Background: PHD2 is the central enzyme that controls hypoxia-inducible factor-α (HIF-α) protein levels.
Results: PHD2 binds a Pro-Xaa-Leu-Glu motif in two HSP90 co-chaperones, and knockdown of one of these, p23, augments hypoxia-induced HIF-1α protein levels.
Conclusion: PHD2 is linked to the HSP90 pathway, facilitating its hydroxylation of HIF-1α.
Significance: This uncovers a new model by which PHD2 controls HIF-1α.
Keywords: Heat Shock Protein, Hsp90, Hypoxia, Hypoxia-inducible Factor (HIF), EGLN1, PHD2, Prolyl Hydroxylase Domain Protein 2
Abstract
Prolyl hydroxylase domain protein 2 (PHD2, also known as Egg Laying Defective Nine homolog 1) is a key oxygen-sensing protein in metazoans. In an oxygen-dependent manner, PHD2 site-specifically prolyl hydroxylates the master transcription factor of the hypoxic response, hypoxia-inducible factor-α (HIF-α), thereby targeting HIF-α for degradation. In this report we show that the heat shock protein 90 (HSP90) co-chaperones p23 and FKBP38 interact via a conserved Pro-Xaa-Leu-Glu motif (where Xaa = any amino acid) in these proteins with the N-terminal Myeloid Nervy and DEAF-1 (MYND)-type zinc finger of PHD2. Knockdown of p23 augments hypoxia-induced HIF-1α protein levels and HIF target genes. We propose that p23 recruits PHD2 to the HSP90 machinery to facilitate HIF-1α hydroxylation. These findings identify a link between two ancient pathways, the PHD:HIF and the HSP90 pathways, and suggest that this link was established concurrent with the emergence of the PHD:HIF pathway in evolution.
Introduction
The central pathway regulating the transcriptional response to hypoxia is the PHD:HIF:von Hippel Lindau protein (VHL)2 pathway (1–3). HIF is a heterodimeric complex consisting of an α and a β subunit. In mammals there are two main α subunits, HIF-1α and HIF-2α, and one β subunit (HIF-1β, also known as the aryl hydrocarbon nuclear translocator). The protein stability of the α subunit is regulated in an oxygen-dependent manner. Under normoxic conditions, PHD constitutively hydroxylates HIF-α on specific prolyl residues. In the case of HIF-1α, the primary site of hydroxylation is Pro-564 (4–6). This provides a binding platform for VHL, a component of an E3 ubiquitin ligase complex that selectively recognizes modified HIF-α and targets it for degradation by the ubiquitin-proteasome pathway (7). Under hypoxic conditions, prolyl hydroxylation of HIF-α is attenuated, allowing the stabilization of HIF-α and activation of a broad range of HIF target genes (2). These genes promote glycolysis, glucose uptake, angiogenesis, and erythropoiesis (3, 8).
Three mammalian PHD isoforms have been identified: PHD1, PHD2, and PHD3 (9, 10). All three shared a conserved prolyl hydroxylase domain. Among the three PHD isoforms, PHD2 is regarded as the central one mediating HIF-α turnover. Knockdown of PHD2 in most cell types is sufficient to induce HIF activity (11, 12). PHD2 is also the most abundant isoform in most cell types (12). Knock-out of the Phd2 gene in mice results in embryonic lethality, whereas Phd1−/− and Phd3−/− mice are viable (13). In humans, patients with germ line PHD2 mutations display erythrocytosis due to dysregulation of the Erythropoietin gene (14–17). Moreover, PHD2 haplotypes are associated with high altitude adaptation in Tibetans (18, 19). PHD2 is distinctive among the three PHDs in that it harbors at its N terminus a domain that is predicted to be a zinc finger of the Myeloid Nervy and DEAF-1 (MYND) type (20).
The PHD:HIF:VHL pathway shows broad conservation in animals (21, 22), and experiments demonstrate that it is functional in the simplest metazoan, Trichoplax adhaerens (21). T. adhaerens contains single isoforms for PHD, HIF-α, and VHL, and intriguingly, its PHD contains a MYND-type zinc finger homologous to that seen in mammalian PHD2. These observations indicate that prolyl hydroxylation is an ancient mechanism to transduce changes in oxygen concentration to changes in cellular function and furthermore suggest that mammalian PHD2 is the isoform most closely related to the single ancestral PHD.
The HSP90 pathway is an even more ancient pathway, showing conservation in bacteria (23). It is centered around the HSP90 chaperone protein. Although it was originally identified as a pathway that is heat-inducible and that promotes protein folding, it is clear that it plays a central role in the maturation of a multitude of client proteins, particularly ones involved in signal transduction and transcription. Indeed, HIF-1α is a client of the HSP90 pathway (24). A salient feature of the HSP90 pathway is that it employs a set of co-chaperones that regulate the activity of HSP90 and can interact with client proteins (25, 26). These co-chaperones show varying degrees of conservation in evolution (27). As one example, the co-chaperone p23, which binds to HSP90 in an ATP-dependent manner and stabilizes a closed conformation of HSP90, shows extensive conservation. It is present, for example, in Saccharomyces cerevisiae. The conservation of various HSP90 co-chaperones raises interesting questions as to the selective pressures that have maintained them in evolution.
The PHDs site-specifically hydroxylate HIF-α on prolyl residues that occur in an LXXLAP motif (where X = any amino acid). In vitro experiments demonstrate that HIF-1α peptides as short as 19 amino acids in length that contain this motif are sufficient to allow for site-specific hydroxylation and subsequent recognition by VHL (5, 28, 29). Although the data imply that the core machinery of the pathway consists of PHD, HIF, and VHL, the extent to which it is integrated with other cellular pathways remains poorly understood. In the present report we identify a direct interaction between the MYND-type zinc finger of PHD2 and a peptide motif in select co-chaperones of the HSP90 pathway, including p23. We provide evidence for the functional importance of this interaction in promoting efficient HIF-α degradation. Taken together, the data establish a link between oxygen-sensing and the HSP90 pathway and suggest that this link emerged early in the evolution of metazoans.
EXPERIMENTAL PROCEDURES
Plasmids
pcDNA5/FRT/TO-FLAG-PHD2 was constructed by subcloning the 1.5-kb HindIII/SphI fragment of pcDNA3-FLAG-PHD2 (30) into the HindIII/SphI site of pcDNA5/FRT/TO. pcDNA3-HA-PHD2 was constructed by subcloning the 1.3-kb BamHI/XbaI fragment of pcDNA3-FLAG-PHD2 into the BamHI/XbaI site of pcDNA3-HA. pcDNA3-HA-PHD2 (1–196) was constructed by subcloning the 0.6-kb BamHI/XhoI fragment of pcDNA3-FLAG-PHD2 (30) into the BamHI/XhoI site of pcDNA3-HA. pcDNA3-HA-PHD2 (130–426) was constructed by digesting pcDNA3-HA-PHD2 with NheI and NotI, blunting the ends with the Klenow fragment of Escherichia coli DNA polymerase I, and then self-ligating. pcDNA3-HA-PHD2 (1–196) C36S/C42S was constructed by PCR-mediated mutagenesis. Then, the 0.7-kb XhoI/XbaI fragment of pcDNA3-FLAG-PHD2 was subcloned into the XhoI/XbaI site of pcDNA3-HA-PHD2 (1–196) C36S/C42S to yield pcDNA3-HA-PHD2 C36S/C42S.
pGEX-PHD2 (1–63) was constructed by subjecting pGEX-PHD2 (1–124) to digestion with NotI, partial digestion with SfoI, blunting with the Klenow fragment of E. coli DNA polymerase I, and self- ligation. pMAL-PHD2 (1–63) was constructed by subcloning the 0.2-kb BamHI/NotI fragment of pGEX-PHD2 (1–63) into the BamHI/NotI site of pMAL-5X-1.
pcDNA3-HA-PHD1 was constructed by subcloning the 1.8-kb BamHI/XbaI fragment of pcDNA3-FLAG-PHD1 (30) into the BamHI/XbaI site of pcDNA3-HA. pcDNA3-HA-PHD3 was constructed by subcloning the 0.8-kb BamHI (partial)/XhoI fragment of pcDNA3-FLAG-PHD3 (30) into the BamHI/XhoI site of pcDNA3-HA.
pcDNA5/FRT/TO-3xFLAG-p23 was constructed by amplifying the coding sequence of pOTB7-p23 (IMAGE clone 2821965) by PCR using the following primers: 5′-GTA CGG ATC CAA ATG CAG CCT GCT TCT GCA AAG-3′ and 5′-GTA CCT CGA GTT ACT CCA GAT CTG GCA T-3′. The 0.5-kb product was digested with BamHI and XhoI and subcloned into the BamHI/XhoI site of pcDNA5/FRT/TO-3xFLAG. pcDNA3-HA-p23 was constructed by subcloning the 0.5-kb BamHI/XhoI coding sequence fragment of pcDNA5/FRT/TO-3xFLAG-p23 into the BamHI/XhoI site of pcDNA3-HA. pcDNA5/FRT/TO-3xFLAG-p23 P157A/L159A/E160A was constructed by amplifying the coding sequence of pcDNA3-HA-p23 using the primers 5′-GTA CGG ATC CAA ATG CAG CCT GCT TCT GCA AAG-3′ and 5′-GTA CCT CGA GCG GCC GCT ACG CCG CAT CTG CCA TTT TTT CAT CAT CAC TGT CTT GTG-3′. The 0.5-kb product was digested with BamHI and XhoI and subcloned into the BamHI/XhoI site of pcDNA5/FRT/TO-3xFLAG.
pcDNA3-HA-FKBP38 was constructed by amplifying the coding sequence of pDONR-FKBP38 (Dana Farber/Harvard Cancer Core clone HsCD000824) with the primers 5′-GTA CGG ATC CAG ATG GCA TCG TGT GCT GAA C-3′ and 5′-GTA TGC GGC CGC TCT AGA TTA GTT CCT GGC AGC GAT GAC CAC AGA GAG TGC CAC ACC CCC CAA G-3′. The 1.2-kb product was digested with BamHI and NotI and subcloned into the BamHI/NotI site of pcDNA3-HA. pcDNA5/FRT/TO-3xFLAG-FKBP38 was constructed by digesting pcDNA3-HA-FKBP38 with BamHI and NotI and then subcloning the 1.2-kb coding sequence into the BamHI/NotI site of pcDNA5/FRT/TO-3xFLAG.
pcDNA3–3xFLAG-FKBP52 was constructed by amplifying the coding sequence of pOTB7-FKBP52 (IMAGE clone 3542330) with the primers 5′-GTA CCT TAA GAT GAC AGC CGA GGA GAT GAA G-3′ and 5′-GTA CCT CGA GCT ATG CTT CTG TCT CCA CCT G-3′. The 1.4-kb product was digested with AflII and XhoI and subcloned into the into AflII/XhoI site of pcDNA3–3xFLAG. pcDNA5/FRT/TO-3xFLAG-FKBP51 was constructed by amplifying the coding sequence of pCMV-SPORT6-FKBP51 (IMAGE clone 4539929) with the primers 5′-GTA CGG ATC CAG ATG ACT ACT GAT GAA GGT GC-3′ and 5′-GTA CCT CGA GTC ATA CGT GGC CCT CAG GTT-3′. The 1.4-kb product was digested with BamHI and XhoI and subcloned into the BamHI/XhoI site of pcDNA5/FRT/TO-3xFLAG.
pcDNA3–3xFLAG-FKBP16 was constructed by amplifying the coding sequence of pOTB7-FKBP16 (IMAGE clone 8143800) with the primers 5′-GTA CGA ATT CAG ATG GCG GAT ATC ATC GCA AG-3′ and 5′-GTA CCT CGA GTC AAT GGG AGA AGA TCC-3′. The 1.0-kb product was digested with EcoRI and XhoI and then subcloned into the EcoRI/XhoI site of pcDNA3–3xFLAG. pcDNA5/FRT/TO-3xFLAG-PPP5C was constructed by amplifying the coding sequence of pCMV-SPORT-PPP5C (IMAGE clone 3459309) with the primers 5′-GTA CGG ATC CAG ATG GCG ATG GCG GAG GGC GAG-3′ and 5′-GTA CCT CGA GTC ACA TCA TTC CTA GCT GCA G-3′. The 1.5-kb product was digested with BamHI and XhoI and then subcloned into the BamHI/XhoI site of pcDNA5/FRT/TO-3xFLAG.
The authenticity of recombinant plasmids was confirmed by sequencing. The (eHRE)3-Luc plasmid, which contains three copies of the hypoxia response element from the human EPO gene enhancer, and pGEX-HIF-1α (531–575) have been described (6, 28). The plasmid pRL-TK, which expresses Renilla luciferase from a minimal thymidine kinase promoter, was obtained from Promega.
Cell Lines
A cell line with doxycycline-inducible expression of FLAG-PHD2 was prepared using pcDNA5/FRT/TO-FLAG-PHD2, HEK293 Flp-In T-Rex cells, and Flp recombinase according the manufacturer's instructions (Invitrogen). These cells are hereafter referred to as HEK293 Flp-In T-Rex/FLAG-PHD2. Stably transfected HEK293 Flp-In T-Rex/FLAG-p23 cells were prepared in an analogous manner using pcDNA5/FRT/TO-3xFLAG-p23.
HeLa and PC3 cells were obtained from ATCC. HEK293FT cells were obtained from Invitrogen. MCF7 and RCC4 cells were kind gifts from Dr. Mark Lemmon (Perelman School of Medicine, University of Pennsylvania) and Dr. Celeste Simon, respectively (Perelman School of Medicine, University of Pennsylvania). All cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. Transfections of plasmids and siRNA were performed using Lipofectamine 2000 and antibiotic-free media. Hypoxia experiments were performed in an In Vivo 200 Hypoxia Work station (Ruskinn Technologies). Sf9 cells were maintained in Sf-900 II serum-free media. 17-AAG was obtained from Cayman Chemical. MG132 was from Sigma.
Mass Spectrometry
HEK293 Flp-In T-Rex and HEK293 Flp-In T-Rex/FLAG-PHD2 cells were grown in 15-cm dishes using light isotope-containing DMEM for the former and heavy isotope (l-[13C6]lysine and l-[13C6,15N4]arginine)-containing DMEM for the latter (SILAC kit from Pierce) (31). The cells were induced with 1 μg/ml doxycycline for 18 h. The cells were then gently washed with PBS, then lysed in 1 ml of buffer A (50 mm Hepes, pH 7.5, 0.5% Triton X-100) supplemented with mammalian protease inhibitor mixture (Sigma). Lysates were clarified by centrifugation at 15,800 × g for 10 min at 4 °C. Equal protein amounts (as determined by the Bio-Rad DC Protein Assay) were then added to 15-μl aliquots of anti-FLAG (M2)-agarose (Sigma) and rocked for 1 h at 4 °C. The resins were washed 3 times with buffer A and then eluted by the addition of 40 μl of a 0.2 μg/μl solution of 3× FLAG peptide. Aliquots of eluates were saved for later Western analysis. Immunoprecipitations of heavy labeled FLAG-p23 were performed in an analogous manner.
Eluates were mixed and subjected to alkylation and trypsin digestion using Filter Aided Sample Preparation (FASP) and Microcon YM-30 filtration units (32) followed by desalting and concentration using C18 StageTips (33). Peptides were separated on 50-cm × 75-μm column packed with 4 μm C12 Jupiter Proteo beads (Phenomenex) that was heated to 50 °C (Phoenix S&T). Nanoflow chromatography was performed at 300 nl/min using a 90-min gradient from 2 to 42% acetonitrile in 0.5% acetic acid. Eluents were electrosprayed for analysis at 2.7 kV with a Phoenix S&T μAutoNano spray source that was coupled to a Thermo Scientific LTQ-Orbitrap hybrid mass spectrometer. Mass spectra (MS) data were acquired in the Orbitrap, and MS/MS fragmentation was performed in the linear ion trap using a top-5 data-dependent MS/MS method. Data were analyzed using MaxQuant software Version 1.2.2.5 (34), and spectra were searched using MaxQuant's included search engine, Andromeda, against the IPI human database Version 3.68. SILAC doublets for Arg-10 and Lys-6 were specified. Oxidation of methionine and proline and acetylation of protein N termini were employed as variable modifications, whereas carbamidomethylation of cysteine was employed as a fixed modification.
Peptide Binding Assays
Peptides corresponding to residues 151–160 of p23 and residues 47–56 of FKBP38 were synthesized by Genscript. p23 peptides were also prepared with the following amino acid substitutions: P157A/L159A/E160A, E160A, E160D, E160Q, L159A, L159I, L159V, D158A, D158P, D158L, P157A, P157S, M156A, M156L, M156V, M156F, M156E, K155A, E154A, D153A, D152A, and D152A/D153A/E154A. An FKBP38 peptide with a P53A/L55A/E56A substitution was prepared. All peptides possessed an N-terminal tyrosine residue to allow spectrophotometric quantitation using a molar absorptivity of 1280 m−1 cm−1 at 280 nm (35), and all were biotinylated at the N terminus.
Peptides (0.5 μg) were prebound to 15-μl aliquots of streptavidin-agarose (Sigma). The resins were incubated with Sf9 lysates containing baculovirus-expressed His6FLAG-PHD2 (30) for 1 h with rocking at 4 °C in buffer B (20 mm Tris, pH 7.6, 150 mm NaCl, 25 mm β-glycerol phosphate, 2 mm pyrophosphate, 10% glycerol, 1% Triton X-100). The resins were washed four times with buffer A and eluted, and the eluates were subjected to SDS-PAGE and Western blotting using anti-FLAG antibody-alkaline phosphatase conjugates (Sigma). Quantification of binding was performed using ImageJ software (National Institutes of Health).
GST Pulldown Assays
GST and GST-HIF-1α (531–575) were purified from E. coli DH5α transformed with pGEX-5X-1 and pGEX-HIF-1α (531–575), respectively, using affinity chromatography on glutathione (GSH)-Sepharose (GE Healthcare). GST or GST-HIF-1α (531–575) prebound to 15 μl of GSH-Sepharose was incubated in buffer C (50 mm Tris, pH 7.5, 100 mm NaCl, 0.5% Triton X-100) with rocking with cellular extracts for 1 h at 4 °C. The resins were washed three times, and eluates were subjected to SDS-PAGE and Western blotting.
Immunoprecipitations
After a wash with PBS, cells were lysed in buffer C supplemented with mammalian protease inhibitor mixture. The lysate was clarified by centrifugation at 15,800 × g for 10 min at 4 °C. These whole cell extracts were then added to 10-μl aliquots of anti-FLAG (M2)-agarose and rocked for 1 h at 4 °C. The resins were washed four times with buffer C and eluted, and the eluates were subjected to SDS-PAGE and Western blotting using anti-HA antibody-alkaline phosphatase conjugates (Sigma). In the case of the VHL capture assays, recombinant FLAG-tagged VHL (in a complex with Elongin B and Elongin C) purified from Sf9 cells (30) was incubated with the extracts for 1 h before immunoprecipitation, and Western blotting was conducted using anti-HIF-1α antibodies (Santa Cruz Biotechnology, H-206). For immunoprecipitation of endogenous PHD2, we employed anti-PHD2 monoclonal antibody 6.9 (36) and protein G-agarose (Invitrogen); for control antibody, we used monoclonal antibody G3A1 (Cell Signaling Technology).
Western Blotting
Western blotting was performed essentially as described (28), except that the secondary antibodies were alkaline phosphatase-conjugated anti-rabbit IgG or anti-mouse IgG (Santa Cruz Biotechnology), and the substrate was CDP-Star (Roche Applied Science). Rabbit polyclonal antibodies to PHD2 were prepared as follows. First, GST-PHD2 (1–63) and MBP-PHD2 (1–63) were purified from E. coli transformed with pGEX-PHD2 (1–63) and pMAL-PHD2 (1–63), respectively, using affinity chromatography on GSH-Sepharose and amylose-agarose (New England Biolabs), respectively. Then polyclonal antibodies to MBP-PHD2 (1–63) were then raised in rabbits and affinity-purified on GST-PHD2 (1–63) coupled to agarose by Covance Research Products. A rabbit monoclonal antibody to PHD2 was obtained from Epitomics (catalog #5859).
Anti-p23 (catalog #2731) and anti-FKBP38 (catalog #5792) were from Epitomics. Anti-HSP90 (clone C45G5) and anti-HSP70 (catalog #4872) were from Cell Signaling. Anti-FKBP16/ARA9 (catalog #GTX110665) was from GeneTex. Mouse monoclonal anti-HIF-1α was from BD Biosciences (clone 54). The E7 monoclonal antibody against β-tubulin developed by Dr. Michael Klymkowsky was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa, Department of Biology, Iowa City, IA 52242.
Real Time PCR
Total RNA was harvested from cells using TRIzol reagent (Invitrogen). Reverse transcription reactions were performed using High Capacity cDNA kits (ABI). Real time PCR reactions were performed on 20-ng equivalents of cDNA using an ABI 7300 Real Time PCR machine and SYBR Green Master mixes. Relative quantification was performed using the ΔΔCt method and 18 S RNA as the endogenous control. The following primer pairs were employed, and in all cases dissociation curve analysis revealed a single peak for each: P23, 5′-CCA AAT GAT TCC AAG CAT AAA AGA-3′ and 5′-GGC CAG ATT CTC CTT TTC GTA A-3′; PHD2, 5′-TGA AGC TGG CGC TCG AGT A-3′ and 5′-CAC ACA GAT GCC GTG CTT GT-3′; CA9, 5′-CGG AAG AAA ACA GTG CCT ATG-A 3′ and 5′-CTT CCT CAG CGA TTT CTT CCA-3′; GLUT1, 5′-TGC TCA TGG GCT TCT CGA A-3′ and 5′-AAG CGG CCC AGG ATC AG-3′; HIF1A, 5′-TTT TAC CAT GCC CCA GAT TCA-3′ and 5′-AGT GCT TCC ATC GGA AGG ACT-3′; 18S, 5′-TCG GAA CTG AGG CCA TGA TT-3′ and 5′-TAG CGG CGC AAT ACG AAT G-3′.
siRNA
siRNAs to p23 (PTGES3_1 (SI02780911) and PTGES3_2 (SI02781170); the former was employed for most experiments) and FKBP38 (FKBP8_5 (SI03035977)) were obtained from Qiagen. siRNA to PHD2 (PHD2-A) has been described (37). The control non-targeting siRNA #2 (D-001210-02) and non-targeting siRNA #3 (D-001210-03) were from Dharmacon (the latter was employed for most experiments). Transfections were performed with siRNA concentrations of 20 nm. Lysates were prepared in buffer C supplemented with mammalian protease inhibitor mixture and clarified by centrifugation at 15,800 × g for 10 min at 4 °C. Protein concentrations were measured using a Bio-Rad DC Protein Assay, and equal amounts were examined by Western blotting.
Luciferase Assays
Luciferase assays were performed on a Wallac LB9507 luminometer using a Dual Luciferase Reporter Assay System (Promega).
Statistical Analysis
Analysis of variance (GraphPad Prism) was employed for statistical analysis. Differences were considered significant when p < 0.05.
RESULTS
PHD2 Interacts with Select HSP90 Co-chaperones
In many signal transduction pathways key enzymes do not act in isolation but, rather, interact with other proteins. We hypothesized that this might be the case with PHD2. To pursue this, we generated a human embryonic kidney (HEK) 293 cell line expressing FLAG-tagged PHD2. Using a stable isotope labeling by amino acids in cell culture (SILAC) approach (31), we subjected this cell line (labeled with heavy isotope) as well as the parental cell line (labeled with light isotope) to immunoprecipitation with anti-FLAG antibodies and eluted with FLAG peptide (Fig. 1A). The eluates were mixed, digested with trypsin, and analyzed by mass spectrometry. As expected, PHD2 displays a very high heavy isotope to light (H/L) isotope ratio (39.5). The group of proteins with the highest degree of enrichment were ones of the HSP90 pathway, including FKBP38 (H/L ratio of 22.5), p23 (19.0), FKBP51 (17.5), FKBP52 (6.3), and HSP90 itself (6.6 for the AA isoform, 6.4 for the AB isoform).
FIGURE 1.
Mass spectrometry identification of PHD2-interacting proteins. A, Flp-In T-Rex HEK293 or Flp-In T-Rex HEK293/FLAG-PHD2 cells were grown in the presence of amino acids with light or heavy labeled lysine/arginine, respectively and subjected to immunoprecipitation (IP) with anti-FLAG antibodies, and the eluates were mixed and subjected to tryptic digest and mass spectrometry. The square and hexagon depict proteins that bind specifically to PHD2, whereas the small circles depict proteins that bind non-specifically to the antibody (or resin). B, HEK293FT cells were transfected with expression vectors for the indicated proteins, lysed, and subjected to immunoprecipitation with anti-FLAG antibodies, and Western blots (WB) were then performed. The positions of HA-PHD2 or molecular weight markers are shown to the right. C, Flp-In T-Rex HEK293 or Flp-In T-Rex HEK293/FLAG-PHD2 cells were induced with doxycycline, lysed, and subjected to immunoprecipitation with anti-FLAG antibodies. The immunoprecipitates were eluted with 3X FLAG peptide. Aliquots of lysate or eluate were then subjected to Western blotting using antibodies against the indicated proteins. Anti-FLAG antibodies were employed to detect FLAG-PHD2. D and E, HEK293FT cells were maintained under normoxia or hypoxia (1% O2 for 4 h) (D) or treated with control or p23 siRNA (E). Ab, antibody. Cells were lysed, incubated with 10 μg of either control or anti-PHD2 monoclonal antibody, and immunoprecipitated with protein G-agarose, and Western blots were then performed.
HSP90 can associate with a multitude of co-chaperones that perform a number of functions, including the recruitment of regulatory or client proteins to the HSP90 machinery (23, 25, 26). Only a few co-chaperones were identified in the SILAC experiment just described. To explore the possibility that select co-chaperones may interact with PHD2, we coexpressed PHD2 with various HSP90 co-chaperones and immunoprecipitated the latter. We found that PHD2 selectively coimmunoprecipitates with p23 and FKBP38 (Fig. 1B, top panel, lanes 2 and 5). In reciprocal coimmunoprecipitations, we confirmed the presence of p23, FKBP38, and HSP90 in PHD2 immunoprecipitates (Fig. 1C).
We next employed a monoclonal antibody that can specifically immunoprecipitate PHD2 but not PHD1 or PHD3 (36). We immunoprecipitated endogenous PHD2 from HEK293FT cells and demonstrate the presence of endogenous p23 in these immunoprecipitates (Fig. 1D, top panel, lane 2). Exposure of cells to 1% O2 did not affect the association (Fig. 1D, top panel, compare lanes 2 and 4). We also knocked down p23 using siRNA in these cells, immunoprecipitated endogenous PHD2, and examined the immunoprecipitates for the presence of HSP90. We found, as expected, that HSP90 coimmunoprecipitates with PHD2 (Fig. 1E, top panel, lane 2). Knockdown of p23 substantially diminishes this coimmunoprecipitation (Fig. 1E, top panel, compare lanes 2 and 3), suggesting that p23 is an important, although not necessarily the exclusive, means by which PHD2 interacts with HSP90.
Other proteins of the HSP90 pathway, such as HSP70, HOP (also known as STIP1), PPP5C, or FKBP16 (also known as AIP or XAP2) as well as others involved in protein folding, such as HSP60, GRP94, HSP73, or GRP75, were not enriched in PHD2 immunoprecipitates, as assessed by Western blotting (Fig. 1, B and C) or SILAC analysis (data not shown). The selective interaction of PHD2 with p23 and FKBP38 is consistent with their relatively high heavy to light isotope ratio in the SILAC experiment.
PHD2 Binds to a PXLE Motif
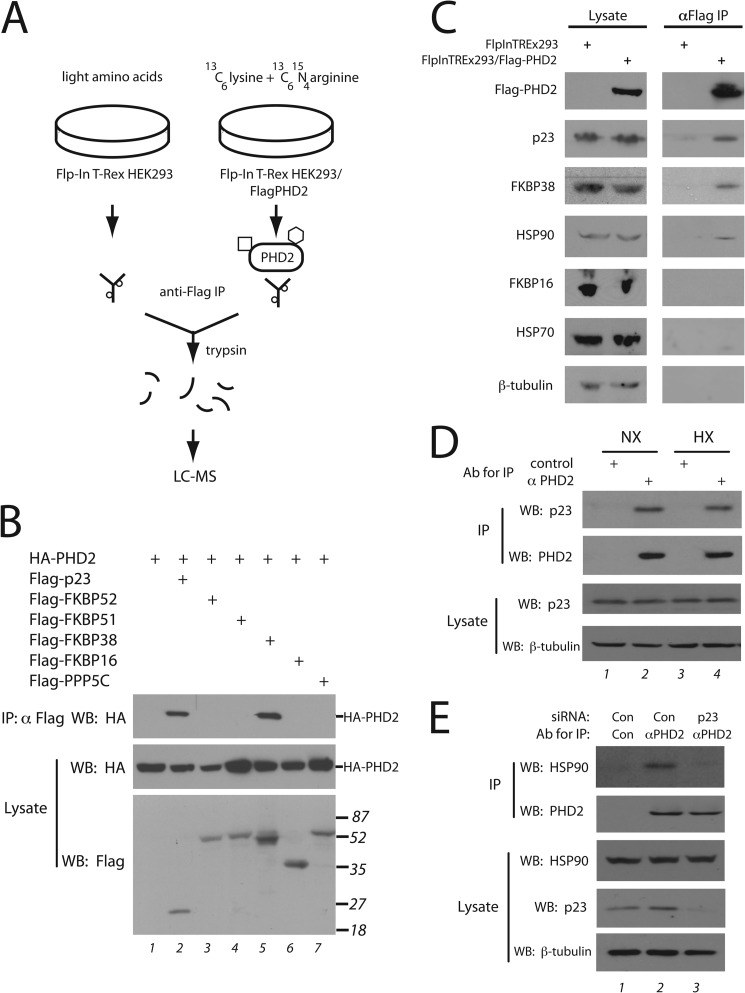
p23 is an evolutionarily conserved co-chaperone that binds to HSP90 in an ATP-dependent manner and plays a critical role in numerous HSP90-dependent processes (25, 38). For certain client proteins, it enters late in the HSP90-dependent maturation cycle. FKBP38 appears to have emerged later in evolution and is a co-chaperone anchored to the endoplasmic reticulum (39). It also appears to have a more specialized function. For example, it has been implicated in the regulation of Bcl-2 (40). The interaction between PHD2 and FKBP38 has been reported previously and was shown to depend on the interaction of PHD2 with an acidic region in FKBP38 comprising residues 37–56 (41). We confirmed this by deleting the N-terminal 57 amino acids of FKBP38 and observing that this abolishes interaction with PHD2 in coimmunoprecipitation assays (data not shown). Inspection of the p23 amino sequence reveals that it possesses an acidic region at its C terminus that shares a PXLE motif with FKBP38 (Fig. 2A). This motif shows strong conservation in p23 through evolution (Fig. 2B).
FIGURE 2.
A conserved PXLE motif in select HSP90 co-chaperones. A, shown is a schematic diagram of p23. Shading denotes the CHORD and Sgt1 (CS) domain, and numbers indicate amino acid number. The C-terminal 20 amino acids of p23 are shown and compared with the indicated residues of FKBP38. B, shown is a comparison p23 sequences from Homo sapiens (Unigene Hs.50425), Mus musculus (Mm.305816), Xenopus. laevis (Xl.14340), Danio rario (Dr.77365), C. elegans (Cel.8134), and Trichoplax adhaerens (XP_002111186). Shading indicates residues conserved in all of the proteins shown. C, HEK293FT were transfected and immunoprecipitated (IP) as in panel B. AAA denotes a P157A/L159A/E160A mutation in p23. WB, Western blot.
We examined the importance of the PXLE motif in p23 by mutating the conserved Pro, Leu, and Glu simultaneously to Ala. Using coimmunoprecipitation assays with full-length proteins, we observed that this mutation abolishes interaction with PHD2 (Fig. 2C, top panel, lanes 3 and 4). We also immobilized a peptide comprising p23 (151–160) on agarose beads (Fig. 3A) and found that this peptide is sufficient to confer interaction with PHD2 (Fig. 3B, top left panel, second lane). In the context of this peptide, the P157A/L159A/E160A mutation abolishes the interaction (third lane). A parallel experiment with an FKBP38 peptide (residues 47–56) containing the PXLE motif shows that this peptide is sufficient to confer interaction with PHD2, which again is abolished by mutation of this motif (Fig. 3B, bottom right panel, last two lanes).
FIGURE 3.
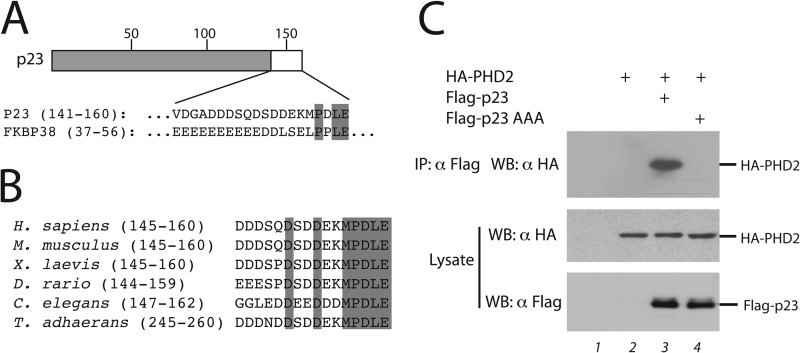
PHD2 binds to a PXLE motif. A, shown is a comparison of p23 residues 151–160 to FKBP38 residues 47–56. The numbers above the sequences correspond to p23 residues. Shading indicates conserved residues of the PXLE motif. B, Sf9 lysates containing His6-FLAG-PHD2 were incubated with p23 peptides immobilized on streptavidin-agarose and washed, and the eluates were examined for the presence of PHD2 by Western blotting using anti-FLAG antibodies. Amino acid substitutions are indicated in each lane. PLE>AAA denotes P157A/L159A/E160A, and DDE>AAA denotes D152A/D153A/E154A. Experiments with FKBP38 peptides are shown in the last two lanes of the bottom right panel. FKBP38 AAA denotes P53A/L55A/E56A. In each panel, peptide was omitted in the first lane. The relative degree of binding for each peptide, normalized to that of wild-type (WT) p23 peptide, is given below each panel.
In further experiments with p23 we found that mutation of Glu-160 (to Ala, Asp, or Gln), Leu-159 (to Ala, Ile, or Val), or Pro-157 (to Ala or Ser) substantially reduces, if not abolishes PHD2 binding, highlighting the stringency of this amino acid motif. In contrast, amino acid substitutions at position 158 (to Ala, Pro, and Leu), which is not conserved between p23 and FKBP38, can be tolerated. Amino acid 156 is a hydrophobic residue (Met), and we found that substitutions to Leu and Val preserve the interaction; indeed, the Leu substitution has a somewhat increased binding. In contrast, substitutions to Ala, Phe, or Glu essentially abolish it, suggesting a preference for select hydrophobic amino acids. Mutations of each of the four amino acids preceding the MPDLE motif preserves the interaction, with the exception of D153A, which substantially diminishes it, as does a triple mutation of the three acidic residues (D152A/D153A/E154A). The importance of Asp at position 153 is likely to be specific to p23, as the corresponding residue in FKBP38 is Leu (Fig. 3A), but may nevertheless point to a necessity for having one or more acidic residues preceding the PXLE motif.
The MYND-type Zinc Finger of PHD2 Binds the PXLE Motif
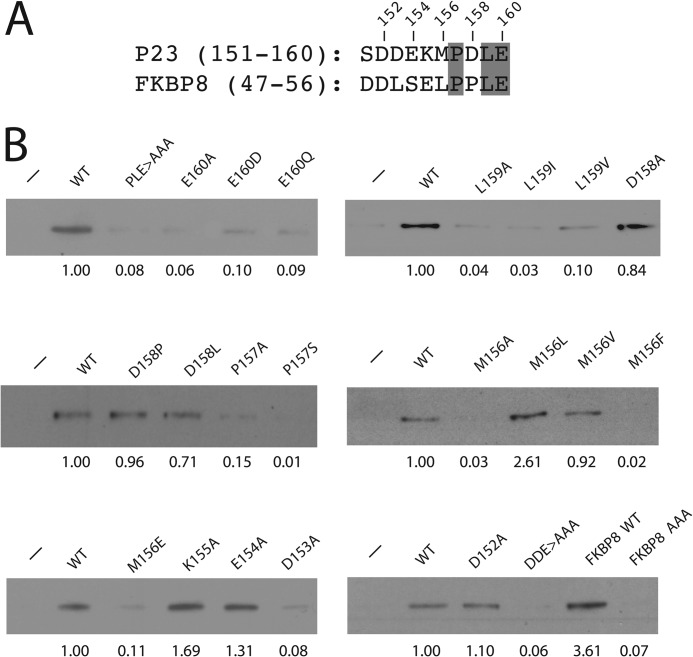
PHD2 is one of three PHD isoforms that has the capacity to hydroxylate HIF-α (9, 10). We coexpressed FLAG-p23 with HA-tagged versions of PHD1, PHD2, or PHD3 and found that only PHD2 interacts with p23 (Fig. 4A, lane 5). A similar result has been observed with FKBP38 (42). The three PHDs share a conserved C-terminal hydroxylase domain but differ at their N termini, raising the possibility that the N terminus of PHD2 mediates this interaction. Indeed, an N-terminal fragment of PHD2 interacts with p23, whereas a C-terminal fragment does not (Fig. 4B, compare lanes 5 and 7). The N terminus of PHD2 is distinctive among the three PHDs in that it is predicted to harbor a MYND-type zinc finger (9, 20) (Fig. 4C). Mutation of predicted zinc-chelating residues (C36S/C42S) in PHD2 abolishes its ability to coimmunoprecipitate with p23 (Fig. 4B, lane 9). This mutation also eliminates the ability of GST-PHD2 (1–124) to bind FKBP38 (data not shown). These results strongly suggest that the integrity of the MYND-type zinc finger is essential for interaction with the PXLE motif.
FIGURE 4.
The MYND-type zinc finger of PHD2 is required for its interaction with p23. A and B, HEK293FT were transfected and immunoprecipitated (IP) as in Fig. 1B. WB, Western blot. C, shown is a schematic diagram of PHD2. Dark shading denotes the N-terminal MYND-type zinc finger (ZF), light shading denotes the C-terminal prolyl hydroxylase (PH) domain, and numbers indicate amino acid number. Shown at the top are amino acids 21–58. Underlining indicates predicted zinc chelating residues. Inverted triangles indicate Cys-36 and Cys-42.
p23 Is Functionally Important for HIF-1α Regulation
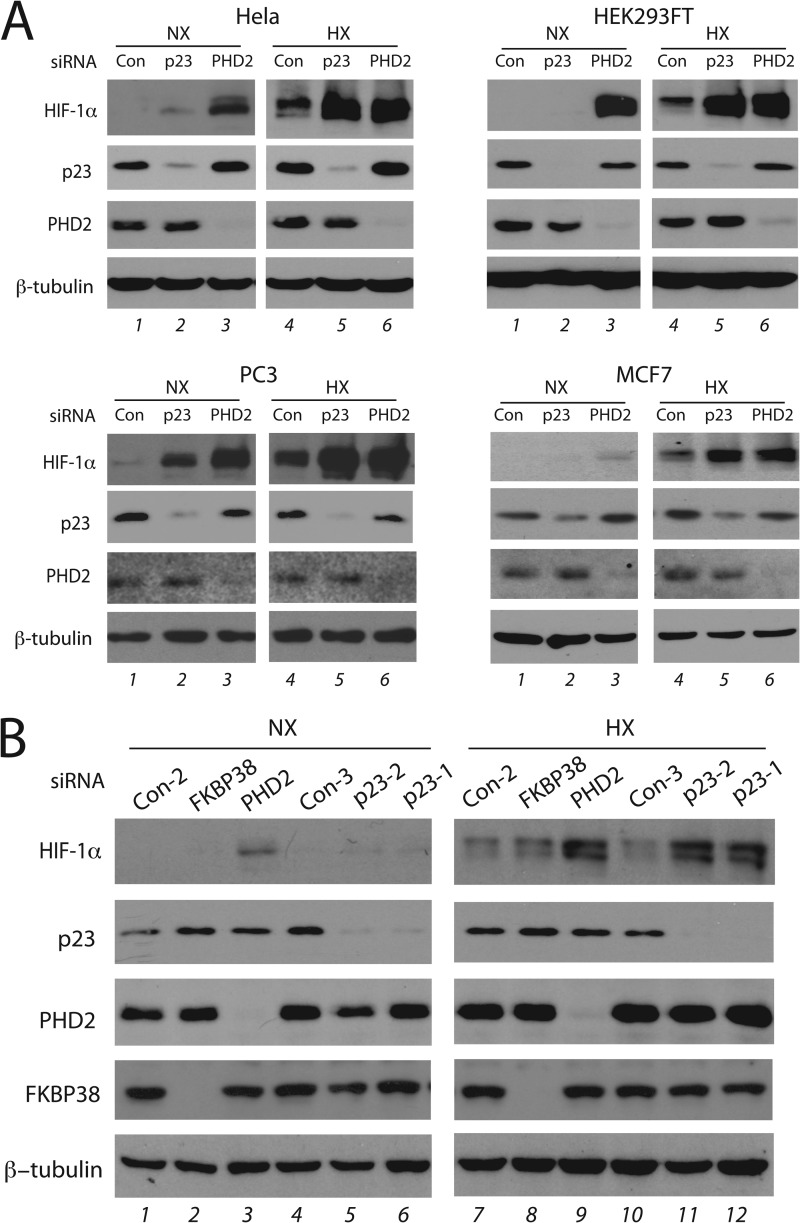
We knocked down p23 or PHD2 using siRNA in HeLa cells and examined the levels of various proteins by Western blotting. Under normoxic conditions we found that p23 knockdown has at best a marginal effect on HIF-1α levels (Fig. 5A, left upper panel, lane 2). We then examined these cells under moderate hypoxia (2% O2), a situation in which PHD2 continues to have activity; indeed, PHD2 knockdown results in a substantial increase in HIF-1α levels (Fig. 5A, left upper panel, compare lanes 4 and 6). Importantly, we found that p23 knockdown augments HIF-1α levels to ones comparable to those seen after PHD2 knockdown (lane 5). Equally significant, this p23 knockdown augmentation of hypoxic HIF-1α levels is seen in a range of other cell types, including HEK293, PC3, and MCF7 cells (compare lanes 4 and 5 in each set of panels). Augmentation of normoxic HIF-1α levels after p23 knockdown was seen in PC3 cells but not in HEK293 and MCF7 cells (compare lanes 1 and 2 in each set of panels). Western blotting indicates that the effects of p23 knockdown cannot be attributed to changes in PHD2 levels (Fig. 5A). Knockdown of p23 using an independent siRNA yields the same effect (Fig. 5B, lanes 11 and 12); in addition, under the conditions of this experiment, we found that FKBP38 knockdown did not have an effect on HIF-1α levels (lanes 2 and 8).
FIGURE 5.
Knockdown of p23 augments HIF-1α protein levels. A, the indicated cell lines were treated with the indicated siRNA and then maintained under normoxia (NX) or subjected to 2% O2 (HX) for 4 h. Cell lysates were prepared, and equal protein amounts were then subjected to Western blotting using antibodies to the indicated proteins. Con = control non-targeting siRNA #3. The p23 siRNA was PTGES3_1. B, HeLa cells were treated with the indicated siRNA and then examined as in A. The p23-1 siRNA was PTGES3_1, and the p23-2 siRNA was PTGES3_2.
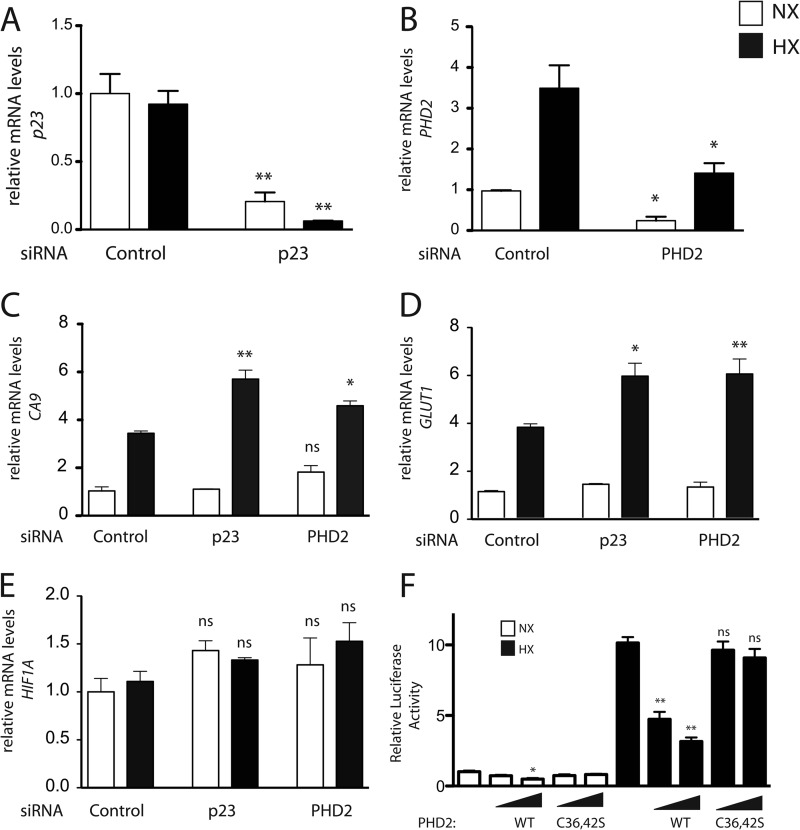
We performed real time PCR on RNA extracted from HeLa cells after p23 or PHD2 siRNA treatment. This analysis confirmed the efficacy of knockdown of p23 and PHD2 (Fig. 6, A and B, respectively). Consistent with the previous results examining HIF-1α protein levels (Fig. 5), knockdown of PHD2 yields an increase in hypoxia-induced mRNA levels of the canonical HIF target genes, CA9 and GLUT1 (Fig. 6, C and D, respectively). Importantly, we found that p23 knockdown also augments message levels of these same targets. For either p23 or PHD2 knockdown, this augmentation cannot be attributed to changes in HIF1A mRNA levels (Fig. 6E).
FIGURE 6.
Knockdown of p23 augments HIF-1α target genes. A–E, HeLa cells were treated with the indicated siRNA for 72 h and then maintained under normoxia (NX) or subjected to 2% O2 (HX) for 16 h. mRNA levels for the indicated genes were measured by real time PCR and normalized to that of 18 S rRNA. Shown are the means ± S.D., n = 3. *, p < 0.05 compared with the respective normoxic or hypoxic control; **, p < 0.01 compared with the respective normoxic or hypoxic control; ns, not significant compared with the respective normoxic or hypoxic control. F, HEK293FT cells were transfected with 50 ng of (eHRE)3-Luc, 50 ng of pRL-TK, and either 5 or 15 ng of either pcDNA3-FLAG-PHD2 or pcDNA3-HA-PHD2 (C36S/C42S). DNA doses were held constant by the addition of pcDNA3. After 8 h, cells were exposed to 1% O2 (HX) for an additional 16 h or maintained under normoxia (NX). Cells were lysed, and luciferase activity was measured and normalized to that of Renilla luciferase. Shown are the means ± S.D., n = 3. *, p < 0.05 compared with the respective normoxic or hypoxic control; **, p < 0.01 compared with the respective normoxic or hypoxic control; ns, not significant compared with the respective normoxic or hypoxic control.
The data support an important role for an interaction between p23 and the zinc finger of PHD2. In a different experiment to assess this, we transfected HEK293FT cells with a luciferase reporter gene with a hypoxia response element reporter gene and constructs for either wild-type or zinc finger-defective (C36S/C42S) PHD2. The cells were exposed to hypoxia, and luciferase activities were measured. We found that hypoxia, as expected, robustly induces activity from this reporter gene (Fig. 6F). Under conditions where wild-type PHD2 effectively inhibits activation of this reporter, zinc finger-defective PHD2 does not. This, therefore, provides further evidence for a functionally significant interaction between p23 and PHD2.
We next generated a HEK293 cell line expressing FLAG-tagged p23 and then immunoprecipitated the p23 in a SILAC experiment analogous to the one that originally identified p23 as a PHD2-interacting protein. By mass spectrometry, we found that p23 displays the highest H/L ratio, 26.9 and that other components of the HSP90 pathway are also enriched, consistent with the well established role of p23 in HSP90 dynamics. These proteins include FKBP51 (H/L ratio of 12.0), PPP5C (6.8), FKBP52 (4.8), HSP90AB1 (3.8), and HSP90AA1 (3.6). Among non-HSP90 pathway proteins, KIF1-binding protein displays the highest H/L ratio (21.6). Strikingly, PHD2 displays the next highest (13.3). Western blotting confirms the presence of endogenous PHD2 in p23 immunoprecipitates in this cell line (data not shown). Therefore, this unbiased SILAC experiment independently supports an interaction between p23 and PHD2.
p23 Is Functionally Important for HIF-1α Hydroxylation
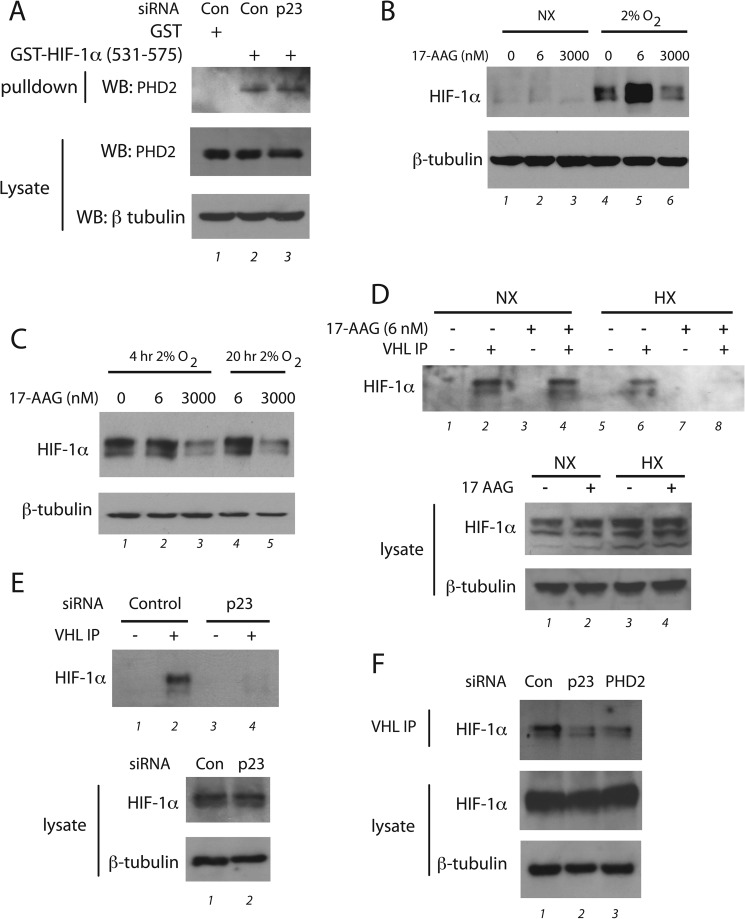
One possible explanation for the present results is that PHD2 itself is a client of the HSP90 pathway, i.e. the HSP90 pathway properly folds PHD2 into a catalytically active conformation. A number of observations argue against this. First, p23 knockdown does not affect PHD2 protein levels (Fig. 5). Second, mutation of the MYND-type zinc finger of PHD2 abolishes its interaction with p23 (Fig. 4B), indicating that at least for this particular domain of PHD2, it is actually the mature folded state, as opposed to an unfolded state, that is essential for interaction. To explore the possibility that p23 may be necessary for folding of the other domain of PHD2, i.e. its catalytic domain, we assessed the interaction of PHD2 with HIF-1α (530–575), which contains the hydroxylacceptor proline, Pro-564. We knocked down p23 in HeLa cells, incubated cellular extracts with GST-HIF-1α (531–575), and assessed PHD2 binding by Western blotting. As shown in Fig. 7A, we did not find any detectable effect of p23 knockdown on PHD2 binding to HIF-1α (530–575), suggesting that p23 is not essential for the folding of the PHD2 catalytic domain.
FIGURE 7.
The PHD2-p23 interaction facilitates prolyl hydroxylation of HIF-1α. A, HeLa cells were treated with control (Con) or p23 siRNA, and cellular extracts were prepared. GST or GST-HIF-1α (531–575) prebound to GSH-agarose was incubated with the extracts and washed, and bound PHD2 was then assessed by Western blotting (WB) using the indicated antibodies. B, PC3 cells were treated with the indicated concentrations of 17-AAG and then immediately subjected to normoxia (NX) or 2% O2 for 4 h. Cell lysates were prepared, and equal protein amounts were subjected to Western blotting using antibodies to the indicated proteins. C, RCC4 cells were treated with the indicated concentrations of 17-AAG and then immediately subjected to normoxia or 2% O2 for 4 or 20 h. Cell lysates were prepared, and equal protein amounts were subjected to Western blotting using antibodies to the indicated proteins. D, RCC4 cells were subjected to either 6 nm 17-AAG, 2% O2 (HX), or both for 20 h. The cells were lysed, and equal protein amounts were incubated with or without recombinant FLAG-VHL, the FLAG-VHL was immunoprecipitated, and proteins were captured by VHL analyzed by Western blotting using antibodies to HIF-1α. Lysates were also analyzed by Western blotting. E, HeLa cells were treated with the indicated siRNA and then subjected to 2% O2 for 4 h in the presence of 10 μm MG132. The cells were lysed, equal protein amounts were incubated with or without recombinant FLAG-VHL, the FLAG-VHL was immunoprecipitated, and proteins were captured by VHL analyzed by Western blotting using antibodies to HIF-1α. Lysates were also analyzed by Western blotting. F, HeLa cells were treated with the indicated siRNA and then subjected to 2% O2 for 4 h in the presence of 10 μm MG132. The cells were lysed, equal protein amounts were incubated with recombinant FLAG-VHL, the FLAG-VHL was immunoprecipitated, and proteins were captured by VHL analyzed by Western blotting using antibodies to HIF-1α. Lysates were also analyzed by Western blotting.
HSP90 inhibitors, such as geldanamycin and 17-N-allylamino-17-demethoxygeldanamycin (17-AAG), are known to produce a biphasic response on HIF-1α levels. At high (μm) concentrations, they diminish HIF-1α levels (24), whereas at low (nm) concentrations, they increase it (43). The former is consistent with HIF-1α being an HSP90 client protein. The latter, based on the present data, might conceivably be due to effects on the PHD2-p23 axis, as it is known that both HSP90 inhibitors and p23 bind to the N terminus of HSP90 (38, 44). To examine this, we treated PC3 cells with 17-AAG and then either maintained cells under normoxia or exposed them to hypoxia. Consistent with previous results, we found diminished hypoxia-induced HIF-1α levels at high (3 μm) 17-AAG concentrations (Fig. 7B, compare lanes 4 and 6). Importantly, we observed increased HIF-1α levels at low (6 nm) 17-AAG concentrations, particularly under hypoxic conditions (lane 5), a situation that mirrors p23 knockdown. We also examined the effect of 17-AAG on HIF-1α levels in RCC4 cells, which lack functional VHL and thereby dissociate prolyl hydroxylation from degradation. Again, consistent with previous results (24), high 17-AAG concentrations diminished HIF-1α levels (Fig. 7C, lanes 3 and 5). Low 17-AAG concentrations, in contrast, have no effect on HIF-1α levels in these cells under either normoxic or hypoxic conditions (lanes 2 and 4), thereby arguing that the effects of low 17-AAG concentrations seen in PC3 cells are not due to effects on HIF-1α translation.
We further employed the RCC4 cells to examine the hydroxylation state of HIF-1α. We treated cells with or without 6 nm 17-AAG in the absence or presence of hypoxia (2% O2) for 20 h. Cells were lysed and mixed with recombinant FLAG-tagged VHL (as a complex with Elongin B and Elongin C) to capture hydroxylated HIF-1α. The FLAG-VHL was then immunoprecipitated, and eluates were then examined for the presence of HIF-1α by Western blotting. Examination of the lysates indicates that neither low dose (6 nm) 17-AAG nor hypoxia had an appreciable effect on HIF-1α levels (Fig. 7D, second panel from bottom), consistent with results of Fig. 7C. In the absence of VHL capture probe, no HIF-1α was detectable in the anti-FLAG immunoprecipitates, whereas in its presence HIF-1α is readily detectable (top panel, compare lanes 1 and 2), confirming the specificity of the interaction and also indicating the presence of a steady state level of HIF-1α hydroxylation in these cells. Under normoxia, we found no difference in the extent of HIF-1α hydroxylation upon 17-AAG treatment (lanes 2 and 4). Under hypoxia, the level of HIF-1α hydroxylation is diminished in control cells yet was still detectable (compare lanes 2 and 6). Importantly, we found that this hydroxylation was further and substantially attenuated by 17-AAG treatment (lane 8). These findings are consistent with low dose 17-AAG inhibiting PHD2-induced hydroxylation of HIF-1α, presumably by interference with p23-mediated recruitment of PHD2 to HSP90.
We pursued this further by employing the VHL capture assay to assess HIF-1α hydroxylation after p23 knockdown in HeLa cells. In these experiments we knocked down p23 and then exposed cells to 2% O2 in the presence of the proteasome inhibitor MG132 to allow stabilization of hydroxylated HIF-1α. Recombinant VHL was then employed to capture hydroxylated HIF-1α, which was then detected by Western blotting. As before, no HIF-1α was detected in the absence of recombinant VHL, confirming the specificity of the capture probe (Fig. 7E, top panel, compare lanes 1 and 2). In control cells, hydroxylated HIF-1α was readily detected, indicating that prolyl hydroxylation occurs and is detectable under these modest hypoxic conditions (lane 2). Strikingly, we found that p23 knockdown results in markedly lower levels of HIF-1α hydroxylation (lane 4). Additional experiments using this same VHL capture approach reveal that the decrease in HIF-1α hydroxylation induced by p23 knockdown is comparable with that induced by PHD2 knockdown (Fig. 7F, top panel, lanes 2 and 3). We conclude that p23 serves to recruit PHD2 to HSP90 to promote optimal HIF-1α hydroxylation.
DISCUSSION
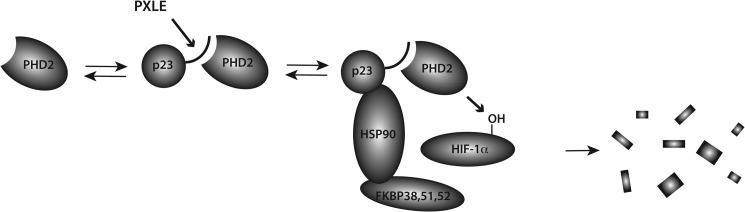
These data provide evidence for a critical link between PHD2 and the HSP90 pathway (Fig. 8). A model consistent with the data is that p23 recruits PHD2 to HSP90 to facilitate HIF-1α hydroxylation. This model is compatible with previous observations that HIF-1α is itself an HSP90 client protein (24) and also with previous work which indicates that p23 enters the HSP90 cycle at a late stage in the maturation cycle of HSP90 client proteins (23). The model would allow for efficient PHD2-mediated hydroxylation, as HIF-1α proceeds through an obligatory HSP90-dependent pathway. The more pronounced effect of p23 knockdown seen under hypoxic conditions would suggest that the coupling to the HSP90 pathway is relatively more important under these conditions.
FIGURE 8.
Model for interaction of PHD2 with HSP90 pathway. PHD2 associates with p23 via a PXLE motif in p23 (denoted by curved line). p23 in turn is known to bind to the N terminus of HSP90. This facilitates PHD2-catalyzed hydroxylation of HIF-1α, which is known to be an HSP90 client protein. Prolyl hydroxylation in turn marks HIF-1α for degradation. The C terminus of HSP90 is known to bind a subset of HSP90 co-chaperones that includes FKBP38, FKBP51, and FKBP52. FKBP38 possesses a PXLE motif and, therefore, provides an independent means for recruiting PHD2 to the HSP90 machinery.
HSP90 co-chaperones are diverse in terms of both structure and function (25, 26). Many HSP90 co-chaperones serve to recruit HSP90 client proteins (23). Prominent examples include the recruitment of protein kinases by Cdc37 (45) and Nod-like receptors by Sgt1 (46, 47). In addition, select co-chaperones have intrinsic catalytic activities. Examples of these include FKBP52, which has peptidyl prolyl isomerase activity, and PPP5C, which has protein phosphatase activity. Our model is different from these previously described uses of co-chaperones in that it proposes that a co-chaperone (p23) recruits an enzyme (PHD2), which post-translationally modifies proteins. The enzymatic activity of PHD2 is clearly essential for its ability to regulate HIF-1α (9, 10, 30), and it will be of interest to determine if the proposed mechanism may also be used to promote the hydroxylation of non-HIF protein targets. In addition, it will be of interest to examine whether there might be hydroxylase-independent functions of PHD2. Interestingly, previous studies on PHD2 point to this possibility (48, 49). It might be noted that the capacity of FKBP52 to serve as a co-chaperone in the glucocorticoid receptor maturation pathway is independent of its peptidyl prolyl isomerase activity (50).
The present studies also identify a functional role for the MYND-type zinc finger of PHD2, namely, interaction with a PXLE motif present in p23 and FKBP38. MYND domains serve as protein-protein interaction motifs in other proteins (51). For example, the MYND domain-containing transcriptional corepressor BS69 binds to a PXLXP motif present in E1A (52), whereas the MYND domain-containing AML1/ETO fusion protein binds to a PPPLI motif in SMRT (53). These motifs share similarity to the PXLE motif to which the PHD2 MYND zinc finger binds. It is conceivable that MYND zinc fingers as a whole bind a core consensus sequence of PXL, with adjacent residues determined in a MYND domain-specific manner; additional studies on other MYND zinc fingers will be necessary to address this possibility. The SMRT PPPLI motif binds in an extended conformation to AML1/ETO (53). The p23 PXLE motif is C-terminal, and studies indicate that it is unstructured (54, 55), raising the possibility that it may also bind in an extended conformation to the PHD2 MYND-type zinc finger.
Taken together, these findings provide evidence for a direct link between two ancient pathways, the oxygen-sensing pathway and the HSP90 pathway. The significance of this link is supported by the strong conservation of the MYND domain of PHD2 through evolution (21, 22). This domain, for example, is present in the single PHD isoform from the simplest animal, T. adhaerens, which possesses both a PXLE motif-containing p23 homologue (Fig. 2B) and a functional PHD:HIF pathway (21). This suggests that the coupling of the PHD:HIF pathway to the HSP90 machinery emerged early in evolution, indeed perhaps concurrent with the appearance of the PHD:HIF pathway itself.
Acknowledgments
We thank Heddy Kerestes for excellent technical assistance, Yu Jin Chung for helpful discussions, Dr. Celeste Simon for the gift of RCC4 cells, Dr. Mark Lemmon for the gift of MCF7 cells, and Dr. David B. Roth and Dr. Mark L. Tykocinski for support.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-GM090301 (to F. S. L. and S. R. M.) and R01-CA153347 (to F. S. L.).
- VHL
- von Hippel Lindau protein
- 17-AAG
- 17-N-allylamino-17-demethoxygeldanamycin
- FKBP
- FK506-binding protein
- HIF
- hypoxia-inducible factor
- HSP
- heat shock protein
- PHD
- prolyl hydroxylase domain protein
- MYND
- Myeloid Nervy and DEAF-1
- SILAC
- stable isotope labeling by amino acids in cell culture
- H/L
- heavy to light ratio.
REFERENCES
- 1. Kaelin W. G., Jr., Ratcliffe P. J. (2008) Oxygen sensing by metazoans. The central role of the HIF hydroxylase pathway. Mol. Cell 30, 393–402 [DOI] [PubMed] [Google Scholar]
- 2. Semenza G. L. (2007) Life with oxygen. Science 318, 62–64 [DOI] [PubMed] [Google Scholar]
- 3. Majmundar A. J., Wong W. J., Simon M. C. (2010) Hypoxia-inducible factors and the response to hypoxic stress. Mol. Cell 40, 294–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ivan M., Haberberger T., Gervasi D. C., Michelson K. S., Günzler V., Kondo K., Yang H., Sorokina I., Conaway R. C., Conaway J. W., Kaelin W. G., Jr. (2002) Biochemical purification and pharmacological inhibition of a mammalian prolyl hydroxylase acting on hypoxia-inducible factor. Proc. Natl. Acad. Sci. U.S.A. 99, 13459–13464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jaakkola P., Mole D. R., Tian Y. M., Wilson M. I., Gielbert J., Gaskell S. J., von Kriegsheim A., Hebestreit H. F., Mukherji M., Schofield C. J., Maxwell P. H., Pugh C. W., Ratcliffe P. J. (2001) Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292, 468–472 [DOI] [PubMed] [Google Scholar]
- 6. Yu F., White S. B., Zhao Q., Lee F. S. (2001) HIF-1α binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc. Natl. Acad. Sci. U.S.A. 98, 9630–9635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Maxwell P. H., Wiesener M. S., Chang G. W., Clifford S. C., Vaux E. C., Cockman M. E., Wykoff C. C., Pugh C. W., Maher E. R., Ratcliffe P. J. (1999) The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399, 271–275 [DOI] [PubMed] [Google Scholar]
- 8. Lendahl U., Lee K. L., Yang H., Poellinger L. (2009) Generating specificity and diversity in the transcriptional response to hypoxia. Nat. Rev. Genet. 10, 821–832 [DOI] [PubMed] [Google Scholar]
- 9. Epstein A. C., Gleadle J. M., McNeill L. A., Hewitson K. S., O'Rourke J., Mole D. R., Mukherji M., Metzen E., Wilson M. I., Dhanda A., Tian Y. M., Masson N., Hamilton D. L., Jaakkola P., Barstead R., Hodgkin J., Maxwell P. H., Pugh C. W., Schofield C. J., Ratcliffe P. J. (2001) C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107, 43–54 [DOI] [PubMed] [Google Scholar]
- 10. Bruick R. K., McKnight S. L. (2001) A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294, 1337–1340 [DOI] [PubMed] [Google Scholar]
- 11. Berra E., Benizri E., Ginouvès A., Volmat V., Roux D., Pouysségur J. (2003) HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1α in normoxia. EMBO J. 22, 4082–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Appelhoff R. J., Tian Y. M., Raval R. R., Turley H., Harris A. L., Pugh C. W., Ratcliffe P. J., Gleadle J. M. (2004) Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J. Biol. Chem. 279, 38458–38465 [DOI] [PubMed] [Google Scholar]
- 13. Takeda K., Ho V. C., Takeda H., Duan L. J., Nagy A., Fong G. H. (2006) Placental but not heart defects are associated with elevated hypoxia-inducible factor α levels in mice lacking prolyl hydroxylase domain protein 2. Mol. Cell. Biol. 26, 8336–8346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Percy M. J., Zhao Q., Flores A., Harrison C., Lappin T. R., Maxwell P. H., McMullin M. F., Lee F. S. (2006) A family with erythrocytosis establishes a role for prolyl hydroxylase domain protein 2 in oxygen homeostasis. Proc. Natl. Acad. Sci. U.S.A. 103, 654–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee F. S., Percy M. J. (2011) The HIF pathway and erythrocytosis. Annu. Rev. Pathol. 6, 165–192 [DOI] [PubMed] [Google Scholar]
- 16. Percy M. J., Furlow P. W., Beer P. A., Lappin T. R., McMullin M. F., Lee F. S. (2007) A novel erythrocytosis-associated PHD2 mutation suggests the location of a HIF binding groove. Blood 110, 2193–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ladroue C., Carcenac R., Leporrier M., Gad S., Le Hello C., Galateau-Salle F., Feunteun J., Pouysségur J., Richard S., Gardie B. (2008) PHD2 mutation and congenital erythrocytosis with paraganglioma. N. Engl. J. Med. 359, 2685–2692 [DOI] [PubMed] [Google Scholar]
- 18. Simonson T. S., Yang Y., Huff C. D., Yun H., Qin G., Witherspoon D. J., Bai Z., Lorenzo F. R., Xing J., Jorde L. B., Prchal J. T., Ge R. (2010) Genetic evidence for high-altitude adaptation in Tibet. Science 329, 72–75 [DOI] [PubMed] [Google Scholar]
- 19. Beall C. M. (2011) Genetic changes in Tibet. High Alt. Med. Biol. 12, 101–102 [DOI] [PubMed] [Google Scholar]
- 20. Taylor M. S. (2001) Characterization and comparative analysis of the EGLN gene family. Gene 275, 125–132 [DOI] [PubMed] [Google Scholar]
- 21. Loenarz C., Coleman M. L., Boleininger A., Schierwater B., Holland P. W., Ratcliffe P. J., Schofield C. J. (2011) The hypoxia-inducible transcription factor pathway regulates oxygen sensing in the simplest animal, Trichoplax adhaerens. EMBO Rep. 12, 63–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rytkönen K. T., Williams T. A., Renshaw G. M., Primmer C. R., Nikinmaa M. (2011) Molecular evolution of the metazoan PHD-HIF oxygen-sensing system. Mol. Biol. Evol. 28, 1913–1926 [DOI] [PubMed] [Google Scholar]
- 23. Taipale M., Jarosz D. F., Lindquist S. (2010) HSP90 at the hub of protein homeostasis. Emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 11, 515–528 [DOI] [PubMed] [Google Scholar]
- 24. Isaacs J. S., Jung Y. J., Mimnaugh E. G., Martinez A., Cuttitta F., Neckers L. M. (2002) Hsp90 regulates a von Hippel Lindau-independent hypoxia-inducible factor-1 α-degradative pathway. J. Biol. Chem. 277, 29936–29944 [DOI] [PubMed] [Google Scholar]
- 25. Echtenkamp F. J., Freeman B. C. (2012) Expanding the cellular molecular chaperone network through the ubiquitous cochaperones. Biochimica et biophysica acta 1823, 668–673 [DOI] [PubMed] [Google Scholar]
- 26. Li J., Soroka J., Buchner J. (2012) The Hsp90 chaperone machinery. Conformational dynamics and regulation by co-chaperones. Biochim. Biophys. Acta 1823, 624–635 [DOI] [PubMed] [Google Scholar]
- 27. Johnson J. L., Brown C. (2009) Plasticity of the Hsp90 chaperone machine in divergent eukaryotic organisms. Cell Stress Chaperones 14, 83–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yu F., White S. B., Zhao Q., Lee F. S. (2001) Dynamic, site-specific interaction of hypoxia-inducible factor-1α with the von Hippel-Lindau tumor suppressor protein. Cancer Res. 61, 4136–4142 [PubMed] [Google Scholar]
- 29. Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., Salic A., Asara J. M., Lane W. S., Kaelin W. G., Jr. (2001) HIFα targeted for VHL-mediated destruction by proline hydroxylation. Implications for O2 sensing. Science 292, 464–468 [DOI] [PubMed] [Google Scholar]
- 30. Huang J., Zhao Q., Mooney S. M., Lee F. S. (2002) Sequence determinants in hypoxia-inducible factor-1α for hydroxylation by the prolyl hydroxylases PHD1, PHD2, and PHD3. J. Biol. Chem. 277, 39792–39800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 32. Winiewski J. R., Zougman A., Nagaraj N., Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 33. Rappsilber J., Mann M., Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 34. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 35. Gill S. C., von Hippel P. H. (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182, 319–326 [DOI] [PubMed] [Google Scholar]
- 36. Huang J. H., Lee F. S., Pasha T. L., Sammel M. D., Karakousis G., Xu G., Fraker D., Zhang P. J. (2010) Cancer Biol. Ther. 9, 303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huang J., Song D., Flores A., Zhao Q., Mooney S. M., Shaw L. M., Lee F. S. (2007) IOP1, a novel hydrogenase-like protein that modulates hypoxia-inducible factor-1α activity. Biochem. J. 401, 341–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ali M. M., Roe S. M., Vaughan C. K., Meyer P., Panaretou B., Piper P. W., Prodromou C., Pearl L. H. (2006) Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature 440, 1013–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Somarelli J. A., Lee S. Y., Skolnick J., Herrera R. J. (2008) Structure-based classification of 45 FK506-binding proteins. Proteins 72, 197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Edlich F., Lücke C. (2011) From cell death to viral replication. The diverse functions of the membrane-associated FKBP38. Curr. Opin. Pharmacol. 11, 348–353 [DOI] [PubMed] [Google Scholar]
- 41. Barth S., Edlich F., Berchner-Pfannschmidt U., Gneuss S., Jahreis G., Hasgall P. A., Fandrey J., Wenger R. H., Camenisch G. (2009) Hypoxia-inducible factor prolyl-4-hydroxylase PHD2 protein abundance depends on integral membrane anchoring of FKBP38. J. Biol. Chem. 284, 23046–23058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Barth S., Nesper J., Hasgall P. A., Wirthner R., Nytko K. J., Edlich F., Katschinski D. M., Stiehl D. P., Wenger R. H., Camenisch G. (2007) The peptidyl prolyl cis/trans isomerase FKBP38 determines hypoxia-inducible transcription factor prolyl-4-hydroxylase PHD2 protein stability. Mol. Cell. Biol. 27, 3758–3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ibrahim N. O., Hahn T., Franke C., Stiehl D. P., Wirthner R., Wenger R. H., Katschinski D. M. (2005) Induction of the hypoxia-inducible factor system by low levels of heat shock protein 90 inhibitors. Cancer Res. 65, 11094–11100 [DOI] [PubMed] [Google Scholar]
- 44. Forafonov F., Toogun O. A., Grad I., Suslova E., Freeman B. C., Picard D. (2008) p23/Sba1p protects against Hsp90 inhibitors independently of its intrinsic chaperone activity. Mol. Cell. Biol. 28, 3446–3456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stepanova L., Leng X., Parker S. B., Harper J. W. (1996) Mammalian p50Cdc37 is a protein kinase-targeting subunit of Hsp90 that binds and stabilizes Cdk4. Genes Dev. 10, 1491–1502 [DOI] [PubMed] [Google Scholar]
- 46. da Silva Correia J., Miranda Y., Leonard N., Ulevitch R. (2007) SGT1 is essential for Nod1 activation. Proc. Natl. Acad. Sci. U.S.A. 104, 6764–6769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mayor A., Martinon F., De Smedt T., Pétrilli V., Tschopp J. (2007) A crucial function of SGT1 and HSP90 in inflammasome activity links mammalian and plant innate immune responses. Nat. Immunol. 8, 497–503 [DOI] [PubMed] [Google Scholar]
- 48. Chan D. A., Kawahara T. L., Sutphin P. D., Chang H. Y., Chi J. T., Giaccia A. J. (2009) Tumor vasculature is regulated by PHD2-mediated angiogenesis and bone marrow-derived cell recruitment. Cancer Cell 15, 527–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bordoli M. R., Stiehl D. P., Borsig L., Kristiansen G., Hausladen S., Schraml P., Wenger R. H., Camenisch G. (2011) Prolyl-4-hydroxylase PHD2- and hypoxia-inducible factor 2-dependent regulation of amphiregulin contributes to breast tumorigenesis. Oncogene 30, 548–560 [DOI] [PubMed] [Google Scholar]
- 50. Riggs D. L., Cox M. B., Tardif H. L., Hessling M., Buchner J., Smith D. F. (2007) Noncatalytic role of the FKBP52 peptidyl-prolyl isomerase domain in the regulation of steroid hormone signaling. Mol. Cell. Biol. 27, 8658–8669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Matthews J. M., Bhati M., Lehtomaki E., Mansfield R. E., Cubeddu L., Mackay J. P. (2009) It takes two to tango. The structure and function of LIM, RING, PHD, and MYND domains. Curr. Pharm. Des. 15, 3681–3696 [DOI] [PubMed] [Google Scholar]
- 52. Ansieau S., Leutz A. (2002) The conserved Mynd domain of BS69 binds cellular and oncoviral proteins through a common PXLXP motif. J. Biol. Chem. 277, 4906–4910 [DOI] [PubMed] [Google Scholar]
- 53. Liu Y., Chen W., Gaudet J., Cheney M. D., Roudaia L., Cierpicki T., Klet R. C., Hartman K., Laue T. M., Speck N. A., Bushweller J. H. (2007) Structural basis for recognition of SMRT/N-CoR by the MYND domain and its contribution to AML1/ETO's activity. Cancer Cell 11, 483–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Weaver A. J., Sullivan W. P., Felts S. J., Owen B. A., Toft D. O. (2000) Crystal structure and activity of human p23, a heat shock protein 90 co-chaperone. J. Biol. Chem. 275, 23045–23052 [DOI] [PubMed] [Google Scholar]
- 55. Weikl T., Abelmann K., Buchner J. (1999) An unstructured C-terminal region of the Hsp90 co-chaperone p23 is important for its chaperone function. J. Mol. Biol. 293, 685–691 [DOI] [PubMed] [Google Scholar]