Background: RHBDD2 is distantly related to rhomboids, membrane-bound proteases.
Results: In retina, RHBDD2 exists as a monomer in all cells throughout life and a homotrimer only in cone outer segments; a mutation in RHBDD2 possibly leads to retinitis pigmentosa.
Conclusion: RHBDD2 plays important roles in development and normal retinal function.
Significance: This is the first characterization of RHBDD2 and its association with retinal disease.
Keywords: Cloning; Gene Expression; Immunohistochemistry; Molecular Cell Biology; Mutagenesis, Site-specific; Retina; Cone Outer Segments; Glycine Zipper; Golgi Localization; Retinitis Pigmentosa
Abstract
The novel rhomboid-like protein RHBDD2 is distantly related to rhomboid proteins, a group of highly specialized membrane-bound proteases that catalyze regulated intramembrane proteolysis. In retina, RHBDD2 is expressed from embryonic stages to adulthood, and its levels show age-dependent changes. RHBDD2 is distinctly abundant in the perinuclear region of cells, and it localizes to their Golgi. A glycine zipper motif present in one of the transmembrane domains of RHBDD2 is important for its packing into the Golgi membranes. Its deletion causes dislodgment of RHBDD2 from the Golgi. A specific antibody against RHBDD2 recognizes two forms of the protein, one with low (39 kDa; RHBDD2L) and the other with high (117 kDa; RHBDD2H) molecular masses in mouse retinal extracts. RHBDD2L seems to be ubiquitously expressed in all retinal cells. In contrast, RHBDD2H seems to be present only in the outer segments of cone photoreceptors and may correspond to a homotrimer of RHBDD2L. This protein consistently co-localizes with S- and M-types of cone opsins. We identified a homozygous mutation in the human RHBDD2 gene, R85H, that co-segregates with disease in affected members of a family with autosomal recessive retinitis pigmentosa. Our findings suggest that the RHBDD2 protein plays important roles in the development and normal function of the retina.
Introduction
Highly specialized membrane-bound proteases, which cleave other membrane proteins within their membrane-spanning segments, have been intensively studied during the last decade. These enzymes are identifiable in nearly all the sequenced genomes of archaea, bacteria, eukaryotic organisms, and mitochondria. They influence diverse processes, including transcriptional control, cellular differentiation, lipid metabolism, mitochondrial membrane remodeling, parasite invasion, and bacterial protein translocation (1–6). Rhomboid proteins, widely conserved polytopic membrane proteins with six or seven transmembrane domains, are members of the intramembrane proteolytic protein family. The rhomboid gene was first discovered in Drosophila and is an important component of the epidermal growth factor (EGF) receptor signaling pathway during development (7–10). Rhomboids directly recognize their substrates in vitro (11), and their conserved Ser and His residues are essential for proteolytic activity (12). Their active site is situated close to the extracellular face of the membrane, which allows them to release effector domains to the cell exterior. Mainly, rhomboids cleave transmembrane segments of type I orientation (N termini outside the cell) (6) and regulate the location and topology of membrane proteins with great precision.
A simplified phylogenetic tree of the rhomboid family shows three major taxonomic groups: the rhomboids, inactive rhomboids (iRhoms),2 and inactive rhomboid homologues (13). The eukaryotic active rhomboids are a major group consisting of presenilin-associated rhomboid-like (PARL)-type rhomboids, which are mitochondrial, and secretases (types A and B), which reside in the secretory pathway.
Rhomboid-like proteins that lack essential catalytic residues are identified as iRhoms. Their characteristic features are a large loop between transmembrane domains 1 and 2 and an extended cytoplasmic N terminus (13, 14). Little is known about their function. Other rhomboid-like homologues that lack catalytic residues but do not cluster with the iRhoms are scattered across evolution.
Rhomboids have been implicated in a variety of human diseases as a result of their distinct functions. As a component of the mechanism of parasitic invasion in toxoplasmosis and malaria, these cell surface molecules obligate human pathogens to invade the host cells by forming irreversible junctures between the plasma membrane of the invading parasite and the host cell (15–17). As another example, the mitochondrial Rhomboid-7 is required to cleave the precursor forms of both Pink1 and Omi, proteins that are mutated in Parkinson disease (18). In addition, an identified mutation in PARL may be associated with insulin resistance and type 2 diabetes (19). Recently, two independent groups reported that iRhom2 is required for tumor necrosis factor release in mice. iRhom2 interacts with TNFα-converting enzyme and regulates shedding of soluble, active TNFα. Thus, iRhom2 may represent an attractive therapeutic target for treating TNFα-mediated diseases (20, 21).
The focus of this study is the novel intramembrane, growth/development-associated, rhomboid-like protein RHBDD2. Here we describe the distribution of Rhbdd2 transcripts in mouse tissues and their developmental expression as well as that of the RHBDD2 protein in retina. We show that two forms of RHBDD2 are present in mouse retina and demonstrate that one of these forms is specifically expressed in cone photoreceptor outer segments. Most importantly, we report a novel recessive missense mutation in the RHBDD2 gene, which maps to the human 7q11 locus, in members of a family affected with autosomal recessive retinitis pigmentosa (arRP). This mutation co-segregates with the disease and links the RHBDD2 gene to the arRP phenotype.
EXPERIMENTAL PROCEDURES
Animals
C57BL/6J mice were obtained from our colonies bred from stock originated at The Jackson Laboratory (Bar Harbor, ME). Mouse eyes were quickly enucleated after death, and the retinas were dissected and frozen. In addition, other tissues were obtained from these animals and immediately frozen. All experiments were conducted in accordance with the approved UCLA Animal Care and Use Committee protocol and the Association for Research in Vision and Ophthalmology statement for the use of animals in ophthalmic and vision research.
Embryo Collection
Pregnant C57BL/6 mice were sacrificed at 12.5, 15.5, and 18.5 days postcoitum, and embryos were removed from the embryonic sac. Theiler staging was used to confirm the phenotype of the 12.5-, 15.5-, and 18.5-day-postcoitum embryos. Embryonic heads were fixed and embedded in OCT for cryosectioning and immunohistochemistry, whereas embryonic eyes were collected for RNA extraction and quantitative PCR analysis. For the former experiments, embryo heads were fixed in 4% paraformaldehyde in 10 mm phosphate-buffered saline (PBS) overnight. The next day, the tissue was rinsed three times with PBS and infiltrated with 30% sucrose in PBS overnight at 4 °C with gentle rotation. The following day, embryonic heads were placed overnight in a 1:1 ratio of 30% sucrose in PBS to OCT. Heads were then embedded in OCT and stored at −80 °C. The embedded tissue was sectioned (10 μm) and used for immunohistochemistry experiments.
RNA Isolation and Northern Blot Analysis
Total RNA was extracted from mouse retinas (TRIzol, Invitrogen). Poly(A+) RNA was obtained with an mRNA purification kit (Oligotex, Qiagen, Valencia, CA). RNA was quantified with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE) and stored at −80 °C. Two micrograms of poly(A+) RNA were electrophoresed on 1.2% denaturing formaldehyde-agarose gels and transferred to a Hybond N+ membrane (Amersham Biosciences). An 821-bp mouse retinal cDNA fragment containing part of the coding region and 3′-UTR was amplified with forward (5′-CTCATCTGACTCCAAGTTATC) and reverse (5′-AGAAGCCCAGGAGCCTCAAGAC) primers from the sequence of Rhbdd2, labeled with [32P]dCTP, and used as a probe to hybridize the retinal and multiple mouse tissue (Ambion, Austin, TX) Northern blots.
In Situ Hybridization
In situ hybridization using 8-μm frozen sections of mouse retina embedded in OCT was performed as described previously (22). The same fragment of the mouse Rhbdd2 cDNA used for Northern blot analysis was subcloned into the pCRII plasmid vector (Invitrogen) for generation of riboprobes. The antisense and sense digoxigenin (DIG)-labeled RNA riboprobes were synthesized with SP6 and T7 RNA polymerases (according to the DIG Labeling kit protocol, Roche Applied Science) and purified by spin columns (NucWay, Ambion). Sections were permeabilized with proteinase K (1 μg/ml). Prehybridization was performed at 70 °C for 30 min in a mixture of 50% formamide, 5× SSC (SSC: 150 mm NaCl, 15 mm sodium citrate, pH 4.5), 50 μg/ml yeast RNA, 50 μg/ml heparin, and 1% SDS. Hybridization occurred overnight at 70 °C in a humidified chamber. Sections were then washed with 5× SSC and 1% SDS in 50% formamide at 70 °C; 2× SSC in 50% formamide at 65 °C; and Tris-buffered saline, 0.1% Tween 20 at room temperature. Sections were incubated overnight at 4 °C with alkaline phosphatase-conjugated sheep anti-DIG antibody (1:200; Roche Applied Science) in 25 mm Tris-HCl, pH 7.5, 140 mm NaCl, 2.7 mm KCl, 1% Tween 20, 1% sheep serum; washed with TBST (140 mm NaCl, 2.7 mm KCl, 25 mm Tris-HCl, pH 7.5, 0.1% Tween 20); and incubated with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate to visualize the mRNA. The reaction was stopped with acidic PBS containing 0.1% Tween 20.
Quantitative Real Time RT-PCR and Statistical Analyses
To analyze the levels of Rhbdd2 transcripts, quantitative PCR was performed on first strand cDNAs. Primers were designed using Primer 3 Internet software (Whitehead Institute, Massachusetts Institute of Technology, Cambridge, MA) and synthesized by Integrated DNA Technology (San Diego, CA). The primer pair for total Rhbdd2 mRNA was: forward, 5′-TCCCTCAGACCTCCTTCCTC and reverse, 5′-GTCAGATGAGGGTGGCAACTC. The primer pair for the long 3′-UTR Rhbdd2 mRNA was: forward, 5′-GATGTGGGCTCTTAGGCAAG and reverse, 5-′ATCTAGGGGCAGTCCATCAG. Total RNA was isolated from embryonic eyes and retinal samples using the RNAqueous 4PCR protocol (Ambion) and treating twice with DNase I. RNA concentrations were measured using the NanoDrop ND-1000 spectrophotometer, and 1 μg of RNA from each age was reverse transcribed. The quantitative PCRs were performed in SYBR Green Master Mix with the corresponding primer sets using a quantitative PCR system (MX3000P, Stratagene, La Jolla, CA). The melting curves of the PCR products were monitored to ensure that a single melting curve was obtained for each of the samples. To analyze the real time PCR data, signals from each sample were normalized to values obtained for β-actin cDNA, which was assayed simultaneously with the experimental samples. Analysis of variance using Monte Carlo bootstrapping was performed to analyze the possibility of a significant time effect in the expression of the total and long 3′-UTR Rhbdd2 RNAs. Post hoc analyses were applied to determine significant changes in the total mRNA levels among the different age points. Bootstrap analysis was used because of the lack of a normal distribution in our data.
Generation of Anti-RHBDD2 Antibody
The RHBDD2 C-terminal peptide containing amino acids 344–360 (LGTPGATGSKESSKVAM) was synthesized and used to immunize rabbits for generation of the anti-RHBDD2 antibody. The obtained antibody (designated 7Rc) was affinity-purified (ProSci Inc., Poway, CA).
Expression Construct
To produce the full-length protein, the full-size Rhbdd2 cDNA was amplified via RT-PCR from mouse retinal RNA using a primer set (5′-GGATCCATGGCGGCCCCGGGCCCCGCGAGT) and (5′-GAATTCCTTAGGGCAT GGCTACCTTGGAAGA) containing the desired restriction enzyme recognition sites (BamHI and EcoRI). The digested and purified PCR product (PCR gel extraction kit, Qiagen) was cloned into the pcDNA4/HisMax C vector between its BamHI/EcoRI cloning sites and sequenced for confirmation. The CMV promoter of this vector drove expression of the RHBDD2 fusion protein with the Xpress epitope at its N terminus.
Site-directed Mutagenesis to Modify the Glycine Zipper Motif
All glycine zipper motif mutant cDNAs were created using site-directed mutagenesis (QuikChange® II site-directed mutagenesis kit, Stratagene) to change the glycines in the zipper to leucines. S185L, G189L, G193L, and G197L are the single leucine substitution mutants (SSMs). In the triple substitution mutant, the Gly-189, Gly-193, and Gly-197 were collectively replaced with leucines. To delete 27 nucleotides from the glycine zipper motif sequence, two overlapping primers were made, each having 36 nucleotides. The forward primer consisted of 9 nucleotides upstream of the motif and 27 nucleotides downstream of the motif. The reverse primer had 27 nucleotides upstream of the motif and 9 nucleotides downstream. The two primers had an overlapping region of 18 nucleotides lacking the 9 amino acid residues of the motif. The PCR mixture was set up according to the QuikChange II XL site-directed mutagenesis kit. Once the PCR was completed, 10 units of DpnI were added, and the reaction mixture was incubated at 37 °C for 1 h to degrade the original plasmid used for PCR. EcoRI and BamHI sites were introduced into the 5′- and 3′-ends of the wild-type and mutant cDNAs to directionally subclone them into the pEGFP-N3 vector (BD Biosciences Clontech). The RHBDD2-EGFP fusion constructs were verified by sequencing. The list of specific primers used in site-directed mutagenesis is presented in Table 1.
TABLE 1.
The sequences of the PCR primers used to obtain the 7R mutants
Mutated codons are in bold. TSM, triple substitution mutant; GZMD, glycine zipper motif (amino acids 189–197) deletion mutant.
| Mutants | Primer name | Primer sequence (5′–3′) |
|---|---|---|
| SSM/S185L | m7R 4F | CAGACCTCCTTCCTCCTTAACGTCTCTGGCCTT |
| m7R 4R | AAGGCCAGAGACGTTAAGGAGGAAGGAGGTCTG | |
| SSM/G189L | m7R 5F | CTCAGTAACGTCTCTCTCCTTTTGATTGGCCTG |
| m7R 5R | CAGGCCAATCAAAAGGAGAGAGACGTTACTGAG | |
| SSM/G193L | m7R 6F | CTCTGGCCTTTTGATTCTCCTGTCCTATGGCCT |
| m7R 6R | AGGCCATAGGACAGGAGAATCAAAAGGCCAGAG | |
| SSM/G197L | m7R 7F | GATTGGCCTGTCCTATCTCCTCACCTACTGCTAC |
| m7R 7R | GTAGCAGTAGGTGAGGAGATAGGACAGGCCAATC | |
| TSM | m7R 8F | CTCTCTCCTTTTGATTCTCCTGTCCTATCTCCT |
| m7R 8R | AGGAGATAGGACAGGAGAATCAAAAGGAGAGAG | |
| GZMD | m7R 9F | AACGTCTCTCTCACCTACTGCTACTCCCTCGACCTC |
| m7R 9R | GTAGGTGAGAGAGACGTTACTGAGGAAGGAGGTCTG |
Transient and Stable Transfections
HEK293 cells obtained from the American Type Culture Collection (Manassas, VA) were grown and transfected using the pcDNA4/HisMax-RHBDD2, each of the pEGFP-N3-RHBDD2 expression constructs, and the PolyFect transfection reagent (Qiagen). All experiments included the pcDNA4/HisMax plasmid without the insert as an internal normalization control. Transiently transfected cells were harvested for analysis of newly synthesized protein. Linearized plasmid was used to achieve stable transfection of HEK293 cells, and positive clones were selected (Zeocin, Invitrogen).
Protein Extraction and Immunoblot Analysis
Nuclear and cytoplasmic protein extracts from transfected cells, mouse retinas, and mouse brains were prepared using nuclear and cytoplasmic extraction reagents (NE-PER, Pierce). For total protein extraction, the harvested cells and tissue samples were lysed with lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 10% (w/v) glycerol, 100 mm NaF, 10 mm EGTA, 1 mm Na3VO4, 1% (w/v) Triton X-100, 5 μm ZnCl2) and the Complete EDTA-free protease inhibitor mixture (Roche Applied Science) and then centrifuged at 10,000 × g for 20 min at 4 °C. Fifty micrograms of the extracted proteins were separated by SDS-PAGE on 7.5% gels (Pierce). Blots were incubated with primary antibody (1:7000 dilution) and secondary anti-rabbit IgG antibodies labeled with alkaline phosphatase (1:5000 dilution; Vector Laboratories). Western blots were visualized with either of two kits (the Amplified-Alkaline Phosphatase kit from Bio-Rad or the enhanced chemiluminescence (ECL) kit from Amersham Biosciences).
Immunostaining of Cultured Cells
Transfected HEK293 cells on coverslips were permeabilized with 100% methanol for 6 min at −20 °C, rinsed three times in PBS, blocked with 3% BSA in PBS containing 0.1% Triton X-100 (PBST) for 45 min, and incubated for 2 h with 7Rc rabbit polyclonal antibody (1:200 dilution) and subsequently for 1 h with fluorescein- or rhodamine-conjugated goat anti-rabbit antibody (1:200; Santa Cruz Biotechnology, Santa Cruz, CA). Next, the transfected cells were incubated for 1 h with a mouse monoclonal anti-Xpress-FITC antibody (1:100 dilution; Invitrogen). The cells were then washed three times in PBST, stained with propidium iodide or 4′,6-diamidino-2-phenylindole (DAPI) for detection of nuclei, and viewed with fluorescence microscopy.
Immunostaining for Golgi markers GM130 and TGN38 was carried out following the same protocol described above using antibodies from Abcam (Cambridge, MA) at 1:200 and 1:500 dilutions, respectively. The co-localization of these Golgi markers and wild-type and mutant RHBDD2 was quantified using the program Olympus Fluoview Version 3.0. Transfected cells were outlined to quantify the degree of overlap. Co-localization was reported as a Pearson's coefficient, which measures how well the pixels from two different color channels fit to a linear relation. The Pearson's coefficient can have values from −1 to 1 with 1 indicating absolute overlap, 0 showing that there is no co-localization, and −1 representing an inverse correlation (relatively high scores on one variable paired with relatively low scores on the other variable).
Immunostaining of Mouse Retinal Sections
All tissue processing, image acquisition, and analysis procedures were as described (23–25). Central sections (10 μm thick; three nonadjacent sections per slide) were located 200–400 μm from the optic nerve. Before immunofluorescence staining, the sections were incubated with the same primary and secondary antibodies described in the previous section. To show the co-localization of RHBDD2 with both cone and Müller cells, sections were also incubated with cone opsin antibodies (short wavelength-sensitive opsin (S opsin) (OS2) and midwavelength-sensitive opsin (M opsin) (COS1) kindly provided by Dr. Agoston Szél, Semmelweis University Medical School, Budapest, Hungary) and glutamine synthetase antibody (1:5000; Sigma-Aldrich G2781, rabbit polyclonal), respectively, with the corresponding secondary antibodies. Confocal images were acquired using a Leica TCS SP2 laser-scanning confocal microscope (Leica Microsystems, Exton, PA). Images were processed using Adobe Photoshop software. The results shown are representative of five separate immunolabeling experiments from three different adult C57BL/6 mouse retinas.
For unknown reasons, retinal sections incubated with GM130 antibody in the conditions used for cultured HEK293 cells showed no results. Therefore, we obtained retinal sections following the fixation times and temperatures described by Kerov et al. (26). Briefly, the enucleated mouse eyes were poked through the cornea with a 21-gauge needle and fixed with 4% paraformaldehyde in PBS for 1 h at 25 °C. After fixation and removal of the cornea and lens, the eyes were hemisected, submersed in a 30% sucrose solution in PBS for 5 h at 48 °C, then embedded in OCT, frozen, and sectioned. The 10-μm sections were incubated for 2 h with 7Rc (1:200 dilution) and GM130 antibody (1:50 dilution) and subsequently for 1 h with Alexa Fluor 488 goat anti-rabbit or Alexa Fluor 568 goat anti-mouse secondary antibodies (1:500; Invitrogen).
Screening the DNA of Patients with Retinal Degenerations for Variants in the RHBDD2 Gene
The DNA of 110 unrelated patients of mixed ethnicities who were diagnosed with various retinal diseases such as cone dystrophy, cone-rod dystrophy, Stargardt disease, and autosomal dominant and recessive retinitis pigmentosa was screened for variants in the RHBDD2 gene. The DNA of 95 control individuals with similar ethnic distribution (56% white, 15% black, 22% Asian, and 7% Hispanic) was also screened for RHBDD2 variants. PCR products resulting from the targeted amplification of RHBDD2 coding sequences were subjected to dideoxy sequencing and analyzed using Sequencher software (Gene Codes Corp., Ann Arbor, MI).
RESULTS
Cloning and Characterization of the Rhbdd2 cDNA
To identify novel cone photoreceptor genes, we carried out representational difference analysis using mRNAs from adult cone degeneration (cd) and normal dog retinas as described previously (27). One of the clones isolated in the screen was predicted to encode a part of a protein containing the rhomboid domain. With this clone as a probe, we then screened a dog retinal cDNA library and isolated an ∼2.0-kb cDNA clone that appeared to have the entire open reading frame of the predicted rhomboid domain-containing protein. Using public database information, the sequence of this clone was found to be highly homologous to sequences of mRNAs from three different species: mouse (GenBankTM accession number BC018360, identified as rhomboid domain-containing 2 (Rhbdd2)), rat (GenBank accession number XM_341058, called rhomboid veinlet-like 7 (Rhbdl7)), and human (GenBank accession numbers AF226732 and BC069017, identified as NPD007 and RHBDD2, respectively). We then isolated the corresponding mouse cDNA from a mouse retinal cDNA library. Because the database mouse mRNA sequence and that of our isolated mouse clone did not have an in-frame termination codon upstream of the first methionine and the N termini of the human AF226732 and rat clones encode predicted proteins that are 45 and 27 amino acids longer than BC018360, respectively, we needed to confirm whether our mouse clone was full length or a fragment. We searched for cDNAs with a longer 5′-end using 5′ RACE (rapid amplification of cDNA ends) (Invitrogen) but were unable to isolate a longer transcript after multiple attempts. Therefore, we concluded that this mRNA contains the complete ORF that encodes a protein of 361 amino acids. We also identified another mouse Rhbdd2 transcript with the same ORF but with a very long 3′-UTR and isolated two human transcripts that are splicing variants produced by insertion of 122- or 126-bp fragments into the first intron of RHBDD2. Both isoforms contain a shorter ORF than that of BC069017 and code for a human protein of 223 instead of 364 amino acids. Using an InterProScan algorithm, we found that RHBDD2, an uncharacterized 39-kDa protein that is highly conserved between species, contains five transmembrane helices; however, TMpred parameters revealed seven possible transmembrane helices.
Distribution of Rhbdd2 mRNA in Mouse Tissues and within the Retina
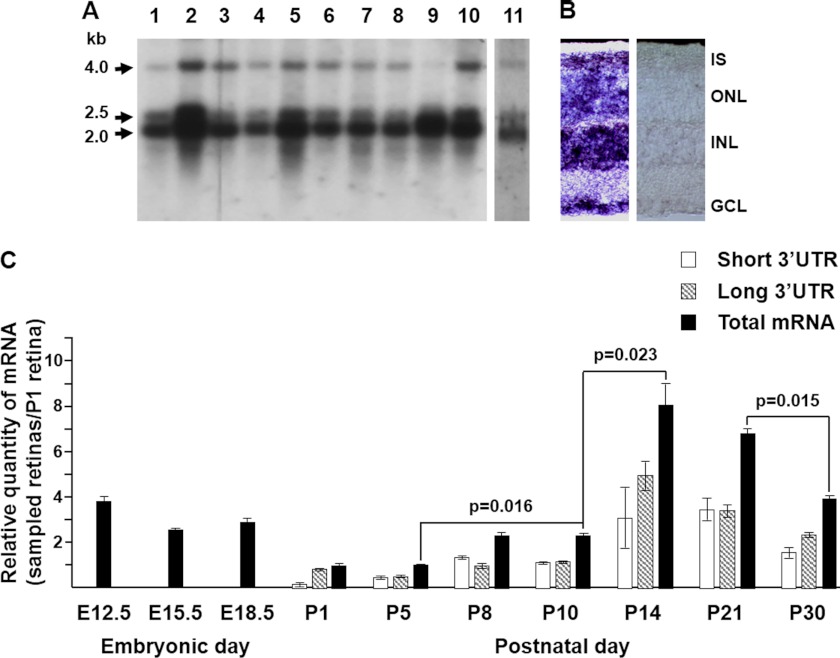
Northern blots from different mouse tissues hybridized with the mouse Rhbdd2 cDNA show a major transcript of ∼2.0 kb in all tissues studied with very intense signals in brain, kidney, testis, and ovary and weaker bands in heart, liver, spleen, embryo, and lung (Fig. 1A). Two other minor transcripts, one of ∼2.5 kb with tissue distribution identical to that of the 2.0-kb band, and a second of ∼4.0 kb found mainly in brain, liver, kidney, and ovary but hardly present in testis, are also observed. The retinal Rhbdd2 mRNA (lane 11) displays the same hybridization pattern (Fig. 1A). The 2.0- and 4.0-kb transcripts are in agreement with the length of the isolated Rhbdd2 cDNAs, but the 2.5-kb mRNA had not been described previously.
FIGURE 1.
Rhbdd2 mRNA expression in mouse tissues and its quantification during retinal development. A, a mouse multitissue Northern blot of mRNAs from heart (lane 1), brain (lane 2), liver (lane 3), spleen (lane 4), kidney (lane 5), embryo (lane 6), lung (lane 7), thymus (lane 8), testis (lane 9), and ovary (lane 10) and a separate blot containing mouse retinal mRNA (lane 11) were probed with a 32P-labeled Rhbdd2 cDNA. Arrows indicate the sizes of the three detected transcripts (2.0, 2.5, and 4 kb), respectively. B, in situ hybridization of Rhbdd2 mRNAs in mouse retina with antisense (left panel) and sense (right panel) DIG-labeled riboprobes. The Rhbdd2 mRNA localized to all the nuclear layers; no signal was detected with the sense probe. C, quantitative RT-PCR amplification of mRNAs from mouse embryonic eyes and postnatal retinas at different times of development. Relative expression of Rhbdd2 is normalized to the β-actin mRNA level. Error bars indicate S.E. IS, inner segment; ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer.
In situ hybridization using a Rhbdd2 antisense riboprobe to assess the expression of Rhbdd2 mRNA in mouse retina shows its presence in cell bodies located in all layers of the retina (Fig. 1B) as well as in the inner segments of photoreceptor cells.
To define the developmental expression pattern of Rhbdd2 mRNA in the mouse retina, we performed qRT-PCR on mRNA from eyes of E12.5, E15.5, and E18.5 embryos and in retinas from mice at birth (P1), P5, P8, P10, P14, P21, and P30 and compared the levels at all ages with that observed at P1 (Fig. 1C, black bars). The obtained data were subjected to repeated one-way analysis of variance to check for a significant effect of time on mRNA expression. Success in the Omnibus test indeed suggested significance in the effect of time on mRNA transcription (F statistic = 7.44). Our results indicate that the amount of transcript at E12.5 is higher than at the more advanced embryonic stages, and it is very similar to that of total mRNA measured in the adult retina at P30 (Fig. 1C). Rhbdd2 mRNA levels after birth (P1) and until P5 are further reduced from those at E15.5 and E18.5 and are not significantly different (p = 0.93). At P8 and P10, Rhbdd2 mRNA levels are double that at P5 (p = 0.016). The strongest expression is observed at P14 (p = 0.023), and it decreases thereafter with a significant change between P21 and P30 (p = 0.015). We also measured by qRT-PCR the levels of the long 3′-UTR transcript at all the different postnatal developmental stages (striped bars) and determined the levels of the short 3′-UTR mRNAs (white bars) by subtraction from the total mRNA. Except for the P1 and P30 samples, both short and long 3′-UTR transcripts seem to contribute similarly to the total amount of Rhbdd2 mRNA.
Expression of the RHBDD2 Protein in Mouse Retina during Development
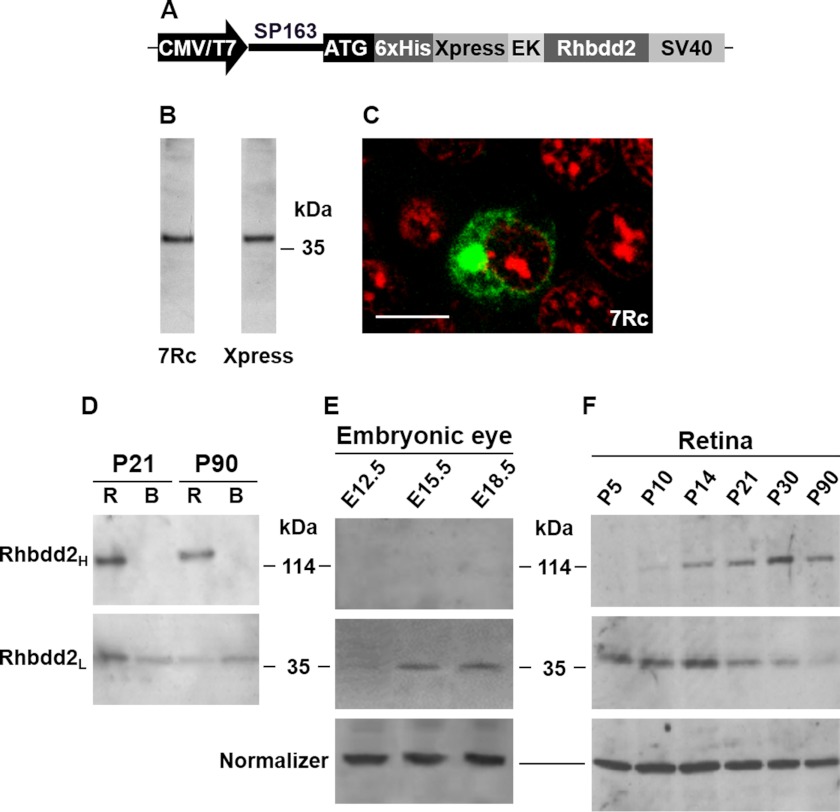
A rabbit antiserum against RHBDD2 (referred to as 7Rc) was generated using a synthetic C-terminal peptide of RHBDD2 (ProSci Inc.). To determine the specificity of this antiserum, we prepared an Xpress-tagged RHBDD2 expression vector in the pcDNA4/HisMax plasmid (Invitrogen), which contained the predicted open reading frame of Rhbdd2 cDNA inserted downstream of the polyhistidine (His6) tag and the Xpress epitope region (Fig. 2A). This vector was transfected into HEK293 cells. Immunoblotting analysis of protein extracts from these cells demonstrated that 7Rc strongly bound a ∼44-kDa protein, also recognized by the Xpress antibody (Fig. 2B). The molecular mass of this protein, ∼5 kDa heavier than that estimated from the RHBDD2 primary structure (39 kDa), may result from the additional 32-amino acid vector sequence that was fused to the N-terminal region of RHBDD2 as well as from possible modification(s) of the protein. In addition, the immunocytochemical localization of RHBDD2 in transfected HEK cells using 7Rc or the Xpress antibody was identical (thus, only 7Rc staining is shown in Fig. 2C). The staining was strong in a distinct perinuclear region, and it resembled a bead necklace around the nucleus. Further confirmation of the specificity of 7Rc for RHBDD2 was obtained by the disappearance of all labeling in RHBDD2-transfected HEK cells incubated with 7Rc that had been preabsorbed with the polypeptide used to generate it.
FIGURE 2.
Expression of the RHBDD2 protein in transfected HEK293 cells and in mouse embryonic eyes, postnatal retina, and brain. A, schematic illustration of the pcDNA4/HisMax RHBDD2 expression construct. The vector contains the SP163 translational enhancer directly upstream of the ATG initiation codon, and the enterokinase (EK) cleavage site preceding the Rhbdd2 cDNA. B, Western blot of a crude protein extract obtained from cultured Rhbdd2-transfected cells using 7Rc (left lane) and the Xpress antibody (right lane). C, immunocytochemical detection of the RHBDD2 protein (green) after transient transfection of the expression vector into HEK293 cells. Propidium iodide was used to visualize the nucleus of the cells (red). Scale bar, 15 μm. D, retinal and brain protein extracts (60 μg each) from P21 and P90 mice were subjected to 8% SDS-PAGE under reducing conditions followed by immunoblotting using 7Rc. The lower panel shows 39-kDa protein bands in both tissue samples. The 117-kDa band identified in retinal extract was absent in brain, indicating tissue specificity (upper panel). R, retina; B, brain. E, expression of RHBDD2 in mouse embryonic eyes during development. Protein extracts (50 μg) were loaded into each lane of a 4–12% gel that was subjected to SDS-PAGE, and results were normalized to α-tubulin. F, Western blot of retinal extracts at different developmental times. The upper and middle blots correspond to the trimeric and monomeric forms of RHBDD2, respectively. Protein levels were normalized to β-actin.
Because the same Rhbdd2 transcripts were observed in all mouse tissues studied, we initially examined the expression of the RHBDD2 protein only in retina and brain. As shown in Fig. 2D, 7Rc recognized the 39-kDa protein band (RHBDD2L; monomer) in both tissues at P21 and P90 and an additional protein band of 117 kDa (RHBDD2H; trimer) only in retinal extracts. We then tested other mouse tissues for RHBDD2H but did not detect it (data not shown).
To determine the expression of RHBDD2 from mouse retina during development, we obtained retinal protein extracts from E12.5, E15.5, and E18.5 embryonic eyes and from retinas of P5, P10, P14, P21, P30, and P90 animals and subjected them to Western blot analyses using 7Rc. Interestingly, the RHBDD2L and RHBDD2H proteins recognized by the 7Rc antibody respond in an opposing manner as mice age. The data indicate that although RHBDD2H is not seen at embryonic stages (Fig. 2E), only increases from P5 to P30, and remains at similar levels at P90 (Fig. 2F), the levels of RHBDD2L increase from E12.5 to E15.5 (Fig. 2E) and are thereafter similar until P14, decrease between this time and P30, and remain the same until P90 (Fig. 2F). Furthermore, the total level of the two retinal RHBDD2 proteins (RHBDD2L plus RHBDD2H) is higher at P14 compared with other ages. This is in agreement with the expression profile of mRNA in developing retina (Fig. 1C).
Golgi Localization of RHBDD2
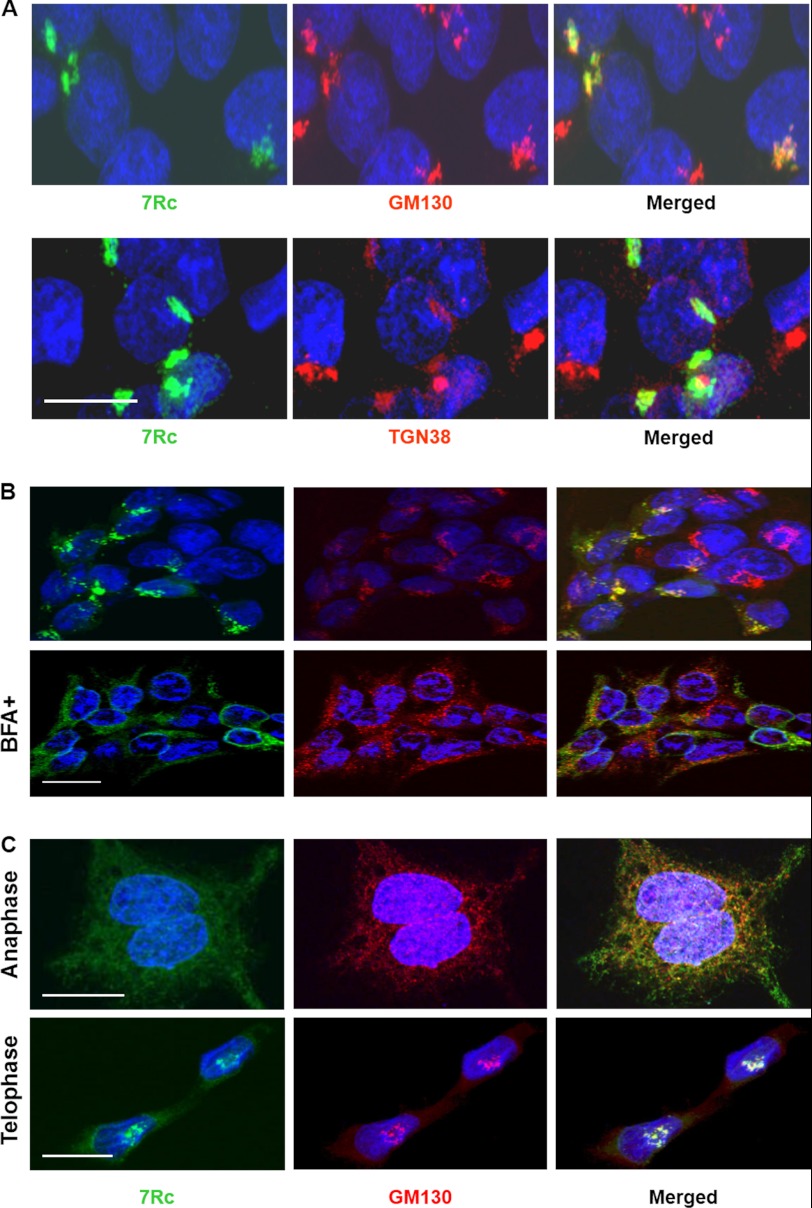
As seen in Fig. 2C, 7Rc strongly labeled the region where the Golgi apparatus resides in Rhbdd2-transfected HEK293 cells. Therefore, we used Golgi markers to analyze the localization of RHBDD2 in this structure of the cells. Fig. 3A shows that the RHBDD2 signal exhibits a high degree of overlap with the signal of the cis-Golgi matrix protein GM130 (Pearson's coefficient, 0.95 ± 0.01; top panel). On the other hand, 7Rc staining appears to overlap to a much lesser extent with that of anti-TGN38, a trans-Golgi marker protein (Pearson's coefficient, 0.32 ± 0.03; bottom panel). These results suggest that the overexpressed RHBDD2 is localized predominantly to the cis-side of the Golgi apparatus and may function in the early stages of the endosomal sorting pathway rather than during the later stages.
FIGURE 3.
Golgi localization of RHBDD2 in transfected HEK293 cells. A, immunocytochemical detection of RHBDD2 and GM130 (top panel) or RHBDD2 and TGN38 (bottom panel) proteins after transient transfection of the Rhbdd2 expression vector into HEK293 cells. The obtained images for each staining were merged. Scale bar, 15 μm. B, Rhbdd2-transfected cells with or without 50 μm BFA were incubated for 30 min at 37 °C before fixation. Cells were stained with 7Rc or anti-GM130 antibody. RHBDD2 co-localization with GM130 is sensitive to the exposure of cells to BFA. C, RHBDD2 and GM130 co-localize during mitosis. RHBDD2-expressing cells were fixed, permeabilized, and double stained with the same antibodies during anaphase and telophase. Scale bars for B and C, 10 μm.
It has been observed before that upon treatment of cells with brefeldin A (BFA), a Golgi-destabilizing agent, the components of the cis-Golgi matrix intersperse in the cytoplasm and appear as puncta (28). Furthermore, other studies have demonstrated that Golgi-specific spectrins associate with Golgi membranes in a BFA-sensitive manner (29). Therefore, as an alternative approach to evaluate the Golgi localization of RHBDD2, we tested its sensitivity to BFA to examine whether BFA treatment would disrupt the association of RHBDD2 with the Golgi complex as in the case of spectrins. Fig. 3B shows that treatment of Rhbdd2-transfected HEK293 cells with BFA for 30 min results in disruption of the stacked Golgi structure, inducing a rapid spreading of RHBDD2, as demonstrated by staining with anti-GM130. This result indicates that RHBDD2 associates with the Golgi membranes and that this association is BFA-sensitive.
The structural integrity of the Golgi apparatus is compromised during mitosis as part of the normal inheritance process. We analyzed the localization of RHBDD2 in the cells during mitosis to determine in which compartment it resides. Exponentially growing and stably transfected HEK293 cells were fixed and double stained with 7Rc and anti-GM130 antibodies. Fig. 3C shows co-localization of RHBDD2 and GM130 even when the Golgi apparatus is dispersed during anaphase (Pearson's coefficient, 0.741 ± 0.008). In telophase, the Golgi apparatus reformed in each of the daughter cells that display a high degree of RHBDD2 and GM130 co-localization (Pearson's coefficient, 0.852 ± 0.008). These results suggest that RHBDD2 associates with Golgi membranes during the cell cycle.
As mentioned above, prediction algorithms revealed that RHBDD2 possesses potential transmembrane domains, one of which, either the fifth or the sixth (depending on the algorithm used), contains a conserved glycine zipper motif. A rigorous analysis of sequence patterns has indicated that the GXXXG (GG4 motif) is the most highly biased sequence motif in naturally occurring transmembrane domains. Its glycine residues are usually separated by three large (Val, Leu, and Ile) or small (Gly, Ala, and Ser) residues in the transmembrane domains (30). Among glycine zipper motifs, the (G/A/S)XXXGXXXG and GXXXGXXX(G/S/T) are the most significant (31). Analysis of RHBDD2 sequences from different species revealed that they have glycine zipper motifs that are highly conserved (Fig. 4). Furthermore, it is interesting to note that the identified sequences contain the most common glycine zippers, which have two or more glycines and in which glycine occupies the central position (31), but that a separate set that includes those RHBDD2 sequences from human, chimpanzee, orangutan, mouse, rat, and other mammals exhibits extended glycine zipper motifs (Fig. 4, top panel); chicken and Xenopus tropicalis also contain these extended motifs. Of interest, some membrane proteins with homooligomeric bundle structures have extended glycine zipper motifs containing four glycine residues (GXXXGXXXGXXXG). It has been reported that mutation of one or more of these conserved glycine residues is in many cases deleterious to function (31).
FIGURE 4.
The glycine zipper motif sequences in the RHBDD2 protein from different species. The glycine zipper packing residues are highlighted in red, indicating the positions of the mutated Ser-185, Gly-189, Gly-193, and Gly-197, respectively.
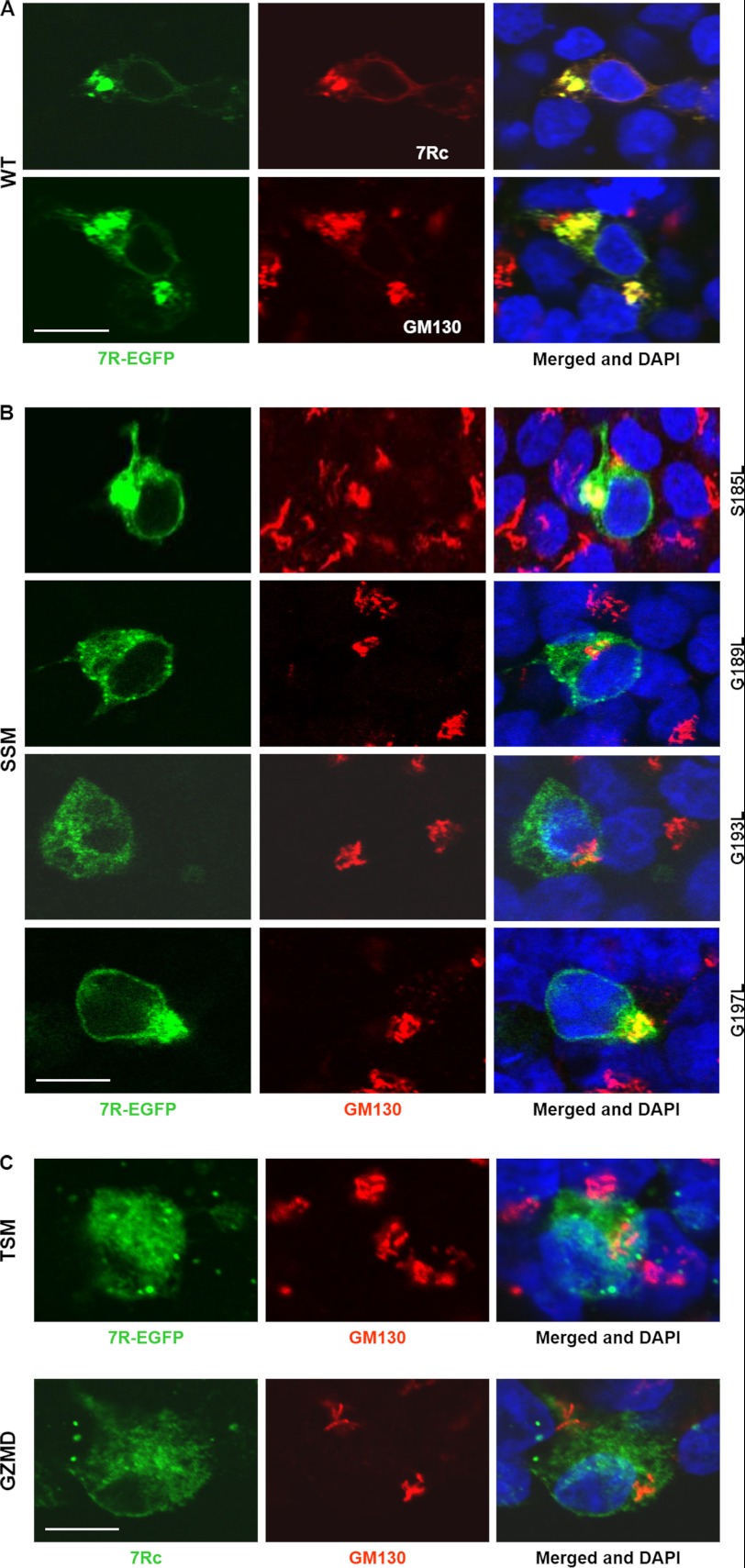
To further analyze whether the glycine zipper motif in RHBDD2 is important for the packing of the protein into membranes, we used the pEGFP-N3 vector (BD Biosciences Clontech) to generate several mutant RHBDD2 proteins fused to the N terminus of EGFP and allow their localization in vivo. The mutant proteins had Ser-185, Gly-189, Gly-193, and Gly-197 individually or collectively (Gly-189, Gly-193, and Gly-197) substituted with leucine, disrupting the glycine zipper packing interface. We also generated a fusion protein that had amino acid residues 189–197 deleted from the RHBDD2 protein. HEK293 cells were then transfected with pEGFP-N3 containing the Rhbdd2 or mutant cDNAs, and the expressed proteins were analyzed either by the green signal from EGFP or by immunocytochemistry using 7Rc and GM130 antibodies. Our results revealed that EGFP fused to RHBDD2 did not affect localization of the RHBDD2 protein (Fig. 5A) and that each of the Ser/Gly-to-Leu SSMs behaved differently (Fig. 5B). Changes in protein distribution as well as the lack of co-localization with GM130 were most notable for the G189L (Pearson's coefficient, 0.22 ± 0.01) and the G193L mutants (Pearson's coefficient, 0.31 ± 0.02). In contrast, the S185L and G197L mutants did not induce significant changes (Pearson's coefficients, 0.70 ± 0.04 and 0.69 ± 0.02, respectively). Surprisingly, RHBDD2 carrying the three Gly-to-Leu substitutions had a stronger co-localization with GM130 (Pearson's coefficient, 0.42 ± 0.01; Fig. 5C, TSM) than the G189L and G193L SSMs. The mutant resulting from the deletion of the amino acid residues 189–197 did not co-localize with GM130 (Pearson's coefficient, 0.14 ± 0.07; Fig. 5C, GZMD), but because we noticed that the green fluorescence excitation of this motif-deleted protein faded very fast, we used 7Rc to detect its localization (Fig. 5C, GZMD). In both cases, a dispersion of RHBDD2 in the cytoplasm was observed, indicating a dislodgment of RHBDD2 from the Golgi apparatus and suggesting that RHBDD2 might be anchored on this organelle. To support this idea, we next fractionated by ultracentrifugation the subcellular organelles/membrane vesicles of retinal tissue using a discontinuous sucrose gradient as described by Gangalum et al. (32). Western blots of the fractions were then incubated with 7Rc and GM130 antibodies. The results indicate that only the S2 fraction (the Golgi membrane fraction) contains an appreciable amount of RHBDD2 protein and that upon recentrifugation of S2 a significant amount of RHBDD2 can be found in the stacked Golgi fraction 3 (Fig. 6A, SGF3), further supporting that RHBDD2 is localized to the Golgi apparatus of retinal cells.
FIGURE 5.
Subcellular localization of RHBDD2 glycine zipper mutants. HEK293 cells were transfected with Rhbdd2-EGFP or Rhbdd2-EGFP mutants and stained with 7Rc or double stained with 7Rc and GM130. The cells were transfected with vector containing a wild-type (WT) Rhbdd2 cDNA together with green fluorescent protein used as a control for experiments and to monitor transfection efficiency (A); S185L, G189L, G193L, and G197L SSMs (B); a G189L, G193L, and G197L triple substitution mutant (TSM) (C); and the glycine zipper motif (amino acids 189–197) deletion mutant (GZMD) (D). Immunofluorescence imaging was performed to verify the expression and distribution of RHBDD2-EGFP mutants. Scale bars, 15 μm.
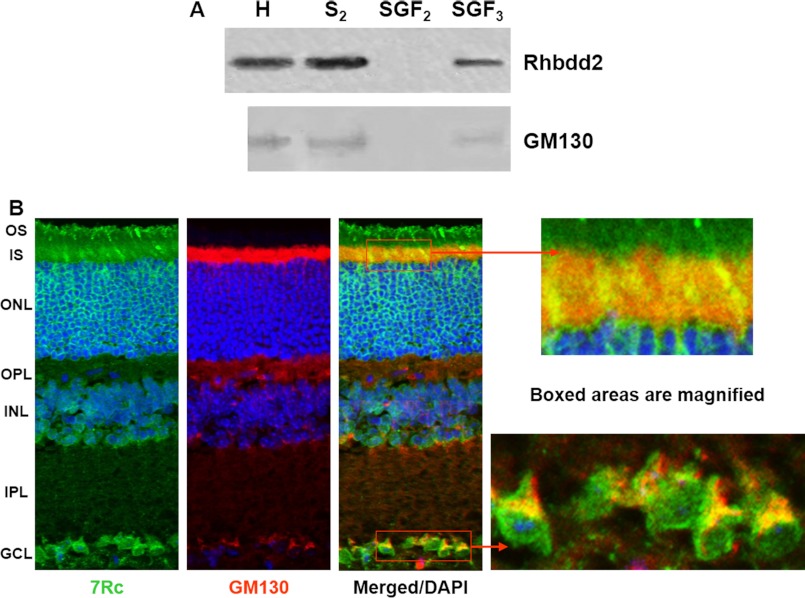
FIGURE 6.
Fractionation of mouse retinal protein extract and co-localization of RHBDD2 with a Golgi marker. A, the postnuclear supernatants were fractionated on discontinuous sucrose gradients. Western blots incubated with 7Rc and GM130 antibody show that the S2 fraction contains a substantial amount of RHBDD2 protein; upon recentrifugation of S2, a significant amount of RHBDD2 is found in the stacked Golgi fraction 3 (SGF3). H, homogenate. B, confocal microscopic images of adult mouse retinal sections stained with 7Rc, GM130 antibody, and DAPI. The merged image illustrates co-localization of these two proteins. The far right panel shows the magnified boxed areas. OS, outer segment; IS, inner segment; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer.
Immunohistochemistry experiments confirmed that RHBDD2 and GM130 co-localize in the retina. Double stained images obtained using antibodies against RHBDD2 and GM130 (Fig. 6B) show the co-localization of these proteins in the inner segment of photoreceptor cells (Pearson's coefficient, 0.48 ± 0.03), mainly in cone inner segments, as well as in the perinuclear region of ganglion cells (a stronger Pearson's coefficient, 0.67 ± 0.05, similar to what was seen in transfected HEK293 cells. In addition, very weak signals were observed on the inner nuclear layer edges bordering the outer plexiform layer and inner plexiform layer.
Specific Pattern of RHBDD2 Immunoreactivity in Developing and Adult Mouse Retina
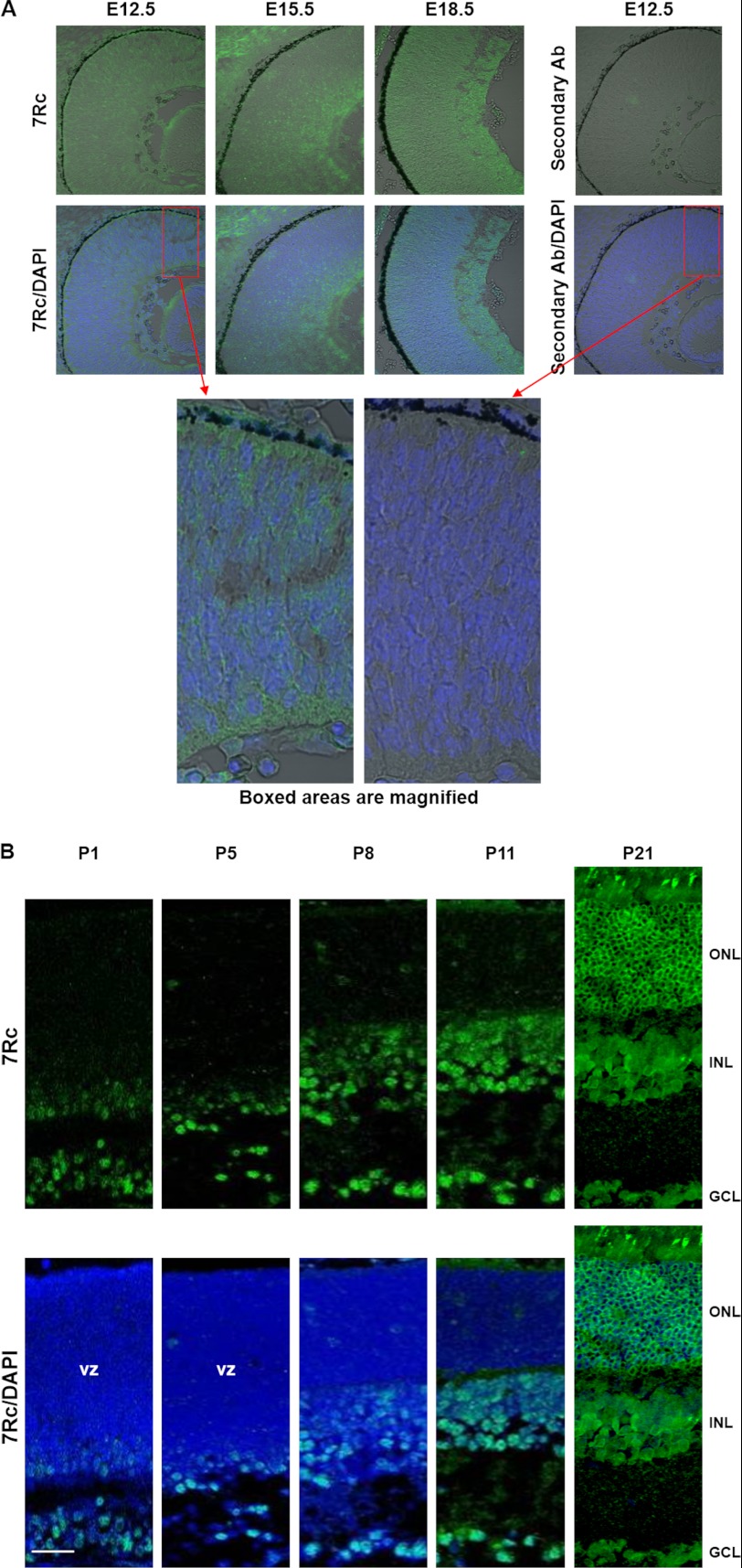
We used immunohistochemistry to follow the expression of RHBDD2 in mouse retina during development from E12.5 to adulthood (Figs. 7 and 8). Our results show that the distribution of RHBDD2 is more complex than our previous in situ hybridization data suggested (Fig. 1B). We detected a weak but clearly positive signal in E10.5 (data not shown), the youngest tissue in which we examined RHBDD2 expression, although it is possible that the RHBDD2-positive cells appear at an even younger age. A stronger signal is observed at E12.5, and the intensity of the staining increases in E15.5 and E18.5 retinas. At these embryonic stages, all cells are positive for RHBDD2 (Fig. 7A). The gradual, age-dependent appearance of the RHBDD2 protein in the layers of the retina begins at birth and continues throughout development (Fig. 7B). At postnatal day 1, a small number of positive cells are observed at the level of the ganglion cell layer (GCL) and in the inner edge of the ventricular zone of the retina. Between P1 and P5, the labeling of RHBDD2 shows a similar spatial pattern, but by P8 to P11, immunostaining has progressively increased and entirely occupies the GCL and inner nuclear layer. However, no significant RHBDD2 immunoreactivity is observed in the photoreceptor cell bodies of the outer nuclear layer (ONL). At P21, RHBDD2 is localized to all nuclear layers of the retina (Fig. 7B).
FIGURE 7.
RHBDD2 immunoreactivity during development of mouse retina. A, at E12.5, E15.5, and E18.5, RHBDD2 is ubiquitously expressed in all retinoblasts, and the intensity of the signal increases with age. The right panel is a section from an E12.5 eye stained only with the secondary antibody (Ab), demonstrating the specificity of the primary antibody signal. The bottom panels show the magnified boxed areas. B, at P1 and P5, RHBDD2 is expressed in some cells of the ventricular zone (vz) and in the GCL. At P8 and P11, immunostaining is clearly detected in the GCL, inner nuclear layer (INL), and in some cells of the ONL. By P21, RHBDD2 is localized to all retinal nuclear layers. In the ONL, RHBDD2 is mainly present around the nuclei of photoreceptors. Scale bar, 20 μm.
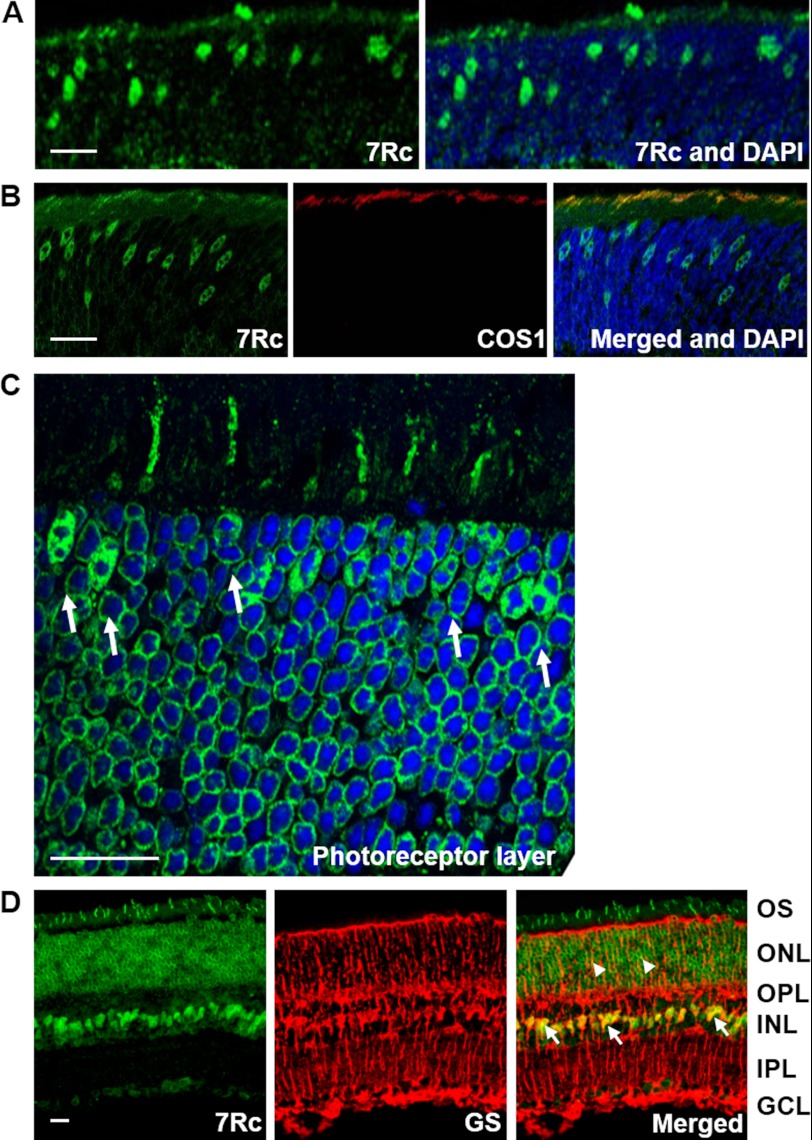
FIGURE 8.
Detection of RHBDD2 expression in young and adult mouse retina sections. A, section of a 12-day-old mouse retina stained with 7Rc-FITC and DAPI, which was used to stain the nuclei. The photoreceptor layer shows strongly stained cone cell nuclei migrating toward the outer edge of ONL and short outer segments. B, a 12-day-old mouse retinal section triple stained with 7Rc tagged with FITC, the cone marker COS1 tagged with Alexa Fluor 647, and DAPI. The merged image shows overlapping RHBDD2 and COS1 staining. C, a magnified image of the ONL from an adult retinal section immunolabeled with 7Rc. The arrows point to the larger, oval nuclei of cones that appear to contain several clumps of irregularly shaped heterochromatin. D, images of an adult retina showing RHBDD2 localization (green), glutamine synthetase (GS) localization (red), and their co-localization in the somas of many Müller glial cells (arrows) and some of their processes (arrowheads). Scale bars, 20 μm. INL, inner nuclear layer; OPL, outer plexiform layer; IPL, inner plexiform layer.
In the ONL, the RHBDD2 signal is primarily present in the cytoplasm of the cell soma adjacent to the plasma membrane of the photoreceptor cells, and it is different from that observed in embryonic and early retinal developmental stage cells where it is localized both in the nucleus and cytoplasm. The consistent perinuclear signal present in the later ages may suggest that compartmentalization is an essential process for the normal functioning of RHBDD2 in more mature retina. Interestingly, a small number of strongly stained RHBDD2 cells with very short outer segments (OS) and an irregular distribution first appear in the outer retina at P12 (Fig. 8A). To determine the identity of these RHBDD2-positive cells, we double labeled the photoreceptors of the mouse retina at P13–14 with both 7Rc and antibodies against S opsin, which is abundant in the cones of the mouse ventral retina, or M opsin, which is concentrated in the dorsal retina cones (33). Our results showed that RHBDD2 immunoreactivity consistently co-localized with S and M opsins in the OS of both types of cones (Fig. 8B, only M opsin is shown).
The characteristics of RHBDD2 staining in the ONL were seen more thoroughly with confocal microscopy images of adult retinal sections. Fig. 8C shows that all photoreceptor cells stained positive for RHBDD2; however, a small number of short OS were stained with 7Rc. These OS corresponded to cells that after differentiation and migration aligned near the outermost region of the ONL; their nuclei contained several clumps of irregularly shaped heterochromatin. The positional and morphological characteristics of these cells are hallmarks of mature cone photoreceptors (34, 35). As also seen in Fig. 8C, RHBDD2 immunostaining was restricted to the cytoplasm surrounding the nuclei, but we were unable to determine whether it was or was not on the nuclear membranes.
In addition to the photoreceptors, the staining pattern of RHBDD2 in adult retinas showed strong RHBDD2 expression in the GCL and inner nuclear layer. In the latter, a small number of cells were more intensively labeled than others. Double labeling with 7Rc and the Müller cell marker glutamine synthetase antibody revealed co-localization of RHBDD2 and glutamine synthetase in these same cells, identifying them as Müller cells (Fig. 8D, arrows). Very weak co-localization also was present on some radial processes of Müller cells in the ONL (arrowheads).
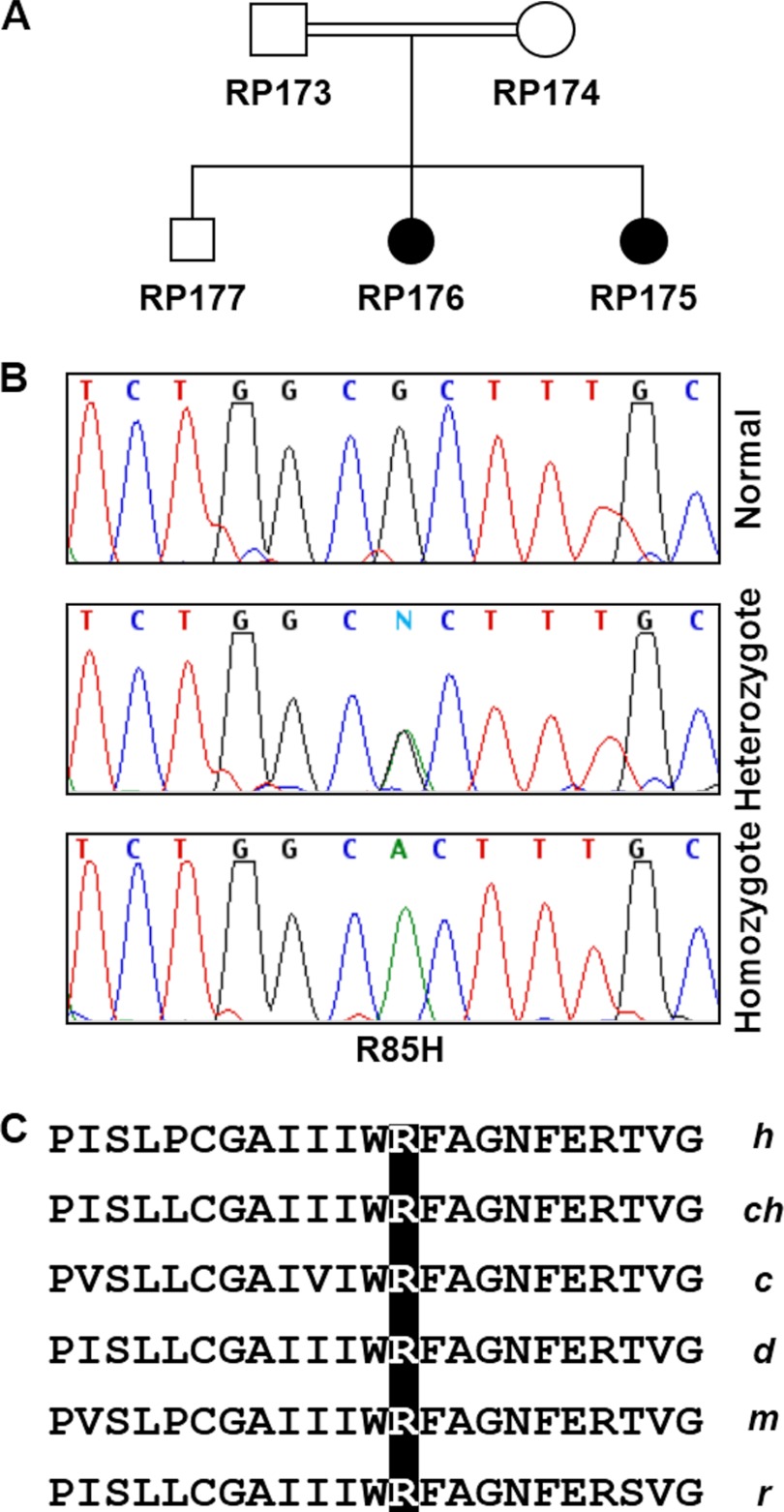
A Missense Mutation in RHBDD2 Co-segregates with Disease in a Family Affected with arRP
A systematic mutational analysis was initiated to determine whether there are disease-causing mutations in the human RHBDD2 gene. We screened the DNA of a small group of patients with different types of retinal degeneration by direct DNA sequence analysis of all exons and exon-intron boundaries and found variants of RHBDD2 in arRP patients. One of them, proband RP176 (Fig. 9A), had a G to A transition in exon 2 that changed arginine to histidine at codon 85 (Fig. 9B). This R85H allele co-segregated with the clinical phenotype in the patient's family. In addition, 95 control individuals (190 chromosomes) did not carry the R85H mutation. Arg-85 is located in the intracellular loop between the second and third predicted transmembrane domains of the RHBDD2 protein and is highly conserved among human, chimpanzee, cow, dog, mouse, rat, and many more species (Fig. 9C). Interestingly, in another unrelated individual, two different RHBDD2 transcripts were identified: a wild-type mRNA and a second transcript containing a 7-nucleotide deletion in exon 2 of RHBDD2 that leads to a frameshift and the creation of a premature stop codon. This individual also had a common single nucleotide polymorphism (SNP) at the −13 position upstream of exon 2. Unfortunately, no other family members are available for testing to determine whether the deletion is linked to any disease phenotype. We excluded the known disease-causing genes PDE6A, PDE6B, RPE65, ABCA4, and TULP1 from all the families studied using linkage analysis. All patients' DNAs were also screened for the CRX, RDS, NR2E3, and RHO genes, and no mutations were found.
FIGURE 9.
R85H mutation in the human RHBDD2 gene. A, pedigree of an arRP family carrying the R85H mutation in the RHBDD2 gene. Filled circles represent the affected members of the family. B, sequences of DNA fragments containing the site of mutation in the RHBDD2 gene. C, Arg-85 is conserved in the RHBDD2 sequence among different species of mammals. h, human; ch, chimpanzee; c, cow; d, dog; m, mouse; r, rat.
DISCUSSION
Herein, we identified a cDNA encoding RHBDD2, a novel rhomboid-like protein, in a screen for mRNAs present in cone photoreceptors. However, we found Rhbdd2 transcripts in all major mouse tissues. The expressed RHBDD2 is an intramembrane protein with a localization restricted to the Golgi apparatus in transfected HEK293 cells. Retina is the only tissue where we have detected two forms of the RHBDD2 protein, RHBDD2L and RHBDD2H, which most likely is an RHBDD2L trimer. Inspection of databases indicates that RHBDD2 is conserved among species.
This study is the first to examine the developmental profile and cellular localization of RHBDD2 in mouse retina. We found that RHBDD2 is expressed in early life (with immunoreactivity observed at E10.5) and becomes widespread and differentially distributed after all the retinal layers have formed. The lowest levels of RHBDD2 were seen at the beginning of postnatal life, and the maximum level was observed at P14.
At P1, RHBDD2 immunoreactivity is strongest in the GCL, and it spreads during development from the inner to the outer retina. In the ONL, the first cells stained by 7Rc are detected at P12 and are somewhat scattered. By P21, RHBDD2 is all over the retina, and it appears in many neuronal and glial Müller cells. Quantification of the expression of the two different forms of RHBDD2 indicated that RHBDD2L minimally changed during development until P14 and then decreased as the age of the mouse progressed, whereas RHBDD2H increased from P5 to P30 and remained close to this level thereafter.
One of the interesting issues concerning RHBDD2 expression is the appearance of homomultimers of this protein. Our results provide evidence that RHBDD2L exists as a monomer in the inner retina neurons and the soma of photoreceptors, whereas RHBDD2H seems to be found exclusively as a polymer in cone OS. Indeed, the levels of RHBDD2H increase precisely at the time in development when the outer segments are elongating. We consider that RHBDD2H is a trimer of RHBDD2L because of its molecular mass and because it is recognized by 7Rc, a specific antibody for both forms of the RHBDD2 protein. RHBDD2H detection is not appreciably altered by increasing mercaptoethanol/DTT, treating with different concentrations of SDS before electrophoresis, or boiling the samples before loading them on the gel. RHBDD2H is present in S and M cone outer segments. It is most likely that the RHBDD2L monomer is conserved between species.
Because the biochemical functions of RHBDD2 have not been determined and establishing them will require a more complete understanding of the protein, currently, we can only speculate that RHBDD2H may be involved in the growth and maturation of the cone OS, whereas RHBDD2L may be an active participant in the early development of retinal cells. The generation of RHBDD2 knock-out mice will provide evidence for the role of RHBDD2 in these processes.
As shown in Figs. 3 and 5, the recombinant RHBDD2 localized to the Golgi apparatus of HEK293 cells. Furthermore, in retinal sections, co-localization of RHBDD2 and the Golgi marker GM130 is clearly seen at the inner segment of photoreceptors and perinuclear region of ganglion cells (Fig. 6B), which is an obvious indication of the existence of RHBDD2 in the Golgi apparatus. This organelle is considered to be a distribution and shipment center for proteins and lipids inside the cell as well as for their export out of the cell. In recent years, discoveries have indicated that the Golgi complex can also be considered as a center of operations where cargo sorting/processing, basic metabolism, signaling, and cell fate decisional processes converge (36). Interestingly, cells overexpressing RHBDD2 had a compacted Golgi similar to that observed in non-transfected cells with normal reticular morphology. While performing transfection experiments with constructs containing different RHBDD2 glycine zipper mutations, we noticed that for some expressed mutant proteins the staining pattern of RHBDD2 was altered, but the Golgi remained intact. Because it is known that glycine zipper motif structures are directly involved in helix interactions, the existence of the GXXXG sequence in RHBDD2 suggests that this motif may not only be involved in the high affinity association of transmembrane helices but also in the association of RHBDD2 with the Golgi apparatus. In addition, our results suggest that the glycine zipper motif may play an important role in the oligomerization of RHBDD2 in OS of photoreceptors. The observation that RHBDD2 localization to the Golgi complex is glycine zipper motif-dependent even though there are no links between the function of RHBDD2 and the Golgi apparatus generates the appealing hypothesis that the formation of the trimer in outer segments of adult retina also relies on the glycine zipper motif.
Our studies also have provided evidence supporting the role of the RHBDD2 gene, which is localized on chromosome 7q11, in the pathogenesis of retinitis pigmentosa, an inherited retinal disease that results in the loss of photoreceptors and is characterized by pigment deposits predominantly in the peripheral retina. To date, 43 genes and 50 loci have been identified for autosomal dominant, autosomal recessive, and X-linked forms of non-syndromic retinitis pigmentosa (RetNet, The Retinal Information Network). We found a homozygous mutation in the RHBDD2 gene, R85H, that co-segregates with disease in a family affected with arRP but that does not exist in 95 controls. No other gene of the several that we investigated showed any mutation in the affected members of this family. Thus, it is possible that the RHBDD2 gene is a new disease-causing gene and that 7q11 is a new locus for arRP. Several studies suggest that arginine residues may form salt bridges with acidic residues to stabilize protein structure, bind cofactors, or interact with DNA (37–39). The observed substitution of His for Arg at position 85 may thus disrupt the structure of the RHBDD2 protein.
Recently, Abba et al. (40) determined that the RHBDD2 mRNA and its expressed protein are significantly elevated in breast carcinomas as compared with normal breast tissue samples or benign breast lesions with overexpression predominantly observed in advanced stages and that silencing of RHBDD2 expression results in a decrease of cell proliferation. The authors showed a strong association between high RHBDD2 expression and decreased overall survival, relapse-free survival, and metastasis-free intervals in patients with primary estrogen receptor-negative breast carcinomas. They suggested that RHBDD2 overexpression behaves as an indicator of poor prognosis and may play a role in facilitating breast cancer progression. Also, they recognized two RHBDD2 alternatively spliced mRNA isoforms expressed in breast cancer cell lines (40). We have found that these two isoforms are also expressed in normal retinal tissue samples. In addition, we identified a third isoform of RHBDD2 from human retinal samples, the rhomboid domain-containing 2, transcript variant 3.
Although Abba et al. (40) present solid mRNA data indicating strong correlation between expression of RHBDD2 transcripts and breast carcinomas, their Western blot and immunohistochemistry results are contrary to our results on RHBDD2. First, the polyclonal antibody that these authors used was raised against peptides that were synthesized based on the RHBDD2 protein sequence (NCBI Reference Sequence NP_065735) corresponding to residues 30–43, 253–266, and 393–406. However, the record for NP_065735 has been permanently removed from the database because this sequence is a nonsense-mediated mRNA decay candidate. Second, these three peptide sequences are encoded by the ORF of human NPD007 mRNA (GenBank accession number AF226732) but are not found in the RHBDD2 protein sequence. NPD007 mRNA codes for a protein with an N terminus 45 amino acids longer than RHBDD2. Peptide 30–43 is located in those 45 amino acids. Also, the NPD007 sequence is missing two nucleotides after position 724 (G and C), which leads to a reading frame change and the creation of a new protein. The 253–266 and 393–406 peptides are located in the diverging sequence downstream of the two missing nucleotides. This is why these peptides are not present in the RHBDD2 sequence. Third, from the Western blot analysis of normal and breast cancer cell lines using their polyclonal antibody, the authors identified a 47-kDa product that they assume is encoded by isoform1 as well as a smaller protein of ∼40 kDa, a product of isoform2. However, the sequences presented by the authors in Fig. 3D of their study (40) contain an ORF encoding 223 amino acids for isoform1 and 364 amino acids for isoform2. Consequently, isoform1 would code for a much smaller protein than that encoded by isoform2. Therefore, it is not clear what proteins the authors are detecting with the polyclonal antibody that they generated. These proteins are definitely different from RHBDD2.
Very recently, a study has been published describing the possible role of RHBDD2 in colorectal cancer progression. The authors indicate that expression of RHBDD2 significantly increases in advanced stages of colorectal cancer. Also, they found a significant increase of RHBDD2 mRNA and protein after treatment with the chemotherapy agent 5-fluorouracil (41).
In conclusion, we have characterized RHBDD2, a ubiquitously expressed transmembrane rhomboid-like protein associated with the Golgi apparatus that exists as a monomer in all retinal cells and as a trimer only in retinal photoreceptor outer segments, mainly in cones. Our genetic findings indicate that a mutation in the RHBDD2 gene may lead to arRP. The association of RHBDD2 abnormalities with diseases like neurodegenerative retinitis pigmentosa and cancer is intriguing. The unknown function of the RHBDD2 protein in mammals together with the undiscovered molecular pathways in which RHBDD2 may participate in human disorders further substantiates future studies to determine the potential role of RHBDD2 in normal cells and tissues.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 EY08285 (to D. B. F.), RO1 ES012482 (to D. A. F.), P30 EY000331 (to the Jules Stein Eye Institute), and T32 EY07024 and P30 EY07551 (to the University of Houston College of Optometry). This work was also supported by Hope for Vision and Oppenheimer Foundation grants (to N. B. A.).
- iRhom
- inactive rhomboid
- E
- embryonic day
- P
- postnatal day
- EGFP
- enhanced green fluorescent protein
- arRP
- autosomal recessive retinitis pigmentosa
- OCT
- optimal cutting temperature medium
- DIG
- digoxigenin
- BFA
- brefeldin A
- GCL
- ganglion cell layer
- ONL
- outer nuclear layer
- OS
- outer segments.
REFERENCES
- 1. Brown M. S., Ye J., Rawson R. B., Goldstein J. L. (2000) Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100, 391–398 [DOI] [PubMed] [Google Scholar]
- 2. Urban S., Freeman M. (2003) Substrate specificity of rhomboid intramembrane proteases is governed by helix-breaking residues in the substrate transmembrane domain. Mol. Cell 11, 1425–1434 [DOI] [PubMed] [Google Scholar]
- 3. Weihofen A., Martoglio B. (2003) Intramembrane-cleaving proteases: controlled liberation of proteins and bioactive peptides. Trends Cell Biol. 13, 71–78 [DOI] [PubMed] [Google Scholar]
- 4. Wolfe M. S., Kopan R. (2004) Intramembrane proteolysis: theme and variations. Science 305, 1119–1123 [DOI] [PubMed] [Google Scholar]
- 5. Ha Y. (2009) Structure and mechanism of intramembrane protease. Semin. Cell Dev. Biol. 20, 240–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Urban S. (2009) Making the cut: central roles of intramembrane proteolysis in pathogenic microorganisms. Nat. Rev. Microbiol. 7, 411–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mayer U., Nüsslein-Volhard C. (1988) A group of genes required for pattern formation in the ventral ectoderm of the Drosophila embryo. Genes Dev. 2, 1496–1511 [DOI] [PubMed] [Google Scholar]
- 8. Bier E., Jan L. Y., Jan Y. N. (1990) Rhomboid, a gene required for dorsoventral axis establishment and peripheral nervous system development in Drosophila melanogaster. Genes Dev. 4, 190–203 [DOI] [PubMed] [Google Scholar]
- 9. Wasserman J. D., Urban S., Freeman M. (2000) A family of rhomboid-like genes: Drosophila rhomboid-1 and roughoid/rhomboid-3 cooperate to activate EGF receptor signaling. Genes Dev. 14, 1651–1663 [PMC free article] [PubMed] [Google Scholar]
- 10. Lee J. R., Urban S., Garvey C. F., Freeman M. (2001) Regulated intracellular ligand transport and proteolysis control EGF signal activation in Drosophila. Cell 107, 161–171 [DOI] [PubMed] [Google Scholar]
- 11. Urban S., Wolfe M. S. (2005) Reconstitution of intramembrane proteolysis in vitro reveals that pure rhomboid is sufficient for catalysis and specificity. Proc. Natl. Acad. Sci. U.S.A 102, 1883–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maegawa S., Ito K., Akiyama Y. (2005) Proteolytic action of GlpG, a rhomboid protease in the Escherichia coli cytoplasmic membrane. Biochemistry 44, 13543–13552 [DOI] [PubMed] [Google Scholar]
- 13. Lemberg M. K., Freeman M. (2007) Functional and evolutionary implications of enhanced genomic analysis of rhomboid intramembrane proteases. Genome Res. 17, 1634–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Freeman M. (2009) Rhomboids: 7 years of a new protease family. Semin. Cell Dev. Biol. 20, 231–239 [DOI] [PubMed] [Google Scholar]
- 15. Srinivasan P., Coppens I., Jacobs-Lorena M. (2009) Distinct roles of Plasmodium rhomboid 1 in parasite development and malaria pathogenesis. PLoS Pathog. 5, e1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Morahan B. J., Sallmann G. B., Huestis R., Dubljevic V., Waller K. L. (2009) Plasmodium falciparum: genetic and immunogenic characterisation of the rhoptry neck protein PfRON4. Exp. Parasitol. 122, 280–288 [DOI] [PubMed] [Google Scholar]
- 17. Cowman A. F., Crabb B. S. (2006) Invasion of red blood cells by malaria parasites. Cell 124, 755–766 [DOI] [PubMed] [Google Scholar]
- 18. Whitworth A. J., Lee J. R., Ho V. M., Flick R., Chowdhury R., McQuibban G. A. (2008) Rhomboid-7 and HtrA2/Omi act in a common pathway with the Parkinson's disease factors Pink1 and Parkin. Dis. Model. Mech. 1, 168–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Walder K., Kerr-Bayles L., Civitarese A., Jowett J., Curran J., Elliott K, Trevaskis J, Bishara N., Zimmet P., Mandarino L., Ravussin E., Blangero J., Kissebah A., Collier G. R. (2005) The mitochondrial rhomboid protease PSARL is a new candidate gene for type 2 diabetes. Diabetologia 48, 459–468 [DOI] [PubMed] [Google Scholar]
- 20. Adrain C., Zettl M., Christova Y., Taylor N., Freeman M. (2012) Tumor necrosis factor signaling requires iRhom2 to promote trafficking and activation of TACE. Science 335, 225–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McIlwain D. R., Lang P. A., Maretzky T., Hamada K., Ohishi K., Maney S. K., Berger T., Murthy A., Duncan G., Xu H. C., Lang K. S., Häussinger D., Wakeham A., Itie-Youten A., Khokha R., Ohashi P. S., Blobel C. P., Mak T. W. (2012) iRhom2 regulation of TACE controls TNF-mediated protection against Listeria and responses to LPS. Science 335, 229–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lerner L. E., Peng G. H., Gribanova Y. E., Chen S., Farber D. B. (2005) Sp4 is expressed in retinal neurons, activates transcription of photoreceptor-specific genes, and synergizes with Crx. J. Biol. Chem. 280, 20642–20650 [DOI] [PubMed] [Google Scholar]
- 23. Saghizadeh M., Gribanova Y., Akhmedov N. B., Farber D. B. (2011) ZBED4, a cone and Müller cell protein in human retina, has a different cellular expression in mouse. Mol. Vis. 17, 2011–2018 [PMC free article] [PubMed] [Google Scholar]
- 24. Johnson J. E., Jr., Perkins G. A., Giddabasappa A., Chaney S., Xiao W., White A. D., Brown J. M., Waggoner J., Ellisman M. H., Fox D. A. (2007) Spatiotemporal regulation of ATP and Ca2+ dynamics in vertebrate rod and cone photoreceptor ribbon synapses. Mol. Vis. 13, 887–919 [PMC free article] [PubMed] [Google Scholar]
- 25. Giddabasappa A., Hamilton W. R., Chaney S., Xiao W., Johnson J. E. (2011) Low-level gestational lead exposure increases retinal progenitor cell proliferation and rod photoreceptor and bipolar cell neurogenesis in mice. Environ. Health Perspect. 119, 71–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kerov V. S., Natochin M., Artemyev N. O. (2005) Interaction of transducin-α with LGN, a G-protein modulator expressed in photoreceptor cells. Mol. Cell. Neurosci. 28, 485–495 [DOI] [PubMed] [Google Scholar]
- 27. Saghizadeh M., Akhmedov N. B., Yamashita C. K., Gribanova Y., Theendakara V., Mendoza E., Nelson S. F., Ljubimov A. V., Farber D. B. (2009) ZBED4, a BED-type zinc-finger protein in the cones of the human retina. Invest. Ophthalmol. Vis. Sci. 50, 3580–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chiu C. F., Ghanekar Y., Frost L., Diao A., Morrison D., McKenzie E., Lowe M. (2008) ZFPL1, a novel ring finger protein required for cis-Golgi integrity and efficient ER-to-Golgi transport. EMBO J. 27, 934–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gough L. L., Fan J., Chu S., Winnick S., Beck K. A. (2003) Golgi localization of Syne-1. Mol. Biol. Cell 14, 2410–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Senes A., Gerstein M., Engelman D. M. (2000) Statistical analysis of amino acid patterns in transmembrane helices: the GxxxG motif occurs frequently and in association with β-branched residues at neighboring positions. J. Mol. Biol. 296, 921–936 [DOI] [PubMed] [Google Scholar]
- 31. Kim S., Jeon T. J., Oberai A., Yang D., Schmidt J. J., Bowie J. U. (2005) Transmembrane glycine zippers: physiological and pathological roles in membrane proteins. Proc. Natl. Acad. Sci. U.S.A. 102, 14278–14283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gangalum R. K., Schibler M. J., Bhat S. P. (2004) Small heat shock protein αB-crystallin is part of cell cycle-dependent Golgi reorganization. J. Biol. Chem. 279, 43374–43377 [DOI] [PubMed] [Google Scholar]
- 33. Szél A., Röhlich P., Caffé A. R., Juliusson B., Aguirre G., Van Veen T. (1992) Unique topographic separation of 2 spectral classes of cones in the mouse retina. J. Comp. Neurol. 325, 327–342 [DOI] [PubMed] [Google Scholar]
- 34. Carter-Dawson L. D., LaVail M. M. (1979) Rods and cones in the mouse retina. I. Structural analysis using light and electron microscopy. J. Comp. Neurol. 188, 245–262 [DOI] [PubMed] [Google Scholar]
- 35. Perkins G. A., Ellisman M. H., Fox D. A. (2003) Three-dimensional analysis of mouse rod and cone mitochondrial cristae architecture: bioenergetic and functional implications. Mol. Vis. 9, 60–73 [PubMed] [Google Scholar]
- 36. Wilson C., Venditti R., Rega L. R., Colanzi A., D'Angelo G., De Matteis M. A. (2011) The Golgi apparatus: an organelle with multiple complex functions. Biochem. J. 433, 1–9 [DOI] [PubMed] [Google Scholar]
- 37. Sundaralingam M., Drendel W., Greaser M. (1985) Stabilization of the central helix of troponin C by intrahelical salt bridges between charged amino acid side chains. Proc. Natl. Acad. Sci. U.S.A. 82, 7944–7947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Llewellyn D. H., Whatley S., Elder G. H. (1993) Acute intermittent porphyria caused by an arginine to histidine substitution (R26H) in the cofactor-binding cleft of porphobilinogen deaminase. Hum. Mol. Genet. 2, 1315–1316 [DOI] [PubMed] [Google Scholar]
- 39. Frigyes D., Alber F., Pongor S., Carloni P. (2001) Arginine-phosphate salt bridges in protein-DNA complex: a Car-Parrinello study. J. Mol. Struct. (Theochem) 574, 39–45 [Google Scholar]
- 40. Abba M. C., Lacunza E., Nunez M. I., Colussi A., Isla-Larrain M., Segal-Eiras A., Croce M. V., Aldaz C. M. (2009) Rhomboid domain containing 2 (RHBDD2): a novel cancer-related gene over-expressed in breast cancer. Biochim. Biophys. Acta 1792, 988–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lacunza E., Canzoneri R., Rabassa M. E., Zwenger A., Segal-Eiras A., Croce M. V., Abba M. C. (2012) RHBDD2: a 5-fluorouracil responsive gene overexpressed in the advanced stages of colorectal cancer. Tumour Biol. 33, 2393–2399 [DOI] [PubMed] [Google Scholar]