Background: The fimbrial tip adhesin CfaE comprises stacked adhesin and pilin domains.
Results: The CfaE adhesin domain mutation G168D loosens the interdomain interface and perturbs only the adjoining pilin domain structure, yet alters CfaE adhesive properties.
Conclusion: The structure-function relationship of CfaE reveals an intimately coupled, bipartite molecular assembly.
Significance: These findings underscore the dynamic nature of host-pathogen interactions.
Keywords: Allosteric Regulation, Bacterial Adhesion, Crystal Structure, Escherichia coli, Molecular Dynamics, Adhesins, Enterotoxigenic Escherichia coli, Fimbriae
Abstract
CfaE, the tip adhesin of enterotoxigenic Escherichia coli colonization factor antigen I fimbriae, initiates binding of this enteropathogen to the small intestine. It comprises stacked β-sandwich adhesin (AD) and pilin (PD) domains, with the putative receptor-binding pocket at one pole and an equatorial interdomain interface. CfaE binding to erythrocytes is enhanced by application of moderate shear stress. A G168D replacement along the AD facing the CfaE interdomain region was previously shown to decrease the dependence on shear by increasing binding at lower shear forces. To elucidate the structural basis for this functional change, we studied the properties of CfaE G168D (with a self-complemented donor strand) and solved its crystal structure at 2.6 Å resolution. Compared with native CfaE, CfaE G168D showed a downward shift in peak erythrocyte binding under shear stress and greater binding under static conditions. The thermal melting transition of CfaE G168D occurred 10 °C below that of CfaE. Compared with CfaE, the atomic structure of CfaE G168D revealed a 36% reduction in the buried surface area at the interdomain interface. Despite the location of this single modification in the AD, CfaE G168D exhibited structural derangements only in the adjoining PD compared with CfaE. In molecular dynamics simulations, the G168D mutation was associated with weakened interdomain interactions under tensile force. Taken together, these findings indicate that the AD and PD of CfaE are conformationally tightly coupled and support the hypothesis that opening of the interface plays a critical modulatory role in the allosteric activation of CfaE.
Introduction
Some cell surface proteins mediate attachment to other cells or matrices, and certain of these adhesins are designed to sustain attachments in the face of mechanical forces. Well studied examples include mammalian carbohydrate-binding selectins governing platelet and leukocyte interactions with the endothelium (1–3) and the type 1 pilus adhesin FimH of uropathogenic Escherichia coli, which attaches to the uroepithelium in a discontinuous flow environment (4). Studies of these systems have been instructive in defining the allosteric mechanisms involved in the formation of so-called catch bonds (2, 5), which strengthen upon application of force. Preliminary studies of the colonization factor antigen I (CFA/I)4 fimbrial tip adhesin CfaE of enterotoxigenic E. coli, an important cause of secretory diarrhea, indicate that it also exhibits catch bond-like adhesion properties under shear stress (6).
FimH of E. coli type 1 pili was the first prokaryotic surface protein shown to form stronger bonds under mechanical force (4). Located distal to the bacterium atop heteropolymeric pilus projections, FimH binds to mannosyl ligands (7, 8). Its atomic structure features two adjoining Ig-like β-sandwich domains, a proximal pilin domain (PD) connecting it to the pilus tip complex, and a distal lectin domain with the mannose-binding pocket at its upper pole (9). A comparison of the crystal structure of high- and low-affinity forms of FimH reveals remarkable conformational differences and provides insights into the allosteric basis of catch bond formation (9, 10). Interdomain interactions appear to regulate the switch from the low- to high-affinity binding state (11, 12). Domain separation, either by application of force or other means, causes elongation and untwisting of the β-strands constituting the lectin domain β-sandwich (10), which in turn constricts the binding pocket and ensnares the ligand. This deformation explains the tenacity of bacterial adhesion to the uroepithelium, countering the forces generated by urine outflow.
Enterotoxigenic E. coli causes secretory diarrhea in humans and livestock by adherence to and colonization of the small intestinal mucosa and subsequent enterotoxin production. CFA/I fimbriae represent an archetypal adhesive structure purported to bind to sialylated protein ligands on the human epithelium while exposed to peristaltic forces (13, 14). CFA/I is composed of CfaE, a tip-localized adhesin, noncovalently interlocked with a helical homopolymeric tract of CfaB pilin subunits through a donor β-strand exchange mechanism (13, 15, 16). Although the primary sequences of CfaE and the type 1 pilus adhesin FimH show nominal pairwise identity (12%), CfaE assumes a cylindrical shape composed of an N-terminal adhesin and C-terminal pilin β-sandwich domain with extensive interdomain interactions (17), resembling the quaternary structure of FimH. The putative ligand-binding pocket of CfaE resides at the pole opposite to where it attaches to the fimbrial stalk and is lined by three Arg residues, all required for binding competence (15, 17, 18). Structure-function studies of CfaE have been facilitated by development of an in cis donor strand complemented CfaE (dscCfaE) variant, which is stabilized with a C-terminal extension comprising a tetrapeptide linker followed by its missing G β-strand, the latter normally being donated in trans by a subjacent CfaB major fimbrial subunit (17).
Both purified CFA/I fimbriae and dscCfaE display shear-enhanced binding to erythrocytes (6), a model cell substrate, by an interaction consistent with catch bond formation. A Gly-to-Asp mutation of adhesin domain (AD) residue 168, positioned at the interdomain interface of CfaE, is purported to disrupt four of the 17 interdomain hydrogen bonds (6, 17). Under conditions of increasing flow, purified CFA/I fimbriae modified to contain CfaE G168D show a pattern of erythrocyte interaction characterized by a shift in activation to the high-affinity state at a 10-fold lower shear stress than exhibited by native CFA/I (6). Above this lowered shear threshold, binding gradually diminishes with the application of increasing force. This suggests that, as with FimH, disruption of the CfaE interdomain interface regulates binding behavior through distal effects on the upper pole of the CfaE AD housing the ligand-binding pocket.
Given the differences in their host niches and the very distant evolutionary relatedness of type 1 pili and CFA/I fimbriae, further structure-function studies of CfaE promise to expand our understanding of the repertoire of mechanisms by which proteins mediate catch bond formation. Toward this end, we engineered the dscCfaE G168D mutant and solved its crystal structure. Here, we report studies of the in vitro binding properties and biophysical state of dscCfaE G168D, its atomic structure, and the results of steered molecular dynamics simulations with application of force along the longitudinal axis of this protein.
EXPERIMENTAL PROCEDURES
Engineering and Purification of dscCfaE G168D and dscCfaE C238S/C326S
The plasmids pET24-dsc19CfaE G168D-His6, and pET24-dsc19cfaE C238S/C326S-His6 were constructed using site-directed mutagenesis (QuikChange, Stratagene) with pET24-dsc19cfaE-His6 as the template (15). Each of these recombinant plasmids, which both include an in-frame C-terminal hexahistidine affinity tag, was transformed into E. coli BL21(DE3) for expression. Cell growth, induction, harvesting, and lysis done as described previously for dscCfaE (15). The lysate was centrifuged at 17,000 × g for 60 min at 4 °C, and the supernatant was loaded onto a HisTrap FF column (GE Healthcare) pre-equilibrated with PBS (pH 7.4). Proteins were eluted using a gradient to 300 mm imidazole over 20 column volumes. Fractions containing target protein were ascertained by SDS-PAGE analysis. These fractions were pooled and diluted 6-fold with buffer containing 20 mm MES (pH 6.0) before loading onto a HiTrap SP column (GE Healthcare). Bound proteins were eluted using a gradient to 500 mm NaCl over 20 column volumes. Fractions containing dsc19CfaE G168D-His6 (hereafter referred to as dscCfaE G168D) were further purified by adding ammonium sulfate to reach 20% saturation before loading onto a phenyl-Sepharose column (GE Healthcare) pre-equilibrated with buffer containing 40% ammonium sulfate and 20 mm Tris (pH 7.7). The protein of interest was eluted using a gradient to 50 mm NaCl and 20 mm Tris (pH 7.7) over 20 column volumes. Purified fractions of dscCfaE G168D were pooled and dialyzed extensively against buffer containing 20 mm Tris (pH 7.7) and 100 mm NaCl. The genetic engineering and purification of dscCfaE R181A and dscCfaE R67A were described previously (15, 17).
Crystallization, Data Collection, Phase Determination, and Refinement of dscCfaE G168D Structure
dscCfaE G168D was crystallized using the hanging drop vapor diffusion method at 15 °C. Initial crystallization screenings were performed with a Mosquito automated solution dispenser (TTP LabTech) using high-throughput screening kits from Hampton Research, Molecular Dimensions, and Emerald BioStructures. The lead conditions were reproduced and optimized with solutions prepared in-house by mixing 1 μl of protein solution (4.4 mg/ml dscCfaE G168D, 20 mm Tris (pH 7.7), and 100 mm NaCl) with 1 μl of reservoir solution (0.1 m sodium citrate (pH 5.8), 1.5 m NaCl, 18% PEG 4000, and 50 mm guanidine chloride). Each drop was equilibrated against 500 μl of reservoir solution. The dscCfaE G168D protein crystals grew to dimensions of 0.6 × 0.4 × 0.1 mm within 1 week. Addition of 30% ethylene glycol was used for cryoprotection. The x-ray diffraction data sets were collected at the Southeast Regional Collaborative Access Team (SER-CAT) beamline at the Advanced Photon Source of the Argonne National Laboratory with a MAR 300 CCD detector. The raw diffraction patterns were indexed, integrated, and scaled using the HKL2000 package (19). The structure was solved by molecular replacement using dscCfaE (Protein Data Bank code 2HB0, chain A) as a phasing template in MOLREP of the CCP4 package (20). The model was refined in REFMAC (21) with 2-fold non-crystallographic symmetry restraints applied and remodeled in Coot (22).
Circular Dichroism Spectroscopy
All samples were prepared at 0.1 mg/ml in 20 mm phosphate (pH 6.1). The thermal denaturation was monitored at 217 nm as the temperature was increased at rate of 80 °C/h. All experiments were performed in a Jasco J-815 CD spectrometer with a 0.1-cm cuvette.
Erythrocyte Adherence Assay
Dynamic flow chamber assays were performed as described previously (6). To assess erythrocyte binding under static conditions, proteins were adsorbed to wells on a 96-well microtiter plate with 100 μl of dscCfaE, dscCfaE G168D, dscCfaE R67A, and dscCfaE R181A at 10, 25, and 50 μg/ml. Each condition was run in duplicate. After incubation at 37 °C for 1 h, the plate was washed three times with 250 μl of PBS. Each well was then blocked by addition of 200 μl of PBS with 0.1% (w/v) BSA at 37 °C for 1 h. After washing the plate three times, prewashed bovine erythrocytes were resuspended at 0.05% (v/v) using PBS (pH 7.4) with 0.5% mannose, and 100 μl of suspension was added per well. After incubation at 37 °C for 1 h, plates were washed three times, and cell dissociation buffer (50 μl/well) was added to release bound erythrocytes. The number of bound erythrocytes was counted using either a hemocytometer or a Countess cell counter (Invitrogen). Each assay was performed three times. Graphical displays were prepared using GraphPad Prism 5.
ELISA
Proteins were coated on a 96-well microtiter plate with 100 μl of dscCfaE, dscCfaE G168D, dscCfaE R67A, and dscCfaE R181A at 2 μg/ml. Each condition was run in duplicate. After incubation at 37 °C for 1 h, the plate was washed three times with 250 μl of PBS. Each well was then blocked by addition of 250 μl of PBS with 5% (v/v) fetal calf serum at 37 °C for 1 h. After washing three times with 250 μl of PBS with 0.05% Tween 20 (PBST), each well was incubated at 37 °C for 1 h with 100 μl of mouse monoclonal antibody P10A7 in a series of 10-fold dilutions. The plate was washed five times with 250 μl of PBST, and each well was incubated at 25 °C for 1 h with 100 μl of horseradish peroxidase-conjugated goat anti-mouse secondary antibodies. After washing three times with 250 μl of PBST, each well was incubated at 25 °C for 20 min with 100 μl of o-phenylenediamine substrates. Absorbances were measured by a plate reader at 450 nm.
Molecular Dynamics Simulations of dscCfaE and dscCfaE G168D under Tensile Force
The dscCfaE (Protein Data Bank 2HB0, chain A) and dscCfaE G168D chain A structures were used to perform molecular dynamics simulations in the program NAMD2 (23) using CHARMM (24), all-hydrogen force field PARAM22 (25), and the TIP3P water model. The proteins were aligned with the longest dimension parallel to the x axis and solvated in a rectangular water box (135 × 80 × 80 Å3) containing 150 mm NaCl, resulting in a system with a total of ∼80,000 atoms. Standard protocols for the non-bonded interactions, temperature, and pressure were described previously (26). For each of the proteins, three 10-ns-long simulations were performed at 27 °C to equilibrate their structures. The conformations obtained at the end of the equilibration runs were resolvated into a rectangular water box (170 × 65 × 65 Å3) to accommodate the extension due to pulling. A constant pulling force was applied at the center of mass of Arg-67, Arg-181, and Arg-182 through a virtual spring with a stiffness of 140 piconewtons/Å and a velocity of 2 Å/ns while the C terminus of the protein was anchored. The pulling simulations were stopped after complete separation of the domains.
RESULTS
Functional Impact of Mutation G168D in CfaE
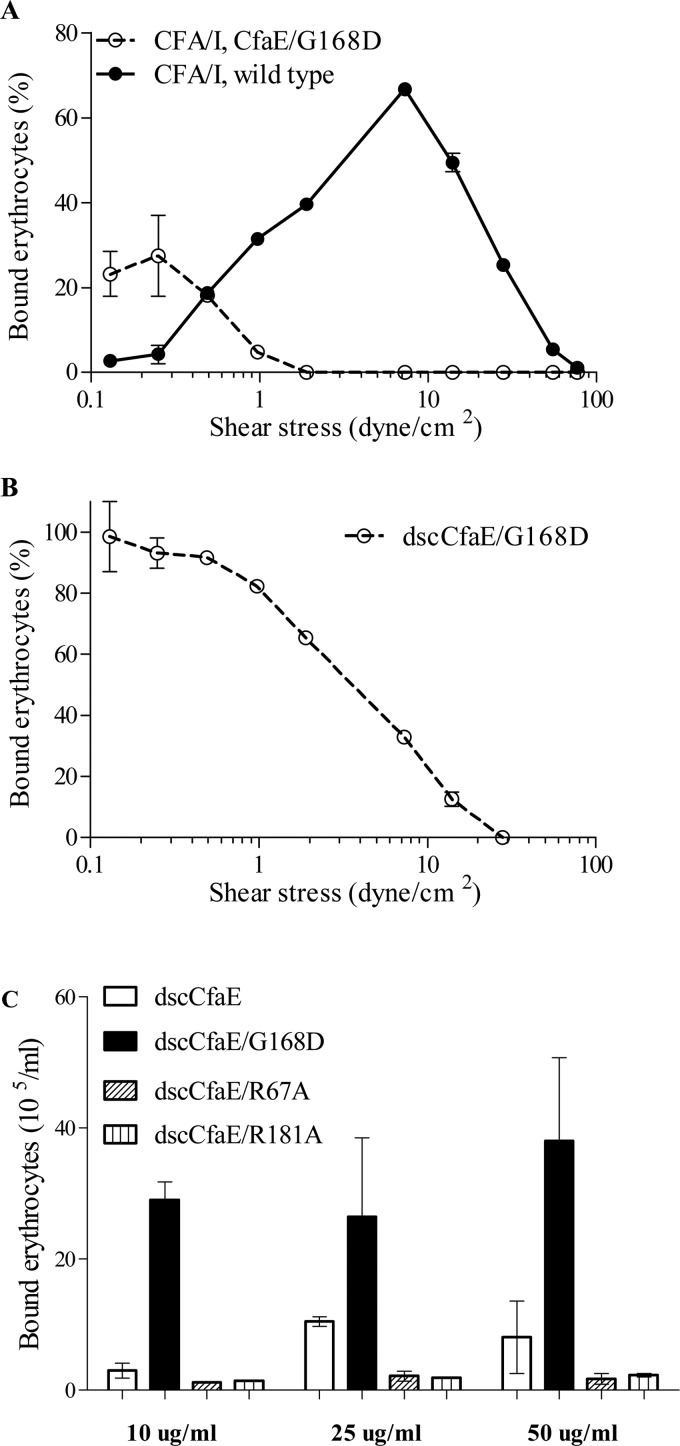
The effects of the G168D mutation in CfaE on erythrocyte adherence were assessed in both a dynamic and static binding assay. In the dynamic flow chamber assay, CFA/I fimbriae harboring CfaE G168D showed a shift in the point of inflection from shear enhancement to shear inhibition to a shear stress level that was lower than that of native CFA/I (Fig. 1A) (6). Restricting our field of view to the tip-localized minor subunit of CFA/I, we studied the recombinant dscCfaE protein and the modified derivative dscCfaE G168D. In the same dynamic assay, dscCfaE G168D showed inflection at a similarly low-shear stress value (Fig. 1B), suggesting that this effect is specifically attributable to the G168D mutation. In a static erythrocyte binding assay, dscCfaE G168D exhibited significantly higher binding than dscCfaE over a range of adhesin concentrations from 10 to 50 μg/ml (Fig. 1C), a finding predicted by the dynamic binding assay by extrapolation to the point of no shear stress. dscCfaE proteins with point mutations in either of two Arg residues (R67A and R181A) in the putative receptor-binding domain were run as controls in this assay, and each showed nominal binding under static conditions (Fig. 1C).
FIGURE 1.
Erythrocyte binding. A, bovine erythrocytes bound to native CFA/I fimbriae and fimbriae with the CfaE G168D mutation under shear stress. B, human erythrocytes bound to recombinant dscCfaE G168D under shear stress. C, erythrocytes bound more strongly to dscCfaE G168D than dscCfaE under static conditions. dscCfaE R67A and dscCfaE R181A served as negative controls and are known to abolish erythrocyte binding. Experiments were reproduced three times, and results from a representative experiment are shown. Error bars represent S.D. of bound erythrocytes.
Reactivity of dscCfaE and dscCfaE G168D to a Receptor-binding Pocket-specific Monoclonal Antibody
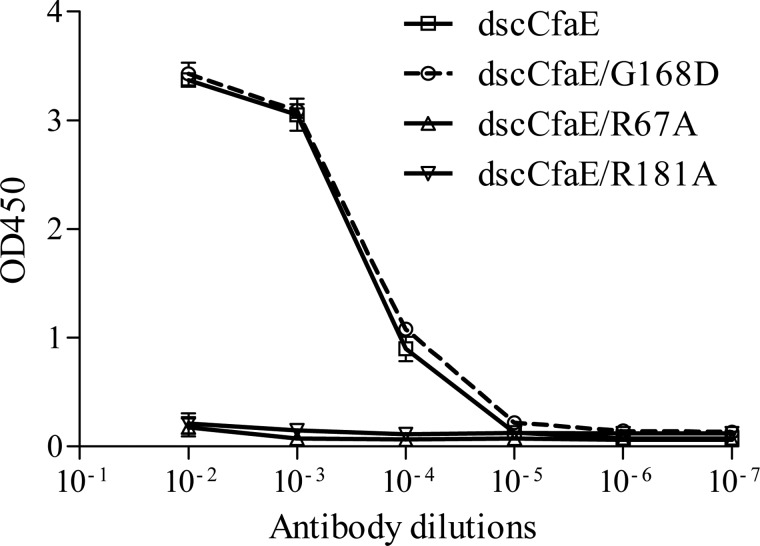
As determined by ELISA, monoclonal antibody P10A7 exhibited binding to dscCfaE but not to either of the two proteins with a point mutation of Arg-67 (dscCfaE R67A) or Arg-181 (dscCfaE R181A) (Fig. 2). This indicates that the binding epitope of P10A7 overlaps the Arg-rich receptor-binding pocket at the upper pole of CfaE. Consequently, we used this monoclonal antibody as an immunological probe to detect conformational shifts of the Arg-67 and Arg-181 side chains in the structure of dscCfaE G168D. In fact, the reactivity of P10A7 was similar for dscCfaE and dscCfaE G168D (Fig. 2). This finding predicts that dscCfaE G168D shares the same conformational epitope in the neighborhood of the receptor-binding pocket in solution.
FIGURE 2.
Monoclonal antibody P10A7 ELISA of different dscCfaE mutants. Error bars represent S.D. of absorbance readings.
Thermal Stability of dscCfaE G168D
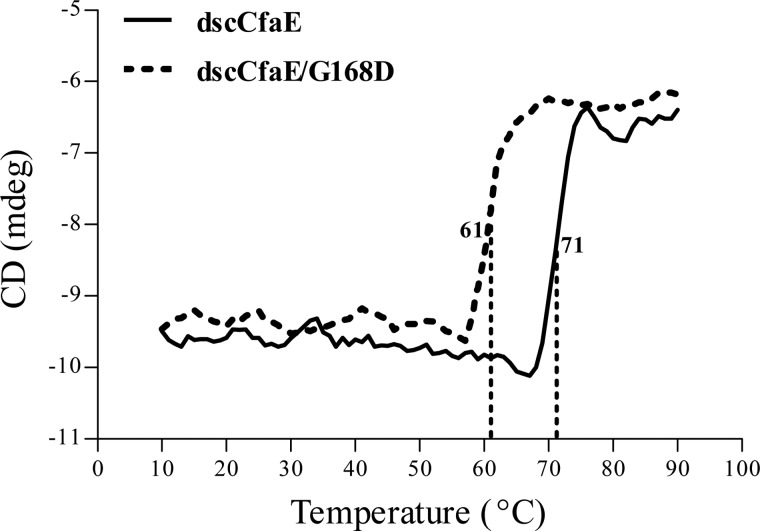
As monitored by CD spectroscopy, the melting temperature of dscCfaE G168D is 61 °C, which is 10 °C lower than that of dscCfaE (Fig. 3). Hence, disturbance of the interdomain interface by the G168D modification of dscCfaE results in an appreciable decrease in thermostability.
FIGURE 3.
Thermal stability of dscCfaE and dscCfaE G168D. The melting temperatures of dscCfaE and dscCfaE G168D were measured by monitoring the secondary structure changes during heating. mdeg, millidegrees.
Determination of the X-ray Crystal Structure of dscCfaE G168D
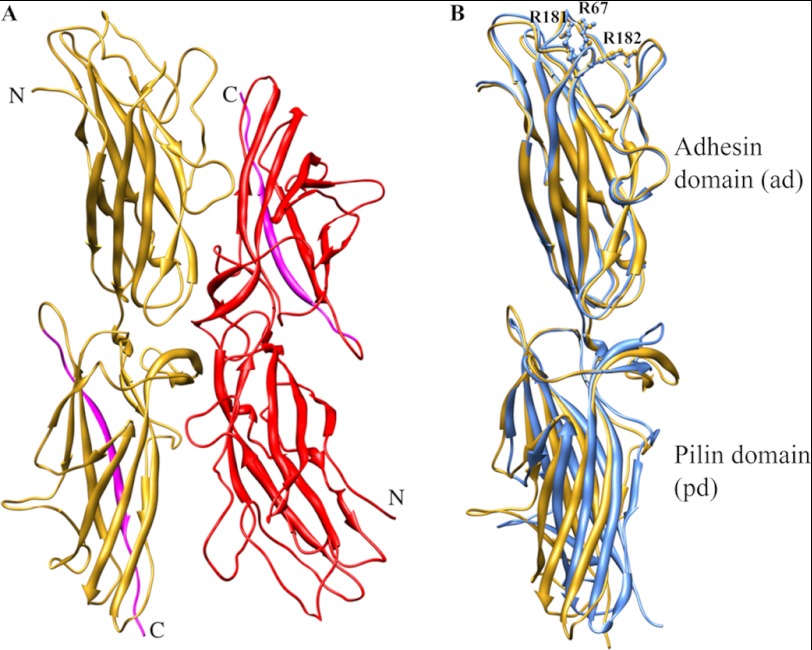
To elucidate the structural basis for the observed functional and biophysical differences between dscCfaE G168D and dscCfaE, we determined the crystal structure of dscCfaE G168D (Protein Data Bank code 3VAC). dscCfaE G168D was purified to homogeneity and crystallized. The structure was determined by molecular replacement using dscCfaE (Protein Data Bank code 2HB0) as a model and refined to 2.6 Å resolution. The data set and refinement statistics are shown in Table 1. Two dscCfaE G168D molecules (chains A and B) are present in one asymmetric unit and are arranged in a head-to-tail orientation (Fig. 4A). dscCfaE G168D chain A has 350 residues visible in the electron density map from Ala-23 to Thr-372, whereas chain B has 354 visible residues from Ala-23 to Asp-376. Approximately 250 water molecules were modeled in the structure. Of the 19 residues composing the extension peptide at the C terminus, only the first 14 residues were modeled for both chains A and B. The modified Asp-168 residue is clearly seen in both chains (supplemental Fig. S1). Chains A and B of dscCfaE G168D are almost identical, with a root mean square deviation (r.m.s.d.) of 0.321 Å (between 350 Cα atoms).
TABLE 1.
Statistics of the x-ray diffraction data set and refinement of the dscCfaE G168D structure
| dscCfaE G168D | |
|---|---|
| Data statistics | |
| Wavelength (Å) | 0.97856 |
| Space group | C2 |
| Cell dimensions | |
| a/b/c (Å) | 104.06/126.36/78.70 |
| β | 100.49° |
| Resolution (Å) | 50–2.6 |
| No. of unique reflections | 30,301 |
| Rmergea | 0.095 (0.310)b |
| Completeness (%) | 98.4 (89.1) |
| Redundancy | 3.6 (2.4) |
| 〈I/σ〈I〉〉 | 13.0 (2.2) |
| Refinement statistics | |
| Rwork | 0.184 |
| Rfreec | 0.234 |
| No. of residues | 712 |
| No. of protein atoms | 5536 |
| No. of non-protein atoms | 249 |
| Wilson B factor (Å2) | 38 |
| Mean B factor (Å2) | 40 |
| r.m.s.d. bond length (Å) | 0.017 |
| r.m.s.d. bond angle | 1.712° |
| Ramachandran plot (%)d | |
| Favored | 97.3 |
| Allowed | 2.7 |
| Disallowed | 0 |
a Rmerge is defined as Σ|Ih,i − 〈Ih〉|/ΣIh,i, where Ih,i is the intensity for the ith observation of a reflection with Miller index h, and 〈Ih〉 is the mean intensity for all measured values of Ih.
b Numbers in parentheses are for the outer resolution shell (2.69–2.60 Å).
c 5% of total reflections were set aside randomly for the Rfree calculation.
FIGURE 4.
A, two dscCfaE G168D molecules (yellow and red) are present in one asymmetric unit in the crystal structure. The donor strands are highlighted in cyan. B, superposition of chain A from both the dscCfaE (blue) and dscCfaE G168D (yellow) structures. The side chains of three arginines (Arg-67, Arg-181, and Arg-182) are shown in ball and stick.
Comparison of dscCfaE G168D and dscCfaE Structures
Because both dscCfaE and dscCfaE G168D have two protein chains in one asymmetric unit that display virtually identical structures, we selected chain A from both structures for comparison. The r.m.s.d. of the Cα atoms in dscCfaE G168D chain A and dscCfaE chain A is 1.879 Å (350 atoms). There are no significant structural changes observed between the N-terminal ADs of these two proteins, inasmuch as the r.m.s.d. of these two domains is 0.386 Å (178 atoms). The three critical Arg residues (Arg-67, Arg-181, and Arg-182) in the AD of both structures are superimposable (Fig. 4B). However, the interdomain G168D mutation did result in spatial rearrangements in the PD (r.m.s.d. of 1.403 Å between 172 atoms) and also at the interface. Not unexpectedly, the buried surface area at the AD-PD interface of dscCfaE G168D was 36% lower than that of dscCfaE, and the number of direct hydrogen bonds at the interface of dscCfaE G168D was reduced by 33% (Table 2 and supplemental Fig. S2).
TABLE 2.
Interdomain interface analysis as measured by buried surface area, hydrogen bonds, and joint and twisting angles between the AD and PD
| Subunit | Domains | Buried surface area | Hydrogen bondsa | Joint angleb | Twisting anglec |
|---|---|---|---|---|---|
| dscCfaE G168D chain A | AD–PD | 444Å2 | 16 (6) | 163.4° | 164.8° |
| dscCfaE chain A | AD–PD | 692Å2 | 23 (9) | 172.6° | 169.5° |
a Direct hydrogen bonds (in parentheses) and indirect hydrogen bonds mediated by one water molecule between two domains were counted.
b The joint angle between the AD and PD is defined between the longest inertial vectors of each domain.
c The twisting angle between AD and PD is based on the transformation matrix obtained from structural alignment between domains that share a common interface. The angle represents a rotation in polar space around an axis to bring two domains into superposition.
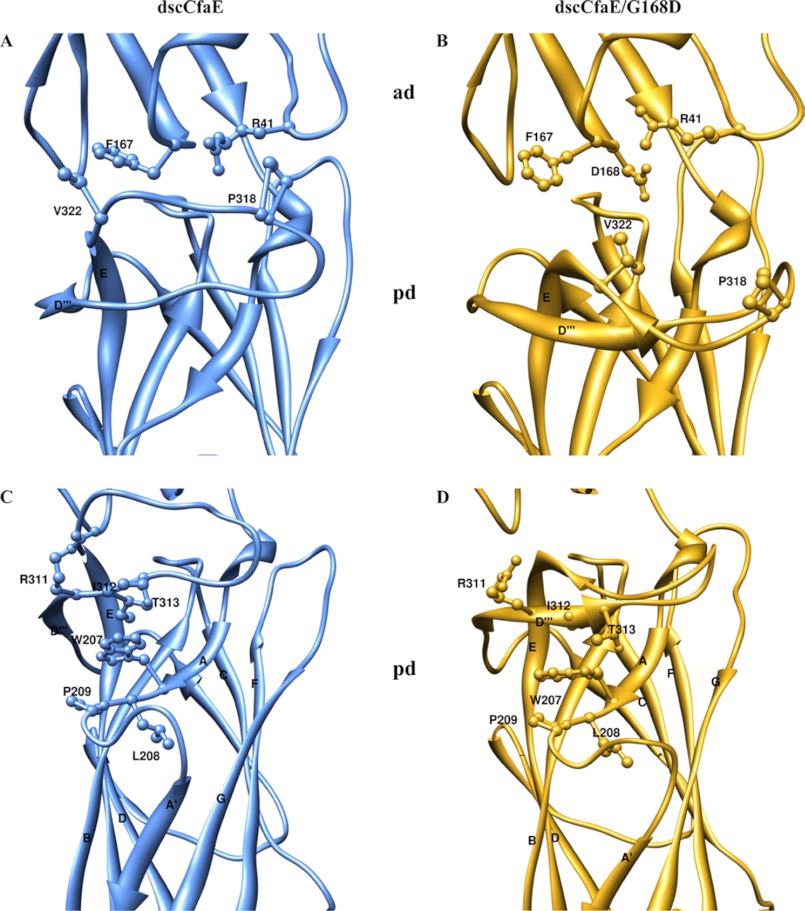
Comparing the overall orientation between the ADs and PDs of dscCfaE and dscCfaE G168D, the joint angle is smaller (9° difference) and the twisting angle is lower (5° difference) for dscCfaE G168D (Table 2) (16). These changes in AD-PD twisting and bending angles in dscCfaE G168D result from a cascade of shifts in the position of the PD β-strands occurring with the introduction of Asp at position 168. This modification results in displacement of the Phe-167 and Arg-41 side chains from their original positions (i.e. in the dscCfaE structure) (Fig. 5, A and B, and supplemental Table S1). In turn, their displaced locations clash with Val-322 and Pro-318 in the loop between the D‴ and E strands in the PD, resulting in a nearly 90° flip in the D‴-E loop (Fig. 5, C and D, and supplemental Table S1). In turn, Arg-311, Ile-312, and Thr-313 in the reoriented D‴-E loop of dscCfaE G168D clash with the original location of Trp-207, resulting in displacement of Leu-208 and Pro-209 in the A-A′ loop and shifting of the A′ strand by 3 Å.
FIGURE 5.
Structural comparison of the interdomain interface of dscCfaE and dscCfaE G168D. A, dscCfaE (blue). B, dscCfaE G168D (yellow). Mutation G168D at the interface caused the steric clashes and reorientation of the highlighted residues. C, dscCfaE (blue). D, dscCfaE G168D (yellow). Reorganization of the cascade of the β-strands in the PD started at the loop between the D‴ and E strands. The E strand in the PD started at Ser-321 in the dscCfaE G168D structure instead of Val-324 as in the dscCfaE structure.
The PD β-strand shifts in dscCfaE G168D are also associated with an increase in the distance between the two sulfur atoms in Cys-238 and Cys-326 located within the PD (from 2.04 Å in dscCfaE to 3.58 Å in dscCfaE G168D) (supplemental Fig. S3, A and B), disrupting the native disulfide bonding of these two residues, which interlink the B and E strands in the CfaE PD structure. To determine whether breakage of this disulfide bond results in the change in thermostability of dscCfaE G168D, we genetically engineered and purified the double mutant dscCfaE C238S/C326S. The thermal melting point of this variant as monitored by CD spectroscopy is 68 °C. Hence, the 10° drop in thermal melting of dscCfaE G168D (see Fig. 3) is only partially attributable to breakage of this bond.
Steered Molecular Dynamics Simulation of dscCfaE and dscCfaE G168D under Tensile Force
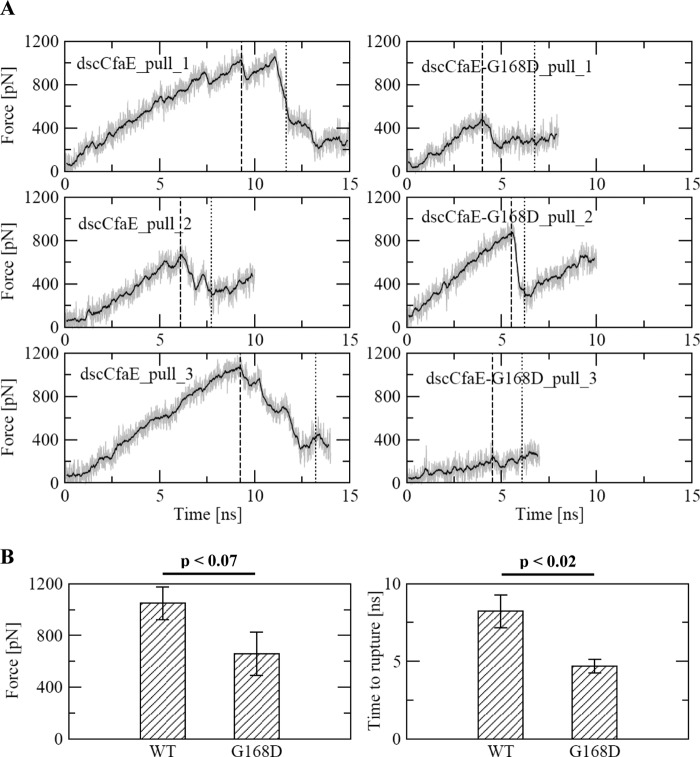
The experimental findings show that the G168D mutation in CfaE results in increased erythrocyte binding under low-shear or static conditions, corresponding to structural alterations that loosen the interdomain interface and alter β-strand packing in the PD but not the AD. Taking this all into account, we hypothesized that this mutation decreases the threshold for complete disarticulation of the AD and PD, which is thought to trigger conformational changes in the ligand-binding domain that toggle it from a low- to high-affinity binding state. To test this hypothesis, we performed molecular dynamics simulations of dscCfaE and dscCfaE G168D in which the center of mass of the putative receptor-binding region in the AD was pulled away from the C terminus of the PD (see “Experimental Procedures”). Because allosterically induced conformational changes in the backbone are not expected to occur in the time scale accessible to molecular dynamics simulations, the analysis presented here focuses on the separation event between the two domains (Fig. 6A). The mean rupture force required to separate the AD and PD was 60% lower for dscCfaE G168D (659 piconewtons) than for dscCfaE (1048 piconewtons) (p < 0.07) (Fig. 6B). The mean rupture time for dscCfaE G168D (4.683 ns) was 75% shorter (p < 0.02) than the rupture time for dscCfaE (8.224 ns) (Fig. 6B). These results suggest that, under tensile force, the AD in dscCfaE G168D more readily separates from the PD than it does in dscCfaE.
FIGURE 6.
Molecular dynamics simulations of dscCfaE and dscCfaE G168D. A, time series of the applied tensile force during the pulling trajectories. The solid lines show the pulling forces averaged over a 200-ps time window. The vertical dashed lines and dotted lines indicate the time points at which the two domains ruptured and completely separated, respectively. Complete separation is defined as the time point at which all hydrogen bonds, which were persistent in the equilibration runs (supplemental Fig. S2), were broken and did not reform for the rest of the run (supplemental Figs. S4 and S5). The simulations were repeated three times for each protein. B, statistical analysis (one-tailed Student's t test) of the rupture force (the highest force measured before complete separation of the domains) and rupture time (time point of the rupture force) during separation of the two domains of dscCfaE and dscCfaE G168D. pN, piconewtons.
DISCUSSION
The functional, biophysical, structural, and molecular dynamics computational studies of dscCfaE G168D presented here reveal the properties of a molecule whose interdomain interface has been partially pried open by insertion of a residue that carries a bulky side chain and breaks several of the hydrogen bonds between the CfaE AD and PD normally formed by the native Gly-168 residue. This modification results in incomplete separation of the domains, impacting its biophysical state as evidenced by a distinct decrease in thermostability. Structurally, there is a diminution in the buried surface area between the two domains, moderate structural shifts in the arrangement of loops and strands in the PD β-sandwich structure, and a small decrease in the joint angle between the AD and PD. There is, however, virtually no conformational disturbance of the AD core or of the apical loops that form its ligand-binding pocket. That this point mutation on the AD interdomain face results largely in structural rearrangements of the adjoining PD illustrates the intimate connectivity of these two domains. The functional consequence of the G168D modification is a downward shift in the force required for shear-enhanced binding of CfaE to erythrocytes and increased erythrocyte binding under static conditions. Taken together with the pulling simulations showing a lower resistance against complete separation of the AD and PD, we postulate that the crystal structure of dscCfaE G168D represents an intermediate state, which facilitates the transition to a high-affinity binding state more easily than the native state in dscCfaE.
The structure of dscCfaE G168D is distinct from the two conformational states that have been reported for the most closely studied catch bond adhesins, FimH (9, 10) and P-selectin (1), in that it exhibits partial disruption of the interdomain interface. Generally, the known structures of FimH and P-selectin fall into one of two different conformations: an extended conformation with complete separation between the lectin and subjacent PD (FimH) or EGF domain (P-selectin), which exhibits high-affinity ligand binding (1, 9), and a compressed conformation featuring contact between the two domains, which exhibits low-affinity ligand binding (10, 27). Although FimH mutants with structural mutations that partially disrupt the interdomain interface have been shown to increase mannose binding, the corresponding crystal structures have not been solved (11). Two previously reported CfaE structures are of variants in which the adhesin has been stabilized by in cis donor strand complementation, namely dscCfaE and dscCfaEB (16, 17). The two-domain CfaE structure in each of these molecules is superimposable (r.m.s.d. of 0.77 Å), with each showing highly conserved interdomain interactions and a large buried surface area (700 Å2). Functional analysis of dscCfaE indicates that it exhibits low-affinity binding to erythrocytes under static or low-shear conditions but can switch to a high-affinity binding state under moderate shear stress (6). The dscCfaE G168D interface mutant exhibits stronger binding to erythrocytes under static or low-shear conditions, as shown in this and previous work (6). Other CfaE interface mutations designed to disrupt interdomain interactions, namely CfaE S244N, or to disrupt the secondary structure of the 3/10-helical interdomain linker, CfaE K201P, have been shown to increase erythrocyte binding of derivative fimbriae at low shear or to increase bacterial binding to Caco-2 intestine-derived tissue culture cells under static conditions (6). We hypothesize that the structure of the dscCfaE G168D mutant is intermediary between the low- and high-affinity allosteric forms, to which the addition of ligand results in a more ready conversion to the high-affinity state than is observed with native CfaE. Consistent with this is our observation that lower shear stress forces are required to enhance binding of CFA/I fimbriae containing CfaE G168D compared with native CfaE. Furthermore, we postulate that CfaE G168D is precluded from assuming the low-affinity conformation that is associated with complete engagement of the interdomain surfaces due to the physical wedge effect of the bulky side chain of Asp-168, which essentially stabilizes what may otherwise represent an unstable intermediate state for native CfaE that occurs transiently in the switch from low- to high-affinity conformation.
Although the findings presented here support the notion that the interdomain interface of CfaE, like that of the other catch bond-forming adhesins FimH and P-selectin, modulates adhesive function, they provide only circumstantial support for a proposed allosteric mechanism. Indeed, if the dscCfaE G168D structure represents an intermediate allosteric form of the adhesin, then more convincing evidence for this mechanism will require solving the structure of a high-affinity allosteric form. Evidence exists to implicate a sialylated glycoprotein as the intestinal receptor for CFA/I fimbriae (14), which presumably interacts with CfaE at its arginine-rich binding pocket. The identification of a sialic acid-containing ligand and its co-crystallization with dscCfaE or dscCfaE G168D might reveal this hypothetical high-affinity allosteric form of the adhesin. Without such proof, it is worth considering alternative explanations for our findings. One possibility is that the G168D modification in CfaE unmasks a second erythrocyte-binding site that exhibits shear-enhanced binding with a lower shear threshold for conversion from catch-to-slip bond formation. One might reason that such an unmasked binding site would be localized to the CfaE PD because the structural differences between dscCfaE and dscCfaE G168D are largely confined to this domain. Evidence against this alternative is the finding that point mutations in any one of the three Arg residues that line the ligand-binding pocket at the apex of the CfaE AD abolish erythrocyte agglutination by the resulting CFA/I-expressing bacteria (17). Another possibility is that the G168D modification results in a change in orientation of the CfaE AD and PD such that it favors multiple simultaneous binding events to erythrocytes by fimbriae or adhesins, as presented in the flow chamber apparatus used here, or by extrapolation to binding of bacteria to the intestinal tract in nature. A theoretical framework has recently been presented whereby multi-fimbriated bacteria that individually bind via slip bonds can exhibit catch bond behavior when working in aggregate (28). However, this does not match well with the minor changes in AD-PD joint and twisting angle orientation, nor does it explain the marked increase in erythrocyte binding of dscCfaE G168D over dscCfaE under static conditions.
The findings presented here and previously (6) suggest that the CfaE interdomain interface plays a role in modulating the function of the distal ligand-binding pocket at the apex of the CfaE AD. Moreover, this is a concept for which there is strong experimental proof for the related bacterial adhesin FimH, as well as other biologically important adhesins. At first glance, however, the juxtaposition of this concept with our finding that the G168D modification in CfaE perturbs the conformation of the PD rather than that of the AD seems counterintuitive. That is to say that the G168D modification leads to an increase in erythrocyte binding to the AD, yet it results in observable structural changes only in the β-strand and loop structure of the PD. This seeming inconsistency is an indicator of the crudeness of our current understanding of the structure-function relationship of the CfaE adhesin and a reminder that this tip-localized protein serves as one working part of CFA/I fimbriae, a polymeric surface organelle that functions as a complex biomachine promoting epithelial adherence (16). In the intestinal environment, enterotoxigenic E. coli CFA/I bacteria travel from the mouth into the small intestine, streaming via mechanical propulsive forces generated by patterned contractions in the smooth muscle wall of this tract. Initial bacterial attachments to the epithelial surface presumably occur via specific CfaE-glycoprotein receptor-ligand interactions (18). Continued peristaltic activity places hydrodynamic drag forces on the bacterium, with transduction of that force through the fimbrial stalk from base to tip, where shear stress is then exerted upon the receptor-ligand interaction. It has recently been shown that the CFA/I fimbrial stalk can absorb some level of external mechanical forces by reversible unwinding of its helically arranged major subunits (29), providing some dampening effect on the forces exerted on the CfaE-ligand bond. For significant drag forces on the bacterium that transduce to CfaE, it would be reasonable to postulate that, in an allosteric model of CfaE catch bond formation, a cascade of events would begin with conformational changes in the PD, the nature of which may mimic the changes we have observed in dscCfaE G168D. With transduction of sufficient force, this would be followed by complete separation of the AD and PD, passing through the normally unstable intermediate of partial separation that we have visualized in the structure of dscCfaE G168D. Extrapolating from experimental findings with FimH, the ultimate effects of this mechanical force would be to alter the conformation of the AD, with these allosteric changes leading from a low- to high-affinity conformation of the ligand-binding pocket, strengthening the bond and resisting dislodgment.
In summary, we propose that dscCfaE G168D displays an allosteric form of CfaE that is intermediate between its low- and high-affinity binding states and that more readily assumes the high-affinity allosteric form with engagement of its cognate ligand under static or low-shear conditions. Further evidence for this model would come from determination of the structure of dscCfaE G168D bound to a suitable ligand. We expect that these structures may also reveal the structural sectors that interconnect the interdomain interface and the outer loops of the AD surrounding the ligand-binding pocket.
Acknowledgments
We thank the staff of the Southeast Regional Collaborative Access Team for assistance for data collection. The molecular dynamics simulations were run on the Trestles supercomputer at the San Diego Supercomputer Center supported by TeraGrid Grant TG-MCB060069N with funding from the National Science Foundation. We are grateful to Daniel Isaac for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 AI050940 (to E. S.) and by the National Cancer Institute Intramural Research Program and the Trans-National Institutes of Health/Food and Drug Administration Intramural Biodefense Program (to D. X.). This work was also supported by United States Army Military Infectious Diseases Research Program Work Unit A1207 and the Henry M. Jackson Foundation for the Advancement of Military Medicine (to S. J. S.). This work was prepared as part of the official duty of S. J. S.

This article contains supplemental Figs. S1–S5 and Table S1.
The atomic coordinates and structure factors (code 3VAC) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- CFA/I
- colonization factor antigen I
- PD
- pilin domain
- dscCfaE
- donor strand complemented CfaE
- AD
- adhesin domain
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1. Somers W. S., Tang J., Shaw G. D., Camphausen R. T. (2000) Insights into the molecular basis of leukocyte tethering and rolling revealed by structures of P- and E-selectin bound to SLeX and PSGL-1. Cell 103, 467–479 [DOI] [PubMed] [Google Scholar]
- 2. Springer T. A. (2009) Structural basis for selectin mechanochemistry. Proc. Natl. Acad. Sci. U.S.A. 106, 91–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Marshall B. T., Long M., Piper J. W., Yago T., McEver R. P., Zhu C. (2003) Direct observation of catch bonds involving cell-adhesion molecules. Nature 423, 190–193 [DOI] [PubMed] [Google Scholar]
- 4. Thomas W. E., Trintchina E., Forero M., Vogel V., Sokurenko E. V. (2002) Bacterial adhesion to target cells enhanced by shear force. Cell 109, 913–923 [DOI] [PubMed] [Google Scholar]
- 5. Thomas W. E., Vogel V., Sokurenko E. (2008) Biophysics of catch bonds. Annu. Rev. Biophys. 37, 399–416 [DOI] [PubMed] [Google Scholar]
- 6. Tchesnokova V., McVeigh A. L., Kidd B., Yakovenko O., Thomas W. E., Sokurenko E. V., Savarino S. J. (2010) Shear-enhanced binding of intestinal colonization factor antigen I of enterotoxigenic Escherichia coli. Mol. Microbiol. 76, 489–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Krogfelt K. A., Bergmans H., Klemm P. (1990) Direct evidence that the FimH protein is the mannose-specific adhesin of Escherichia coli type 1 fimbriae. Infect. Immun. 58, 1995–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Abraham S. N., Sun D., Dale J. B., Beachey E. H. (1988) Conservation of the d-mannose-adhesion protein among type 1 fimbriated members of the family Enterobacteriaceae. Nature 336, 682–684 [DOI] [PubMed] [Google Scholar]
- 9. Choudhury D., Thompson A., Stojanoff V., Langermann S., Pinkner J., Hultgren S. J., Knight S. D. (1999) X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science 285, 1061–1066 [DOI] [PubMed] [Google Scholar]
- 10. Le Trong I., Aprikian P., Kidd B. A., Forero-Shelton M., Tchesnokova V., Rajagopal P., Rodriguez V., Interlandi G., Klevit R., Vogel V., Stenkamp R. E., Sokurenko E. V., Thomas W. E. (2010) Structural basis for mechanical force regulation of the adhesin FimH via finger trap-like β-sheet twisting. Cell 141, 645–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Aprikian P., Tchesnokova V., Kidd B., Yakovenko O., Yarov-Yarovoy V., Trinchina E., Vogel V., Thomas W., Sokurenko E. (2007) Interdomain interaction in the FimH adhesin of Escherichia coli regulates the affinity to mannose. J. Biol. Chem. 282, 23437–23446 [DOI] [PubMed] [Google Scholar]
- 12. Aprikian P., Interlandi G., Kidd B. A., Le Trong I., Tchesnokova V., Yakovenko O., Whitfield M. J., Bullitt E., Stenkamp R. E., Thomas W. E., Sokurenko E. V. (2011) The bacterial fimbrial tip acts as a mechanical force sensor. PLoS Biol. 9, e1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Anantha R. P., McVeigh A. L., Lee L. H., Agnew M. K., Cassels F. J., Scott D. A., Whittam T. S., Savarino S. J. (2004) Evolutionary and functional relationships of colonization factor antigen I and other class 5 adhesive fimbriae of enterotoxigenic Escherichia coli. Infect. Immun. 72, 7190–7201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bartus H., Actor P., Snipes E., Sedlock D., Zajac I. (1985) Indications that the erythrocyte receptor involved in enterotoxigenic Escherichia coli attachment is a sialoglycoconjugate. J. Clin. Microbiol. 21, 951–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Poole S. T., McVeigh A. L., Anantha R. P., Lee L. H., Akay Y. M., Pontzer E. A., Scott D. A., Bullitt E., Savarino S. J. (2007) Donor strand complementation governs intersubunit interaction of fimbriae of the alternate chaperone pathway. Mol. Microbiol. 63, 1372–1384 [DOI] [PubMed] [Google Scholar]
- 16. Li Y. F., Poole S., Nishio K., Jang K., Rasulova F., McVeigh A., Savarino S. J., Xia D., Bullitt E. (2009) Structure of CFA/I fimbriae from enterotoxigenic Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 106, 10793–10798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li Y. F., Poole S., Rasulova F., McVeigh A. L., Savarino S. J., Xia D. (2007) A receptor-binding site as revealed by the crystal structure of CfaE, the colonization factor antigen I fimbrial adhesin of enterotoxigenic Escherichia coli. J. Biol. Chem. 282, 23970–23980 [DOI] [PubMed] [Google Scholar]
- 18. Baker K. K., Levine M. M., Morison J., Phillips A., Barry E. M. (2009) CfaE tip mutations in enterotoxigenic Escherichia coli CFA/I fimbriae define critical human intestinal binding sites. Cell. Microbiol. 11, 742–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Otwinowski Z., Minor M. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 20. Collaborative Computational Project Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 21. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 22. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 23. Kalé L., Skeel R., Bhandarkar M., Brunner R., Gursoy A., Krawetz N., Phillips J., Shinozaki A., Varadarajan K., Schulten K. (1999) NAMD2: greater scalability for parallel molecular dynamics. J. Comput. Phys. 151, 283–312 [Google Scholar]
- 24. Brooks B. R., Brooks C. L., 3rd, Mackerell A. D., Jr., Nilsson L., Petrella R. J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., Caflisch A., Caves L., Cui Q., Dinner A. R., Feig M., Fischer S., Gao J., Hodoscek M., Im W., Kuczera K., Lazaridis T., Ma J., Ovchinnikov V., Paci E., Pastor R. W., Post C. B., Pu J. Z., Schaefer M., Tidor B., Venable R. M., Woodcock H. L., Wu X., Yang W., York D. M., Karplus M. (2009) CHARMM: the biomolecular simulation program. J. Comput. Chem. 30, 1545–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. MacKerell A., Bashford D., Bellott M., Dunbrack R., Evanseck J., Field M., Fischer S., Gao J., Guo H., Ha S., Joseph-McCarthy D., Kuchnir L., Kuczera K., Lau F., Mattos C., Michnick S., Ngo T., Nguyen D., Prodhom B., Reiher W., Roux B., Schlenkrich M., Smith J., Stote R., Straub J., Watanabe M., Wiorkiewicz-Kuczera J., Yin D., Karplus M. (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. 102, 3586–3616 [DOI] [PubMed] [Google Scholar]
- 26. Interlandi G., Thomas W. (2010) The catch bond mechanism between von Willebrand factor and platelet surface receptors investigated by molecular dynamics simulations. Proteins 78, 2506–2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Graves B. J., Crowther R. L., Chandran C., Rumberger J. M., Li S., Huang K. S., Presky D. H., Familletti P. C., Wolitzky B. A., Burns D. K. (1994) Insight into E-selectin/ligand interaction from the crystal structure and mutagenesis of the lec/EGF domains. Nature 367, 532–538 [DOI] [PubMed] [Google Scholar]
- 28. Björnham O., Axner O. (2010) Catch-bond behavior of bacteria binding by slip bonds. Biophys. J. 99, 1331–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Andersson M., Björnham O., Svantesson M., Badahdah A., Uhlin B. E., Bullitt E. (2012) A structural basis for sustained bacterial adhesion: biomechanical properties of CFA/I pili. J. Mol. Biol. 415, 918–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]