Background: During endochondral ossification, cells in the perichondrium give rise to osteoblast precursors.
Results: Bone morphogenetic protein (BMP) interacted with hedgehog (Hh) to enhance osteogenesis, whereas in the absence of Hh, BMP enhanced ectopic chondrogenesis in the perichondrium.
Conclusion: Hh alters the function of BMP to specify perichondrial cells into osteoblasts.
Significance: This provides an insight into signaling network in osteogenesis.
Keywords: Bone Morphogenetic Protein (BMP), Chondrocytes, Development, Hedgehog, Osteoblasts, Lineage Specification, Perichondrium
Abstract
Specification of progenitors into the osteoblast lineage is an essential event for skeletogenesis. During endochondral ossification, cells in the perichondrium give rise to osteoblast precursors. Hedgehog (Hh) and bone morphogenetic protein (BMP) are suggested to regulate the commitment of these cells. However, properties of perichondrial cells and regulatory mechanisms of the specification process are still poorly understood. Here, we investigated the machineries by combining a novel organ culture system and single-cell expression analysis with mouse genetics and biochemical analyses. In a metatarsal organ culture reproducing bone collar formation, activation of BMP signaling enhanced the bone collar formation cooperatively with Hh input, whereas the signaling induced ectopic chondrocyte formation in the perichondrium without Hh input. Similar phenotypes were also observed in compound mutant mice, where signaling activities of Hh and BMP were genetically manipulated. Single-cell quantitative RT-PCR analyses showed heterogeneity of perichondrial cells in terms of natural characteristics and responsiveness to Hh input. In vitro analyses revealed that Hh signaling suppressed BMP-induced chondrogenic differentiation; Gli1 inhibited the expression of Sox5, Sox6, and Sox9 (SRY box-containing gene 9) as well as transactivation by Sox9. Indeed, ectopic expression of chondrocyte maker genes were observed in the perichondrium of metatarsals in Gli1−/− fetuses, and the phenotype was more severe in Gli1−/−;Gli2−/− newborns. These data suggest that Hh-Gli activators alter the function of BMP to specify perichondrial cells into osteoblasts; the timing of Hh input and its target populations are critical for BMP function.
Introduction
Specification of progenitors into a certain lineage is an essential event for organogenesis. In mammals, appendicular and axial skeletons develop through endochondral ossification, one of the two ossification processes in vertebrates (1). In an early phase of the process, two distinct cell populations arise shortly after the condensation of mesenchymal cells: cartilage-forming chondrocytes and perichondrial cells (1). Sox9 (SRY box-containing gene 9) has been shown to be essential for the mesenchymal condensations and subsequent cartilage formation (2, 3). Perichondrial cells constitute the perichondrium, a thin layer of fibroblastic cells surrounding the cartilage mold. During normal development, a population of the perichondrial cells is specified into an osteoblast lineage; they differentiate into osteoblasts through several stages of precursors, contributing to formation of the bone collar, a predecessor of cortical bones (1, 4). Runx2 (Runt-related transcription factor) and Osx (osterix), two essential transcription factors for osteoblast differentiation, are known to sequentially regulate the differentiation program (5–7).
Several lines of evidence have suggested that Hh,3 Wnt, and BMP signaling pathways act as a switch for the program in perichondrial cells and that a bipotential cell population exists in the perichondrium (8–10). We and others have clarified crucial roles of Hh signaling in an early commitment and differentiation of osteoblasts in the perichondrium (8, 11–13). Defects in Wnt/β-catenin signaling as well as the absence of Hh input lead to a complete lack of osteoblasts, allowing perichondrial cells to adopt a chondrocyte lineage (8, 9, 14, 15). On the other hand, forced activation of BMP signaling has been shown to cause ectopic chondrocytes in the perichondrium (16). Although Wnt/β-catenin signaling acts downstream of Hh signaling in the specification of osteoblasts in the perichondrium (9), the hierarchy or interaction of BMP and Hh signaling pathways in the specification has not been clarified. In addition, there have been no data that advance our understanding of the characteristics of perichondrial cells at the single-cell level.
The present study was aimed to elucidate molecular mechanisms underlying the specification of an osteoblast lineage in perichondrial cells, with a particular focus on the interaction between Hh and BMP signaling pathways, through an organ culture system and mouse genetics as well as biochemical approaches. In addition, to gain insight into the specification process, we investigated the expression of osteoblast and chondrocyte marker genes in primary perichondrial cells at the single-cell level.
EXPERIMENTAL PROCEDURES
Animals
Ptch1tm1Mps (Ptch1+/−), Tg(Prrx1-cre)1Cjt (Prx1-Cre), and Ihhtm1Amc (Ihh+/−) mice were obtained from The Jackson Laboratory, wild-type (WT) C57BL/6J mice were from Charles River Japan, and CAG-LoxP-caBMPr1a, Gli1+/−, and Gli2+/− mice were generated as described previously (17–19). All experiments were performed in accordance with the protocol approved by the Animal Care and Use Committee of The University of Tokyo.
Reagents
Smoothened agonist (SAG) was purchased from Calbiochem; cyclopamine was from Biomol; LiCl and dorsomorphin were from Sigma-Aldrich; and IWP2 was from Tocris Bioscience. recombinant human TGFβ1 (rhTGFβ1) from R&D Systems; recombinant human BMP2 (rhBMP2) was provided by Astellas Pharma, Inc.
Metatarsal Organ Culture
Embryonic metatarsals were dissected as described previously (20). For the osteogenic culture, each metatarsal was cultured in 0.25% FBS/high glucose Dulbecco's modified Eagle's medium (Sigma-Aldrich) containing 1% penicillin/streptomycin, 50 μg/ml ascorbic acid phosphate, 10 mm β-glycerophosphate, and 0.1 μm dexamethasone.
Cell Culture
C3H10T1/2 cells were obtained from the RIKEN Cell Bank. Perichondrial cells were isolated as described previously (11). Plasmid transfection was performed using FuGENE HD (Roche).
Single-cell Analyses
Primary perichondrial cells treated with DMSO or SAG (1 μm) for 5 days were collected by a capillary tip after trypsin treatment. cDNA libraries from a single-cell were prepared by using oligo(dT)30-immobilized beads and gene expressions were quantified by qPCR as described previously (21). Hierarchical clustering was performed with Mathematica software (version 7.0, Wolfram Research, Inc.). The detailed procedures and primer sequences are described in supplemental “Experimental Procedures.”
qRT-PCR
Total RNA extraction, reverse-transcription, and qPCR were performed as described previously (11). The primer sequences are available upon request.
Histology
The procedures for von Kossa staining, safranin O staining, and in situ hybridization have been described previously (11, 20). Images were taken using an Axio Imajor A1 microscope (Carl Zeiss).
Luciferase Assay
Cells were plated onto 24-well plates and transfected with 0.4 μg of DNA mixture containing the test reporter plasmids, control reporter plasmids encoding Renilla luciferase, and effector plasmids. A Dual-Luciferase assay was performed as described previously (11).
ChIP
ChIP was performed with a One-Day ChIP kit (Diagenode). To shear genomic DNA, a Shearing ChIP kit (Diagenode) was used according to the manufacturer's instructions. The primer sequences are available upon request.
Statistical Analysis
The means of groups were compared by analysis of variance, and the significance of differences was determined by post hoc testing using Tukey's method.
RESULTS
Development of an Organ Culture System That Enables the Observation of Bone Collar Formation in a Near in Vivo Setting
With the ultimate aim of defining the roles of signaling pathways in osteogenesis in the perichondrium, we set out to establish an organ culture system that enabled us to analyze bone collar formation in endochondral ossification ex vivo. We found that a metatarsal organ culture using an osteogenic medium reproduced bone collar formation in synchronization with cartilage mineralization, which had not yet been achieved by the culture methods reported previously (20). To verify that it reliably reproduced phenotypes that were observed in mice with mutations in Hh, BMP, or Wnt signaling, we histologically analyzed metatarsals cultured with activators and inhibitors of the Hh, BMP, or Wnt signaling pathway. We used SAG, an Hh signaling activator; cyclopamine, an Hh signaling inhibitor; rhBMP2; dorsomorphin, a BMP signaling inhibitor (22); lithium chloride (LiCl), a glycogen synthase kinase-3 (GSK3) inhibitor; and IWP2, an inhibitor of Wnt processing and secretion (23).
SAG induced ectopic bone collar formation in the perichondrium and inhibited cartilage mineralization; cyclopamine blocked bone collar formation and accelerated cartilage mineralization (Fig. 1A). rhBMP2 did not induce ectopic bone collar formation but enhanced chondrogenesis in the growth plate; dorsomorphin did not affect osteogenesis but inhibited chondrogenesis (Fig. 1, A and B). LiCl promoted bone collar formation in synchronization with cartilage mineralization; IWP2 inhibited bone collar formation (supplemental Fig. S1A). These observations were in line with those obtained from gain- and loss-of-function mouse models for the Hh, BMP, and Wnt signaling pathways (8, 9, 12, 14–16, 22, 24, 25).
FIGURE 1.
Effects of the manipulation of Hh and BMP signaling on bone collar formation and ectopic chondrocyte formation in the perichondrium. A and B, von Kossa staining (A) and safranin O staining (B) of representative sections obtained from metatarsals cultured with DMSO, SAG (1 μm), cyclopamine (Cyc; 5 μm), rhBMP2 (100 ng/ml), dorsomorphin (Dor; 5 μm), or a combination of these agents for 7 days. C, in situ hybridization for Col2a1 of representative sections obtained from metatarsals cultured with cyclopamine (5 μm), rhBMP2 (100 ng/ml), or the combination of cyclopamine and rhBMP2 for 7 days. Higher magnification of the boxed areas in the left panels is shown in the right panels. Scale bars, 100 μm.
The integrity of the organ culture system was further verified and investigated in terms of the signaling hierarchy of Hh and Wnt and the interaction of Hh and TGFβ during the commitment of the osteoblast lineage in the perichondrium. Hh signaling is known to be required prior to the requirement of Wnt signaling in osteoblast differentiation (9). In the cultured metatarsals, Osx was expressed in the SAG-induced ectopic bone collar. Although IWP2 inhibited SAG-induced formation of ectopic bone collar, ectopic Osx expression was maintained (supplemental Fig. S1B). As for the loss-of-function of Hh signaling, cyclopamine alone suppressed both bone collar formation and Osx expression in the perichondrium. LiCl did not reverse the cyclopamine-induced suppression of Osx expression and bone collar formation in the perichondrium (supplemental Fig. S1B). TGFβ signaling is reported to be involved in the proliferation of perichondrial cells and osteoblast differentiation (26, 27). In the cultured metatarsals in the present study, rhTGFβ1 increased the thickness of the perichondrium and inhibited the SAG-induced ectopic bone collar formation (supplemental. Fig. S2). These data not only confirmed the previous reports but also suggest that TGFβ signaling inhibits Hh-induced osteogenesis. Thus, pharmacological manipulation of this organ culture system appeared to enable easier and less time-consuming investigation of the signaling network and hierarchy in the perichondrium than genetic approaches.
Hh Signaling-mediated Alteration of the BMP Function in the Perichondrium from Chondrogenic to Osteogenic
By taking advantage of the present organ culture system described above, we next attempted to clarify the signaling network between Hh and BMP in the perichondrium; we examined the effects of combined treatments of activators and inhibitors of Hh and BMP signaling on osteogenesis. Combined treatment of SAG and rhBMP2 increased the thickness but not the region of the bone collar compared with SAG alone (Fig. 1A). On the other hand, dorsomorphin, a BMP inhibitor, suppressed SAG-induced ectopic bone collar formation (Fig. 1A). Regarding the loss of function of Hh signaling, cyclopamine inhibited bone collar formation in organ culture (Fig. 1A), whereas it did not alter the expressions of BMP ligands including Bmp2, Bmp4, Bmp6, and Bmp7 in primary perichondrial cells (supplemental Fig. S3). Interestingly, rhBMP2 did not reverse the cyclopamine-induced suppression of bone collar formation (Fig. 1A). Instead, it induced safranin O-positive chondrocyte-like cells in the perichondrium when combined with cyclopamine, suggesting that ectopic chondrocytes were formed upon the treatment (Fig. 1B). In situ hybridization for Col2a1 (type II collagen α1 chain) revealed that cyclopamine alone induced ectopic Col2a1 expression in the perichondrium, and combined treatment with cyclopamine and rhBMP2 expanded the Col2a1-expressing region (Fig. 1C).
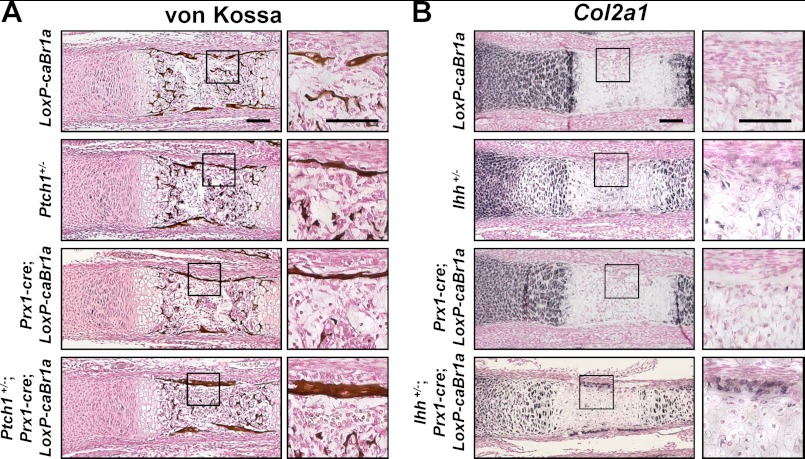
To confirm the interactions between Hh and BMP signaling in vivo, we analyzed E18.5 metatarsals of Ptch1+/−;Prx1-cre;CAG-LoxP-caBmpr1a mice, in which both Hh and BMP signaling were up-regulated in limb mesenchymal cells. The thickness of the bone collar was increased in Ptch1+/−;Prx1-cre;CAG-LoxP-caBmpr1a mice (Fig. 2A), which was consistent with the findings in the metatarsals cultured with the combination of SAG and rhBMP2 (Fig. 1A). In Ptch1+/− or Prx1-cre;CAG-LoxP-caBmpr1a mice, as well as CAG-LoxP-caBmpr1a (control) mice, bone collar formation remained normal. We also analyzed E17.5 metatarsals of Ihh+/−;Prx1-cre;CAG-LoxP-caBmpr1a mice, in which Hh signaling was down-regulated and BMP signaling was up-regulated in limb mesenchymal cells. One out of three mutants of Ihh+/−;Prx1-cre;CAG-LoxP-caBmpr1a showed ectopic Col2a1 expression in the perichondrium (Fig. 2B), which was consistent with the findings in the metatarsals cultured with a combined treatment of cyclopamine and rhBMP2 (Fig. 1B). The other two mutants as well as Ihh+/− and Prx1-cre;CAG-LoxP-caBmpr1a mice did not show such phenotypes. Overall, the findings on the in vivo genetic approach as well as pharmacological manipulation in the ex vivo organ culture suggested that BMP signaling acts as an accelerator for both osteogenesis and chondrogenesis in the perichondrium, after Hh-dependent lineage specification into osteoblasts or chondrocytes has taken place.
FIGURE 2.
Effects of the manipulation of Hh and BMP signaling pathways on bone collar formation and ectopic chondrocyte formation in the perichondrium in vivo. A, von Kossa staining of representative sections of metatarsals obtained from CAG-LoxP-caBmpr1a (LoxP-caBr1a), Ptch1+/−, Prx1-cre;CAG-LoxP-caBmpr1a (Prx1-cre;LoxP-caBr1a), and Ptch1+/−;Prx1-cre1;CAG-LoxP-caBmpr1a (Ptch1+/−;Prx1-cre;LoxP-caBr1a) mice (E18.5). Higher magnification of the boxed areas in the left panels is shown in the right panels. Scale bars, 100 μm. B, in situ hybridization for Col2a1 of representative sections of metatarsals obtained from CAG-LoxP-caBmpr1a (LoxP-caBr1a), Ihh+/−, Prx1-cre;CAG-LoxP-caBmpr1a (Prx1-cre;LoxP-caBr1a), and Ihh+/−;Prx1-cre1;CAG-LoxP-caBmpr1a (Ptch1+/−;Prx1-cre;LoxP-caBr1a) mice (E17.5). Higher magnification of the boxed areas in the left panels is shown in the right panels. Scale bars, 100 μm.
Heterogeneity of Perichondrial Cells
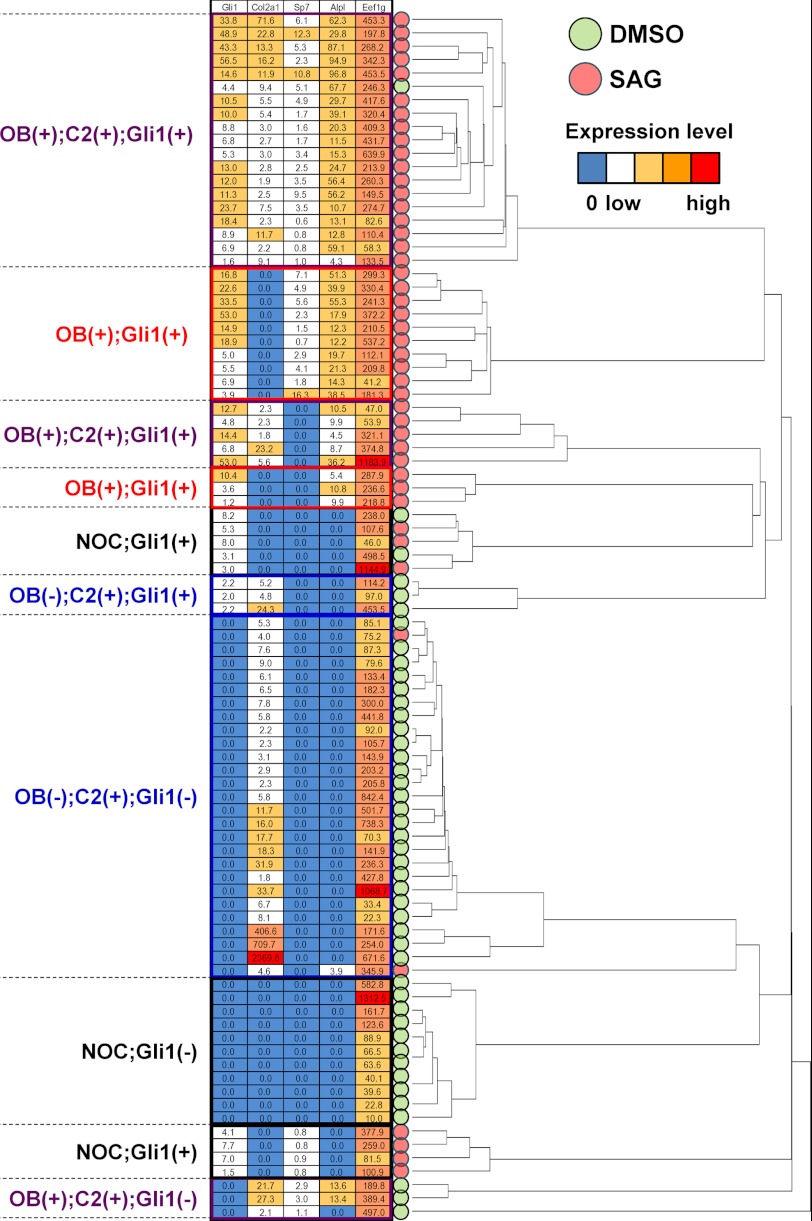
The different phenotypes in the perichondrium upon different combined stimuli in the organ culture suggest the heterogeneity of perichondrial cells. To gain insight into the heterogeneity, we analyzed the expression of osteoblast and chondrocyte marker genes in primary perichondrial cells at the single-cell level by quantitative RT-PCR (qRT-PCR) using a magnetic dT primer (21), focusing on their characteristics in nature as well as their properties acquired in response to Hh input.
The expressions of five genes were examined in 90 cells treated with SAG (activation of Hh signaling, 44 cells) or DMSO (control, 46 cells): Alpl, an early marker of osteoblast differentiation; Sp7 (osterix), a key transcription factor for osteoblast differentiation; Col2a1, a chondrocytic marker; Gli1, a readout of Hh signaling activation; and Eef1g, a housekeeping gene as a control. Clustering analysis on the qRT-PCR data classified the 90 cells into several clusters based on their expression patterns for these five genes (Fig. 3). Most of the DMSO-treated cells exhibited either of two features, chondrocyte-like phenotypes characterized by Col2a1 expression or non-osteochondrogenic phenotypes characterized by the lack of osteoblast and chondrocyte marker gene expression (NOC state). SAG-treated cells showed more heterogeneity in terms of their differentiation states, although the activation of Hh signaling was evidenced by up-regulation of Gli1 in these cells. Some of the cells still maintained an NOC state, but others expressed osteoblast marker genes, Alpl and Sp7, or both osteoblast and chondrocyte marker genes. Thus, the perichondrium likely consists of a heterogenous cell population with different properties and different levels of responsiveness to differentiation stimuli.
FIGURE 3.
Heterogeneity of perichondrial cells. Hierarchical clustering of gene expression in each of 90 single-cell samples obtained from primary perichondrial cells treated with DMSO (46 samples) or SAG (44 samples) for 5 days. The mRNA expression levels of the osteoblast marker genes and of Col2a1 and Gli1 were determined by qRT-PCR analysis. The cluster dendrogram indicates the four cellular subtypes: those expressing the osteoblast (OB) marker genes and the Col2a1, OB(+);C2(+) (shown in purple); those expressing osteoblast marker genes but not Col2a1, OB(+);C2(−) (shown in red); those expressing Col2a1 but not the osteoblast marker genes, OB(−);C2(+) (shown in blue); and those expressing neither osteoblast marker genes nor Col2a1, NOC (shown in black). Information on Gli1 expression is also presented for each of the above.
Hh Signaling Suppresses BMP-induced in Vitro Chondrogenic Differentiation, the Expression of Sox Trio, and Transactivation by Sox9
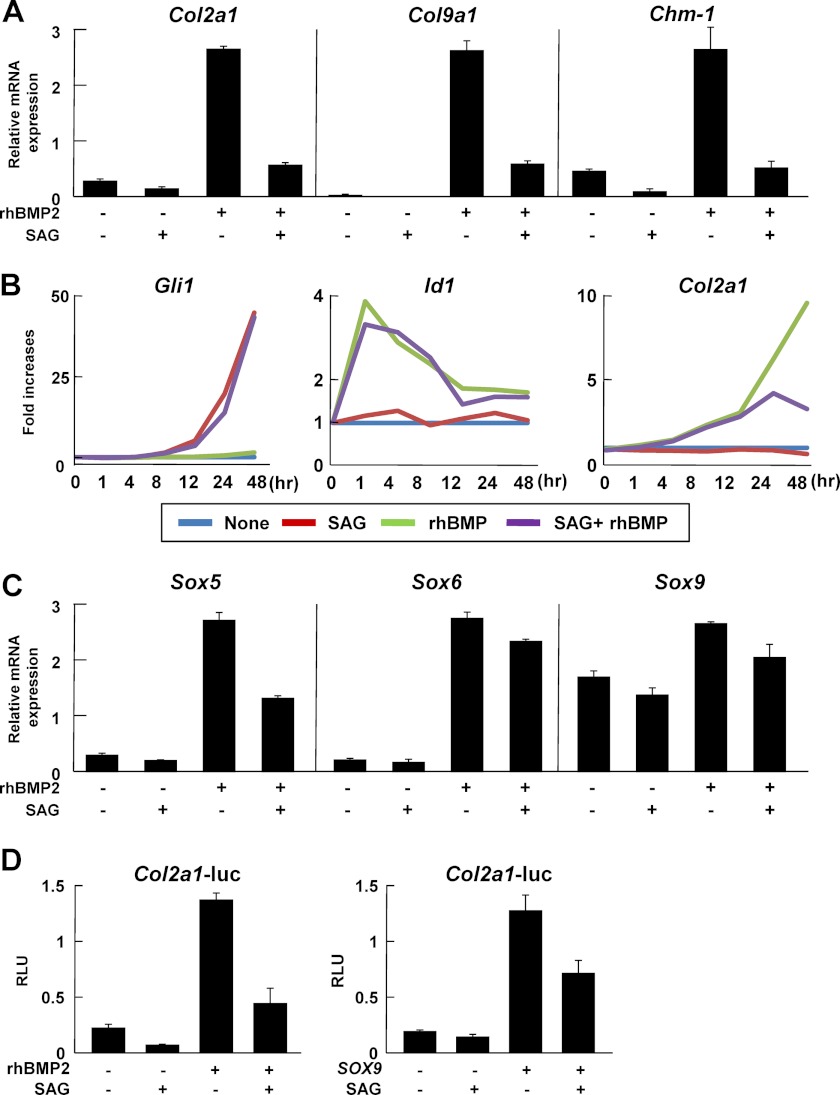
Given that both Hh and BMP signaling are activated in the perichondrium during endochondral ossification (28), the data shown in Figs. 1 and 2 led us to investigate whether Hh signaling had a negative impact on the chondrogenic activity of BMP signaling in the perichondrium. We considered that such an impact, if present, would arise via either of two mechanisms: a direct interaction between Hh and BMP signaling, or an indirect one, by which Hh signaling regulated chondrogenesis-related factors acting downstream of BMP signaling. We initially investigated the former possibility using the C3H10T1/2 cell line, which has been reported to differentiate into both chondrogenic and osteogeneic cells (29). SAG suppressed rhBMP2-induced mRNA expression of chondrocyte marker genes including Col2a1, Col9a1 (type IX collagen α1 chain), and chondromodulin 1 (Chm1) (Fig. 4A). Time course analysis of gene expression revealed that the suppression was synchronized with the up-regulation of Gli1, a readout of Hh signaling (Fig. 4B). However, the following observations suggested that there was no direct interaction or signal cross-talk between Hh and BMP signaling in the regulation of chondrocyte phenotypes (Fig. 4B). First, there was a time lag between expression of Id1, a readout of BMP signaling, and Col2a1 expression in response to rhBMP2 treatment, which indicated that activation of BMP signaling indirectly up-regulated Col2a1 as suggested in previous reports (30, 31). Second, the expression of target genes of either signaling was not altered by the addition of their counterparts (SAG versus SAG+rhBMP2 in Gli1 expression; rhBMP2 versus SAG+rhBMP2 in Id1 expression).
FIGURE 4.
Effects of Hh signaling on BMP signaling-induced expression of the Sox trio and transactivation by Sox9 in vitro. A and B, mRNA expression of chondrocyte marker genes determined by qRT-PCR analysis in C3H10T1/2 cells. Cells were treated with rhBMP2 (100 ng/ml), SAG (1 μm), or the combination of rhBMP2 and SAG for 2 days (A) or various periods up to 48 h (B). C, mRNA expressions of Sox 5, 6, and 9 determined by qRT-PCR analysis in C3H10T1/2 cells. Cells were treated with rhBMP2 (100 ng/ml), SAG (1 μm), or the combination of rhBMP2 and SAG for 2 days. D, luciferase analysis using reporter constructs containing a human COL2A1 regulatory region (+285 to +2450) in C3H10T1/2 cells. In the left panel, cells were incubated for 12 h after transfection with the reporter construct, followed by exposure to rhBMP2 (100 ng/ml), SAG (1 μm), or the combination of rhBMP2 and SAG for 48 h. In the right panel, cells were transfected with the reporter construct and Sox9, followed by exposure to SAG (1 μm) for 48 h (right panel). RLU, relative light units. For A, C, and D, data are expressed as the means ± S.D. of triplicate wells, and representative data of independent experiments are shown.
BMP signaling has been shown to regulate chondrogenesis through Sox5, Sox6, and Sox9 (Sox trio)-dependent mechanisms, and a BMP-responsive element in the Col2a1 region corresponded to a Sox9 binding site (+2429 to +2435) in the first intron of the Col2a1 gene (30–34). Therefore, we tested the second possibility, i.e. the existence of an indirect interaction between Hh and BMP signaling, by examining the effects of Hh signaling on the expression or function of Sox transcription factors. We found that SAG suppressed rhBMP2-induced mRNA expression of Sox5, Sox6, and Sox9 (Fig. 4C). A luciferase assay using the reporter construct containing a Sox9-responsive element in the COL2A1 region (+285 to +2450; COL2A1-Luc) showed that SAG inhibited Sox9-mediated reporter activity as well as rhBMP2-mediated activity (Fig. 4D). These data suggest that Hh signaling suppresses BMP signaling-mediated chondrogenesis, and that this effect occurs at least in part through inhibition of both the expression of the Sox trio and the transactivation by Sox9.
Gli Activators Are Involved in the Hh-mediated Suppression of Chondrogenesis in the Perichondrium
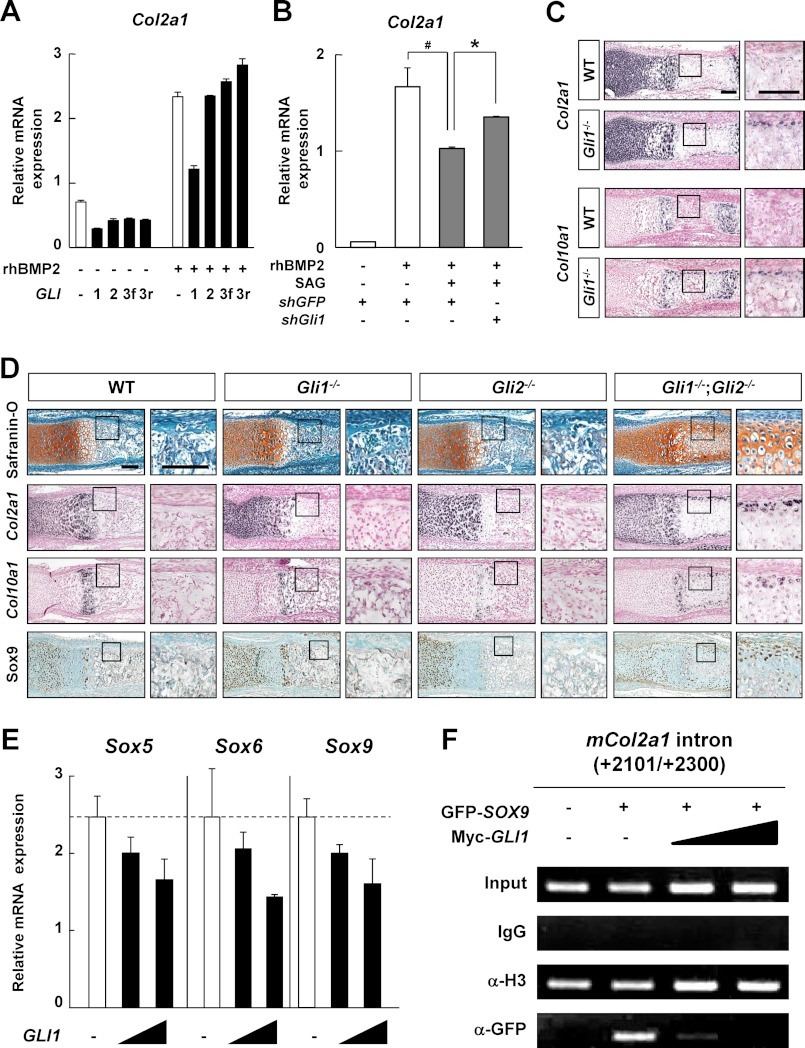
We next attempted to elucidate which molecules contribute to the inhibitory effect of Hh signaling on chondrogenic activity of the BMP-Sox trio, focusing on the transcriptional machinery of Hh-Gli signaling. In C3H10T1/2 cells, Gli1 overexpression suppressed the rhBMP2-induced Col2a1 mRNA expression, whereas this suppression was not observed in response to the overexpressions of Gli2, the full-length form of Gli3, or the repressor form of Gli3 (Fig. 5A). The suppressive effects of Gli1 on chondrogenesis were confirmed by gain-of-function analyses of primary perichondrial cells (supplemental Fig. S2A) and loss-of-function analyses using short interfering RNA for Gli1 (shGli1) in C3H10T1/2 cells (Fig. 5B).
FIGURE 5.
Involvement of Gli transcription factors in the Hedgehog-mediated inhibition of chondrogenesis in the perichondrium. A and B, Col2a1 mRNA expression determined by qRT-PCR analysis in C3H10T1/2 cells. After being transfected with the indicated plasmids, cells were incubated for 12 h, followed by exposure to rhBMP2 (100 ng/ml) (A), or the combination of rhBMP2 and SAG (1 μm) (B) for 48 h. GLI3f, the full-length form of GLI3; GLI3r, the repressor form of GLI3. *, p < 0.05; #, p < 0.05. C and D, in situ hybridization (Col2a1 and Col10a1), safranin O staining, and immunohistochemistry for Sox9 of the representative sections of metatarsals obtained from the indicated mouse embryos (E17.5) (C) or indicated mouse newborns (D). Higher magnification of the boxed areas is shown at the right. Scale bars, 100 μm. E, mRNA expressions of endogenous Sox5, Sox6, and Sox9 determined by qRT-PCR analysis in C3H10T1/2 cells. Cells were transfected with GLI1 (0.1 or 1 μg) and cultured for 48 h. F, ChIP assay in the perichondrial cells transfected with the indicated plasmids. ChIP was performed with an anti-GFP antibody (Sox9), IgG (negative control), or an anti-acetyl histone H3 antibody (positive control). For A, B, and E, data are expressed as the means ± S.D. of triplicate wells, and representative data of independent experiments are shown.
We found no bone collar formation in the metatarsals of Gli1−/− and Gli2−/− mice at E17.5, and the phenotype was more severe in the Gli1−/−;Gli2−/− mice than in either the Gli1−/− or Gli2−/− mice at postnatal day 0 (11). The in vitro data obtained so far led us to examine whether Gli1-mediated suppression of chondrogenesis was associated with the phenotypes in the perichondrium of Gli1−/− or Gli1−/−;Gli2−/− mice. Indeed, ectopic expression of both Col2a1 and Col10a1 (type X collagen α1 chain) was observed in metatarsals of the Gli1−/− perichondrium at E17.5 (Fig. 5C). Moreover, deposition of cartilaginous matrix as well as ectopic expression of Col2a1, Col10a1, and Sox9 were observed in the Gli1−/−;Gli2−/− perichondrium at postnatal day 0 (Fig. 5D). In addition, overexpression of both Gli1 and Gli2 significantly inhibited rhBMP2-induced Col2a1 expression compared with that of Gli1 alone (supplemental Fig. 6S). Thus, Gli activators may have a negative impact on chondrogenesis in the perichondrium during endochondral ossification.
Finally, we investigated the molecular mechanism underlying the suppression of chondrogenic differentiation by the Hh-Gli1 pathways. Gli1 overexpression suppressed the mRNA expressions of the Sox trio in C3H10T1/2 cells (Fig. 5E). A luciferase assay using COL2A1-Luc revealed that Gli1 inhibited the transactivation by Sox9 (supplemental Fig. S3B). We also examined the effects of Gli1 on the Sox9-dependent induction of COL2A1 mRNA using the HEK293 cell line, since this cell line is known to express COL2A1 mRNA upon transfection of the Sox trio (35); we found that Gli1 overexpression inhibited the COL2A1 mRNA expression induced by the Sox trio in a dose-dependent manner (supplemental Fig. S2C), without any changes in the protein localization of exogenous Sox9 (supplemental Fig. S2D). In addition, a chromatin immunoprecipitation (ChIP) assay revealed that Gli1 overexpression inhibited the recruitment of Sox9 onto the Sox9 binding site in the COL2A1 first intron in a dose-dependent manner in perichondrial cells (Fig. 5F). Thus, Gli1 suppresses the expressions of the Sox trio and the recruitment of Sox9 to the Col2a1 enhancer region, and this suppression may underlie the negative impact of Hh signaling on chondrogenic activity of the BMP-Sox trio.
DISCUSSION
This study has three major findings. First, BMP signaling interacted with Hh signaling to enhance the bone collar formation, whereas in the absence of Hh input, BMP enhanced ectopic chondrocyte formation in the perichondrium. Second, perichondrial cells showed heterogeneity in terms of natural characteristics and responsiveness to differentiation stimuli. Third, Hh signaling suppressed BMP signaling-induced chondrogenic differentiation, possibly by the Gli1-mediated inhibition of expression of the Sox trio and transactivation by Sox9.
Our organ culture system using osteogenic medium enabled us to observe the bone collar formation of endochondral ossification. The ex vivo cultures of fetal long bones reported previously had not fully reproduced endochondral ossification because they lacked bone collar formation (20). As indicated in this study, our system would offer clues to understanding the network of multiple signaling pathways that regulate the differentiation of perichondrial cells, in a near in vivo setting and in a simple manner, prior to proceeding to genetic approaches. Although it is important to keep in mind the potential off-target effects of stimuli in our organ culture system, the system would enable easy manipulation of different signaling pathways at different time points.
BMP signaling is known to function as an osteo-chondrogenic factor with positive impacts on both osteogenesis and chondrogenesis during endochondral ossification (36). Rodda et al. (9) raised the possibility that Hh signaling altered the responsiveness of osteoblast progenitors to a BMP input in the perichondrium. Our findings support this possibility and further demonstrate how BMP signaling interacts with Hh signaling to induce osteoblast differentiation in the perichondrium; BMP signaling is not likely to be involved in specification of the osteoblast lineage, but it clearly has a positive impact on osteogenesis in the committed osteoblast precursors. Given that BMP signaling enhanced chondrogenesis in the perichondrium in the absence of Hh input, and Hh signaling suppressed BMP-mediated chondrocyte differentiation in vitro, we infer that the osteo-chondrogenic function of BMP is directed by Hh signaling in the perichondrium.
We were not able to discern the reasons why only one of the three Ihh+/−;Prx1-cre;CAG-LoxP-caBmpr1a mutants showed ectopic Col2a1 expression in the perichondrium (Fig. 2A). We infer that penetrance may underlie the phenotype (37). In addition, these data support the threshold concept for ectopic chondrocyte formation in the perichondrium, where the positive action of BMP and the inhibitory action of Hh may compete with each other in chondrogenesis. The balance between the two signals would determine a threshold for the ectopic chondrocyte formation; a low level of Hh signaling may decrease the threshold, resulting in chondrogenesis in the perichondrium upon the activation of BMP signaling. Moreover, single-cell analyses showed that among cells that expressed Gli1 upon Hh input, not all, but some populations expressed osteoblast marker genes. These data are consistent with the notion that Hh signaling is necessary but not sufficient to induce bone formation (8). Hence, the timing of Hh input and its target population is critical for the osteogenic function of BMP signaling.
In this study, we performed the first examination of the populations of perichondrial cells at a single-cell level. Our data showed that cells stimulated by an osteogenic reagent showed heterogeneity in terms of their differentiation state. Most cells treated with DMSO were either cells expressing Col2a1 or NOC cells. In response to Hh input, the majority of cells expressed osteoblast marker genes. There is a possibility that the Col2a1-positive cells observed in the DMSO group were derived from cartilage due to contamination during the isolation procedure. If the contamination occurred, the chondrocytes might have kept their phenotypes or newly acquired osteoblast phenotypes upon Hh input. However, we did not observe a chondrocyte-like population in the SAG-treated group. Furthermore, chondrocytes are not likely to differentiate into osteoblasts upon Hh input because it was previously reported that no ectopic osteoblasts were observed in cartilage-forming regions in chondrocytes overexpressing constitutive active Smoothened (38) or ablation of Patched1 (24). Therefore, we assume that little, if any, contamination of chondrocytes occurred during our isolation procedure and that the Col2a1-positive cells or NOC cells residing in the perichondrium give rise to cells expressing osteoblast marker genes upon Hh input. Col2a1-expressing cells are known to exist in the perichondrium and contribute to the osteoblast linage (10), which supports this assumption. Indeed, several lines of study have shown that gene manipulation by Col2a1-Cre mice caused phenotypes characterized by osteogenesis in the perichondrium (8, 15, 24). Unlike the Col2a1-expressing cells mentioned above, the properties of NOC cells remain to be clarified, although it is possible that some of these cells are caused by the contamination of fibroblasts, or that they are a progenitor-like undifferentiated population contributing to the maintenance of a progenitor pool in the perichondrium and/or to osteoblast formation.
The Hh-Gli1 pathway inhibited chondrocyte differentiation, possibly by inhibition of the expression of the Sox trio and transactivation by Sox9. Given that Sox9 is required for the expressions of Sox5 and Sox6 (3) and Sox9 transcription is suggested to be controlled by Sox9 itself (39), the initial events in this context may be an inhibition of the recruitment of Sox9 into its own regulatory regions by Gli1. However, a recent paper showed that there were conserved Gli-binding sites near the Sox motif in the Sox5 and Sox6 regions (40), which raised the possibility that an inhibitory action by Gli1 might occur in these regions. This study, however, does not address how Gli1 inhibits the recruitment of Sox9 onto DNA. Further studies including genome-wide analyses may be needed to clarify whether competition between Gli1 and Sox9 occurs in association with DNA binding or through epigenetic regulation. On the other hand, Zhao et al. (41) reported that Hh-Gli2 signaling is a mediator of Bmp2 expression, which may not concur with our data. In our analyses, Hh agonist did not alter the expression of Id1, a target gene of BMP signaling (Fig. 4B), and the Hh inhibitor did not inhibit Bmp2 expression (supplemental Fig. S3). The discrepancy may be due to differences in the cells used between the Zhao et al. (41) study and our work; they used C2C12, a myoblast cell line, 2T3, a calvaria osteoblast cell line, and primary calvaria osteoblasts to examine the effects of Hh-Gli2 signaling on endogenous Bmp2 mRNA expression, whereas we used C3H10T1/2 and primary perichondrial cells. We believe that clarifying the distinct roles of Hh and BMP signaling in each cell type will be important contributions to our understanding of bone development and clinical applications.
In conclusion, this study demonstrated that the osteo-chondrogenic function of BMP was directed toward osteogenesis by Hh-Gli activators in the perichondrium. Perichondrial cells showed various properties upon Hh input, which may underlie the Hh-mediated alteration of BMP function in perichondrial cells. Previous reports described interactions between Hh and BMP signaling toward chondrogenesis in limb bud mesenchymal cells (42, 43), which appear to be inconsistent with our findings. We hypothesize that the discrepancy may be caused by the difference of cells used between these studies and our work. The target populations of Hh and BMP signaling, or other signaling pathways interacting with them, may be different between the limb bud and the perichondrium. In addition, cellular characteristics may be distinct between the limb bud and perichondrium, in terms of multipotency and responsiveness to stimuli for lineage specifications. Sox9-positive cells in the limb bud were reported to differentiate into various skeletal cells, including chondrocytes, osteoblasts, synovial cells, and tendon cells (44). However, the differentiation potential of perichondrial cells was restricted to an osteoblast or chondrocyte lineage in vivo (8). Although the distinct characteristics and signaling networks remain to be clarified, our study provides insights into the molecular basis of the signaling network between Hh and BMP in the perichondrium as well as the natural characteristics of perichondrial cells themselves. Studies including single-cell tracking and genome-wide analyses may lead to further understanding of the signaling network determining the fates of perichondrial cells and to clinical application of such knowledge to the treatment of bone defect-related diseases.
Acknowledgments
We thank Dr. G. Yamada and Astellas Pharma, Inc. for providing experimental materials.
This work was supported by grants-in-aid for scientific research from the Japan Society for the Promotion of Science, as well as by the Center for NanoBio Integration, the Center for Medical System Innovation, and the Funding Program for World Leading Innovative R&D on Science and Technology (FIRST program).

This article contains supplemental “Experimental Procedures,” Table S1, and Figs. S1–S6.
- Hh
- hedgehog
- BMP
- bone morphogenetic protein
- qPCR
- quantitative PCR
- Runx2
- runt-related transcription factor 2
- Osx
- osterix
- Ihh
- Indian hedgehog
- rhBMP2
- recombinant human BMP2
- DMSO
- dimethyl sulfoxide
- E17
- embryonic day 17
- Sox9
- SRY box-containing gene 9
- Ptch1
- patched 1
- caBMPr1a
- constitutively active form of Bmpr1a
- SAG
- Smoothened agonist
- rhTGFβ
- recombinant human TGFβ
- GSK3
- glycogen synthase kinase-3
- Col2a1
- type II collagen α1 chain
- Col9a1
- type IX collagen α1 chain
- Col10a1
- type X collagen α1 chain
- Chm1
- chondromodulin 1.
REFERENCES
- 1. Kronenberg H. M. (2003) Developmental regulation of the growth plate. Nature 423, 332–336 [DOI] [PubMed] [Google Scholar]
- 2. Bi W., Deng J. M., Zhang Z., Behringer R. R., de Crombrugghe B. (1999) Sox9 is required for cartilage formation. Nat. Genet. 22, 85–89 [DOI] [PubMed] [Google Scholar]
- 3. Akiyama H., Chaboissier M. C., Martin J. F., Schedl A., de Crombrugghe B. (2002) The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 16, 2813–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Colnot C., Lu C., Hu D., Helms J. A. (2004) Distinguishing the contributions of the perichondrium, cartilage, and vascular endothelium to skeletal development. Dev. Biol. 269, 55–69 [DOI] [PubMed] [Google Scholar]
- 5. Nakashima K., Zhou X., Kunkel G., Zhang Z., Deng J. M., Behringer R. R., de Crombrugghe B. (2002) The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 108, 17–29 [DOI] [PubMed] [Google Scholar]
- 6. Long F. (2012) Building strong bones: molecular regulation of the osteoblast lineage. Nat. Rev. Mol. Cell Biol. 13, 27–38 [DOI] [PubMed] [Google Scholar]
- 7. Komori T., Yagi H., Nomura S., Yamaguchi A., Sasaki K., Deguchi K., Shimizu Y., Bronson R. T., Gao Y. H., Inada M., Sato M., Okamoto R., Kitamura Y., Yoshiki S., Kishimoto T. (1997) Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89, 755–764 [DOI] [PubMed] [Google Scholar]
- 8. Long F., Chung U. I., Ohba S., McMahon J., Kronenberg H. M., McMahon A. P. (2004) Ihh signaling is directly required for the osteoblast lineage in the endochondral skeleton. Development 131, 1309–1318 [DOI] [PubMed] [Google Scholar]
- 9. Rodda S. J., McMahon A. P. (2006) Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development 133, 3231–3244 [DOI] [PubMed] [Google Scholar]
- 10. Maes C., Kobayashi T., Selig M. K., Torrekens S., Roth S. I., Mackem S., Carmeliet G., Kronenberg H. M. (2010) Osteoblast precursors, but not mature osteoblasts, move into developing and fractured bones along with invading blood vessels. Dev. Cell 19, 329–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hojo H., Ohba S., Yano F., Saito T., Ikeda T., Nakajima K., Komiyama Y., Nakagata N., Suzuki K., Takato T., Kawaguchi H., Chung U. I. (2012) Gli1 protein participates in Hedgehog-mediated specification of osteoblast lineage during endochondral ossification. J. Biol. Chem. 287, 17860–17869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. St-Jacques B., Hammerschmidt M., McMahon A. P. (1999) Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 13, 2072–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Joeng K. S., Long F. (2009) The Gli2 transcriptional activator is a crucial effector for Ihh signaling in osteoblast development and cartilage vascularization. Development 136, 4177–4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hill T. P., Später D., Taketo M. M., Birchmeier W., Hartmann C. (2005) Canonical Wnt/β-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev. Cell 8, 727–738 [DOI] [PubMed] [Google Scholar]
- 15. Day T. F., Guo X., Garrett-Beal L., Yang Y. (2005) Wnt/β-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev. Cell 8, 739–750 [DOI] [PubMed] [Google Scholar]
- 16. Kobayashi T., Lyons K. M., McMahon A. P., Kronenberg H. M. (2005) BMP signaling stimulates cellular differentiation at multiple steps during cartilage development. Proc. Natl. Acad. Sci. U.S.A. 102, 18023–18027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Park H. L., Bai C., Platt K. A., Matise M. P., Beeghly A., Hui C. C., Nakashima M., Joyner A. L. (2000) Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development 127, 1593–1605 [DOI] [PubMed] [Google Scholar]
- 18. Mo R., Freer A. M., Zinyk D. L., Crackower M. A., Michaud J., Heng H. H., Chik K. W., Shi X. M., Tsui L. C., Cheng S. H., Joyner A. L., Hui C. (1997) Specific and redundant functions of Gli2 and Gli3 zinc finger genes in skeletal patterning and development. Development 124, 113–123 [DOI] [PubMed] [Google Scholar]
- 19. Kamiya N., Ye L., Kobayashi T., Mochida Y., Yamauchi M., Kronenberg H. M., Feng J. Q., Mishina Y. (2008) BMP signaling negatively regulates bone mass through sclerostin by inhibiting the canonical Wnt pathway. Development 135, 3801–3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hojo H., Yano F., Ohba S., Igawa K., Nakajima K., Komiyama Y., Kan A., Ikeda T., Yonezawa T., Woo J. T., Takato T., Nakamura K., Kawaguchi H., Chung U. I. (2010) Identification of oxytetracycline as a chondrogenic compound using a cell-based screening system. J. Bone Miner. Metab. 28, 627–633 [DOI] [PubMed] [Google Scholar]
- 21. Taniguchi K., Kajiyama T., Kambara H. (2009) Quantitative analysis of gene expression in a single cell by qPCR. Nat. Methods 6, 503–506 [DOI] [PubMed] [Google Scholar]
- 22. Yu P. B., Hong C. C., Sachidanandan C., Babitt J. L., Deng D. Y., Hoyng S. A., Lin H. Y., Bloch K. D., Peterson R. T. (2008) Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat. Chem. Biol. 4, 33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen B., Dodge M. E., Tang W., Lu J., Ma Z., Fan C. W., Wei S., Hao W., Kilgore J., Williams N. S., Roth M. G., Amatruda J. F., Chen C., Lum L. (2009) Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat. Chem. Biol. 5, 100–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mak K. K., Chen M. H., Day T. F., Chuang P. T., Yang Y. (2006) Wnt/β-catenin signaling interacts differentially with Ihh signaling in controlling endochondral bone and synovial joint formation. Development 133, 3695–3707 [DOI] [PubMed] [Google Scholar]
- 25. Bandyopadhyay A., Tsuji K., Cox K., Harfe B. D., Rosen V., Tabin C. J. (2006) Genetic analysis of the roles of BMP2, BMP4, and BMP7 in limb patterning and skeletogenesis. PLoS Genet. 2, e216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matsunobu T., Torigoe K., Ishikawa M., de Vega S., Kulkarni A. B., Iwamoto Y., Yamada Y. (2009) Critical roles of the TGF-β type I receptor ALK5 in perichondrial formation and function, cartilage integrity, and osteoblast differentiation during growth plate development. Dev. Biol. 332, 325–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen G., Deng C., Li Y. P. (2012) TGF-β and BMP signaling in osteoblast differentiation and bone formation. Int. J. Biol. Sci. 8, 272–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Retting K. N., Song B., Yoon B. S., Lyons K. M. (2009) BMP canonical Smad signaling through Smad1 and Smad5 is required for endochondral bone formation. Development 136, 1093–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Date T., Doiguchi Y., Nobuta M., Shindo H. (2004) Bone morphogenetic protein-2 induces differentiation of multipotent C3H10T1/2 cells into osteoblasts, chondrocytes, and adipocytes in vivo and in vitro. J. Orthop. Sci. 9, 503–508 [DOI] [PubMed] [Google Scholar]
- 30. Fernández-Lloris R., Viñals F., López-Rovira T., Harley V., Bartrons R., Rosa J. L., Ventura F. (2003) Induction of the Sry-related factor SOX6 contributes to bone morphogenetic protein-2-induced chondroblastic differentiation of C3H10T1/2 cells. Mol. Endocrinol. 17, 1332–1343 [DOI] [PubMed] [Google Scholar]
- 31. Chimal-Monroy J., Rodriguez-Leon J., Montero J. A., Gañan Y., Macias D., Merino R., Hurle J. M. (2003) Analysis of the molecular cascade responsible for mesodermal limb chondrogenesis: Sox genes and BMP signaling. Dev. Biol. 257, 292–301 [DOI] [PubMed] [Google Scholar]
- 32. Yoon B. S., Ovchinnikov D. A., Yoshii I., Mishina Y., Behringer R. R., Lyons K. M. (2005) Bmpr1a and Bmpr1b have overlapping functions and are essential for chondrogenesis in vivo. Proc. Natl. Acad. Sci. U.S.A. 102, 5062–5067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zehentner B. K., Dony C., Burtscher H. (1999) The transcription factor Sox9 is involved in BMP-2 signaling. J. Bone Miner. Res. 14, 1734–1741 [DOI] [PubMed] [Google Scholar]
- 34. Pizette S., Niswander L. (2000) BMPs are required at two steps of limb chondrogenesis: formation of prechondrogenic condensations and their differentiation into chondrocytes. Dev. Biol. 219, 237–249 [DOI] [PubMed] [Google Scholar]
- 35. Ikeda T., Kamekura S., Mabuchi A., Kou I., Seki S., Takato T., Nakamura K., Kawaguchi H., Ikegawa S., Chung U. I. (2004) The combination of SOX5, SOX6, and SOX9 (the SOX trio) provides signals sufficient for induction of permanent cartilage. Arthritis Rheum. 50, 3561–3573 [DOI] [PubMed] [Google Scholar]
- 36. Wan M., Cao X. (2005) BMP signaling in skeletal development. Biochem. Biophys. Res. Commun. 328, 651–657 [DOI] [PubMed] [Google Scholar]
- 37. Nadeau J. H. (2001) Modifier genes in mice and humans. Nat. Rev. Genet. 2, 165–174 [DOI] [PubMed] [Google Scholar]
- 38. Long F., Zhang X. M., Karp S., Yang Y., McMahon A. P. (2001) Genetic manipulation of hedgehog signaling in the endochondral skeleton reveals a direct role in the regulation of chondrocyte proliferation. Development 128, 5099–5108 [DOI] [PubMed] [Google Scholar]
- 39. Sekido R., Lovell-Badge R. (2008) Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature 453, 930–934 [DOI] [PubMed] [Google Scholar]
- 40. Leung V. Y., Gao B., Leung K. K., Melhado I. G., Wynn S. L., Au T. Y., Dung N. W., Lau J. Y., Mak A. C., Chan D., Cheah K. S. (2011) SOX9 governs differentiation stage-specific gene expression in growth plate chondrocytes via direct concomitant transactivation and repression. PLoS Genet. 7, e1002356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhao M., Qiao M., Harris S. E., Chen D., Oyajobi B. O., Mundy G. R. (2006) The zinc finger transcription factor Gli2 mediates bone morphogenetic protein 2 expression in osteoblasts in response to hedgehog signaling. Mol. Cell. Biol. 26, 6197–6208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Murtaugh L. C., Chyung J. H., Lassar A. B. (1999) Sonic hedgehog promotes somitic chondrogenesis by altering the cellular response to BMP signaling. Genes Dev. 13, 225–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lopez-Rios J., Speziale D., Robay D., Scotti M., Osterwalder M., Nusspaumer G., Galli A., Holländer G. A., Kmita M., Zeller R. (2012) GLI3 constrains digit number by controlling both progenitor proliferation and. Dev. Cell 22, 837–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Akiyama H., Kim J. E., Nakashima K., Balmes G., Iwai N., Deng J. M., Zhang Z., Martin J. F., Behringer R. R., Nakamura T., de Crombrugghe B. (2005) Osteo-chondroprogenitor cells are derived from Sox9 expressing precursors. Proc. Natl. Acad. Sci. U.S.A. 102, 14665–14670 [DOI] [PMC free article] [PubMed] [Google Scholar]