Background: Acyl-CoA synthetase 1 (ACSL1) promotes inflammatory effects in macrophages, but its regulation and biological role remain largely unknown.
Results: Multiple inflammatory pathways contribute to ACSL1 induction, and this induction allows for phospholipid turnover in activated macrophages.
Conclusion: The regulation and function of ACSL1 differ substantially in macrophages and insulin target tissues.
Significance: These findings indicate a novel role for ACSL1 in innate immunity.
Keywords: Cytokines/Interferon, Fatty Acid Metabolism, Lipopolysaccharide (LPS), Macrophages, Phospholipid Turnover, Acyl-CoA Synthetase, Gram-negative Bacteria
Abstract
The enzyme acyl-CoA synthetase 1 (ACSL1) is induced by peroxisome proliferator-activated receptor α (PPARα) and PPARγ in insulin target tissues, such as skeletal muscle and adipose tissue, and plays an important role in β-oxidation in these tissues. In macrophages, however, ACSL1 mediates inflammatory effects without significant effects on β-oxidation. Thus, the function of ACSL1 varies in different tissues. We therefore investigated the signals and signal transduction pathways resulting in ACSL1 induction in macrophages as well as the consequences of ACSL1 deficiency for phospholipid turnover in LPS-activated macrophages. LPS, Gram-negative bacteria, IFN-γ, and TNFα all induce ACSL1 expression in macrophages, whereas PPAR agonists do not. LPS-induced ACSL1 expression is dependent on Toll-like receptor 4 (TLR4) and its adaptor protein TRIF (Toll-like receptor adaptor molecule 1) but does not require the MyD88 (myeloid differentiation primary response gene 88) arm of TLR4 signaling; nor does it require STAT1 (signal transducer and activator of transcription 1) for maximal induction. Furthermore, ACSL1 deletion attenuates phospholipid turnover in LPS-stimulated macrophages. Thus, the regulation and biological function of ACSL1 in macrophages differ markedly from that in insulin target tissues. These results suggest that ACSL1 may have an important role in the innate immune response. Further, these findings illustrate an interesting paradigm in which the same enzyme, ACSL1, confers distinct biological effects in different cell types, and these disparate functions are paralleled by differences in the pathways that regulate its expression.
Introduction
Acyl-CoA synthetases (ACSLs)4 esterify long chain free fatty acids, converting them to intracellular fatty acyl-CoAs. Esterification is a requisite step for subsequent metabolic fates of fatty acids, including β-oxidation, neutral lipid synthesis, or incorporation into phospholipids. Five distinct ACSL subtypes encoded by different genes have been identified in mammals, and they exhibit overlapping yet distinct patterns of tissue distribution, subcellular localization, and substrate selectivity (1, 2). ACSL1 esterifies long chain fatty acids (C:14–C:22) and is enriched in liver, heart, and adipose tissue (3–5). Functionally, ACSL1 has been implicated in pathways of neutral lipid synthesis and both mitochondrial and peroxisomal β-oxidation in these tissues (6, 7). A pivotal function for ACSL1 in β-oxidation in heart and adipose tissue recently has been verified by mouse models with conditional ACSL1 deficiency in these tissues (8, 9). Notably, this role in β-oxidation initially was suggested by the finding that Acsl1 is a peroxisome proliferator-activated receptor-α (PPARα) and PPARγ target gene (6, 10–13).
ACSL1 also is expressed in monocytes and macrophages, and we have recently demonstrated that ACSL1 expression is induced in classically activated (M1) macrophages and in monocytes and macrophages from mouse models of type 1 diabetes (14). ACSL1-deficient macrophages exhibit reduced expression of inflammatory cytokines, including IL-1β and TNFα under diabetic conditions (14), further implicating ACSL1 in the innate immune response. In contrast to adipocytes, ACSL1-deficient macrophages do not exhibit reduced β-oxidation, suggesting that ACSL1 may play a functionally distinct role in macrophages. However, its role in macrophage biology is poorly understood, and the macrophage-specific mechanisms by which it is regulated are unknown. Delineating these regulatory pathways could generate insights into the functional significance of ACSL1 in macrophages and thereby contribute to our understanding of the cellular response to pathogen and other inflammatory stimuli.
In the present study, we demonstrate that macrophages exhibit a distinct profile of ACSL1 regulation. In contrast to other studied cell types, macrophage ACSL1 expression is induced by the proinflammatory molecules LPS, IFN-γ, and TNFα as well as in vitro infection with the Gram-negative pathogens Escherichia coli and Salmonella typhimurium. Further, Acsl1 is not a PPAR target gene in macrophages but is rather regulated by multiple inflammatory signaling pathways, including TLR4 and its adaptor protein TRIF and c-Jun N-terminal kinase (JNK). ACSL1 induction after LPS stimulation predominantly depends on TLR4-TRIF-mediated signaling, but E. coli can induce its expression by alternative pathways. Whereas JNK signaling contributes to ACSL1 expression after IFN-γ stimulation, it is not required for LPS- or E. coli-mediated ACSL1 induction. Finally, ACSL1 deficiency inhibits LPS-stimulated turnover of several phospholipid species. These findings collectively suggest a novel and distinct regulation of ACSL1 in macrophages and an important role for ACSL1 in inflammation and the innate immune response.
EXPERIMENTAL PROCEDURES
Mice
Male and female adult (12–18 weeks of age) mice on the C57BL6/J background were used for all experiments. Mice were fed a regular chow diet. Mice with myeloid-specific ACSL1 deficiency have been described previously (14), as have mice with double JNK1/JNK2 (Mapk8/Mapk9) deficiency (15) and myeloid cell-targeted JNK1/JNK2 deficiency (16), TLR4 (Tlr4) deficiency (17), MyD88 (Myd88) deficiency (18), and TRIF (Ticam1) deficiency (19). All experiments were performed in accordance with an approved University of Washington Institutional Animal Care and Use Committee protocol.
Bone Marrow-derived Macrophage (BMDM) Isolation and Culture
Bone marrow was harvested from the femurs and tibias of mice 10–14 weeks of age. Bone marrow was flushed with RPMI medium through a 26-gauge needle and then centrifuged at 150 × g for 5 min. The supernatant was aspirated, and bone marrow-derived cells (BMDCs) were suspended in red blood cell lysis buffer (15 mm NH4Cl4, 0.5 mm KHCO3, 18.6 mm Na2EDTA) for 5 min. The reaction was quenched with PBS, and samples again were centrifuged at 150 × g for 5 min. The supernatant was aspirated, and the pellet was resuspended in RPMI medium. Live BMDCs were counted using trypan blue stain to exclude dead cells and were then plated at a concentration of 2 × 106 cells/well in 6-well plates. BMDCs were cultured in RPMI 1640 medium (11.2 mm glucose) with 7% heat-inactivated fetal bovine serum (FBS), 30% L929 cell-conditioned medium as a source of M-CSF, and 1% penicillin, streptomycin, and fungizone. Cells were differentiated in the presence of M-CSF for 7 days, and the medium was changed every 48 h.
For stimulation/activation, media were changed on day 6 to low glucose DMEM (5.5 mm) with 1% non-essential amino acids (Invitrogen), 7% FBS, 30% L929 cell-conditioned medium, and antibiotics. BMDMs were variably stimulated with LPS (E. coli 055:B5; Sigma-Aldrich), ultrapure LPS (E. coli 0111:B4; List Biological Laboratories, Inc., Campbell, CA), recombinant murine TNFα (PeproTech, Inc., Rocky Hill, NJ), recombinant murine IFN-γ (eBioscience Inc., San Diego, CA), rosiglitazone, the PPARδ agonist GW 0742 (Sigma-Aldrich), or the highly specific PPARα agonist CP900691 (a generous gift from Pfizer Research and Development) as described under “Results.” For protein kinase inhibitor studies, macrophages were incubated in the presence of the indicated inhibitor for 30 min prior to LPS stimulation. The following inhibitors were employed: the p38 MAPK inhibitor LY2228820, the mitogen-activated protein kinase kinase 1 inhibitor PD98059 (both from Selleckchem, Houston, TX), and the inhibitor of κB kinase β (IKKβ) inhibitor SC-514 (Cayman Chemical, Ann Arbor, MI).
Peritoneal Macrophage Harvest
Mice were administered an intraperitoneal injection of thioglycollate (BD Biosciences), as described previously (14). Five days later, a midline incision was made in the abdomen with dissection of skin to expose the peritoneum. A 26-gauge needle attached to a syringe filled with RPMI medium was inserted into the peritoneum, and peritoneal fluid was collected through lavage. Cells were pelleted by centrifugation (5 min at 150 × g) and plated. After 2 h of incubation in RPMI medium with 10% serum, the medium was removed, and attached cells were washed with PBS. Fresh medium was added, and cells were treated as described under “Results.”
RNA Extraction and Real-time PCR
Cell lysates were suspended in RLT lysis buffer (Qiagen Inc., Valencia, CA) with β-mercaptoethanol (10 μl/ml RLT buffer). Total RNA was isolated using RNeasy isolation kits (Qiagen) according to the manufacturer's instructions. DNA digest was performed subsequently with DNase I treatment (1 μg/sample; Thermo Scientific, Glen Burnie, MD) followed by a 30-min incubation at 37 °C. The reaction was quenched with EDTA through a 10-min incubation at 67 °C. For each sample, 100–500 ng of RNA was processed to cDNA using RevertAid RT, RiboLock RT inhibitor, 5× reaction buffer, dNTP, and random primers (Thermo Scientific). cDNA was prepared in a 20-μl reaction mixture to a final concentration of 5 ng/μl.
RT-PCR was performed using the SYBR Green I detection system (Applied Biosystems, Inc., Carlsbad, CA). Each 20-μl sample of reaction mixture comprised 10–20 ng of cDNA and gene-specific primers at a final concentration of 2.5 nm. All samples were run in duplicate in an Applied Biosystems 7900 HT thermocycler with cycling conditions as follows: 48 °C for 30 min, 95 °C for 10 min, and 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Data were normalized to Rn18s mRNA expression and shown as -fold increase over control. The following primer sets were used for real-time PCR: ACSL1 (Acsl1) (sense, 5′-GCGGAGGAGAATTCTGCATAGAGAA-3′; antisense, 5′-ATATCAGCACATCATCTGTGGAAG-3′); IL-1β (Il1b) (sense, 5′- GGGCTGCTTCCAAACCTTTG-3′; antisense, 5′-TGATACTGCCTGCCTGAAGCTC-3′); CD36 (Cd36) (sense, 5′- GCAAAGAAGGAAAGCCTGTG-3′; antisense, 5′-CCAATGGTCCCAGTCTCATT-3′); CPT1α (Cpt1a) (sense, 5′-CCAGGCTACAGTGGGACATT-3′; antisense, 5′-GACTTGCCCATGTCCTTGT-3′); IL-6 (Il6) (sense, 5′-TAGTCCTTCCTACCCCAATTTCC-3′; antisense, 5′-TTGGTCCTTAGCCACTCCTTC-3′); TNFα (Tnfa) (sense, 5′-CCCTCACACTCAGATCATCTTCT-3′; antisense, 5′-GCTACGACGTGGGCTACAG-3′); p38α MAPK (Mapk14) (sense, 5′-GAGAAGATGCTCGTTTTGGACT-3′; antisense, 5′-GGACTGGTCATAAGGGTCAGC-3′); IL-12p40 (Il12b) (sense, 5′-TGGTTTGCCATCGTTTTGCTG-3′; antisense, 5′-ACAGGTGAGGTTCACTGT-3′); c-Jun (Jun) (sense, 5′-ACTGGGACCTTCTCACGTC-3′; antisense, 5′-GGTCGGTGTAGTGGTGATGT-3′); COX2 (Ptgs2) (sense, 5′-TGAGCAACTATTCCAAACCAGC-3′; antisense, 5′-GCACGTAGTCTTCGATCACTAT-3′); IFNβ (Ifnb) (sense, 5′-AGCTCCAAGAAAGGACGAACA-3′; antisense, 5′-GCCCTGTAGGTGAGGTTGAT-3′); MyD88 (Myd88) (sense, 5′-TCATGTTCTCCATACCCTTGGT-3′; antisense, 5′-AAACTGCGAGTGGGGTCAG-3′). Resulting Ct values were normalized to Rn18s, and the ΔΔCt method was then used to express values as -fold over control samples. All samples were run in at least duplicates, and statistical analysis was performed on 2−ΔCt values.
Western Blot
Cultured macrophages were suspended in Western lysis buffer (25 mm Tris-HCl, 2 mm EDTA, 10 mm Na2SO4, 150 mm NaCl, 50 mm NaF, 1% Triton X-100) containing protease inhibitors (Complete Protease Inhibitor Mixture, Santa Cruz Biotechnology, Inc., Santa Cruz, CA). The lysate was centrifuged (12,000 × g, 5 min), and the supernatant was collected. Protein concentration was quantified by a BCA assay, and equal amounts of protein extract (5–50 μg) were loaded onto 10% SDS-polyacrylamide gels. Protein then was transferred overnight to Immobilon-P transfer membranes (Millipore, Billerica, MA). Membranes were washed and blocked for 1 h with 5% milk in Tris-buffered saline solution with 1% Tween 20 (TBST). Membranes then were incubated overnight at 4 °C in the appropriate primary antibody (anti-ACSL1 at 1:1000, anti-phospho-JNK/SAPK Thr-183/Tyr-185 at 1:1000, anti-total JNK/SAPK at 1:1000, anti-phospho-p38 MAPK Thr-180/Tyr-182 at 1:1000, anti-total p38α MAPK at 1:1000, anti-phospho-IKKα/β Ser-176/180 at 1:1000, anti-total IKKβ at 1:1000, anti-phospho-STAT1 Tyr-701 at 1:1000, anti-total STAT1 at 1:1000, and anti-β-actin at 1:10,000; Cell Signaling Technology (Danvers, MA)). Membranes were washed in TBST with 1% Tween and incubated for 1 h at room temperature in horseradish peroxidase (HRP)-conjugated anti-rabbit or anti-mouse secondary antibody. Membranes again were washed in TBST with 1% Tween and developed using Immobilon Chemoluminescent HRP substrate (Millipore). Densitometry was performed using ImageJ software.
Macrophage Infection
E. coli (XL-1 Blue, Stratagene) and S. typhimurium (strain SL1344) (20) were cultured overnight in Luria-Burtani broth. Prior to infection, macrophages were changed to antibiotic-free medium. Bacteria were counted using optic densitometry, and 15 bacteria/macrophage were added to each well. Plates were centrifuged at 100 × g for 5 min. After 60 min of incubation, 1% gentamicin was added to the medium. After 6 h, cells were washed with PBS and suspended in RLT lysis buffer.
Chromatin Immunoprecipitation (ChIP) Assay
BMDCs were cultured and differentiated in the presence of M-CSF as described above. On day 7 of culture, macrophages were stimulated with LPS (5 ng/ml). At the designated time points, chromatin cross-linking was achieved through formaldehyde fixation for 15 min at room temperature with a final concentration of 1.5% formaldehyde. The reaction was quenched with 150 mm glycine before the cells were scraped and collected. Samples were washed with PBS and suspended in 150 μl of immunoprecipitation buffer (150 mm NaCl, 50 mm Tris-HCl, 5 mm EDTA, 0.5% Nonidet P-40, 1% Triton X-100, pH 7.5) with phosphatase (P2850 and P8340, Sigma) and protease (P5726, Sigma) inhibitors. Chromatin shearing was performed in a Bioruptor (Diagenode, Philadelphia, PA) at high intensity for 30 min with 30-s on-off cycles.
The ChIP assay was performed using the matrix ChIP method described elsewhere (21). Briefly, 96-well plates were incubated overnight with 0.2 mg of protein A in 100 μl of PBS/well. Wells were washed with 200 μl of PBS and then blocked for 1 h at room temperature with 200 μl of blocking buffer (IP buffer with 5% BSA and 100 μg/ml sheared salmon sperm DNA). Next, wells were incubated with antibody diluted in blocking buffer (2.5 μl/100 μl of buffer for polymerase II (Pol II), 0.5 μl/100 μl of buffer for c-Jun) for 1 h at room temperature. Chromatin samples (2 μl/well) were added, and the plate was floated in an ultrasonic water bath (1 h, 4 °C) to promote antibody binding. The wells then were washed three times with immunoprecipitation buffer (200 μl/wash) followed by three washes with TE buffer (200 μl/wash; 10 mm Tris-HCl, 1 mm EDTA, pH 7.0). Antibody-bound DNA was then eluted in 100 μl/well elution buffer (25 mm Tris, 1 mm EDTA, 1% IP buffer). Elution was performed in a thermal cycler (15 min at 55 °C, 15 min at 95 °C). RT-PCR was performed with 2 μl of eluted sample/well with 2 μl of SYBR Green master mix and appropriate primers at a final primer concentration of 0.25 μm and a total reaction volume of 4 μl. Primer sets were designed through use of the Primer-BLAST program. RT-PCR was run in 384-well Optical Reaction Plates (Applied Biosystems) using the 7900HT real-time PCR system and SDS software. All PCRs were run in triplicates. Data were normalized to Ct values for input wells, which contained eluted sample but not antibody. The RNA polymerase II (Pol II carboxyl terminus domain) and c-Jun antibodies were obtained from Santa Cruz Biotechnology, Inc. For PCR, the following primer sets were used: Acsl1 promoter −320 bp (sense, 5′-GTGACCTTAGTGGCTTTTGCTT-3′; antisense, 5′-GCTCCAGTCCTTCCCCTC-3′); Acsl1 promoter, −1250 bp (sense, 5′-TCTGCCCTTCAGTTTCCTGT-3′; antisense, 5′-GCTCCCAGCATTAGATGGAA-3′); Acsl1 promoter −1800 bp (sense, 5′-CCCTGAGAAAGGGTCATTCA-3′; antisense, 5′-AGTCTGAGGCTCCTTGTCCA-3′); Acsl1 promoter, −2700 bp (sense, 5′-CCTTTGCAGACCTCAGGAAC-3′; antisense, 5′-GCAACCACACACCAGAAATG-3′); Acsl1 transcription start site +200 bp (sense, 5′-GATGTCTGTGTTTTCTGTGGTCTT-3′; antisense, 5′-ATCCTGCCTTGGGTGACTT-3′); intergenic region −4 kb (sense, 5′-CAAACTGAGGCAATGCTGAA-3′; antisense, 5′-GGATTTCCTGGACGTTTTGA-3′).
Generation of JNK1/2 (Mapk8/Mapk9)-, p38α MAPK-, and STAT1-deficient Macrophages
In the Mapk8flox/flox9−/− mice, a Cre-ERT fusion protein that is sensitive to 4-hydroxytamoxifen was used to knock out Mapk8 (22). On day 3 of culture, BMDMs were incubated with 10 nm tamoxifen for 24 h, generating a double knock-out through knockdown of JNK1 (Mapk8) in Mapk9-knock-out mice. Macrophages from mice with myeloid cell-targeted JNK1/JNK2 deficiency were also used (16). Cells were washed with medium three times and stimulated with LPS or IFN-γ 72 h later.
p38α MAPK and STAT1 deficiency were achieved using two sequences of siRNA (Mapk14 siRNA A (s77114; reference sequences NM_001168508.1, NM_001168513.1, NM_001168514.1, and NM_011951.3) and B (s77113; reference sequences NM_001168508.1, NM_001168513.1, NM_001168514.1, and NM_011951.3; Stat1 siRNA A (s74444; reference sequence NM_009283.3) and B (s74445; reference sequence NM_009283.3); Ambion® Silencer Select, Invitrogen). Ambion® Silencer Negative Control 2 siRNA was used as a control for all experiments. To increase the efficiency of siRNA delivery, electroporation was performed, using the Amaxa Mouse Macrophage Nucleofector kit (Lonza Group Ltd.) and the Amaxa Nucleofector I electroporator (Lonza Group Ltd). Peritoneal macrophages were harvested 5 days after intraperitoneal thioglycollate injection and maintained in a single-cell suspension in RPMI. Cells were centrifuged for 5 min at 150 × g, and after aspiration of the supernatant, 2 million cells were resuspended in 100 μl of nucleofector solution with 30 pmol of siRNA and added to a cuvette for electroporation. The samples were electroporated in the Amaza Nucleofector I device using program Y-01. After electroporation, cells were plated in 12-well plates and incubated for 2 h in RPMI medium with 1% penicillin, streptomycin. The wells then were aspirated to remove non-adherent cells, and the remaining macrophages were maintained for 4 days in RPMI medium with 1% penicillin, streptomycin. The first 3 days after transfection, the media were also supplemented with 10% serum, which was removed 24 h prior to stimulation with LPS. After 4 days, macrophages were treated with ultrapure LPS (10 ng/ml) or E. coli. All cells were harvested in RW1 lysis buffer (Macherey-Nagel) 6 h after LPS stimulation, and mRNA was isolated using the Nucleospin RNA II kit according to the manufacturer's instructions. Knockdown of Mapk14 and Stat1 mRNA and induction of Acsl1 were verified by quantitative PCR. To verify efficiency of the knockdown at the protein level, additional samples were harvested in Western lysis buffer after 4 days of culture and 5 μg of protein from each condition were analyzed by Western blot.
Acyl-CoA Analysis
Long-chain acyl-CoA species were quantified by liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS), as described previously (14) in wild type and ACSL1-deficient BMDMs stimulated with LPS or vehicle for 24 h.
Phospholipid and Ceramide Analysis
Wild type and ACSL1-deficient BMDMs were stimulated for 12 h with LPS or left as unstimulated controls. In some experiments, 10 μm [d8]arachidonic acid (5Z,8Z,11Z,14Z-eicosatetraenoic-5,6,8,9,11,12,14,15-d8 acid; item number 10007277, Cayman Chemical) was added concurrently with LPS. Lipids were extracted by the method of Bligh and Dyer (23) in the presence of internal standards, including 1,2-dieicosanoyl-sn-glycero-3-phosphocholine and N-heptadecanoyl ceramide. Extracted lipids were resuspended and diluted in methanol/chloroform (4:1 by volume) prior to analysis by direct infusion electrospray ionization-mass spectrometry using a Thermo Fisher TSQ Quantrum Ultra® instrument. Samples were analyzed as sodiated adduct positive ions and deprotonated negative ions using tandem mass spectrometry (24, 25). For phosphatidylcholine (PC), neutral loss scanning of 59.1 was monitored at a collision energy of −28 eV in positive ion mode. Neutral loss scanning of 256.2 was performed for ceramide in the negative ion mode at a collision energy of 32 eV (26). Individual molecular species were quantified by comparing the ion intensity of individual molecular species with that of the appropriate internal standards following corrections for type I and type II 13C isotope effects (25). Specific molecular species of arachidonylated and [d8]arachidonylated plasmenylethanolamine and phosphatidylinositol were compared using selected reaction monitoring of the precursor ion and fragment fatty acid ion in negative ion mode using a collision energy of 25 eV. These selected reaction monitorings included 16:0–20:4 plasmenylethanolamine (722 → 303), 16:0-[d8]20:4 plasmenylethanolamine (730 → 311), 18:0–20:4 plasmenylethanolamine (750 → 303), 18:0-[d8]20:4 plasmenylethanolamine (758 → 311), 18:1–20:4 plasmenylethanolamine (748 → 303), 18:1-[d8]20:4 plasmenylethanolamine (756 → 311), 18:0–20:4 phosphatidylinositol (885 → 303), and 18:0-[d8]20:4 phosphatidylinositol (893 → 311).
Statistical Analysis
Statistical analyses were performed using GraphPad Prism 5 software (GraphPad, Inc., La Jolla, CA). All experiments were performed at least three times in independent experiments. Comparisons between two groups were made with an unpaired Student's t test, and comparisons among three or more groups were made with one-way or two-way analysis of variance and Tukey, Bonferroni, or Neuman-Keuls post-test. Results are expressed as mean values, and error bars indicate S.E. For all analyses, a p value of <0.05 was considered statistically significant.
RESULTS
ACSL1 Expression in Macrophages Is Induced by Proinflammatory Molecules and Infection with Gram-negative Bacteria
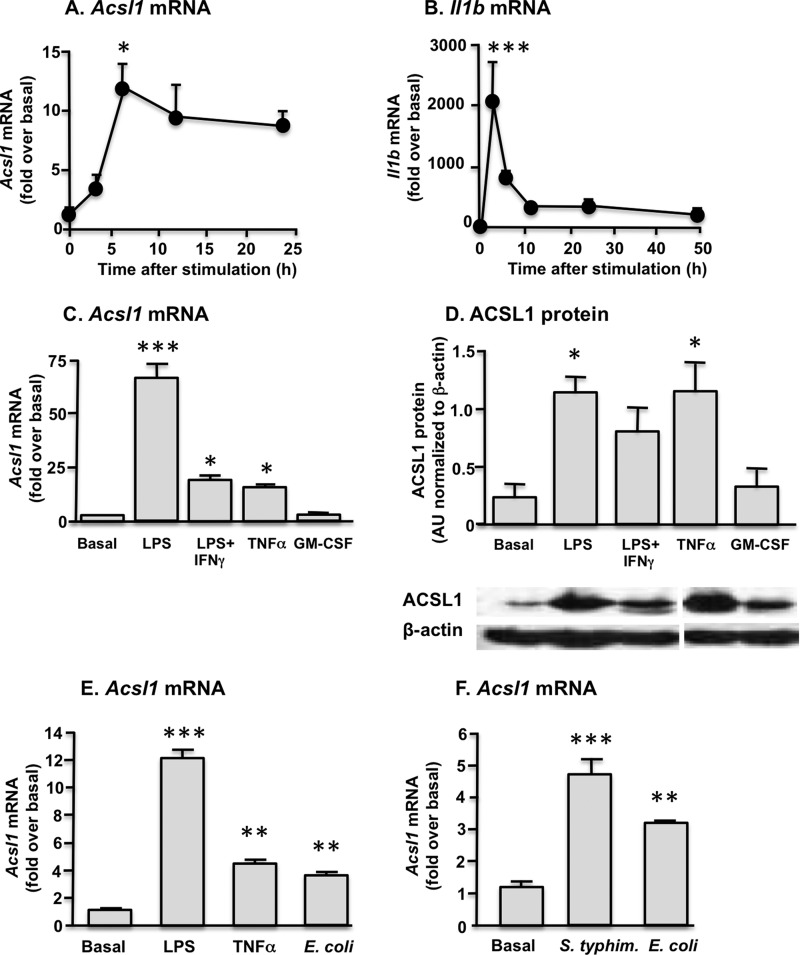
In order to gain insight into the stimuli and intracellular signal transduction pathways responsible for ACSL1 induction in macrophages, we first treated BMDMs with several different cytokines and LPS. LPS (5 ng/ml) plus IFN-γ (12 ng/ml) induced a marked increase in Acsl1 mRNA levels, which peaked 6 h after stimulation and remained elevated for at least 24 h (Fig. 1A). This response differed from that of Il1b mRNA induction, which was much more rapid and transient (Fig. 1B). Of the inflammatory mediators evaluated, LPS, LPS + IFN-γ (5 ng/ml and 12 ng/ml, respectively), and TNFα (20 ng/ml) significantly increased Acsl1 mRNA. In contrast, GM-CSF (10 ng/ml) did not induce Acsl1 (Fig. 1C). Similar results were obtained when ACSL1 protein levels were quantified by Western blot analysis (Fig. 1D).
FIGURE 1.
ACSL1 is induced by inflammatory stimuli in macrophages. A, significant induction of ACSL1 expression was evident by 6 h after macrophage stimulation with LPS + IFN-γ. B, in contrast, IL-1β (Il1b) expression peaked within 1 h of stimulation and rapidly declined thereafter. Significant induction of ACSL1 expression occurred after macrophage treatment with LPS, LPS + IFN-γ, and TNF-α but not GM-CSF. ACSL1 expression was comparable at the mRNA (C) and protein (D) levels. Samples in D were run on the same gel. E, macrophage exposure to LPS, TNFα, or E. coli significantly induced Acsl1 expression. F, exposure to the Gram-negative pathogens E. coli and S. typhimurium generated comparable induction of Acsl1 expression. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus basal or time 0. Error bars, S.E.
Because these findings suggest a role for ACSL1 in macrophage responses to pathogen, we examined ACSL1 expression in macrophages subsequent to acute bacterial infection. Macrophages were incubated with E. coli (15 bacteria/macrophage) for 6 h. A significant induction of Acsl1 mRNA was observed after exposure to E. coli, and the magnitude of this induction was comparable with that seen with direct TNFα stimulation but lower than that of LPS (Fig. 1E). To replicate these findings and to determine whether induction of ACSL1 was specific to E. coli or represented a more general pathogen response, we next incubated macrophages for 6 h with either E. coli or S. typhimurium. As with E. coli infection, significant induction of Acsl1 mRNA levels was observed after exposure to S. typhimurium (Fig. 1F). Together, these results demonstrate that ACSL1 is readily induced by inflammatory stimuli and Gram-negative bacteria in macrophages.
ACSL1 Expression in Macrophages Is Not Induced by PPAR Agonists
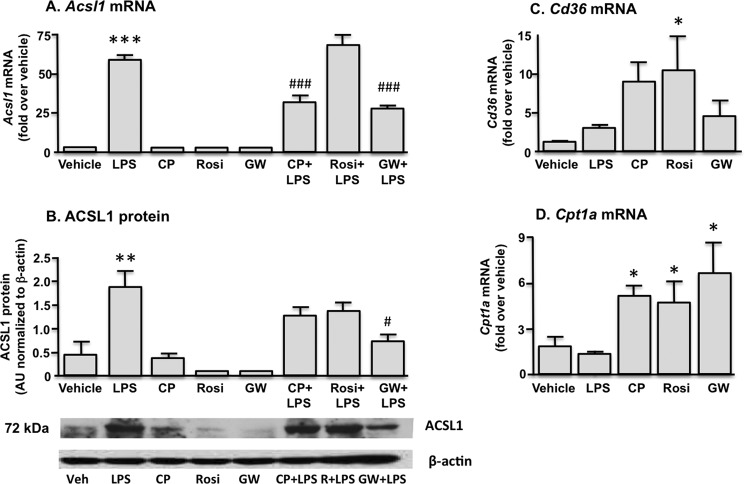
Because Acsl1 is a known transcriptional target of PPARα and PPARγ in other tissues (6, 10–13), we next examined the effect of PPAR agonists on macrophage ACSL1 expression. Macrophages were treated with either LPS, PPAR subtype-specific agonists, or LPS and a PPAR agonist in combination. None of the PPAR agonists induced ACSL1 expression (Fig. 2A). Notably, both the PPARα agonist CP900691 (10 μm) (27) and the PPARδ agonist GW 0742 (100 μm) exerted a suppressive effect on LPS-induced ACSL1 expression. The PPAR agonist-mediated suppression of the LPS effect was statistically significant at the mRNA expression level for both agonists and at the protein level for GW 0742 (Fig. 2B). A suppressive effect was not observed with the PPARγ agonist rosiglitazone (25 μm). Cd36 and Cpt1a mRNA levels were measured by real-time PCR to verify induction of known PPAR target genes (Fig. 2, C and D). Thus, in contrast to skeletal muscle and adipose tissue, PPAR agonists do not induce ACSL1 in macrophages but rather suppress the induction of ACSL1 by LPS.
FIGURE 2.
Macrophage stimulation with the PPARα agonist CP900691 (CP), the PPARγ agonist rosiglitazone (Rosi), or the PPARδ agonist GW 0742 (GW) does not induce ACSL1 expression. Treatment with the PPARα and PPARδ agonists partially attenuated LPS-mediated increases in ACSL1. Similar findings were observed at the mRNA (A) and protein (B) levels. Effective PPAR activation was verified by increased expression of the known PPAR target genes Cd36 (C) and Cpt1a (D). *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus vehicle control. #, p < 0.05; ###, p < 0.001 versus LPS-stimulated samples. Error bars, S.E.
LPS Induces ACSL1 through a TLR4-dependent and in Part TRIF-dependent Pathway in Macrophages
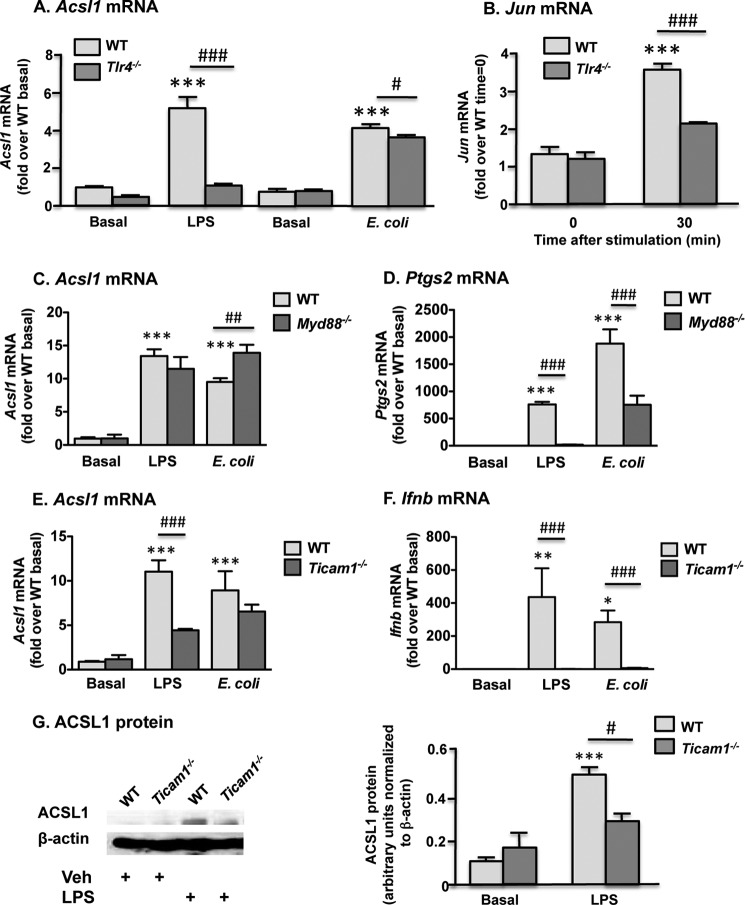
LPS is a known ligand for TLR4, which stimulates cellular responses through two principal downstream pathways. Thus, among the genes regulated by LPS, some depend on MyD88 signaling, whereas others require signaling through TRIF (28). To verify whether LPS and E. coli induce ACSL1 expression through TLR4 signaling, we first stimulated BMDMs derived from TLR4-deficient (Tlr4−/−) mice with LPS and E. coli. As anticipated, Tlr4−/− macrophages exhibited no increase in ACSL1 expression after LPS treatment, whereas E. coli-induced ACSL1 expression was minimally but significantly reduced by TLR4 deficiency (Fig. 3A). Neither LPS- nor E. coli-induced Acsl1 mRNA levels were reduced by MyD88 deficiency; rather, Acsl1 expression was higher in Myd88−/− macrophages subsequent to E. coli exposure, indicating a possible suppressive role for MyD88-dependent signal transduction (Fig. 3C). These findings thus suggested that the TRIF pathway may be predominantly responsible for LPS induction of ACSL1. Control experiments confirmed that the BMDMs from MyD88-deficient mice were devoid of Myd88 mRNA and exhibited a reduced ability to induce COX2 (Ptgs2) expression (Fig. 3D) (data not shown), a known MyD88-regulated gene (29). We next analyzed ACSL1 induction in BMDMs from TRIF-deficient (Ticam1−/−) mice. TRIF deficiency markedly attenuated LPS-induced ACSL1 expression at both the mRNA and protein levels (Fig. 3, E and G). Modest induction of ACSL1 nonetheless persisted after LPS stimulation in the Ticam1−/− macrophages, and no decrement in ACSL1 expression was observed after exposure to E. coli, thus underscoring the redundancy of pathways involved in ACSL1 regulation. To verify the inhibition of TRIF-dependent signaling in Ticam1−/− macrophages, Ifnb expression also was assessed and exhibited virtually complete abrogation after both LPS and E. coli stimulation (Fig. 3F). For these experiments, ultrapure LPS was employed to minimize the possibility of stimulating non-TLR4-dependent pathways.
FIGURE 3.
ACSL1 is induced by LPS through a TRIF-dependent pathway in macrophages. A, LPS- but not E. coli-mediated ACSL1 induction was completely abolished in TLR4-deficient macrophages. B, decreased Jun expression was apparent in Tlr4−/− macrophages subsequent to LPS stimulation. MyD88 deficiency did not attenuate ACSL1 expression after LPS or E. coli stimulation (C) but resulted in pronounced suppression of COX2 (Ptgs2) induction (D). E, TRIF-deficient macrophages exhibited reduced levels of Acsl1 mRNA after treatment with LPS but not E. coli. F, induction of Ifnb was completely abolished in Ticam1−/− macrophages. G, reduced ACSL1 expression was evident at the protein level in TRIF-deficient macrophages 24 h after LPS stimulation. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus WT basal control. #, p < 0.05; ##, p < 0.01; ###, p < 0.001 versus LPS-stimulated or E. coli-stimulated control, as indicated. Error bars, S.E.
Several Redundant Signaling Pathways Downstream of TLR4 Are Likely to Contribute to ACSL1 Induction in Macrophages
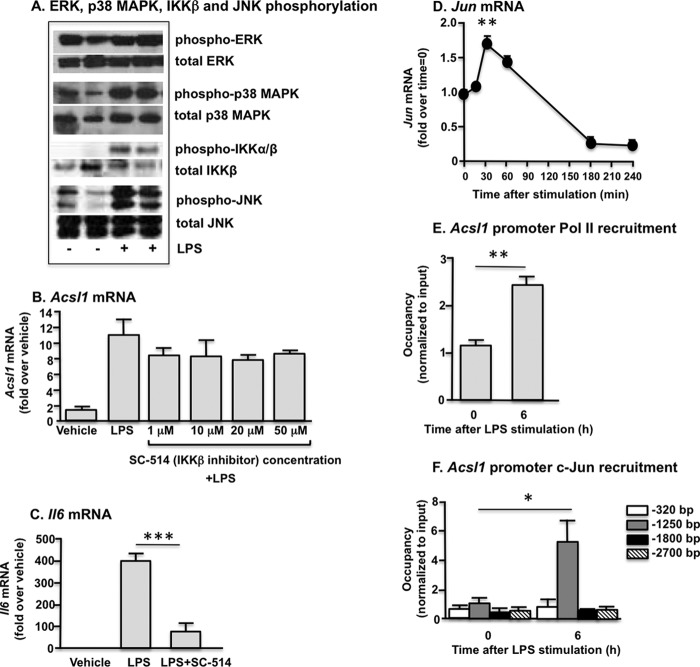
To further delineate the signaling pathways involved in LPS-induced ACSL1 expression, we analyzed some of the known mediators downstream of TRIF (JNK, p38 MAPK, ERK1/2, and IKKβ) in the macrophage LPS response (30). As expected, these pathways all were activated by LPS (Fig. 4A).
FIGURE 4.
LPS stimulates multiple pathways in macrophages. A, macrophage stimulation with LPS induces phosphorylation of the ERK1/2, JNK1/2, and p38 MAPKs and IKKβ. B, the IKKβ inhibitor SC-514 did not affect LPS-mediated induction of Acsl1 expression. C, inhibition of NF-κB activity with SC-514 was demonstrated by significant suppression of LPS-induced Il6 expression. D, after LPS stimulation, a significant increase in Jun expression was evident at 30 min, with subsequent decline to below base-line expression by 3 h. E, significant recruitment of Pol II to the Acsl1 TSS was evident 6 h after LPS stimulation. F, LPS treatment enhanced c-Jun binding to the Acsl1 promoter at a site 1250 bp upstream of the TSS. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus the indicated treatment group or versus time 0 (D). Error bars, S.E.
The role of IKK activation in LPS-induced ACSL1 expression was addressed first. The IKKβ inhibitor SC-514 had no effect on LPS-induced Acsl1 mRNA levels at concentrations up to 50 μm, whereas it markedly inhibited LPS induction of Il6 mRNA (Fig. 4, B and C). The MEK1 inhibitor PD98059 also had no effect on LPS-induced Acsl1 mRNA levels (data not shown). We therefore focused our studies on the JNK and p38 MAPK pathways, which mediate inflammatory effects in macrophages (16, 31–33).
Both JNK and p38 MAPK can signal through the transcription factor activator protein-1 (AP-1) (33, 34), and the Acsl1 promoter contains several AP-1 consensus sequences (TGACTCA). Thus, AP-1 appeared promising as an initial candidate among transcription factors that could be implicated in ACSL1 expression. The AP-1 heterodimer contains Jun proteins, so we first investigated LPS-mediated regulation of Jun. A significant increase in Jun expression was evident 30 min after LPS stimulation, prior to appreciable induction of ACSL1 (Fig. 4D). Further, diminished Jun expression was observed in Tlr4−/− macrophages in association with the abrogation of LPS-induced ACSL1 expression (Fig. 3B). To investigate the possibility that LPS induces AP-1 recruitment to the Acsl1 promoter, we performed chromatin immunoprecipitation (ChIP) in tandem with RT-PCR.
Bone marrow-derived cells were isolated and differentiated in the presence of M-CSF. Macrophages were stimulated with LPS (5 ng/ml) and harvested at 0, 1, 3, 6, 9, and 24 h. Chromatin was immunoprecipitated with an RNA Pol II antibody, and RT-PCR subsequently was performed to quantify binding at a region 200 bp downstream of the Acsl1 transcription start site (TSS). A time course of Pol II binding near the TSS recapitulated our mRNA time course data; Pol II binding increased within 1 h of LPS treatment and peaked at 3–6 h after stimulation (data not shown), further suggesting that LPS increases Acsl1 mRNA levels through a transcriptionally mediated effect. We therefore used the 0 and 6 h time points for additional experiments aimed to investigate the role of AP-1 in LPS-induced ACSL1 expression.
Chromatin was isolated from macrophages prior to and 6 h after stimulation with LPS (5 ng/ml). ChIP was repeated with a Pol II antibody to verify induction of Acsl1 transcription and to optimize chromatin volume. Significantly increased Pol II binding again was observed near the Acsl1 TSS (+200 bp) (Fig. 4E), whereas no enhanced binding was evident at a negative control site selected in an intergenic region 4 kb upstream of the Acsl1 gene (data not shown).
We examined recruitment of the AP-1 constituent c-Jun to the Acsl1 promoter at four potential AP-1 binding sites. Six hours after LPS stimulation, enhanced c-Jun binding was evident at a region 1250 bp upstream of the TSS (Fig. 4F). In contrast, no change in c-Jun binding was observed at the −320, −1800, or −2700 bp regions. These results suggest that LPS-induced AP-1 activation may be partly responsible for ACSL1 induction.
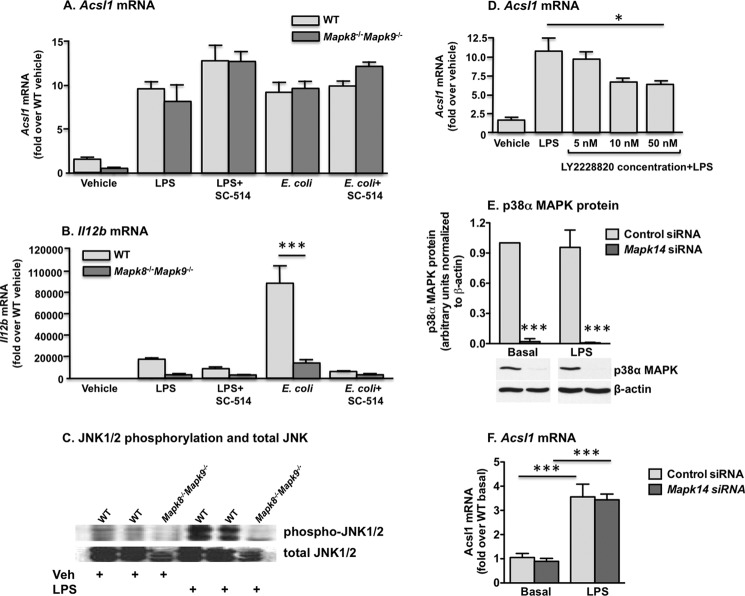
We next sought to determine the potential roles of the JNK and p38 MAPK pathways in ACSL1 induction. We first examined the expression profile of ACSL1 in macrophages deficient in JNK1 (Mapk8) and JNK2 (Mapk9), the JNK subtypes present in macrophages (31). No attenuation of ACSL1 expression in Mapk8−/−Mapk9−/− double deficient macrophages was observed subsequent to LPS or E. coli stimulation (Fig. 5A). Importantly, combined JNK1 and JNK2 deficiency dramatically inhibited LPS- and E. coli-induced IL-12p40 (Il12b) induction, consistent with previous results (Fig. 5B) (35). Western blot analysis was performed to verify JNK1/2 deficiency (Fig. 5C).
FIGURE 5.
Deficiency of JNK1/2 signaling in macrophages does not attenuate ACSL1 expression subsequent to LPS or E. coli exposure, as compared with WT macrophages. A, no effect on ACSL1 expression was evident with co-administration of the IKKβ inhibitor SC-514. B, reduced signaling through JNK1/2- and IKK-mediated pathways was confirmed by expression levels of IL-12p40 (Il12b). JNK deficiency was confirmed at the protein level by Western blot. C, a representative blot. D, pretreatment with the p38 MAPK inhibitor LY2228820 partially but significantly reduced LPS-stimulated ACSL1 expression in macrophages. Macrophage transfection with p38α MAPK (Mapk14) siRNA reduced p38α MAPK protein levels (E) but did not attenuate LPS-induced ACSL1 expression (F). *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus the indicated treatment group. Error bars, S.E.
Although our studies with protein kinase inhibitors did not support NF-κB-mediated regulation of ACSL1, the Acsl1 promoter has a putative NF-κB binding site; thus, genetic deletion of both JNK subtypes potentially could induce ACSL1 through an alternative, LPS-responsive pathway. To investigate this possibility, we also examined Acsl1 expression in Mapk8−/−Mapk9−/− macrophages after pretreatment with the IKKβ inhibitor SC-514. Again, no reduction in LPS- or E. coli-induced ACSL1 expression was observed. Thus, reducing JNK-mediated pathways alone or in combination with IKKβ inhibition is not sufficient to inhibit TLR4-mediated ACSL1 expression.
We next investigated the role of p38 MAPK because p38 MAPK is induced by LPS and also can activate AP-1 (33). In macrophages, p38α and p38δ are the predominant isoforms, whereas the p38β isoform is not readily detected (36). We first targeted the p38 MAPK isoform using the pharmacological inhibitor LY2228820. LY2228820 partially inhibited ACSL1 induction after LPS stimulation (Fig. 5D). We next employed an siRNA approach to specifically target p38α MAPK. Using electroporation, we observed robust (>90%) reduction of p38α MAPK protein (Fig. 5E). However, despite nearly complete p38α MAPK knockdown, no decrement in ACSL1 expression was evident after LPS stimulation (Fig. 5F). These results therefore collectively suggest that several redundant signaling pathways downstream of TLR4 are likely to contribute to ACSL1 induction in macrophages.
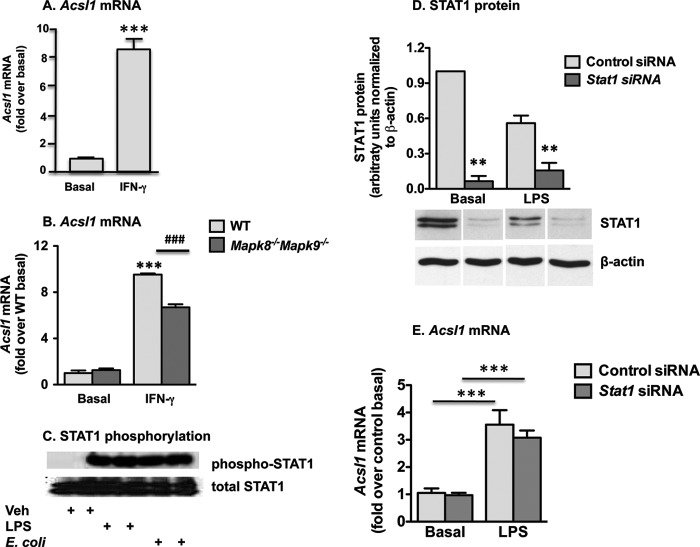
IFN-γ Mediates ACSL1 Induction in Macrophages in Part through JNK1/JNK2
IFN-γ is a key cytokine that induces inflammatory macrophage activation. We therefore investigated whether IFN-γ induces ACSL1. Indeed, a significant induction of Acsl1 mRNA was obvious in BMDMs 6 h after stimulation with IFN-γ (Fig. 6A). IFN-γ signaling stimulates MAPK-dependent pathways, in part through inhibition of MAPK phosphatase-1 (37), so we again examined ACSL1 expression in Mapk8−/−Mapk9−/− macrophages, this time subsequent to IFN-γ treatment. Notably, the absence of JNK1/2 significantly reduced Acsl1 induction in response to IFN-γ (Fig. 6B), suggesting that JNK1/2 signaling plays a more prominent part in ACSL1 induction by IFN-γ, compared with that induced by LPS or E. coli.
FIGURE 6.
IFN-γ induces ACSL1 expression partly through JNK. A, IFN-γ treatment significantly induced ACSL1 expression in macrophages. B, Acsl1 expression was diminished in Mapk8−/−Mapk9−/− macrophages subsequent to treatment with IFN-γ. C, robust increases in STAT1 phosphorylation were evident 2 h after macrophage stimulation with LPS or E. coli. D, macrophage transfection with Stat1-targeting siRNA generated >70% knockdown of STAT1 protein (basal and LPS data derive from two different blots, but control siRNA and Stat1 siRNA lanes are from the same blot for basal and LPS-stimulated samples, respectively, and β-actin blots for control siRNA-treated samples are the same as in Fig. 5E because these samples were run on the same gels). However, STAT1 knockdown did not reduce LPS-stimulated Acsl1 expression (E). **, p < 0.01 versus control siRNA; ***, p < 0.001 versus basal; ###, p < 0.001 versus IFN-γ-stimulated WT. Error bars, S.E.
Array studies have recently shown that Acsl1 is among the IFN-γ-induced genes that require STAT1 signaling in macrophages (38). TLR4-TRIF signaling results in type I IFN production (39), which, like IFN-γ, signals through STAT1. Thus, TLR4-TRIF might induce ACSL1 expression through an autocrine mechanism involving type I IFN release and STAT1 activation. Accordingly, we next assessed STAT1 phosphorylation after macrophage stimulation with LPS or E. coli and found robust increases in phosphorylated (Tyr-710) STAT1 subsequent to either treatment (Fig. 6C).
Again using an siRNA approach, we investigated the effect of STAT1 knockdown on Acsl1 expression subsequent to LPS stimulation. With cell electroporation to optimize siRNA delivery, we achieved substantial (>70%) knockdown of STAT1 protein under basal and LPS-stimulated conditions (Fig. 6D). Nonetheless, inhibition of STAT1 signaling yielded no significant reduction in LPS-induced Acsl1 expression (Fig. 6E).
Together, these findings show that ACSL1 is induced by multiple inflammatory signaling pathways in macrophages. Moreover, different inflammatory signals appear to induce ACSL1 expression through discrete signaling mechanisms characterized by redundancy in the implicated pathways.
ACSL1 Is Required for LPS-mediated Phospholipid Turnover in Activated Macrophages
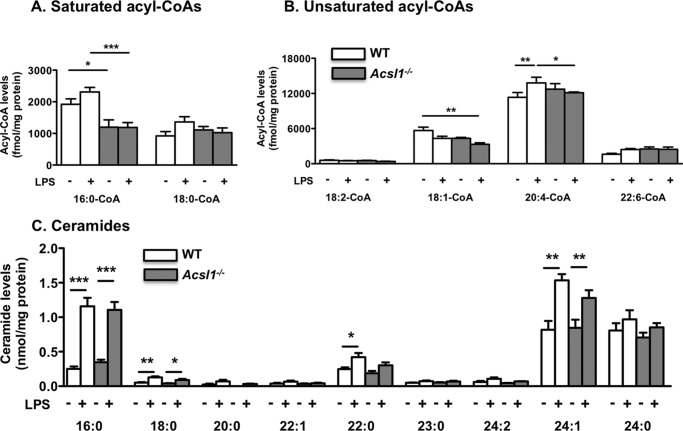
In order to gain insight into ACSL1 function in LPS-stimulated macrophages, we investigated its potential role in modulating LPS-mediated effects on acyl-CoA species, ceramides, and phospholipids in BMDMs harvested from wild type and myeloid-specific ACSL1-deficient mice, described previously (14). Of the saturated acyl-CoAs measured, 16:0-CoA was more abundant than 18:0-CoA (Fig. 7A). Among the unsaturated acyl-CoAs, 20:4-CoA was the most abundant species, consistent with results in thioglycollate-elicited peritoneal macrophages (14). ACSL1 deficiency resulted in a significant reduction in 16:0-CoA levels under both basal and LPS-stimulated conditions and a reduction in 18:1-CoA levels under LPS-stimulated conditions (Fig. 7, A and B). LPS had no significant effect on any of the acyl-CoA species analyzed except for 20:4-CoA, in which a small but significant increase was observed (Fig. 7B). ACSL1 deficiency prevented this LPS-mediated rise in 20:4-CoA levels. LPS treatment resulted in a marked increase in several ceramide species 12 h after stimulation, including 16:0-, 18:0-, 22:0-, and 24:1-ceramide, but ACSL1 deficiency had no significant impact on these LPS-mediated effects at this time point (Fig. 7C).
FIGURE 7.
ACSL1 deficiency leads to reduced 16:0-CoA, 18:1-CoA, and 20:4-CoA levels in LPS-stimulated macrophages but does not affect ceramide levels. ACSL1-deficient BMDMs and WT controls were stimulated with LPS or vehicle for 24 h, and saturated and unsaturated acyl-CoA species were measured by LC-ESI-MS/MS (A and B). Ceramides were measured by direct infusion ESI-MS, as described in detail under “Experimental Procedures.” C, ceramide molecular species in ACSL1-deficient BMDMs and WT controls stimulated with LPS for 12 h. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus the indicated treatment group; white bars, WT macrophages; gray bars, ACSL1-deficient macrophages. Error bars, S.E.
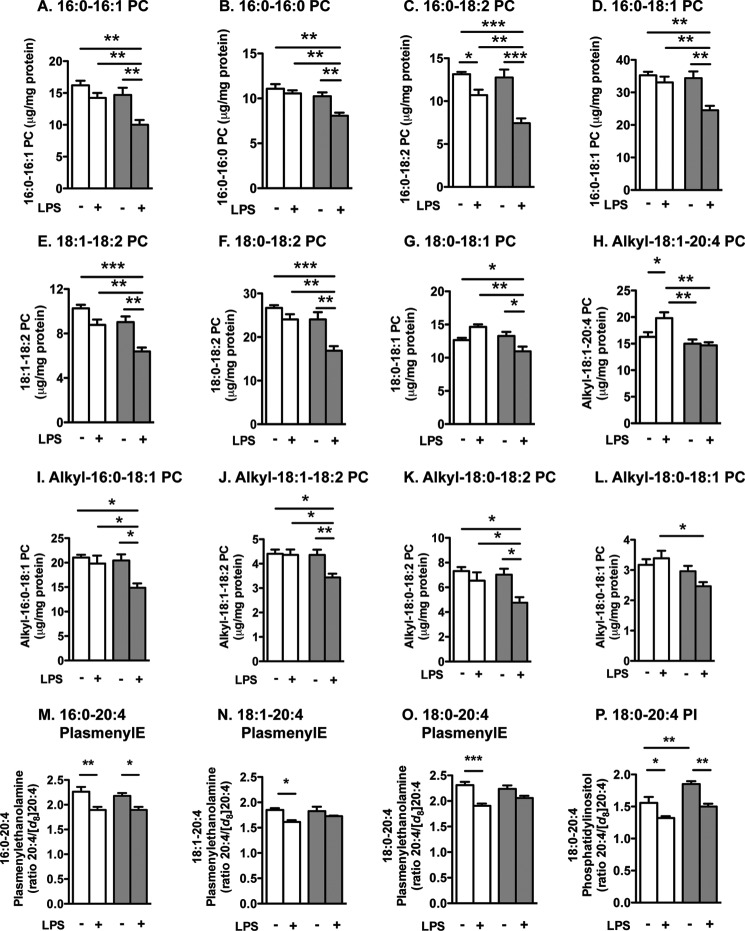
However, ACSL1 deficiency had significant effects on PC species under LPS-stimulated conditions. Thus, in LPS-stimulated BMDMs, ACSL1 deficiency significantly reduced levels of several PC species, including 16:0–16:1 PC, 16:0–16:0 PC, 16:0–18:2 PC, 16:0–18:1 PC, 18:1–18:2 PC, 18:0–18:2 PC, 18:0–18:1 PC, alkyl-18:1–20:4 PC, alkyl-16:0–18:1 PC, alkyl-18:1–18:2 PC, alkyl-18:0–18:2 PC, and alkyl-18:0–18:1 PC (Fig. 8, A–L). Interestingly, ACSL1 deficiency had no significant effect or a much smaller effect on these phospholipids in cells under basal conditions.
FIGURE 8.
ACSL1 deficiency leads to reduced turnover of many phospholipid species and reduced arachidonic acid turnover in phospholipids. ACSL1-deficient BMDMs and WT controls were stimulated with LPS or vehicle for 12 h. PC, plasmenylethanolamine (PlasmenylE), and PI were measured by direct infusion ESI-MS as described in detail under “Experimental Procedures.” A–L, PC molecular species were quantified. M–P, turnover of 20:4 in plasmenylethanolamine and PI molecular species was evaluated by incubating the macrophages in the presence of 10 μm deuterated 20:4 during the 12 h. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus the indicated treatment group; white bars, WT macrophages; gray bars, ACSL1-deficient macrophages. Error bars, S.E.
LPS stimulates 20:4-CoA synthesis and metabolism in macrophages, and we previously have demonstrated that ACSL1 deficiency results in reduced 20:4-CoA levels in thioglycollate-elicited peritoneal macrophages (14); therefore, we next investigated turnover of 20:4-containing phosphatidylethanolamine and phosphatidylinositol (PI) by measuring the ratio of non-deuterated 20:4 versus deuterated 20:4 in phosphatidylethanolamine and PI. In wild type BMDMs, LPS caused a significant reduction in this ratio (Fig. 8, M–P), indicating an increased turnover. This accelerated turnover was inhibited by ACSL1 deficiency for 18:1–20:4 and 18:0–20:4 plasmenylethanolamine (Fig. 8, N–O). However, the effect of LPS was not attenuated by ACSL1 deficiency for 16:0–20:4 plasmenylethanolamine or 18:0–20:4 PI (Fig. 8, M and P). Finally, ACSL1 deficiency significantly reduced turnover of 20:4 in 18:0–20:4 PI under basal conditions (Fig. 8P). Together, these results indicate that ACSL1 deficiency leads to reduced 16:0-CoA, 18:1-CoA, and 20:4-CoA levels in LPS-stimulated macrophages and that LPS-induced turnover of a number of phospholipids containing these fatty acids is impaired by ACSL1 deficiency.
DISCUSSION
In the present study, we demonstrate that ACSL1 expression in macrophages is induced by acute exposure to Gram-negative bacterial pathogens and the proinflammatory molecules LPS, IFN-γ, and TNFα. In contrast to its regulation in other cell types, Acsl1 is not a PPAR transcriptional target in macrophages but rather is a downstream target of multiple signal transduction pathways, with the TLR4-TRIF pathway specifically implicated in LPS-induced expression and JNK1/JNK2 implicated in IFN-γ-induced ACSL1. In contrast, E. coli-mediated induction of ACSL1 is only partly dependent on TLR4, and neither TRIF- nor JNK-dependent signaling is required for ACSL1 expression after macrophage stimulation with E. coli. These findings collectively underscore the cell- and stimulus-specific regulation of ACSL1 and suggest a novel role for this enzyme in the innate immune response. Further, the apparent redundancy of the pathways implicated in ACSL1 regulation supports the biological importance of ACSL1 in activated macrophages.
An emerging research area of interest is the differential regulation of gene expression in different tissues due to chromatin accessibility and histone marks. For example, PPARγ regulates different genes in macrophages and adipocytes due to differences in binding regions marked with repressive histone modifications (40). It is likely that the absence of ACSL1 induction with PPAR agonists in macrophages is due to repressive histone modifications of the Acsl1 promoter in these cells, whereas adipocytes and myocytes exhibit different modification profiles. Indeed, histone deacetylase 3 has recently been shown to regulate the inflammatory program in macrophages (41).
The initial suggestion that ACSL1 plays a role in innate immunity derives from our observations that a combination of LPS and IFN-γ induces ACSL1 expression in macrophages and, further, that ACSL1-deficient macrophages exhibit diminished cytokine generation (14). Here we demonstrate that ACSL1 induction by LPS requires signaling through TLR4 and the adaptor protein TRIF but not MyD88. A number of redundant signaling pathways appear to be involved in inducing ACSL1 downstream of TLR4-TRIF. Thus, inhibiting or knocking down IKK, ERK1/2, JNK1/2, or p38α MAPK was inefficient in reducing LPS-induced ACSL1 expression, at least at the time of maximal stimulation (6 h). However, ChIP-PCR data supported the involvement of JNK and/or p38 MAPK pathways in Acsl1 transcriptional regulation because increased recruitment of c-Jun was observed at a specific putative AP-1 binding site located on the Acsl1 promoter. This c-Jun recruitment was concurrent with enhanced Pol II binding at the TSS following LPS stimulation, strongly implicating AP-1 as a contributing transcription factor in LPS-induced ACSL1 expression. Further supporting the role of AP-1 in ACSL1 regulation, simultaneous treatment of macrophages with LPS and IFN-γ appeared to attenuate LPS induction of ACSL1, and IFN-γ has been shown to inhibit LPS-induced AP-1 activity in human monocytes (42). p38 MAPK isoforms are known regulators of AP-1 and can activate constituent AP-1 proteins, including ATF2 (43). p38α MAPK also induces Jun expression through the transcription factor MEF2c (44), and LPS-stimulated increases in Jun transcript precede those in Acsl1, lending additional albeit indirect support for p38 MAPK-AP-1 mediated regulation of Acsl1 transcription. Findings derived from use of the pharmacological p38 MAPK inhibitor LY2228820 supported this putative signaling pathway, but experiments employing siRNA clearly demonstrated that p38α MAPK signaling is not required for maximal LPS-induced ACSL1 expression. We cannot exclude a role for p38δ MAPK in ACSL1 regulation. Thus, p38 MAPK may contribute to ACSL1 regulation, but the importance of additional pathways appears likely.
Because JNK is also an upstream regulator of AP-1, we further interrogated this signaling pathway. JNK exists in three distinct subtypes encoded by separate genes, each with its respective isoforms generated through alternative splicing (31). Two of these subtypes, JNK1 and JNK2, are expressed in macrophages and exert discrete biological effects (45–47). Accordingly, we used macrophages deficient in both JNK1 and JNK2 to evaluate the role of JNK signaling in ACSL1 regulation. Similar to our results with p38 MAPK, the nearly complete absence of JNK-dependent signaling yielded no attenuation of LPS-induced ACSL1 expression. Another possibility is that TLR4-TRIF-induced ACSL1 induction is due to IRF3 (interferon regulatory transcription factor 3). However, there is also remarkable redundancy in pathways utilizing IRF family transcription factors. For example, macrophages deficient in IRF3, IRF5, and IRF7 are still able to produce IFN-β following viral infection (48).
Isolated IFN-γ stimulation produces robust induction of ACSL1, which elsewhere has been shown to be contingent on STAT1 signaling (38). Accordingly, we also examined the role of STAT1 in mediating LPS-induced ACSL1 expression. Although we achieved robust knockdown of STAT1 through an siRNA strategy, no attenuation of LPS-induced ACSL1 expression was observed, arguing against a pivotal role for STAT1 in ACSL1 regulation downstream of TLR4-TRIF signaling. Interestingly, however, JNK-deficient macrophages exhibited reduced ACSL1 expression exclusively after IFN-γ but not LPS treatment, underscoring the stimulus-dependent nature of ACSL1 regulation. Notably, too, selective inhibition of any of the examined pathways, with the exception of a small effect of TLR4 deficiency, failed to attenuate ACSL1 expression after macrophage exposure to E. coli. Thus, additional work is needed to determine the signals responsible specifically for Gram-negative pathogen-mediated ACSL1 induction.
ACSL1 expression increased after macrophage exposure to both E. coli and S. typhimurium, suggesting this enzyme may be induced broadly in response to Gram-negative bacteria rather than specific, individual pathogens. The functional role of ACSL1 in macrophages nonetheless has remained unclear because esterification is a critical, preliminary step for fatty acid utilization in a variety of cellular functions. In adipocytes and hepatocytes, ACSL1 principally generates fatty acyl-CoAs that serve as substrate for β-oxidation and triglyceride synthesis (7, 8). In contrast, macrophages do not rely predominantly on mitochondrial fatty acid oxidation for ATP generation, and exposure to bacterial pathogens or LPS further skews macrophage energy metabolism toward glycolysis (49). Moreover, previous work has demonstrated that ACSL1-deficient macrophages do not exhibit reduced production of β-oxidation metabolites but do show significantly reduced arachidonoyl-CoA levels, which probably contributes to the reduced production of certain prostaglandin species we observed (14). Thus, ACSL1 appears to play a fundamentally different role in macrophages from that in other cell types. To investigate the biological role of ACSL1 in macrophages, we examined changes in acyl-CoAs, ceramides, and phospholipids in ACSL1-deficient macrophages.
The functional significance of LPS-induced ACSL1 expression was investigated by thorough analysis of changes in acyl-CoAs, ceramides, and phospholipids in wild type and ACSL1-deficient macrophages. Together, these findings indicated that ACSL1 promotes synthesis of 16:0-CoA, 18:1-CoA, and 20:4-CoA in LPS-stimulated macrophages, but LPS conferred minimal effects on these CoA species, with only a small stimulatory effect on 20:4-CoA levels at the time point studied. In contrast, LPS caused a marked increase in several ceramide species, none of which was affected by ACSL1 deficiency. This absence of effect on ceramide production occurred despite the fact that ACSL1-deficient macrophages exhibited decreased levels of 16:0-CoA, the precursor for de novo ceramide biosynthesis. Ceramides previously have been shown to mediate the synergistic effects of LPS and fatty acids on enhanced cytokine production in stimulated macrophages (50). The present findings therefore argue against reduced ceramide synthesis as a mechanism underlying the anti-inflammatory effects of ACSL1 deficiency. Conversely, ACSL1 deficiency significantly reduced the turnover of several phospholipid species containing 16:0 and 20:4 acyl chains in LPS-stimulated macrophages. This finding is likely to explain the reduced prostaglandin E2 generation in ACSL1-deficient macrophages observed previously (14), and it also suggests that ACSL1 deficiency might negatively regulate signaling pathways downstream of other phospholipases in addition to phospholipase A2. In aggregate, these data therefore indicate that the cellular capacity to re-esterify fatty acids may prove a key, rate-limiting step in the generation of certain bioactive lipids in stimulated macrophages.
Collectively, these findings demonstrate a distinct profile of ACSL1 regulation in macrophages and suggest in parallel a highly cell-specific functional role for this enzyme. Future investigation will reveal whether this mode of regulation of ACSL1 proves wholly unique to macrophages or is shared with other cell types. Future studies also are necessary to determine how ACSL1-dependent flux in phospholipid species contributes functionally to host defense and other facets of innate immune activity.
Acknowledgments
CP900691 was a generous gift from Pfizer, Inc. Acyl-CoAs were analyzed through the Molecular Phenotyping Core, Michigan Nutrition and Obesity Center (supported by National Institutes of Health Grant P30 DK089503).
This work was supported, in whole or in part, by National Institutes of Health Grants HL062887, HL092969, HL097365 (to K. E. B.); 6K12HD053984 (to K. B. R.); AI062859 (to K. D. S.); HL074214 and HL111906 (to D. A. F.); DK082841 (to S. P.); and DK083310 and R37 DK45978 (to K. B.). This work was also supported by Juvenile Diabetes Research Foundation Grant 42-2009-779 (to K. B.). Part of the study was supported by the Diabetes Research Center at the University of Washington (National Institutes of Health Grant DK017047). This work was also supported by NIAID, National Institutes of Health, contract HHSN272200700038C, “Systems Approach to Immunity and Inflammation.”
- ACSL
- acyl-CoA synthetase
- PPAR
- peroxisome proliferator-activated receptor
- BMDM
- bone marrow-derived macrophage
- BMDC
- bone marrow-derived cell
- Pol
- polymerase
- ESI
- electrospray ionization
- PC
- phosphatidylcholine
- PI
- phosphatidylinositol
- TSS
- transcription start site.
REFERENCES
- 1. Mashek D. G., Li L. O., Coleman R. A. (2006) Rat long-chain acyl-CoA synthetase mRNA, protein, and activity vary in tissue distribution and in response to diet. J. Lipid Res. 47, 2004–2010 [DOI] [PubMed] [Google Scholar]
- 2. Suzuki H., Kawarabayasi Y., Kondo J., Abe T., Nishikawa K., Kimura S., Hashimoto T., Yamamoto T. (1990) Structure and regulation of rat long-chain acyl-CoA synthetase. J. Biol. Chem. 265, 8681–8685 [PubMed] [Google Scholar]
- 3. Lobo S., Wiczer B. M., Bernlohr D. A. (2009) Functional analysis of long-chain acyl-CoA synthetase 1 in 3T3-L1 adipocytes. J. Biol. Chem. 284, 18347–18356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. de Jong H., Neal A. C., Coleman R. A., Lewin T. M. (2007) Ontogeny of mRNA expression and activity of long-chain acyl-CoA synthetase (ACSL) isoforms in Mus musculus heart. Biochim. Biophys. Acta 1771, 75–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fujimoto Y., Onoduka J., Homma K. J., Yamaguchi S., Mori M., Higashi Y., Makita M., Kinoshita T., Noda J., Itabe H., Takanoa T. (2006) Long-chain fatty acids induce lipid droplet formation in a cultured human hepatocyte in a manner dependent of acyl-CoA synthetase. Biol. Pharm. Bull. 29, 2174–2180 [DOI] [PubMed] [Google Scholar]
- 6. Coleman R. A., Lewin T. M., Van Horn C. G., Gonzalez-Baró M. R. (2002) Do long-chain acyl-CoA synthetases regulate fatty acid entry into synthetic versus degradative pathways? J. Nutr. 132, 2123–2126 [DOI] [PubMed] [Google Scholar]
- 7. Li L. O., Ellis J. M., Paich H. A., Wang S., Gong N., Altshuller G., Thresher R. J., Koves T. R., Watkins S. M., Muoio D. M., Cline G. W., Shulman G. I., Coleman R. A. (2009) Liver-specific loss of long chain acyl-CoA synthetase-1 decreases triacylglycerol synthesis and β-oxidation and alters phospholipid fatty acid composition. J. Biol. Chem. 284, 27816–27826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ellis J. M., Li L. O., Wu P. C., Koves T. R., Ilkayeva O., Stevens R. D., Watkins S. M., Muoio D. M., Coleman R. A. (2010) Adipose acyl-CoA synthetase-1 directs fatty acids toward β-oxidation and is required for cold thermogenesis. Cell Metab. 12, 53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ellis J. M., Mentock S. M., Depetrillo M. A., Koves T. R., Sen S., Watkins S. M., Muoio D. M., Cline G. W., Taegtmeyer H., Shulman G. I., Willis M. S., Coleman R. A. (2011) Mouse cardiac acyl coenzyme a synthetase 1 deficiency impairs fatty acid oxidation and induces cardiac hypertrophy. Mol. Cell. Biol. 31, 1252–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rakhshandehroo M., Hooiveld G., Müller M., Kersten S. (2009) Comparative analysis of gene regulation by the transcription factor PPARα between mouse and human. PLoS One 4, e6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schoonjans K., Watanabe M., Suzuki H., Mahfoudi A., Krey G., Wahli W., Grimaldi P., Staels B., Yamamoto T., Auwerx J. (1995) Induction of the acyl-coenzyme A synthetase gene by fibrates and fatty acids is mediated by a peroxisome proliferator response element in the C promoter. J. Biol. Chem. 270, 19269–19276 [DOI] [PubMed] [Google Scholar]
- 12. Schoonjans K., Staels B., Grimaldi P., Auwerx J. (1993) Acyl-CoA synthetase mRNA expression is controlled by fibric-acid derivatives, feeding and liver proliferation. Eur. J. Biochem. 216, 615–622 [DOI] [PubMed] [Google Scholar]
- 13. Martin G., Schoonjans K., Lefebvre A. M., Staels B., Auwerx J. (1997) Coordinate regulation of the expression of the fatty acid transport protein and acyl-CoA synthetase genes by PPARα and PPARγ activators. J. Biol. Chem. 272, 28210–28217 [DOI] [PubMed] [Google Scholar]
- 14. Kanter J. E., Kramer F., Barnhart S., Averill M. M., Vivekanandan-Giri A., Vickery T., Li L. O., Becker L., Yuan W., Chait A., Braun K. R., Potter-Perigo S., Sanda S., Wight T. N., Pennathur S., Serhan C. N., Heinecke J. W., Coleman R. A., Bornfeldt K. E. (2012) Diabetes promotes an inflammatory macrophage phenotype and atherosclerosis through acyl-CoA synthetase 1. Proc. Natl. Acad. Sci. U.S.A. 109, E715–E724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Das M., Sabio G., Jiang F., Rincón M., Flavell R. A., Davis R. J. (2009) Induction of hepatitis by JNK-mediated expression of TNF-α. Cell 136, 249–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Han M. S., Jung D. Y., Morel C., Lakhani S. A., Kim J. K., Flavell R. A., Davis R. J. (2013) JNK expression by macrophages promotes obesity-induced insulin resistance and inflammation. Science 339, 218–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hoshino K., Takeuchi O., Kawai T., Sanjo H., Ogawa T., Takeda Y., Takeda K., Akira S. (1999) Cutting edge. Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide. Evidence for TLR4 as the Lps gene product. J. Immunol. 162, 3749–3752 [PubMed] [Google Scholar]
- 18. Hawn T. R., Smith K. D., Aderem A., Skerrett S. J. (2006) Myeloid differentiation primary response gene (88)- and Toll-like receptor 2-deficient mice are susceptible to infection with aerosolized Legionella pneumophila. J. Infect. Dis. 193, 1693–1702 [DOI] [PubMed] [Google Scholar]
- 19. Yamamoto M., Sato S., Hemmi H., Hoshino K., Kaisho T., Sanjo H., Takeuchi O., Sugiyama M., Okabe M., Takeda K., Akira S. (2003) Role of adaptor TRIF in the MyD88-independent Toll-like receptor signaling pathway. Science 301, 640–643 [DOI] [PubMed] [Google Scholar]
- 20. Hoiseth S. K., Stocker B. A. (1981) Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291, 238–239 [DOI] [PubMed] [Google Scholar]
- 21. Flanagin S., Nelson J. D., Castner D. G., Denisenko O., Bomsztyk K. (2008) Microplate-based chromatin immunoprecipitation method, matrix ChIP. A platform to study signaling of complex genomic events. Nucleic Acids Res. 36, e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Das M., Jiang F., Sluss H. K., Zhang C., Shokat K. M., Flavell R. A., Davis R. J. (2007) Suppression of p53-dependent senescence by the JNK signal transduction pathway. Proc. Natl. Acad. Sci. U.S.A. 104, 15759–15764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bligh E. G., Dyer W. J. (1959) A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 24. Ford D. A., Monda J. K., Brush R. S., Anderson R. E., Richards M. J., Fliesler S. J. (2008) Lipidomic analysis of the retina in a rat model of Smith-Lemli-Opitz syndrome. Alterations in docosahexaenoic acid content of phospholipid molecular species. J. Neurochem. 105, 1032–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Han X., Gross R. W. (2005) Shotgun lipidomics. Electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom. Rev. 24, 367–412 [DOI] [PubMed] [Google Scholar]
- 26. Han X. (2002) Characterization and direct quantitation of ceramide molecular species from lipid extracts of biological samples by electrospray ionization tandem mass spectrometry. Anal. Biochem. 302, 199–212 [DOI] [PubMed] [Google Scholar]
- 27. Wagner J. D., Shadoan M. K., Zhang L., Ward G. M., Royer L. J., Kavanagh K., Francone O. L., Auerbach B. J., Harwood H. J., Jr. (2010) A selective peroxisome proliferator-activated receptor α agonist, CP-900691, improves plasma lipids, lipoproteins, and glycemic control in diabetic monkeys. J. Pharmacol. Exp. Ther. 333, 844–853 [DOI] [PubMed] [Google Scholar]
- 28. Beutler B., Hoebe K., Du X., Ulevitch R. J. (2003) How we detect microbes and respond to them. The Toll-like receptors and their transducers. J. Leukoc. Biol. 74, 479–485 [DOI] [PubMed] [Google Scholar]
- 29. Suram S., Gangelhoff T. A., Taylor P. R., Rosas M., Brown G. D., Bonventre J. V., Akira S., Uematsu S., Williams D. L., Murphy R. C., Leslie C. C. (2010) Pathways regulating cytosolic phospholipase A2 activation and eicosanoid production in macrophages by Candida albicans. J. Biol. Chem. 285, 30676–30685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pålsson-McDermott E. M., O'Neill L. A. (2004) Signal transduction by the lipopolysaccharide receptor, Toll-like receptor-4. Immunology 113, 153–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Davis R. J. (2000) Signal transduction by the JNK group of MAP kinases. Cell 103, 239–252 [DOI] [PubMed] [Google Scholar]
- 32. Kang Y. J., Chen J., Otsuka M., Mols J., Ren S., Wang Y., Han J. (2008) Macrophage deletion of p38α partially impairs lipopolysaccharide-induced cellular activation. J. Immunol. 180, 5075–5082 [DOI] [PubMed] [Google Scholar]
- 33. Schieven G. L. (2005) The biology of p38 kinase. A central role in inflammation. Curr. Top. Med. Chem. 5, 921–928 [DOI] [PubMed] [Google Scholar]
- 34. Bogoyevitch M. A., Boehm I., Oakley A., Ketterman A. J., Barr R. K. (2004) Targeting the JNK MAPK cascade for inhibition. Basic science and therapeutic potential. Biochim. Biophys. Acta 1697, 89–101 [DOI] [PubMed] [Google Scholar]
- 35. Ma W., Gee K., Lim W., Chambers K., Angel J. B., Kozlowski M., Kumar A. (2004) Dexamethasone inhibits IL-12p40 production in lipopolysaccharide-stimulated human monocytic cells by down-regulating the activity of c-Jun N-terminal kinase, the activation protein-1, and NF-κB transcription factors. J. Immunol. 172, 318–330 [DOI] [PubMed] [Google Scholar]
- 36. Hale K. K., Trollinger D., Rihanek M., Manthey C. L. (1999) Differential expression and activation of p38 mitogen-activated protein kinase α, β, γ, and δ in inflammatory cell lineages. J. Immunol. 162, 4246–4252 [PubMed] [Google Scholar]
- 37. Comalada M., Lloberas J., Celada A. (2012) MKP-1. A critical phosphatase in the biology of macrophages controlling the switch between proliferation and activation. Eur. J. Immunol. 42, 1938–1948 [DOI] [PubMed] [Google Scholar]
- 38. Maloney N. S., Thackray L. B., Goel G., Hwang S., Duan E., Vachharajani P., Xavier R., Virgin H. W. (2012) Essential cell-autonomous role for interferon (IFN) regulatory factor 1 in IFN-γ-mediated inhibition of norovirus replication in macrophages. J. Virol. 86, 12655–12664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sato S., Sugiyama M., Yamamoto M., Watanabe Y., Kawai T., Takeda K., Akira S. (2003) Toll/IL-1 receptor domain-containing adaptor inducing IFN-β (TRIF) associates with TNF receptor-associated factor 6 and TANK-binding kinase 1 and activates two distinct transcription factors, NF-κB and IFN-regulatory factor-3, in the Toll-like receptor signaling. J. Immunol. 171, 4304–4310 [DOI] [PubMed] [Google Scholar]
- 40. Lefterova M. I., Steger D. J., Zhuo D., Qatanani M., Mullican S. E., Tuteja G., Manduchi E., Grant G. R., Lazar M. A. (2010) Cell-specific determinants of peroxisome proliferator-activated receptor γ function in adipocytes and macrophages. Mol. Cell. Biol. 30, 2078–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mullican S. E., Gaddis C. A., Alenghat T., Nair M. G., Giacomin P. R., Everett L. J., Feng D., Steger D. J., Schug J., Artis D., Lazar M. A. (2011) Histone deacetylase 3 is an epigenomic brake in macrophage alternative activation. Genes Dev. 25, 2480–2488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ho H. H., Antoniv T. T., Ji J. D., Ivashkiv L. B. (2008) Lipopolysaccharide-induced expression of matrix metalloproteinases in human monocytes is suppressed by IFN-γ via superinduction of ATF-3 and suppression of AP-1. J. Immunol. 181, 5089–5097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cuenda A., Rousseau S. (2007) p38 MAP-kinases pathway regulation, function and role in human diseases. Biochim. Biophys. Acta 1773, 1358–1375 [DOI] [PubMed] [Google Scholar]
- 44. Han J., Jiang Y., Li Z., Kravchenko V. V., Ulevitch R. J. (1997) Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature 386, 296–299 [DOI] [PubMed] [Google Scholar]
- 45. Utsugi M., Dobashi K., Ono A., Ishizuka T., Hisada T., Koga Y., Shimizu Y., Kawata T., Matsuzaki S., Aoki H., Kamide Y., Mori M. (2010) JNK1 and JNK2 differently regulate IL-12 production in THP-1 macrophage cells. Cytokine 51, 127–131 [DOI] [PubMed] [Google Scholar]
- 46. Sánchez-Tilló E., Comalada M., Xaus J., Farrera C., Valledor A. F., Caelles C., Lloberas J., Celada A. (2007) JNK1 Is required for the induction of Mkp1 expression in macrophages during proliferation and lipopolysaccharide-dependent activation. J. Biol. Chem. 282, 12566–12573 [DOI] [PubMed] [Google Scholar]
- 47. Zhu J., Krishnegowda G., Gowda D. C. (2005) Induction of proinflammatory responses in macrophages by the glycosylphosphatidylinositols of Plasmodium falciparum. The requirement of extracellular signal-regulated kinase, p38, c-Jun N-terminal kinase and NF-κB pathways for the expression of proinflammatory cytokines and nitric oxide. J. Biol. Chem. 280, 8617–8627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lazear H. M., Lancaster A., Wilkins C., Suthar M. S., Huang A., Vick S. C., Clepper L., Thackray L., Brassil M. M., Virgin H. W., Nikolich-Zugich J., Moses A. V., Gale M., Jr., Früh K., Diamond M. S. (2013) IRF-3, IRF-5, and IRF-7 coordinately regulate the type I IFN response in myeloid dendritic cells downstream of MAVS signaling. PLoS Pathog. 9, e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ruiz-García A., Monsalve E., Novellasdemunt L., Navarro-Sabaté A., Manzano A., Rivero S., Castrillo A., Casado M., Laborda J., Bartrons R., Díaz-Guerra M. J. (2011) Cooperation of adenosine with macrophage Toll-4 receptor agonists leads to increased glycolytic flux through the enhanced expression of PFKFB3 gene. J. Biol. Chem. 286, 19247–19258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schwartz E. A., Zhang W. Y., Karnik S. K., Borwege S., Anand V. R., Laine P. S., Su Y., Reaven P. D. (2010) Nutrient modification of the innate immune response. A novel mechanism by which saturated fatty acids greatly amplify monocyte inflammation. Arterioscler. Thromb. Vasc. Biol. 30, 802–808 [DOI] [PubMed] [Google Scholar]