Background: The expanded C9orf72 (GGGGCC)n repeat causes amyotrophic lateral sclerosis and frontotemporal dementia.
Results: The r(GGGGCC)n repeat forms stable, tract length-dependent unimolecular and multimolecular RNA G-quadruplexes.
Conclusion: G-quadruplex formation by the r(GGGGCC)n repeats may contribute to normal and pathogenic functions of C9orf72.
Significance: Understanding the RNA structures formed by this repeat may have implications for how repeat expansion causes disease.
Keywords: Amyotrophic Lateral Sclerosis (Lou Gehrig Disease), Neurodegenerative Diseases, Nucleotide Repeat Disease, RNA Structure, RNA-Protein Interaction, ALS-FTD, C9ORF72, G-quadruplex, Multimolecular, Translation
Abstract
Certain DNA and RNA sequences can form G-quadruplexes, which can affect promoter activity, genetic instability, RNA splicing, translation, and neurite mRNA localization. Amyotrophic lateral sclerosis and frontotemporal dementia were recently shown to be caused by expansion of a (GGGGCC)n·(GGCCCC)n repeat in the C9orf72 gene. Mutant r(GGGGCC)n-containing transcripts aggregate in nuclear foci possibly sequestering repeat-binding proteins, suggesting a toxic RNA pathogenesis. We demonstrate that the r(GGGGCC)n RNA but not the C-rich r(GGCCCC)n RNA forms extremely stable uni- and multimolecular parallel G-quadruplex structures (up to 95 °C). Multimolecular G-quadruplex formation is influenced by repeat number and RNA concentration. MBNL1, a splicing factor that is sequestered in myotonic dystrophy patients by binding to expanded r(CUG)n repeat hairpins, does not bind the C9orf72 repeats, but the splicing factor ASF/SF2 can bind the r(GGGGCC)n repeat. Because multimolecular G-quadruplexes are enhanced by repeat length, RNA-RNA interactions facilitated by G-quadruplex formation at expanded repeats might influence transcript aggregation and foci formation in amyotrophic lateral sclerosis-frontotemporal dementia cells. Tract length-dependent G-quadruplex formation by the C9orf72 RNA should be considered when assessing the role of this repeat in C9orf72 gene activity, protein binding, transcript foci formation, and translation of the C9orf72 product, including the noncanonical repeat-associated non-ATG translation (RAN translation) into pathologic dipeptide repeats, as well as any oligonucleotide repeat-based therapy.
Introduction
Amyotrophic lateral sclerosis (ALS),4 often referred to as Lou Gehrig disease, is a fatal neurodegenerative disorder (1). ALS is part of a spectrum of disorders including frontotemporal dementia (FTD) (2). It was recently reported that an expansion of a (GGGGCC)n·(GGCCCC)n repeat within a noncoding region of C9orf72 could cause ALS and FTD (3, 4). Unaffected individuals have 2–19 repeats, whereas affected individuals have 250–1600 repeats and can show symptoms with as few as 20–22 repeats (5). The C9orf72 transcript with the expanded repeat was shown to aberrantly aggregate into RNA nuclear foci (4). This suggested that ALS-FTD may share a toxic RNA pathogenic path with myotonic dystrophy whose r(CUG)n repeat-containing RNA sequesters MBNL1 proteins bound to the expanded repeat. The C9orf72 repeat expansion is the most frequent cause of inherited ALS-FTD (3, 4).
Expansion of gene-specific repeats is the causative mutation of numerous neurological and neuromuscular diseases including myotonic dystrophy type 1 (DM1) fragile X type A, fragile X tremor ataxia syndrome (FXTAS), Huntington disease, and many spinocerebellar ataxias (6). ALS-FTD is the most recent member of the DNA repeat expansion diseases. There are multiple mechanisms through which an expanded repeat tract can cause disease. Expansion of a (CTG)n·(CAG)n repeat in the myotonic dystrophy protein kinase (DMPK) gene results in the formation of long, stable RNA hairpin structures formed by the expanded r(CUG)n repeat, which binds to and sequesters the muscleblind-like (MBNL) family of splicing regulators to nuclear foci (7, 8). Loss of MBNL results in mis-splicing of its many target RNAs, leading to pathogenesis (8). Expansion of the (CGG)n·(CCG)n repeat in the FMR1 gene can lead to three distinct syndromes, fragile X mental retardation, primary ovarian insufficiency, and fragile X-associated ataxia, where the latter is thought be mediated through sequestration of RNA-binding proteins bound to a toxic CGG-containing RNA found in nuclear foci in FXTAS brains (9). Expansions of coding CAG repeats, as occur in Huntington disease and several spinocerebellar ataxias, lead to toxic polyglutamine proteins, but some polyglutamine diseases are also thought to have toxic CAG RNAs (10). An underlying feature of all repeat diseases is the expansion of an (often bidirectionally) transcribed repeat tract that has the potential to form unusual DNA and RNA structures (6, 8, 11).
The G-rich nature of the ALS-FTD repeat makes it a candidate for secondary structure formation (6). G-rich DNA and RNAs can form unusual structures called G-quadruplexes in vitro as well as in vivo (see Fig. 1) (12–14). In G-quadruplexes, four guanines interact through Hoogsteen bonding in a planar configuration around a central monovalent cation to form G-quartets (see Fig. 1, A–C) (12, 13). Guanine residues can interact with other guanines of the same nucleic acid molecule to form unimolecular G-quadruplexes (see Fig. 1B) or with guanines from separate molecules to form multimolecular G-quadruplexes (see Fig. 1C) (12, 13). G-quadruplexes are associated with biological processes including genetic instability, telomere regulation, gene regulation, immunoglobulin class switch recombination, splicing, and RNA translation regulation (15–19).
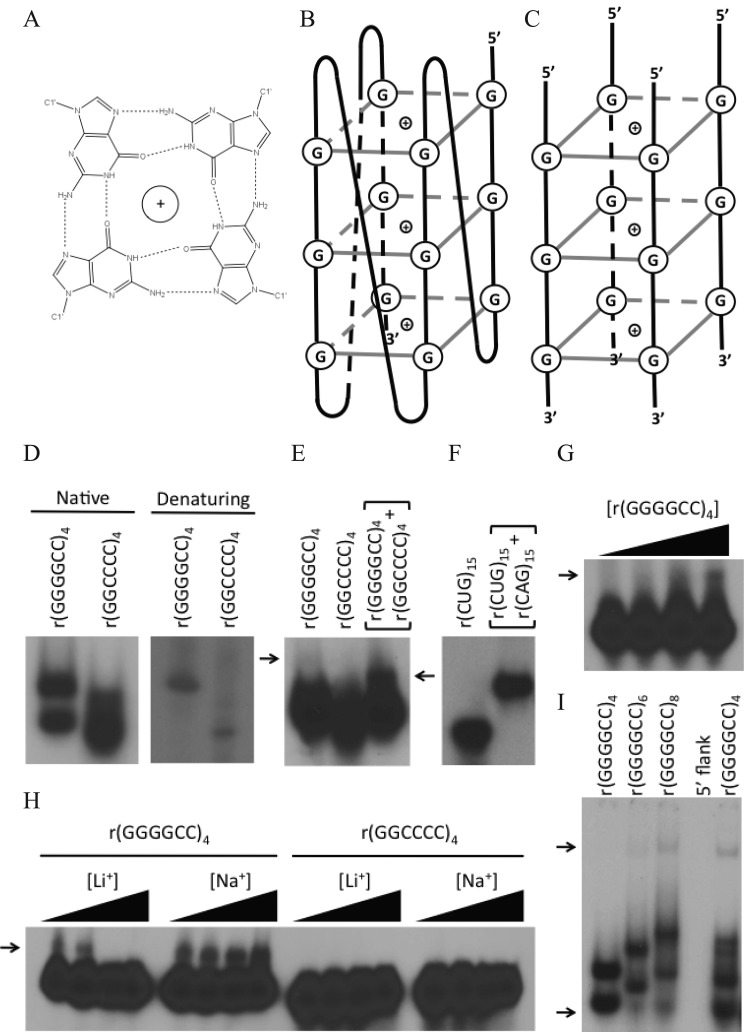
FIGURE 1.
Electrophoretic migration of the ALS-FTD RNA repeats. A, schematic of G-quartet with 4 guanine residues interacting through Hoogsteen bonding (dashed lines), stabilized by a potassium or sodium ion. B, three G-quartets are shown to form a unimolecular parallel G-quadruplex where guanines interact with each other within the same molecule. C, three G-quartets are shown to form a multimolecular G-quadruplex from four nucleic acid molecules. Other configurations are also possible. D, migration of [γ-32P]ATP end-labeled RNA (frozen and thawed at room temperature) in native 6% polyacrylamide gel or in 6% denaturing 8 m urea sequencing gel. E, migration of [γ-32P]ATP end-labeled r(GGGGCC)4 or r(CCCCGG)4 samples or a mixture of the two (heated at 95 °C and cooled at room temperature for 5 h) on a 6% polyacrylamide gel. F, migration of [γ-32P]ATP end-labeled sense strand r(CUG)15 or labeled sense strand with unlabeled antisense strand r(CAG)15 oligonucleotides in a 6% polyacrylamide gel. G, RNA concentration effect on the slow migrating r(GGGGCC)4 product. 100 fmol of [γ-32P]ATP end-labeled r(GGGGCC)4 oligonucleotides were heated at 95 °C with 0, 25, 50, or 100 pmol of unlabeled r(GGGGCC)4 and incubated at room temperature for 5 h, transferred to 4 °C, and stored overnight. Samples were subsequently chilled on ice for 1 h prior to electrophoresis in an 8% native polyacrylamide gel. The slow migrating species is indicated by an arrow. H, cation effect on the slow migrating r(GGGGCC)4 product. [γ-32P]ATP end-labeled RNA oligonucleotides were heated at 95 °C and incubated at room temperature for 1 h with increasing amounts of LiCl and NaCl (0, 10, 100, and 1000 mm). Slow migrating species are indicated by arrows. I, repeat length and flanking sequence effect on structure formation. [γ-32P]ATP end-labeled r(GGGGCC)n (n = 4, 6, or 8) repeat units or n = 4 + 15 nucleotides of genomic flank (r(AGGAGUCGCGCGCUA)r(GGGGCC)4) in 100 mm KCl were electrophoretically separated on 6% native polyacrylamide. Arrows indicate the fast and slow migrating species present in the n = 6- and 8-repeat RNAs.
Here we report formation of extremely stable uni- and multimolecular G-quadruplex structures in the RNA sequences of the sense ALS-FTD r(GGGGCC)n repeat, but not the complementary C-rich strand. G-quadruplex formation and complexity are affected by the number of repeats as well as the flanking sequence. Neither r(GGGGCC)n nor r(GGCCCC)n repeats are able to bind MBNL1 protein, but r(GGGGCC)n can bind the ASF/SF2 splicing regulator. Our findings suggest that G-quadruplex formation by the r(GGGGCC)n repeat tract from C9orf72 may shed light on normal and pathogenic roles of the repeat.
EXPERIMENTAL PROCEDURES
RNA Oligonucleotide Synthesis and Labeling
Oligonucleotides (Invitrogen) were heated to 95 °C and placed on ice. Oligonucleotides were end-labeled using [γ-32P]ATP. Equal sample amounts, based on Cerenkov counting (10,000 cpm/sample), were electrophoresed on 6 or 8% polyacrylamide gels at 120 V for 60 min. All oligonucleotide sequences used in this study are indicated in the figures.
Circular Dichroism (CD) Spectroscopy
CD analysis in a Jasco-J-815 spectrometer used 310 μl of RNA in a 1-mm cuvette. CD melting experiments used 1 ml of sample in a 1-cm cuvette. Oligonucleotides were heated to 95 °C and cooled at room temperature in buffer containing 10 mm K2HPO4 and 80 mm KCl (pH 7.5) (total [K+] = 100 mm) unless otherwise stated. An average of three CD scans, over the wavelength range of 330–200 nm, was acquired at a scan rate of 20 nm min−1 with an 8-s response time. CD spectra were corrected for buffer contributions. Data were collected at 25 °C unless otherwise stated.
Protein Purification
MBNL1 N-terminal protein construct (pGEX-6P-1-MBNL1-N; residues 1–283) was kindly provided by Dr. Maurice Swanson (20). Protein purification was carried out as described previously (20).
Electrophoretic Mobility Shift Assays
Reactions were carried out in a 10-μl volume containing ∼10 fmol of [γ-32P]ATP end-labeled RNA (10,000 cpm) with the indicated amounts of either MBNL1 or ASF/SF2 (Abcam). MBNL1 reactions contained 50 mm Tris-HCl (pH 7.6), 100 mm NaCl, 5% glycerol, 1 mm DTT, 100 ng/μl tRNA, and 1 μg/μl BSA. ASF/SF2 reactions contained 10 mm Hepes (pH 7.4) 200 mm KCl, 20 mm NaCl, 0.05% Nonidet, 1 mm DTT, 10% glycerol, 100 ng/μl tRNA, and 1 μg/μl BSA. Reactions were incubated at room temperature for 30 min, loaded onto 6% native polyacrylamide gels, electrophoresed at 120 V for 1 h in 0.5× Tris borate-EDTA, dried, and autoradiographed.
RESULTS
The C9orf72 Repeat RNA Forms Electrophoretically Slow Migrating Structures
To determine whether the r(GGGGCC)n and r(GGCCCC)n repeats had the potential to form alternative structures, RNA oligonucleotides were assessed for their electrophoretic migration on native polyacrylamide gels. Frozen, [γ-32P]ATP end-labeled oligonucleotides were thawed at room temperature and electrophoresed on native or denaturing polyacrylamide gels. Autoradiography revealed two distinct products for r(GGGGCC)4 on a native gel but only one product on the denaturing gel, suggestive of unusual structure formation (Fig. 1D). The C-rich repeat did not display slow migrating products to the same degree (Fig. 1D). Notably, mfold predicts that r(GGGGCC)4 and r(GGCCCC)4 sequences will assume hairpins, but other structure prediction software such as EuQuad predicts G-quadruplex formation of G-rich sequences. Hairpin formation may compete with or contribute to G-quadruplex or other structure formation.
When the oligonucleotides were first heated to 95 °C for 5 min and allowed to cool for 5 h to room temperature, we observed a reduced amount of the slower migrating r(GGGGCC)4 product (Fig. 1E, top arrow). This suggests that the slower migrating species may be an intermolecular G-quadruplex that was denatured at the high temperature that did not have sufficient time to reform during cooling. Multimolecular complexes form very slowly. These putative multimolecular r(GGGGCC) structures may be sensitive to freezing as demonstrated for other G-quadruplex-forming sequences (21). Denaturation and renaturation of the r(GGGGCC)4 RNA in the presence of its complementary r(GGCCCC)4 RNA resulted in only a very small proportion of double-stranded, shifted species (Fig. 1E, lower arrow), suggesting that the vast majority of the unshifted species were in a highly stable, alternative structural state that was not available for regular Watson-Crick base pairing. The slower migrating r(GGGGCC)4 product is distinct from the double-stranded r(GGGGCC)4·r(GGCCCC)4. In contrast, when a DM1-associated r(CUG)15 hairpin is denatured and renatured in the presence of its complementary sequence, there is complete formation of a double-stranded, shifted species (Fig. 1F). Collectively, these observations indicate that the r(GGGGCC)4 RNA adopts unimolecular and multimolecular G-quadruplexes where the slower migrating products are in a multimolecular form and the faster migrating products are in unimolecular form but may also contain a proportion of stable hairpins as predicted by mfold.
Slow Migrating r(GGGGCC)4 Structure Is Sensitive to RNA Concentration
Multimolecular G-quadruplexes are dependent upon molecular concentration. To further confirm the multimolecularity of the r(GGGGCC)4 species, we performed denaturation/renaturation experiments at increasing RNA concentrations (Fig. 1G). We observed a concentration-dependent increase in the slower migrating r(GGGGCC)4 species (Fig. 1G). This is strong evidence that multimolecular G-quadruplexes are formed.
Ion Dependence of the Slow Migrating r(GGGGCC)4 Structure
Characteristically, G-quadruplexes are stabilized by potassium or sodium ions and destabilized by lithium (12, 13) (Fig. 1, A–C). The slower migrating electrophoretic species we observe formed in the presence of potassium ions, supporting its identity as a multimolecular G-quadruplex (Fig. 1, D–G). Toward confirming this, we assessed the effects of lithium or sodium ions (Fig. 1H). Lithium concentrations disfavored the slow migrating r(GGGGCC)4 species, whereas sodium enhanced it; both observations are consistent with the formation of a multimolecular G-quadruplex (Fig. 1H). No slow migrating species were observed for r(GGCCCC)4 (Fig. 1H).
Repeat Length and Flanking Sequence Affect Structure Formation
Repeat tract length is an important determinant of disease severity for repeat diseases including ALS-FTD (6). Increasing repeat numbers can affect G-quadruplex formation by telomere repeats and slipped DNAs and R-loops by (CAG)/(CTG) and (CGG)/(CCG) repeats (22–26). Here we analyzed increasing r(GGGGCC)n repeat lengths of n = 4, 6, and 8 units (Fig. 1I). We observed a slow migrating species for 6 and 8 r(GGGGCC)n repeats as we did for 4 repeats, but we also observed additional faster and slower migrating species with the 6 and 8 repeats not evident in the 4-repeat RNA (Fig. 1I, arrows). These products were similar to faster and slower migrating G-quadruplex species reported for telomere oligonucleotides with varying repeat numbers (22). The formation of additional and distinct electrophoretic species is consistent with the formation of uni- and multimolecular G-quadruplexes with increasing repeat length (Fig. 1I). These oligonucleotides displayed only a single species on denaturing gels, confirming the altered migration on native gels to be due to secondary structure (data not shown). Thus, increased numbers of molecules of the r(GGGGCC)4 (Fig. 1G), as well as increased repeat numbers per molecule (n = 4, 6, and 8 repeats) (Fig. 1I) increased the formation of multimolecular G-quadruplexes.
It was of interest to determine whether the r(GGGGCC)4 repeat formed G-quadruplexes in the context of C9orf72 as sequences immediately adjacent to G-quadruplex-forming sequences can cause structural alterations (27). Inclusion of 15 nucleotides of the C9orf72 gene immediately upstream of the repeat tract increased the heterogeneity of products (Fig. 1I). Multiple species are evident following native gel electrophoresis that are not present on denaturing gels (data not shown). This suggests that the r(GGGGCC)n quadruplex may be stabilized in numerous conformations in the context of the C9orf72 transcript.
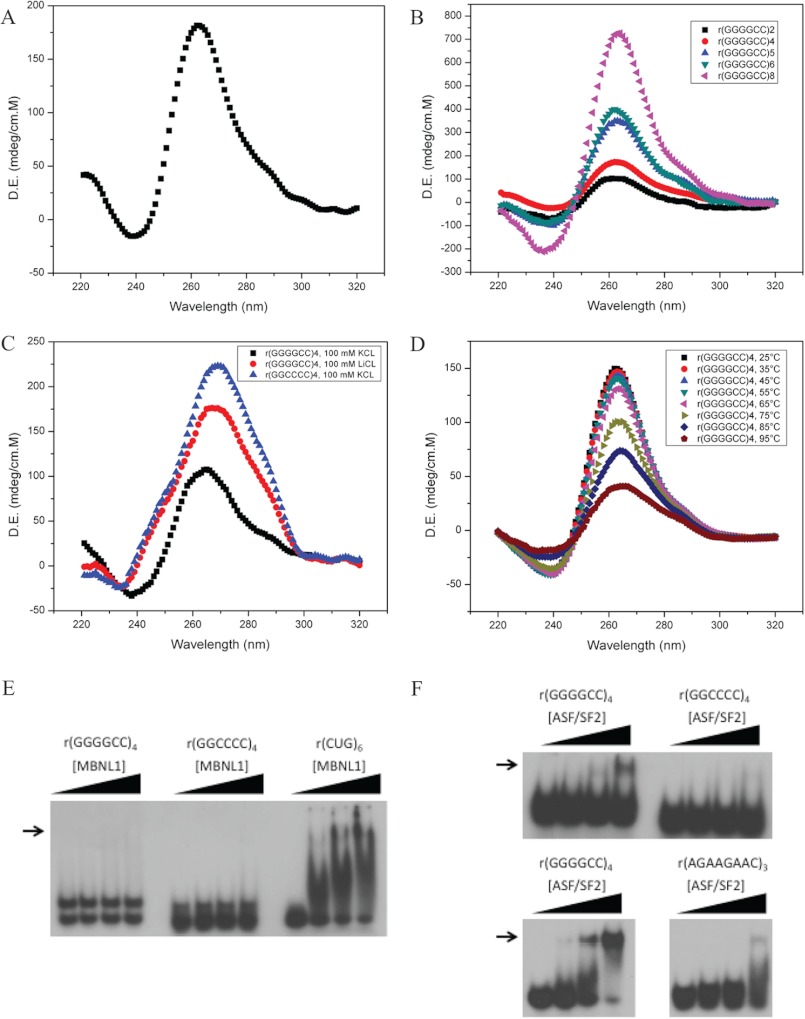
Circular Dichroism Shows That the r(GGGGCC)n Repeat Forms a Parallel G-quadruplex
CD spectroscopy is a biophysical means to assess G-quadruplex formation in RNA (28, 29). CD analysis of the r(GGGGCC)4 repeat in potassium buffer revealed a positive peak around 260 nm and a negative peak around 240 nm (Fig. 2A). These spectral features are characteristic of a parallel RNA G-quadruplex (28, 29). We observed similar CD spectra for repeat lengths of r(GGGGCC)2, -5, -6, and -8, which collectively account for ∼90% of nonexpanded repeat lengths in the population (4) (Fig. 2B). Notably, there was a repeat tract length-dependent increase in G-quadruplex-associated CD signal (higher 260 nm peak and 240 nm negative peak), further supporting repeat tract length dependence of G-quadruplex formation. The spectrum was potassium ion-dependent, characteristic of G-quadruplexes; spectra obtained in lithium revealed a shift in the spectrum, from 260 nm toward 270 nm, as well as a shift in the negative peak at 240 nm, resembling the A-form of RNA hairpins predicted by mfold (Fig. 2C). CD analysis of the antisense r(GGCCCC)4 repeat in potassium revealed a positive peak at 270 nm and no negative peak at 240 nm, indicating that r(GGCCCC)4 did not form aG-quadruplex (Fig. 2C). Again, the peak closer to 270 nm and lack of the 240 nm negative peak are more typical of A-form RNA as described previously for r(CUG)n hairpins (29) and may be similar to the mfold hairpin predictions. Thus, the CD spectra provide further support of G-quadruplex formation by r(GGGGCC)n repeats.
FIGURE 2.
CD spectra and protein binding of the ALS-FTD RNA repeats. A, CD spectrum of 5 μm r(GGGGCC)4 repeat in 100 mm potassium buffer. D.E., Delta Epsilon. mdeg, millidegrees. B, CD spectrum of 5 μm r(GGGGCC)2–8 repeats in 100 mm potassium buffer C, CD spectrum of 5 μm r(GGGGCC)4 or r(GGCCCC)4 in 100 mm KCL or 100 mm LiCl. D, CD spectrum of 6.7 μm r(GGGGCC)4 repeat in 100 mm potassium buffer at increasing temperatures. E, human MBNL1 EMSA with the indicated [γ-32P]ATP end-labeled RNA oligonucleotides (10 fmol) and 0, 1, 5, and 10 pmol of purified human MBNL1 N-terminal protein containing both sets of zinc fingers. RNA-protein complex is indicated by the arrow. F, human ASF/SF2 EMSA with the indicated [γ-32P]ATP end-labeled RNA oligonucleotides. The upper panel uses 10 fmol of RNA and 0, 1, 5, or 10 pmol of full-length human ASF/SF2 (Abcam). The lower panel uses 5 fmol of RNA and 0, 5, 10, or 20 pmol of full-length human ASF/SF2 (Abcam). RNA-protein complex is indicated by the arrow.
r(GGGGCC)4 G-quadruplex Exhibits Unusually High Thermostability
Other sequences that form G-quadruplexes show large variability in their thermal stability. To test the thermal stability of the structures formed by the r(GGGGCC)4 repeat, we measured the CD spectrum at increasing temperatures (Fig. 2D). There was a clear temperature-dependent reduction in the CD signal at higher temperatures. Surprisingly, although the CD signal decreased, the CD data did not indicate the complete dissociation of the G-quadruplex even at 95 °C (Fig. 2D). Furthermore, the repeat could not be melted even by lowering the potassium concentration to 10 mm in UV melting experiments (data not shown). Such extreme thermostability has previously been observed for G-rich quadruplex-forming sequences (30).
The r(GGGGCC)4 Repeat Does Not Bind MBNL1 but Can Bind ASF/SF2 Splicing Factor in Vitro
Structured repeat-containing RNA such as hairpins formed by pathogenic expansions of r(CUG)n repeats are targets for the splicing regulator muscleblind-like 1 (MBNL1). MBNL1 specifically interacts with r(CUG)n repeat hairpins (7, 31). To determine whether MBNL1 could recognize the structured C9orf72 RNA repeats, we performed electrophoretic mobility shift assays with purified human MBNL1 protein and radiolabeled RNA (Fig. 2E). MBNL1 did not bind either the G-rich or the C-rich repeats under conditions that it bound an r(CUG)6 repeat (Fig. 2E). This suggests that MBNL1 displays a binding preference for r(CUG)n hairpins but not r(GGGGCC)n G-quadruplexes.
G-quadruplexes in RNA have been associated with regulating alternative splicing by serving as an exonic splice enhancer (32, 33). DeJesus-Hernandez et al. (4) reported three variant splice isoforms of the C9orf72 transcript where the (GGGGCC)n repeat is situated in the promoter (v1) or the first intron between two noncoding exons (v2,3). To determine whether a splicing regulator could specifically recognize the r(GGGGCC) repeat, we performed a candidate search in ESEfinder (34), which identified the ASF/SF2 essential splicing regulator. As predicted ASF/SF2 bound the r(GGGGCC)4 repeat but not the r(GGCCCC)4 repeat (Fig. 2F). We compared binding of ASF/SF2 to r(GGGGCC)4 versus a previously reported ASF/SF2 SELEX-identified consensus sequence, r(AGAAGAAC), which when present in three copies could function as a strong splicing enhancer (35). Under identical conditions, ASF/SF2 bound both sequences with some preference for r(GGGGCC)4 over r(AGAAGAAC)3 (Fig. 2F).
DISCUSSION
In this study we provide evidence of G-quadruplex formation by the G-rich C9orf72 RNA repeat. We confirm a recent study demonstrating unimolecular G-quadruplex formation of an r(GGGGCC)4 sequence and extend this finding (36). We used oligonucleotide models having repeat lengths present in the nonaffected/nonmutant C9orf72 gene; the nonaffected population has 2–19 (GGGGCC)n units, and the affected population can have n = 20–1600 units. We demonstrate that: 1) this repeat adopts extremely stable uni- and multimolecular G-quadruplexes that are stabilized by potassium and sodium but not lithium; 2) structure formation and complexity are affected by the RNA concentration, repeat length, and flanking sequence; and 3) neither of the repeats is bound by the r(CUG)n hairpin-binding MBNL1, but can be bound in vitro by the ASF/SF2 splicing regulator.
Repeat tract length is positively correlated with disease severity in repeat expansion diseases (6). For example, in the case of DM1, inherited expansions of >100 repeats can progress as the individual ages to lengths of several thousand (CTG)n repeats, with increasing disease severity and progression for longer expansions (6). Transcription of these repeats results in toxic RNAs containing the expanded r(CUG)n repeats that fold into hairpins that sequester MBNL1 proteins into ribonuclear foci, preventing their normal activities (8). ALS-FTD patients are thought to follow the same toxic RNA pathogenesis as DM1 because they have long (GGGGCC)n expansions that, like DM1, can form ribonuclear foci, suggesting that the mutant C9orf72 transcript may be a structured toxic RNA possibly bound by proteins (3, 4). The fact that we observe that r(GGGGCC)n repeats form multimeric G-quadruplexes, whose formation is sensitive to repeat length, suggests that intermolecular G-quadruplex interactions might contribute to RNA foci formation.
G-quadruplex formation by r(GGGGCC)n repeats but not r(GGCCCC)n repeats may have implications upon their ability to be translated into aggregating dipeptide repeats in brains of patients with C9orf72 expansions (37, 38) through repeat-associated non-ATG translation (RAN translation) (39), as initially observed for trinucleotide repeat diseases by Ranum and colleagues (40). Ranum and colleagues (40) proposed that formation of hairpin structures by the expanded r(CAG)n and r(CUG)n repeats was required to initiate RAN translation. If RAN translation occurs from the opposite strand r(GGCCCC)n, it may do so through a distinct structure as we did not observe G-quadruplex formation with this sequence. It should be noted that both the C-rich and the G-rich repeat strands can potentially form hairpins as predicted by mfold, and thus, determining how each of these structures influences RAN translation leading to cellular dipeptide repeat aggregates is an important question requiring further investigation.
Proteins that recognize G-quadruplex structures may also contribute to ribonuclear aggregation. It is important to determine which proteins are binding these expanded repeats and how they influence aggregation in ALS-FTD patients harboring expanded repeats.
We found that MBNL1 does not bind the sense nor the antisense RNA repeat, suggesting that ALS-FTD may not involve MBNL1 sequestration. We cannot definitively rule out the possibility that MBNL1 may be able to interact with very long pathogenic r(GGGGCC)n repeats or be associated with ribonuclear foci via an indirect association, as in FXTAS (41). Because ALS-FTD is suspected to involve toxic RNA pathogenesis, as in DM1, the identification of proteins that bind to the expanded C9orf72 r(GGGGCC)n repeat is of great interest.
G-quadruplex formation within the repeat may contribute to the normal function of the C9orf72 gene transcript. G-quadruplexes can affect alternative splicing (33, 42). The (GGGGCC)n repeat lies within the noncoding region of the C9orf72 gene, which can be alternatively spliced to three isoforms (3, 4). The relative abundance of these splice isoforms in ALS-FTD patient tissues and cells showed a 50% reduction of v1 relative to v2 and v3 in individuals bearing an expanded repeat but not in individuals with an unexpanded repeat (4). Although preliminary, these results suggest that the expanded (GGGGCC)n repeat may potentially autoregulate C9orf72 splicing (4). Our observation that the r(GGGGCC)n repeat can assume a G-quadruplex opens the question as to whether such structures may affect C9orf72 splicing outcomes, as occurs for other genes (33, 42). We found that the splicing regulator ASF/SF2 bound in vitro to the C9orf72 repeat, as predicted based on sequence specificity (34). The role, if any, of ASF/SF2 in C9orf72 transcript metabolism including the effects of G-quadruplex formation is unknown. More research is necessary to validate the link of C9orf72 splicing with repeat expansion, the role of RNA structure, and the proteins involved.
Considerable attention has been given toward therapeutically targeting certain DNA and RNA G-quadruplex-forming sequences to modulate disease (43–45). Various small molecules have been used to target the G-quadruplex-forming sequences, to either enhance or perturb structure formation to modulate the maintenance of telomere lengths or gene activity (43–45). If the formation of RNA foci is playing a toxic role in ALS-FTD, and their formation is exacerbated in part by intermolecular multimerization of G-quadruplexes, then disruption of these foci may have beneficial outcomes. Application of G-quadruplex-interacting small molecules may modulate ALS-FTD pathogenesis.
An additional consideration of the ability of C9orf72 repeats to form G-quadruplexes is that any antisense oligonucleotide repeat-based therapeutic approach must consider the structures that the oligonucleotides will tend to assume. Notably, the formation of G-quadruplexes by GGGGCC repeats may influence the ability to form a complementary hybrid as the formation of hairpins by CTG and CAG repeats did not override hybrid formation by morpholino-based therapies for DM1 (46).
Acknowledgment
We thank Dr. Maurice Swanson for providing human MBNL1 bacterial expression constructs.
This work was supported by grants from the Amyotrophic Lateral Sclerosis Association of America (to C. E. P.), the Canadian Institutes of Health Research (CIHR) to (C. E. P.), and the Natural Science and Engineering Research Council of Canada (to R. B. M.).
- ALS
- amyotrophic lateral sclerosis
- FTD
- frontotemporal dementia
- FXTAS
- fragile X tremor ataxia syndrome
- RAN
- repeat-associated non-ATG
- DM1
- myotonic dystrophy type 1
- MBNL
- muscleblind-like.
REFERENCES
- 1. Rowland L. P., Shneider N. A. (2001) Amyotrophic lateral sclerosis. N. Engl. J. Med. 344, 1688–1700 [DOI] [PubMed] [Google Scholar]
- 2. Graff-Radford N. R., Woodruff B. K. (2007) Frontotemporal dementia. Semin. Neurol. 27, 48–57 [DOI] [PubMed] [Google Scholar]
- 3. Renton A. E., Majounie E., Waite A., Simón-Sánchez J., Rollinson S., Gibbs J. R., Schymick J. C., Laaksovirta H., van Swieten J. C., Myllykangas L., Kalimo H., Paetau A., Abramzon Y., Remes A. M., Kaganovich A., Scholz S. W., Duckworth J., Ding J., Harmer D. W., Hernandez D. G., Johnson J. O., Mok K., Ryten M., Trabzuni D., Guerreiro R. J., Orrell R. W., Neal J., Murray A., Pearson J., Jansen I. E., Sondervan D., Seelaar H., Blake D., Young K., Halliwell N., Callister J. B., Toulson G., Richardson A., Gerhard A., Snowden J., Mann D., Neary D., Nalls M. A., Peuralinna T., Jansson L., Isoviita V. M., Kaivorinne A. L., Hölttä-Vuori M., Ikonen E., Sulkava R., Benatar M., Wuu J., Chiò A., Restagno G., Borghero G., Sabatelli M., Heckerman D., Rogaeva E., Zinman L., Rothstein J. D., Sendtner M., Drepper C., Eichler E. E., Alkan C., Abdullaev Z., Pack S. D., Dutra A., Pak E., Hardy J., Singleton A., Williams N. M., Heutink P., Pickering-Brown S., Morris H. R., Tienari P. J., Traynor B. J. (2011) A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. DeJesus-Hernandez M., Mackenzie I. R., Boeve B. F., Boxer A. L., Baker M., Rutherford N. J., Nicholson A. M., Finch N. A., Flynn H., Adamson J., Kouri N., Wojtas A., Sengdy P., Hsiung G. Y., Karydas A., Seeley W. W., Josephs K. A., Coppola G., Geschwind D. H., Wszolek Z. K., Feldman H., Knopman D. S., Petersen R. C., Miller B. L., Dickson D. W., Boylan K. B., Graff-Radford N. R., Rademakers R. (2011) Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gómez-Tortosa E., Gallego J., Guerrero-López R., Marcos A., Gil-Neciga E., Sainz M. J., Díaz A., Franco-Macías E., Trujillo-Tiebas M. J., Ayuso C., Pérez-Pérez J. (2013) C9ORF72 hexanucleotide expansions of 20–22 repeats are associated with frontotemporal deterioration. Neurology 80, 366–370 [DOI] [PubMed] [Google Scholar]
- 6. Pearson C. E., Nichol Edamura K., Cleary J. D. (2005) Repeat instability: mechanisms of dynamic mutations. Nat. Rev. Genet. 6, 729–742 [DOI] [PubMed] [Google Scholar]
- 7. Miller J. W., Urbinati C. R., Teng-Umnuay P., Stenberg M. G., Byrne B. J., Thornton C. A., Swanson M. S. (2000) Recruitment of human muscleblind proteins to (CUG)n expansions associated with myotonic dystrophy. EMBO J. 19, 4439–4448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shin J., Charizanis K., Swanson M. S. (2009) Pathogenic RNAs in microsatellite expansion disease. Neurosci. Lett. 466, 99–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Willemsen R., Levenga J., Oostra B. A. (2011) CGG repeat in the FMR1 gene: size matters. Clin. Genet. 80, 214–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wojciechowska M., Krzyzosiak W. J. (2011) CAG repeat RNA as an auxiliary toxic agent in polyglutamine disorders. RNA Biol. 8, 565–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Batra R., Charizanis K., Swanson M. S. (2010) Partners in crime: bidirectional transcription in unstable microsatellite disease. Hum. Mol. Genet. 19, R77–R82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Neidle S. (2009) The structures of quadruplex nucleic acids and their drug complexes. Curr. Opin. Struct. Biol. 19, 239–250 [DOI] [PubMed] [Google Scholar]
- 13. Bugaut A., Balasubramanian S. (2012) 5′-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res. 40, 4727–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Biffi G., Tannahill D., McCafferty J., Balasubramanian S. (2013) Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 5, 182–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lopes J., Piazza A., Bermejo R., Kriegsman B., Colosio A., Teulade-Fichou M. P., Foiani M., Nicolas A. (2011) G-quadruplex-induced instability during leading-strand replication. EMBO J. 30, 4033–4046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Piazza A., Boulé J. B., Lopes J., Mingo K., Largy E., Teulade-Fichou M. P., Nicolas A. (2010) Genetic instability triggered by G-quadruplex interacting Phen-DC compounds in Saccharomyces cerevisiae. Nucleic Acids Res. 38, 4337–4348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wu Y., Brosh R. M., Jr. (2010) G-quadruplex nucleic acids and human disease. FEBS J. 277, 3470–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lipps H. J., Rhodes D. (2009) G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 19, 414–422 [DOI] [PubMed] [Google Scholar]
- 19. Melko M., Bardoni B. (2010) The role of G-quadruplex in RNA metabolism: involvement of FMRP and FMR2P. Biochimie 92, 919–926 [DOI] [PubMed] [Google Scholar]
- 20. Yuan Y., Compton S. A., Sobczak K., Stenberg M. G., Thornton C. A., Griffith J. D., Swanson M. S. (2007) Muscleblind-like 1 interacts with RNA hairpins in splicing target and pathogenic RNAs. Nucleic Acids Res. 35, 5474–5486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhai Q., Deng M., Xu L., Zhang X., Zhou X. (2012) Promoting the formation of tetramolecular G-quadruplexes under freezing condition. Bioorg. Med. Chem. Lett. 22, 1142–1145 [DOI] [PubMed] [Google Scholar]
- 22. Pedroso I. M., Duarte L. F., Yanez G., Burkewitz K., Fletcher T. M. (2007) Sequence specificity of inter- and intramolecular G-quadruplex formation by human telomeric DNA. Biopolymers 87, 74–84 [DOI] [PubMed] [Google Scholar]
- 23. Abu-Ghazalah R. M., Macgregor R. B., Jr. (2009) Structural polymorphism of the four-repeat Oxytricha nova telomeric DNA sequences. Biophys. Chem. 141, 180–185 [DOI] [PubMed] [Google Scholar]
- 24. Víglaský V., Bauer L., Tlucková K. (2010) Structural features of intra- and intermolecular G-quadruplexes derived from telomeric repeats. Biochemistry 49, 2110–2120 [DOI] [PubMed] [Google Scholar]
- 25. Reddy K., Tam M., Bowater R. P., Barber M., Tomlinson M., Nichol Edamura K., Wang Y. H., Pearson C. E. (2011) Determinants of R-loop formation at convergent bidirectionally transcribed trinucleotide repeats. Nucleic Acids Res. 39, 1749–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pearson C. E., Sinden R. R. (1996) Alternative structures in duplex DNA formed within the trinucleotide repeats of the myotonic dystrophy and fragile X loci. Biochemistry 35, 5041–5053 [DOI] [PubMed] [Google Scholar]
- 27. Arora A., Nair D. R., Maiti S. (2009) Effect of flanking bases on quadruplex stability and Watson-Crick duplex competition. FEBS J. 276, 3628–3640 [DOI] [PubMed] [Google Scholar]
- 28. Kumari S., Bugaut A., Huppert J. L., Balasubramanian S. (2007) An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 3, 218–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sobczak K., Michlewski G., de Mezer M., Kierzek E., Krol J., Olejniczak M., Kierzek R., Krzyzosiak W. J. (2010) Structural diversity of triplet repeat RNAs. J. Biol. Chem. 285, 12755–12764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Merkina E. E., Fox K. R. (2005) Kinetic stability of intermolecular DNA quadruplexes. Biophys. J. 89, 365–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kino Y., Mori D., Oma Y., Takeshita Y., Sasagawa N., Ishiura S. (2004) Muscleblind protein, MBNL1/EXP, binds specifically to CHHG repeats. Hum. Mol. Genet. 13, 495–507 [DOI] [PubMed] [Google Scholar]
- 32. Didiot M. C., Tian Z., Schaeffer C., Subramanian M., Mandel J. L., Moine H. (2008) The G-quartet containing FMRP binding site in FMR1 mRNA is a potent exonic splicing enhancer. Nucleic Acids Res. 36, 4902–4912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fisette J. F., Montagna D. R., Mihailescu M. R., Wolfe M. S. (2012) A G-rich element forms a G-quadruplex and regulates BACE1 mRNA alternative splicing. J. Neurochem. 121, 763–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cartegni L., Wang J., Zhu Z., Zhang M. Q., Krainer A. R. (2003) ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 31, 3568–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tacke R., Manley J. L. (1995) The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 14, 3540–3551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fratta P., Mizielinska S., Nicoll A. J., Zloh M., Fisher E. M., Parkinson G., Isaacs A. M. (2012) C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci. Rep. 2, 1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mori K., Weng S. M., Arzberger T., May S., Rentzsch K., Kremmer E., Schmid B., Kretzschmar H. A., Cruts M., Van Broeckhoven C., Haass C., Edbauer D. (February 7, 2013) The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 10.1126/science.1232927 [DOI] [PubMed] [Google Scholar]
- 38. Ash P. E. A., Bieniek K. F., Gendron T. F., Caulfield T., Lin W.-L., DeJesus-Hernandez M., van Blitterswijk M. M., Jansen-West K., Paul J. W., 3rd, Rademakers R., Boylan K. B., Dickson D. W., Petrucelli L. (2013) Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77, 639–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pearson C. E. (2011) Repeat associated non-ATG translation initiation: one DNA, two transcripts, seven reading frames, potentially nine toxic entities! PLoS Genet. 7, e1002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zu T., Gibbens B., Doty N. S., Gomes-Pereira M., Huguet A., Stone M. D., Margolis J., Peterson M., Markowski T. W., Ingram M. A., Nan Z., Forster C., Low W. C., Schoser B., Somia N. V., Clark H. B., Schmechel S., Bitterman P. B., Gourdon G., Swanson M. S., Moseley M., Ranum L. P. (2011) Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl. Acad. Sci. U.S.A. 108, 260–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sellier C., Rau F., Liu Y., Tassone F., Hukema R. K., Gattoni R., Schneider A., Richard S., Willemsen R., Elliott D. J., Hagerman P. J., Charlet-Berguerand N. (2010) Sam68 sequestration and partial loss of function are associated with splicing alterations in FXTAS patients. EMBO J. 29, 1248–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Marcel V., Tran P. L., Sagne C., Martel-Planche G., Vaslin L., Teulade-Fichou M. P., Hall J., Mergny J. L., Hainaut P., Van Dyck E. (2011) G-quadruplex structures in TP53 intron 3: role in alternative splicing and in production of p53 mRNA isoforms. Carcinogenesis 32, 271–278 [DOI] [PubMed] [Google Scholar]
- 43. Siddiqui-Jain A., Grand C. L., Bearss D. J., Hurley L. H. (2002) Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 99, 11593–11598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Neidle S. (2010) Human telomeric G-quadruplex: the current status of telomeric G-quadruplexes as therapeutic targets in human cancer. FEBS J. 277, 1118–1125 [DOI] [PubMed] [Google Scholar]
- 45. Faudale M., Cogoi S., Xodo L. E. (2012) Photoactivated cationic alkyl-substituted porphyrin binding to g4-RNA in the 5′-UTR of KRAS oncogene represses translation. Chem. Commun. (Camb.) 48, 874–876 [DOI] [PubMed] [Google Scholar]
- 46. Wheeler T. M., Sobczak K., Lueck J. D., Osborne R. J., Lin X., Dirksen R. T., Thornton C. A. (2009) Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science 325, 336–339 [DOI] [PMC free article] [PubMed] [Google Scholar]