Background: The platform protein PilC and/or the alignment subcomplex PilMNOP may coordinate type IV pilus (T4P) extension/retraction ATPases.
Results: Loss of PilC in a retraction-deficient background abolished assembly. C-terminal mutations were permissive for assembly but not twitching motility.
Conclusion: PilC is essential for pilus biogenesis and twitching motility.
Significance: Platform proteins are conserved essential components of T4P and type II secretion machines.
Keywords: Bacterial Adhesion, Membrane Proteins, Microbiology, Molecular Motors, Pseudomonas aeruginosa, Twitching Motility, Type II Secretion System, Type IV Pili
Abstract
A systematic genetic analysis was performed to identify the inner membrane proteins essential for type IV pilus (T4P) expression in Pseudomonas aeruginosa. By inactivating the retraction aspect of pilus function, genes essential for T4P assembly were discriminated. In contrast to previous studies in the T4P system of Neisseria spp., we found that components of the inner membrane subcomplex consisting of PilMNOP were not essential for surface pilus expression, whereas the highly conserved inner membrane protein PilC was essential. Here, we present data that PilC may coordinate the activity of cytoplasmic polymerization (PilB) and depolymerization (PilT) ATPases via their interactions with its two cytoplasmic domains. Using in vitro co-affinity purification, we show that PilB interacts with the N-terminal cytoplasmic domain of PilC. We hypothesized that PilT similarly interacts with the PilC C-terminal cytoplasmic domain. Overexpression of that domain in the wild-type protein reduced twitching motility by ∼50% compared with the vector control. Site-directed mutagenesis of conserved T4P-specific residues in the PilC C-terminal domain yielded mutant proteins that supported wild-type pilus assembly but had a reduced capacity to support twitching motility, suggesting impairment of putative PilC-PilT interactions. Taken together, our results show that PilC is an essential inner membrane component of the T4P system, controlling both pilus assembly and disassembly.
Introduction
Type IV pili (T4P)5 are long helical fibers composed of pilin subunits and are expressed on the surfaces of a wide variety of Gram-positive, Gram-negative, and archaeal species (1, 2). T4P assembly systems are evolutionarily and functionally related to the type II secretion (T2S) system, which is responsible for the extrusion of folded proteins and protein complexes from the periplasm of Gram-negative bacteria (3–5). Both machineries are thought to function through the reversible polymerization of pilin subunits into long helical T4P or short T2S pseudopili. T4P are divided into type IVa (T4aP) and type IVb (T4bP) subclasses. Both subclasses are associated with adherence and cell-cell aggregation, but T4aP also mediate a form of flagellum-independent locomotion called “twitching” motility (6, 7). The forces that drive twitching arise from the depolymerization of surface-attached pili and consequent translocation of the cell body toward the point of pilus attachment (8–11).
Four interdependent subcomplexes are proposed to form the apparatus responsible for T4P and T2S biogenesis and function (7). The outer membrane subcomplex consists of an oligomeric secretin, through which pili or secreted proteins exit the cell; in some systems, the secretin is associated with a pilotin controlling its localization and/or oligomerization (12). There are two inner membrane subcomplexes, the motor and alignment subcomplexes. The motor subcomplex is highly conserved across T4P and T2S systems and is proposed to be composed of the inner membrane platform protein PilC, associated cytoplasmic ATPases, and potentially the prepilin peptidase (7). The alignment subcomplex may link the outer membrane secretin and inner membrane motor subcomplexes and potentially participate in secretin gating dynamics. In Pseudomonas aeruginosa and other T4aP-expressing bacteria, the alignment subcomplex consists of the cytoplasmic actin-like protein PilM, which binds to the short cytoplasmic N terminus of the inner membrane protein PilN (13, 14). In turn, PilN forms heterodimers with the inner membrane protein PilO, and PilNO interacts with the inner membrane lipoprotein PilP via the latter's unstructured N terminus (15). Interaction of the C-terminal β-domain of PilP and the PilQ secretin completes the trans-envelope network.6 The T4bP and T2S system motor subcomplexes typically have a single ATPase, and the proteins forming the alignment subcomplexes are different in sequence and number than those of T4aP (7, 16). The final and most dynamic subcomplex is the pilus/pseudopilus, composed of major and minor subunits (17, 18).
In current models of pilus function, pilins are first secreted into the inner membrane by the Sec system (19, 20) and then polymerized by the assembly system through the action of a hexameric ATPase (PilB in P. aeruginosa), which undergoes large conformational changes upon ATP hydrolysis (21, 22). The resulting mechanical energy is thought to be transferred via an inner membrane protein(s) to drive pilin subunits into the base of the growing fiber during polymerization (23). Pilus depolymerization may result from reversal of this process using a second, retraction ATPase (PilT in P. aeruginosa) (24). A third PilT-like ATPase in P. aeruginosa (PilU) has an as-yet undefined role in retraction (25). In recent years, components of both the motor and alignment subcomplexes in the T4P and T2S systems have been suggested to transduce the conformational changes of the ATPases (26, 27), making it challenging to propose a unifying model of fiber assembly and function.
In the P. aeruginosa T4aP system, the platform protein PilC was proposed to be essential for surface pilus expression and twitching motility based on the phenotypes of pilC mutants (28). Similar studies of platform proteins in the T4bP and T2S systems have also supported a critical role in assembly (23, 29). However, the equivalent platform protein PilG in the T4aP system of Neisseria meningitidis was reported to be dispensable for pilus assembly (26, 30), as mutation of pilG in a retraction-deficient background resulted in wild-type surface piliation and adhesive properties. Use of retraction-deficient backgrounds permits the differentiation of components that are essential for assembly from those that antagonize pilus retraction (31). In N. meningitidis, the PilMNOP alignment subcomplex was found to be essential for pilus assembly in a retraction-deficient strain. PilMNOP was proposed to function as a “scaffold” to connect pilin subunits embedded in the inner membrane with the cytoplasmic ATPases (26). Although studies in Myxococcus, Thermus, and Pseudomonas suggested that alignment subcomplex proteins are important for pilus expression and twitching motility (14, 32–34), none examined the phenotypes of retraction-deficient double mutants (35).
Because PilC homologs (but not PilMNOP homologs) are highly conserved across T4P and T2S systems, which are thought to function in a similar manner, we hypothesized that PilC would be an essential component of the P. aeruginosa T4P system. Our data support roles for PilC (but not PilMNOP) in T4P polymerization and depolymerization. We demonstrate interaction of the N-terminal cytoplasmic domain of PilC with PilB by stability studies and co-affinity purification. Although the C-terminal cytoplasmic domain of PilC could not be purified in soluble form, its overexpression in the wild type led to defects in twitching motility. Site-directed mutagenesis of residues conserved in platform proteins from T4P systems, but not T2S systems (which lack a retraction ATPase), recapitulated a retraction-deficient phenotype without affecting pilus assembly or ATPase levels, showing that the extension and retraction functions of PilC are separable. These data support a central role for PilC in T4P function and allow us to propose a model in which alternate interactions of PilB-PilC and PilT-PilC direct pilus assembly and disassembly, respectively.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Plasmids
Strains and plasmids used in this study are listed in supplemental Table 1. Plasmids were transformed via electroporation or heat shock into chemically competent cells.
Twitching Motility Assays
Twitching assays were performed as described previously (36). Briefly, single colonies were stab-inoculated to the bottom of 1% LB agar plates in triplicate. The plates were incubated at 37 °C for 36 h. Post-incubation, the agar was carefully removed, and adherent bacteria were stained with 1% crystal violet, followed by a wash with tap water to remove unbound dye. The areas of twitching zones were measured using NIH ImageJ software. At least three experiments were performed.
Analysis of Sheared Surface Proteins
Surface pili and flagella were analyzed as described previously (36). Briefly, strains of interest were streaked in a grid-like pattern on LB agar plates and incubated at 37 °C for ∼14 h. Using glass cover-slips, the cells were gently scraped from the plates and resuspended in 4.5 ml of 1× PBS (pH 7.4). Surface appendages were sheared by vortexing the cell suspensions for 30 s. Subsequently, the suspensions were transferred to 1.5-ml Eppendorf tubes, and the cells were pelleted by centrifugation at 11,688 × g for 5 min at room temperature. The supernatant was transferred to fresh tubes and recentrifuged at 11,688 × g for 20 min at room temperature to remove the remaining cells. The supernatant was transferred to a new tube, and 0.1 volume of each 5 m NaCl and 30% (w/v) polyethylene glycol (molecular weight range, ∼8000) was added. Soluble proteins were precipitated on ice for 90 min. The precipitated proteins were collected by centrifugation at 11, 688 × g for 30 min at 4 °C. The supernatant was discarded, and the tubes were recentrifuged for 3 min. The remaining supernatant was discarded, and the pellet was resuspended in 150 μl of 1× SDS sample buffer (80 mm Tris (pH 6.8), 5.3% (v/v) 2-mercaptoethanol, 10% (v/v) glycerol, 0.02% (w/v) bromphenol blue, and 2% (w/v) SDS). The samples were boiled for 10 min and separated on 15% SDS-polyacrylamide gels. Proteins were visualized with Coomassie Brilliant Blue (Sigma).
Preparation of Whole Cell Lysates
Overnight cultures were grown in LB broth with appropriate antibiotics where indicated to A600 = 0.6, and then cells from a 1-ml aliquot were collected by centrifugation at 11,688 × g for 2 min in a microcentrifuge. The pellet was resuspended in 100 μl of 1× SDS sample buffer and boiled for 10 min. The whole cell lysate samples were subsequently subjected to Western blot analysis.
Western Blot Analysis
Whole cell lysate samples were separated on 15% SDS-polyacrylamide gels at 80–120 V. Samples were transferred to nitrocellulose membranes at 225 mA for 1 h. The membranes were blocked in 5% (w/v) skim milk in 1× PBS (pH 7.0) for 1 h, followed by incubation in rabbit antisera specific for the proteins of interest (34, 37, 38) for 18 h at 4 °C. The anti-PilC antibody was generated using a peptide antigen corresponding to residues 81–102 of P. aeruginosa PilC (in the first cytoplasmic domain) to immunize rabbits (Cedarlane Laboratories, Burlington, Ontario, Canada). The membranes were washed three times with 1× PBS for 5 min and incubated in alkaline phosphatase-conjugated goat anti-rabbit IgG secondary antibody (Bio-Rad) following the manufacturer's protocol. The membranes were washed three times with 1× PBS for 5 min and visualized with alkaline phosphatase developing reagent (Bio-Rad) following the manufacturer's protocol.
Expression and Purification of the N-terminal Domain of PilC
The pET28b::pilC(1–173) (N-terminal His6 tag) expression construct was transformed into Escherichia coli BL21(DE3) cells. One-ml trials were performed to identify optimal expression conditions. Induction with 1 mm isopropyl 1-thio-β-d-galactopyranoside at A = 0.6 for 3 h led to high level expression. Expression experiments were subsequently scaled up to 1 liter. Western blot analysis using anti-His and anti-PilC antibodies confirmed the expression of the N-terminal domain (NTD) of PilC and the in-frame incorporation of the purification tag. The cell pellet was resuspended in lysis buffer containing 1% (w/v) lauryldimethylamine oxide, 10 ng/ml DNase and RNase, and 100 mg/liter benzamidine prior to lysis via French press and centrifuged at 48,258 × g to separate the clarified lysate from the unlysed cells. The lysate was collected and applied to 10-ml nickel-nitrilotriacetic acid (Ni-NTA) columns containing 1 ml of Ni-NTA resin. The column was subsequently washed with 20 ml of Ni+ buffer (20 mm Tris (pH 8.0) and 250 mm KCl) and 10 ml of Ni+ buffer containing 15, 30, 150, and 300 mm imidazole to elute the NTD of PilC-His6.
Circular Dichroism Spectroscopy of the NTD of PilC
Circular dichroism spectroscopy was used to examine the secondary structure composition of the purified NTD of PilC. Briefly, the NTD was expressed and purified as described above. The 150 and 300 mm imidazole elution fractions were collected and dialyzed overnight in 1× PBS at 4 °C. The protein sample was adjusted to final concentrations of 0.45 and 0.23 μg/ml prior to the assay. Absorbance was measured at intervals of 1 nm from 190 to 260 nm, with a data collection time of 3 s. The temperature of the sample chamber was set to 25 °C. Absorbance values were transferred to Microsoft Excel, and the data were normalized, averaged, and converted to molar ellipticity ([θ]) using the following formula: [θ] = (absorbance/1000)/((concentration (mg/ml)/molecular mass (Da)/1000) × no. of amino acids). Control spectra containing buffer alone were collected and subtracted from the sample spectra to account for background absorbance.
In Vitro Co-affinity Purification
The NTD of PilC-His6 was expressed and purified as described above. The 150 and 300 mm imidazole elution fractions were collected and dialyzed overnight in 1× PBS at 4 °C. The dialyzed protein was concentrated to 2 μg/ml and incubated overnight with 1 ml of Ni-NTA resin at 4 °C. The PAK strain of P. aeruginosa was streaked out on a 1.5% LB agar plate in a grid-like fashion and grown overnight at 37 °C. The cells were harvested by gentle scraping with a sterile coverslip and resuspended in 1× PBS containing 100 mg/liter benzamidine and 10 ng/ml DNase and RNase. The cells were sonicated on ice with alternating 10-s on and 10-s off periods for a total on time of 2 min. The cell lysates were centrifuged at 3000 × g for 15 min to separate unlysed cells. The supernatant was mixed with purified bait protein plus Ni-NTA resin for 2 h at 4 °C. The mixture was then applied to a pre-equilibrated gravity flow 10-ml nickel column, and the flow-through was collected. Washes and elution fractions of 10 ml were collected: 1× PBS, 5 mm imidazole elution, and 300 mm imidazole elution. All fractions were used for SDS-PAGE and Western blot analysis as described above.
In Vivo Arabinose-induced Overexpression of the C-terminal Domain
The C-terminal domain (CTD; residues 240–379) of PilC was cloned into the arabinose-inducible vector pBADGr (37). The pBADGr::pilC(240–379) construct was electroporated into the WT PAK strain and subjected to growth assays, sheared surface protein preparations, and twitching motility assays as described above.
Site-directed Mutagenesis
Site-directed mutagenesis of PilC was performed using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol using the oligonucleotides listed in supplemental Table 2. The pUCP20Gm::pilC plasmid was used as the template DNA, with an annealing temperature of 55 °C and an extension time of 16 min for 18 cycles. Site-directed mutagenesis reactions were digested with 1 μl of 10 units/μl DpnI (Fermentas) for 2 h in a 37 °C water bath. Digestion reactions were then transformed into chemically competent E. coli DH5α cells, which were subsequently grown overnight at 37 °C on 1.5% LB agar plates supplemented with the appropriate antibiotics. Mutations were verified by DNA sequencing (McMaster Institute for Molecular Biology and Biotechnology (MOBIX), Hamilton, Ontario, Canada). Mutant constructs were electroporated into PAK pilC and pilC-pilT mutant strains, and transformants were selected by plating onto 1.5% LB agar supplemented with the appropriate antibiotics. Phenotypes were analyzed using a twitching motility assay and sheared surface pilin preparations.
RESULTS
PilC Is Essential and PilMNOP Is Dispensable for Pilus Biogenesis
Surface piliation of single and retraction-deficient double mutants in pilC and pilMNOP was examined. As reported previously, single mutants in pilMNOP lack surface pili (34). However, pilMNOP-pilT double mutants had surface pili, although the levels were reduced to ∼15–20% of the pilT control (Fig. 1). In contrast, both pilC and pilC-pilT mutants lacked surface pili (Fig. 1). Complementation of the mutant strains with the relevant genes restored the corresponding single mutant phenotypes, and Western blot analysis of whole cell lysate samples confirmed expression of the proteins in trans (data not shown). These results suggest that PilC (but not PilMNOP) is essential for T4aP biogenesis in P. aeruginosa.
FIGURE 1.
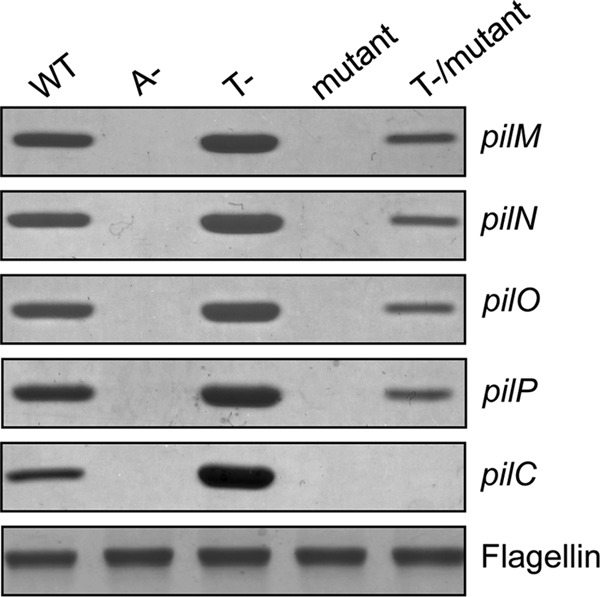
Platform protein PilC is essential for surface pilus expression in P. aeruginosa. Sheared surface proteins from the WT PAK strain, A− (pilA::FRT), T− (pilT::FRT), mutant (ΔpilM, pilN::FRT, pilO::FRT, pilP::FRT, pilC::FRT), and T−/mutant were separated on a 15% SDS-polyacrylamide gel and stained with Coomassie Blue. The flagellin band was used as a loading control.
The First Start Site of pilC Is Used in Vivo
When creating the complementation construct for pilC, we noted that the annotated sequences of pilC from two closely related P. aeruginosa strains (PAK and PAO1) have different predicted starts, making the PAO1 PilC protein 32 residues shorter than the PAK protein (Fig. 2A). Alignment of PAK and PAO1 PilC with other platform proteins (data not shown) suggested that the longer product was more likely to be correct. Western blot analyses with anti-PilC sera showed that both strains express PilC with a mass corresponding to the first start site (Fig. 2B). Interestingly, twitching motility assays revealed that a truncated version of PilC expressed from the second predicted start site could restore twitching to ∼75% of wild-type levels (Fig. 2C). Together, these data suggest that the first start of PilC is used in vivo and that the first 32 amino acids are required for full function.
FIGURE 2.
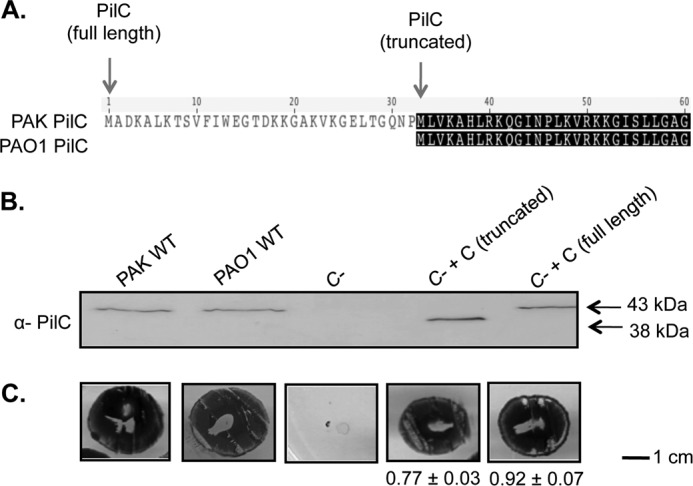
The first start site of pilC is used in vivo. A, sequences of the PilC proteins from P. aeruginosa strains PAO1 and PAK were aligned using Geneious (Biomatters). The PAK protein is predicted to be 32 residues longer than the PAO1 protein. B, Western blot analyses using an anti-PilC antibody of whole cell lysates from PAK and PAO1, the PAK pilC mutant, and the mutant complemented with the truncated and full-length versions of pilC. For both wild-type strains, the first start site is used in vivo. C, twitching motility of the strains in B. The truncated form of PilC complements motility, but not to the same extent as the full-length protein. Numbers are percent of wild-type PAK (n = three assays, each with three replicates).
PilB Levels Are Reduced in pilC Mutants
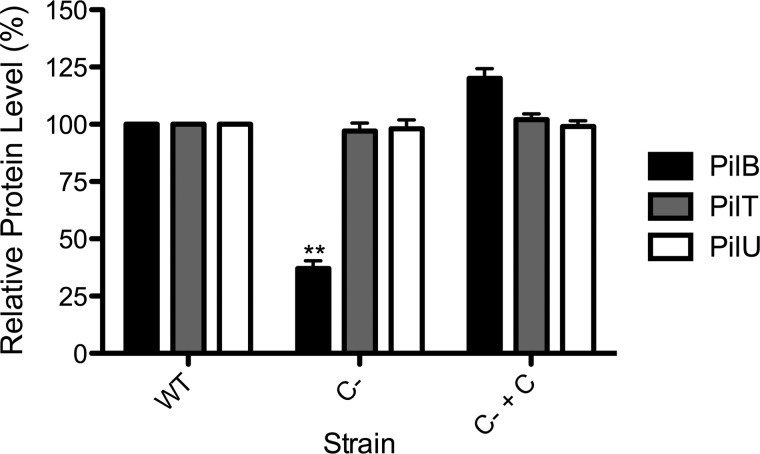
We showed previously that the normally bipolar ATPase PilB is delocalized in the absence of PilC (39). Western blot analyses with specific antisera showed that PilB levels were reduced to ∼37% (p < 0.01) of wild-type levels in the pilC background, whereas the levels of PilT and PilU were unchanged (Fig. 3), suggesting that loss of PilC specifically affects PilB stability. Furthermore, the levels of the ATPases in double mutants lacking pilC plus any of pilM/N/O/P were comparable to those of the pilC single mutant (data not shown). The reduction in PilB levels was not due to polar effects, as complementation with pilC in trans restored PilB levels. In fact, expression of PilC from the multicopy vector tended to increase PilB levels compared with wild-type levels (Fig. 3), although the increase was not statistically significant. Thus, PilC is important for both the localization and stability of PilB.
FIGURE 3.
Intracellular levels of PilB are reduced in the absence of PilC. Whole cell lysates of the WT PAK strain, C− (pilC::FRT), and C− + C (pilC::FRT + pUCP20Gm::pilC) were probed with anti-PilB, anti-PilT, or anti-PilU antiserum. Densitometry (NIH ImageJ) was performed on three independent Western blots, and the results were averaged. The levels of PilB, PilT, and PilU in the wild-type strain were set to 100%. **, p < 0.01.
The Cytoplasmic NTD of PilC Pulls Down PilB
Previous studies in the T4bP and T2S systems provided both indirect and direct evidence of interactions between the NTD of the platform protein and the polymerization ATPase (23, 27, 40, 41). To test for interaction between the NTD of PilC and PilB in P. aeruginosa, we performed in vitro co-affinity purification. Using the previously established boundaries of the NTD from orthologous platform proteins (42, 43) and predictions generated by the TMHMM algorithm (44), we generated a construct expressing a soluble NTD fragment of P. aeruginosa PilC. Circular dichroism spectroscopy was used to confirm its predicted α-helical secondary structure (data not shown). Purified PilB was initially tested for interactions with the PilC NTD by co-affinity purification, but problems with PilB aggregation prompted us to use whole cell lysates instead. We elected to use wild-type instead of pilC mutant whole cell lysates for this experiment due to the significantly reduced amount of PilB present in the pilC background (Fig. 3), despite the competition of full-length PilC for PilB. After incubation with whole cell lysates from wild-type P. aeruginosa PAK, the NTD and any bound untagged partners were purified by nickel affinity chromatography. Western blot analyses showed that PilB was found in the bound fraction at the highest concentration of imidazole (Fig. 4) only when the PilC NTD was present. The retraction ATPases PilT and PilU were not detected in the final elution fraction using specific antisera (data not shown), suggesting that they do not interact with this domain of PilC. Together, the results support the hypothesis that PilB interacts with the NTD of PilC.
FIGURE 4.
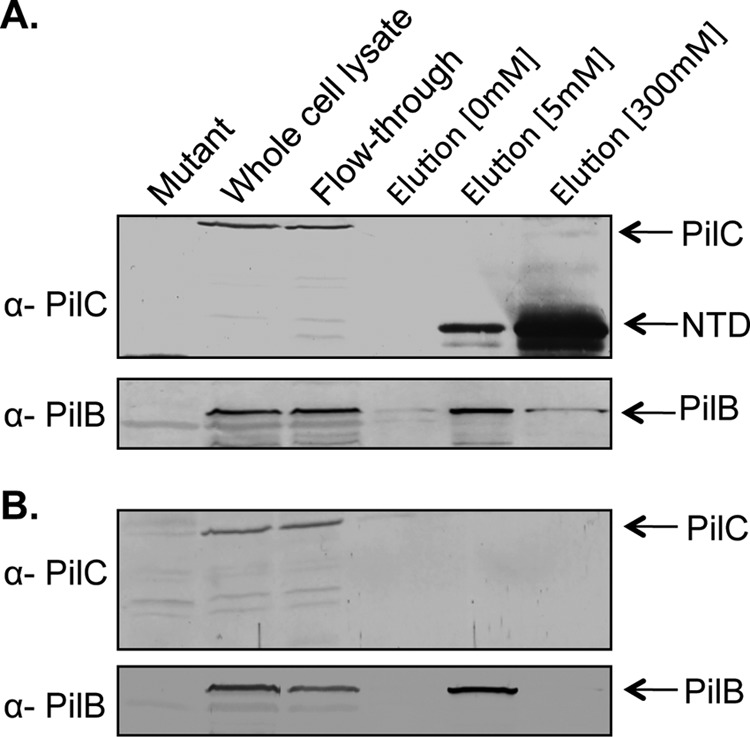
In vitro co-affinity purification of the NTD of PilC and the ATPase PilB. A, the purified NTD of PilC was incubated with whole cell lysates of the PAK strain and then repurified by nickel affinity chromatography as described under “Experimental Procedures.” Whole cell lysates, the column flow-through, and imidazole wash fractions (0, 5, and 300 mm) were probed with anti-PilC or anti-PilB antiserum. Full-length PilC present in the lysates did not bind the column. A fraction of PilB was retained in the 300 mm imidazole fraction. B, when the NTD of PilC was not included, PilB did not appear in the 300 mm imidazole elution fraction.
The Cytoplasmic CTD of PilC Functions in Pilus Retraction
P. aeruginosa PilC is predicted to have two cytoplasmic domains, with 21% amino acid sequence identity (34% similarity) to one another and similar predicted secondary structure. Abendroth et al. (42) proposed previously that based on such similarities, the cytoplasmic domains of the PilC ortholog EpsF and its relatives were likely to have similar folds. We hypothesized that although the NTD of PilC likely interacts with PilB, its CTD may interact with PilT. A previous study in the T4bP system of E. coli showed that overexpression of a peptide from the C-terminal region of its PilC ortholog (BfpE) caused phenotypes consistent with the loss of pilus retraction (40). To establish the role of the PilC CTD in the T4aP system of P. aeruginosa, residues 240–379 of PilC were first expressed in trans in the wild type using an arabinose-inducible vector. Twitching motility was reduced in a dose-dependent manner to approximately half that of the vector control at 0.5% arabinose (p < 0.001), the highest concentration tested (Fig. 5), without effects on growth or pilus biogenesis (data not shown). These data suggest that the CTD could have a role in pilus retraction.
FIGURE 5.
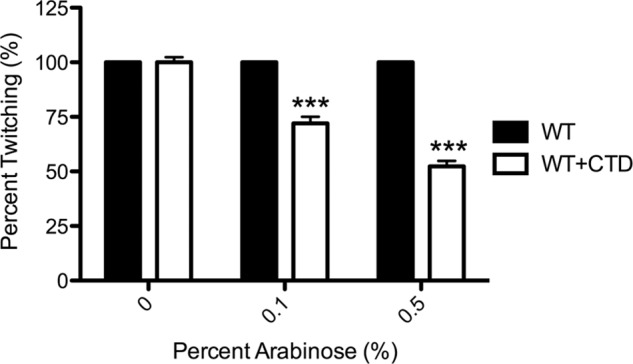
Expression of the PilC CTD in trans reduces twitching motility. A DNA fragment encoding the CTD of PilC was cloned into the arabinose-inducible vector pBADGr and introduced into the PAK strain. Colonies were stab-inoculated in triplicate onto 1% agar containing 0, 0.1, or 0.5% l-arabinose; twitching zones were visualized by crystal violet staining; and the areas were measured using NIH ImageJ. The amount of twitching at 0% arabinose was set to 100% (n = three assays, each with three replicates). ***, p < 0.001.
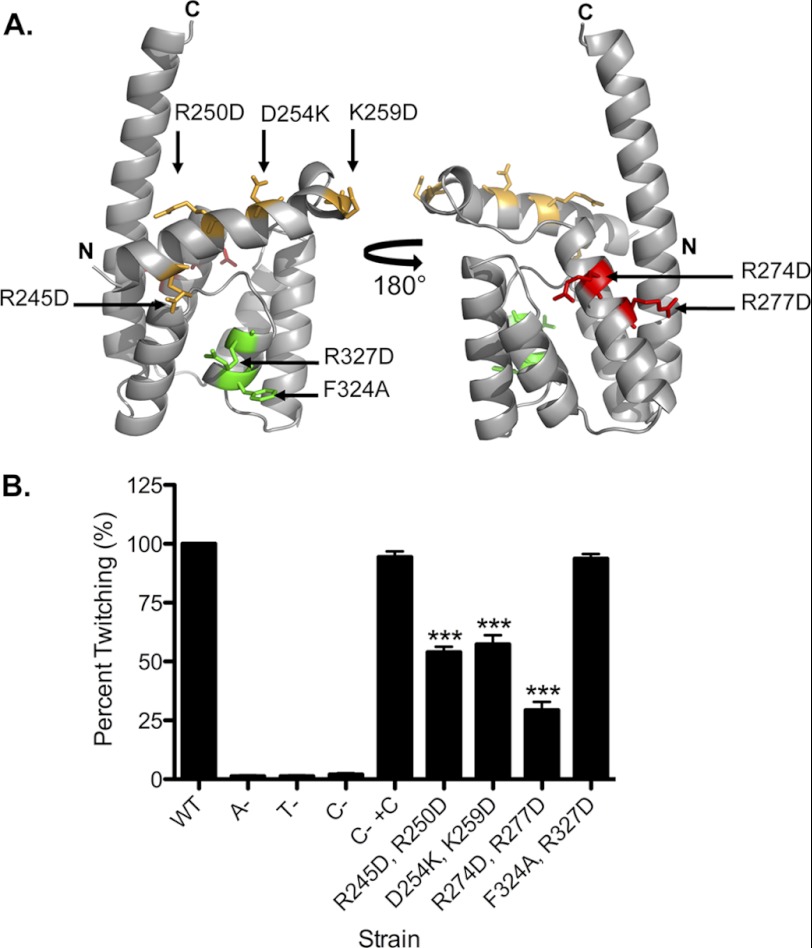
Next, we considered which residues of the CTD might be important for twitching motility. First, the CTDs of platform proteins from a number of T4P and T2S systems were aligned (data not shown), and conserved residues were identified. Because the T2S system and most T4bP systems lack a retraction ATPase, we reasoned that residues conserved in platform proteins across the three groups were more likely to have structural roles. Instead, we selected residues that were conserved particularly in T4aP platform proteins. We mapped them onto a model of the P. aeruginosa CTD generated using the structural prediction algorithm I-TASSER (45), selecting a subset of those predicted to be surface-exposed and thus potentially available for interaction with other components (Fig. 6A). To facilitate rapid screening, residues were mutated in pairs to the opposite charge or to Ala in the case of Phe-324. The ability of each mutant protein to restore T4P production (a measure of pilus assembly) and twitching (a measure of disassembly) was tested by complementation of the pilC mutant. All CTD mutations reduced twitching motility (p < 0.001) with the exception of F324A/R327D, which gave wild-type levels of twitching (Fig. 6B). These results show that only a subset of conserved residues in the T4aP CTD are important for twitching. Although all mutations reduced the detectable levels of PilC protein, the amount of surface pili in the mutant strains was comparable to that in the pilC mutant complemented with wild-type PilC (Fig. 7, A and B). Together with the observation that the F324A/R327D mutant has wild-type motility, the reduced levels of PilC in the point mutants appeared sufficient for normal pilus assembly. Thus, reductions in motility observed for specific mutants were more likely to be due to retraction defects. Western blot analyses showed that the intracellular levels of PilT, PilU, and PilA in each of the mutants were equivalent to those of the strain complemented with wild-type PilC (Fig. 7B), consistent with the data in Fig. 3 showing that PilT and PilU levels were unaffected in the pilC mutant background. Therefore, the decreases in twitching motility were not due to decreased PilT or PilU stability.
FIGURE 6.
Specific mutations in the CTD of PilC reduce twitching motility. A, I-TASSER model of the CTD of P. aeruginosa PilC using the crystal structure of the T. thermophilus PilC NTD (Protein Data Bank code 2WHN) as a template. The residues selected for mutagenesis are labeled: those in green have no effect on twitching, those in yellow have a moderate effect, and those in red have the most pronounced impact, as quantified in B. B, strains were stab-inoculated in triplicate onto 1% agar twitching plates and incubated for 36 h as described under “Experimental Procedures.” Twitching zones were visualized by crystal violet staining, and the areas were measured using NIH ImageJ. The strains used were WT PAK, A− (pilA::FRT), T−(pilT::FRT), C− (pilC::FRT), C− + C (pilC::FRT + pUCP20Gm::pilC), and the pilC::FRT mutant complemented with pUCP20Gm::pilC point mutants encoding the indicated alterations. Wild-type motility was set to 100% (n = three assays, each with three replicates). ***, p < 0.001.
FIGURE 7.
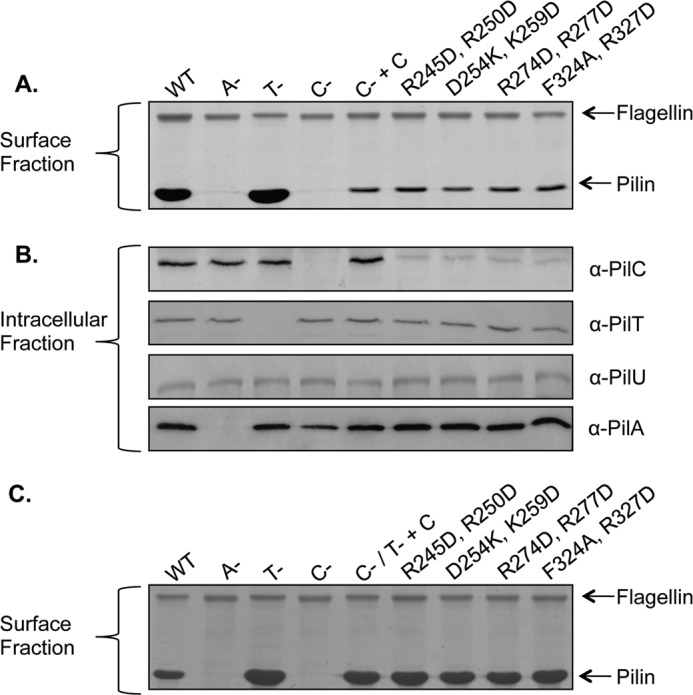
PilC CTD mutations do not affect T4P assembly. A, sheared surface proteins from each of the strains indicated were separated on a 15% SDS-polyacrylamide gel, followed by Coomassie Blue staining. The flagellin band was used as a loading control. All strains are on the PAK background: WT, A− (pilA::FRT), T− (pilT::FRT), C− (pilC::FRT), C− + C (pilC::FRT + pUCP20Gm::pilC), and the pilC::FRT mutant complemented with site-directed mutants of pUCP20Gm::pilC encoding the indicated alterations. B, immunoblot analysis of whole cell lysates probed with anti-PilC, anti-PilT, anti-PilU, or anti-PilA antiserum. C, sheared surface proteins from the pilC::FRT-pilT::FRT double mutant transformed with CTD site-directed mutant constructs were separated on a 15% SDS-polyacrylamide gel, followed by Coomassie Blue staining. The flagellin band was used as a loading control.
To specifically examine the assembly capacity of mutant PilC proteins, each of the constructs was introduced into the pilC-pilT double mutant. Examination of sheared surface proteins showed that complementation with any of the CTD mutants resulted in hyperpiliated phenotypes that were indistinguishable from that of the double mutant complemented with wild-type pilC (Fig. 7C). These data confirm that point mutations in the CTD of PilC do not affect pilus assembly and support the idea that pilus extension and retraction are independent processes that rely on separate domains of PilC. Together, the results suggest that the NTD of PilC mediates pilus assembly through interactions with PilB and that the CTD of PilC is involved in T4P retraction, potentially through interactions with PilT and/or PilU.
DISCUSSION
Aside from the pilins, homologs of the platform protein PilC and the polymerization ATPase PilB are the most highly conserved components among T2S and T4P systems in bacteria and archaea, suggesting that they form the basic motor unit. Here, we have provided evidence that PilC is essential for T4P polymerization and depolymerization through its potential associations with the cytoplasmic ATPases PilB and PilT/PilU.
Components of the PilMNOP subcomplex are conditionally dispensable for T4P assembly in P. aeruginosa, although the amount of pili recovered in retraction-deficient backgrounds is reduced. PilMNOP is proposed to align the motor and outer membrane subcomplexes to ensure that pili assembled at the periplasmic face of the inner membrane are directed through the periplasm and peptidoglycan layer (46) to the secretin for egress. In their absence, pilus assembly is inefficient (∼15–20% of control), and those fibers that are assembled are likely rapidly retracted. Mounting evidence that components of the alignment subcomplex and the periplasmic regions of the secretin can interact with pilins (47, 48)6 suggests that PilMNOP might increase the local concentration of membrane-bound subunits near the complex, increasing polymerization efficiency and limiting diffusion of newly disassembled subunits away from the site of assembly. We showed previously that loss of PilMNOP decreases secretin levels (34), which could partly explain the ∼80–85% reduction in the amount of surface piliation observed in double mutants relative to the pilT control (Fig. 1).
The hexameric T4P and T2S ATPases are composed of monomers with an N-terminal head domain attached to a C-terminal body domain by a flexible linker. The ATPase active site is formed at the interface between two subunits, and ATP hydrolysis causes an ∼15–65° rotation of the head domain relative to the body (21, 22, 49). An unidentified membrane protein(s) is thought to transduce ATPase movements to associated pilins, driving them into a growing fiber or back into the membrane during T4P polymerization and depolymerization, respectively (4, 35). Our data provide evidence that PilC fulfills this role. One or more of its transmembrane segments could interact transiently with pilins whose hydrophobic N termini are fully or partly buried in the membrane, whereas its cytoplasmic domains could interact with the ATPases to transduce the forces generated upon ATP hydrolysis.
The genetic conservation and linkage of pilB and pilC homologs across T4P and T2S systems support the functional association of the two gene products. Together, our localization (39), stability, and co-purification data support interaction of the NTD of PilC and the assembly ATPase PilB. Similarly, the hypothesis that the CTD of PilC participates in the depolymerization of T4P is supported by the data showing that its overexpression decreased wild-type twitching motility without affecting pilus assembly (Fig. 5). Although we speculate that this effect was caused by titration of PilT and/or PilU away from full-length PilC, we were unable to formally test those interactions due insolubility of the PilC CTD in vitro.
Instead, site-directed mutagenesis revealed key residues in the CTD of PilC involved in T4P retraction. These residues were predicted to have surface-exposed charged or hydrophobic side chains that could be important for interactions with the ATPase(s). Mutations R245D/R250D, D254K/K259D, and R247D/R277D reduced twitching motility (Fig. 7). The first two pairs of residues are close to the predicted membrane interface and might affect movements of the CTD (and adjacent transmembrane segments) induced by ATPase conformational changes that are important for pilin depolymerization and thus motility. The third pair of residues is predicted to be farther from the membrane and might contribute to ATPase interactions. In contrast, mutation of residues predicted to be distal to the membrane (F324A/R327D), including the potentially solvent-exposed Phe-324, did not affect twitching motility. This region was suggested to be important for dimerization of the NTD in the crystal structure of Thermus thermophilus PilC (43). However, it is unclear if these residues are functionally equivalent in the CTD. More accurate structural information is necessary before the exact role of these residues can be clarified.
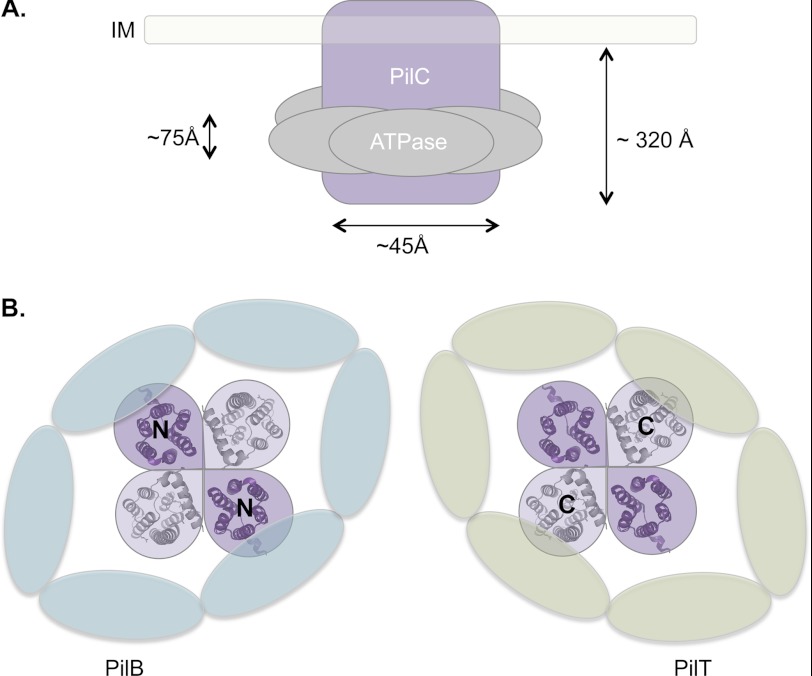
Although some models have been proposed, the precise stoichiometry of PilC and the ATPases has yet to be experimentally determined. Previous studies in the T4aP systems of Neisseria and Thermus suggested that PilC is a dimer and possibly a dimer of dimers or a tetramer (43, 50). Our data suggest that the ATPase hexamers may interact with independent cytoplasmic domains of PilC, perhaps by encircling them. However, a tetrameric arrangement of PilC (with four NTDs and four CTDs, each predicted to be ∼20–25 Å at the widest point) is too large to fit the experimentally determined inner diameter of the ATPases at ∼25–50 Å (22, 51, 52). Instead, we suggest that PilC is more likely arranged as a dimer, which is in agreement with a T2S system homolog (39). We speculate that this arrangement, with two NTDs and two CTDs together inside the lumen of a hexameric ATPase (Fig. 8) might allow necessary and potentially alternate contacts of the cytoplasmic domains of PilC with two different ATPases in their active asymmetric configuration (22).
FIGURE 8.
Model for PilC-ATPase interactions. A, approximate dimensions of the cytoplasmic domains of the inner membrane (IM) protein PilC and a hexameric ATPase (22, 50). B, model of the NTD of PilC (purple; Protein Data Bank code 2WHN) interacting with the hexameric ATPase PilB and of the CTD (gray; I-TASSER prediction using code 2WHN as a template) interacting with the hexameric ATPase PilT in an asymmetric conformation.
The data presented here suggest that the platform protein PilC and the ATPases PilB and PilT/PilU compose the minimal motor of the T4aP system, driving pilus assembly and disassembly. A second inner membrane subcomplex, PilMNOP, is required for efficient pilus assembly. Additional studies are required to further substantiate the proposed interactions of PilC with the retraction ATPases; however, these findings are an important step forward in defining the elements that are essential for T4P function in P. aeruginosa.
Acknowledgments
We thank Hanjeong Harvey, Lisa Wong, and Santina Colalillo for generation of pilC and pilM mutants.
This work was supported in part by Operating Grant MOP 93585 from the Canadian Institutes of Health Research (to L. L. B. and P. L. H.).

This article contains supplemental Tables 1 and 2.
Tammam, S., Sampaleanu, L. M., Koo, J., Manoharan, K., Daubaras, M., Burrows, L. L., and Howell, P. L. (2013) PilMNOPQ from the Pseudomonas aeruginosa type IV pilus system form a transenvelope protein interaction network that interacts with PilA. J. Bacteriol., in press.
- T4P
- type IV pilus/pili
- T2S
- type II secretion
- NTD
- N-terminal domain
- Ni-NTA
- nickel-nitrilotriacetic acid
- CTD
- C-terminal domain
- FRT
- Flp recombinase target.
REFERENCES
- 1. Pelicic V. (2008) Type IV pili: e pluribus unum? Mol. Microbiol. 68, 827–837 [DOI] [PubMed] [Google Scholar]
- 2. Albers S. V., Pohlschröder M. (2009) Diversity of archaeal type IV pilin-like structures. Extremophiles 13, 403–410 [DOI] [PubMed] [Google Scholar]
- 3. Hobbs M., Mattick J. S. (1993) Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol. Microbiol. 10, 233–243 [DOI] [PubMed] [Google Scholar]
- 4. Ayers M., Howell P. L., Burrows L. L. (2010) Architecture of the type II secretion and type IV pilus machineries. Future Microbiol. 5, 1203–1218 [DOI] [PubMed] [Google Scholar]
- 5. Peabody C. R., Chung Y. J., Yen M. R., Vidal-Ingigliardi D., Pugsley A. P., Saier M. H., Jr. (2003) Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology 149, 3051–3072 [DOI] [PubMed] [Google Scholar]
- 6. Mattick J. S. (2002) Type IV pili and twitching motility. Annu. Rev. Microbiol. 56, 289–314 [DOI] [PubMed] [Google Scholar]
- 7. Burrows L. L. (2012) Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu. Rev. Microbiol. 66, 493–520 [DOI] [PubMed] [Google Scholar]
- 8. Bradley D. E. (1972) Evidence for the retraction of Pseudomonas aeruginosa RNA phage pili. Biochem. Biophys. Res. Commun. 47, 142–149 [DOI] [PubMed] [Google Scholar]
- 9. Semmler A. B., Whitchurch C. B., Mattick J. S. (1999) A re-examination of twitching motility in Pseudomonas aeruginosa. Microbiology 145, 2863–2873 [DOI] [PubMed] [Google Scholar]
- 10. Craig L., Pique M. E., Tainer J. A. (2004) Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2, 363–378 [DOI] [PubMed] [Google Scholar]
- 11. Skerker J. M., Berg H. C. (2001) Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. U.S.A. 98, 6901–6904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Koo J., Burrows L. L., Howell P. L. (2012) Decoding the roles of pilotins and accessory proteins in secretin escort services. FEMS Microbiol. Lett. 328, 1–12 [DOI] [PubMed] [Google Scholar]
- 13. Sampaleanu L. M., Bonanno J. B., Ayers M., Koo J., Tammam S., Burley S. K., Almo S. C., Burrows L. L., Howell P. L. (2009) Periplasmic domains of Pseudomonas aeruginosa PilN and PilO form a stable heterodimeric complex. J. Mol. Biol. 394, 143–159 [DOI] [PubMed] [Google Scholar]
- 14. Karuppiah V., Derrick J. P. (2011) Structure of the PilM-PilN inner membrane type IV pilus biogenesis complex from Thermus thermophilus. J. Biol. Chem. 286, 24434–24442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tammam S., Sampaleanu L. M., Koo J., Sundaram P., Ayers M., Chong P. A., Forman-Kay J. D., Burrows L. L., Howell P. L. (2011) Characterization of the PilN, PilO, and PilP type IVa pilus subcomplex. Mol. Microbiol. 82, 1496–1514 [DOI] [PubMed] [Google Scholar]
- 16. Roux N., Spagnolo J., de Bentzmann S. (2012) Neglected but amazingly diverse type IVb pili. Res. Microbiol. 163, 659–673 [DOI] [PubMed] [Google Scholar]
- 17. Giltner C. L., Habash M., Burrows L. L. (2010) Pseudomonas aeruginosa minor pilins are incorporated into the type IV pilus. J. Mol. Biol. 398, 444–461 [DOI] [PubMed] [Google Scholar]
- 18. Winther-Larsen H. C., Wolfgang M., Dunham S., van Putten J. P., Dorward D., Løvold C., Aas F. E., Koomey M. (2005) A conserved set of pilin-like molecules controls type IV pilus dynamics and organelle-associated functions in Neisseria gonorrhoeae. Mol. Microbiol. 56, 903–917 [DOI] [PubMed] [Google Scholar]
- 19. Francetic O., Buddelmeijer N., Lewenza S., Kumamoto C. A., Pugsley A. P. (2007) Signal recognition particle-dependent inner membrane targeting of the PulG pseudopilin component of a type II secretion system. J. Bacteriol. 189, 1783–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Arts J., van Boxtel R., Filloux A., Tommassen J., Koster M. (2007) Export of the pseudopilin XcpT of the Pseudomonas aeruginosa type II secretion system via the signal recognition particle-Sec pathway. J. Bacteriol. 189, 2069–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Misic A. M., Satyshur K. A., Forest K. T. (2010) P. aeruginosa PilT structures with and without nucleotide reveal a dynamic type IV pilus retraction motor. J. Mol. Biol. 400, 1011–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Satyshur K. A., Worzalla G. A., Meyer L. S., Heiniger E. K., Aukema K. G., Misic A. M., Forest K. T. (2007) Crystal structures of the pilus retraction motor PilT suggest large domain movements and subunit cooperation drive motility. Structure 15, 363–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Py B., Loiseau L., Barras F. (2001) An inner membrane platform in the type II secretion machinery of Gram-negative bacteria. EMBO Rep. 2, 244–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wolfgang M., Lauer P., Park H. S., Brossay L., Hébert J., Koomey M. (1998) PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol. Microbiol. 29, 321–330 [DOI] [PubMed] [Google Scholar]
- 25. Whitchurch C. B., Mattick J. S. (1994) Characterization of a gene, pilU, required for twitching motility but not phage sensitivity in Pseudomonas aeruginosa. Mol. Microbiol. 13, 1079–1091 [DOI] [PubMed] [Google Scholar]
- 26. Carbonnelle E., Helaine S., Nassif X., Pelicic V. (2006) A systematic genetic analysis in Neisseria meningitidis defines the Pil proteins required for assembly, functionality, stabilization and export of type IV pili. Mol. Microbiol. 61, 1510–1522 [DOI] [PubMed] [Google Scholar]
- 27. Arts J., de Groot A., Ball G., Durand E., El Khattabi M., Filloux A., Tommassen J., Koster M. (2007) Interaction domains in the Pseudomonas aeruginosa type II secretory apparatus component XcpS (GspF). Microbiology 153, 1582–1592 [DOI] [PubMed] [Google Scholar]
- 28. Nunn D., Bergman S., Lory S. (1990) Products of three accessory genes, pilB, pilC, and pilD, are required for biogenesis of Pseudomonas aeruginosa pili. J. Bacteriol. 172, 2911–2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ramer S. W., Schoolnik G. K., Wu C. Y., Hwang J., Schmidt S. A., Bieber D. (2002) The type IV pilus assembly complex: biogenic interactions among the bundle-forming pilus proteins of enteropathogenic Escherichia coli. J. Bacteriol. 184, 3457–3465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tønjum T., Freitag N. E., Namork E., Koomey M. (1995) Identification and characterization of pilG, a highly conserved pilus-assembly gene in pathogenic Neisseria. Mol. Microbiol. 16, 451–464 [DOI] [PubMed] [Google Scholar]
- 31. Wolfgang M., van Putten J. P., Hayes S. F., Dorward D., Koomey M. (2000) Components and dynamics of fiber formation define a ubiquitous biogenesis pathway for bacterial pili. EMBO J. 19, 6408–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nudleman E., Wall D., Kaiser D. (2006) Polar assembly of the type IV pilus secretin in Myxococcus xanthus. Mol. Microbiol. 60, 16–29 [DOI] [PubMed] [Google Scholar]
- 33. Rumszauer J., Schwarzenlander C., Averhoff B. (2006) Identification, subcellular localization and functional interactions of PilMNOWQ and PilA4 involved in transformation competency and pilus biogenesis in the thermophilic bacterium Thermus thermophilus HB27. FEBS J. 273, 3261–3272 [DOI] [PubMed] [Google Scholar]
- 34. Ayers M., Sampaleanu L. M., Tammam S., Koo J., Harvey H., Howell P. L., Burrows L. L. (2009) PilM/N/O/P proteins form an inner membrane complex that affects the stability of the Pseudomonas aeruginosa type IV pilus secretin. J. Mol. Biol. 394, 128–142 [DOI] [PubMed] [Google Scholar]
- 35. Wolfgang M., Park H. S., Hayes S. F., van Putten J. P., Koomey M. (1998) Suppression of an absolute defect in type IV pilus biogenesis by loss-of-function mutations in pilT, a twitching motility gene in Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. U.S.A. 95, 14973–14978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kus J. V., Tullis E., Cvitkovitch D. G., Burrows L. L. (2004) Significant differences in type IV pilin allele distribution among Pseudomonas aeruginosa isolates from cystic fibrosis (CF) versus non-CF patients. Microbiology 150, 1315–1326 [DOI] [PubMed] [Google Scholar]
- 37. Chiang P., Sampaleanu L. M., Ayers M., Pahuta M., Howell P. L., Burrows L. L. (2008) Functional role of conserved residues in the characteristic secretion NTPase motifs of the Pseudomonas aeruginosa type IV pilus motor proteins PilB, PilT and PilU. Microbiology 154, 114–126 [DOI] [PubMed] [Google Scholar]
- 38. Harvey H., Habash M., Aidoo F., Burrows L. L. (2009) Single residue changes in the C-terminal disulfide-bonded loop of the Pseudomonas aeruginosa type IV pilin influence pilus assembly and twitching motility. J. Bacteriol. 191, 6513–6524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chiang P., Habash M., Burrows L. L. (2005) Disparate subcellular localization patterns of Pseudomonas aeruginosa type IV pilus ATPases involved in twitching motility. J. Bacteriol. 187, 829–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Milgotina E. I., Lieberman J. A., Donnenberg M. S. (2011) The inner membrane subassembly of the enteropathogenic Escherichia coli bundle-forming pilus machine. Mol. Microbiol. 81, 1125–1127 [DOI] [PubMed] [Google Scholar]
- 41. Py B., Loiseau L., Barras F. (1999) Assembly of the type II secretion machinery of Erwinia chrysanthemi: direct interaction and associated conformational change between OutE, the putative ATP-binding component and the membrane protein OutL. J. Mol. Biol. 289, 659–670 [DOI] [PubMed] [Google Scholar]
- 42. Abendroth J., Mitchell D. D., Korotkov K. V., Johnson T. L., Kreger A., Sandkvist M., Hol W. G. (2009) The three-dimensional structure of the cytoplasmic domains of EpsF from the type 2 secretion system of Vibrio cholerae. J. Struct. Biol. 166, 303–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Karuppiah V., Hassan D., Saleem M., Derrick J. P. (2010) Structure and oligomerization of the PilC type IV pilus biogenesis protein from Thermus thermophilus. Proteins 78, 2049–2057 [DOI] [PubMed] [Google Scholar]
- 44. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 45. Roy A., Kucukural A., Zhang Y. (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Scheurwater E. M., Burrows L. L. (2011) Maintaining network security: how macromolecular structures cross the peptidoglycan layer. FEMS Microbiol. Lett. 318, 1–9 [DOI] [PubMed] [Google Scholar]
- 47. Gray M. D., Bagdasarian M., Hol W. G., Sandkvist M. (2011) In vivo cross-linking of EpsG to EpsL suggests a role for EpsL as an ATPase-pseudopilin coupling protein in the type II secretion system of Vibrio cholerae. Mol. Microbiol. 79, 786–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Georgiadou M., Castagnini M., Karimova G., Ladant D., Pelicic V. (2012) Large-scale study of the interactions between proteins involved in type IV pilus biology in Neisseria meningitidis: characterization of a subcomplex involved in pilus assembly. Mol. Microbiol. 84, 857–873 [DOI] [PubMed] [Google Scholar]
- 49. Savvides S. N. (2007) Secretion superfamily ATPases swing big. Structure 15, 255–257 [DOI] [PubMed] [Google Scholar]
- 50. Collins R. F., Saleem M., Derrick J. P. (2007) Purification and three-dimensional electron microscopy structure of the Neisseria meningitidis type IV pilus biogenesis protein PilG. J. Bacteriol. 189, 6389–6396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yamagata A., Tainer J. A. (2007) Hexameric structures of the archaeal secretion ATPase GspE and implications for a universal secretion mechanism. EMBO J. 26, 878–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Herdendorf T. J., McCaslin D. R., Forest K. T. (2002) Aquifex aeolicus PilT, homologue of a surface motility protein, is a thermostable oligomeric NTPase. J. Bacteriol. 184, 6465–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]