Background: Small GTPase Rab17 regulates dendritic morphogenesis of hippocampal neurons, but its activation mechanism is completely unknown.
Results: Expression of Rabex-5 in mouse hippocampal neurons promoted dendritic localization of Rab17, and Rabex-5 knockdown resulted in inhibition of neurite morphogenesis.
Conclusion: Rabex-5 regulates neurite morphogenesis by activating Rab5 and Rab17.
Significance: Our findings revealed a crucial role of Rabex-5 and its targets in neuronal development.
Keywords: Guanine Nucleotide Exchange Factor (GEF), Membrane Trafficking, Neurobiology, Rab, Small GTPases
Abstract
Small GTPase Rab17 has recently been shown to regulate dendritic morphogenesis of mouse hippocampal neurons; however, the exact molecular mechanism of Rab17-mediated dendritogenesis remained to be determined, because no guanine nucleotide exchange factor (GEF) for Rab17 had been identified. In this study we screened for the Rab17-GEF by performing yeast two-hybrid assays with a GDP-locked Rab17 mutant as bait and found that Rabex-5 and ALS2, both of which were originally described as Rab5-GEFs, interact with Rab17. We also found that expression of Rabex-5, but not of ALS2, promotes translocation of Rab17 from the cell body to the dendrites of developing mouse hippocampal neurons. The shRNA-mediated knockdown of Rabex-5 or its known downstream target Rab5 in hippocampal neurons inhibited morphogenesis of both axons and dendrites, whereas knockdown of Rab17 affected dendrite morphogenesis alone. Based on these findings, we propose that Rabex-5 regulates neurite morphogenesis of hippocampal neurons by activating at least two downstream targets, Rab5, which is localized in both axons and dendrites, and Rab17, which is localized in dendrites alone.
Introduction
Rab-type small GTPases are conserved membrane trafficking proteins in all eukaryotes, and they mediate various steps in membrane trafficking, including vesicle budding, vesicle movement, vesicle docking to specific membranes, and vesicle fusion (1, 2). Rabs function as a molecular switch by cycling between two nucleotide-bound states, a GDP-bound inactive state and a GTP-bound active state. In general, Rabs are activated by specific guanine nucleotide exchange factors (GEFs),3 which promote the release of GDP from Rab and binding of GTP to Rab (3), and the activated Rabs are then inactivated by GTPase-activating proteins (GAPs) or spontaneously inactivated by their intrinsic GTPase activity, either of which terminates the cycle (3, 4). Therefore, the identification and characterization of these Rab regulators, especially of GEFs, is crucial to understanding the spatiotemporal regulation of Rab GTPase activation.
A variety of putative Rab-GEFs have been identified during the past decade, and they have largely been classified into four groups based on the similarities between their putative Rab-GEF domains or their structures (3), i.e. DENN (differentially expressed in normal and neoplastic cells) domains (5), Sec2 domains (6, 7), VPS9 (vacuolar protein sorting 9) domains (8), and multimeric GEFs, including a TRAPP complex (9), Hps1-Hps4 (10), and Mon1-Ccz1 (11). The DENN domain-containing proteins constitute the largest group of these putative Rab-GEFs, and the in vitro targets of most of them have recently been identified. For example, DENN/MADD/Rab3-GEP exhibits GEF activity toward Rab3 and Rab27 (12, 13), DENND1/connecdenn/RME-4 toward Rab35 (14, 15), DENND2 toward Rab9 (16), and DENND4 toward Rab10 (16, 17). One of the other three groups, the VPS9 domain-containing proteins, activate Rab5/Ypt51p subfamily GTPases (8, 18), whereas the Sec2 domain-containing proteins, Sec2p and Rabin8, activate Sec4p and Rab8, respectively (6, 7). Despite the increasing numbers of Rab-GEFs that have been identified, specific GEFs for about half of the mammalian Rabs remain unknown.
Rab17 is one of the Rab isoforms whose specific and physiological GEFs have not been identified. Rab17 was originally described as an epithelial cell-specific Rab that regulates polarized trafficking (19, 20), but the results of our previous study indicated that Rab17 is also expressed in mouse brain and that it regulates dendrite morphogenesis and postsynaptic development of hippocampal neurons (21). Because Rab17 is the only dendrite-specific Rab protein, unraveling the activation mechanism of Rab17 is crucial to a better understanding the molecular mechanism of dendrite outgrowth and branching. In this study we screened for Rab17-GEFs by using a GDP-locked Rab17 mutant as bait and identified Rabex-5 (22) and ALS2 (amyotrophic lateral sclerosis 2) (23), both of which were originally described as Rab5-GEFs, as putative Rab17-GEFs. We found that Rabex-5, but not ALS2, is required for stage-dependent movement of Rab17 protein from the cell body to the dendrites of mouse hippocampal neurons. We also showed that knockdown of Rabex-5 inhibited morphogenesis of both axons and dendrites in developing neurons. We discuss the possible functions of Rabex-5 in neurite morphogenesis of hippocampal neurons based on our findings.
EXPERIMENTAL PROCEDURES
Antibodies
The following antibodies used in this study were obtained commercially: anti-c-Myc (9E10) mouse monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA), anti-actin mouse monoclonal antibody (ABM, Richmond, Canada), anti-neurofilament-H mouse monoclonal antibody (American Research Products, Belmont, MA), anti-MAP2 chick polyclonal antibody (Millipore Corp., Billerica, MA), anti-GFP rabbit polyclonal antibody (MBL, Nagoya, Japan), horseradish peroxidase (HRP)-conjugated anti-FLAG tag (M2) mouse monoclonal antibody and M2-conjugated agarose beads (Sigma), HRP-conjugated anti-T7 tag antibody (Novagen, Darmstadt, Germany), and Alexa-Fluor 488/594/633-conjugated anti-mouse/rabbit/chick IgG goat antibody (Invitrogen). Anti-Rab17 rabbit polyclonal antibody and anti-GFP guinea pig polyclonal antibody were prepared as described previously (21). Anti-Rabex-5 rabbit polyclonal antibody was raised against glutathione S-transferase (GST)-Rabex5-C (amino acid residues (aa) 208–491) and affinity purified by exposure to antigen-bound Affi-Gel 10 beads (Bio-Rad) as described previously (24).
Plasmid Construction
cDNAs encoding the human DENN domain-containing proteins, mouse VPS9 domain-containing proteins, and mouse Sec2 domain-containing proteins were amplified from Marathon-Ready adult human or mouse brain and/or testis cDNA (Clontech-Takara Bio Inc., Shiga, Japan) by performing PCR with the following specific oligonucleotides as described previously (25): DENN/MADD/Rab3-GEP (aa 1–570), DENND1A (aa 1–380), DENND1B (aa 5–410), DENND1C (aa 1–424), DENND2A (aa 551–1009), DENND2B (aa 669–1237), DENND2C (aa 392–881), DENN2D (aa 1–471), DENND3 (aa 24–435), DENND4A (aa 139–660), DENND4B (aa 99–650), DENND4C (aa 1–420), DENND5A (aa 1–599), DENND5B (aa 18–600), DENND6A (aa 1–605), DENND6B (aa 1–585), Sbf1/MTMR5 (aa 1–467), Sbf2/MTMR13 (aa 1–585), Varp (aa 1–450), Rabex-5 (aa 208–491), Gapex-5 (aa 1281–1437), Rin1 (aa 446–763), Rin2 (aa 593–858), Rin3 (aa 713–980), ALS2 (aa 1485–1651), ALS2-cl (aa 764–947), RINL (aa 314–563), Rabin3 (aa 1–428), and GRAB (aa 1–384). The sequences of the oligonucleotides used are available from the authors on request. Purified PCR products were directly inserted into the pGEM-T Easy vector (Promega, Madison, WI) and verified with an automated sequencer. The cDNAs were excised from the pGEM-T Easy vector with appropriate restriction enzymes and then subcloned into the pGAD-C1 vector (26). pGBD-C1 vector harboring constitutive negative (CN or GDP-fixed) or constitutive active (CA, GTP-fixed) mutants of Rab1–43 lacking the C-terminal geranylgeranylation site was prepared as described previously (27). cDNAs encoding mouse Rabex-5 (aa 1–491), Rabex-5-C (aa 208–491), ALS2 (aa 1–1651), and ALS2-C (aa 1485–1651) were similarly produced by conventional PCR techniques (see Fig. 1D). A Rabex-5 mutant carrying an Asp-to-Ala mutation at amino acid positions 313 (D313A) and an ALS2 mutant (D1593A) were prepared by using conventional PCR techniques and mutagenic oligonucleotides as described previously (28) (see Fig. 1D). The cDNA of each of the Rabex-5 (or ALS2) constructs was subcloned into the pGAD-C1 vector, pEF-T7 vector (25), pEF-Myc vector (21), pEGFP-C1 vector (Clontech-Takara Bio Inc.), and/or pGEX-4T-3 vector (GE Healthcare). The pSilencer-EGFP vector (21) or pSilencer 2.1-U6 neo (Ambion, Austin, TX) encoding a mouse Rabex-5-shRNA (short hairpin RNA) (19-base target site 5′-GTTCAAGACATTGTTGAGA-3′), Rab5A-shRNA (19-base target site 5′-GCACAGTCCTATGCAGATG-3′), Rab5B/C-shRNA (19-base target site 5′-GTTTGAGATCTGGGACACA-3′), Rab21-shRNA (19-base target site 5′-TTTACTACCGAGATTCGAA-3′), and ALS2-shRNA (21-base target site 5′-GAACTCTTGCAAGATTTGTCA-3′) was constructed as described previously (21).
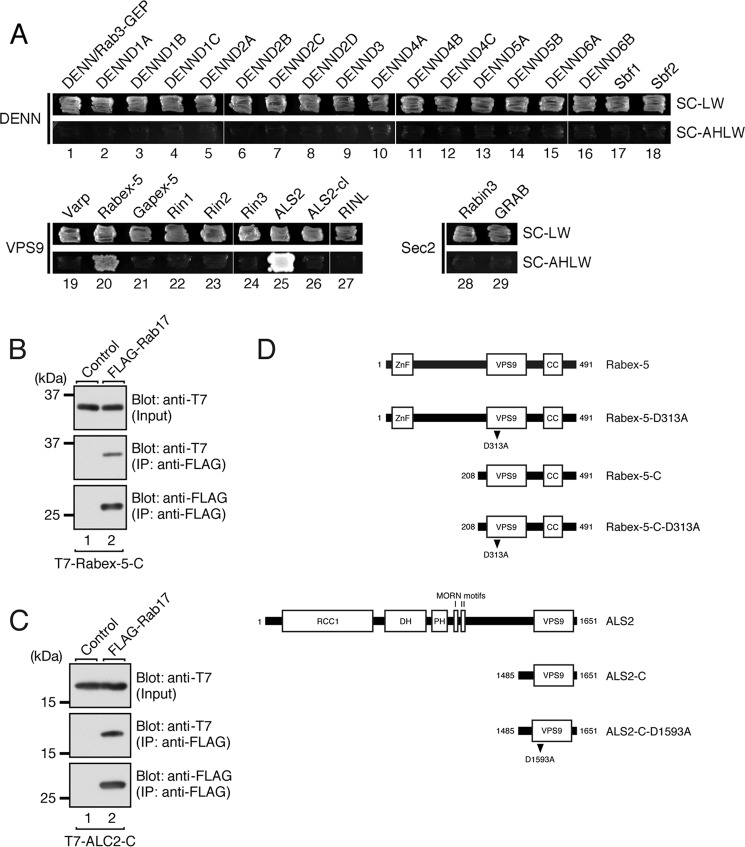
FIGURE 1.
Identification of Rabex-5 and ALS2 as putative Rab17-GEFs. A, screening of Rab-GEFs by yeast two-hybrid assays with the GDP-locked CN form of Rab17 as bait is shown. Yeast cells containing pGBD plasmid expressing Rab17(CN) and pGAD-C1 plasmid expressing each of the DENN, VPS9, or Sec2 domain-containing proteins were streaked on SC-LW (top panels) and SC-AHLW (selection medium; bottom panels) and incubated at 30 °C for 1 day and for 1 week, respectively. Note that only two of the 29 putative Rab-GEFs tested, Rabex-5 (lane 20) and ALS2 (lane 25), bound Rab17(CN). B and C, Rabex-5 (B) and ALS2 (C) interact with Rab17 in the presence of 1 mm GDP. The T7-tagged VPS9 domain of Rabex-5 (Rabex-5-C) or VPS9 domain of ALS2 (ALS2-C) and FLAG-tagged Rab17 were co-expressed in COS-7 cells, and they were analyzed for associations by coimmunoprecipitation assays with anti-FLAG tag antibody-conjugated agarose beads. Input means volume of the reaction mixture used for immunoprecipitation (top panels). Immunoprecipitated (IP) FLAG-Rab17 (bottom panels) and coimmunoprecipitated T7-Rabex-5-C or T7-ALS2-C (middle panels) were detected with HRP-conjugated anti-FLAG tag antibody and HRP-conjugated anti-T7 tag antibody, respectively. The positions of the molecular mass markers (in kilodaltons) are shown on the left. D, shown is a schematic representation of the Rabex-5 constructs and ALS2 constructs used in this study. Rabex-5 (aa 1–491) contains a zinc finger domain (ZnF), VPS9 domain, and coiled-coiled domain (CC). Rabex-5-C (aa 208–491) contains the VPS9 domain and CC domain. ALS2 (aa 1–1651) contains a RCC1 domain, DH domain, PH domain, MORN motifs I and II, and VPS9 domain (see Ref. 38). ALS2-C (aa 1485–1651) contains only the VPS9 domain. GEF activity-deficient VPS9 mutants, Rabex-5-D313A and ALS2-D1593A, are shown.
pEGFP-C1-Rabex-5SR (an shRNA-resistant Rabex-5 mutant) and pEGFP-C1-Rabex-5SR-D313A were produced by using the same method as described previously (21) and the following mutagenic oligonucleotides (substituted nucleotides are shown in italics): Rabex-5-SR-5′ primer, 5′-CGGGATTGCCAAGGAGGTACAGGATATAGTAGAAAAATACCCACTGGAG-3′, and Rabex-5-SR-3′ primer, 5′-CTCCAGTGGGTATTTTTCTACTATATCCTGTACCTCCTTGGCAATCCCG-3′. pEF-FLAG-Rab17, pEF-Myc-Rab17, pEF-Myc-Rab17-Q77L, pEGFP-C1-Rab17, pEGFP-C1-Rab17-Q77L, pmCherry-C1-Rab17, and pSilencer-EGFP-Rab17 were prepared as described previously (21). pEF-FLAG-Rab5A, pEF-FLAG-Rab5B, pEF-FLAG-Rab5C, and pEF-FLAG-Rab21 were also prepared as described previously (29, 30). The cDNAs of Rab5A, Rab17, and Rab21 were subcloned into the pMyc vector (31). pmStrawberry-C1 was also prepared as described previously (32).
Yeast Two-hybrid Assays
The yeast strain, medium, culture conditions, and transformation protocol used are described in James et al. (26). The materials used for the two-hybrid assay in this study were: yeast strain pJ69-4A, a synthetic complete medium lacking leucine and tryptophan (SC-LW medium: 0.67% yeast nitrogen base without amino acids, 2% glucose, 2% Bacto agar, 0.02% adenine, 0.01% uracil, 0.01% histidine, 0.015% lysine, and 0.01% methionine), and a synthetic complete medium lacking adenine, histidine, leucine, and tryptophan (SC-AHLW) as the selection medium.
Hippocampal Neuron Culture and Transfection
Mouse hippocampal neuronal cultures were prepared essentially as described previously (33). In brief, hippocampi were dissected from embryonic day 16.5 mice and dissociated with 0.25% trypsin (Invitrogen). The cells were plated at a density of 3–6 × 104 cells/ml onto coverglasses in a 6-well plate or glass-bottom dishes (35-mm dish, MatTek Corp., Ashland, MA) coated with poly-l-lysine hydrobromide (Nacalai Tesque, Kyoto, Japan). The cells were maintained in MEM containing B27® supplements, 1% fetal bovine serum, and 0.5 mm glutamine (Invitrogen). Plasmid DNAs were transfected into neurons at 2–4 days in vitro (DIV) by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Other Cell Cultures and Transfections
COS-7 cells and Neuro2A cells were cultured in Dulbecco's modified Eagle's medium (Wako Pure Chemical Industries, Osaka, Japan) supplemented with 10% fetal bovine serum, 50 units/ml penicillin, and 50 units/ml streptomycin. The cells were plated onto a 6-well plate. Plasmid DNAs were transfected into COS-7 cells and Neuro2A cells by using Lipofectamine Plus (Invitrogen) and Lipofectamine 2000, respectively, each according to the manufacturer's instructions.
Immunocytochemistry
Neurons were fixed for 10 min at room temperature with 4% paraformaldehyde (Wako Pure Chemicals Industries) and 4% sucrose in 0.1 m sodium phosphate buffer. After permeabilizing the cells with 0.1% Triton X-100 for 1 min, they were blocked with the blocking buffer (10% fetal bovine serum in phosphate-buffered saline) for 1 h. The cells were then immunostained for 1 h with anti-MAP2 chick antibody (1/1000 dilution), anti-neurofilament-H mouse antibody (1/500 dilution), anti-Tau mouse antibody (1/100 dilution), anti-Myc mouse antibody (1/500 dilution), anti-Rab17 rabbit antibody (1/500 dilution), anti-GFP rabbit antibody (1/1000 dilution), and/or anti-GFP guinea pig antibody (1/1000 dilution), after which they were incubated for 1 h at room temperature with Alexa-Fluor 488/594/633-labeled secondary IgG (1/5000 dilution). The cells were examined for fluorescence with a confocal laser-scanning microscope (Fluo-view 1000, Olympus, Tokyo, Japan), and the images were processed with Adobe Photoshop software (CS4). Fluorescent intensity was quantified with ImageJ software (Version 1.42q; National Institutes of Health).
Quantification of Rab Proteins Translocated into the Axon or Dendrites of Hippocampal Neurons
To quantify the signals of each Rab protein in the entire neuron (cell body, dendrites, and axon) or just the axon region or dendrite region, image thresholds were set to exclude pixels that did not fall over the cell body, the axon, or the dendrites, and after subtracting the background intensity values from each image before quantification, the integrated fluorescence intensity of Rab proteins in the entire neuron or just the axon (or dendrite) region was obtained. The proportion (%) of each Rab protein in the axon (or the dendrites) was calculated by dividing the fluorescence intensity of the axon (or dendrite) region by the fluorescence intensity of the entire neuron (i.e. total fluorescence intensity).
Morphometric Analyses of the Neurites of Hippocampal Neurons
Neurites were morphometrically analyzed as described previously with slight modifications (21). Hippocampal neurons were fixed and then subjected to immunocytochemistry with antibodies against GFP, MAP2, and neurofilament-H (or Tau). All quantitative analyses were carried out based on immunostaining of the morphometric marker GFP. Dendrites and axons were identified based on the presence of a dendrite-specific marker (MAP2) and an axon-specific marker (neurofilament-H or Tau), respectively. Total dendrite length, total dendrite branch tip numbers, total axon length, and total axon branch tip numbers were determined manually by using the NeuronJ (Version 1.1.0) (34) plug-in to the ImageJ software program.
Statistical Analyses
The results shown in Figs. 2C, 3F, 4D, 5 (F and I), 6 (B–F, H, and J), 7(C and D), and 8 (E–H) are presented as the means and S.E. Values were compared by means of Student's unpaired t test. A p value of <0.05 was considered statistically significant. All statistical analyses were performed on data from three independent experiments, and the data from a representative experiment are shown here.
FIGURE 2.
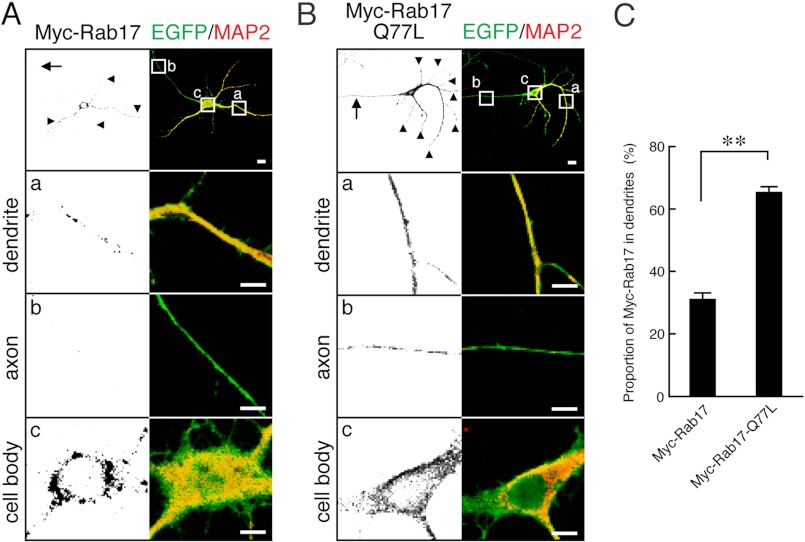
The constitutive active mutant of Rab17 preferentially localized in the dendrites of hippocampal neurons. A and B, shown are typical images of Rab17-expressing neurons and Rab17-Q77L-expressing neurons. At 4 DIV hippocampal neurons were transfected with vectors encoding EGFP and Myc-tagged Rab17 (A) or Myc-tagged Rab17-Q77L (B), and at 7 DIV the neurons were fixed and subjected to immunocytochemistry with antibodies against GFP (green), Myc (black), and MAP2 (a dendrite marker; red). The arrows and arrowheads point to axons and dendrites, respectively. The bottom three panels (a–c) are magnified views of the boxed areas in the top right panels. Bars, 10 μm. C, shown is quantification of the proportions of Myc-Rab17 (n = 20) and Myc-Rab17-Q77L (n = 20) in the dendrites shown in A and B. The proportion (%) of dendrite-localized Myc-Rab17 was calculated by dividing the dendrite-specific Myc-Rab17 fluorescence intensity by the total Myc-Rab17 fluorescence intensity. Note that active Rab17 was preferentially localized in the dendrites. **, p < 0.0025.
FIGURE 3.
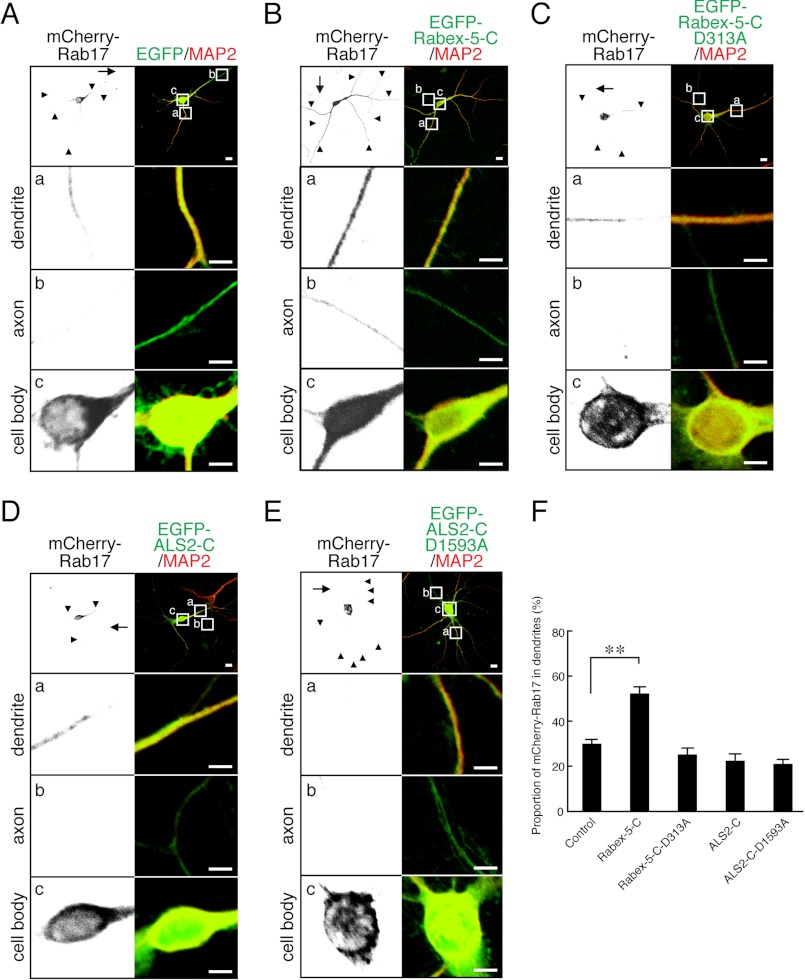
The VPS9 domain of Rabex-5, but not of ALS2, promoted translocation of recombinant Rab17 to the dendrites of hippocampal neurons. A–E, shown are typical images of mCherry-Rab17 in Rabex-5-C-coexpressing or EGFP-ALCS-2-C-coexpressing neurons. At 4 DIV hippocampal neurons were transfected with vectors encoding mCherry-tagged Rab17 and EGFP (A), EGFP-Rabex-5-C (B), EGFP-Rabex-5-C-D313A (C), EGFP-ALS2-C (D), or EGFP-ALS2-C-D1593A (E), and at 7 DIV the neurons were fixed and subjected to immunocytochemistry with antibodies against GFP (green), Rab17 (black), and MAP2 (a dendrite marker; red). The arrows and arrowheads point to axons and dendrites, respectively. The bottom three panels (a–c) are magnified views of the boxed areas in the top right panels. Bars, 10 μm. F, shown is quantification of the proportion of mCherry-Rab17 in the dendrites in the presence of EGFP (n = 20), EGFP-Rabex-5-C (n = 20), EGFP-Rabex-5-C-D313A (n = 10), EGFP-ALS2-C (n = 10), or EGFP-ALS2-C-D1593A (n = 10) shown in A–E. The proportion (%) of dendrite-localized mCherry-Rab17 was calculated by dividing the dendrite-specific mCherry-Rab17 fluorescence intensity by the total mCherry-Rab17 fluorescence intensity. Note that EGFP-Rabex-5-C, but not ALS2-C, promoted translocation of mCherry-Rab17 from the cell body to the dendrites. **, p < 0.0025.
FIGURE 4.
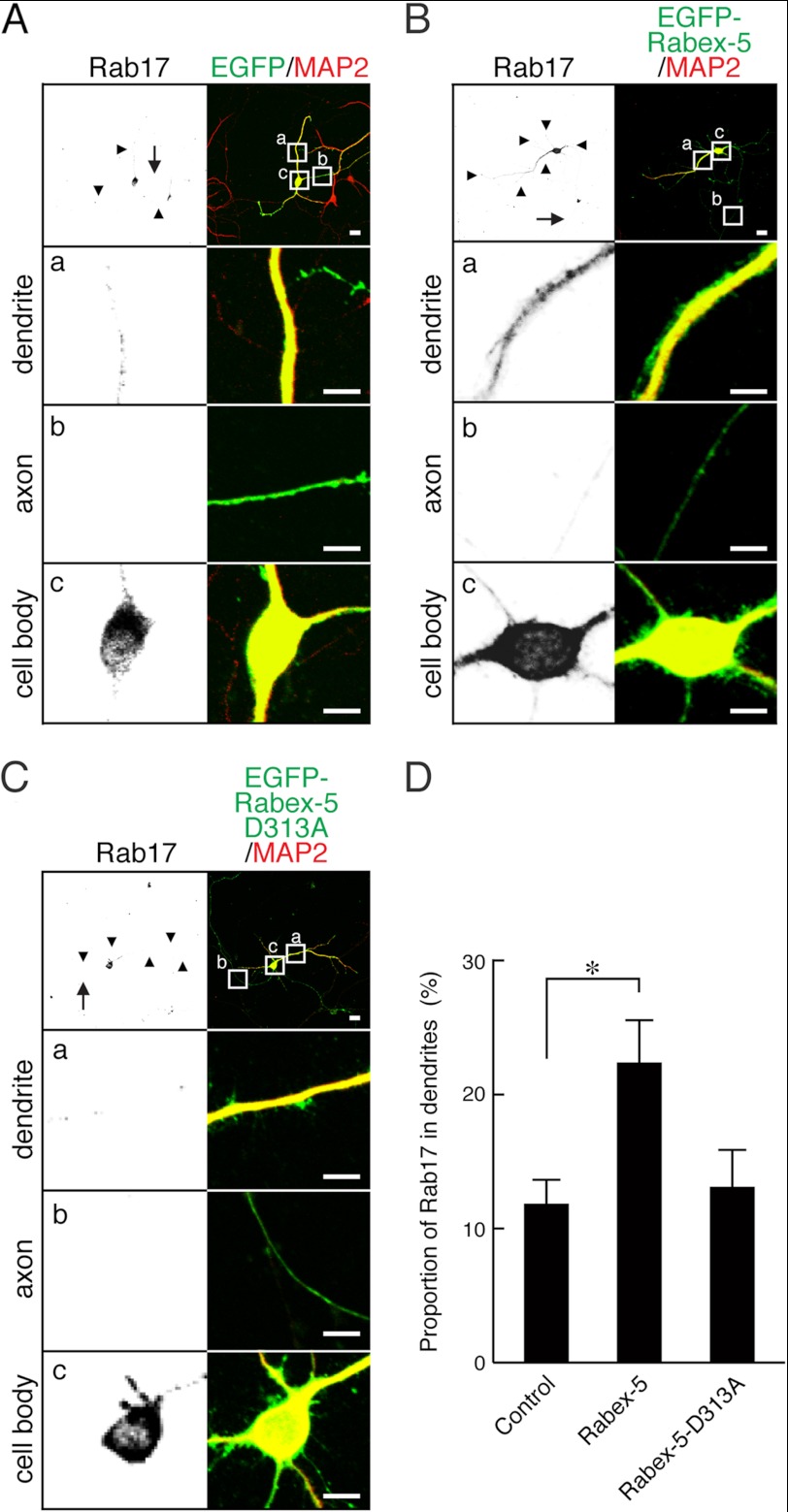
Rabex-5 promotes translocation of endogenous Rab17 to the dendrites of hippocampal neurons. A–C, shown are typical images of endogenous Rab17 protein in Rabex-5-expressing or EGFP-Rabex-5-D313A-expressing neurons. At 4 DIV hippocampal neurons were transfected with a vector encoding EGFP (A), EGFP-Rabex-5 (B), or EGFP-Rabex-5-D313A (C), and at 12 DIV the neurons were fixed and subjected to immunocytochemistry with antibodies against GFP (green), Rab17 (black), and MAP2 (red). The arrows and arrowheads point to axons and dendrites, respectively. The bottom three panels (a–c) are magnified views of the boxed areas in the top right panels. Bars, 10 μm. D, shown is quantification of the proportion of endogenous Rab17 protein in the dendrites in the presence of EGFP (n = 20), EGFP-Rabex-5 (n = 20), or EGFP-Rabex-5-D313A (n = 12) shown in A–C. The proportion (%) of Rab17 in the dendrites was calculated by dividing the dendrite-specific Rab17 fluorescence intensity by the total Rab17 fluorescence intensity. Note that EGFP-Rabex-5, but not its GEF activity-deficient mutant (D313A), promoted translocation of Rab17 from the cell body to the dendrites. *, p < 0.025.
FIGURE 5.
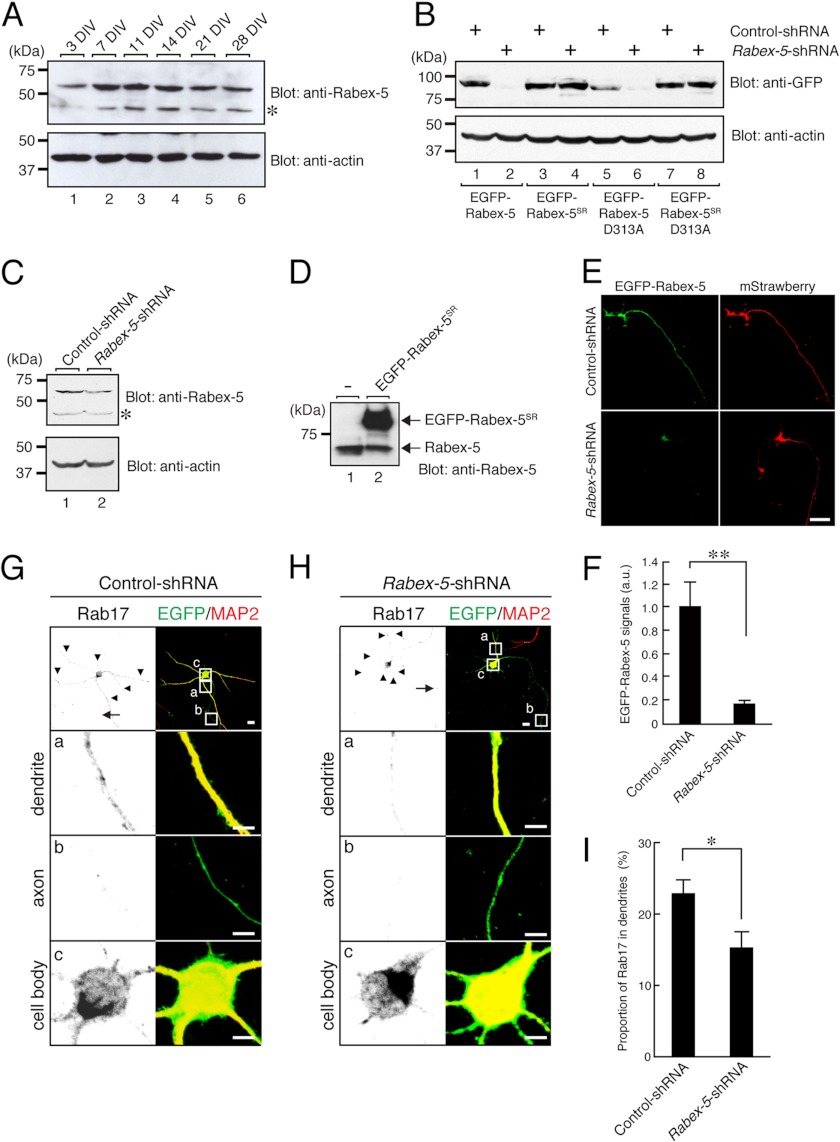
Rabex-5 is required for the dendritic localization of Rab17 in hippocampal neurons. A, Rabex-5 is expressed in primary mouse hippocampal neuron cultures. Lysates of mouse hippocampal neurons at 3, 7, 11, 14, 21, or 28 DIV were analyzed by immunoblotting with anti-Rabex-5 antibody (upper panel) and anti-actin antibody (lower panel). The asterisks indicate nonspecific bands. B and C, knockdown efficiency of Rabex-5-shRNA is shown. B, Neuro2A cells were transfected with a vector encoding control-shRNA (lanes 1, 3, 5, and 7) or Rabex-5-shRNA (lanes 2, 4, 6, and 8) together with pEGFP-C1-Rabex-5 (lanes 1 and 2), pEGFP-C1-Rabex-5SR (lanes 3 and 4), pEGFP-C1-Rabex-5-D313A (lanes 5 and 6), or pEGFP-C1-Rabex-5-D313ASR (lanes 7 and 8). Two days after transfection the cells were lysed and subjected to immunoblot analysis with anti-GFP antibody (upper panel) and anti-actin antibody (lower panel). C, Neuro2A cells were transfected with a vector encoding control-shRNA (lane 1) or Rabex-5-shRNA (lane 2). Two days after transfection the cells were lysed and subjected to immunoblot analysis with anti-Rabex-5 antibody (upper panel) and anti-actin antibody (lower panel). The asterisk indicates nonspecific bands. Note that the level of expression of endogenous Rabex-5 in Neuro2A cells was reduced in the presence of Rabex-5-shRNA. D, shown is a comparison of the protein expression level of EGFP-Rabex-5SR and endogenous Rabex-5 in Neuro2A cells. Neuro2A cells were transfected with pEGFP-C1-Rabex-5SR (lane 2) or nothing (lane 1). Two days after transfection the cells were lysed and subjected to immunoblot analysis with anti-Rabex-5 antibody. The positions of the molecular mass markers (in kilodaltons) are shown on the left. Note that the expression level of EGFP-Rabex-5SR is approximately 5 times higher than that of endogenous Rabex-5 in Neuro2A cells. E and F, shown are typical images of EGFP-Rabex-5-expressing neurons in the presence and absence of Rabex-5-shRNA. At 3 DIV hippocampal neurons were transfected with a vector encoding control-shRNA or Rabex-5-shRNA together with EGFP-Rabex-5 and mStrawberry, and at 5 DIV the neurons were fixed and subjected to immunocytochemistry with antibodies against GFP (green). Bar, 10 μm. F, shown is quantification of the EGFP-Rabex-5 of the control-shRNA-transfected neurons (n = 20) or Rabex-5-shRNA-transfected neurons (n = 20) shown in E. a.u., arbitrary units. G and H, shown are typical images of Rab17 in the Rabex-5-shRNA-transfected neurons. At 4 DIV hippocampal neurons were transfected with a vector encoding control-shRNA (G) or Rabex-5-shRNA (H), and at 14 DIV the neurons were fixed and subjected to immunocytochemistry with antibodies against GFP (green), Rab17 (black), and MAP2 (blue). The arrows and arrowheads point to axons and dendrites, respectively. The bottom three panels (a–c) are magnified views of the boxed areas in the top right panels. Bars, 10 μm. I, shown is quantification of the proportion of Rab17 in the dendrites of control-shRNA-transfected neurons (n = 20) and Rabex-5-shRNA-transfected neurons (n = 20) shown in G and H. The proportion (%) of Rab17 in the dendrites was calculated by dividing the dendrite-specific Rab17 fluorescence intensity by the total Rab17 fluorescence intensity. Note that translocation of Rab17 to the dendrites was significantly reduced after knockdown of Rabex-5. **, p < 0.0025.
FIGURE 6.
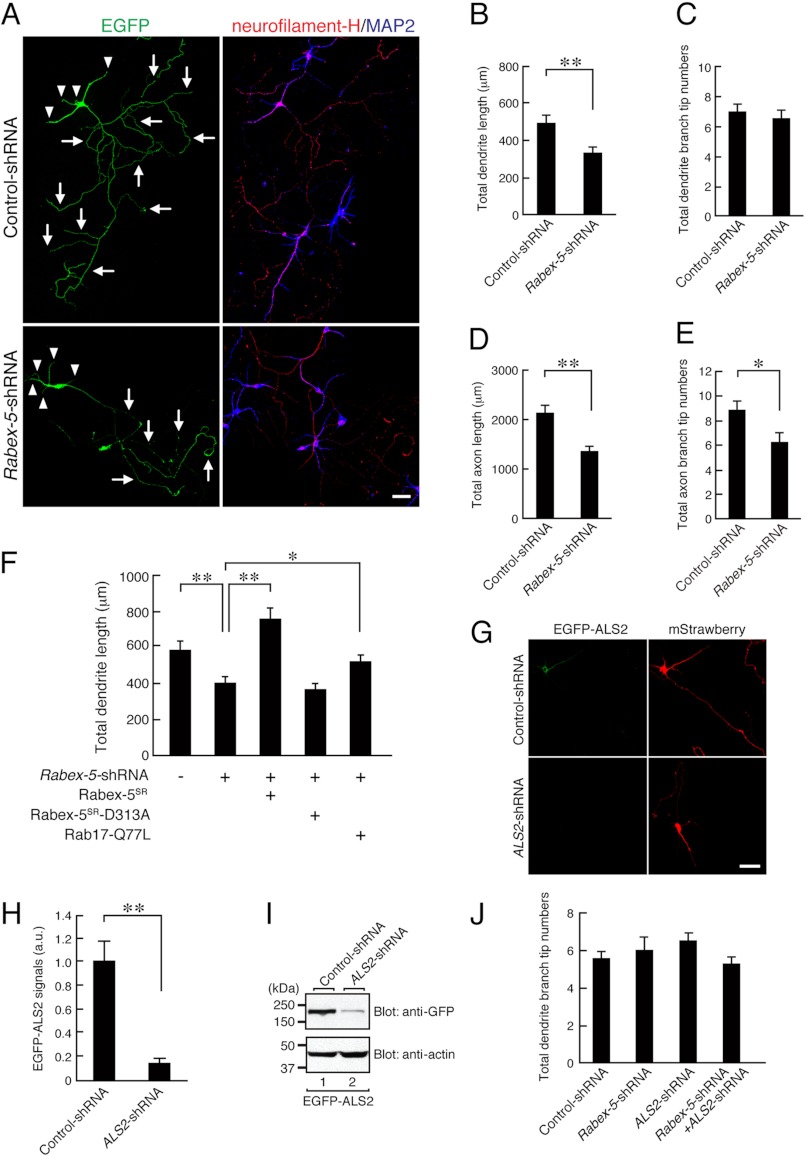
Rabex-5 regulates axon and dendrite morphogenesis in hippocampal neurons. A, shown are typical images of Rabex-5 knockdown neurons. At 4 DIV hippocampal neurons were transfected with a vector encoding EGFP and control-shRNA (upper panels) or Rabex-5-shRNA (lower panels), and at 11 DIV the neurons were fixed and subjected to immunocytochemistry with antibodies against GFP, neurofilament-H (an axon marker; red), and MAP2 (blue). The arrows and arrowheads point to axons and dendrites, respectively. Bar, 50 μm. B–E, shown is quantification of the total dendrite length (B), total dendrite branching tip numbers (C), total axon length (D), and total axon branching tip numbers (E) of the control neurons (n = 20), and Rabex-5 knockdown neurons (n = 20). Note that the total dendrite length, total axon length, and total axon branching tip numbers of the Rabex-5 knockdown neurons were significantly lower than in the control cells. **, p < 0.0025. F, shown is quantification of the total dendrite length of the control neurons (n = 32), Rabex-5 knockdown neurons (n = 35), EGFP-Rabex-5SR-expressing Rabex-5 knockdown neurons (n = 32), EGFP-Rabex-5-D313ASR-expressing Rabex-5 knockdown neurons (n = 34), and EGFP-Rab17-Q77L-expressing Rabex-5 knockdown neurons (n = 35). Note that the reduction in total dendrite length of the Rabex-5 knockdown neurons was rescued by re-expression of Rabex-5SR or by expression of Rab17-Q77L. **, p < 0.0025; *, p < 0.025. G, shown are typical images of EGFP-ALS2-expressing neurons in the presence and absence of ALS2-shRNA. At 3 DIV hippocampal neurons were transfected with a vector encoding control-shRNA or ALS2-shRNA together with EGFP-ALS2 and mStrawberry, and at 5 DIV the neurons were fixed and subjected to immunocytochemistry with antibodies against GFP (green). Bar, 10 μm. H, shown is quantification of the EGFP-ALS2 of control-shRNA-transfected neurons (n = 20) and Rabex-5-shRNA-transfected neurons (n = 20) shown in G. a.u., arbitrary units. I, COS7 cells were transfected with a vector encoding EGFP-ALS2 together with control-shRNA or ALS2-shRNA. Two days after transfection the cells were lysed and subjected to immunoblot analysis with anti-GFP antibody (upper panel) and anti-actin antibody (lower panel). The positions of the molecular mass markers (in kilodaltons) are shown on the left. J, shown is quantification of the total dendrite branching tip numbers of the control neurons (n = 20), ALS2 knockdown neurons (n = 20), Rabex-5 knockdown neurons (n = 20), and Rabex-5 and ALS2 double-knockdown neurons (n = 20). Note that the total dendrite branching tip numbers of neither the ALS2 knockdown neurons nor the Rabex-5 and ALS2 double-knockdown neurons were altered in comparison with the control neurons.
FIGURE 7.
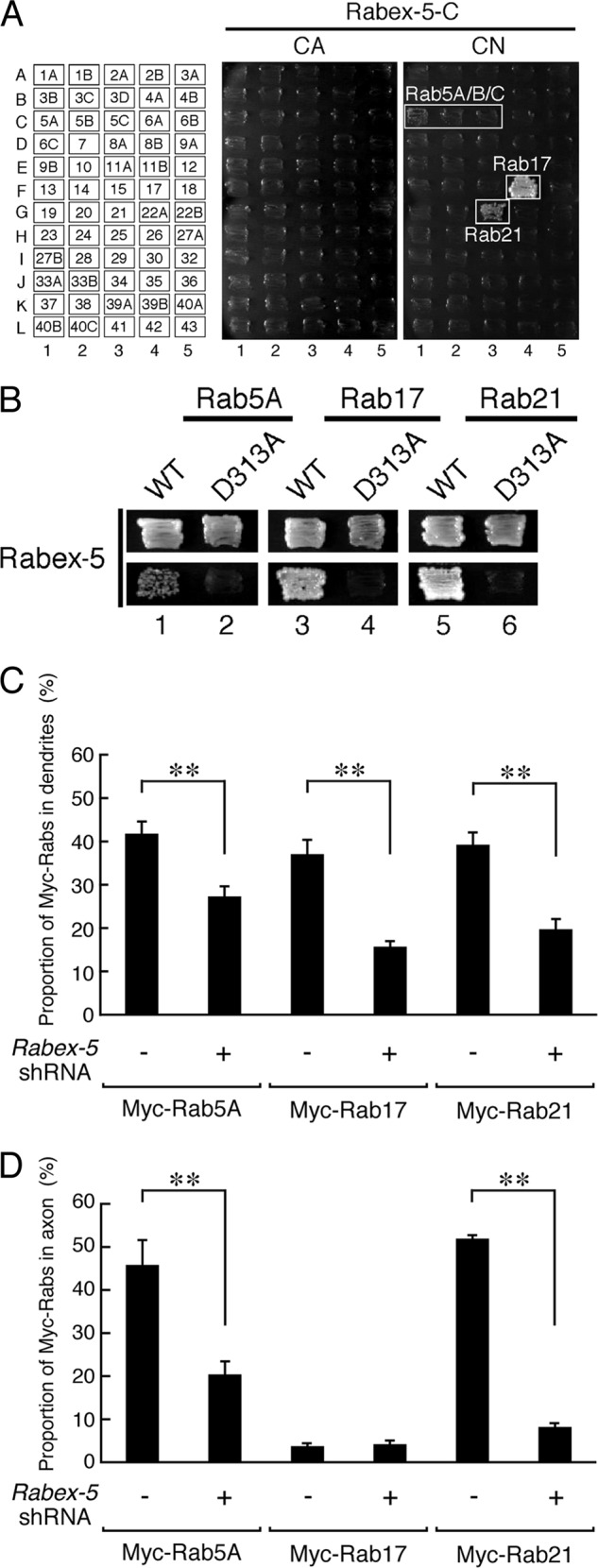
Rabex-5 promotes translocation of Rab5 and Rab21 to the axon and dendrites of hippocampal neurons. A, shown is Rab binding specificity of Rabex-5 as revealed by yeast two-hybrid panels. Yeast cells containing pGBD plasmid expressing CA or CN mutants of Rab (positions indicated in the left panels) and pGAD plasmid expressing Rabex-5-C protein were streaked on SC-AHLW and incubated at 30 °C for 1 week. Positive patches are boxed. Note the specific interactions between Rabex-5-C and the CN (GDP-fixed) form of Rab5A, Rab5B, Rab5C, Rab17, and Rab21. B, yeast cells containing pGBD plasmid expressing GDP-locked CN forms of Rab5A, Rab17, and Rab21 and pGAD plasmid expressing the VPS9 domain of Rabex-5 or of Rabex-5-D313A were streaked on an SC-LW plate (upper panels) or SC-AHLW plate (lower panels) and incubated at 30 °C. Note that the GEF activity-deficient Rabex5-D313A mutant completely abrogated the interaction with Rab5A, Rab17, and Rab21. C and D, shown is quantification of the proportions of Myc-Rab5A, Myc-Rab17, and Myc-Rab21 in the dendrites (C) and axon (D) of control shRNA-transfected neurons (n = 20) and Rabex-5-shRNA-transfected neurons (n = 20). At 4 DIV hippocampal neurons were transfected with a vector encoding control shRNA or Rabex-5-shRNA together with Myc-Rab5A, Myc-Rab17, or Myc-Rab21, and at 11 DIV the neurons were fixed and subjected to immunocytochemistry with antibodies against GFP, Myc, and MAP2. The proportions (%) of Myc-Rab5A, Myc-Rab17, and Myc-Rab21 in the axon and dendrites were calculated by dividing the axon- or dendrite-specific Myc-Rabs fluorescence intensity by the total Myc-Rabs fluorescence intensity. Note that translocation of Myc-Rab5A and Myc-Rab21 to the axon and dendrites was significantly reduced after knockdown of Rabex-5. **, p < 0.0025.
FIGURE 8.
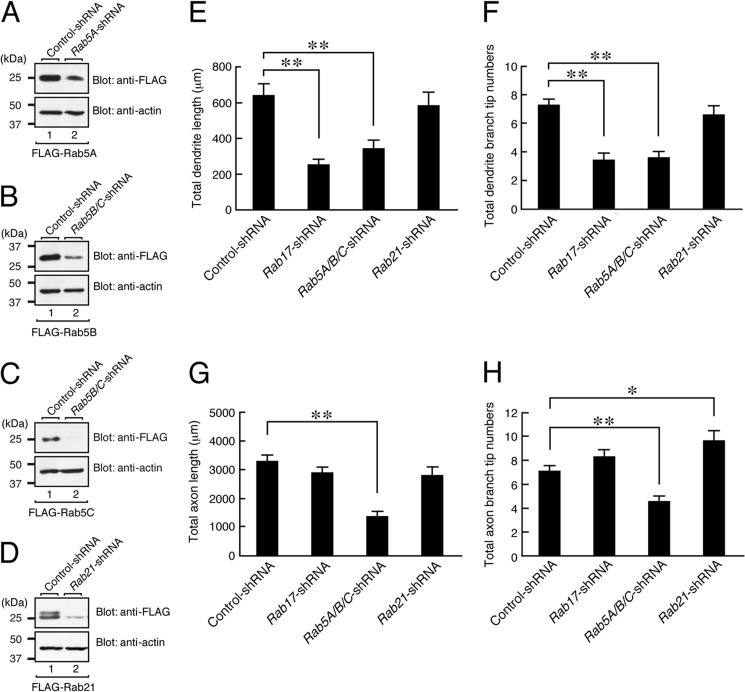
Rab5 and Rab17, but not Rab21, are required for the control of neurite morphogenesis of hippocampal neurons. A–D, COS7 cells were transfected with a vector encoding FLAG-Rab5A together with control-shRNA or Rab5A-shRNA (A), FLAG-Rab5B/C together with control-shRNA or Rab5B/C-shRNA (B and C), or FLAG-Rab21 together with control-shRNA or Rab21-shRNA (D). Two days after transfection the cells were lysed and subjected to immunoblot analysis with anti-FLAG tag antibody (upper panels) and anti-actin antibody (lower panels). The positions of the molecular mass markers (in kilodaltons) are shown on the left. The knockdown efficiency of each shRNA was also evaluated by expressing EGFP-Rab in hippocampal neurons (Rab5A-shRNA, 46.5 ± 17.7% reduction of EGFP-Rab5A; Rab5B/C-shRNA, 69.0 ± 9.5% reduction of EGFP-Rab5B and 84.4 ± 2.7% reduction of EGFP-Rab5C; Rab17-shRNA, 78.8 ± 7.8% reduction of EGFP-Rab17; Rab21-shRNA, 91.2 ± 2.1% reduction of EGFP-Rab21 in comparison with the control shRNA). E–H, shown is quantification of the total dendrite length (E), total dendrite branching tip numbers (F), total axon length (G), and total axon branching tip numbers (H) of the control neurons (n = 21), Rab17 knockdown neurons (n = 20), Rab5A/B/C knockdown neurons (n = 21), and Rab21-knockdown neurons (n = 21). At 4 DIV hippocampal neurons were transfected with a vector encoding EGFP and control-shRNA, Rab17-shRNA, Rab5A-shRNA/Rab5B/C-shRNA, or Rab21-shRNA, and at 11 DIV the neurons were fixed and subjected to immunocytochemistry with antibodies against GFP, neurofilament-H, and MAP2. Note that the dendrite length and branching tip numbers of the Rab17 knockdown neurons were significantly lower than in the control cells, whereas the length and branching tip numbers of both the axon and dendrites of the Rab5A/B/C knockdown neurons were significantly lower than in the control cells. **, p < 0.0025; *, p < 0.025.
RESULTS
Screening for Rab17-GEFs by Yeast Two-hybrid Assays with a GDP-locked Rab17 Mutant as Bait
To identify a Rab17-GEF molecule(s) that functions in neurons, we focused on previous reports showing that Rab-GEFs often physically interact with their substrate GDP-Rabs, e.g. DENND1/RME-4 interacts with Rab35 (14), ALS2 with Rab5 (23), Rin1 with Rab5 (35), Varp with Rab21 (36), and Rabin8 with Rab8 (7). We, therefore, performed yeast two-hybrid assays to test for possible interactions between putative GEF domains and a GDP-locked, constitutive negative (CN) form of Rab17 (Rab17-T33N). As shown in Fig. 1A, we screened 18 DENN domains (DENN, DENND1A–C, DENND2A–D, DENND3, DENND4A–C, DENND5A–B, DENND6A–B, and Sbf1–2), 9 VPS9 domains (Varp, Rabex-5, Gapex-5, Rin1–3, ALS2, ALS-cl, and RINL), and 2 Sec2 domains (Rabin3 and GRAB) by yeast two-hybrid assays and succeeded in identifying two candidates, Rabex-5 and ALS2 (Fig. 1, A, lanes 20 and 25, and D), as Rab17-GEFs. It should be noted that Rabex-5 has recently been reported to exhibit Rab17-GEF activity in vitro (16), thereby validating our screening procedure. The interaction between GDP-Rab17 and the VPS9 domain of Rabex-5 (Rabex-5-C) or ALS2 (ALS2-C) was confirmed by co-immunoprecipitation assays in COS-7 cells (Fig. 1, B and C). We, therefore, selected Rabex-5 and ALS2 for subsequent analysis as candidates for the Rab17-GEF(s) in mouse hippocampal neurons.
The VPS9 Domain of Rabex-5, but Not of ALS2, Activates Rab17 in Hippocampal Neurons
We next investigated whether these candidate proteins actually act as a Rab17 activator in cultured mouse hippocampal neurons. We focused on the stage-dependent difference in Rab17 localization in hippocampal neurons as a means of monitoring activation of Rab17. We previously showed that Rab17 was detected only in the cell body at an early stage (3 DIV, 6.0 ± 1.7% of Rab17; 7 DIV, 6.4 ± 2.3% of Rab17 in the dendrites) but that some of the Rab17 had translocated from the cell body to the dendrites at a later stage (11 DIV, 11.6 ± 1.2% of Rab17 in the dendrites) (21). We hypothesized that the differences in the localization of Rab17 during neuronal development are attributable to the difference in ratio of GTP-Rab17 to GDP-Rab17. If activation of Rab17, i.e. an increase in GTP-Rab17, is required for Rab17 to translocate to dendrites, a constitutive active form of Rab17 (Rab17-Q77L) should be preferentially localized in the dendrites rather than in the cell body. As expected, at 7 DIV the wild-type Rab17 was found to be mainly localized in the cell body, and only a small portion of Rab17 was detected in the dendrites (31.3 ± 1.9% of Myc-Rab17 was in the dendrites, and this value was higher than the proportion of endogenous Rab17 in the dendrites described above; the discrepancy may be caused by the lower expression level of endogenous Rab17 and by the low quality of our anti-Rab17 antibody, which does not sufficiently detect the low level expression of endogenous Rab17 protein in the dendrites) (Fig. 2A and C). By contrast, a large proportion of the Rab17-Q77L was localized in the dendrites (65.9 ± 1.6% of Rab17-Q77L was in the dendrites) (Fig. 2, B and C). Interestingly, however, a small proportion of the Myc-Rab17-Q77L signals was also detected in the axon, in contrast to the lack of wild-type Myc-Rab17 signals in the axon (Fig. 2, A and B, panels b, left). These results strongly supported our hypothesis that Rab17 is translocated from the cell body to the dendrites in a GTP-dependent manner.
If Rabex-5 and ALS2 actually have the ability to activate Rab17 in neurons, forced overexpression of their GEF domains, i.e. enhanced GFP (EGFP)-tagged Rabex-5-C and ALS-2-C, in hippocampal neurons should promote translocation of Rab17 from the cell body to the dendrites. As expected, at 7 DIV the proportion of dendrite-localized Rab17 was significantly higher in the EGFP-Rabex-5-C-expressing neurons than it was in the control neurons (52.3 ± 2.9% of mCherry-Rab17 in the dendrites of the Rabex-5-C-expressing neurons versus 30.0 ± 1.9% of mCherry-Rab17 in the dendrites of control neurons) (Fig. 3, A, B, and F). Interestingly, some mCherry-Rab17 signals were also detected in the axon of Rabex-5-C-expressing cells (Fig. 3B, panel b, left), the same as in the Rab17-Q77L-expressing cells (Fig. 2B, panel b, left). This Rabex-5-C-dependent translocation of Rab17 to dendrites must have been caused by the GEF activity of the VPS9 domain of Rabex-5, because a GEF activity-deficient mutant of Rabex-5-C-D313A (18) had no effect on the Rab17 distribution (21.8 ± 2.5% of mCherry-Rab17 in the dendrites of Rabex-5-C-D313A-expressing neurons) (Fig. 3, C and F). By contrast, neither the wild-type ALS-2-C nor a GEF activity-deficient mutant of ALS-2-C-D1593A altered the Rab17 distribution (Fig. 3, D–F). These results indicated that the VPS9 domain of Rabex-5, but not of ALS2, has the ability to activate Rab17 in hippocampal neurons.
To further confirm that endogenous Rab17 protein is also activated by Rabex-5, we overexpressed full-length Rabex-5 in hippocampal neurons and evaluated its effect on the subcellular localization of endogenous Rab17 protein by immunocytochemistry with the anti-Rab17-specific antibody. The results showed that at 12 DIV the proportion of endogenous Rab17 protein localized in the dendrites was 11.8 ± 1.9% in the control EGFP-expressing cells (Fig. 4, A and D) and significantly higher (21.8 ± 2.5%) in the Rabex-5-expressing neurons (Fig. 4, B and D). Interestingly, in contrast to the mCherry-Rab17- and EGFP-Rabex-5-C-expressing cells (Fig. 3B, panel b, left), no endogenous Rab17 signals were observed in the axon of EGFP-Rabex-5-expressing cells (Fig. 4B, panel b, left).
Rabex-5 Is Required for Translocation of Rab17 to the Dendrites of Hippocampal Neurons
To determine whether endogenous Rabex-5 protein also regulates the dendritic localization of Rab17 in hippocampal neurons, we evaluated the impact of knockdown of endogenous Rabex-5. First, we investigated whether Rabex-5 is actually expressed in cultured mouse hippocampal neurons by immunoblotting with a specific antibody against Rabex-5. As shown in Fig. 5A, endogenous Rabex-5 protein was easily detected in cultured hippocampal neurons, and the level of Rabex-5 protein seemed to peak at 7–14 DIV. We then knocked down Rabex-5 with a specific shRNA, Rabex-5-shRNA (Fig. 5, B and C), which dramatically decreased the protein expression level of EGFP-Rabex-5 (77.2 ± 3.0% reduction in comparison with the control) in hippocampal neurons (Fig. 5, E and F). We found that Rabex-5 knockdown resulted in a marked reduction in the proportion of Rab17 that had been translocated to the dendrites of the hippocampal neurons at 14 DIV (22.9 ± 1.9% of endogenous Rab17 in the dendrites of the control neurons versus 15.3 ± 2.2% of endogenous Rab17 in the dendrites of the Rabex-5 knockdown neurons) (Fig. 5, G–I). These results enabled us to conclude that Rabex-5 is required for the translocation of Rab17 to the dendrites of hippocampal neurons.
Rabex-5 Regulates Dendrite and Axon Morphogenesis of Hippocampal Neurons
Because we previously showed that Rab17 specifically regulates dendrite morphogenesis of hippocampal neurons, we proceeded to investigate the involvement of Rabex-5 in neurite morphogenesis of hippocampal neurons. In contrast to the knockdown of Rab17, however, Rabex-5 knockdown resulted in a marked reduction in the total dendrite length, total axon length, and number of axon branches (to 72.3 ± 8.3, 61.8 ± 5.7, and 76.9 ± 7.7%, respectively, of the control), but it had no effect on the number of dendrite branches (Fig. 6, A–E). These effects could not have been caused by an off-target effect of shRNA, because the reduction in the total dendrite length in Rabex-5 knockdown neurons (to 69.0 ± 5.7% of the control) was completely rescued by re-expression of shRNA-resistant Rabex-5SR (Fig. 5, B and D) (to 129.5 ± 10.1% of the control neurons) but not of a GEF activity-deficient Rabex-5SR-D313A mutant (Fig. 6F). Moreover, we found that the reduction in total dendrite length of the Rabex-5 knockdown neurons was partially rescued by co-expression of Rab17-Q77L, i.e. forced activation of Rab17 (to 89.0 ± 5.9% of the control) (Fig. 6F), indicating that Rabex-5 contributes to Rab17-mediated dendrite outgrowth.
Because knockdown of Rabex-5 did not affect the number of dendrite branches, we next investigated the possible involvement of ALS2, another GDP-Rab17-binding protein (Fig. 1), in dendrite branching by knocking down its expression with a specific shRNA (Fig. 6, G–I). Although the ALS2-shRNA we prepared dramatically decreased the protein expression level of EGFP-ALS2 in the hippocampal neurons (80.7 ± 1.5% reduction in comparison with the control) (Fig. 6H), neither knockdown of ALS2 alone nor double knockdown of Rabex-5 and ALS2 affected the number of dendrite branches of hippocampal neurons (Fig. 6J). The same results were obtained with two independently prepared ALS2-shRNAs (data not shown). Based on these findings, ALS2 is unlikely to regulate Rab17-mediated dendrite branching.
Rabex-5 Is Also Required for the Neuritic Localization of Rab5 and Rab21 in Hippocampal Neurons
Different contributions of Rabex-5 and Rab17 to neurite morphogenesis (i.e. Rabex-5 to dendrite outgrowth and axon morphogenesis and Rab17 to dendrite morphogenesis) suggests that Rabex-5 activates other Rab family proteins besides Rab17 during the course of neurite morphogenesis. To pursue this possibility, we performed yeast two-hybrid assays with a panel of 60 different CA (GTP-locked) and CN (GDP-locked) Rabs to identify additional target Rabs of Rabex-5 (36). The results of the Rab-family-wide analysis showed that Rabex-5 specifically interacted with the CN forms of Rab5A/B/C, Rab17, and Rab21 (Fig. 7A), which was consistent with the known GEF activity of Rabex-5 in vitro (16, 18). In addition, the D313A mutation of Rabex-5 completely abrogated the binding of Rabex-5 to Rab5A, Rab17, and Rab21 in yeast two-hybrid assays (Fig. 7B). Next, we investigated whether Rabex-5 also regulates the localization of Rab5 and Rab21, in addition to the localization of Rab17, in hippocampal neurons. The results showed that Rabex-5 knockdown resulted in a marked reduction in the proportions of Myc-tagged Rab5A, Rab17, and Rab21 that had been translocated to the dendrites (Myc-Rab5A, 34.7 ± 5.2% reduction; Myc-Rab17, 57.6 ± 4.6% reduction; Myc-Rab21, 49.8 ± 5.1% reduction in comparison with the control) (Fig. 7C) and to the axon (Myc-Rab5A, 55.5 ± 8.8% reduction; Myc-Rab21, 84.2 ± 1.6% reduction in comparison with the control) (Fig. 7D). These results indicated that Rabex-5 also functions as an upstream regulator of Rab5 and Rab21.
Rab5 and Rab17 Regulate Neurite Morphogenesis of Hippocampal Neurons Differently
Last, we investigated the involvement of Rab5A/B/C and Rab21, other putative downstream targets of Rabex-5, in neurite morphogenesis of hippocampal neurons by using specific shRNAs (Fig. 8, A–D). Consistent with our previous finding (21), knockdown of Rab17 resulted in a marked reduction in both total dendrite length and the number of dendrite branches (to 39.7 ± 4.4 and 47.4 ± 3.1%, respectively, of the control) (Fig. 8, E and F), but total axon length and the number of axon branches were unaffected (Fig. 8, G and H). By contrast, knockdown of all three Rab5 isoforms (Rab5A/B/C) resulted in a marked reduction in total dendrite length, the number of dendrite branches, total axon length, and the number of axon branches (to 53.7 ± 7.0, 49.7 ± 5.2, 41.8 ± 5.0, and 64.3 ± 6.0%, respectively, of the control) (Fig. 8, E–H). Interestingly, however, knockdown of Rab21 did not reduce total neurite length or the total number of neurite branches (Fig. 8, E–H). The same results were obtained when we used another site of Rab21-shRNA (data not shown). These results indicated that Rab5 and Rab17, but not Rab21, are likely to function as downstream targets of Rabex-5 during neurite morphogenesis of hippocampal neurons.
DISCUSSION
We previously showed that Rab17 regulates dendrite morphogenesis and postsynaptic development of hippocampal neurons (21). However, the spatiotemporal regulation of Rab17 activation remained completely unknown, because no Rab17-GEF that functions in hippocampal neurons had been identified. In the present study we succeeded in identifying Rabex-5 and ALS2 as candidate Rab17-GEFs by means of yeast two-hybrid assays (Fig. 1A). Several lines of evidence indicated that Rabex-5, but not ALS2, is likely to function as a Rab17-GEF in mouse hippocampal neurons: (i) overexpression of the GEF domain of Rabex-5, but not of ALS2, in hippocampal neurons promoted the dendritic localization of Rab17 (Figs. 3 and 4), (ii) knockdown of endogenous Rabex-5, but not of ALS2, in hippocampal neurons decreased the dendritic localization of Rab17 (Fig. 5, H and I), (iii) the total dendrite length of Rabex-5 knockdown neurons was partially restored by forced activation of Rab17, i.e. expression of Rab17-Q77L (Fig. 6F). Based on our findings together with the previous report that Rabex-5 exhibited Rab17-GEF activity in vitro (16), we concluded that Rabex-5 is an upstream activator of Rab17 in mouse hippocampal neurons.
However, Rabex-5 cannot be the sole Rab17-GEF in hippocampal neurons because the phenotypes of Rabex-5 knockdown neurons and Rab17 knockdown neurons differed with respect to dendrite branching (Figs. 6C and 8F), although both exhibited reduced total dendrite length (Figs. 6B and 8E). We speculate that Rab17 is differently activated by two GEFs, i.e. by Rabex-5 and by an as yet unidentified GEF, in the following manner; activation of Rab17 by Rabex-5 specifically regulates dendrite outgrowth, whereas activation of Rab17 by the as yet unidentified GEF promotes dendrite branching. One plausible candidate for the unidentified GEF is ALS2, which also binds the GDP-locked Rab17 (Fig. 1). However, the GEF domain of ALS2 was unable to promote dendritic localization of Rab17 in hippocampal neurons (Fig. 3, D and E), and knockdown of ALS2 alone (or double knockdown of ALS2 and Rabex-5) in hippocampal neurons had no effect on the number of their dendrite branches (Fig. 6J), indicating that ALS2 is not involved in the dendrite branching step in hippocampal neurons. Although ALS2 does not contribute to dendrite outgrowth or branching, it is still possible that ALS2 activates Rab17 at the spine, because Rab17 is required for postsynaptic development (21), and ALS2 has been reported to localize at the spine (37). At any rate, further extensive studies are necessary to determine whether ALS2 possesses in vitro Rab17-GEF activity and functions as a Rab17-GEF in hippocampal neurons. Because ALS2 was originally identified from ALS patients and hereditary spastic paraplegia (HPS) motor neuron disease patients (38–40), it will also be interesting to investigate the relationship between Rab17 and these diseases in motor neurons in the future.
Our findings also indicated that Rab17 is not the sole target of Rabex-5, because defects in axon morphogenesis were observed in Rabex-5 knockdown neurons alone (Figs. 6, D and E, and 8, G and H). Although several in vitro target Rabs of Rabex-5, Rab5, Rab17, Rab21, and Rab22, have been reported (16, 18, 41) and at least three of them interacted with Rabex-5 in yeast two-hybrid assays (Fig. 7, A and B), the observation that the defects in axon morphogenesis of Rabex-5 knockdown neurons were phenocopied by Rab5A/B/C knockdown in the present study (Figs. 6, D and E, and 8, G and H) indicated that Rab5 is the most likely target of Rabex-5 during axon morphogenesis of hippocampal neurons. However, activation of Rab5 (and also Rab17) at the dendrite branching step is unlikely to be mediated by Rabex-5, because Rabex-5 is not involved in dendrite branching (Fig. 6C). Another type of Rab5-GEF, e.g. Rin (35), or Gapex-5/RME-6 (42) may be involved in this process.
Although involvement of two other target Rabs, Rab21 and Rab22, in neurite outgrowth of PC12 cells has been reported (41, 43), knockdown of neither Rab21 nor Rab22A/B with specific shRNAs had virtually any effect on neurite morphogenesis under our experimental conditions (Fig. 8, E–H).4 The lack of effect of our Rab21-shRNA is unlikely to be caused by insufficient knockdown of Rab21, because the same shRNA strongly inhibited forskolin-induced dendrite formation of melanocytes (44). In contrast to our finding, however, it has been reported that knockdown of Varp, a Rab21-specific GEF (45), in hippocampal neurons reduced total axon length at 4 DIV (43). This discrepancy may be attributable to the difference in assay protocols, 4 DIV in Ref. 43 and 11 DIV in this study. We speculate that Rab21 and Varp function at an early stage of axon outgrowth.
In summary, we demonstrated that Rabex-5 functions as a Rab17-GEF that regulates Rab17 localization. We also showed that Rabex-5 and Rab5 are required for both axon morphogenesis and dendrite morphogenesis. Based on our findings, we propose that Rabex-5 activates at least two distinct Rabs, Rab5 and Rab17, and that the activated Rabs cooperatively mediate dendritic morphogenesis of hippocampal neurons.
Acknowledgments
We thank Megumi Aizawa for technical assistance and members of the Fukuda Laboratory for valuable discussions.
This work was supported in part by grants-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, and Technology (MEXT) of Japan (to Y. M. and M. F.) and by a grant from the Daiichi-Sankyo Foundation of Life Science (to M. F.).
Y. Mori and M. Fukuda, unpublished observations.
- GEF
- guanine nucleotide exchange factor
- aa
- amino acid residues
- ALS
- amyotrophic lateral sclerosis
- CA/CN
- constitutive active/negative
- DENN
- differentially expressed in normal and neoplastic cells
- DIV
- days in vitro
- EGFP
- enhanced GFP
- SR
- shRNA-resistant
- VPS9
- vacuolar protein sorting 9.
REFERENCES
- 1. Fukuda M. (2008) Regulation of secretory vesicle traffic by Rab small GTPases. Cell. Mol. Life Sci. 65, 2801–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stenmark H. (2009) Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10, 513–525 [DOI] [PubMed] [Google Scholar]
- 3. Barr F., Lambright D. G. (2010) Rab GEFs and GAPs. Curr. Opin. Cell Biol. 22, 461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fukuda M. (2011) TBC proteins. GAPs for mammalian small GTPase Rab? Biosci. Rep. 31, 159–168 [DOI] [PubMed] [Google Scholar]
- 5. Marat A. L., Dokainish H., McPherson P. S. (2011) DENN domain proteins. Regulators of Rab GTPases. J. Biol. Chem. 286, 13791–13800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Walch-Solimena C., Collins R. N., Novick P. J. (1997) Sec2p mediates nucleotide exchange on Sec4p and is involved in polarized delivery of post-Golgi vesicles. J. Cell Biol. 137, 1495–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hattula K., Furuhjelm J., Arffman A., Peränen J. (2002) A Rab8-specific GDP/GTP exchange factor is involved in actin remodeling and polarized membrane transport. Mol. Biol. Cell 13, 3268–3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carney D. S., Davies B. A., Horazdovsky B. F. (2006) Vps9 domain-containing proteins. Activators of Rab5 GTPases from yeast to neurons. Trends Cell Biol. 16, 27–35 [DOI] [PubMed] [Google Scholar]
- 9. Cai Y., Chin H. F., Lazarova D., Menon S., Fu C., Cai H., Sclafani A., Rodgers D. W., De La Cruz E. M., Ferro-Novick S., Reinisch K. M. (2008) The structural basis for activation of the Rab Ypt1p by the TRAPP membrane-tethering complexes. Cell 133, 1202–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gerondopoulos A., Langemeyer L., Liang J.-R., Linford A., Barr F. A. (2012) BLOC-3 mutated in Hermansky-Pudlak syndrome is a Rab32/38 guanine nucleotide exchange factor. Curr. Biol. 22, 2135–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nordmann M., Cabrera M., Perz A., Bröcker C., Ostrowicz C., Engelbrecht-Vandré S., Ungermann C. (2010) The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr. Biol. 20, 1654–1659 [DOI] [PubMed] [Google Scholar]
- 12. Wada M., Nakanishi H., Satoh A., Hirano H., Obaishi H., Matsuura Y., Takai Y. (1997) Isolation and characterization of a GDP/GTP exchange protein specific for the Rab3 subfamily small G proteins. J. Biol. Chem. 272, 3875–3878 [DOI] [PubMed] [Google Scholar]
- 13. Mahoney T. R., Liu Q., Itoh T., Luo S., Hadwiger G., Vincent R., Wang Z.-W., Fukuda M., Nonet M. L. (2006) Regulation of synaptic transmission by RAB-3 and RAB-27 in Caenorhabditis elegans. Mol. Biol. Cell 17, 2617–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sato M., Sato K., Liou W., Pant S., Harada A., Grant B. D. (2008) Regulation of endocytic recycling by C. elegans Rab35 and its regulator RME-4, a coated-pit protein. EMBO J. 27, 1183–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Allaire P. D., Marat A. L., Dall'Armi C., Di Paolo G., McPherson P. S., Ritter B. (2010) The Connecdenn DENN domain. A GEF for Rab35 mediating cargo-specific exit from early endosomes. Mol. Cell 37, 370–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yoshimura S., Gerondopoulos A., Linford A., Rigden D. J., Barr F. A. (2010) Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J. Cell Biol. 191, 367–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sano H., Peck G. R., Kettenbach A. N., Gerber S. A., Lienhard G. E. (2011) Insulin-stimulated GLUT4 protein translocation in adipocytes requires the Rab10 guanine nucleotide exchange factor Dennd4C. J. Biol. Chem. 286, 16541–16545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Delprato A., Merithew E., Lambright D. G. (2004) Structure, exchange determinants, and family-wide rab specificity of the tandem helical bundle and Vps9 domains of Rabex-5. Cell 118, 607–617 [DOI] [PubMed] [Google Scholar]
- 19. Hunziker W., Peters P. J. (1998) Rab17 localizes to recycling endosomes and regulates receptor-mediated transcytosis in epithelial cells. J. Biol. Chem. 273, 15734–15741 [DOI] [PubMed] [Google Scholar]
- 20. Zacchi P., Stenmark H., Parton R. G., Orioli D., Lim F., Giner A., Mellman I., Zerial M., Murphy C. (1998) Rab17 regulates membrane trafficking through apical recycling endosomes in polarized epithelial cells. J. Cell Biol. 140, 1039–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mori Y., Matsui T., Furutani Y., Yoshihara Y., Fukuda M. (2012) Small GTPase Rab17 regulates dendritic morphogenesis and postsynaptic development of hippocampal neurons. J. Biol. Chem. 287, 8963–8973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Horiuchi H., Lippé R., McBride H. M., Rubino M., Woodman P., Stenmark H., Rybin V., Wilm M., Ashman K., Mann M., Zerial M. (1997) A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell 90, 1149–1159 [DOI] [PubMed] [Google Scholar]
- 23. Otomo A., Hadano S., Okada T., Mizumura H., Kunita R., Nishijima H., Showguchi-Miyata J., Yanagisawa Y., Kohiki E., Suga E., Yasuda M., Osuga H., Nishimoto T., Narumiya S., Ikeda J.-E. (2003) ALS2, a novel guanine nucleotide exchange factor for the small GTPase Rab5, is implicated in endosomal dynamics. Hum. Mol. Genet. 12, 1671–1687 [DOI] [PubMed] [Google Scholar]
- 24. Fukuda M., Mikoshiba K. (1999) A novel alternatively spliced variant of synaptotagmin VI lacking a transmembrane domain. Implications for distinct functions of the two isoforms. J. Biol. Chem. 274, 31428–31434 [DOI] [PubMed] [Google Scholar]
- 25. Fukuda M., Kanno E., Mikoshiba K. (1999) Conserved N-terminal cysteine motif is essential for homo- and heterodimer formation of synaptotagmins III, V, VI, and X. J. Biol. Chem. 274, 31421–31427 [DOI] [PubMed] [Google Scholar]
- 26. James P., Halladay J., Craig E. A. (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fukuda M., Kobayashi H., Ishibashi K., Ohbayashi N. (2011) Genome-wide investigation of the Rab binding activity of RUN domains. Development of a novel tool that specifically traps GTP-Rab35. Cell Struct. Funct. 36, 155–170 [DOI] [PubMed] [Google Scholar]
- 28. Fukuda M., Kojima T., Aruga J., Niinobe M., Mikoshiba K. (1995) Functional diversity of C2 domains of synaptotagmin family. Mutational analysis of inositol high polyphosphate binding domain. J. Biol. Chem. 270, 26523–26527 [DOI] [PubMed] [Google Scholar]
- 29. Kuroda T. S., Fukuda M., Ariga H., Mikoshiba K. (2002) The Slp homology domain of synaptotagmin-like proteins 1–4 and Slac2 functions as a novel Rab27A binding domain. J. Biol. Chem. 277, 9212–9218 [DOI] [PubMed] [Google Scholar]
- 30. Itoh T., Satoh M., Kanno E., Fukuda M. (2006) Screening for target Rabs of TBC (Tre-2/Bub2/Cdc16) domain-containing proteins based on their Rab-binding activity. Genes Cells 11, 1023–1037 [DOI] [PubMed] [Google Scholar]
- 31. Kobayashi H., Fukuda M. (2012) Rab35 regulates Arf6 activity through centaurin-β2 (ACAP2) during neurite outgrowth. J. Cell Sci. 125, 2235–2243 [DOI] [PubMed] [Google Scholar]
- 32. Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N. G., Palmer A. E., Tsien R. Y. (2004) Improved monomeric red, orange, and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22, 1567–1572 [DOI] [PubMed] [Google Scholar]
- 33. Furutani Y., Matsuno H., Kawasaki M., Sasaki T., Mori K., Yoshihara Y. (2007) Interaction between telencephalin and ERM family proteins mediates dendritic filopodia formation. J. Neurosci. 27, 8866–8876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Meijering E., Jacob M., Sarria J.-C. F., Steiner P., Hirling H., Unser M. (2004) Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A 58, 167–176 [DOI] [PubMed] [Google Scholar]
- 35. Tall G. G., Barbieri M. A., Stahl P. D., Horazdovsky B. F. (2001) Ras-activated endocytosis is mediated by the Rab5 guanine nucleotide exchange activity of RIN1. Dev. Cell 1, 73–82 [DOI] [PubMed] [Google Scholar]
- 36. Tamura K., Ohbayashi N., Maruta Y., Kanno E., Itoh T., Fukuda M. (2009) Varp is a novel Rab32/38-binding protein that regulates Tyrp1 trafficking in melanocytes. Mol. Biol. Cell 20, 2900–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lai C., Xie C., McCormack S. G., Chiang H.-C., Michalak M. K., Lin X., Chandran J., Shim H., Shimoji M., Cookson M. R., Huganir R. L., Rothstein J. D., Price D. L., Wong P. C., Martin L. J., Zhu J. J., Cai H. (2006) Amyotrophic lateral sclerosis 2-deficiency leads to neuronal degeneration in amyotrophic lateral sclerosis through altered AMPA receptor trafficking. J. Neurosci. 26, 11798–11806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hadano S., Hand C. K., Osuga H., Yanagisawa Y., Otomo A., Devon R. S., Miyamoto N., Showguchi-Miyata J., Okada Y., Singaraja R., Figlewicz D. A., Kwiatkowski T., Hosler B. A., Sagie T., Skaug J., Nasir J., Brown R. H., Jr., Scherer S. W., Rouleau G. A., Hayden M. R., Ikeda J.-E. (2001) A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nat. Genet. 29, 166–173 [DOI] [PubMed] [Google Scholar]
- 39. Yang Y., Hentati A., Deng H. X., Dabbagh O., Sasaki T., Hirano M., Hung W. Y., Ouahchi K., Yan J., Azim A. C., Cole N., Gascon G., Yagmour A., Ben-Hamida M., Pericak-Vance M., Hentati F., Siddique T. (2001) The gene encoding alsin, a protein with three guanine-nucleotide exchange factor domains, is mutated in a form of recessive amyotrophic lateral sclerosis. Nat. Genet. 29, 160–165 [DOI] [PubMed] [Google Scholar]
- 40. Gros-Louis F., Meijer I. A., Hand C. K., Dubé M.-P., MacGregor D. L., Seni M.-H., Devon R. S., Hayden M. R., Andermann F., Andermann E., Rouleau G. A. (2003) An ALS2 gene mutation causes hereditary spastic paraplegia in a Pakistani kindred. Ann. Neurol. 53, 144–145 [DOI] [PubMed] [Google Scholar]
- 41. Wang L., Liang Z., Li G. (2011) Rab22 controls NGF signaling and neurite outgrowth in PC12 cells. Mol. Biol. Cell 22, 3853–3860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sato M., Sato K., Fonarev P., Huang C.-J., Liou W., Grant B. D. (2005) Caenorhabditis elegans RME-6 is a novel regulator of RAB-5 at the clathrin-coated pit. Nat. Cell Biol. 7, 559–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Burgo A., Sotirakis E., Simmler M.-C., Verraes A., Chamot C., Simpson J. C., Lanzetti L., Proux-Gillardeaux V., Galli T. (2009) Role of Varp, a Rab21 exchange factor and TI-VAMP/VAMP7 partner, in neurite growth. EMBO Rep. 10, 1117–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ohbayashi N., Yatsu A., Tamura K., Fukuda M. (2012) The Rab21-GEF activity of Varp, but not its Rab32/38 effector function, is required for dendrite formation in melanocytes. Mol. Biol. Cell 23, 669–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang X., He X., Fu X. Y., Chang Z. (2006) Varp is a Rab21 guanine nucleotide exchange factor and regulates endosome dynamics. J. Cell Sci. 119, 1053–1062 [DOI] [PubMed] [Google Scholar]