6, H2B K123R, and Bre1Δ50 mutant strains (Figs. 1, B, C, and E, and 2, B and C). Taken together, our results support that the non-RING domain of Bre1p plays a crucial role to promote histone H2B ubiquitylation via its interaction with Paf1C and Rad6p in addition to its repressive function as schematically shown in Fig. 8.
It has been recently demonstrated that deletion of the RING domain of Bre1p does not alter its interaction with Rad6p (19). Furthermore, as discussed above, Paf1C interacts with the non-RING domain of Bre1p, and its recruitment is not impaired in the absence of the RING domain of Bre1p (Fig. 4F). Thus, deletion of the RING domain of Bre1p does not direct the degradation or disassembly of the entire complex (Fig. 8). The complete deletion of BRE1 would dissociate the complex as Rad6p and Paf1C would loose their interaction with Bre1p (Fig. 8). However, the individual subunits following disintegration of the complex do not display residual activity toward histone H2B ubiquitylation as several previous studies (15, 16, 19) and our current studies (Fig. 1H) have demonstrated a dramatic loss of histone H2B ubiquitylation in the Δbre1 strain. Thus, reversal of the defect of RNA polymerase II association with GAL1 caused by the loss of the RING domain in the Δbre1 strain is not mediated by the residual activity of the individual component, but rather a repression domain of Bre1p as schematically shown in Fig. 8.
As discussed above, the complete deletion of BRE1 lowers the recruitment of Paf1C to GAL1 (Fig. 4B) and the absence of Paf1C impairs the association of RNA polymerase II with GAL1 as well as transcription (Fig. 4, D and E). Based on these results, it is expected to observe an impairment of RNA polymerase II association with GAL1, and transcription in the absence of Bre1p. However, we find that RNA polymerase II association with GAL1 is not impaired in the Δbre1 strain (Fig. 1, D and F). Consistently, GAL1 transcription is not decreased in the absence of Bre1p (Fig. 1, E and G). These results support the presence of a repression domain within the non-RING domain of Bre1p, as discussed above. Such a domain might be exhibiting its repressive role by interacting with another factor(s). The loss of interaction of yet another unknown factor(s) with Bre1p in the Δbre1 strain might have reversed the defect in RNA polymerase II association and transcription in the absence of the RING domain and interactions of Bre1p with Rad6p and Paf1C in Δbre1, as schematically shown in Fig. 8. Intriguingly, protein-protein interaction studies (18, 54–56) revealed the interaction of Bre1p with histone deacetylase, a transcriptional repressor. Thus, histone deacetylase might be repressing transcription via its interaction with Bre1p. Such repression is relieved in the absence of Bre1p, and hence the defect in RNA polymerase II association with GAL1 in the absence of the RING domain (or histone H2B ubiquitylation) is reversed in the null mutation of BRE1 (Fig. 8). Alternatively, Bre1p might be interacting with other transcriptional repressor(s). However, these possibilities remain to be further elucidated. Nonetheless, our data demonstrate that a non-RING domain of Bre1p has a repressive role, and thus, the complete deletion of BRE1 reverses the defect in transcription and RNA polymerase II association phenotypes caused by the loss of H2B ubiquitylation or the RING domain (or interactions with Rad6p and Paf1C), as schematically shown in Fig. 8.
Recently, the homologue of Bre1p in humans has been shown to repress the expression of the proto-oncogenes via inhibition of the recruitment of TFIIS that promotes transcription (48). Based on these recent results, it is expected that Bre1p in yeast might be exhibiting its repressive role by inhibiting the recruitment of TFIIS. However, our data reveal that Bre1p does not alter the recruitment of TFIIS in yeast (Fig. 3, B and C). Thus, the function of the homologue of Bre1p in humans in repressing the expression of proto-oncogenes via TFIIS appears to be gene-specific or more complex as compared with yeast.
In summary, we find that the RING domain of Bre1p has a stimulatory role in targeted histone H2B ubiquitylation for enhanced transcription as well as RNA polymerase II association with the active gene (Fig. 8). On the other hand, the non-RING domain of Bre1p has both the stimulatory and repressive roles in regulating the association of RNA polymerase II with the active gene and hence transcription (Fig. 8). The stimulatory function of the non-RING domain of Bre1p is mediated via its interaction with Rad6p and Paf1C, which promotes H2B ubiquitylation (Fig. 8). Although the function of Bre1p in transcription stimulation is well known via histone H2B ubiquitylation, this study unravels for the first time a new repressive role of the non-RING domain of Bre1p in regulating the association of RNA polymerase II with active gene and hence transcription (Fig. 8). Such a repressive role appears to be confined within the first ∼200 amino acids at the N-terminal of Bre1p. This domain of Bre1p may be exhibiting its repressive role by decreasing the histone H2B ubiquitylation activity. However, we rule out this possibility as a recent study has implicated the interaction of this domain with Rad6p for histone H2B ubiquitylation (19). Consistently, deletion of this domain has been shown to impair histone H2B ubiquitylation (19). Furthermore, removal of this domain does not alter Bre1p homomeric complex formation (19). Thus, the first ∼200 amino acid domain of Bre1p is likely to play its repressive role via a different mechanism. It has been recently demonstrated that the 1–210-amino acid domain of Bre1p is ubiquitylated by Rad6p (19), and such ubiquitylation may play a repressive role in association of RNA polymerase II and transcription. Alternatively, this domain may also play a repressive role in association of RNA polymerase II via histone deacetylation, because Bre1p has been shown to interact with histone deacetylase (18, 54–56), as discussed above. However, these possibilities remain to be further elucidated. Nonetheless, this study demonstrates for the first time a new repressive role of Bre1p in regulation of RNA polymerase II association with active gene and hence transcription. Such a repressive role of Bre1p is likely to play a crucial function to slow down elongating RNA polymerase II for allowing histone H2B ubiquitylation and subsequent chromatin reassembly in maintaining productive transcription, because previous studies have implicated histone H2B ubiquitylation in chromatin reassembly in the wake of elongating RNA polymerase II (10, 11). Thus, the repressive role of Bre1p appears to be physiologically linked to its stimulatory function in histone H2B ubiquitylation, chromatin dynamics, and transcription.
Acknowledgments
We thank Mary Ann Osley, Ali Shilatifard, David Stillman, and Judith K. Davie for yeast strains; and Shivani Malik for critical reading of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant 1R15GM088798-01 (to S. R. B.), Grant-in-aid 10GRNT4300059 from the American Heart Association (Greater Midwest Affiliate), a Mallinckrodt Foundation grant, and Excellence in Academic Medicine awards of the Southern Illinois University School of Medicine.
- Paf1
- polymerase II-associated factor 1
- MMS
- methyl methanosulfonate
- HU
- hydroxy urea.
REFERENCES
- 1. Bhaumik S. R., Smith E., Shilatifard A. (2007) Covalent modifications of histones during development and disease pathogenesis. Nat. Struct. Mol. Biol. 14, 1008–1016 [DOI] [PubMed] [Google Scholar]
- 2. Shukla A., Chaurasia P., Bhaumik S. R. (2009) Histone methylation and ubiquitination with their cross-talk and roles in gene expression and stability. Cell Mol. Life Sci. 66, 1419–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Malik S., Bhaumik S. R. (2010) Mixed lineage leukemia: histone H3 lysine 4 methyltransferases from yeast to human. FEBS J. 277, 1805–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shilatifard A. (2006) Chromatin modifications by methylation and ubiquitination. Implications in the regulation of gene expression. Annu. Rev. Biochem. 75, 243–269 [DOI] [PubMed] [Google Scholar]
- 5. Selth L. A., Sigurdsson S., Svejstrup J. Q. (2010) Transcript elongation by RNA polymerase II. Annu. Rev. Biochem. 79, 271–293 [DOI] [PubMed] [Google Scholar]
- 6. Strahl B. D., Allis C. D. (2000) The language of covalent histone modifications. Nature 403, 41–45 [DOI] [PubMed] [Google Scholar]
- 7. Bradbury E. M. (1992) Reversible histone modifications and the chromosome cell cycle. Bioessays 14, 9–16 [DOI] [PubMed] [Google Scholar]
- 8. Shukla A., Bhaumik S. R. (2007) H2B-K123 ubiquitination stimulates RNAPII elongation independent of H3-K4 methylation. Biochem. Biophys. Res. Commun. 359, 214–220 [DOI] [PubMed] [Google Scholar]
- 9. Xiao T., Kao C. F., Krogan N. J., Sun Z. W., Greenblatt J. F., Osley M. A., Strahl B. D. (2005) Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol. Cell. Biol. 25, 637–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fleming A. B., Kao C. F., Hillyer C., Pikaart M., Osley M. A. (2008) H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol. Cell. 31, 57–66 [DOI] [PubMed] [Google Scholar]
- 11. Batta K., Zhang Z., Yen K., Goffman D. B., Pugh B. F. (2011) Genome-wide function of H2B ubiquitylation in promoter and genic regions. Genes Dev. 25, 2254–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tanny J. C., Erdjument-Bromage H., Tempst P., Allis C. D. (2007) Ubiquitylation of histone H2B controls RNA polymerase II transcription elongation independently of histone H3 methylation. Genes Dev. 21, 835–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pavri R., Zhu B., Li G., Trojer P., Mandal S., Shilatifard A., Reinberg D. (2006) Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125, 703–717 [DOI] [PubMed] [Google Scholar]
- 14. Vitaliano-Prunier A., Menant A., Hobeika M., Géli V., Gwizdek C., Dargemont C. (2008) Ubiquitylation of the COMPASS component Swd2 links H2B ubiquitylation to H3K4 trimethylation. Nat. Cell Biol. 10, 1365–1371 [DOI] [PubMed] [Google Scholar]
- 15. Hwang W. W., Venkatasubrahmanyam S., Ianculescu A. G., Tong A., Boone C., Madhani H. D. (2003) A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell 11, 261–266 [DOI] [PubMed] [Google Scholar]
- 16. Wood A., Krogan N. J., Dover J., Schneider J., Heidt J., Boateng M. A., Dean K., Golshani A., Zhang Y., Greenblatt J. F., Johnston M., Shilatifard A. (2003) Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell 11, 267–274 [DOI] [PubMed] [Google Scholar]
- 17. Song Y. H., Ahn S. H. (2010) A Bre1-associated protein, large 1 (Lge1), promotes H2B ubiquitylation during the early stages of transcription elongation. J. Biol. Chem. 285, 2361–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pan X., Ye P., Yuan D. S., Wang X., Bader J. S., Boeke J. D. (2006) A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell 124, 1069–1081 [DOI] [PubMed] [Google Scholar]
- 19. Kim J., Roeder R. G. (2009) Direct Bre1-Paf1 complex interactions and RING finger-independent Bre1-Rad6 interactions mediate histone H2B ubiquitylation in yeast. J. Biol. Chem. 284, 20582–20592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Murén E., Oyen M., Barmark G., Ronne H. (2001) Identification of yeast deletion strains that are hypersensitive to brefeldin A or monensin, two drugs that affect intracellular transport. Yeast 18, 163–172 [DOI] [PubMed] [Google Scholar]
- 21. Jentsch S., McGrath J. P., Varshavsky A. (1987) The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature 329, 131–134 [DOI] [PubMed] [Google Scholar]
- 22. Sung P., Prakash S., Prakash L. (1988) The RAD6 protein of Saccharomyces cerevisiae polyubiquitinates histones, and its acidic domain mediates this activity. Genes Dev. 2, 1476–1485 [DOI] [PubMed] [Google Scholar]
- 23. Robzyk K., Recht J., Osley M. A. (2000) Rad6-dependent ubiquitination of histone H2B in yeast. Science 287, 501–514 [DOI] [PubMed] [Google Scholar]
- 24. Giannattasio M., Lazzaro F., Plevani P., Muzi-Falconi M. (2005) The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J. Biol. Chem. 280, 9879–9886 [DOI] [PubMed] [Google Scholar]
- 25. Chernikova S. B., Razorenova O. V., Higgins J. P., Sishc B. J., Nicolau M., Dorth J. A., Chernikova D. A., Kwok S., Brooks J. D., Bailey S. M., Game J. C., Brown J. M. (2012) Deficiency in mammalian histone H2B ubiquitin ligase Bre1 (Rnf20/Rnf40) leads to replication stress and chromosomal instability. Cancer Res. 72, 2111–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Walter D., Matter A., Fahrenkrog B. (2010) Bre1p-mediated histone H2B ubiquitylation regulates apoptosis in Saccharomyces cerevisiae. J. Cell Sci. 123, 1931–1939 [DOI] [PubMed] [Google Scholar]
- 27. Game J. C., Chernikova S. B. (2009) The role of RAD6 in recombinational repair, checkpoints and meiosis via histone modification. DNA Repair 8, 470–482 [DOI] [PubMed] [Google Scholar]
- 28. Wyce A., Xiao T., Whelan K. A., Kosman C., Walter W., Eick D., Hughes T. R., Krogan N. J., Strahl B. D., Berger S. L. (2007) H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Mol. Cell 27, 275–288 [DOI] [PubMed] [Google Scholar]
- 29. Sun Z. W., Allis C. D. (2002) Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418, 104–108 [DOI] [PubMed] [Google Scholar]
- 30. Mutiu A. I., Hoke S. M., Genereaux J., Liang G., Brandl C. J. (2007) The role of histone ubiquitylation and deubiquitylation in gene expression as determined by the analysis of an HTB1(K123R) Saccharomyces cerevisiae strain. Mol. Genet. Genomics 277, 491–506 [DOI] [PubMed] [Google Scholar]
- 31. Henry K. W., Wyce A., Lo W. S., Duggan L. J., Emre N. C., Kao C. F., Pillus L., Shilatifard A., Osley M. A., Berger S. L. (2003) Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 17, 2648–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schulze J. M., Hentrich T., Nakanishi S., Gupta A., Emberly E., Shilatifard A., Kobor M. S. (2011) Splitting the task. Ubp8 and Ubp10 deubiquitinate different cellular pools of H2BK123. Genes Dev. 25, 2242–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ng H. H., Dole S., Struhl K. (2003) The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J. Biol. Chem. 278, 33625–33628 [DOI] [PubMed] [Google Scholar]
- 34. Wood A., Schneider J., Dover J., Johnston M., Shilatifard A. (2003) The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 278, 34739–34742 [DOI] [PubMed] [Google Scholar]
- 35. Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 36. Sikorski R. S., Hieter P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tsukuda T., Fleming A. B., Nickoloff J. A., Osley M. A. (2005) Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature 438, 379–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee S. E., Moore J. K., Holmes A., Umezu K., Kolodner R. D., Haber J. E. (1998) Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94, 399–409 [DOI] [PubMed] [Google Scholar]
- 39. Bhaumik S. R., Green M. R. (2002) Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol. Cell. Biol. 22, 7365–7371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bhaumik S. R., Green M. R. (2003) Interaction of Gal4p with components of transcription machinery in vivo. Methods Enzymol. 370, 445–454 [DOI] [PubMed] [Google Scholar]
- 41. Shukla A., Stanojevic N., Duan Z., Sen P., Bhaumik S. R. (2006) Ubp8p, a histone deubiquitinase whose association with SAGA is mediated by Sgf11p, differentially regulates lysine 4 methylation of histone H3 in vivo. Mol. Cell. Biol. 26, 3339–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bhaumik S. R., Raha T., Aiello D. P., Green M. R. (2004) In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 18, 333–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Malik S., Shukla A., Sen P., Bhaumik S. R. (2009) The 19S proteasome subcomplex establishes a specific protein interaction network at the promoter for stimulated transcriptional initiation in vivo. J. Biol. Chem. 284, 35714–35724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Malik S., Chaurasia P., Lahudkar S., Durairaj G., Shukla A., Bhaumik S. R. (2010) Rad26p, a transcription-coupled repair factor, is recruited to the site of DNA lesion in an elongating RNA polymerase II-dependent manner in vivo. Nucleic Acids Res. 38, 1461–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Peterson C. L., Kruger W., Herskowitz I. (1991) A functional interaction between the C-terminal domain of RNA polymerase II and the negative regulator SIN1. Cell 64, 1135–1143 [DOI] [PubMed] [Google Scholar]
- 46. Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Struhl K. (2001) Current Protocols in Molecular Biology, Wiley, New York [Google Scholar]
- 47. Trujillo K. M., Osley M. A. (2012) A role of H2B ubiquitylation in DNA replication. Mol. Cell 48, 734–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shema E., Kim J., Roeder R. G., Oren M. (2011) RNF20 inhibits TFIIS-facilitated transcriptional elongation to suppress pro-oncogenic gene expression. Mol. Cell. 42, 477–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Costa P. J., Arndt K. M. (2000) Synthetic lethal interactions suggest a role for the Saccharomyces cerevisiae Rtf1 protein in transcription elongation. Genetics 156, 535–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mueller C. L., Jaehning J. A. (2002) Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol. Cell. Biol. 22, 1971–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pokholok D. K., Hannett N. M., Young R. A. (2002) Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell 9, 799–809 [DOI] [PubMed] [Google Scholar]
- 52. Krogan N. J., Kim M., Ahn S. H., Zhong G., Kobor M. S., Cagney G., Emili A., Shilatifard A., Buratowski S., Greenblatt J. F. (2002) RNA polymerase II elongation factors of Saccharomyces cerevisiae. A targeted proteomics approach. Mol. Cell. Biol. 22, 6979–6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Squazzo S. L., Costa P. J., Lindstrom D. L., Kumer K. E., Simic R., Jennings J. L., Link A. J., Arndt K. M., Hartzog G. A. (2002) The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 21, 1764–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., Sevier C. S., Ding H., Koh J. L., Toufighi K., Mostafavi S., Prinz J., St Onge R. P., VanderSluis B., Makhnevych T., Vizeacoumar F. J., Alizadeh S., Bahr S., Brost R. L., Chen Y., Cokol M., Deshpande R., Li Z., Lin Z. Y., Liang W., Marback M., Paw J., San Luis B. J., Shuteriqi E., Tong A. H., van Dyk N., Wallace I. M., Whitney J. A., Weirauch M. T., Zhong G., Zhu H., Houry W. A., Brudno M., Ragibizadeh S., Papp B., Pál C., Roth F. P., Giaever G., Nislow C., Troyanskaya O. G., Bussey H., Bader G. D., Gingras A. C., Morris Q. D., Kim P. M., Kaiser C. A., Myers C. L. (2010) The genetic landscape of a cell. Science 327, 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Collins S. R., Miller K. M., Maas N. L., Roguev A., Fillingham J., Chu C. S., Schuldiner M., Gebbia M., Recht J., Shales M., Ding H., Xu H., Han J., Ingvarsdottir K., Cheng B., Andrews B., Boone C., Berger S. L., Hieter P., Zhang Z., Brown G. W., Ingles C. J., Emili A., Allis C. D., Toczyski D. P., Weissman J. S., Greenblatt J. F., Krogan N. J. (2007) Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446, 806–810 [DOI] [PubMed] [Google Scholar]
- 56. Lin Y. Y., Qi Y., Lu J. Y., Pan X., Yuan D. S., Zhao Y., Bader J. S., Boeke J. D. (2008) A comprehensive synthetic genetic interaction network governing yeast histone acetylation and deacetylation. Genes Dev. 22, 2062–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]