Background: Pregnancy-associated plasma protein-A (PAPP-A) is known to regulate insulin-like growth factor (IGF) bioavailability by specific IGF binding protein proteolysis.
Results: Early developmental delay resulting from zebrafish papp-a knockdown can be rescued by wild-type or proteolytically inactive zebrafish Papp-a.
Conclusion: Papp-a has functionality independent of its proteolytic activity.
Significance: This is the first report of non-proteolytic PAPP-A function with important implications for understanding the biology of PAPP-A.
Keywords: Development, Insulin-like Growth Factor (IGF), Metalloprotease, Proteolytic Enzymes, Zebrafish, Developmental Delay, Pregnancy-associated Plasma Protein A
Abstract
Pregnancy-associated plasma protein-A (PAPP-A) is a large metalloproteinase specifically cleaving insulin-like growth factor (IGF) binding proteins, causing increased IGF bioavailability and, hence, local regulation of IGF receptor activation. We have identified two highly conserved zebrafish homologs of the human PAPP-A gene. Expression of zebrafish Papp-a, one of the two paralogs, begins during gastrulation and persists throughout the first week of development, and analyses demonstrate highly conserved patterns of expression between adult zebrafish, humans, and mice. We show that the specific knockdown of zebrafish papp-a limits the developmental rate beginning during gastrulation without affecting the normal patterning of the embryo. This phenotype is different from those resulting from deficiency of Igf receptor or ligand in zebrafish, suggesting a function of Papp-a outside of the Igf system. Biochemical analysis of recombinant zebrafish Papp-a demonstrates conservation of proteolytic activity, specificity, and the intrinsic regulatory mechanism. However, in vitro transcribed mRNA, which encodes a proteolytically inactive Papp-a mutant, recues the papp-a knockdown phenotype as efficiently as wild-type Papp-a. Thus, the developmental phenotype of papp-a knockdown is not a consequence of lacking Papp-a proteolytic activity. We conclude that Papp-a possesses biological functions independent of its proteolytic activity. Our data represent the first evidence for a non-proteolytic function of PAPP-A.
Introduction
Many lines of evidence suggest the metalloproteinase pregnancy-associated plasma protein A (PAPP-A, pappalysin-1)2 as a local regulator of insulin-like growth factor (IGF) bioavailability. Under physiological conditions, the vast majority of IGF is found in complex with one of six IGF binding proteins (IGFBP-1 through -6) (1), and for signaling to occur, IGF must be released to activate the IGF receptor (IGF-IR) (2, 3). PAPP-A specifically cleaves insulin-like growth factor binding proteins IGFBP-4 and -5 with high efficiency (4, 5), resulting in increased IGF bioavailability because of reduced affinity for IGF of the cleavage fragments (6). PAPP-A retains this activity when associated with the cell surface (7), and, therefore, catalytic amounts of PAPP-A have the potential to efficiently and locally promote IGF receptor activation (4–6).
The PAPP-A knockout mouse phenocopies the IGF-II null mouse in producing viable and fertile offspring 60% the size of the wild type at birth (8–10). The postnatal phenotype can be rescued by disruption of IGF-II imprinting (11) or partially by crossing the PAPP-A knockout mouse with the IGFBP-4 knockout mouse (12).
The PAPP-A knockout mouse strongly suggests a requirement for PAPP-A in normal development in utero, and in humans, PAPP-A is an established diagnostic marker of several complications of human pregnancy. Low maternal serum PAPP-A in the first trimester correlates with increased risk of adverse fetal conditions, including small-for-gestational-age pregnancies, early fetal loss, preterm birth, stillbirth, and low birth weight at term (13–16). In humans, circulating maternal PAPP-A derives from the placental syncytiotrophoblast (17), which secretes extremely high levels of PAPP-A (18), indicating that placental malfunction contributes to these complications of pregnancy. However, in mice, the placenta is not a source of PAPP-A (19), and the PAPP-A knockout phenotype is, therefore, caused by the absence of PAPP-A in the embryo proper. Currently, knowledge about PAPP-A in the developing embryo is scarce, and in particular, the consequences of embryonic PAPP-A deficiency during the earliest stages of development are unknown.
To explore the function of PAPP-A in early development, we established the zebrafish as a model organism to study PAPP-A function. The zebrafish was selected because of the rapid and external development of its transparent embryo, allowing real-time investigation of embryonic phenotypes. Importantly, the use of a non-placental vertebrate organism eliminates placental contributions to embryonic development. Furthermore, the core components of the IGF system are known to be conserved between vertebrate species, and the suitability of the zebrafish as an animal model for this system is well established (20–23).
We identified the zebrafish papp-a gene, characterized the phenotype of papp-a knockdown in zebrafish embryos, and analyzed zebrafish Papp-a biochemically. We present data showing conservation of the proteolytic activity of Papp-a in the IGF system and that Papp-a, independent of this activity, is required to maintain a normal rate of early embryonic development.
EXPERIMENTAL PROCEDURES
Animals
Zebrafish were fed twice daily and kept at 28.5 °C on a 14 h light/10 h dark cycle. Embryos were obtained by natural crosses, reared in E3 buffer (5 mm NaCl, 0.17 mm KCl, 0.33 mm MgSO4, 10−5% (w/w) methylene blue, 2 mm Hepes (pH 7.0)), and staged according to Kimmel et al. (24). For phenotype quantification and documentation, live embryos were sedated in tricaine (150 ng/ml) (Aldrich) in E3.
Sequence Analysis
Human preproPAPP-A (Q13219) was blasted against the Zv8 assembly of the zebrafish genomic database using TBLASTN. Synteny analysis was performed in the basis of zebrafish genome assembly Zv8 and human genome assembly GRCh37. A phylogenetic tree of PAPP-A and PAPP-A2 was constructed by the neighbor-joining method with protein Poisson distances using the MEGA4 software. Gaps in the amino acid sequences as aligned by CLUSTAL X were excluded from the phylogenetic construction. The reliability of the estimated tree was evaluated by the bootstrap method with 1000 replications.
Cloning, Sequencing, and Mutagenesis
Total RNA was isolated from embryos, adults, or adult tissues using TRI reagent (Molecular Research Centre, Inc.) according to the recommendations of the manufacturer. RNA quality was assessed by denaturing RNA gel electrophoresis using the FlashGel system (Lonza), and RNA content was quantified by spectroscopy. Random hexamer-primed cDNA was reverse-transcribed from zebrafish total RNA using the Thermoscript RT-PCR system for first-strand cDNA synthesis (Invitrogen).
For all PCR reactions, KOD Hot Start DNA polymerase (Novagen) was used. DNA encoding full-length zebrafish preproPapp-a was obtained by single PCR using primers 5′-CTTGGTTGGTGTTGAACACGC-3′ and 5′-TGGAAAGGCCCTCCTATAAGC-3′. The PCR product was gel-purified using the QIAquick gel extraction kit (Qiagen), ligated into the pSC-B vector using the StrataClone Blunt PCR cloning kit (Stratagene) (pzfPapp-aSC-B), and sequenced. The coding sequence was ligated into the pcDNA3.1/myc-His(+)A (Invitrogen) expression vector as three fragments using internal original BamHI and XbaI restriction sites and introduced flanking restriction sites. A 5′ KpnI site was introduced using primer set 5′-AAAAAGGTACCCAAATCCCACTCATCCATTGACCA-3′ and 5′-AGCCACATCTTCATCCGATGAG-3′, allowing for ligation using KpnI and BamHI. In the 3′ end, high guanine-cytosine content immediately downstream from the stop codon compromised primer design. To circumnavigate this, the 3′ fragment was excised using XbaI and KpnI and ligated into pcDNA3.1(−) (Invitrogen), creating the construct pzfPA3′cDNA3.1(−). Using this construct as a template, PCR with the primer set 5′-TAATACGACTCACTATAGGG-3′ and 5′-TTTTTTGGGCCCGGCTAAGCCGATGGA-3′ destroyed the original KpnI site and the stop codon and introduced the 3′ ApaI site for ligation into pcDNA3.1/myc-His(+)A.
The active site mutant E495A was constructed by overlap extension PCR producing an A to C mutation in the desired glutamate codon using the primer pairs 5′-TATTATGATCACGGGGACTGCTGCAA-3′ and 5′-CAATCTCGTGGATCATGGTGTGAG-3′ and 5′-TCACACCATGATCCACGCGATTG-3′ and 5′-TTGCACTGGTTTGAGCAGCCATC-3′. The PCR product was ligated into pzfPapp-a-cDNA3.1/Myc-His(+)A using BamHI and XbaI. The full-length early stop codon variant Y70stop was constructed by overlap extension PCR producing a TAC to TAA mutation using the primer pairs 5′-TAATACGACTCACTATAGGG-3′ and 5′-GCATTTATCTTAGAGACCTCC-3′ and 5′-GGAGGTCTCTAAGATAAATGC-3′ and 5′-CGAAGGGTTTAGCACAATCC-3′. The PCR product was ligated into pzfPapp-a-cDNA3.1/Myc-His(+)A using KpnI and BamHI. All constructs were verified by sequencing.
Expression Analysis
Zebrafish papp-a specific primer set 5′-CCGACGATTACAGAACACCA-3′ and 5′-CGAAGGGTTTAGCACAATCC-3′ and beta-actin primer set 5′-CACGAGACCACCTTCAACT-3′ and 5′-ATCCAGACGGAGTATTTGC-3′) were used for RT-PCR. The optimal PCR cycle number was determined experimentally as 34 cycles for papp-a and 28 cycles for beta-actin using mixed-sex adult zebrafish cDNA as a template.
Whole mount in situ hybridization was performed as described previously (25). Two non-overlapping zebrafish papp-a probe templates were constructed from cDNA using the primer sets 5′-CCGACGATTACAGAACACCA-3′ and 5′-CGAAGGGTTTAGCACAATCC-3′ (nucleotide 1229–1774) and 5′-GCCCATTTGCACTTGGCTCTTATGC-3′ and 5′-GCTGTGTTGGAGGTGATGGTGGTGA-3′ (nucleotide 163-687). The Pax2a probe was constructed from cDNA using the primer set 5′-CTTCTAACAGGCACATCCCAT-3′ and 5′-CATTAACCCTCACTAAAGGGAACTATCCGTTCAAAGCCCG-3′ (nucleotide 1–419 of NM_131184.2). Other probes used were ntla (cb240), myogenin (cb553), emx3 (eu682), and eng2a (eu759).3
Whole Mount Immunohistochemistry
Embryos were fixed in 4% PFA in PBS for 1 h of shaking at room temperature. Fixed embryos were washed three times for 5 min each time in PBT (PBS + 1% Triton X-100), blocked in PBT + 10% goat serum for 1 h at room temperature, washed three times for 5 min each time in PBT, incubated with Ab-F59 cell supernatant (Developmental Studies Hybridoma Bank, Santa Cruz Biotechnology, Inc.) diluted 1:10 in PBT + 1% goat serum overnight at 4 °C, washed three times for 5 min each time in PBT, incubated with Alexa Fluor 488 goat anti-mouse IgG (Invitrogen) diluted 1:400 in PBT + 1% goat serum for 3 h at room temperature, and washed overnight in PBT with three buffer changes. Labeled embryos were transferred to glycerol for storage and imaging.
Gene-specific Knockdown and Overexpression
Microinjection volumes were calibrated by performing 10 injections into a 0.5-μl microcapillary tube (Drummond Microcaps), measuring the amount of liquid using a ruler, and calculating the volume per injection. 5 nl of injection mixture was microinjected into the center of the yolk of zebrafish zygotes. Sequence-specific morpholinos (Gene Tools, LLC) were designed from the experimentally determined zebrafish papp-a cDNA sequence and the identified database genomic sequences. One morpholino (sMO, TTAAGCAAACCAAACCTCCGATAAC) was designed to anneal in the border region of exon 1 and intron 1 to cause aberrant splicing of the pre-mRNA. The specific efficiency of inhibition of zebrafish papp-a pre-mRNA splicing was assessed by analytic PCR using the primers 5′-CCGACGATTACAGAACACCA-3′ and 5′-CGAAGGGTTTAGCACAATCC-3′) on cDNA prepared from total RNA extracted from injected embryos. A translation-inhibiting morpholino (tMO, AAGTTACAAAAGCCTGTCAAGACGC) was designed to anneal to the 5′ untranslated region. A standard control morpholino (Gene Tools, LLC, CCTCTTACCTCAGTTACAATTTATA) was used for control experiments. All knockdown experiments were repeated systematically with coinjection of a p53 morpholino (GCGCCATTGCTTTGCAAGAATTG) (26) to verify that knockdown phenotypes were not dependent on nonspecific p53 activation. For phenotyping by whole mount in situ hybridization, sMO was coinjected with 5 ng/embryo of standard zebrafish p53 morpholino.
Capped mRNA was synthesized using the mMessage mMachine T7 ULTRA Kit (Ambion, Inc). Plasmid templates were linearized with NaeI and NdeI for in vitro transcription. Upon synthesis, the reaction was incubated with TURBO DNase (Ambion) followed by RNA purification using the RNeasy MinElute cleanup kit (Qiagen). RNA quality was assessed by denaturing RNA gel electrophoresis using the FlashGel system (Lonza), and RNA concentration was assessed spectrometrically by absorbance at 260 nm.
Recombinant Protein Expression and Purification
HEK293T cells (293tsA1609neo) were maintained in high-glucose DMEM supplemented with 10% fetal bovine serum, 2 mm glutamine, non-essential amino acids, and gentamicin (Invitrogen). Transient transfections were carried out by calcium phosphate coprecipitation (27). Conditioned medium was harvested and cleared by centrifugation 48 h after transfection.
For N-terminal sequence analysis, recombinant myc-His-tagged zebrafish Papp-a was partially purified from culture supernatants of transfected cells by nickel affinity on Chelating-Sepharose Fast Flow (Amersham Biosciences), followed by immunoprecipitation on protein G-Sepharose 4 Fast Flow (Amersham Biosciences) saturated with anti-c-myc (9E10) monoclonal antibody. Recombinant IGF binding proteins -1 through -6 were expressed, purified, and radio-labeled as described previously (28).
Biochemical Analysis
Western blotting was performed as described previously (28) using 9E10 anti-c-myc-conditioned medium diluted 1:100 and HRP-conjugated rabbit anti-mouse (P0260, DAKO) diluted 1:2000. The N-terminal sequence of purified zebrafish Papp-a and IGF binding proteins was determined by Edman degradation. Briefly, cleavage reactions were separated by SDS-PAGE and blotted onto PVDF membranes. The membranes were stained with Coomassie Blue, and bands of interest were excised and analyzed (3–11 pmol) on an Applied Biosystems 491 protein sequencer.
Proteolytic assays were conducted as described previously (28). Briefly, conditioned serum-containing medium harvested from HEK293T cells transiently transfected with pzfPapp-a-cDNA3.1/myc-His(+)A was added to 125I-labeled substrate preincubated with or without molar excess of human IGF-II (Bachem). Reactions were incubated at 28 °C and stopped by the addition of EDTA, followed by separation by SDS-PAGE and visualization by autoradiography.
Statistics
Raw data were subjected to the extreme studentized deviate method for detection of outliers (p < 0.05). Statistical analysis was performed using GraphPad Prism version 5.00 for Windows (GraphPad Software, San Diego, CA). Data were plotted as mean ± S.D. Means were compared by one-way analysis of variance analysis with Tukey's multiple comparison test.
Microscopy and Imaging
Live embryos were observed and documented on an Olympus IX71 microscope equipped with an Olympus DP71 camera. Embryos stained by in situ hybridization were assessed and documented on an Olympus SZX16 stereomicroscope equipped with an Altra20 camera and a Photonic LED transmitted light stage. Fluorescent images were captured with a Zeiss Axiocam MRm camera mounted on a Zeiss Axio Observer.D1 stand with a Colibri LED light source.
Analysis of Papp-a′
This paragraph details experiments carried out on Papp-a′. The preceding paragraphs detail experiments carried out on Papp-a. DNA encoding full-length zebrafish preproPapp-a′ was obtained by PCR using primers 5′-ATGCTCCGCTCACTTCAAC-3′ and 5′-TGCGCTCATGCTTTCTTTCA-3′ on cDNA reverse-transcribed from total RNA extracted from adult zebrafish of the SAT strain. The PCR product was gel-purified using the QIAquick gel extraction kit (Qiagen), ligated into the pJET1.2 vector using the CloneJET PCR cloning kit (Fermentas), and sequenced. Using this construct as template, the open reading frame was amplified by PCR using primers 5′-GTCGGATCCACATGAAAGTTTGGACTTTTCTCCAGTG-3′ and 5′-TGAGCGGCCGCGAGCCAAGAGAGTGTAAATGCTCA-3′ for the introduction of a 5′ BamHI restriction site, a 3′ NotI restriction site, and mutation of the stop codon. The PCR product was cloned into the BamHI/NotI sites of pcDNA3.1/myc-His(+)A. Recombinant expression and proteolytic assays were carried out as for Papp-a described above. For quantitative reverse transcription real-time PCR, total RNA extracted from staged embryos using TRI reagent was DNase-treated and purified using RNase-free DNaseI (Qiagen) and the RNeasy mini kit (Qiagen). Reactions were set up using Brilliant III Ultra Fast SYBR Green QRT-PCR Master Mix (Agilent Technologies). Primer pairs 5′-CCCAGCTTGTGTTACTTCTACG-3′ and 5′-CCTTCGACTGACAGCTCTTT-3′, 5′-AAAGAGGAGGGCGTTCAAG-3′ and 5′-TGCAGCGGATCACATTAGAG-3′, and 5′-GCCTTCACCCCAGAGAAAGG-3′ and 5′-CCGGTTTGGATTTACATGTTG-3′ were used for the amplification of papp-a, papp-a′, and β-2-microglobulin (b2m), respectively. Cycling conditions were as follows: 10 min at 50 °C, 3 min at 95 °C followed by 40 cycles of 20 s at 95 °C and 20 s at 58 °C in a Stratagene Mx3005P (Agilent Technologies) with MxPro QPCR software. Data were analyzed by the Livak method (29). For papp-a′ knockdown, embryos were injected with 5 ng of splice-site targeted morpholino (i3e4, CTAGATAGGCCCTGGAGTCACAGAT or e1i1, AATCACAAAACTGTTCCTACCAGCT) or 2.5 ng of translation-inhibiting ATG morpholino (TTTATCCAAGAACACAGGAGGTGGA).
RESULTS
The Zebrafish Genome Encodes Papp-a, an Ortholog of Human PAPP-A
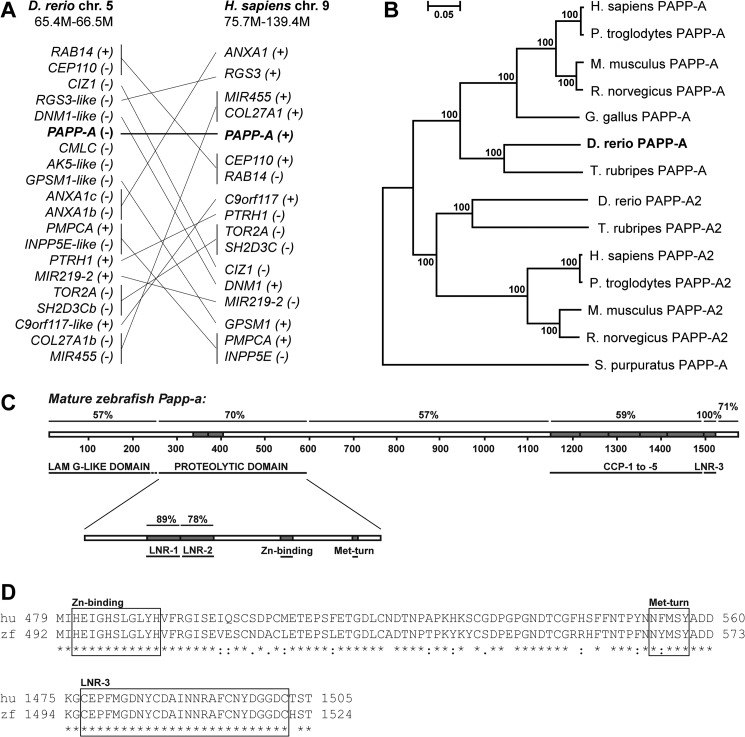
Using the polypeptide sequence of human PAPP-A with TBLASTN on zebrafish genome assembly Zv8, papp-a spanning 190 kb on zebrafish chromosome 5 was identified. The regions immediately surrounding papp-a in zebrafish, and PAPP-A on human chromosome 9, show little conservation. However, 17 of the 19 identified genes located within ∼1.1 Mb of zebrafish papp-a have orthologs on human chromosome 9 (Fig. 1A), suggesting that although gene shuffling has occurred, the zebrafish papp-a and human PAPP-A genes are true orthologs. Phylogenetic analysis of PAPP-A and its only known homolog, PAPP-A2, from selected species groups zebrafish Papp-a in the PAPP-A clade (Fig. 1B), substantiating that the identified genomic sequence is indeed zebrafish papp-a. The cDNA was cloned from total adult zebrafish mRNA and sequenced.
FIGURE 1.
Sequence analysis. A, zebrafish papp-a is orthologous to human PAPP-A. The genes are located on zebrafish chromosome 5 and human chromosome 9, respectively. 17 of 19 genes located within 1.1 Mb of zebrafish papp-a are orthologous to genes on human chromosome 9. B, phylogenetic analysis of PAPP-A and its only known homolog, PAPP-A2. The phylogenetic tree was constructed by the neighbor-joining method with protein Poisson distances. Bootstrap percentages (from 1000 iterations) are shown. The scale bar indicates evolutionary distance in arbitrary units. Prepro sequences were used for alignment, and gaps were excluded from the analysis. Database sequences used with no modifications were human (Homo sapiens) PAPP-A and PAPP-A2 (Q13219 and Q9BXP8), mouse (Mus musculus) PAPP-A and PAPP-A2 (Q8R4K8 and NP_001078845), and rat (Rattus norvegicus) PAPP-A2 (XP_001073420). Chimpanzee PAPP-A was in silico-translated from XR_024483.1. Sea urchin (Strongylocentrotus purpuratus) and chicken (Gallus gallus) PAPP-A (XM_790504 and XM_415522.2) were identified by TBLASTN with the human PAPP-A against the purple sea urchin build 2 genome database, and Gallus_gallus-2.1, respectively. Exon 1 of the chicken gene was extended manually by 24 nucleotides following alignment to human PAPP-A. Zebrafish (Danio rerio) and chimpanzee (Pan troglodytes) PAPP-A2 (NW_001512945.1 and NW_001229605.1) were identified from the Pan_troglodytes-2.1 and zebrafish Zv7 genomic assemblies, respectively, by TBLASTN with the human PAPP-A2 sequence as query. The rat PAPP-A sequence is a correction of EMD10508 obtained by adding the N- and C-terminal sequences identified by TBLASTN with the human query. Fugu (Takifugu rubripes) PAPP-A (scaffold 44) and PAPP-A2 (scaffold 1318) sequence was obtained by using zebrafish Papp-a and zebrafish Papp-a2 as queries in TBLASTN versus the fugu genome. C, schematic representation of mature zebrafish Papp-a primary structure indicating overall structural and functional conservation to human PAPP-A. Domains and their sequence identities to human PAPP-A are shown, and functional motifs, modules, and amino acid residue numbers are indicated. LamG-like, laminin G-like. D, upper panel, clustalW alignment showing high identity of the partial proteolytic domain sequence of human and zebrafish Papp-a, including the elongated zinc-binding site and the Met-turn (zebrafish amino acid residues 492–573). Lower panel, clustalW alignment showing complete identity of human and zebrafish LNR-3 located in the very C terminus (zebrafish amino acid residues 1494–1524). Motifs are boxed and labeled.
Sequence alignment of the prepro forms of human PAPP-A (1627 residues) and zebrafish Papp-a (1624 residues) showed 60% overall identities. Of 84 cysteine residues in human PAPP-A (30), 83 are conserved in zebrafish Papp-a, suggesting conservation of the disulfide structure and overall fold. Notably, recognized domains and sequence motifs are highly conserved (Fig. 1, C and D). Within the proteolytic domain, the metzincin hallmark elongated zinc binding consensus is 100% conserved, and a single conservative substitution is found in the structurally important Met-turn. The three LNR modules are also highly conserved. In particular, the C-terminal LNR-3, which functions as a specificity-determining exosite in human PAPP-A (31), is 100% conserved over its 26-residue sequence stretch (Fig. 1D). Also, CCP-3, which mediates cell surface association in human PAPP-A (7), shows conservation of identified glucosaminoglycan-binding basic residues (32) and 72% overall sequence identity. Taken together, this indicates structural and functional conservation between zebrafish and human PAPP-A.
Papp-a Is Expressed during Early Development, and Expression Is Conserved between Human, Mouse, and Zebrafish
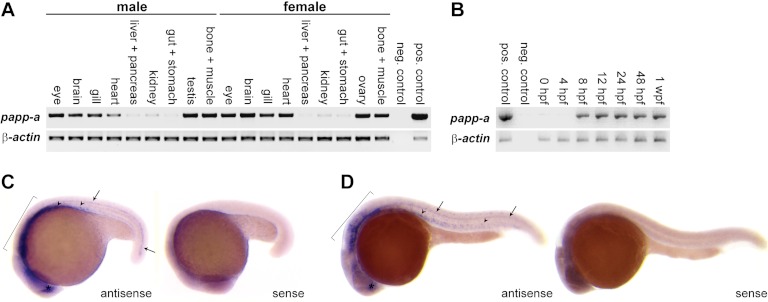
Zebrafish papp-a mRNA was detected in all adult tissues analyzed, with the highest levels in eyes, brain, gills, heart, testes, ovaries, bone, and muscle and the lowest levels in gut, kidney, liver, and pancreas (Fig. 2A). In agreement with these data, PAPP-A-encoding mRNA was detected previously in all human tissues analyzed (18), and in mice, expression was characterized as ubiquitous, with the highest levels in brain, heart, bone, kidney, and lung (8). In whole zebrafish embryos and larvae, papp-a mRNA was detected from mid-gastrulation (8 h post-fertilization (hpf)) throughout the first week of development (Fig. 2B), and whole mount in situ hybridization demonstrated tissue-specific expression in the developing myotomes, ventral neural tube, hindbrain, and forebrain (Fig. 2, C and D).
FIGURE 2.
Zebrafish papp-a expression. A, reverse transcription PCR analysis of papp-a mRNA reveals expression in all adult tissues at varying levels. Negative control (neg. control): no template; positive control (pos. control), plasmid template. B, reverse transcription-PCR analysis of papp-a mRNA reveals expression from 8 hpf throughout the first week of development. Negative control, no template; positive control, plasmid template. C and D, whole mount in situ hybridization showing papp-a mRNA expression at 18 (C) and 24 (D) hpf in developing myotomes (arrowheads), ventral neural tube (arrows), hindbrain (brackets), and forebrain (asterisks). Identical expression patterns were obtained with two non-overlapping antisense probes with no signals detected from either complementary sense probe. Insets, negative controls (sense probe). wpf, weeks post fertilization.
Knockdown of papp-a Causes Early General Developmental Delay
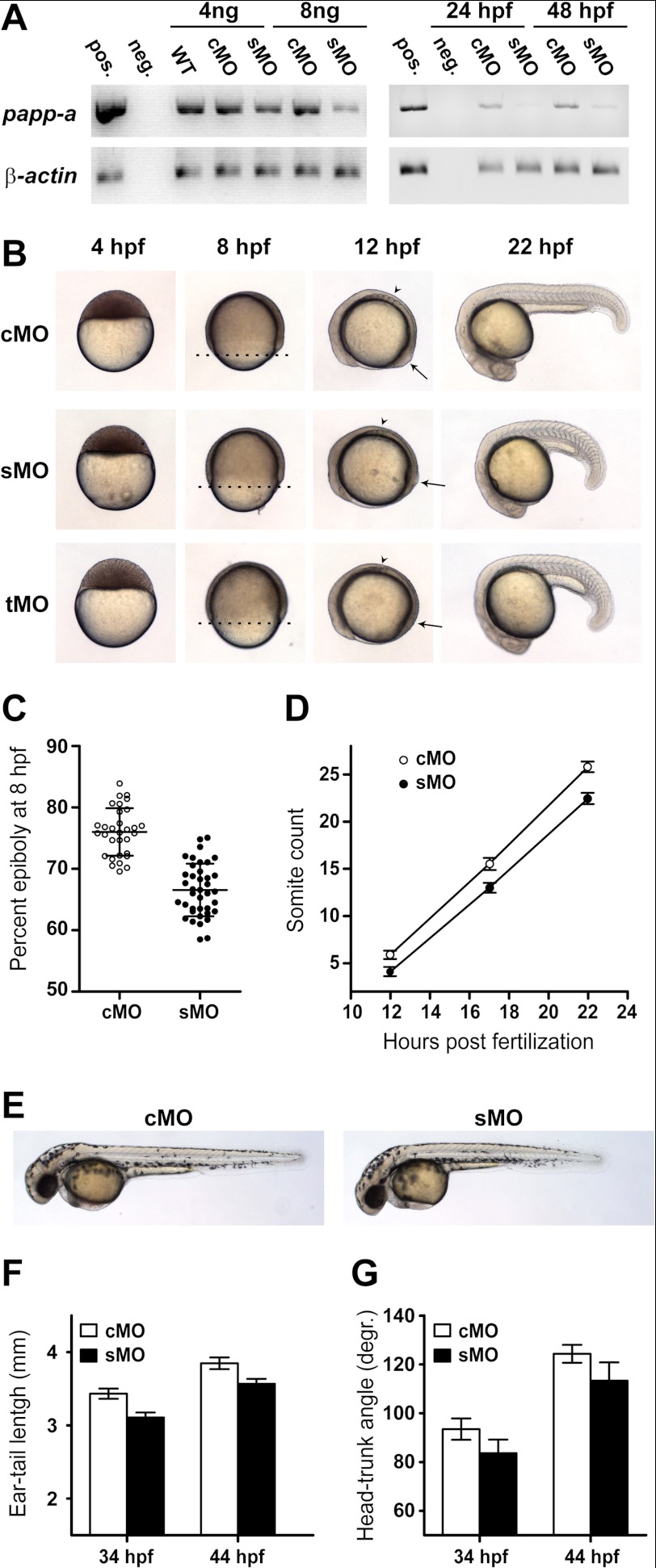
Knockdown experiments were performed using morpholino antisense technology (33). Microinjection of a papp-a-targeted morpholino oligonucleotide (sMO), designed to interfere with normal pre-mRNA splicing, caused a dose-dependent reduction of correctly spliced papp-a mRNA, efficient until at least 48 hpf (Fig. 3A). Four hours post-fertilization, sMO-injected embryos were morphologically indistinguishable from embryos injected with a control morpholino (cMO) (Fig. 3B). By 8 h post-fertilization, approximately halfway through the gastrulation period (5–10 hpf) of development, control embryos averaged 76% epiboly, whereas epiboly had only progressed to 67% in knockdown embryos (Fig. 3, B and C). By 12 hpf, an average of 5.9 and 4.1 somites had developed in control and knockdown embryos, respectively. By 22 hpf, with average somite counts of 25.8 and 22.5, respectively, the difference was even more pronounced (Fig. 3, B and D). The lag in development was significant (p < 0.001) at all stages. At 44 hpf, knockdown embryos showed visibly reduced body length compared with control embryos (Fig. 3E). This difference in size was significant at 34 as well as 44 hpf (p < 0.001) (Fig. 3F). However, quantification of the developmental stage by head-trunk angle (Fig. 3G) showed a significant (p < 0.001) developmental delay, accounting at least in part for the apparent growth deficiency. Also, at 44 hpf, no morphological abnormalities were evident (Fig. 3E), and no excessive mortality was observed during the first 5 days of development.
FIGURE 3.
papp-a knockdown. papp-a knockdown was performed by micro-injecting zygotes with an sMO targeted for the border between exon 1 and intron 1 of the papp-a pre-mRNA. This was predicted to cause inclusion of the 34-kb intron 1 in the spliced mRNA, resulting in a truncated transcript encoding 60 residues of the 256 residue N-terminal LamG-like domain and 20 residues encoded by intron 1. Additionally, a tMO designed to anneal to the 5′ untranslated region of the papp-a transcript was used. A standard cMO was used as negative control. A, the papp-a-targeted sMO efficiently knocks down papp-a expression. Reverse transcription PCR analysis of mRNA levels in response to cMO and sMO injection shows efficient papp-a knockdown from injection of 8 ng of sMO by 24 hpf (left panel). Knockdown is efficient at 24 and 48 hpf (right panel). Pos., positive; neg., negative. B, papp-a knockdown reduces the rate of gastrulation and segmentation. Phenotypes of control (upper panel) and papp-a knockdown (middle and lower panel) embryos at 4, 8, 12, and 22 hpf. Lateral views, animal pole to the top (4 hpf) or anterior to the left (8, 12, and 22 hpf). The dotted lines at 8 hpf show the progression of epiboly. Note the number of somites (arrowheads) and the position of the tail bud (arrows) at 12 hpf. In all of numerous experiments, the penetrance of this phenotype consistently exceeded 95% of the injected embryos. 8 ng of cMO, sMO, or tMO was used in all experiments. C, quantification of epiboly progression 8 hpf. The percent epiboly at 8 hpf was calculated as the percentage of yolk covered by cells as measured along the animal/vegetal axis. n(cMO) = 31, and n(cMO) = 39. Data points are plotted as mean ± S.D. D, quantification of segmentation progression in control and knockdown embryos. Somite counts at 12 hpf (n(cMO) = 20 and n(sMO) = 17), 17 hpf (n(cMO) = 15 and n(sMO) = 23), and 22 hpf (n(cMO) = 15 and n(sMO) = 22) are plotted as mean ± S.D. E, representative control and knockdown embryos 44 hpf. F, ear-tail length of control and knockdown embryos 34 hpf (n(cMO) = 20 and n(sMO) = 25) and 44 hpf (n(cMO) = 20 and n(sMO) = 22). G, head-trunk angle of control and knockdown embryos 34 hpf (n(cMO) = 20, and n(sMO) = 26) and 44 hpf (n(cMO) = 19, and n(sMO) = 24). degr., degrees.
We conclude that the knockdown of papp-a limits the developmental rate in the zebrafish embryo not only during gastrulation but also during the segmentation period (10–24 hpf) and that the effect remains prominent during the pharyngula period (24–48 hpf). Developmental delay was observed in epiboly, somitogenesis, and straightening of the body axis. Also, overall morphology and developmental hallmarks, including optic placodes, otic placodes, and heartbeat, were delayed correspondingly (data not shown), supporting a general delay phenotype. A similar lag in development resulted from the injection of a papp-a-targeted translation-inhibiting morpholino (tMO) (Fig. 3B). For groups injected with either sMO or tMO, penetrance consistently exceeded 95% in all of many independent experiments. No malformations or necrosis were observed in either of the injected groups.
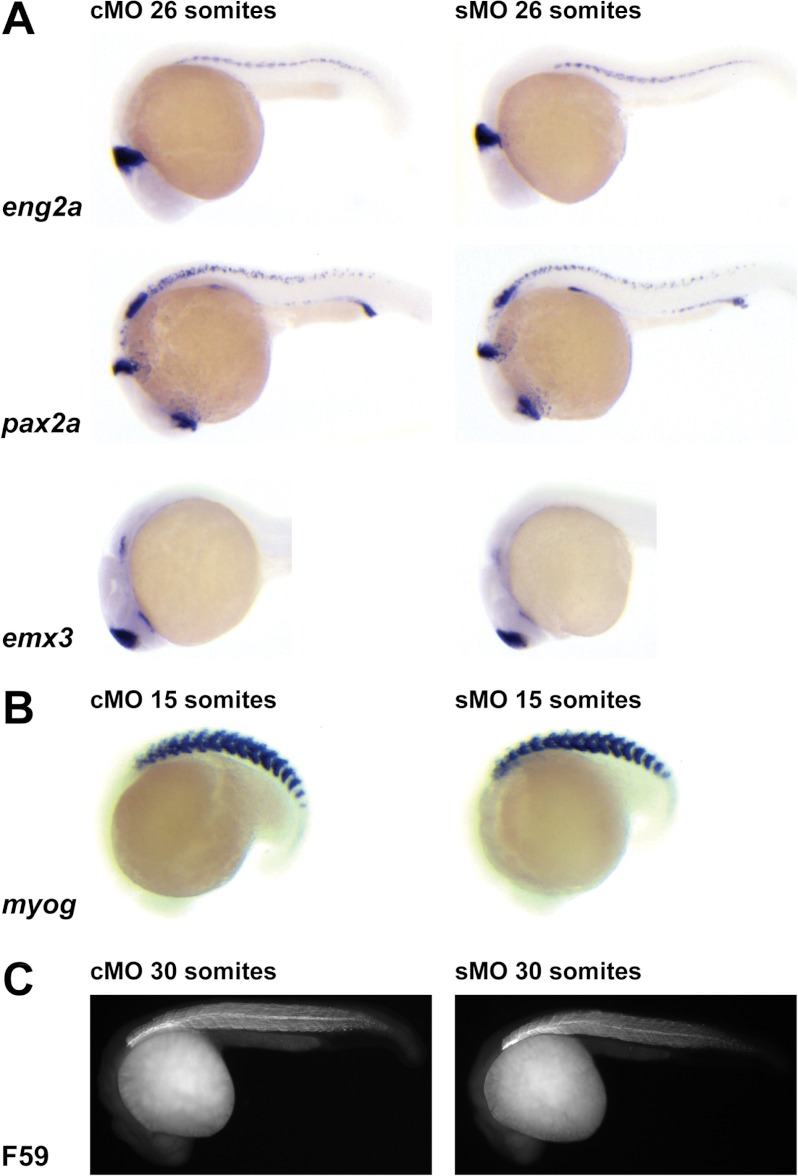
To evaluate the effect of Papp-a deficiency on embryonic patterning, whole mount in situ hybridization for selected marker genes was performed on stage-matched embryos (Fig. 4). In the central nervous system, normal expression of eng2a (midbrain-hindbrain boundary), pax2a (optic stalk and hindbrain and spinal cord neurons), and emx3 (hypothalamus and telencephalon) was observed in knockdown embryos (Fig. 4A). Knockdown of papp-a caused no detectable abnormalities of muscle or myotome as assessed by whole mount in situ hybridization for myogenin (Fig. 4B) and F59 immunostaining of muscle (C). Furthermore, wild-type expression of eng2a in muscle pioneers and of pax2a in the pronephric ducts, otic vesicles, and thyroid primordium was observed (Fig. 4A). Taken together, these data suggest that papp-a knockdown causes a general developmental delay without affecting normal patterning of the zebrafish embryo.
FIGURE 4.
Effect of papp-a knockdown on marker gene expression. The anterior is shown to the left, and the dorsal at the top. Embryos of the same developmental stage are shown. A, papp-a knockdown does not affect patterns of marker gene emx3, eng2a, and pax2a expression, as shown by in situ hybridization. B and C, muscle development is not affected by papp-a knockdown as revealed by in situ staining for myogenin (myog) expression (B) and immunostaining of muscle fibers (F59) (C). Embryos shown are representative of 15 to 30 embryos/group.
Interestingly, the developmental delay, beginning between 4 and 8 h post-fertilization, occurs much earlier than the delays resulting from knockdown in zebrafish of igf-II or igf-Ir, which begin between 12 and 24 hpf and by 16 hpf, respectively (20, 23, 34). The delay resulting from papp-a knockdown is therefore different from the Igf-related delay.
Zebrafish Papp-a Is Conserved as a Specific IGF Binding Protein Protease
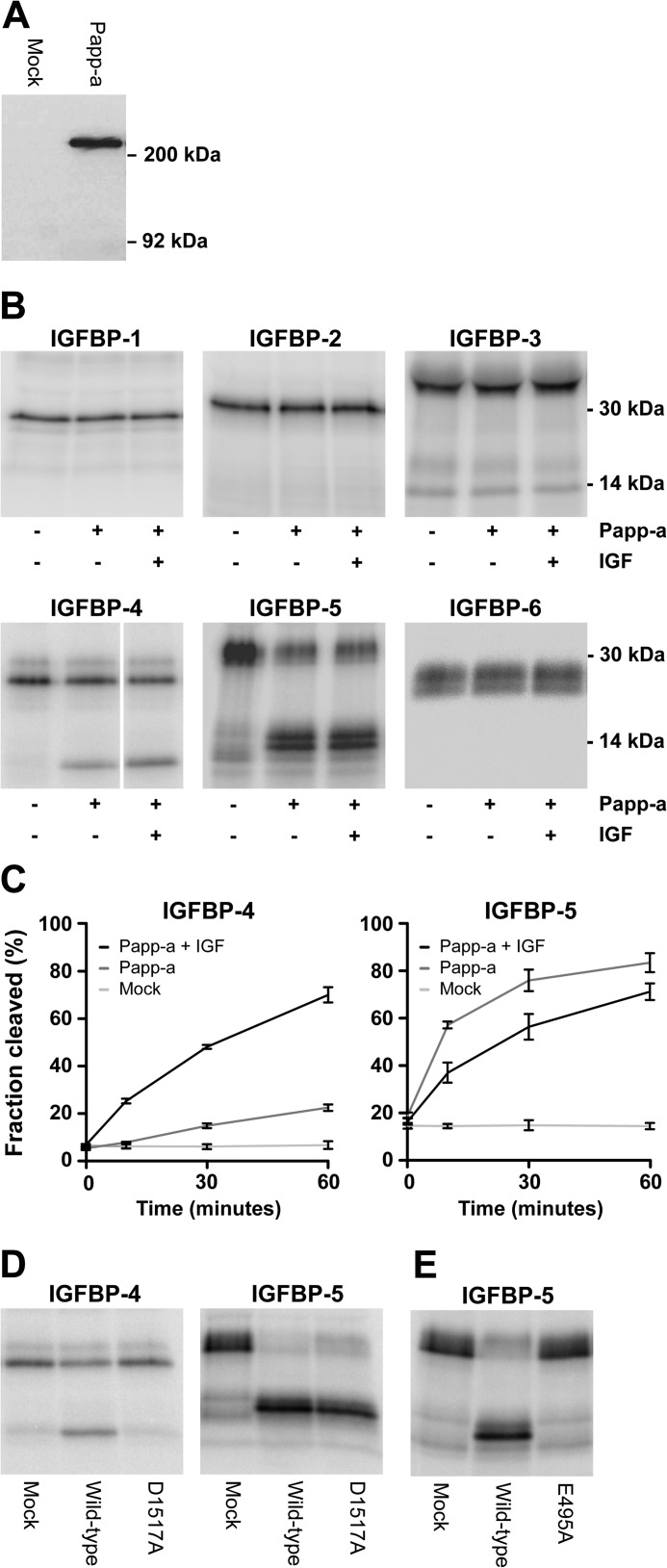
This phenotypic indication of Papp-a function outside of the Igf system prompted the question of whether the proteolytic function of zebrafish Papp-a is conserved. PAPP-A is an unusually large (400 kDa) proteolytic enzyme composed of multiple modules, and cleavage of IGFBP-4 requires homodimerization in trans of the mature polypeptide and is dependent on the distantly located LNR-3 module as a substrate-binding exosite (35). To address this question of functional conservation, recombinant zebrafish Papp-a was expressed in HEK293T cells for biochemical characterization. Expression was confirmed by Western blotting (Fig. 5A), and the N-terminal sequence of the secreted protein was identified by Edman degradation as Ser61-Leu-Pro-Gly-Ile, demonstrating processing into a 1564-residue mature zebrafish Papp-a polypeptide.
FIGURE 5.
Proteolytic activity, specificity, and mechanistic conservation of Papp-a. A, Western blot detection of reduced recombinant myc-His tagged zebrafish Papp-a in medium from transfected HEK293T cells. Zebrafish Papp-a migrates as a monomer of ∼200 kDa. B, assays for proteolytic cleavage of the six human IGF binding proteins by Papp-a in the absence or presence of human IGF-II. For each IGFBP, reactions were analyzed on the same gel in adjacent lanes, except for IGFBP-4, where one lane was excised. Each experiment was performed three times or more with similar results. C, time course proteolytic assays of Papp-a cleavage of IGFBP-4 and IGFBP-5 in the presence or absence of IGF-II. Reactions containing conditioned medium from mock-transfected HEK293T cells were used as negative controls. The plotted values are means of three independent experiments. Error bars indicate the mean ± S.D. D, mutation of Asp-1517 to alanine (Papp-a(D1517A)) specifically abolishes IGFBP-4 cleavage but has no effect on cleavage of IGFBP-5. E, mutation to alanine of active site glutamate residue Glu-495 (Papp-a(E495A)) abrogates the proteolytic activity of zebrafish Papp-a. Cleavage of IGFBP-4 was in the presence of IGF-II. The experiments of D and E were performed three times with similar results.
To determine whether zebrafish Papp-a is capable of cleaving IGF-binding proteins, proteolytic assays were performed on the full panel of human IGFBPs (Fig. 5B). No activity was detected toward IGFBP-1, -2, -3, and -6, whereas IGFBP-4 and IGFBP-5 were cleaved, generating the expected two bands of approximately equal size. For determination of cleavage sites, reactions of IGFBP-4 and -5 were separated by SDS-PAGE, blotted onto a PVDF membrane, and subjected to Edman degradation. The N-terminal sequences of the comigrating IGFBP-4 fragments were determined to be Asp/Lys-Glu/Val, corresponding to Asp1-Glu of the mature, uncleaved IGFBP-4 N terminus and Lys136-Val of the C-terminal fragment. The faster migrating IGFBP-5 fragment yielded the sequence Lys144-Phe-Val-Gly. Thus, zebrafish Papp-a cleaves IGFBP-4 N-terminal to Lys136, and IGFBP-5 N-terminal to Lys144. These cleavage sites are identical to those resulting from cleavage by human PAPP-A (5, 36).
Proteolytic activity toward IGFBP-4 and -5 was further analyzed in time course assays (Fig. 5C). Similar to human PAPP-A (5), zebrafish Papp-a cleavage of IGFBP-4 was potentiated markedly by the presence of IGF-II, IGFBP-5 cleavage was slightly inhibited by IGF-II, and zebrafish Papp-a cleaved IGFBP-5 more efficiently than IGFBP-4. To further assess functional conservation, the variant Papp-a(D1517A), corresponding to human PAPP-A LNR-3 variant D1499A, which is inactive toward IGFBP-4 while maintaining its activity toward IGFBP-5 (37), was constructed and analyzed. Papp-a(D1517A) was found to be able to cleave IGFBP-5 but not -4 (Fig. 5D), suggesting that the unusual LNR-3-based exosite, a part of the intrinsic regulatory mechanism of human PAPP-A proteolytic activity, is also highly conserved functionally in zebrafish. Finally, a conserved glutamate residue within the elongated zinc-binding site of the proteolytic domain is strictly required for proteolytic activity of human PAPP-A (28). Substitution of the corresponding residue in zebrafish Papp-a with alanine (Papp-a(E495A)) completely abolished zebrafish Papp-a proteolytic activity (Fig. 5E). Together, these data strongly support the functional and mechanistic conservation of zebrafish Papp-a as a specific IGF-binding protein protease.
Papp-a Functions Independently of Its Proteolytic Activity
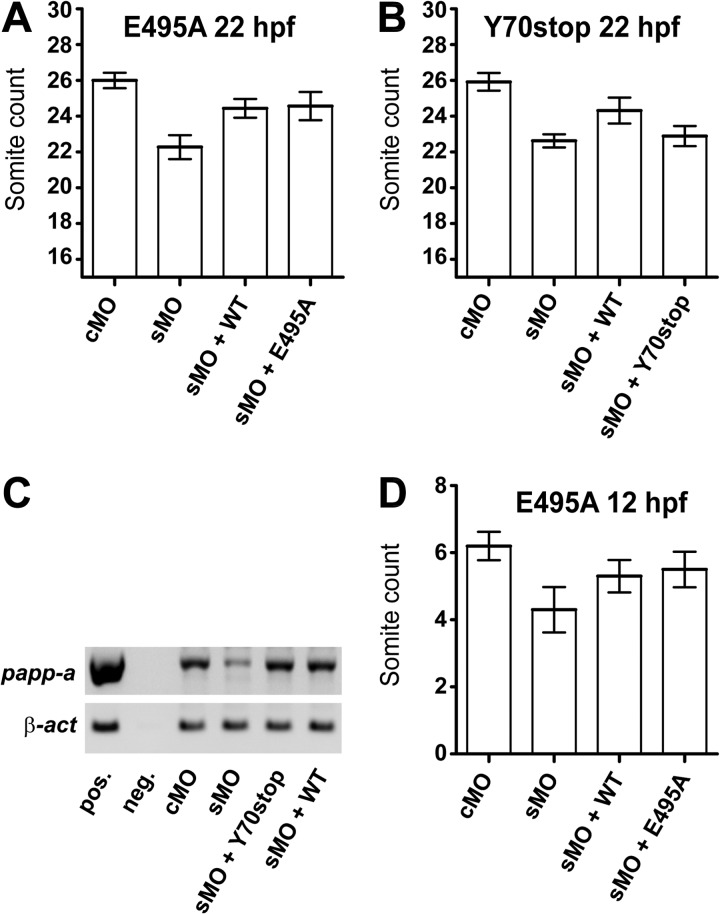
To address the apparent conflict between the late onset of developmental delay caused by the absence of Igf-II or Igf-Ir and the much earlier onset of delay caused by the absence of Papp-a, we carried out experiments where rescue of the papp-a knockdown phenotype was attempted by coinjection of the morpholino with in vitro transcribed zebrafish papp-a mRNA. Significant rescue (p < 0.001) of the delayed development at 22 hpf resulted from coinjecting sMO with zebrafish papp-a mRNA (Fig. 6A). These experiments substantiate that the morpholino-induced phenotype is indeed the result of specific papp-a knockdown and, importantly, that it provides a tool that allows us to probe for specific functions of Papp-a in vivo by attempting rescue with mutated variants of the papp-a mRNA.
FIGURE 6.
Proteolytic activity of Papp-a in vivo. The ability of wild-type and mutant Papp-a to rescue knockdown phenotypes assessed by developmental staging. A, wild-type Papp-a and the proteolytically inactive Papp-a(E495A) variant rescue papp-a knockdown-induced developmental delay 22 hpf significantly (p < 0.001) and with similar efficiency but not to the level of the control (p < 0.001). n(cMO = 12), n(sMO) = 18, n(sMO + WT) = 9, and n(sMO + E495A) = 7. B, wild-type zebrafish Papp-a, but not the full-length early stop codon zebrafish papp-a mRNA variant, Papp-a(Y70stop), rescues developmental delay 22 hpf. n(cMO = 13), n(sMO) = 13, n(sMO + WT) = 18, and n(sMO + Y70stop) = 15. C, Papp-a(Y70stop), differing from wild-type papp-a mRNA by a single nucleotide substitution only, shows stability similar to wild-type zebrafish Papp-a by semi-quantitative RT-PCR 24 h after injection. D, wild-type Papp-a and the proteolytically inactive Papp-a(E495A) variant rescue papp-a knockdown-induced developmental delay 12 hpf significantly (p < 0.01) and with similar efficiency, but not to the level of the control (p < 0.01). n(cMO) = 10, n(sMO) = 10, n(sMO + WT) = 10, and n(sMO + E495A) = 10. Error bars indicate ± S.D.
Surprisingly, equally efficient and significant rescue (p < 0.001) was observed by coinjecting the morpholino with mRNA encoding the proteolytically inactive zebrafish Papp-a variant Papp-a(E495A) compared with the wild-type protein (Fig. 6A). To substantiate that this rescue is caused by the encoded protein and not by unspecific interactions of the morpholino with the injected mRNA itself, we designed a full-length early stop codon variant of the zebrafish papp-a mRNA Papp-a(Y70stop), differing from the wild-type by a single-nucleotide substitution only. Using the Papp-a(Y70stop) mRNA, the developmental delay resulting from papp-a knockdown could not be rescued (Fig. 6B). To exclude transcript instability as the cause of the lacking ability to rescue, we verified that the single nucleotide substitution in the 5-kb mRNA did not alter its stability in vivo compared with the wild-type mRNA (Fig. 6C). This firmly establishes that the observed rescue using mRNA encoding zebrafish wild-type Papp-a or Papp-a(E495A) is an effect of the protein encoded by the injected mRNA, not the transcript itself.
These experiments suggest that the developmental phenotype of papp-a knockdown is a consequence of elimination of Papp-a protein, not its proteolytic activity, and, therefore, that Papp-a possesses biological functions that are not mediated by IGFBP proteolysis. This is true not only by 22 hpf but also by 12 hpf (p < 0.01) (Fig. 6D), before the onset of igf-II and igf-Ir knockdown delay phenotypes, further suggesting that Papp-a functions independently of its proteolytic activity through a mechanism that involves neither Igf-II nor Igf-Ir in early zebrafish development.
A Second PAPP-A Homolog, Papp-a′
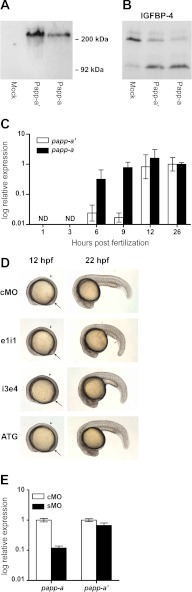
Toward the end of the work presented here, we became aware of the existence of a zebrafish Papp-a paralog in a more recent assembly of the zebrafish genome (Zv9). Although our conclusions above are not compromised by the existence of this additional gene, we carried out similar analyses to assess whether the presence of this paralog, which we designate Papp-a′, has a limiting effect on the severity of the phenotype we have documented here for Papp-a. The Papp-a′ cDNA was cloned from adult zebrafish and sequenced. Sequence alignment of the prepro forms showed 73% identity between Papp-a′ and Papp-a and 58% identity between Papp-a′ and human PAPP-A. Between Papp-a′ and Papp-a, 85 cysteine residues are conserved, and functional domains and motifs show a high degree of conservation (not shown). Recombinantly expressed Papp-a′ showed electrophoretic migration similar to Papp-a (Fig. 7A) and similar proteolytic activity (B). Interestingly, analysis by quantitative RT-PCR revealed that although papp-a expression is abundant from 6 hpf (early gastrulation), only traces of papp-a′ mRNA are present before 12 hpf (onset of segmentation) and, therefore, not present at the onset of the papp-a knockdown phenotype (Fig. 7C).
FIGURE 7.
A second PAPP-A homolog: Biochemistry, expression, and in vivo analysis of Papp-a′. A, Western blot detection of reduced recombinant myc-His tagged zebrafish Papp-a′ in medium from transfected HEK293T cells. Similar to Papp-a, Papp-a′ migrates as a monomer of ∼200 kDa. B, assay for proteolytic cleavage of human IGFBP-4 by Papp-a′ and Papp-a in the presence of human IGF-II. C, quantitative RT-PCR analysis of papp-a′ and papp-a expression levels during early development. Each group contained 20 embryos. Data were normalized to β-2-microglobulin and to the expression level at 26 hpf. The levels of both papp-a′ and papp-a were below the detection limit of our assay at 1 and 3 hpf. ND, not detected. D, phenotypes of control (upper panel) and papp-a knockdown (three lower panels) embryos at 12 and 22 hpf. Views are anterior to the left. Note the number of somites (arrowheads) and position of the tail bud (arrows) at 12 hpf. 5 ng of cMO, 5 ng of splice-site targeting morpholino (e1i1 or i3e4), or 2.5 ng of translation-inhibiting morpholino (ATG) was used. E, quantitative RT-PCR analysis of papp-a′ and papp-a expression levels in papp-a knockdown embryos 24 hpf. Knockdown of papp-a caused a 10-fold (p < 0.001) reduction of papp-a transcript levels, whereas the level of papp-a′ transcript was similar in control and knockdown embryos. Expression levels in knockdown embryos were normalized to β-2-microglobulin and to expression levels in control-injected embryos.
Knockdown of papp-a′ using either of three non-overlapping morpholinos did not cause a delay in developmental rate (Fig. 7D). A slight kink of the tail, however, was apparent 22 hpf, resulting from injection of either of the three morpholinos (Fig. 7D). Finally, we assessed the effect of knockdown of papp-a on papp-a′ expression. No compensatory up-regulation of papp-a′ was observed (Fig. 7E).
DISCUSSION
We have found that the zebrafish genome encodes two homologs of human PAPP-A, Papp-a and Papp-a′, both highly conserved in sequence stretches of identified function. Biochemical analyses of the recombinant zebrafish proteins revealed conservation of proteolytic specificity and mechanism. In addition, the zebrafish papp-a mRNA expression pattern closely resembles the profiles of human and murine PAPP-A, further suggesting suitability of the zebrafish as a vertebrate model for the analysis of PAPP-A function in vivo.
Knockdown of papp-a caused a decrease in the developmental rate of the zebrafish embryo. However, a developmental delay was not observed upon knockdown of papp-a′, in agreement with its relatively late onset of expression. We note that the phenotype of papp-a knockdown embryos was not limited in severity by a hypothetical, compensatory up-regulation of papp-a′.
The general developmental delay caused by the absence of Papp-a begins during gastrulation and increases in severity throughout the segmentation period. Igf-II or igf-Ir knockdown embryos similarly display developmental delays, but with onset well into the segmentation period (20, 23). Thus, the papp-a knockdown phenotype is different from the igf-II and igf-Ir knockdown phenotypes, further substantiated by our finding that the papp-a knockdown phenotype can be rescued by mRNA encoding either wild-type zebrafish Papp-a or a proteolytically inactive variant. These data suggest that the function of Papp-a required for maintaining a normal developmental rate is not potentiation of Igf signaling by Igfbp proteolysis, thereby indicating a function of Papp-a outside of the Igf system. Notably, these data provide the first evidence of non-proteolytic functions of Papp-a.
Given the multidomain structure of the large Papp-a protein, it is not surprising to find that it harbors proteolysis-independent functionality. Biochemical functions have currently been assigned to the proteolytic domain comprising less than a fourth of the protein sequence, two of the five CCP modules (cell association), and the short C terminus-containing LNR-3 (substrate specificity) of human PAPP-A (Fig. 1C). The N-terminal laminin-G-like domain and the central region of ∼550 residues (30) have not yet been assigned any function.
The dwarfism phenotype of the PAPP-A knockout mouse, with its proportional reduction in organ weights, suggests that normal embryonic growth is disrupted prior to organogenesis beginning at E10. Also, a developmental delay of the PAPP-A deficient mouse is observed at E12.5 (8). Thus, the murine phenotype is in accordance with the developmental delay during gastrulation and segmentation in the zebrafish. Importantly, the early growth deficiency of the IGF-II knockout mouse beginning around E9.5 is not accompanied by a developmental delay, exemplified by an unaltered rate of somitogenesis (38). This indicates that similar to our observations in zebrafish embryos, PAPP-A might function independently of its proteolytic activity during early embryonic development in mice. Furthermore, we observed no morphological abnormalities or patterning defects in the embryonic zebrafish, in accordance with the apparent lack of anatomical abnormalities in the PAPP-A knockout mouse.
Later in the embryonic development, at E18.5 and postnatally, double knockout of PAPP-A and IGFBP-4 is reported to result in partial alleviation of the growth deficit of the PAPP-A knockout mouse (12). This experiment strongly suggests that during these later stages, the proteolytic activity of PAPP-A is in fact involved in the potentiation of growth. Our biochemical analysis of zebrafish Papp-a revealed that proteolytic activity, specificity, and the intrinsic regulatory mechanism are conserved between zebrafish and human PAPP-A. Even the highly unusual function of LNR-3 as a specificity-determining exosite (35) required for IGFBP-4 cleavage is conserved, as demonstrated by analysis of mutated zebrafish Papp-a.
Although IGFBP-4 is often considered inhibitory, it is believed to create a pericellular reservoir of bound IGF, which can only become active as a result of PAPP-A cleavage of IGFBP-4 (6). Because PAPP-A is bound to the cell surface, this will occur in close proximity to the IGF receptor and, hence, results in its activation. In the absence of IGFBP-4, IGF is sequestered by other IGFBPs that cannot be cleaved by PAPP-A. Importantly, the dependence on IGF for PAPP-A cleavage of IGFBP-4 is key to this mechanism. Curiously, an ortholog of IGFBP-4 has not yet been identified in the zebrafish genome, but the functional conservation of Papp-a strongly suggest that such an IGF-dependent substrate is present in zebrafish. Because PAPP-A substrate cleavage is not based on recognition of a simple cleavage motif (39), it is not possible to predict whether any of the 10 currently identified zebrafish IGFBPs might be the functional equivalent of mammalian IGFBP-4.
The PAPP-A knockout mouse suggests a requirement for PAPP-A in the embryo proper of normal mammalian development, similar to our observations in embryonic zebrafish. Assuming conservation between vertebrates, some of the fetal growth-related complications in human pregnancies, where the maternal circulating level of PAPP-A is found to be reduced, may be caused by PAPP-A deficiency in the embryo proper. Such conservation implies similar non-proteolytic PAPP-A function in human development. In support of such non-proteolytic function in human pregnancy is the extremely high level of PAPP-A expression in the human placenta, where PAPP-A mRNA is one of the most abundant transcripts (18). In addition, the 50,000-fold increase in circulating PAPP-A during pregnancy would not be expected if PAPP-A functions only by means of its enzymatic activity to increase IGF receptor stimulation.
In conclusion, this work represents the first evaluation of PAPP-A function from the time of fertilization throughout early vertebrate development. We have presented evidence suggesting that zebrafish Papp-a is required for a normal early developmental rate during gastrulation and segmentation, independent of its proteolytic activity. In agreement with its late onset of expression, we find that its paralog, Papp-a′, is not required to maintain a normal developmental rate. Although our data suggest that the proteolytic activity of Papp-a does not contribute to the early regulation of the developmental rate, the functional conservation of the Papp-a proteolytic mechanism indicates that this is involved in other processes in zebrafish, in agreement with data obtained in other organisms.
Note Added in Proof
Papp-a has recently been designated Pappab by ZFIN. Papp-a′ has recently been designated Pappaa by ZFIN (40).
This work was supported by grants from the Danish Council for Independent Research (Medical Sciences, and Technology and Production Sciences), by the Novo Nordic Foundation, by the Karen Elise Jensen Foundation, and by the Danish National Research Infrastructure Program.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) HE999622, HF572951.
B. Thisse, S. Pflumio, M. Fürthauer, B. Loppin, V. Heyer, A. Degrave, R. Woehl, A. Lux, T. Steffan, X. Q. Charbonnier, and C. Thisse, unpublished data.
- PAPP-A
- pregnancy-associated plasma protein A
- IGF
- insulin-like growth factor
- IGFBP
- IGF binding protein
- hpf
- hours post-fertilization
- sMO
- splice site-targeted morpholino
- tMO
- translation-inhibiting morpholino
- cMO
- control morpholino
- E10
- embryonic day 10
- CCP
- complement control protein repeat
- LNR
- Lin12-Notch repeat.
REFERENCES
- 1. Firth S. M., Baxter R. C. (2002) Cellular actions of the insulin-like growth factor binding proteins. Endocr. Rev. 23, 824–854 [DOI] [PubMed] [Google Scholar]
- 2. Bunn R. C., Fowlkes J. L. (2003) Insulin-like growth factor binding protein proteolysis. Trends Endocrinol. Metab. 14, 176–181 [DOI] [PubMed] [Google Scholar]
- 3. Forbes B. E., McCarthy P., Norton R. S. (2012) Insulin-like growth factor binding proteins. A structural perspective. Front. Endocrinol. 3, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lawrence J. B., Oxvig C., Overgaard M. T., Sottrup-Jensen L., Gleich G. J., Hays L. G., Yates J. R., 3rd, Conover C. A. (1999) The insulin-like growth factor (IGF)-dependent IGF binding protein-4 protease secreted by human fibroblasts is pregnancy-associated plasma protein-A. Proc. Natl. Acad. Sci. U.S.A. 96, 3149–3153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Laursen L. S., Overgaard M. T., Søe R., Boldt H. B., Sottrup-Jensen L., Giudice L. C., Conover C. A., Oxvig C. (2001) Pregnancy-associated plasma protein-A (PAPP-A) cleaves insulin-like growth factor binding protein (IGFBP)-5 independent of IGF. Implications for the mechanism of IGFBP-4 proteolysis by PAPP-A. FEBS Lett. 504, 36–40 [DOI] [PubMed] [Google Scholar]
- 6. Laursen L. S., Kjaer-Sorensen K., Andersen M. H., Oxvig C. (2007) Regulation of insulin-like growth factor (IGF) bioactivity by sequential proteolytic cleavage of IGF binding protein-4 and -5. Mol. Endocrinol. 21, 1246–1257 [DOI] [PubMed] [Google Scholar]
- 7. Laursen L. S., Overgaard M. T., Weyer K., Boldt H. B., Ebbesen P., Christiansen M., Sottrup-Jensen L., Giudice L. C., Oxvig C. (2002) Cell surface targeting of pregnancy-associated plasma protein A proteolytic activity. Reversible adhesion is mediated by two neighboring short consensus repeats. J. Biol. Chem. 277, 47225–47234 [DOI] [PubMed] [Google Scholar]
- 8. Conover C. A., Bale L. K., Overgaard M. T., Johnstone E. W., Laursen U. H., Füchtbauer E. M., Oxvig C., van Deursen J. (2004) Metalloproteinase pregnancy-associated plasma protein A is a critical growth regulatory factor during fetal development. Development 131, 1187–1194 [DOI] [PubMed] [Google Scholar]
- 9. DeChiara T. M., Efstratiadis A., Robertson E. J. (1990) A growth-deficiency phenotype in heterozygous mice carrying an insulin-like growth factor II gene disrupted by targeting. Nature 345, 78–80 [DOI] [PubMed] [Google Scholar]
- 10. Liu J. P., Baker J., Perkins A. S., Robertson E. J., Efstratiadis A. (1993) Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r). Cell 75, 59–72 [PubMed] [Google Scholar]
- 11. Bale L. K., Conover C. A. (2005) Disruption of insulin-like growth factor-II imprinting during embryonic development rescues the dwarf phenotype of mice null for pregnancy-associated plasma protein-A. J. Endocrinol. 186, 325–331 [DOI] [PubMed] [Google Scholar]
- 12. Ning Y., Schuller A. G., Conover C. A., Pintar J. E. (2008) Insulin-like growth factor (IGF) binding protein-4 is both a positive and negative regulator of IGF activity in vivo. Mol. Endocrinol. 22, 1213–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fox N. S., Chasen S. T. (2009) First trimester pregnancy associated plasma protein-A as a marker for poor pregnancy outcome in patients with early-onset fetal growth restriction. Prenat. Diagn. 29, 1244–1248 [DOI] [PubMed] [Google Scholar]
- 14. Kirkegaard I., Henriksen T. B., Uldbjerg N. (2011) Early fetal growth, PAPP-A and free β-hCG in relation to risk of delivering a small-for-gestational age infant. Ultrasound Obstet. Gynecol. 37, 341–347 [DOI] [PubMed] [Google Scholar]
- 15. Peterson S. E., Simhan H. N. (2008) First-trimester pregnancy-associated plasma protein A and subsequent abnormalities of fetal growth. Am. J. Obstet. Gynecol. 198, e43–45 [DOI] [PubMed] [Google Scholar]
- 16. Smith G. C., Stenhouse E. J., Crossley J. A., Aitken D. A., Cameron A. D., Connor J. M. (2002) Early-pregnancy origins of low birth weight. Nature 417, 916. [DOI] [PubMed] [Google Scholar]
- 17. Bonno M., Oxvig C., Kephart G. M., Wagner J. M., Kristensen T., Sottrup-Jensen L., Gleich G. J. (1994) Localization of pregnancy-associated plasma protein-A and colocalization of pregnancy-associated plasma protein-A messenger ribonucleic acid and eosinophil granule major basic protein messenger ribonucleic acid in placenta. Lab. Invest. 71, 560–566 [PubMed] [Google Scholar]
- 18. Overgaard M. T., Oxvig C., Christiansen M., Lawrence J. B., Conover C. A., Gleich G. J., Sottrup-Jensen L., Haaning J. (1999) Messenger ribonucleic acid levels of pregnancy-associated plasma protein-A and the proform of eosinophil major basic protein. Expression in human reproductive and nonreproductive tissues. Biol. Reprod. 61, 1083–1089 [DOI] [PubMed] [Google Scholar]
- 19. Søe R., Overgaard M. T., Thomsen A. R., Laursen L. S., Olsen I. M., Sottrup-Jensen L., Haaning J., Giudice L. C., Conover C. A., Oxvig C. (2002) Expression of recombinant murine pregnancy-associated plasma protein-A (PAPP-A) and a novel variant (PAPP-Ai) with differential proteolytic activity. Eur. J. Biochem. 269, 2247–2256 [DOI] [PubMed] [Google Scholar]
- 20. Schlueter P. J., Peng G., Westerfield M., Duan C. (2007) Insulin-like growth factor signaling regulates zebrafish embryonic growth and development by promoting cell survival and cell cycle progression. Cell Death Differ. 14, 1095–1105 [DOI] [PubMed] [Google Scholar]
- 21. Zhong Y., Lu L., Zhou J., Li Y., Liu Y., Clemmons D. R., Duan C. (2011) IGF binding protein 3 exerts its ligand-independent action by antagonizing BMP in zebrafish embryos. J. Cell Sci. 124, 1925–1935 [DOI] [PubMed] [Google Scholar]
- 22. Zou S., Kamei H., Modi Z., Duan C. (2009) Zebrafish IGF genes. Gene duplication, conservation and divergence, and novel roles in midline and notochord development. PLoS ONE 4, e7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. White Y. A., Kyle J. T., Wood A. W. (2009) Targeted gene knockdown in zebrafish reveals distinct intraembryonic functions for insulin-like growth factor II signaling. Endocrinology 150, 4366–4375 [DOI] [PubMed] [Google Scholar]
- 24. Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F. (1995) Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310 [DOI] [PubMed] [Google Scholar]
- 25. Kajimura S., Aida K., Duan C. (2005) Insulin-like growth factor-binding protein-1 (IGFBP-1) mediates hypoxia-induced embryonic growth and developmental retardation. Proc. Natl. Acad. Sci. U.S.A. 102, 1240–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Robu M. E., Larson J. D., Nasevicius A., Beiraghi S., Brenner C., Farber S. A., Ekker S. C. (2007) p53 activation by knockdown technologies. PLoS Genet. 3, e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pear W. S., Nolan G. P., Scott M. L., Baltimore D. (1993) Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. U.S.A. 90, 8392–8396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Boldt H. B., Overgaard M. T., Laursen L. S., Weyer K., Sottrup-Jensen L., Oxvig C. (2001) Mutational analysis of the proteolytic domain of pregnancy-associated plasma protein-A (PAPP-A). Classification as a metzincin. Biochem. J. 358, 359–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔ C(T)) Method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 30. Overgaard M. T., Sorensen E. S., Stachowiak D., Boldt H. B., Kristensen L., Sottrup-Jensen L., Oxvig C. (2003) Complex of pregnancy-associated plasma protein-A and the proform of eosinophil major basic protein. Disulfide structure and carbohydrate attachment. J. Biol. Chem. 278, 2106–2117 [DOI] [PubMed] [Google Scholar]
- 31. Mikkelsen J. H., Gyrup C., Kristensen P., Overgaard M. T., Poulsen C. B., Laursen L. S., Oxvig C. (2008) Inhibition of the proteolytic activity of pregnancy-associated plasma protein-A by targeting substrate exosite binding. J. Biol. Chem. 283, 16772–16780 [DOI] [PubMed] [Google Scholar]
- 32. Weyer K., Overgaard M. T., Laursen L. S., Nielsen C. G., Schmitz A., Christiansen M., Sottrup-Jensen L., Giudice L. C., Oxvig C. (2004) Cell surface adhesion of pregnancy-associated plasma protein-A is mediated by four clusters of basic residues located in its third and fourth CCP module. Eur. J. Biochem. 271, 1525–1535 [DOI] [PubMed] [Google Scholar]
- 33. Nasevicius A., Ekker S. C. (2000) Effective targeted gene “knockdown” in zebrafish. Nat. Genet. 26, 216–220 [DOI] [PubMed] [Google Scholar]
- 34. Schlueter P. J., Royer T., Farah M. H., Laser B., Chan S. J., Steiner D. F., Duan C. (2006) Gene duplication and functional divergence of the zebrafish insulin-like growth factor 1 receptors. FASEB J. 20, 1230–1232 [DOI] [PubMed] [Google Scholar]
- 35. Weyer K., Boldt H. B., Poulsen C. B., Kjaer-Sorensen K., Gyrup C., Oxvig C. (2007) A substrate specificity-determining unit of three Lin12-Notch repeat modules is formed in trans within the pappalysin-1 dimer and requires a sequence stretch C-terminal to the third module. J. Biol. Chem. 282, 10988–10999 [DOI] [PubMed] [Google Scholar]
- 36. Conover C. A., Durham S. K., Zapf J., Masiarz F. R., Kiefer M. C. (1995) Cleavage analysis of insulin-like growth factor (IGF)-dependent IGF-binding protein-4 proteolysis and expression of protease-resistant IGF-binding protein-4 mutants. J. Biol. Chem. 270, 4395–4400 [DOI] [PubMed] [Google Scholar]
- 37. Boldt H. B., Kjaer-Sorensen K., Overgaard M. T., Weyer K., Poulsen C. B., Sottrup-Jensen L., Conover C. A., Giudice L. C., Oxvig C. (2004) The Lin12-notch repeats of pregnancy-associated plasma protein-A bind calcium and determine its proteolytic specificity. J. Biol. Chem. 279, 38525–38531 [DOI] [PubMed] [Google Scholar]
- 38. Burns J. L., Hassan A. B. (2001) Cell survival and proliferation are modified by insulin-like growth factor 2 between days 9 and 10 of mouse gestation. Development 128, 3819–3830 [DOI] [PubMed] [Google Scholar]
- 39. Laursen L. S., Overgaard M. T., Nielsen C. G., Boldt H. B., Hopmann K. H., Conover C. A., Sottrup-Jensen L., Giudice L. C., Oxvig C. (2002) Substrate specificity of the metalloproteinase pregnancy-associated plasma protein-A (PAPP-A) assessed by mutagenesis and analysis of synthetic peptides: substrate residues distant from the scissile bond are critical for proteolysis. Biochem. J. 367, 31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bradford Y., Conlin T., Dunn N., Fashena D., Frazer K., Howe D. G., Knight J., Mani P., Martin R., Moxon S. A., et al. (2011) ZFIN: enhancements and updates to the Zebrafish Model Organism Database. Nucleic Acids Res. 39, D822–D829 [DOI] [PMC free article] [PubMed] [Google Scholar]